Highlight
MLP43 interacts with SnRK2.6 and ABF1 and functions as a positive regulator in ABA and drought stress responses through modulating gene expression, ROS production and primary metabolite profiles.
Key words: ABA signal reconstitution, abscisic acid, drought stress, metabolite profile, MLP43, reactive oxygen species, SnRK2.6.
Abstract
Drought stress is one of the disadvantageous environmental conditions for plant growth and reproduction. Given the importance of abscisic acid (ABA) to plant growth and abiotic stress responses, identification of novel components involved in ABA signalling transduction is critical. In this study, we screened numerous Arabidopsis thaliana mutants by seed germination assay and identified a mutant mlp43 (major latex protein-like 43) with decreased ABA sensitivity in seed germination. The mlp43 mutant was sensitive to drought stress while the MLP43-overexpressed transgenic plants were drought tolerant. The tissue-specific expression pattern analysis showed that MLP43 was predominantly expressed in cotyledons, primary roots and apical meristems, and a subcellular localization study indicated that MLP43 was localized in the nucleus and cytoplasm. Physiological and biochemical analyses indicated that MLP43 functioned as a positive regulator in ABA- and drought-stress responses in Arabidopsis through regulating water loss efficiency, electrolyte leakage, ROS levels, and as well as ABA-responsive gene expression. Moreover, metabolite profiling analysis indicated that MLP43 could modulate the production of primary metabolites under drought stress conditions. Reconstitution of ABA signalling components in Arabidopsis protoplasts indicated that MLP43 was involved in ABA signalling transduction and acted upstream of SnRK2s by directly interacting with SnRK2.6 and ABF1 in a yeast two-hybrid assay. Moreover, ABA and drought stress down-regulated MLP43 expression as a negative feedback loop regulation to the performance of MLP43 in ABA and drought stress responses. Therefore, this study provided new insights for interpretation of physiological and molecular mechanisms of Arabidopsis MLP43 mediating ABA signalling transduction and drought stress responses.
Introduction
Abiotic stresses greatly affect plant growth and crop production. To date, plants have evolved many mechanisms to adapt and survive against these stresses, which include developmental, morphological, physiological and biochemical strategies. Plant hormones play essential roles in promoting and mediating these defence responses (Peleg and Blumwald, 2011). Abscisic acid (ABA) is regarded as a key signal involved in regulating the response of plants to various stresses, and particularly in regulating drought stress responses when plants experience water deficit (Cutler et al., 2010; Lee and Luan, 2012). ABA production increases radically under drought stress conditions and stimulates stomatal closure, changing the expression of various osmotic stress-responsive genes (Kim et al., 2010). In recent years, significant research progress has been made in studies using plants bearing gene mutations involved in hormone-biosynthetic and signalling transduction pathways, and to the identification of the ABA receptors PYR/PYL through chemical genetic approaches (Park et al., 2009; Ma et al., 2009; Hubbard et al., 2010). Given the importance of ABA to plant physiology and development, identification of novel components involved in ABA signalling transduction is critical.
Major latex protein (MLP) was first identified from the latex of the opium poppy (Papaver somniferum) (Nessler et al., 1990; Nessler and Burnett, 1992). The orthologues of MLP, called MLP-like proteins (MLP), were later found in Arabidopsis, soybean and tobacco (Aggelis et al., 1997; Wu et al., 2008). MLP proteins belong to the Bet v 1 family, also known as the pathogenesis related 10 (PR10)-like protein family, which contains proteins with low sequence similarity, but shares a similar three-dimensional structure (Radauer et al., 2008). The Bet v 1 family is divided into 11 subfamilies, including MLP, PR10, cytokinin receptors and plant polyketide cyclase-like subfamilies (Radauer et al., 2008). Most of the Bet v 1 proteins function through binding ligands, such as cytokinins, brassinolides and secondary metabolites and trigger downstream signal transduction (Koistinen et al., 2005; Radauer et al., 2008). In Arabidopsis, the ABA receptor RCAR/PYR/PYL family of START proteins share amino acid and structural similarity with Bet v 1 proteins (Ma et al., 2009; Park et al., 2009). Previous studies indicated that MLPs are down-regulated by blue light in the cry1 loss-of-function mutant (Phee et al., 2007). Two AtMLPs (At2g01520 and At2g01530) could be induced by cis-cinnamic acid and promotes vegetative growth and delays flowering (Guo et al., 2011). One report showed that MLPs were related to the translocation of hydrophobic compounds in the Cucurbitaceae family, which could contribute to decreasing the hydrophobic contamination of fruit (Inui et al., 2013). Three MLPs, existing as duplicated gene pairs, were significantly down-regulated by oxidative stress through whole-genome DNA microarrays analysis, indicating that MLP might be involved in plants stress responses (Kim et al., 2005). Interestingly, a study by Chen and Dai (2010) indicated that overexpression of MLP in Arabidopsis leads to salt stress insensitivity. The homologues of the MLP gene family are differentially expressed in various tissues; however, the molecular mechanism of MLPs in abiotic stress response is still elusive.
In this study, we screened numerous Arabidopsis T-DNA insertion mutants through seed germination assay, and identified the mutant mlp43 with decreased ABA sensitivity in seed germination. The drought stress responses of the mlp43 mutant and MLP43-overexpressed transgenic plants were also examined. The tissue-specific expression patterns and subcellular localization of MLP43 were detected. Physiological and biochemical analysis indicated that MLP43 was involved in ABA- and drought-stress responses in Arabidopsis through modulating primary metabolite profiling and gene expression. Reconstitution analysis of ABA signalling component assay in Arabidopsis protoplasts and direct interaction assay indicated that MLP43 functions as a positive regulator in ABA signalling transduction and could directly interact with SnRK2.6 and ABF1 in yeast.
Materials and methods
Plant materials and growth conditions
Arabidopsis thaliana ecotype Columbia (Col-0) was used in this study and all transgenic plants were generated in a Col-0 background. The line of mlp43 mutant (Salk_109337 and Salk_033347) was obtained from the Arabidopsis Biological Resource Center (ABRC; http://arabidopsis.org/abrc/) and identified by PCR. Primers used are listed in Supplementary Table S1 at JXB online. The mutants of ost1-1 (Mustilli et al., 2002), aba1-1 (Koornneef et al., 1982) and abi1-1 (Koornneef et al., 1984) are derived from the Arabidopsis accession Landsberg erecta (Ler). The pyr1/pyl1/4 triple mutant (Park et al., 2009) is derived from the Arabidopsis accession Col-0. Seeds were surface sterilized and sown on Murashige and Skoog (MS) agar plates containing full-strength MS salts, 0.8% (w/v) agar, and 1% (w/v) sucrose. Germination assay was performed with or without ABA on MS plates (Sigma, A1049). The seeds were stratified at 4ºC for 4 d in darkness and then transferred to growth chamber with 16h/8h light/dark cycle at 23ºC, or were directly sown in soil after stratification under the same conditions.
Plasmids construction and transgenic plant generation
MLP43 cDNA was amplified using the primers MLP43-F and MLP43-R and cloned into Pro35S::GFP to generate Pro35S::MLP43-GFP fusion construct. The fused protein was then digested by XhoI/SacI and inserted into a binary vector pBA to generate pBA-MLP43-GFP construct. Promoter fragment of MLP43 (-1769bp before start code ATG) was amplified using primers ProMLP43-F and ProMLP43-R and subsequently cloned into the pBI101 to generate ProMLP43::GUS construct. The plasmids used in reconstituted ABA signaling pathway have been described by Lu et al. (2013). The detail information of plasmids used in yeast two hybrid (Y2H) assay is listed in Supplementary Table S2. The primers used for plasmids construction are listed in Supplementary Table S3. The mutated ABI5 was generated previously through site mutation (Wang et al., 2013). All transgenic plants were generated by introducing Agrobacterium tumefaciens (strain GV3101) carrying the corresponding plasmids through floral dip-mediated infiltration into Col-0 background (Clough and Bent, 1998). Complementary transgenic plants of Com-3 were generated by introducing a pBA-MLP43-GFP construct into mlp43 mutant background.
GUS histochemical analysis and subcellular localization
GUS signals were detected according to the method described by Jefferson et al. (1987). Plants were pretreated with or without 100 μM ABA for 3h, and then immersed in 90% acetone for 30min. After incubation in the GUS staining solution (0.5mg/ml X-Gluc, 50mM PBS, pH 7.0; 5.0mM potassium ferricyanide, 5.0mM potassium ferrocyanide, 0.1% Triton X-100) at 37ºC over night, the stained plants were washed with 70% ethanol overnight. Images were taken with an inverted microscope (SMZ1500, Nikon). For subcellular localization of MLP43, we transformed the plasmids into Arabidopsis rosette leaves (Col-0) by bombardment. Images were taken with an inverted microscope (TE2000U, Nikon) equipped with cool CCD (CoolSNAP HQ2, Roper Scientific). GFP fluorescence was acquired with 488nm excitation, and the chloroplast auto-fluorescence was acquired with 543nm excitation.
Drought treatment, water loss analysis, and stomatal aperture measurement
For measurement of drought tolerance, water was withheld from 14-day-old plants comparable in size and growing in pots. After 21 d of drought treatment, the plants were re-watered. Survival rates were determined and the plants were photographed 2 d after re-watering. For measurement of water loss from detached leaves, the rosette leaves were detached from four-week-old plants and weighed at the indicated times. To analyse stomatal apertures, we incubated rosette leaves in a solution containing 50mM KCl, 10mM CaCl2 and 10mM MES (pH 6.15) for 3h under light condition. ABA was then added to the solution to a final concentration of 50 μM. Stomatal apertures were then measured after 30min and 1h of ABA treatment, respectively. For each experimental repeat, at least 50 stomata were counted and measured by photoshop software. The values of stomatal width to length ratios (V) were defined as: V≥0.5, open; 0.5 >V≥0.25, partially open; and V<0.25, closed.
Comparison of EL, ROS contents and antioxidant enzymes activities
EL was determined from the detached aerial parts of drought-stressed plants with the indicated time points. The detailed procedure was performed as described by Wang et al. (2013). Superoxide radicals (O−) and hydrogen peroxide (H2O2) were detected by nitroblue tetrazolium chloride (NBT) staining and 3,3’-diaminobenzidine (DAB) staining, respectively, as described previously (Ramel et al., 2009). Two-week-old seedlings were grown in soil, and then withheld water for the number of days indicated. Quantification of H2O2 content was determined using the method described by Hu et al. (2012). The activities of antioxidant enzymes were measured after drought treatment application followed by the procedure described by Wang et al. (2013).
Gene expression analysis by real-time quantitative RT-PCR
Two-week-old plants were treated as indicated phytohormones or abiotic stresses. Total RNA was extracted using a plant RNA purification kit (Tiangen, Beijing, China). Equal amounts of RNA were used for reverse transcription with ReverTra Ace-α-TM (TOYOBO, Tokyo, Japan) according to the manufacturer’s instructions. The primers used in qRT-PCR were designed using web tool GenScript (https://www.genscript.com/ssl-bin/app/primer). The primers used for qRT-PCR experiment are listed in Supplementary Table S4.
Metabolite profiling by gas chromatograph time-of-flight mass spectrometry
Plant samples for metabolite profiling were withheld water for two weeks from the 15th day after sowing in the growth chamber, and then all seedlings were harvested and immediately frozen in liquid nitrogen. The experimental procedure for extract preparation was performed as described previously (Lisec et al., 2006). The total extracts were quantified by performing chromatography on GC-TOF MS (Agilent 7890A/5975C, USA). The detailed procedure has been described by Shi et al. (2014), with the data representing the mean values of three independent experimental repeats. The results were analysed using the Cluster 3.0 program (http://bonsai.hgc.jp/~mdehoon/software/cluster/) and visualized using Java Treeview (http://jtreeview.sourceforge.net/).
Transient expression assay
Mesophyll protoplasts were isolated from four-week-old Col-0 plants according to the methods described previously (Yoo et al., 2007). All of the plasmids used in this assay were prepared through purification with caesium chloride/ethidium bromide (Sambrook et al., 1989). For the luciferase assay, protoplasts were harvested after 12-h incubation under light conditions at 23ºC with or without stimuli (50 μM ABA). The activities of LUC and GUS were measured with the GloMax-Multi Jr Single Tube Multimode Reader (Promega, USA). All experiments were repeated at least three times.
Yeast two hybrid (Y2H) assay
The Y2H assay was carried out according to the instructions for the Matchmaker GAL4-based two-hybrid system (Clontech). The full-length sequence of MLP43 was cloned and inserted into pGADT and pGBKT, respectively. Another two homologous genes, MLP34 and MLP168, were cloned and inserted into pGBKT. We cloned ABI5, SnRK2.2, SnRK2.3, PYL1/2/5/9/13 and ABI1 and fused them into the pGADT vector, respectively. The genes of SnRK2.6, SnRK2.8, SnRK2.10, PYR1, ABF1 and ABF3 were cloned and fused into the pGBKT vector, respectively. The restriction sites and primer sequences for the plasmids used in this assay have been provided in Supplementary Tables S2 and S3, respectively.
Results
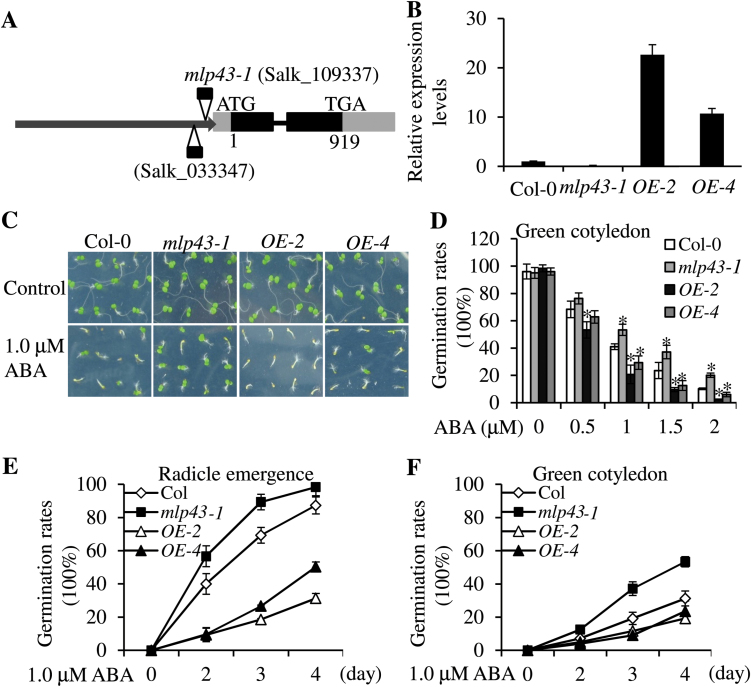
The mlp43 mutant was insensitive to ABA during seed germination
To discover other novel regulators and to expand ABA signalling networks during seed germination and abiotic drought stresses, we screened various T-DNA insertion mutants purchased from the ABRC (http://www.arabidopsis.org/) on MS medium containing 1.0 μM ABA during seed germination. The mutant mlp43-1 (Salk_109337) showing insensitivity to ABA was selected for further anlysis. T-DNA was inserted into the 5’-UTR of MLP43 (Fig. 1A). Transgenic plants of overexpressed MLP43 (OE-2 and OE-4) was generated by introducing Pro35S::MLP43-GFP plasmids into Col-0 plants. The relative expression level of MLP43 was verified by qRT-PCR (Fig. 1B). The expression levels of MLP43-GFP fusion protein was detected using anti-GFP antibody through western-blot assay (Supplementary Fig. S1A). In the absence of ABA, no obvious differences were observed in germination rates between Col-0 and the mlp43-1 mutant (Fig. 1C, D). However, the mlp43 mutant showed significantly decreased ABA sensitivity on the 1.0 μM ABA plate, while overexpressed MLP43 enhanced ABA sensitivity during seed germination (Fig. 1C, D). Moreover, the kinetics of germination time of Col-0, mlp43-1, and MLP43 OE were compared in the presence of 1.0 μM ABA by analysing the percentages of emerged radicles and open green cotyledons (Fig. 1E, F). On the 4th day after stratification, about 80% and 25% of Col-0 emerged with radicles and green cotyledons, respectively. However, the corresponding percentages of mlp43-1 were more than 95% and 50%. Overexpressed MLP43 transgenic plants showed much lower germination rates than Col-0 plants (Fig. 1E, F). We also examined the ABA sensitivity with another T-DNA insertion Salk line mlp43-2 (Salk_033347) in the seed germination assay. As indicated in Supplementary Fig. S1, this mutant line harbours almost null expression levels of MLP43 and was insensitive to ABA in seed germination. The ABA sensitivity of root growth was also examined after ABA treatment and the results indicated that no significant differences in primary root growth and lateral root number were observed among Col-0, mlp43-1, and MLP43 OE seedlings (Supplementary Fig. S2A, B). Taken together, MLP43 might function as a positive regulator in ABA response during seed germination.
Fig. 1.
MLP43 was involved in ABA responses in seed germination assay. Asterisk symbols (*) indicate P<0.05 (Student’s t-test). (A) Diagram showing of the T-DNA insertion site of mlp43-1 mutant. (B) Relative expression levels of MLP43 in the mlp43-1 mutant and overexpressed transgenic plants. Expression levels of β-ACTIN8 represent the internal control. Data represent the means ±SEs of three replicated experiments. (C) Seeds growing on MS medium with or without 1.0 μM ABA for 5 d after stratification. Both overexpressed transgenic plants (OE-2 and OE-4) showed a hypersensitive response to ABA. Photographs were taken to document the phenotypes. (D) Germination rates of green cotyledons with the indicated ABA application. Germination rates (%) were scored 5 d after stratification. Data represent means ±SEs of three replicated experiments (n>60 for each experiment). (E) Germination rates of radicle emergence in 1.0 μM ABA plates at the indicated time points. Data represent the means ±SEs of three replicated experiments (n>60 for each experiment). (F) Germination rates of green cotyledons in 1.0 μM ABA plates at the indicated time points. Data represent the means ±SEs of three replicated experiments (n>60 for each experiment). (This figure is available in colour at JXB online.)
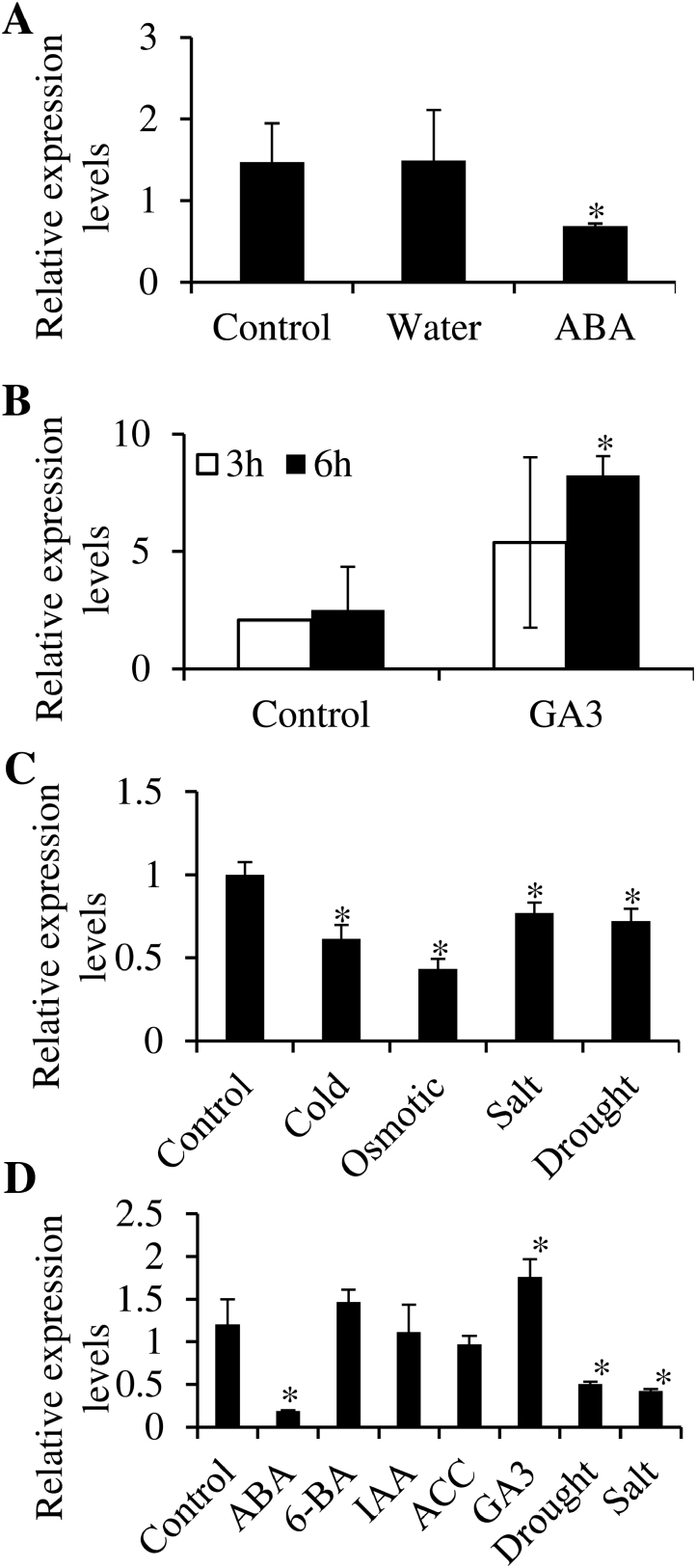
Modulation of MLP43 expression by ABA and abiotic stress treatments
Based on publicly available microarray data (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi; Winter et al., 2007), the MLP43 transcript was inhibited by ABA (30 μM) after 24h of treatment in seeds, but induced by gibberellic acid 3 (GA3, 5 μM) after 3 and 6h of treatment, respectively (Fig. 2A, B). The effects of abiotic stresses on MLP43 transcripts were further analysed. Interestingly, cold (4ºC), osmotic stress (300mM d-mannitol), salt (150mM NaCl) and drought (air dry) negatively regulated MLP43 expression (Fig. 2C). To further confirm the effects of phytohormones and abiotic stresses on MLP43 expression, qRT-PCR was applied to examine relative expression levels of MLP43. The results indicated that MLP43 was actually inhibited by ABA (50 μM, 3h), drought (water withheld for two weeks) and salt (NaCl, 100mM for 3h) treatment, respectively. No significant changes were observed after cytokinin (6-BA), auxin (IAA) or ethylene (ACC) treatment except that GA3 treatment slightly up-regulated MLP43 expression (Fig. 2D). These results showed that ABA and drought treatment decreased MLP43 transcription, which were consistent with the public microarray data.
Fig. 2.
ABA and abiotic stresses affected MLP43 expression. Data represent the means ±SEs of three replicated experiments. Asterisk symbols (*) indicate P<0.05 (Student’s t-test). (A) Publicly available microarray data illustrating the expression level of MLP43 was inhibited by 30 μM ABA treatment for 24h (http://www.bar.utoronto.ca/NASCArrays/index.php?ExpID=183), but was (B) induced by gibberellic acid 3 in seeds (GA3, 5 μM, treated for different time courses) (http://www.bar.utoronto.ca/NASCArrays/index.php?ExpID=184). (C) Publicly available microarray data illustrating the effect of abiotic stress treatment (cold, osmotic, salt and drought) on expression level of MLP43 (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi).
(D) Effects of hormones and abiotic stresses on MLP43 expression using qRT-PCR. ABA (10 μM, 3h), 6-BA (1 μM, 3h), IAA (5 μM, 3h), ACC (5 μM, 3h), GA3 (10 μM, 3h), drought (water withheld for two weeks), salt (NaCl, 100mM, 3h). The values of gene expression levels of Col-0 seedlings without any treatment were taken as ‘1’. Expression levels of β-ACTIN8 represent the internal control.
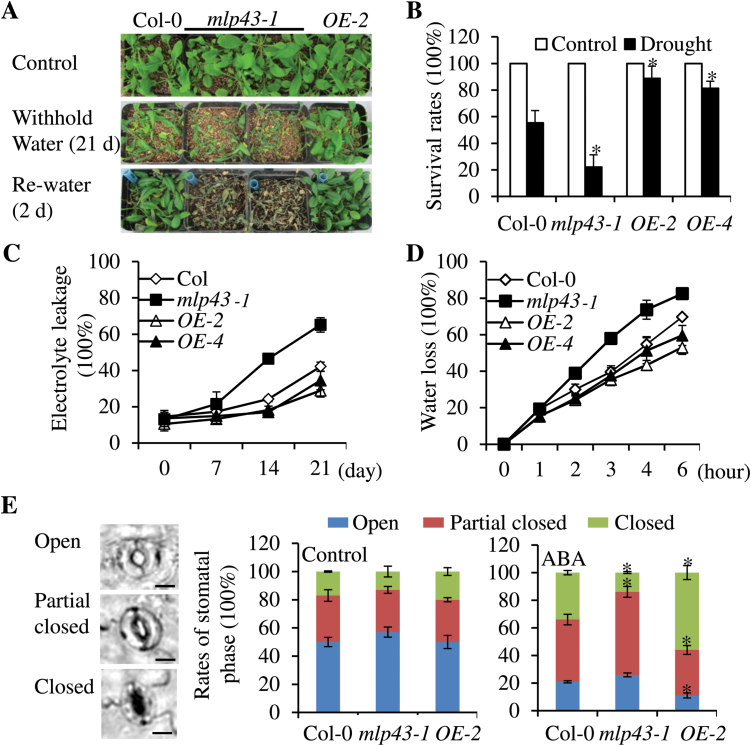
Overexpressed MLP43 confers enhanced drought tolerance
As the mlp43 mutant showed significantly decreased ABA sensitivity in seed germination, and drought treatment inhibited MLP43 expression (Figs 1, 2), the responses to drought stress of the mlp43-1 mutant and MLP43-overexpressed plants were further examined. Two-week-old plants grown under normal conditions were withheld water for three weeks, and then rewatered for 2 d. The results indicated that only 20% of mlp43-1 mutants but more than 80% of MLP43 OE plants recovered from wilting after rehydration, while the survival rate of Col-0 was about 58% (Fig. 3A, B). The EL test showed that the mlp43-1 mutant exhibited a significantly higher EL percentage than Col-0, but MLP43 OE plants showed a lower EL percentage relative to Col-0 (Fig. 3C). Transpirational water loss from detached leaves of 4-week-old plants were further compared at room temperature with a humidity of ~50–60%. Much higher water loss rates were detected in the mlp43-1 mutant compared with Col-0, while MLP43 OE plants showed lower water loss rates compared to Col-0 (Fig. 3D).
Fig. 3.
MLP43 involvement in drought stress responses. (A) Two-week-old plants were well watered (control) or deprived of water for 21 d and then rewatered. The photos were taken two days after rewatering. (B) Survival rates after the drought treatment in panel A. Data represent the means ±SEs of three replicated experiments (n>20 for each genotype, P<0.05). (C) Electrolyte leakage comparison at the indicated time points after drought treatment applied. Data represent the means ±SEs of three replicated experiments (n=15 for each genotype). (D) Transpirational water loss from well-watered detached leaves of four-week-old plants. Water loss rates are indicated as the percentage of the initial fresh weight (% FW). Results are shown as the means ±SEs of three replicated experiments (n=15 for each genotype). (E) Stomatal aperture under ABA treatment. The stomata phases are defined as shown in left panel. Scale bar, 5 μm. The rates of different stomata phases are calculated with or without ABA (30 μM) treatment for 30min. The data represent the means of four replicated experiments (n>50, P<0.05). (This figure is available in colour at JXB online.)
ABA-induced stomatal closure is usually responsible for plant adaptation to drought stress. We defined the stomatal phases as open, partially open and closed, according to the ratios of width to length of stomatal aperture (Fig. 3E). Under normal conditions, the rates of different stomatal phases were comparable among wildtype (Col-0), mlp43-1 and MLP43 OE. However, after ABA treatment for 1h, the rates of closed stomata were significantly increased in Col-0, especially in MLP43 OE. In contrast, the mlp43-1 mutant exhibited fewer closed stomata and more partially closed stomata than those in Col-0 and MLP43 OE (Fig. 3E). Together, these results suggested that the improved drought tolerance of MLP43 OE was associated with an increased sensitivity to ABA-induced stomatal closure in MLP43 OE. The complementary overexpressed transgenic plants generated by introducing Pro35S::MLP43-GFP into mlp43-1 were further assessed for drought resistance. The results indicated that the hypersensitivity of mlp43-1 mutant to drought stress could be restored by MLP43 overexpression (Supplementary Fig. S3).
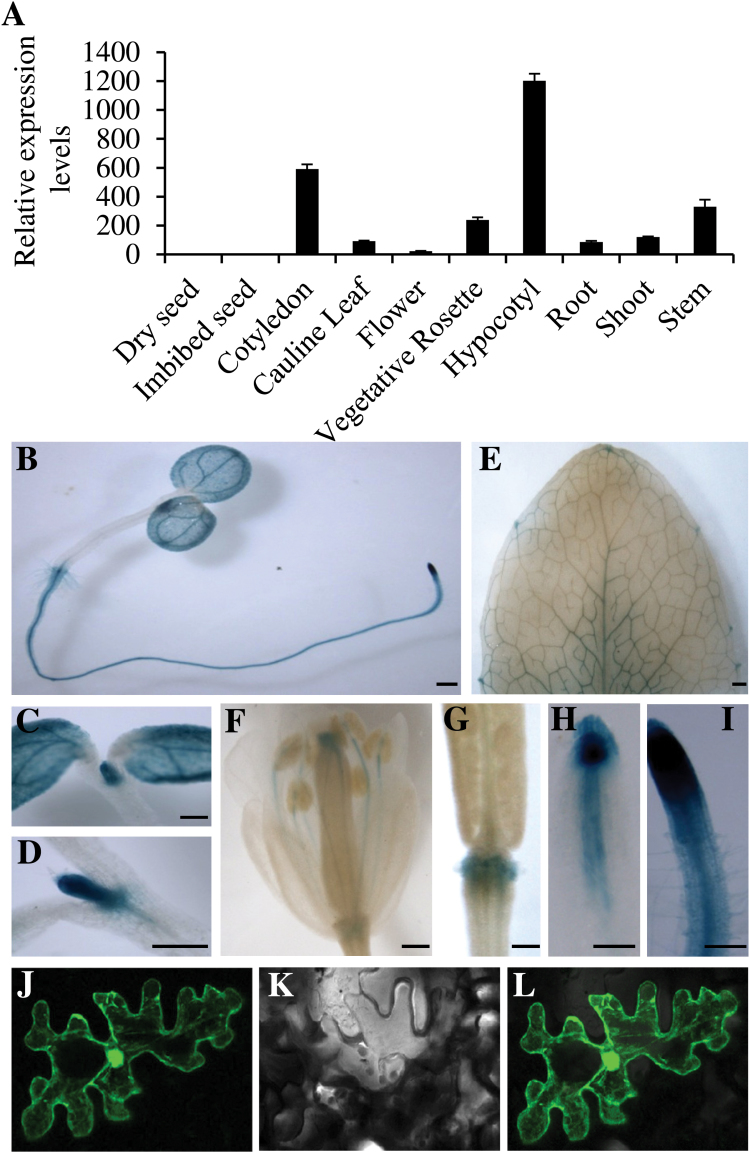
Tissue-specific expression patterns and subcellular localization of MLP43
Tissue-specific expression patterns of MLP43 were firstly analysed based on the microarray data available in public resources (Fig. 4A; http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi; Winter et al., 2007). The results indicated that MLP43 preferentially expressed in cotyledon and hypocotyls in Arabidopsis, while lower expression levels could be detected in roots, shoots, stems and vegetative rosette leaves (Fig. 4A). Almost no expression patterns could be detected either in dry seed or in imbibed seeds (Fig. 4A). To further confirm the specific expression patterns of MLP43 in Arabidopsis, the promoter sequence of MLP43 (−1769 bps upstream of start code ATG) was cloned and a transgenic plant carrying plasmid ProMLP43::GUS was generated. GUS histochemical signals were analysed in various tissues (Fig. 4B–I). There were predominant GUS signals in cotyledons, primary roots and apical meristems (Fig. 4B–D, H, I). GUS signals also could be detected in rosette leaves, flowers and the abscission zone (Fig. 4E–G) whereas no GUS signals had been examined in hypocotyls, which was inconsistent with the microarray data. We transiently expressed the Pro35S::MLP43-GFP fusion construct into epidermal cells of Arabidopsis rosette leaves by bombardment and observed the subcellular localization of the MLP43-GFP fusion protein under a confocal fluorescence microscope. The results indicated that MLP43 were localized in nucleus and cytoplasm in Arabidopsis epidermal cells (Fig. 4J–L).
Fig. 4.
Tissue-specific expression patterns of MLP43. (A) Publicly available microarray data illustrating the tissue specific expression levels of MLP43 (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi). (B)–(I) Analysis of tissue-specific expression patterns by GUS histochemical staining. Scale bar, 0.5mm. (J)–(L) Subcellular localization of MLP43-GFP fusion protein drived by the 35S promoter. Bar, 10 μM. Photos taken under cofocal microscope J, GFP; K, bright field; L, overlay. (This figure is available in colour at JXB online.)
Mutation of MLP43 increased ROS production and modulated primary metabolites after drought treatment
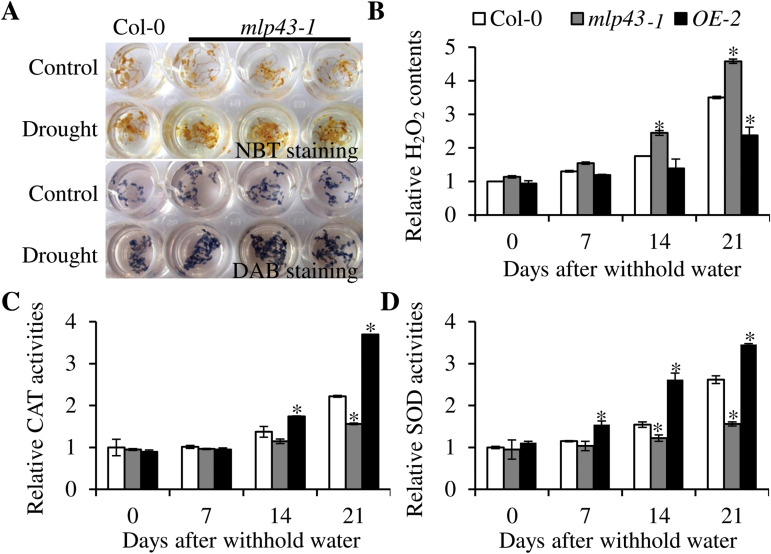
ABA and drought stress trigger ROS accumulation, and a ROS detoxification mechanism is necessary to enable plant survival under drought stress (Smirnoff, 1993). To characterize whether ROS accumulation was altered in the mlp43-1 mutant, we determined H2O2 and O− production by DAB and NBT staining, respectively. The mlp43-1 mutant exhibited a dark colour relative to Col-0 with both DAB and NBT staining after air drought stress treatment, indicating more accumulation of H2O2 and O− in the mlp43-1 mutant (Fig. 5A). We also measured the H2O2 content in the young seedlings, and found that drought stress induced rapid accumulation of H2O2 after 14 d of drought treatment in all genotypes. However, the H2O2 level was significantly higher in the mlp43-1 mutant plants, but lower in MLP43 OE when compared with Col-0 plants (Fig. 5B). Next, the enzymatic activities of SOD and CAT were measured under the same drought conditions. The results indicated that no significant differences of CAT activity were observed among three genotypes after drought treatment for 7 d, however, SOD activities were 1.5-fold higher in MLP43 OE plants than wildtype (Col-0) and the mlp43-1 mutant, respectively (Fig. 5C, D). Moreover, both SOD and CAT activities showed a significant decrease in mlp43-1 and increase in MLP43 OE relative to Col-0 plants after drought treatment for 14 d and 21 d, respectively.
Fig. 5.
MLP43 enhanced drought stress tolerance through inhibiting ROS production and promoting antioxidant enzyme activities. (A) Visualization of superoxide radical and hydrogen peroxide detected by NBT and DAB staining using two-week-old MS-grown plants subjected to subsequent treatment with or without air dry for 30min. Comparison of (B) H2O2 content, (C) CAT and (D) SOD activity in Col-0, mlp43 mutant and overexpressed transgenic plant MLP43 OE. Two-week-old plants were well watered and then withheld water for the indicated number of days. The relative contents of H2O2 and the activity of CAT and SOD were calculated as fold-changes compared with the Col-0 control. Data represent the means ±SEs of three replicated experiments (n=10; P<0.01). (This figure is available in colour at JXB online.)
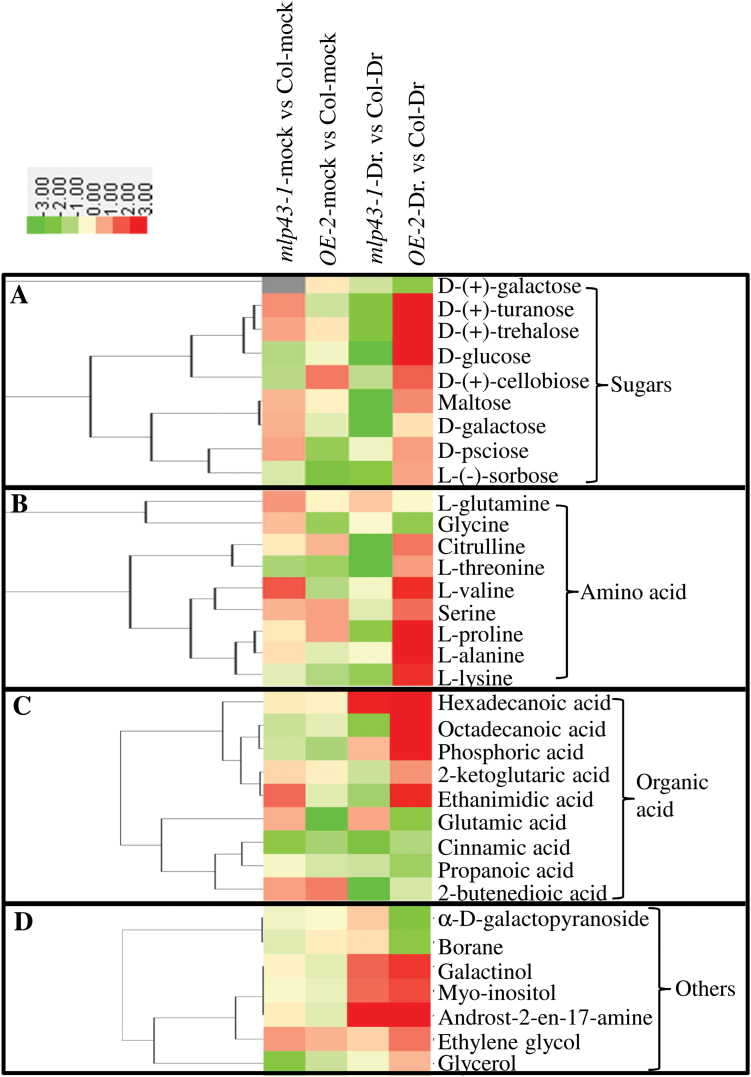
To elucidate the metabolic responses of MLP43 to drought stress treatment, GC-TOF-MS was applied to test whether the primary metabolite profiling was modified by MLP43 overexpression. The results indicated that 34 metabolites including 9 carbohydrates, 9 amino acids, 9 organic acids and 7 other derivatives were reproducibly identified (Fig. 6; Supplementary Table S5). Compared with wildtype (Col-0), most of the detected metabolites showed <2-fold differences in MLP43 ectopic expression lines without drought treatment. However, the contents of most of the examined carbohydrates and amino acids were significantly decreased in the mlp43-1 mutant, but increased in MLP43 OE transgenic plants after drought treatment (Fig. 6). Except D-(+)-galactose, all carbohydrates, including maltose, sorbose, cellobiose, turanose, glucose, psicose and galactose, decreased in the mlp43-1 mutant but increased in MLP43 OE when compared to Col-0 under the withheld water condition (Fig. 6). A similar change was observed for citrulline, threonine, valine, serine, proline, alanine and lysine, with the exception of glutamine and glycine (Fig. 6). In addition, hexadecanoic acid, galactinol, myo-inositol and androst-2-en-17-amine increased, while cinnamic acid, propanoic acid and 2-butenedioic acid decreased either in mlp43-1 mutants or in MLP43 transgenic plants under drought treatment conditions.
Fig. 6.
Comparisons of metabolite profiling under control and drought treatment in Col-0, mlp43-1 and MLP43 OE plants. Thirty-four compounds that could reproducibly be detected by GC-TOF MS in all three genotypic plants were analyzed by hierarchical clustering. The resulting tree was analyzed using CLUSTER 3.0 and shown using Java Treeview. (This figure is available in colour at JXB online.)
Transcriptional alterations of ABA- and drought-responsive genes by MLP43
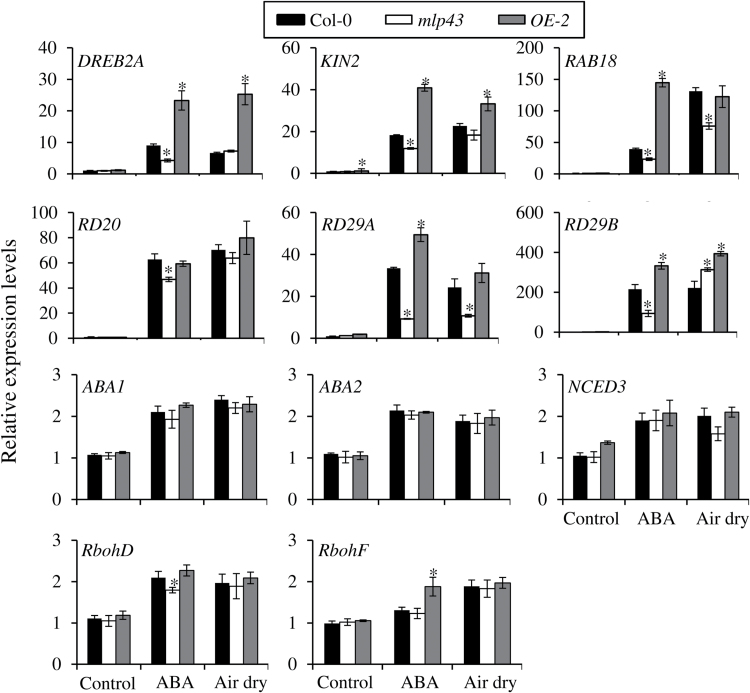
To further determine the role of MLP43 in the ABA signalling pathway and drought stress responses, we assessed the expression patterns of genes involved in ABA- and drought-responsive processes. Two-week-old seedlings were pretreated with 50 μM ABA for 3h or air dried for 6h, respectively, after which gene expression levels were analysed by qRT-PCR. We compared the expression levels of several ABA- and/or abiotic stress-responsive genes, including DREB2A, KIN2, RAB18, RD20, RD29A and RD29B. All of them showed extensively increased transcriptional levels after ABA and air-dry treatments in all three genotypes (Fig. 7). However, the fold changes of DREB2A, KIN2, RAB18 and RD29A were significantly lower in the mlp43-1 mutant, but higher in the MLP43 OE plants compared to those in Col-0. ABA treatment significantly enhanced the transcription of above ABA- and drought-responsive genes in MLP43 OE plants, which was consistent with the ABA hypersensitivity of MLP43 OE in seed germination and stomatal aperture responses. Dehydration stress usually promotes ABA production and actives ABA signal transduction (Chan, 2012), and our results substantiate this, with ABA and air-dry treatments inducing similar expression patterns of the above detected genes (Fig. 6). In addition, we also examined expression level changes of three ABA biosynthesis genes, ABA1, ABA2 and NCED3. The results indicated that there were no significant differences among the three genotypes after ABA or air-dry treatments. As MLP43 modulated ROS production, we also examined the relative expression levels of RbohD and RbohE encoding NADPH oxidases which promote ROS production especially after ABA and drought treatment (Torres et al., 2002; Miller et al., 2010). Results revealed no significant differences except that RbohE showed a relatively higher expression level in the presence of ABA (Fig. 7, last panel). Based on the above results, we concluded that the overexpression of MLP43 enhanced plant drought stress adaptation through an ABA-dependent manner.
Fig. 7.
Relative expression levels of drought- and ABA-responsive genes in mlp43-1 mutant and MLP43 OE plants. The two-week-old plants were subjected to ABA treatment (50 μM) for 3h and air dried for 6h, respectively. The expression levels of the selected drought- and ABA-responsive genes were analysed quantitatively by qRT-PCR. Gene expression levels were analysed relative to the corresponding gene expression levels in Col-0 plants. The values of gene expression levels of Col-0 seedlings without ABA treatment were taken as ‘1’. Expression levels of β-ACTIN8 represent the internal control. The data represent means ±SEs of three reproducible experiments (P<0.01).
MLP43 positively modulated ABA responses in reconstituted ABA signalling pathway
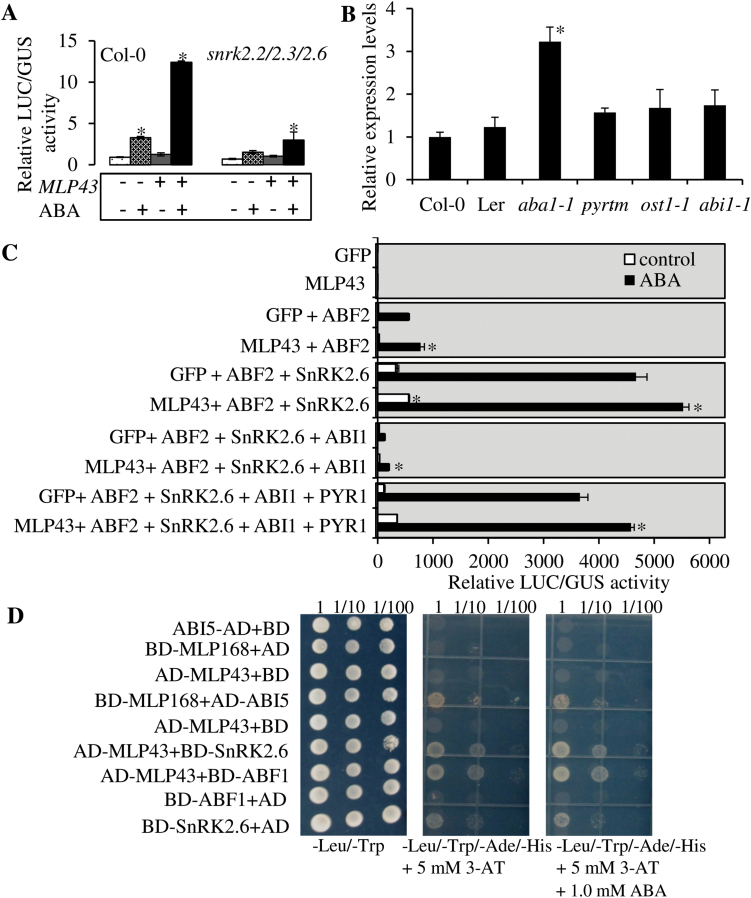
To elucidate how MLP43 modulated the expression of downstream targets of the ABA signalling pathway, we performed a transient expression assay in Arabidopsis mesophyll protoplasts extracted from Col-0 and snrk2.2/2.3/2.6, respectively, using ProRD29A::LUC as reporter and ProUBQ::GUS as internal control, and Pro35S::GFP as empty vector control. The results indicated that transient overexpression of MLP43 significantly enhanced the transcriptional activity of RD29A promoter after ABA treatment in Col-0 but not in snrk2.2/2.3/2.6 (Fig. 8A). We then checked expression patterns of MLP43 in mutants of primary ABA signalling components, including the pyr1/pyl1/pyl4 triple mutant (pyrtm), ost1-1, abi1-1 and the ABA-deficient mutant aba1-1. When compared with wildtype (Col-0 and Ler), the expression level of MLP43 was up-regulated in all the examined mutants, especially in the ABA-deficient mutant aba1-1 with ~3-fold increases at the transcriptional level (Fig. 8B). In order to gain more clues about the effect of MLP43 in the ABA signalling pathway, we reconstituted the ABA signalling pathway in wildtype (Col-0) protoplasts (Fig. 8C). As expected, co-transformation of ABF2 and SnRK2.6 could extensively activate RD29A::LUC activity and PYR1 inhibited ABI1 activity and then enabled expression of the ABA-dependent transcription of RD29A::LUC (Fig. 8C). However, in the presence of MLP43 overexpression, the activity of RD29A::LUC significantly increased, especially after ABA treatment when compared with empty vector control (Fig. 8C).
Fig. 8.
MLP43 functioned as a positive regulator in the reconstituted ABA signalling pathway. The data represent means ±SEs of five reproducible experiments (P<0.01). (A) Relative activities (LUC/GUS) in co-transformation with empty vector (Pro35S::GFP) or MLP43 (Pro35S::MLP43-GFP) in Col-0 and snrk2.2/2.3/2.6 with or without ABA (50 μM) treatment for 12h, respectively. (B) Relative expression levels of MLP43 in the primary ABA signal components related gene mutants using qRT-PCR. The value of gene expression levels of Col-0 seedlings was taken as ‘1’. Expression levels of β-ACTIN8 represent the internal control. (C) Relative activities (LUC/GUS) in the reconstituted ABA signal pathway. Co-transformation of MLP43 enhanced the relative activities (LUC/GUS). ABF2, SnRK2.6, ABI1 and PYR1 were co-transformed as effectors, respectively. (D) Interactions between MLP43 and SnRK2.6 or ABF1 using the yeast two-hybrid assay. The homologous gene MLP168 could interact with wildtype ABI5 in yeast.
We further examined the interactions between MLP43 and the key ABA signal components. First, the phylogenetic relationships among nine MLP homologous genes were analysed based on the full length amino acid sequences (Supplementary Fig. S4). Another two MLPs (MLP34 and MLP168), which shared differencial homology with MLP43, were selected for the yeast two-hybrid assay together with MLP43. The results indicated that MLP43 could interact with SnRK2.6 and ABF1 in the yeast two-hybrid assay independent of ABA (Fig. 8D; Supplementary Fig. S5). However, we did not detect any interactions between MLP34 or MLP168 and the key ABA signal components, except the interaction between MLP168 and wildtype ABI5 protein (Supplementary Fig. S5A). Interestingly, mutated proteins carrying single or triple phosphoamino acid mutations (S42A, S145A and T201A) in ABI5 could eliminate its interactions with MLP168 (Supplementary Fig. S5A). Taken together, the above results indicated that MLP43 might function as a positive regulator through interacting with SnRK2.6 and ABFs in ABA signalling responses, and ABA could negatively regulate the expression of MLP43 as a complementary mechanism to modulate the function of MLP43 in vivo.
Discussion
In this study, the function of MLP43 was characterized during plant stress responses. Both MLP43 and ABA receptors PYL/PYR belong to the Bet v 1 family and share similar protein structure. We speculated that MLP43 might be involved in the ABA signalling pathway. The efficiency of water loss is responsible for plant tolerance to drought stress, and rapid water loss leads to greater drought sensitivity. As indicated, the rosette leaf water loss of the mlp43-1 mutant was significantly more rapid than wildtype (Col-0) under the same drought stress conditions, resulting in higher electrolyte leakage and lower survival rates (Fig. 3). The above results were consistent with ABA insensitivity of mlp43 in seed germination. As we know, ABA and GA3 antagonistically regulate seed germination (Lee et al., 2002; Piskurewicz et al., 2008). Interestingly, MLP43 expression was significantly inhibited by ABA, but promoted by GA3 (Fig. 2A). Moreover, drought and salt treatments also negatively regulated MLP43 expression. We also analysed the 1769bp promoter sequence of MLP43 and found two ACGT-containing ABRE-like elements in the promoter region, located at −1291bp to −1294bp and −1088bp to −1092bp, respectively. Interestingly, a functional G-box (CACGTG) was also identified in the MLP43 promoter, located at −603bp to −608bp close to the ATG start codon of MLP43. The occurrence of both an ABRE and G-box motif in the MLP43 promoter is consistent with previous studies that indicated that ABRE/G-box elements were usually co-recognized by ABA responsive transcription factors (Menkens et al., 1995; Shen and Ho, 1995; Ho et al., 1999).
To further elucidate the mechanism of MLP43 in mediating ABA and drought responses, the comparisons of expression levels of ABA- and drought-responsive genes were performed. As expected, the expression levels of DREB2A, KIN2, RAB18, RD29A and RD29B were up-regulated by ABA treatment, but showed lower expression levels in mlp43-1 and higher expression levels in MLP43 OE when compared with those in Col-0 after ABA treatment. These results indicated that MLP43 enhanced ABA signal transduction and drought stress tolerance through modulating downstream targets of ABA and drought stress responses. In addition, expression levels of ABA biosynthesis-related genes, such as ABA1, ABA2 and NCED3 showed no significant changes in mlp43 and MLP43 OE lines indicating that MLP43 modulated ABA responses independent of the ABA biosynthesis pathway. However, MLP43 expression was up-regulated in the ABA receptor triple mutant pyr1/ pyl1/pyl4, as well as in ost1-1, abi1-1 (dominant negative mutant) and aba1-1 mutants, when compared with those in wildtype Col-0 and Ler (Fig. 8B). In other words, disturbance of ABA signalling transduction or ABA biosynthesis blocked the inhibitory effect of ABA on MLP43 transcription, indicating that the above key ABA signal components are important inter-mediators for the negative regulation of ABA on MLP43 expression. Moreover, the re-constituted ABA signal pathway confirmed that transient over-expression of MLP43 in protoplasts enhanced ABA responses by determining the reporter RD29A::LUC activity (Fig. 8C). However, mutation in SnRK2.2/2.3/2.6 eliminated the positive effect of MLP43 on ABA responses in the transient expression assay, which provides evidence that MLP43 functions up-stream of SnRK2s in the ABA signal pathway (Fig. 8A).
Furthermore, the interactions between MLP43 and SnRK2.6 or ABFs could be detected in the yeast two-hybrid assay (Fig. 8D; Supplementary Fig. S5), which provided direct evidence for the involvement of MLP43 in the ABA signalling pathway. In the core ABA signalling pathway, PYL/PYRs interact with PP2Cs in the present of ABA to form PYL/PYR-PP2C complexes, which in turn inhibits the activity of the PP2Cs in an ABA-dependent manner, allowing activation of SnRK2s (Ma et al., 2009; Park et al., 2009). To date, no direct interactions between ABA receptors PYL/PYRs and SnRK2s have been reported. Therefore, even though MLPs and the ABA receptor RCAR/PYR/PYL family of START proteins share amino acid and structural similarity at the protein level, MLP43 does not interact with ABI1, a key negative regulator of PP2Cs in the ABA signalling pathway. We speculated that MLPs might function as positive ABA regulators through direct regulation of downstream SnRK2s or ABF activities. Solid evidence through the yeast two-hybrid assay verified that MLP43 interacts with SnRK2.6 and ABF1 (Fig. 8D). However, the MLP homologous genes might be functionally diverse in their ABA responses as there were no interactions between MLP34/MLP168 and other ABA signalling components, except that MLP168 could interact with ABI5 in yeast (Supplementary Fig. 5). Presence or absence of ABA had no different effect on the interactions between MLPs and SnRK2/ABFs, indicating the interaction between MLP43 and SnRK2.6 or ABF1 is ABA independent. Therefore, our study provides experimental evidence to elucidate how MLP43 functions as a positive regulator in ABA signalling and drought stress responses.
Accumulating evidence indicates that ABA-enhanced water stress tolerance is associated with induction of antioxidant defence systems, including ROS-scavenging enzymes such as SOD, CAT and APX (Mittler et al., 2011). ROS are small molecules generated during development and in response to stress, and function as eukaryotic intracellular second messengers (Finkel, 1998; Mittler et al., 2011). In Arabidopsis, AtrbohD and AtrbohE encode NADPH oxidases which catalyse NADPH into NADP and ROS especially after ABA and drought treatment (Torres et al., 2002; Kwak et al., 2003; Miller et al., 2010). Our results showed that mutation of MLP43 increased ROS accumulation under drought stress conditions (Fig. 5A), indicating the involvement of MLP43 in ROS-mediated drought responses. However, MLP43 did not modulate RbohD and RbohF transcript levels according to the qRT-PCR results, which indicated that MLP43 preferably regulated ROS scavenging but not production (Fig. 7). The further accumulation of excessive ROS causes more severe oxidative damage to plant cells and stimulates antioxidase activities (Mittler, 2002). SOD and CAT are two important antioxidases that scavenge excessive ROS to prevent cell damage. We determined significant increases of both SOD and CAT activities in MLP43-overexpressed transgenic plants (Fig. 5C, D), which suggests that MLP43 played a role in scavenging the excessive ROS production under dehydration to enhance drought tolerance.
During plant abiotic stress responses, compatible solutes are important in helping plants balance external osmotic pressure and maintain cellular function of macromolecules. In this study, we further examined primary metabolic profiles using GC-TOF MS. The results indicated several carbohydrates and amino acids accumulated in MLP43 plants (Fig. 6). The increased amounts of alanine and glutamic acid in MLP43 OE plants during drought stress treatment might regulate photosynthesis intensity (Bocian et al., 2015). Moreover, overexpression of MLP43 resulted in accumulation of free proline, maltose, trehalose and glucose, which in turn contributed to conferred drought tolerance (Garg et al., 2002). Galactinol has a positive effect in protecting plants from drought stress and oxidative damage (Taji et al., 2002; Nishizawa et al., 2008). In our study, galactinol showed significant increases after drought treatment, especially in MLP43 OE plants (Fig. 6). Moreover, overexpressed GhMLP led to a two-fold increase in flavonoid contents, which suggested that MLP might be involved in flavonoid metaolism (Chen and Dai, 2010). All these results indicated that MLP43 modulated primary and secondary metabolic profiles, which might be contributed to increased abiotic stress tolerance.
In this study, we dissected the functions of MLP43 in ABA signals and drought stress responses. First, ABA and drought stress treatments inhibited the expression of MLP43, but this inhibition was disturbed in the primary ABA signal components mutants. Second, MLP43 overexpression enhanced ABA responses in seed germination and improved drought stress tolerance by modulating ABA- and/or drought-responsive gene expression, ROS homeostasis and primary metabolic profiling. Third, MLP43 promoted RD29A::LUC activity in the reconstituted ABA signalling pathway, and that was dependent upon SnRK2.2/2.3/2.6. Finally, there were direct interactions between MLP43 and SnRK2.6 or ABF1 in the yeast two-hybrid assay. Collectively, we concluded that MLP43 functioned as a positive regulator upstream of SnRK2s, and ABA down-regulated MLP43 expression as a negative feedback loop, so regulating drought stress responses.
Supplementary data
Supplementary data is available at JXB online.
Supplementary Fig. S1. Phenotypic analysis of another MLP43 T-DNA insertion line mlp43-2 (Salk_033347).
Supplementary Fig. S2. Comparison of primary root length and lateral roots number among Col-0, mlp43-1, MLP OE plants after ABA treatment.
Supplementary Fig. S3. Complementary over-expression of MLP43 into the mlp43-1 mutant (Com-3) showed drought-tolerance in soil.
Supplementary Fig. S4. Phylogenetic relationships of nine AtMLP genes.
Supplementary Fig. S5. Yeast two-hybrid assay to detect the interactions between MLP43/MLP34/MLP168 and the key components in the ABA signalling pathway.
Supplementary Table S1. The primers used for identification of the mlp43 mutant (Salk_109337 and Salk_033347).
Supplementary Table S2. Detailed information of plasmid construction in this study.
Supplementary Table S3. The primers used for plasmid construction in this study.
Supplementary Table S4. The primers used for real-time quantitative RT-PCR (qRT-PCR) in this study.
Supplementary Table S5. The detailed list of alterations in metabolite profiling under control and drought treatment in Col-0, mlp43-1 and MLP OE plants.
Acknowledgments
The mlp43 mutant was provided by the Arabidopsis Biological Resource Center (ABRC). We thank Dr Jian-Kang Zhu (Purdue University) for kindly providing ost1-1, abi1-1, aba1, pyr1/pyl1/2/4, and snrk2/3/6 mutant seeds. We thank Dr Pingfang Yang for help with analysis by qRT-PCR. This research was supported by the National Natural Science Foundation of China (grant no. 31370302), the ‘Hundred Talents Program’ (54Y154761O01076 and 29Y329631O0263) (ZC), and by the National Natural Science Foundation of China (grant no. 31300233) (YW). We are grateful to the editor and reviewers for their comments and suggestions.
Glossary
Abbreviations:
- ABA
abscisic acid
- ABRE
ABA-responsive element
- APX
ascorbateperoxidase
- CAT
catalase
- DAB
3,3’-diaminobenzidine
- EL
electrolyte leakage
- GA3
gibberellic acid 3
- GC-TOF MS
gas chromatograph time-of-flight mass spectrometry
- MLP43
major latex protein-like 43
- MS
Murashige and Skoog
- NBT
nitoblue terazolium chloride
- PR10
pathogenesis-related 10
- qRT-PCR
real-time quantitative RT-PCR
- ROS
reactive oxygen species
- SnRK2
SUCROSE NONFERMENTING1-RELATED SUBFAMILY2
- SOD
superoxide dismutase
- START
star-related lipid-transfer domain.
References
- Aggelis A, John I, Karvouni Z, Grierson D. 1997. Characterization of two cDNA clones for mRNAs expressed during ripening of melon (Cucumis melo L) fruits. Plant Molecular Biology 33, 313–322. [DOI] [PubMed] [Google Scholar]
- Bocian A, Zwierzykowski Z, Rapacz M, Koczyk G, Ciesiolka D, Kosmala A. 2015. Metabolite profiling during cold acclimation of Lolium perenne genotypes distinct in the level of frost tolerance. Journal of Applied Genetics . Doi 10. 1007/s13353-015-0293-6. [DOI] [PubMed] [Google Scholar]
- Chan Z. 2012. Expression profiling of ABA pathway transcripts indicates crosstalk between abiotic and biotic stress responses in Arabidopsis . Genomics 100, 110–115. [DOI] [PubMed] [Google Scholar]
- Chen J-Y, Dai X-F. 2010. Cloning and characterization of the Gossypium hirutum major latex protein gene and functional analysis in Arabidopsis thaliana . Planta 231, 861–873. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agro-bacterium-mediated transformation of Arabidopsis thaliana . The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. 2010. Abscisic acid: emergence of a core signaling network. Annual Review of Plant Biology 61, 651–679. [DOI] [PubMed] [Google Scholar]
- Finkel T. 1998. Oxygen radicals and signaling. Current Opinion in Cell Biology 10, 248–253. [DOI] [PubMed] [Google Scholar]
- Garg AK, Kim JK, Owens TG, Ranwala AP, Do Choi Y, Kochian LV, Wu RJ. 2002. Trehalsoe accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proceedings of the National Academy of Science, USA 99, 15898–15903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D, Wong WS, Xu WZ, Sun FF, Qing DJ, Li N. 2011. Cis-cinnamic acid-enhanced 1 gene plays a role in regulation of Arabidopsis bolting. Plant Molecular Biology 75, 481–495. [DOI] [PubMed] [Google Scholar]
- Ho T, Asada M, Kowyama Y, Hattori T. 1999. ACGT-containing abscisic acid response element (ABRE) and coupling element 3 (CE3) are functionally equivalent. The Plant Journal 19, 679–689. [DOI] [PubMed] [Google Scholar]
- Hu L, Li H, Pang H, Fu J. 2012. Responses of antioxidant gene, protein and enzymes to salinity stress in two genotypes of perennial ryegrass (Lolium perenne) differing in salt tolerance. Journal of Plant Physiology 169, 149–156. [DOI] [PubMed] [Google Scholar]
- Hubbard KE, Nishimura N, Hitomi K, Getzoff ED, Schroeder JI. 2010. Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes & Development 24, 1695–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inui H, Sawada M, Goto J, Yamazaki K, Kodama N, Tsuruta H. 2013. A major latex-like protein is a key factor in crop contamination by persistent organic pollutants. Plant Physiology 161, 2128–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. The EMBO Journal 20, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Yu Y, Snesrud EC, Moy LP, Linford LD, Haas BJ. 2005. Transcriptional divergence of the duplicated oxidative stress-responsive genes in the Arabidopsis genome. The Plant Journal 41, 212–220. [DOI] [PubMed] [Google Scholar]
- Kim TH, Bohmer M, Hu H, Nishimura N, Schroeder JI. 2010. Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annual Review of Plant Biology 61, 561–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koistinen KM, Soininen P, Venӓlӓinen TA, Hӓyrinen J, Laatikainen R, Perӓkylӓ M, Tervahauta AI, Kӓrenlampi SO. 2005. Birch PR-10c interacts with several biologically important ligands. Phytochemistry 66, 2524–2533. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Jorna ML, Brinkhorst-van der Swan DLC, Karssen CM. 1982. The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellins sensitive lines of Arabidopsis thaliana (L.) Heynh. Theoretical and Applied Genetics 61, 385–393. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Reuling G, Karssen CM. 1984. The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana . Physiologia Plantarum 61, 377–383. [Google Scholar]
- Kwak J, Mori IC, Pei ZM, Leonhardt N, Torres MA, Dang JL, Bloom RE, Bodde S, Jones JD, Schroeder JI. 2003. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis . The EMBO Journal 22, 2623–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Cheng H, King KE, Wang W, He Y, Hussain A, Lo J, Harberd NP, Peng J. 2002. Gibberellin regulates Arabidopsis seed germination via RGL2, a GA/RGA-like gene whose expression is up-regulated following imbibitions. Genes & Development 16, 646–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Luan S. 2012. ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant, Cell & Environment 35, 53–60. [DOI] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. 2006. Gas chromatography mass spectrometry-based metabolite profiling in plants. Nature Protocol 1, 387–396. [DOI] [PubMed] [Google Scholar]
- Lu Y, Chen X, Wu Y, Wang Y, He Y, Wu Y. 2013. Directly transforming PCR-amplified DNA fragment into plant cells is a versatile system that facilitates the transient expression assay. Plos ONE 8, e57171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. 2009. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. [DOI] [PubMed] [Google Scholar]
- Menkens AE, Schindler U, Casshmore AR. 1995. The G-box: a ubiquitous regulatory DNA elements in plants bound by the CBF family of bZIP proteins. Trends in Biochemistry Science 20, 506–510. [DOI] [PubMed] [Google Scholar]
- Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R. 2010. Reactive oxygen species homeostasis and signaling during drought and salinity stresses. Plant Cell & Environment 33, 453–467. [DOI] [PubMed] [Google Scholar]
- Mittler R. 2002. Oxidative stress, antioxidants and stress tolerance. Trends in Plant Science 7, 405–410. [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, Van Breusegem F. 2011. ROS signaling: the new wave? Trends in Plant Science 16, 300–309. [DOI] [PubMed] [Google Scholar]
- Mustilli A-C, Merlot S, Vavasseur A, Fenzi F, Giraudat J. 2002. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14, 3089–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nessler CL, Burnett RJ. 1992. Organization of the major latex protein gene family in opium poppy. Plant Molecular Biology 20, 749–752. [DOI] [PubMed] [Google Scholar]
- Nessler CL, Kurz WGW, Pelcher LE. 1990. Isolation and analysis of the major latex protein genes of opium poppy. Plant Molecular Biology 15, 951–953. [DOI] [PubMed] [Google Scholar]
- Nishizawa A, Yabuta Y, Shigeoka S. 2008. Galatinol and raffinose constitute a novel function to protect plants from oxidase damage. Plant Physiology 147, 1251–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H. 2009. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START protein. Science 324, 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg Z, Blumwald E. 2011. Hormone balance and abiotic stress tolerance in crop plants. Current Opinion in Plant Biology 14, 290–295. [DOI] [PubMed] [Google Scholar]
- Phee BK, Park S, Cho JH, Jeon JS, Bhoo SH, Hahn TR. 2007. Comparative proteomic analysis of blue light signaling components in the Arabidopsis crytochrome 1 mutant. Molecular Cells 23, 154–160. [PubMed] [Google Scholar]
- Piskurewicz U, Jikumaru Y, Kinoshita N, Nambara E, Kamiya Y, Lopez-Molina L. 2008. The gibberellic acid singaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 20, 2729–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radauer C, Lackner P, Breiteneder H. 2008. The Bet v 1 fold: an ancient, versatile scaffold for binding of large, hydrophobic ligands. BMC Evolutionary Biology 8, 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel F, Sulmon C, Bogard M, Gouesbet G. 2009. Differential patterns of reactive oxygen species and antioxidative mechanisms during atrazine injury and surose-induced tolerance in Arabidopsis thaliana plantlets. BMC Plant Biology 9, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual , 2nd edn Cold Spring Harbor: Cold Spring Harbor Labortory Press. [Google Scholar]
- Shen Q, Ho T. 1995. Functional dissection of an abscisic acid (ABA)-inducible gene reveals two independent ABA-responsive complexes each containing a G-box and a novel cis-acting element. Plant Cell 7, 295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Ye T, Zhong B, Liu X, Chan Z. 2014. Comparative proteomic and metabolomic analyses reveal mechanisms of improved cold stress tolerance in bermudagrass (Cynodon dactylon (L). Pers.) by exogenous calcium. Journal of Integrative Plant Biology 56, 1064–1079. [DOI] [PubMed] [Google Scholar]
- Smirnoff N. 1993. The roles of active oxygen in the response of plants to water deficit and desiccation. New Phytologist 125, 27–58. [DOI] [PubMed] [Google Scholar]
- Taji T, Ohsumi C, Luchi S, Seki M, Kasuga M, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K. 2002. Important roles of drought- and cold- inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana . The Plant Journal 29, 417–426. [DOI] [PubMed] [Google Scholar]
- Torres MA, Dang JL, Jones JD. 2002. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proceedings of the National Academy of Science, USA 8, 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li L, Ye T, Lu Y, Chen X, Wu Y. 2013. The inhibitory effect of ABA on floral transition is mediated by ABI5 in Arabidopsis . Journal of Experimental Botany 64, 675–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yang L, Zheng Z, Grumet R, Loescher W, Zhu J-K, Yang P, Hu Y, Chan Z. 2013. Transcriptomic and physiological variations of three Arabidopsis ecotypes in response to salt stress. Plos ONE . 8, e69036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. 2007. An ‘electronic fluorescent pictograph’ browser for exploring and analyzing large-scale biological data sets. PloS ONE 2, e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FZ, Lu TC, Shen Z, Wang BC, Wang HX. 2008. N-terminal acetylation of two major latex proteins from Arabidopsis thaliana using electrospray ionization tandem mass spectrometry. Plant Molecular Biology Reporter 26, 88–97. [Google Scholar]
- Yoo SD, Cho YH, Sheen J. 2007. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Natural Protocol 2, 1565–1572. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.