Highlight
Strigolactones monitor lateral root development in a spatiotemporal manner by an interplay with cytokinin.
Key words: Arabidopsis thaliana, cytokinin signaling, lateral root development, polar auxin transport, strigolactones.
Abstract
Strigolactones are important rhizosphere signals that act as phytohormones and have multiple functions, including modulation of lateral root (LR) development. Here, we show that treatment with the strigolactone analog GR24 did not affect LR initiation, but negatively influenced LR priming and emergence, the latter especially near the root–shoot junction. The cytokinin module ARABIDOPSIS HISTIDINE KINASE3 (AHK3)/ARABIDOPSIS RESPONSE REGULATOR1 (ARR1)/ARR12 was found to interact with the GR24-dependent reduction in LR development, because mutants in this pathway rendered LR development insensitive to GR24. Additionally, pharmacological analyses, mutant analyses, and gene expression analyses indicated that the affected polar auxin transport stream in mutants of the AHK3/ARR1/ARR12 module could be the underlying cause. Altogether, the data reveal that the GR24 effect on LR development depends on the hormonal landscape that results from the intimate connection with auxins and cytokinins, two main players in LR development.
Introduction
Strigolactones (SLs) are phytohormones that affect lateral branching of the shoot (Gomez-Roldan et al., 2008; Umehara et al., 2008) and many other processes, such as photomorphogenesis, drought tolerance, leaf senescence, and secondary growth, among others (Woo et al., 2001; Snowden et al., 2005; Shen et al., 2007, 2012; Tsuchiya et al., 2010; Agusti et al., 2011; Bu et al., 2014). In the rhizosphere, SLs influence interactions of the host plant with neighboring organisms, such as root-parasitic plants, mycorrrhizal fungi, and rhizobia (for review, see Xie et al., 2010; Rasmussen et al., 2013a). The root system architecture itself is also affected by SLs, because SLs influence adventitious root development, main root growth, root hair development, and lateral root (LR) development (Kapulnik et al., 2011a, 2011b; Ruyter-Spira et al., 2011; Mayzlish-Gati et al., 2012; Rasmussen et al., 2012, 2013a; Sun et al., 2014). The ontogenesis of LRs consists of several successive steps that are highly regulated (reviewed by Péret et al., 2009). The first step is priming of the LR that occurs in the xylem pole pericycle (XPP) cells in the basal meristem zone of the root tip. These primed XPP cells, also designated prebranch sites, have acquired the developmental program to become an LR. As the root grows, the primed XPP cells enter the elongation zone, where they undergo asymmetric cell division, a process designated LR initiation. Through further well controlled division patterns, an LR primordium will be formed that will ultimately develop into a typical dome-shaped primordium that will pierce through the main root and will form an emerged LR.
Regarding LR development, addition of the SL analog GR24 was found to reduce the LR density (LRD), because of a diminished LR initiation and LR outgrowth (Koltai et al., 2010; Kapulnik et al., 2011b; Ruyter-Spira et al., 2011). In Arabidopsis thaliana, mutants in the F-box protein MORE AXILLARY GROWTH2 (MAX2) are perturbed in SL perception and display higher LRDs than the wild-type (WT) plants (Kapulnik et al., 2011b; Kohlen et al., 2011; Ruyter-Spira et al., 2011). When MAX2 function was restored specifically in the root endodermis of max2 mutants, their insensitivity could be partially complemented (Koren et al., 2013). SLs are perceived by an α/β-hydrolase, DWARF14 (D14), that binds and hydrolyzes SLs and plays a central role in downstream signaling activation (Hamiaux et al., 2012; Zhao et al., 2013). In petunia (Petunia hybrida) and rice (Oryza sativa), D14 interacts with MAX2/D3, a nuclear-localized F-box protein that participates in the Skp-Cullin-F-box (SCF) complexes and, thus, can mediate the ubiquitin-dependent degradation of signaling proteins (Hamiaux et al., 2012; Zhao et al., 2013).
The interaction of SLs with auxins and cytokinins in regulation of shoot lateral branching has been thoroughly studied mainly in pea (Pisum sativum) and Arabidopsis (for a review, see Stirnberg et al., 2010; Cheng et al., 2013; Rasmussen et al., 2013a). Indeed, SL biosynthesis and signaling are intimately connected with auxin transport regulation (Foo et al., 2005; Bennett et al., 2006; Brewer et al., 2009; Ferguson and Beveridge, 2009; Hayward et al., 2009; Crawford et al., 2010; Koltai et al., 2010; Shinohara et al., 2013; Pandya-Kumar et al., 2014). The application of GR24 reduces the basipetal auxin transport and the accumulation of PIN-FORMED1 (PIN1) in the plasma membrane of xylem parenchyma cells in the shoot in a MAX2-dependent manner (Crawford et al., 2010). Moreover, in buds, SLs promote PIN1 endocytosis through a clathrin-dependent mechanism that occurs independently of de novo protein synthesis (Shinohara et al., 2013). In pea, SLs have been demonstrated to act also independently of auxin (Brewer et al., 2015). Interestingly, SLs could inhibit shoot lateral branching only when a competing auxin source was available (Crawford et al., 2010; Liang et al., 2010). The auxin landscape also influences the SL control on branching, because the negative effect on shoot lateral branching disappeared and even became positive when the auxin homeostasis was changed (Shinohara et al., 2013). In buds, SLs and cytokinins are known to interact antagonistically and locally (Dun et al., 2012; Zhang et al., 2010; Hu et al., 2014), probably through their common target, BRANCHED1 (BRC1) in Arabidopsis (Minakuchi et al., 2010; Braun et al., 2012; Dun et al., 2012).
Also in the root, the interaction of SLs with auxins has been investigated. PIN1, PIN3, and PIN7 protein levels are reduced upon prolonged treatment with GR24 (Ruyter-Spira et al., 2011). Additionally, during GR24-induced root hair elongation, PIN2 abundance increases at the apical plasma membrane of epidermal cells, suggesting that SLs affect PIN2 endocytosis and endosomal trafficking via actin dynamics in a MAX2-dependent manner (Pandya-Kumar et al., 2014). The inhibitory effect of GR24 on LR development can be reverted to an induction rather than a reduction of LRD by applying a high dose of auxin, or under low phosphate conditions that may increase the auxin sensitivity (Pérez-Torres et al., 2008; Ruyter-Spira et al., 2011). These observations suggest that, just as for branching, changes in the auxin landscape could modulate the impact of GR24 (Ruyter-Spira et al., 2011).
Cytokinins are also well known to influence the root architecture (reviewed in Vanstraelen and Benková, 2012). Cytokinin signaling negatively affects LR development by impinging on PIN-dependent auxin transport (Laplaze et al., 2007; Bishopp et al., 2011; Marhavý et al., 2011, 2014; Bielach et al., 2012; Chang et al., 2013; Moreira et al., 2013). Interaction of SLs with cytokinins during LR development has been poorly studied, but max2-1 mutants have been reported to have a reduced sensitivity to the synthetic cytokinin 6-benzylaminopurine (BAP) (Koren et al., 2013).
Here, LR priming as well as outgrowth are shown to be modulated by treatment with GR24, the latter in a spatiotemporal manner, mainly affecting the emergence of LRs, which are closest to the root–shoot junction. In addition, the ARABIDOPSIS HISTIDINE KINASE3 (AHK3)/ARABIDOPSIS RESPONSE REGULATOR1 (ARR1)/ARR12 cytokinin signaling module interacts with SLs to affect LR development, probably through changes in polar auxin transport. Altogether, the results place the SL action on LR development in the auxin landscape context via cross-talk mechanisms with cytokinin signaling.
Materials and methods
Plant material and growth conditions
The pin7-1 mutant from Arabidopsis thaliana (L.) Heyhn. is in Landsberg erecta (Ler) background, whereas the other lines described are in Columbia-0 (Col-0) background. The plant material used has been described previously: ahk2-2, cre1-12, and ahk3-3 (Higuchi et al., 2004); ahk2;ahk3, ahk2;ahk4, and ahk3;ahk4 (Riefler et al., 2006); arr1, arr12, and arr1;arr12 (Mason et al., 2005); arr3;arr4;arr5;arr6 and arr3;arr4;arr5;arr6;arr8;arr9 (To et al., 2004); pin1-613 (Bennett et al., 2006); 35S:PIN1-GFP (Růžička et al., 2007); pin3-3 (Friml et al., 2002); pin5-3 (Mravec et al., 2009); pin7-1 (Friml et al., 2003); shy2-24 (Tian and Reed, 1999); proAHK3:GUS (Higuchi et al., 2004); proPIN1:GUS and pGATA23:NLS-GFP-GUS (De Rybel et al., 2010); and YUCCA1-D (Zhao et al., 2001).
Seeds were surface-sterilized for 5min in 70% (v/v) ethanol, 0.05% (v/v) SDS solution, then incubated in 95% (v/v) ethanol for 5min, and plated on half-strength Murashige and Skoog (½MS) medium [1% (w/v) sucrose and 0.8% (w/v) agar]. Plants were stratified at 4 °C for 2 d, transferred to a growth chamber at 21 °C (16-h light/8-h dark photoperiod). A racemic mixture of GR24 was supplemented to the growth medium at the start of the experiment and plants were grown for the indicated time. All the experiments were repeated three times. Chemical compounds were added in the following concentrations, except indicated otherwise: 1 μM GR24 and 0.1 μM 1-N-naphthylphthalamic acid (NPA).
Phenotypic analysis and statistics
After 9 d of growth, LRs were counted under a binocular S4E microscope (Leica Microsystems) and root length was measured with ImageJ (http://rsb.info.nih.gov/ij). Both values were used to calculate the LRD. For the decapitation experiments, seedlings were grown for 6 d, whereafter the shoot was removed as described (Forsyth and Van Staden, 1981). The bottom part was transferred to ½MS medium with or without 1 µM GR24. For the complementation with indole-3-acetic acid (IAA), agar blocks (0.5cm3) containing solidified growth medium with and without 10 µM IAA were added to the decapitated site and the LRD was analyzed 5 d later. Replicate means were subjected to statistics by analysis of variance (ANOVA; SAS Institute Inc., Cary, NC, USA).
Stage determination of GATA23 expression analysis
pGATA23:NLS-GFP-GUS seeds were put on medium supplemented with 1 µM GR24 or with the same volume of acetone as control and were stratified for 2 d at 4 °C. Seedlings were grown vertically under continuous white light at 21 °C. At 4 d after germination (DAG), half the seedlings were harvested for analysis, whereas for the remaining seedlings, the position of the main root tip was marked and the plates were transferred back to the growth room. At 9 DAG, the root parts above the mark were harvested. Samples were stained with β-glucuronidase (GUS), cleared as described (Malamy and Benfey, 1997), and analyzed under the microscope (see below). To calculate the percentage of initiated sites, the average of initiations at 9 DAG was divided by the average sites present at 4 DAG. Likewise for the calculations of the percentage of emerged sites, the average of emerged LRs at 9 DAG was divided by that of the sites present at 4 DAG.
Histochemical analysis of GUS activity
Whole seedlings were stained in multiwell plates as described (Jefferson et al., 1987). Samples were cleared as described (Malamy and Benfey, 1997) and were analyzed by a differential interference contrast BX51 microscope (Olympus). Alternatively, samples were mounted directly in chloral hydrate solution (chloral hydrate:water:glycerol, 8:3:1) and microscopically analyzed.
RNA isolation, quantitative RT-PCR, and statistical analysis of PIN1 expression
Arabidopsis pPIN1:GUS seeds were sown on ½MS medium with or without 1 µM GR24. Seeds were stratified for 2 d at 4 °C and then grown in vertical position at 21 °C (16-h light/8-h dark photoperiod). After 7 d, root material was harvested and flash-frozen in liquid nitrogen. The region between the root–shoot junction and the first emerged LR was harvested separately from the remaining root system. Approximately 100 seedlings were used for each treatment and the experiment was repeated three times.
RNA preparation, cDNA synthesis, real-time quantitative (q)RT-PCR, and statistical analysis of expression profiling were done as described (Rasmussen et al., 2013b). The primers used are the following: PIN1_forward GGCATGGCTATGTTCAGTCTTGGG and PIN1_reverse ACGGCAGGTCCAACGACAAATC; ACTIN_forward CGCCATCCAAGCTGTTCTC and ACTIN_reverse TCACGTCCAGCAAGGTCAAG.
Accession numbers
The Arabidopsis Genome Initiative locus identifiers for the genes characterized in this study are: AHK3 (AT1G27320), SHY2 (AT1G04240), PIN1 (AT1G73590), PIN7 (AT1G23080), and YUCCA1 (AT4G32540). Germplasm identification numbers for the seeds are: ahk2 (ahk2-2tk), ahk3-3 (SALK_069269), cre1-12 (SALK_048970), ahk2;ahk3 (ahk2-5ahk3-7), ahk2;ahk4 (ahk2-5cre1-12), ahk3;ahk4 (ahk3-7;cre1-2), arr1-2 (N6368), arr12-1 (CS6978), arr1;arr12 (arr1-3;arr12-1), pin1-613 (SALK_047613), and pin5-3 (SALK_021738).
Results
GR24 reduces lateral rooting in Arabidopsis by affecting LR emergence, especially near the root–shoot junction in a MAX2-dependent manner
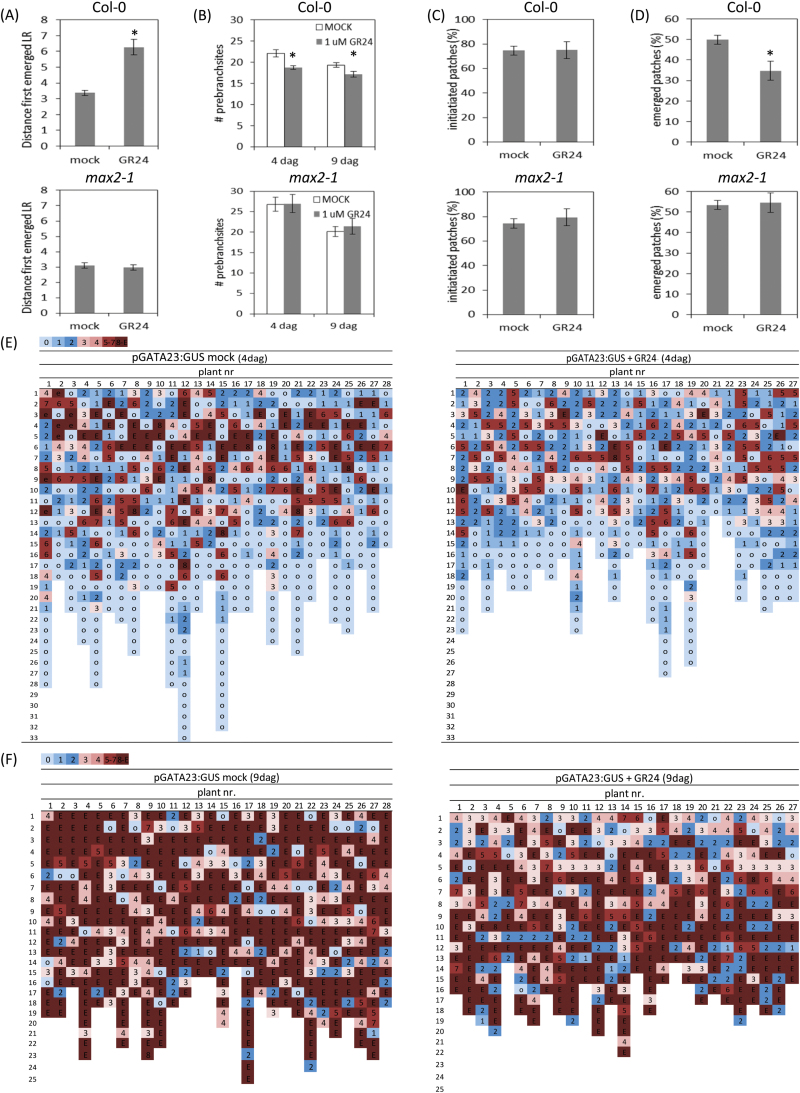
The overall MAX2-dependent reduction in LRD caused by GR24 application had already been reported (Kapulnik et al., 2011b; Kohlen et al., 2011; Ruyter-Spira et al., 2011), but phenotypical insights into this event are still lacking. Upon GR24 treatment, the first emerged LR had an altered position and this effect was abolished in the max2-1 mutant. When plants were grown without GR24 (mock), the distance from the hypocotyl to the first emerged LR was on average 3.37mm, whereas when grown in the presence of GR24 it increased to 6.27mm in WT plants (Fig. 1A).
Fig. 1.
Effect of exogenous GR24 on LR development near the root–shoot junction.
(A) Distance to the first emerged LR in Col-0 (top) and max2-1 (bottom). (B) Total number of prebranch sites under mock (white bars) and GR24 treatment (gray bars), 4 and 9 DAG in Col-0 (top) and max2-1 (bottom). (C) Percentage of initiated patches under mock and 1 μM GR24 treatments in Col-0 (top) and max2-1 (bottom) at 9 DAG. (D) Percentage of emerged patches under mock and GR24 treatment in Col-0 (top) and max2-1 (bottom). (A–D) Data presented are means ± standard error (SE) of three biological repeats (n>20). *P<0.001, according to the Student’s t-test. (E, F) Stages of LR primordia via GATA23:GUS staining in Col-0 under mock (left) and GR24 treatment (right) at 4 DAG (E) and 9 DAG (F). All events, possibly leading to emerged LRs, were scored in individual plants, color-coded, and for each plant, vertically ordered from the closest to the hypocotyl (up) downward to the meristem (down). The root fragments used for analysis were comparable in length. Data of one representative experiment are shown. The experiments were repeated three times with similar results.
To understand this effect, the LR development was spatiotemporally followed, with specific focus on the upper root zone. Therefore, the expression of the early LR marker GATA23 that indicates prebranch sites (De Rybel et al., 2010) was used and combined with the staging of the LR primordia (Malamy and Benfey, 1997), in both WT and max2-1 plants, under mock and GR24 treatments (Fig. 1E, F). As such, all sites in which an LR could develop were visualized from the root–shoot junction down to the root meristem at 4 DAG (Fig. 1E; Supplementary Fig. S1A at JXB online). The progression in LR development was subsequently analyzed at 9 DAG (Fig. 1F; Supplementary Fig. S1B at JXB online) to obtain a spatiotemporal view of how the LR primordium development was affected by GR24 treatment. Fewer GATA23-marked sites were observed at 9 DAG than at 4 DAG, implying that not all primed sites developed into an LR primordium. When the number of LR sites between mock and GR24-grown plants was compared, slightly, but significantly, fewer sites were counted upon GR24 treatment, both at 4 and 9 DAG (Fig. 1B), indicating that GR24 treatment reduced the total number of prebranch sites in WT, but not in max2-1, seedlings (Fig. 1B). Concerning initiated patches (see Materials and Methods), mock and GR24-grown roots of both WT and max2-1 seedlings did not differ, suggesting that GR24 had no effect on LR initiation, once the prebranch site had been formed (Fig. 1C). GR24 treatment also affected LR outgrowth (Kapulnik et al., 2011b; Kohlen et al., 2011; Ruyter-Spira et al., 2011). When the percentage of emerged patches was calculated, significantly fewer sites were counted on GR24-grown roots than on control roots, but again not on max2-1 roots (Fig. 1D). Interestingly, when the emergence pattern was analyzed at 9 DAG (Fig. 1F), the LR outgrowth inhibition was most pronounced at positions 1–8, corresponding to the LR primordia closest to the root–shoot junction, but did not occur in the max2-1 mutant (see Supplementary Fig. S1B at JXB online). These data indicate that mainly the first formed LR primordia, thus those near the root–shoot junction, do not develop when plants are grown in the presence of GR24 and that this effect depends on MAX2.
The cytokinin signaling components AHK3, ARR1, and ARR12 mediate the effect of GR24 on LR development
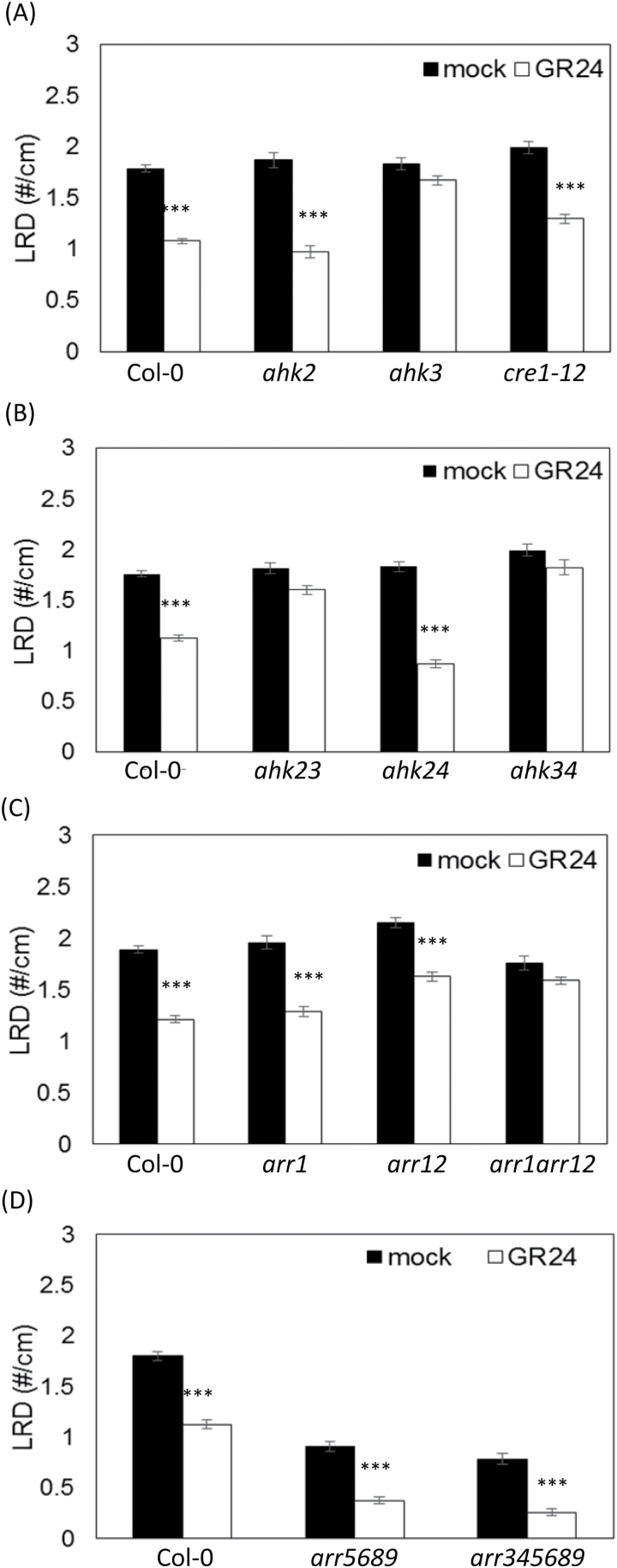
Both cytokinins and SLs have been described as negative regulators of LR development in Arabidopsis (Benková et al., 2003; Li et al., 2006; Laplaze et al., 2007; Kapulnik et al., 2011b; Ruyter-Spira et al., 2011). Therefore, the link between the GR24-mediated LRD reduction and the cytokinin-mediated LRD inhibition was investigated in further detail. First, the LRD of several cytokinin signaling mutants, single and higher-order mutants affected in the cytokinin receptors CYTOKININ RESPONSE1 (CRE1)/AHK4, AHK2, and/or AHK3 (see Materials and Methods) was examined upon treatment with 1 µM GR24 (Fig. 2A, B). For all tested genotypes, GR24 treatment did not significantly affect the main root length (see Supplementary Fig. S2 at JXB online). For Col-0, cre1/ahk4, and ahk2, the LRD was significantly reduced upon GR24 treatment, but not for the ahk3 mutant (Fig. 2A). In the double cytokinin receptor mutant ahk2;ahk4, the LRD decreased significantly upon GR24 treatment, whereas no significant changes in LRD were observed for ahk2;ahk3 and ahk3;ahk4 between mock and GR24 treatment (Fig. 2B). Taken together, these data show that in mutants specifically affected in one member of the cytokinin receptor family, i.e. AHK3 (ahk3, ahk2;ahk3, and ahk3;ahk4), the GR24 impact on LRD was abolished, whereas other cytokinin receptor mutants responded as WT plants. The AHK3 expression was unaffected by GR24 treatment (see Supplementary Fig. S3 at JXB online).
Fig. 2.
Effects of GR24 on cytokinin perception and signaling mutants. LRD of single cytokinin receptor mutants (A), double cytokinin receptor mutants (B), B-type response regulators ARR1, ARR12, and ARR1;ARR12 (C), and mutants in higher-order A-type response regulators (D) upon GR24 treatment. Data presented are means ± SE of three biological repeats (n>20). ***P<0.001, according to ANOVA mixed-model statistical analyses.
These observations prompted the investigation of the downstream signaling components of the cytokinin perception machinery. As the B-type response regulators ARR1 and ARR12 are involved in mediating the AHK3-dependent effects in the root elongation zone (Dello Ioio et al., 2007, 2008), the GR24 impact on the LRD was tested in mutants of these response regulators. The single mutants arr1 and arr12 displayed a sensitivity to GR24 similar to that of Col-0 (Fig. 2C), but the double mutant arr1;arr12 did not, indicating that both ARRs need to be disrupted to interfere with the GR24 effect on LR development (Fig. 2C).
Having established that AHK3, ARR1, and ARR12 are involved in the GR24-mediated reduction of LRD, we analyzed whether mutants affected in A-type response regulators would affect the GR24-mediated LRD reduction. Therefore, the sensitivity was tested of higher-order A-type ARR mutants to GR24, because these negative regulators of the cytokinin response are known to act redundantly in root architecture control (To et al., 2004; Zhang et al., 2011). The arr5;arr6;arr8;arr9 and arr3;arr4;arr5;arr6;arr8;arr9 mutants showed a significant increase in sensitivity to GR24: LRD decreased by 37% in WT and by 58% and 67% in arr5;arr6;arr8;arr9 and arr3;arr4;arr5;arr6;arr8;arr9, respectively (Fig. 2D). Hence, these data support the hypothesis that an altered cytokinin responsiveness can enhance (A-type ARRs) or repress (B-type ARRs or AHK3) the GR24 effect on LR development. Taken together, these experiments demonstrate that specific cytokinin signaling components are needed for the GR24 action on LR development.
The modified sensitivity to GR24 of ahk3/arr1;arr12/shy2 mutants is due to changes in the auxin landscape
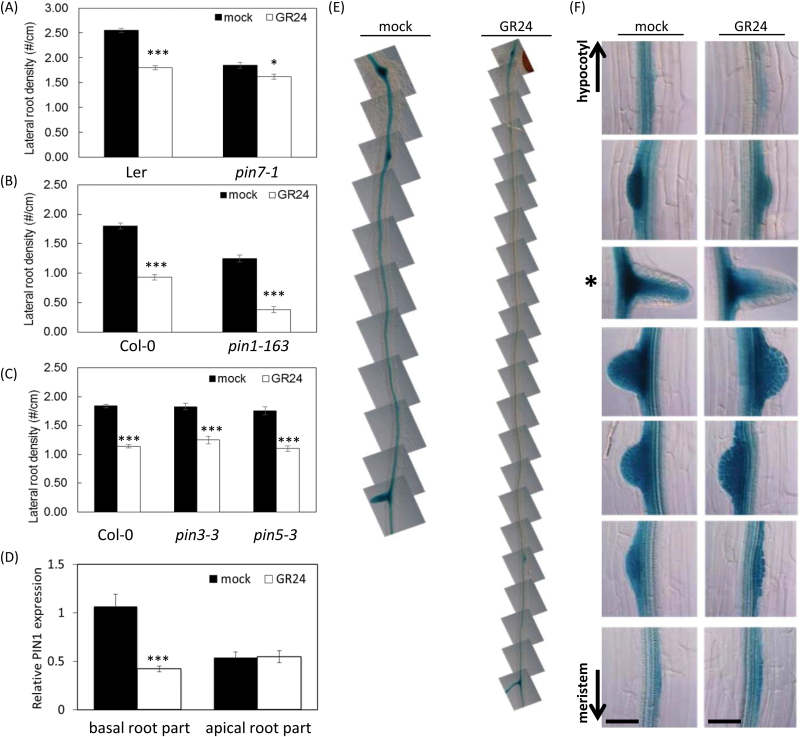
The AHK3/ARR1/ARR12 cytokinin signaling pathway has been shown to act upstream of SHORT HYPOCOTYL2 (SHY2) to control root differentiation (Dello Ioio et al., 2007, 2008) and, additionally, the shy2 loss-of-function mutant to be insensitive to GR24 as well (Koren et al., 2013). To elucidate why mutants in the AHK3/ARR1/ARR12/SHY2 module are affected in their GR24 sensitivity, the GR24 phenotype of different pin mutants was examined, because SHY2 specifically represses PIN1, PIN3, PIN5, and PIN7 and cytokinin treatment downregulates PIN1 and PIN3, but upregulates PIN7 expression (Dello Ioio et al., 2007; Růžička et al., 2009). First, the GR24 effect on LRD of mutations in PIN1, PIN3, PIN5, or PIN7 was analyzed. The decrease in LRD of the pin7 mutants was only minor upon GR24 treatment, indicating that mutations in PIN7 reduced the root sensitivity to GR24 (Fig. 3A); however, the LRD reduction of the pin1-613 mutants was significantly higher than that in WT plants (Fig. 3B). For the pin3-3 and pin5-3 mutants, the LRD did not differ from that of WT plants (Fig. 3C).
Fig. 3.
Interrelation between the polar auxin transport and the GR24 effect on LR development. (A–C) LRD of pin7-1, pin1-613, pin3-3, and pin5-3 mutants compared with WT grown in the presence or absence of GR24. Data presented are means ± SE of three biological repeats (n>20). (D) Relative PIN1 expression in 5-d-old seedlings under mock and GR24 treatment as determined by qRT-PCR. Material was harvested separately from the upper part (old, above the first emerged LR) and the lower part (young) of the root. ***P<0.001, *P<0.05, according to ANOVA mixed-model statistical analyses.(E) pPIN1:GUS expression patterns of plants grown with and without GR24, 7 d after growth. Frames until the first emerged LR are shown. (F) Expression of PIN1 with pPIN1:GUS plants during different stages of LR development under mock and GR24 treatment. The panels indicated by the asterisk display the first emerged LR and those above the asterisk correspond to the LR primordia near the root–shoot junction. Scale bars=40 µm.
Hence, these results provide the first genetic evidence that the LR response to exogenous GR24 is modulated by interference with the polar auxin transport through PIN1 and, to a lesser extent, PIN7. Previously, prolonged, but not short, GR24 treatments had been demonstrated to influence the expression of PIN1, PIN3, and PIN7 in the root meristem, but the expression in root parts other than the meristem had not been assessed (Ruyter-Spira et al., 2011; Shinohara et al., 2013). Therefore, the GR24 effect was investigated on the transcription of PIN1 in the mature root, at the hypocotyl–root junction, where LR emergence is most affected by the GR24 treatment (Fig. 3). The GR24 impact on PIN1 expression was analyzed after 7 d of growth of pPIN1:GUS seedlings. Interestingly, PIN1 expression was affected in a spatial way because, especially closest to the shoot, the expression in the vasculature was lower upon GR24 treatment than under mock conditions (Fig. 3E, F). This observation was confirmed by analyzing the PIN1 gene expression by qRT-PCR of roots grown either in the presence or the absence of GR24 and by assessing the mature versus younger regions of the root (Fig. 3D). Moreover, PIN1 expression was also lower in the developing LRs from the upper part of GR24-treated plants than that of mock-grown roots, in contrast to developing LRs at younger stages, i.e. near the root meristem (Fig. 3F).
Thus far, our data demonstrate that mutations in the AHK3/ARR1/ARR12 cytokinin signaling module and in the auxin transport genes PIN1 and PIN7 affect the root sensitivity to GR24, and that GR24 influences auxin homeostasis by downregulating the expression of PIN1 near the shoot–root junction, in agreement with the decreased PIN protein levels in the root upon prolonged treatments with high concentrations of GR24 (Ruyter-Spira et al., 2011).
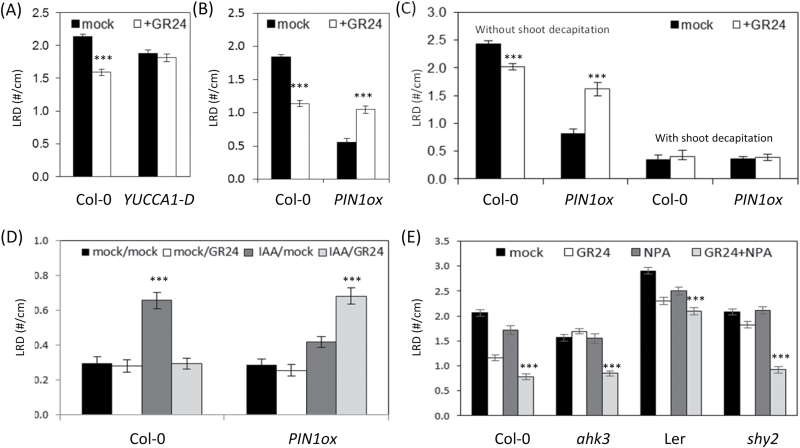
To further investigate how the auxin environment alters the GR24 effect, the GR24 response was examined in plants that overexpressed YUCCA with concomitantly increased free auxin levels (Zhao et al., 2001). The LRD of YUCCA1-D plants did not decrease upon GR24 treatment, indicating that enhanced endogenous auxin levels also modulate the GR24 response in roots (Fig. 4A). Also 35S:PIN1-overexpressing plants that have highly increased frequencies of root primordia with retarded growth were analyzed (Benková et al., 2003). The typical GR24-mediated decrease in LRD was no longer visible, but rather an increase in LRD (Fig. 4B). Moreover, when the foliar auxin source that determines the outgrowth potential of LRs (Bhalerao et al., 2002; Ljung et al., 2005) was removed by decapitation after 6 d of growth and when these plants were subsequently treated with GR24 for 5 d, the effects disappeared on both the PIN1-overpressing lines (increase in LRD) and the WT (decrease in LRD), indicating that shoot-derived auxin is important for the GR24 responses in roots (Fig. 4D). Application of IAA in these experiments (see Materials and Methods) revealed that shoot-derived auxin mediated the effect, because it complemented the phenotype of decapitated plants (Fig. 4D). Altogether, the functional data demonstrate that shoot-derived auxin controls the effect of GR24 on lateral rooting in Arabidopsis, as previously hypothesized (Ruyter-Spira et al., 2011).
Fig. 4.
Dependence of GR24 action on the plant auxin status. (A) LRD of WT and YUCCA-overexpressing (YUCCA1-D) plants, grown with and without GR24. (B) LRD of WT and PIN1-overexpressing (PIN1ox) plants, grown with and without GR24. (C) LRD of Col-0 and 35S:PIN1 (PIN1ox) plants with and without shoot decapitation, grown in the presence or absence of GR24. (D) LRD of Col-0 and PIN1ox plants with decapitation and with and without apically applied IAA grown in the presence or absence of GR24. Mock/mock: decapitated plants grown in the absence of GR24 and without applied IAA; mock/+GR24: decapitated plants grown in the presence of GR24 and without applied IAA; IAA/mock: decapitated plants grown in the absence of GR24 and with apically applied IAA; IAA/+GR24: decapitated plants grown in the presence of GR24 and with apically applied IAA. (E) LRD of Col-0, ahk3, Ler and shy2-24 mutants upon treatment with mock, GR24, NPA, or NPA+GR24. Data presented are means ± SE of three biological repeats (n>20). ***P<0.001, according to ANOVA mixed-model statistical analyses.
All mutants with GR24-insensitive root responses, i.e. ahk3, arr1;arr12, and shy2-24, display enhanced PIN1 expression (Dello Ioio et al., 2007, 2008; Zhang et al., 2011) that might cause their insensitivity toward GR24. This hypothesis was tested by applying low concentrations (100nM) of NPA, a polar auxin transport inhibitor (Himanen et al., 2002). The LRD response was analyzed under mock and GR24 treatment after 9 d of growth (Fig. 4E). Addition of this low concentration of NPA had no impact on the LRD (Fig. 4E) and had no clear effect on PIN1 expression in the main root, although a slight increase in PIN1 gene expression was observed in the root tip (see Supplementary Fig. S4 at JXB online). However, when the ahk3 and shy2-24 mutants were grown on plates supplemented with NPA as well as GR24, the LRD was lower than that of roots grown under mock conditions or supplemented with GR24 or NPA alone, implying that treatment with NPA rendered the mutant plants responsive to GR24 again. For Col-0, no additional effect was seen when the roots were treated with both NPA and GR24.
Discussion
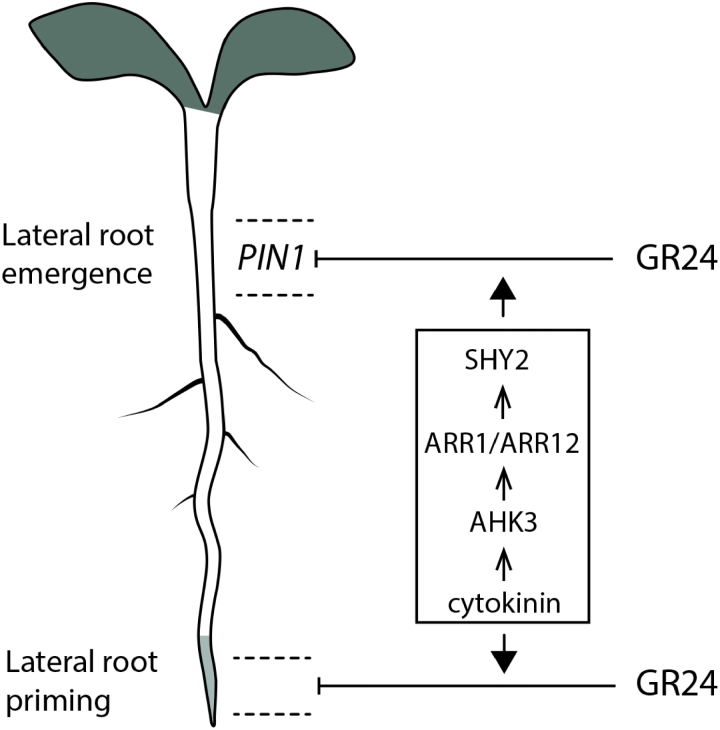
Several aspects of the root system architecture are modulated by SLs (for reviews, see Cheng et al., 2013; Rasmussen et al., 2013a; Koltai, 2014). Here, GR24 was found to control LR development spatiotemporally and to interplay with cytokinin that, just like SLs, regulates LR development. A summarizing model is presented (Fig. 5).
Fig. 5.
Working model on the interaction of cytokinins with the SL analog GR24 to control LR development. GR24 treatment results in inhibition of LR emergence, mainly, but not exclusively, near the root–shoot junction and, to a minor extent, in inhibition of LR priming in the root meristem zone. In the root region near the root–shoot junction, this LR emergence inhibition coincides with a spatial downregulation of PIN1 expression by GR24 treatment. The cytokinin module that signals via AHK3, through the response regulators ARR1/ARR12, and ultimately to SHY2, influences the effect of GR24 on LR development. Mutants in this pathway are insensitive to GR24, probably due to their reported increased PIN1 levels, because reduction of the auxin flux by NPA treatment renders the mutants sensitive again to GR24.
The method established to build a developmental map of all possible initiated LRs combines the GATA23 marker gene for induction of prebranching sites, i.e. pericycle-derived LR founder cells that are predestined to start cell division for LR development and LR positioning (Malamy and Benfey, 1997; De Rybel et al., 2010). Together with the determination of the position of each event along the main root, a precise developmental map provides location and developmental stage of each LR event, thereby revealing that the main effect of GR24 on the development of LRs concerns their emergence. This observation concurs with previously published work, although the proposed specific interruption at stage V of LR development was not detected (Ruyter-Spira et al., 2011).
On the 9-DAG map, the LRs were mainly, but not exclusively, situated close to the root–shoot junction that no longer emerged under GR24 treatment. Accordingly, the distance between the hypocotyl–shoot junction and the first emerged LR was longer in GR24-grown roots than in control roots. This MAX2-dependent effect concurs with its essential function in SL signaling. Hence, GR24 might affect specifically the emergence of the LRs that develop first and are positioned in the older part of the root. This spatiotemporal effect was also seen on the PIN1 expression pattern in the root. Although the reason for this effect still needs to be investigated, the disappearance of the SL receptor might be the underlying cause, because GR24 treatment reduces D14 protein abundance in roots (Chevalier et al., 2014).
Additionally, a small, but significant, decrease in prebranch sites was visible, whereas GR24 had no appreciable impact on LR initiation. The previously detected GR24 effect on LR initiation (Kapulnik et al., 2011b; Ruyter-Spira et al., 2011) might be due to an impact on prebranching. Prebranch sites are established by a periodic oscillation of auxin concentrations accompanied by fluctuations in specific gene expression (De Smet et al., 2007; Moreno-Risueno et al., 2010). This oscillating pattern has been found to be mediated by a carotenoid compound, distinct from SLs (Van Norman et al., 2014). In agreement with the data presented, the max2 mutants also exhibited an increased LR capacity (Van Norman et al., 2014). It would be interesting to analyze whether GR24, as a mimic of SLs or related compounds, modulates the periodic oscillation of auxin to cause the small effect on prebranching. Furthermore, independently of SLs, at 9 DAG, fewer LR events are observed on the same main root part than at 4 DAG, possibly indicating that not all primed sites develop into LRs.
Cytokinins have been identified as endogenous repressors of LR development in a close interplay with auxin (Benková et al., 2003; Li et al., 2006; Laplaze et al., 2007). Here, the GR24 effect on LR development required the functional cytokinin receptor AHK3, but not AHK2 and AHK4/CRE1. The dependence on AHK3 and not on AHK4 is remarkable, because AHK4 has been implicated in LR patterning along the main root (Marhavý et al., 2011), whereas AHK3 and the two immediately downstream B-type response regulator genes, ARR1 and ARR12, play an important role in determining the root meristem size (Dello Ioio et al., 2007, 2008). Also in the experimental setup, the double mutant arr1;arr12 had no LR response toward GR24, implying that the same cytokinin module (AHK3/ARR1/ARR12) that determines the root meristem differentiation also governs the GR24 action on LR development. AHK3 is involved in meristem differentiation by transcriptional control of the auxin-induced SHY2/IAA3 gene (Dello Ioio et al., 2007, 2008). The typical reduction in lateral rooting upon GR24 treatment was indeed not seen in the shy2-24 loss-of-function mutants (Koren et al., 2013), supporting the hypothesis that the AHK3/ARR1/ARR12 module acts through SHY2 to result in GR24 insensitivity.
The AHK3/ARR1/ARR12/SHY2 module negatively influences PIN1/PIN3/PIN5/PIN7 expression (Dello Ioio et al., 2007, 2008), whereas cytokinin treatment downregulates PIN1/PIN3/PIN5, but upregulates PIN7 expression (Laplaze et al., 2007; Růžička et al., 2009). These changes in PIN gene expression and their consequences on polar auxin transport might be the underlying cause for the GR24 insensitivity of the mutants. Several PIN mutants had a modified sensitivity to GR24: pin3 and pin5 mutants still displayed a reduced LR development upon GR24 treatment, whereas pin7 mutants were only slightly responsive to GR24, and pin1-613 mutants were hypersensitive, in agreement with the opposite influence of cytokinins on their expression. In addition, treatment of ahk3 and shy2-24 with NPA made them sensitive again to GR24. Hence, the changes in PIN gene expression, such as the PIN1 overexpression observed in these mutants (Dello Ioio et al., 2007, 2008; Zhang et al., 2011) with an enhanced polar auxin transport as a result, might be the reason that GR24 does not reduce the LRD in these mutants.
Moreover, the data support the central role of auxin transport for SL action. Based on exogenous auxin and phosphate level modulation, the auxin content in roots has been shown to determine its responsiveness toward GR24 (Ruyter-Spira et al., 2011). Indeed, endogenous overproduction of auxin via overexpression of YUCCA could make LR development unresponsive to GR24. As auxin is well known to positively regulate its own efflux from cells, PIN1 internalization in the YUCCA1-D mutant was reduced, resulting in the accumulation of PIN1 on the plasma membrane (Paciorek et al., 2005), an observation fitting the theory that mutants with an enhanced PIN1 expression are insensitive to GR24. Interestingly, PIN1-overexpressing plants no longer displayed a reduced LRD when treated with GR24, but an opposite phenotype with an increased LRD. The difference in phenotypes between the plants overexpressing YUCCA1-D and PIN1 is intriguing, but might be due to differences in the severity of PIN1 accumulation. Also in the shoot, depending on the auxin transport landscape, GR24 could have positive or negative effects on the shoot lateral branching by depleting PIN1 from the membranes of xylem parenchyma cells of inflorescence stems (Shinohara et al., 2013). In addition, GR24 has been shown to have a different impact on LR development that depends on the growth conditions: inhibition under sufficient and induction under low phosphate conditions or with exogenous IAA (Ruyter-Spira et al., 2011). Hence, PIN1 overexpression has an effect on GR24 responses similar to that of phosphate-limiting conditions: an increase, rather than a decrease, in LRD.
In conclusion, the data presented imply that GR24 regulates LR development in a spatiotemporal manner with the strongest effect on emergence of the first developed LR positioned close to the root–shoot junction. This effect is tightly integrated into the auxin–cytokinin network that rules the root architecture, with the polar auxin transport capacity as a central player on which both cytokinin and GR24 act.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Stages of LR primordia via pGATA23:GUS staining in max2-1 under mock and GR24 treatment at 4 and 9 DAG.
Fig. S2. Main root lengths of WT and cytokinin receptor and signal transduction mutants under mock and GR24 treatment.
Fig. S3. pAHK3:GUS expression patterns of LR primordia at different developmental stages under mock and GR24 treatment.
Fig. S4. pPIN1:GUS expression pattern after treatment with 0.1 µM NPA around the root–shoot junction (left) and the root meristem zone (right).
Acknowledgments
We thank Eva Benková and Jiří Friml (Institute of Science and Technology – Austria) for providing mutant and transgenic lines, Martine De Cock and Tom Viaene for help with the manuscript, and Karel Spruyt for support with the figures. This work was supported by the European Cooperation on Science and Technology (COST action FA1206 “Strigolactones: Biological Roles and Applications”). LJ is indebted to the China Scholarship Council for a predoctoral fellowship; CM and BMG are the recipients of a predoctoral and postdoctoral fellowship from the “Bijzonder Onderzoeksfonds” of Ghent University, respectively; and CDC and SD are a predoctoral and a postdoctoral fellow of the Research Foundation-Flanders, respectively.
Glossary
Abbreviations:
- AHK
ARABIDOPSIS HISTIDINE KINASE
- ARR
ARABIDOPSIS RESPONSE REGULATOR
- BAP
6-benzylaminopurine
- BES
BRASSINOSTEROID INSENSITIVE--EMS-SUPPRESSOR
- BRC
BRANCHED
- CRE
CYTOKININ RESPONSE
- D14
DWARF14
- DAG
days after germination
- EMS
ethyl methanesulfonate
- GUS
β-glucuronidase
- IAA
indole-3-acetic acid
- LR
lateral root
- LRD
lateral root density
- MAX
MORE AXILLARY GROWTH
- NPA
1-N-naphthylphthalamic acid
- PIN
PIN-FORMED
- SCF
Skp-Cullin-F-box
- SHY
SHORT HYPOCOTYL
- SL
strigolactone
- WT
wild-type
- XPP
xylem pole pericycle.
References
- Agusti J, Herold S, Schwarz M, et al. 2011. Strigolactone signaling is required for auxin-dependent stimulation of secondary growth in plants. Proceedings of the National Academy of Sciences, USA 108, 20242–20247 [Erratum Proceedings of the National Academy of Sciences, USA 109, 14277]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J. 2003. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602. [DOI] [PubMed] [Google Scholar]
- Bennett T, Sieberer T, Willett B, Booker J, Luschnig C, Leyser O. 2006. The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Current Biology 16, 553–563. [DOI] [PubMed] [Google Scholar]
- Bhalerao RP, Eklöf J, Ljung K, Marchant A, Bennett M, Sandberg G. 2002. Shoot-derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. Plant Journal 29, 325–332. [DOI] [PubMed] [Google Scholar]
- Bielach A, Duclercq J, Marhavý P, Benková E. 2012. Genetic approach towards the identification of auxin-cytokinin crosstalk components involved in root development. Philosophical Transactions of the Royal Society B-Biological Sciences 367, 1469–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishopp A, Benková E, Helariutta Y. 2011. Sending mixed messages: auxin-cytokinin crosstalk in roots. Current Opinion in Plant Biology 14, 10–16. [DOI] [PubMed] [Google Scholar]
- Braun N, de Saint Germain A, Pillot J-P, et al. 2012. The pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching. Plant Physiology 158, 225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer PB, Dun EA, Ferguson BJ, Rameau C, Beveridge CA. 2009. Strigolactone acts downstream of auxin to regulate bud outgrowth in pea and Arabidopsis. Plant Physiology 150, 482–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer PB, Dun EA, Gui R, Mason MG, Beveridge CA. 2015. Strigolactone inhibition of branching independent of polar auxin transport. Plant Physiology 168, 1820–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu Q, Lv T, Shen H, et al. 2014. Regulation of drought tolerance by the F-box protein MAX2 in Arabidopsis. Plant Physiology 164, 424–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Ramireddy E, Schmülling T. 2013. Lateral root formation and growth of Arabidopsis is redundantly regulated by cytokinin metabolism and signalling genes. Journal of Experimental Botany 64, 5021–5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Ruyter-Spira C, Bouwmeester H. 2013. The interaction between strigolactones and other plant hormones in the regulation of plant development. Frontiers in Plant Science 4, 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier F, Nieminen K, Sánchez-Ferrero JC, Rodríguez ML, Chagoyen M, Hardtke CS, Cubas P. 2014. Strigolactone promotes degradation of DWARF14, an α/β hydrolase essential for strigolactone signaling in Arabidopsis . Plant Cell 26, 1134–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford S, Shinohara N, Sieberer T, Williamson L, George G, Hepworth J, Müller D, Domagalska MA, Leyser O. 2010. Strigolactones enhance competition between shoot branches by dampening auxin transport. Development 137, 2905–2913. [DOI] [PubMed] [Google Scholar]
- De Rybel B, Vassileva V, Parizot B, et al. 2010. A novel Aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Current Biology 20, 1697–1706. [DOI] [PubMed] [Google Scholar]
- De Smet I, Tetsumura T, De Rybel B, et al. 2007. Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis . Development 134, 681–690. [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Scaglia Linhares F, Scacchi E, Casamitjana-Martinez E, Heidstra R, Costantino P, Sabatini S. 2007. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Current Biology 17, 678–682. [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, Morita MT, Aoyama T, Costantino P, Sabatini S. 2008. A genetic framework for the control of cell division and differentiation in the root meristem. Science 322, 1380–1384. [DOI] [PubMed] [Google Scholar]
- Dun EA, de Saint Germain A, Rameau C, Beveridge CA. 2012. Antagonistic action of strigolactone and cytokinin in bud outgrowth control. Plant Physiology 158, 487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson BJ, Beveridge CA. 2009. Roles for auxin, cytokinin, and strigolactone in regulating shoot branching. Plant Physiology 149, 1929–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo E, Bullier E, Goussot M, Foucher F, Rameau C, Beveridge CA. 2005. The branching gene RAMOSUS1 mediates interactions among two novel signals and auxin in pea. Plant Cell 17, 464–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsyth C, Van Staden J. 1981. The effects of root decapitation on lateral root formation and cytokinin production in Pisum sativum. Physiologia Plantarum 51, 375–379. [Google Scholar]
- Friml J, Wiśniewska J, Benková E, Mendgen K, Palme K. 2002. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis . Nature 415, 806–809. [DOI] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G. 2003. Efflux-dependent auxin gradients establish the apical–basal axis of Arabidopsis . Nature 426, 147–153. [DOI] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, et al. 2008. Strigolactone inhibition of shoot branching. Nature 455, 189–194. [DOI] [PubMed] [Google Scholar]
- Hamiaux C, Drummond RSM, Janssen BJ, Ledger SE, Cooney JM, Newcomb RD, Snowden KC. 2012. DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Current Biology 22, 2032–2036. [DOI] [PubMed] [Google Scholar]
- Hayward A, Stirnberg P, Beveridge C, Leyser O. 2009. Interactions between auxin and strigolactone in shoot branching control. Plant Physiology 151, 400–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, Pischke MS, Mähönen AP, et al. 2004. In planta functions of the Arabidopsis cytokinin receptor family. Proceedings of the National Academy of Sciences, USA 101, 8821–8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen K, Boucheron E, Vanneste S, de Almeida Engler J, Inzé D, Beeckman T. 2002. Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell 14, 2339–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Yamauchi T, Yang J, et al. 2014. Strigolactone and cytokinin act antagonistically in regulating rice mesocotyl elongation in darkness. Plant and Cell Physiology 55, 30–41. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO Journal 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapulnik Y, Resnick N, Mayzlish-Gati E, Kaplan Y, Wininger S, Hershenhorn J, Koltai H. 2011a. Strigolactones interact with ethylene and auxin in regulating root-hair elongation in Arabidopsis . Journal of Experimental Botany 62, 2915–2924. [DOI] [PubMed] [Google Scholar]
- Kapulnik Y, Delaux P-M, Resnick N, et al. 2011b. Strigolactones affect lateral root formation and root-hair elongation in Arabidopsis . Planta 233, 209–216. [DOI] [PubMed] [Google Scholar]
- Kohlen W, Charnikhova T, Liu Q, Bours R, Domagalska MA, Beguerie S, Verstappen F, Leyser O, Bouwmeester H, Ruyter-Spira C. 2011. Strigolactones are transported through the xylem and play a key role in shoot architectural response to phosphate deficiency in nonarbuscular mycorrhizal host Arabidopsis. Plant Physiology 155, 974–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltai H. 2014. Receptors, repressors, PINs: a playground for strigolactone signaling. Trends in Plant Science 19, 727–733. [DOI] [PubMed] [Google Scholar]
- Koltai H, Dor E, Hershenhorn J, et al. 2010. Strigolactones’ effect on root growth and root-hair elongation may be mediated by auxin-efflux carriers. Journal of Plant Growth Regulation 29, 129–136. [Google Scholar]
- Koren D, Resnick N, Mayzlish Gati E, Belausov E, Weininger S, Kapulnik Y, Koltai H. 2013. Strigolactone signaling in the endodermis is sufficient to restore root responses and involves SHORT HYPOCOTYL 2 (SHY2) activity. New Phytologist 198, 866–874. [DOI] [PubMed] [Google Scholar]
- Laplaze L, Benkova E, Casimiro I, et al. 2007. Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell 19, 3889–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Mo X, Shou H, Wu P. 2006. Cytokinin-mediated cell cycling arrest of pericycle founder cells in lateral root initiation of Arabidopsis . Plant and Cell Physiology 47, 1112–1123. [DOI] [PubMed] [Google Scholar]
- Liang J, Zhao L, Challis R, Leyser O. 2010. Strigolactone regulation of shoot branching in chrysanthemum (Dendranthema grandiflorum). Journal of Experimental Botany 61, 3069–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljung K, Hull AK, Celenza J, Yamada M, Estelle M, Normanly J, Sandberg G. 2005. Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell 17, 1090–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. 1997. Organization and cell differentiation in lateral roots of Arabidopsis thaliana . Development 124, 33–44. [DOI] [PubMed] [Google Scholar]
- Marhavý P, Bielach A, Abas L, et al. 2011. Cytokinin modulates endocytic trafficking of PIN1 auxin efflux carrier to control plant organogenesis. Developmental Cell 21, 796–804. [DOI] [PubMed] [Google Scholar]
- Marhavý P, Duclercq J, Weller B, Feraru E, Bielach A, Offringa R, Friml J, Schwechheimer C, Murphy A, Benková E. 2014. Cytokinin controls polarity of PIN1-dependent auxin transport during lateral root organogenesis. Current Biology 24, 1031–1037. [DOI] [PubMed] [Google Scholar]
- Mason MG, Mathews DE, Argyros DA, Maxwell BB, Kieber JJ, Alonso JM, Ecker JR, Schaller GE. 2005. Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis . Plant Cell 17, 3007–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayzlish-Gati E, De-Cuyper C, Goormachtig S, et al. 2012. Strigolactones are involved in root response to low phosphate conditions in Arabidopsis. Plant Physiology 160, 1329–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minakuchi K, Kameoka H, Yasuno N, et al. 2010. FINE CULM1 (FC1) works downstream of strigolactones to inhibit the outgrowth of axillary buds in rice. Plant and Cell Physiology 51, 1127–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira S, Bishopp A, Carvalho H, Campilho A. 2013. AHP6 inhibits cytokinin signaling to regulate the orientation of pericycle cell division during lateral root initiation. PLoS ONE , 8, e56370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Risueno MA, Van Norman JM, Moreno A, Zhang J, Ahnert SE, Benfey PN. 2010. Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329, 1306–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mravec J, Skůpa P, Bailly A, et al. 2009. Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 459, 1136–1140. [DOI] [PubMed] [Google Scholar]
- Paciorek T, Zažímalová E, Ruthardt N, et al. 2005. Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435, 1251–1256. [DOI] [PubMed] [Google Scholar]
- Pandya-Kumar N, Shema R, Kumar M, et al. 2014. Strigolactone analog GR24 triggers changes in PIN2 polarity, vesicle trafficking and actin filament architecture. New Phytologist 202, 1184–1196. [DOI] [PubMed] [Google Scholar]
- Péret B, De Rybel B, Casimiro I, Benková E, Swarup R, Laplaze L, Beeckman T, Bennett MJ. 2009. Arabidopsis lateral root development: an emerging story. Trends in Plant Science in 14, 399–408. [DOI] [PubMed] [Google Scholar]
- Pérez-Torres C-A, López-Bucio J, Cruz-Ramírez A, Ibarra-Laclette E, Dharmasiri S, Estelle M, Herrera-Estrella L. 2008. Phosphate availability alters lateral root development in Arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. Plant Cell 20, 3258–3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen A, Mason MG, De Cuyper C, et al. 2012. Strigolactones suppress adventitious rooting in Arabidopsis and pea. Plant Physiology 158, 1976–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen A, Depuydt S, Goormachtig S, Geelen D. 2013a. Strigolactones fine-tune the root system. Planta 238, 615–626. [DOI] [PubMed] [Google Scholar]
- Rasmussen A, Heugebaert T, Matthys C, Van Deun R, Boyer F-D, Goormachtig S, Stevens C, Geelen D. 2013b. A fluorescent alternative to the synthetic strigolactone GR24. Molecular Plant 6, 100–112. [DOI] [PubMed] [Google Scholar]
- Riefler M, Novak O, Strnad M, Schmülling T. 2006. Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18, 40–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruyter-Spira C, Kohlen W, Charnikhova T, et al. 2011. Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: another belowground role for strigolactones? Plant Physiology 155, 721–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Růžička K, Ljung K, Vanneste S, Podhorská R, Beeckman T, Friml J, Benková E. 2007. Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19, 2197–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Růžička K, Šimášková M, Duclercq J, Petrášek J, Zažímalová E, Simon S, Friml J, Van Montagu MCE, Benková E. 2009. Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proceedings of the National Academy of Sciences, USA 106, 4284–4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Luong P, Huq E. 2007. The F-Box protein MAX2 functions as a positive regulator of photomorphogenesis in Arabidopsis. Plant Physiology 145, 1471–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Zhu L, Bu Q-Y, Huq E. 2012. MAX2 affects multiple hormones to promote photomorphogenesis. Molecular Plant 5, 750–762. [DOI] [PubMed] [Google Scholar]
- Shinohara N, Taylor C, Leyser O. 2013. Strigolactone can promote or inhibit shoot branching by triggering rapid depletion of the auxin efflux protein PIN1 from the plasma membrane. PLoS Biology 11, e1001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden KC, Simkin AJ, Janssen BJ, Templeton KR, Loucas HM, Simons JL, Karunairetnam S, Gleave AP, Clark DG, Klee HJ. 2005. The Decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. Plant Cell 17, 746–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg P, Ward S, Leyser O. 2010. Auxin and strigolactones in shoot branching: intimately connected? Biochemical Society Transactions 38, 717–722. [DOI] [PubMed] [Google Scholar]
- Sun H, Tao J, Liu S, Huang S, Chen S, Xie X, Yoneyama K, Zhang Y, Xu G. 2014. Strigolactones are involved in phosphate- and nitrate-deficiency-induced root development and auxin transport in rice. Journal of Experimental Botany 65, 6735–6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q, Reed JW. 1999. Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 126, 711–721. [DOI] [PubMed] [Google Scholar]
- To JPC, Haberer G, Ferreira FJ, Deruère J, Mason MG, Schaller GE, Alonso JM, Ecker JR, Kieber JJ. 2004. Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16, 658–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya Y, Vidaurre D, Toh S, Hanada A, Nambara E, Kamiya Y, Yamaguchi S, McCourt P. 2010. A small-molecule screen identifies new functions for the plant hormone strigolactone. Nature Chemical Biology 6, 741–749. [DOI] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, et al. 2008. Inhibition of shoot branching by new terpenoid plant hormones. Nature 455, 195–200. [DOI] [PubMed] [Google Scholar]
- Van Norman JM, Zhang J, Cazzonelli CI, Pogson BJ, Harrison PJ, Bugg TDH, Chan KX, Thompson AJ, Benfey PN. 2014. Periodic root branching in Arabidopsis requires synthesis of an uncharacterized carotenoid derivative. Proceedings of the National Academy of Sciences, USA 111, E1300–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanstraelen M, Benková E. 2012. Hormonal interactions in the regulation of plant development. Annual Review of Cell and Developmental Biology 28, 463–487. [DOI] [PubMed] [Google Scholar]
- Woo HR, Chung KM, Park J-H, Oh SA, Ahn T, Hong SH, Jang SK, Nam HG. 2001. ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 13, 1779–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Yoneyama K, Yoneyama K. 2010. The strigolactone story. Annual Review of Phytopathology 48, 93–117. [DOI] [PubMed] [Google Scholar]
- Zhang S, Li G, Fang J, et al. 2010. The interactions among DWARF10, auxin and cytokinin underlie lateral bud outgrowth in rice. Journal of Integrative Plant Biology 52, 626–638. [DOI] [PubMed] [Google Scholar]
- Zhang W, To JPC, Cheng C-Y, Schaller GE, Kieber JJ. 2011. Type-A response regulators are required for proper root apical meristem function through post-transcriptional regulation of PIN auxin efflux carriers. Plant Journal 68, 1–10. [DOI] [PubMed] [Google Scholar]
- Zhao L-H, Zhou XE, Wu Z-S, et al. 2013. Crystal structures of two phytohormone signal-transducing α/β hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14. Cell Research 23, 436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser X, Cashman JR, Cohen JD, Weigel D, Chory J. 2001. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291, 306–309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.