Abstract
Background
Variations in the alleles for the alcohol metabolizing enzymes have been shown to influence risk for alcohol dependence. One variant, ADH1B*3, is observed almost exclusively in populations of African ancestry and has been shown to be associated with reduced rates of alcohol dependence. We conducted an alcohol challenge study to test whether ADH1B*3 is associated with differences in subjective and physiological response to alcohol.
Method
We administered a moderate dose of alcohol (0.72g/kg for males, 0.65g/kg for females) to a sample of African American young adults (n = 91; ages 21–26). Participants were genotyped for ADH1B, as well as additional polymorphisms that might contribute to alcohol response. Breath alcohol concentration, self-reported sedation and stimulation, and pulse rate were assessed prior to alcohol administration and for 2.5 hours following administration.
Results
ADH1B*3 was associated with higher levels of sedation and a sharper increase in pulse rate immediately following alcohol consumption.
Conclusions
These findings suggest that the lower rates of alcohol dependence in those with ADH1B*3 alleles may be due to differences in alcohol response, particularly increased sedation.
Introduction
Genetic differences in the alcohol metabolizing enzymes have been shown to make a significant contribution to alcohol dependence risk (Luczak et al., 2006; Li, 2000). One hypothesized mechanism by which these genetic differences influence alcohol use disorders is through differences in response to alcohol. Considerable research has demonstrated that individual differences in response to alcohol are genetically influenced (Heath et al., 1999) and related to risk for heavy alcohol use and alcohol dependence (e.g., Conrod et al., 2001; Schuckit & Smith, 2001).
Alleles for the alcohol metabolizing enzymes have been found to occur at different frequencies in different ethnic groups. A variant of one of the alcohol dehydrogenase genes, ADH1B*3, has been found almost exclusively in populations of African ancestry (Osier et al., 2002) and has been shown to be associated with lower rates of alcohol dependence (Edenberg et al., 2006; Wall et al., 2003). The present study is the first to test subjective and physiological response to alcohol as a potential mechanism of the protective effect of this allele.
ADH and ALDH Polymorphisms and Alcohol Dependence Risk
Following consumption, alcohol is primarily metabolized by alcohol dehydrogenase (ADH) enzymes in the liver, which oxidize ethanol into acetaldehyde. There are seven known genes that code for ADH enzymes. The majority of research has examined the class I ADH genes (ADH1A, ADH1B, ADH1C). Two of these (ADH1B and ADH1C) have been found to exhibit variants that encode for enzymes with different kinetic properties (Edenberg, 2007). Acetaldehyde is, in turn, broken down into acetate by aldehyde dehydrogenase (ALDH). There are also several molecular forms of ALDH. The gene for mitochondrial ALDH (ALDH2) has been found to have two allele variants and three variants have been identified in the promoter region of the cytosolic isoform ALDH1A1. Genes that encode for ADH enzymes that more rapidly metabolize alcohol, or for ALDH enzymes that are less efficient in the breakdown of acetaldehyde, have been found to be associated with lower risk for alcohol dependence and heavy drinking. The hypothesized mechanism for this protection is higher transient levels of acetaldehyde, which can produce a stronger response to alcohol, including a “flushing reaction” (see Wall, 2005 for a review). Although acetaldehyde levels are difficult to directly measure in humans, animal models have demonstrated that faster elimination of alcohol, due in part to high ADH activity, can create a short-term acetaldehyde “burst” which is associated with reduced alcohol intake (Quintanilla et al., 2007).
Significant variability in frequencies of ALDH alleles have been observed across ethnic groups. ALDH2*2 alleles have been found at moderate rates (20–50%) in samples of northeast Asian heritage (Goedde et al., 1992), and have been found to be associated with lower risk for the development of alcohol dependence (Luczak et al., 2006). Ethnic differences also have been identified in the frequency of the three variants in the ALDH1A1 promoter region: ALDH1A1*1, ALDH1A1*2, and ALDH1A1*3. ADLH1A1*2 alleles have been found at low frequencies (<4%) in Asian, Jewish, Caucasian, Native American, and African American samples; ADLH1A1*3 alleles have been identified at low frequency (≈3%) in samples of Native American and African ancestry (Ehlers et al., 2004; Moore et al., 2007; Spence et al., 2003). There is some evidence that ALDH1A*3 is associated with alcohol dependence in African Americans (Spence et al, 2003), while the association between ADLH1A1*2 and alcohol dependence is mixed (Ehlers et al., 2004; Moore et al., 2007; Spence et al., 2003). The effect of ALDH1A1 variants on alcohol metabolism and alcohol response is unclear.
Studies of ADH1C variants have demonstrated decreased risk for alcohol dependence in those homozygous for ADH1C*1 compared to those with at least one ADH1C*2 allele in Asian populations (Higuchi et al., 1996; Shen et al., 1997). Modest associations between ADH1C*2 and increased alcohol dependence risk have also been demonstrated in European populations (Whitfield, 1997). Studies of linkage disequilibrium have indicated that the association between ADH1C*2 and alcohol dependence may be due instead to variants of ADH1B in some ethnic groups (Osier et al. 1999), although other studies have demonstrated independent effects of ADH1C*2 (Luo et al., 2006). It may be that ADH1C*2 has a larger effect on alcohol dependence in populations where ADH1B*2 is infrequent, such as Europeans and African Americans (Whitfield, 1997).
ADH1B*2 alleles have been identified at high frequencies (>60%) in east-Asian populations and at low frequencies (<13%) in European and North African populations (Osier et al., 2002). Meta-analytic studies have indicated a significant association between ADH1B*2 alleles and reduced risk for alcohol dependence in both Asian (Luczak et al., 2006) and European samples (Whitfield, 2002).
The ADH1B*3 allele has been found primarily in people of African descent (Bosron & Li, 1987) from almost all regions of Africa (Osier et al., 2002). This allele has also been identified at low frequency (≈ 6%) in certain Native American groups (Wall et al., 1997), although this may be due to population admixture. Up to one-third of African Americans possess an ADH1B*3 allele (Ehlers et al., 2001; Li et al., 2001; McCarver et al., 1997). In African Americans, the presence of at least one ADH1B*3 allele has been associated with reduced drinking behavior (Ehlers et al., 2001) and lower risk for alcohol dependence (Ehlers et al., 2007; Edenberg et al., 2006; Luo et al., 2006). ADH1B*3 has also been found to be negatively associated with family history of alcohol dependence (Ehlers et al., 2001) and associated with decreased risk of alcohol-related birth defects (McCarver et al., 1997). ADH1B*3 alleles have also been associated with reduced likelihood of alcohol dependence and heavy consumption in Native Americans (Wall et al., 2003).
ADH1B*3 and Response to Alcohol in African Americans
Epidemiological data suggest that, compared to Caucasians, African Americans have lower lifetime prevalence of alcohol use disorders (Breslau et al., 2006). African American adolescents show slower increases in rates of drinking (Warheit et al., 1996) and have higher abstention rates (Substance Abuse and Mental Health Services Administration, 2003). African American adolescents (Wallace et al., 2003; Bachman et al., 1991) and college students (O’Malley & Johnston, 2002) also engage in less heavy drinking than Caucasians.
The current study was designed to test the association between ADH1B polymorphisms and alcohol response in an African American sample. Prior studies have demonstrated that ADH1B*3 alleles are associated with faster elimination of alcohol (Thomasson et al., 1995), and in vitro studies support greater ethanol-oxidizing activity in enzymes encoded by ADH1B*3 compared to ADH1B*1 (Lee et al., 2004). However, as Scott and Taylor (2007) have noted, research on alcohol metabolism in African-Americans is limited. To date, no study has examined differences in response to alcohol as the mechanism for the protective effect of this allele.
The current study utilized an alcohol challenge paradigm to test subjects with and without ADH1B*3 alleles for differences in their subjective and physiological response to alcohol. We administered a moderate dose of alcohol, designed to produce a peak breath alcohol concentration (BrAC) of approximately 0.075 to 0.080%, to a sample of African American young adults (ages 21–26). Measures of alcohol response (including self-reported sedation, stimulation, and pulse rate) were assessed at baseline, every 15 minutes during the first hour following ingestion, and every 30 minutes for 90 minutes thereafter. Participants were genotyped for polymorphisms of ADH1B, ADH1C, and the ALDH1A1 promoter region.
We hypothesized that participants with at least one ADH1B*3 allele would exhibit a stronger response to alcohol, indicated by increased sedation, stimulation, and pulse rate following consumption, than those with two ADH1B*1 alleles. Although ADH1B*2 has been shown to be associated with lower risk for alcohol dependence as well (Whitfield, 2002), this variant is rare in populations of African ancestry (Osier et al, 2002). ADH1C*2 has also been found to be associated with increased alcohol dependence risk (Whitfield, 1997), and is thought to be associated with slower metabolism of alcohol (Edenberg, 2007). We tested whether subjects with ADH1C*2 alleles exhibited lower response to alcohol than those with ADH1C*1. As it is unclear whether ADH1B and ADH1C exert independent effects on alcohol metabolism (Luo et al., 2006; Osier et al., 1999), analyses were conducted separately for ADH1B and ADH1C.
Materials and Methods
Participants
The full sample consisted of 116 young adult African Americans. The sample was 42% male and had a mean age of 21.9 years (SD = 1.15, range 21–26). While all study participants were of African descent, 10% of the sample described themselves as mixed race, and 3% identified their ethnicity as Hispanic. For those reporting mixed race, study inclusion criteria required that they have at least one parent of African ancestry. The majority of the sample (77%) had some college education and 20% reported being college graduates. Participants were required to be between the ages of 21 and 26 years and to be current drinkers. Participants were required to report at least one drinking episode in the past six months where their consumption was consistent with the amount they would receive in the present study (e.g., 3–4 drinks for a 165 pound male). Participants were excluded if they were currently abstaining from alcohol because of an alcohol use disorder, had significant medical or psychiatric illness (e.g., psychotic disorders, past head injury with loss of consciousness > 5 minutes), or were currently taking medication for which use of alcohol is contraindicated.
Procedures
Study procedures were approved by the University of Missouri and Washington University Institutional Review Boards, and written informed consent was obtained prior to each session. Participants were recruited from the University of Missouri, the city of Columbia, Missouri, and the surrounding area. Fliers were placed at various locations on campus and at local businesses. Potential participants were screened by phone to determine eligibility for the study. Participants who met eligibility criteria were scheduled for an in-person interview conducted by a trained research assistant in a private office. Participants also completed questionnaire measures, including self-report measures of alcohol use. Buccal brush samples were taken from each participant for genotyping. Participants received $40 for their participation in the interview.
Participants were scheduled for an alcohol challenge session approximately one week after the initial interview. Participants were given an information packet prior to their laboratory appointment, which instructed them to refrain from alcohol for 24 hours before the session and to refrain from other drug use for 48 hours. They were instructed to refrain from eating or drinking caffeinated or dairy beverages for 8 hours prior to their session (starting at 12 midnight the prior evening). Participants arrived at the laboratory at 8:00 a.m. A questionnaire was administered to verify compliance with pre-session instructions. A breath alcohol test was used to verify abstinence from alcohol. Females were given a urine pregnancy test and excluded from the study if they tested positive. A standard low-fat breakfast (bagel and juice) was provided.
The alcohol administration and assessment were conducted in a private office, with a semi-recumbent chair, separate from that used for interviews. This office was equipped with a vital signs monitor and computer. Participants were assessed prior to beverage consumption, in 15 minute intervals for the first hour following consumption, and 30 minute intervals thereafter (i.e., 15, 30, 45, 60, 90, 120, and 150 minutes). Between 8:30 and 9:00 a.m., baseline measures were taken. At 9:00 a.m., participants received an alcoholic beverage. Participants received a dose of alcohol equivalent to 0.72g/kg of alcohol for males and 0.65g/kg for females. This dose was designed to produce a peak BrAC of approximately 0.075 to 0.080 mg% for both males and females (Sher & Walitzer, 1986). The alcohol drinks were made using 50% alcohol (vodka), in 20% solution with non-caffeinated soda (tonic). Beverages were consumed over a 15-minute period. At approximately noon, each participant was provided lunch.
To minimize risk, the following procedures outlined in the NIAAA Recommended Council Guidelines on Ethyl Alcohol Administration in Human Experimentation were used (NIAAA, 2005). Participants were not allowed to leave the laboratory until their observable behavior had returned to normal and until their BrAC fell below 0.02mg%. Each participant was also required to travel home by taxi (provided by the study), or with a friend. Participants were required to state in writing that he or she would not drive a car or operate other machinery for three hours after leaving the laboratory. They were reimbursed $100 for participation in the session.
Measures
Alcohol Use Behavior
Participant alcohol involvement was assessed through both interview (i.e., the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA-II; Bucholz et al., 1994)) and self-report (e.g., Drinking Styles Questionnaire (DSQ: Smith et al., 1995)). Self-report of past month quantity and frequency of alcohol use are included in the current analyses.
BrAC
Breath alcohol readings were taken using a breathalyzer device (Intoximeters, Inc.) at baseline and at all measurement points after consumption of the beverage.
Subjective Feelings of Intoxication
Subjective feelings of intoxication were evaluated at baseline and at all measurement points following beverage consumption, using the Biphasic Alcohol Effects Scale (BAES: Martin et al., 1993). This measure assesses separate sedating and stimulating effects of alcohol on both the ascending and descending limb of the blood alcohol curve (Earleywine & Erblich, 1996).
Pulse Rate
Pulse rate was obtained using an automatic vital signs measurement system at baseline and at all measurement points after consumption of the beverage.
Genotyping
Buccal brush samples were sent to Washington University School of Medicine for genotype analysis. The ADH1B haplotypes were determined by separate PCR amplifications (primers from Osier et al, 2002) of the regions around exon 3 (Arg48His; rs1229984) and exon 9 (Arg370Cys; rs2066702) followed by restriction endonuclease digests (with Msl I and Alw NI, respectively). No samples were heterozygous at both sites, so the haplotypes ADH1B*1 (48Arg-370Arg), ADH1B*2 (48His-370Arg), and ADH1B*3 (48Arg-370Cys) were clearly indicated.
Likewise, the ADH1C haplotypes were determined by separate PCR amplifications of the regions around exon 6 (Arg272Gln; rs1693482; PCR primers from sequencing protocol of ss8819648) and exon 8 (Ile350Val; rs698; primers from Osier et al, 2002), followed by restriction endonuclease digests (with Alu I and Ssp I, respectively). These sites are known to be in strong linkage disequilibrium, and, indeed, all samples were either homozygous for both Arg (exon 6) and Ile (exon 8), homozygous for both Gln (exon 6) and Val (exon 8), or were heterozygous at both sites. Thus, haplotypes ADH1C*1 (272Arg-350Ile) and ADH1C*2 (272Gln-350Val) were determined.
The ALDH1A1 promoter region polymorphism (rs8187866; alleles ALDH1A*1, ALDH1A*2 (17 base-pair deletion) and ALDH1A1*3 (3 base-pair insertion)) was genotyped on the ABI 310 Genetic Analyzer using the A4-forward and A4-reverse primers of Spence and colleagues (2003), with a 5′-FAM-fluorescent dye added to the A4-forward primer. The size estimates for the alleles using the Applied Biosystems Genescan-500 size standard were about 6 basepairs larger (198, 215, 218 bp vs. 192, 209, 212 bp) than Spence and colleagues (2003).
Results
Genotype Frequencies and Descriptive Statistics
Table 1 presents ADH1B and ADH1C genotype frequencies for the full sample. Thirty-four percent of the sample had at least one ADH1B*3 allele, while 25% had at least one ADH1C*2 allele. Hardy-Weinberg equilibrium was not significant for ADH1B (rs2066702; χ2 (1, N = 116) = 0.39, ns; rs1229984; χ2 (1, N = 116) = 0.02, ns) or ADH1C, χ2 (1, N = 116) = 0.07, ns; Rodriguez, Gaunt & Day, 2009). Chi-square analyses did not indicate a significant association between ADH1B and ADH1C genotype frequencies (χ2 (6, N = 116) = 7.27, p = .30). The distribution of genotypes did not differ across gender or ethnic group (African American, mixed race, Hispanic/Latino).
Table 1.
ADH1B and ADH1C Genotype Frequencies.
| ADH1B Genotype | Full Sample (N = 116) | ADH1C*1/*1 (n = 85, 73%) | ADH1C*1/*2 (n = 29, 25%) | ADH1C*2/*2 (n = 2, 2%) |
|---|---|---|---|---|
| ADH1B*1/*1 | 76 (66%) | 50 (43%) | 24 (21%) | 2 (2%) |
| ADH1B*1/*2 | 2 (2%) | 2 (2%) | 0 | 0 |
| ADH1B*1/*3 | 33 (28%) | 28 (24%) | 5 (4%) | 0 |
| ADH1B*3/*3 | 5 (4%) | 5 (4%) | 0 | 0 |
|
| ||||
| Final Sample (n = 91) | ADH1C*1/*1 (n = 67, 74%) | ADH1C*1/*2 (n = 22, 24%) | ADH1C*2/*2 (n = 2, 2%) | |
|
| ||||
| ADH1B*1/*1 | 60 (66%) | 39 (43%) | 19 (21%) | 2 (2%) |
| ADH1B*1/*3 | 26 (29%) | 23 (25%) | 3 (3%) | 0 |
| ADH1B*3/*3 | 5 (5%) | 5 (6%) | 0 | 0 |
Note: Values are number of participants or percent of the full or final sample (in parentheses).
Two participants had one ADH1B*2 allele, rare in samples of African ancestry. Seven participants were heterozygous for ALDH1A*2 (ALDH1A*1/*2) and four were heterozygous for ALDH1A*3 (ALDH1A*1/*3). ADH1B*2 has been shown to influence alcohol response (Cook et al., 2005; Duranceaux et al., 2006), while the influence of ALDH1A*2 and ALDH1A*3 alleles on response to alcohol is uncertain. Participants with these alleles (n = 13) were excluded from study analyses testing the effect of ADH1B and ADH1C on alcohol response1.
Thirteen participants became ill (vomited) following alcohol consumption. Chi-square analyses indicated that those who became ill did not differ from the remainder of the sample in ADH1B or ADH1C status. These participants also did not differ in gender, weight, and past month frequency of alcohol consumption. However, there was a trend for those who became ill to report a lower quantity of past month alcohol consumption (t (114) = 1.87, p = .06; mean drinks per occasion: 3.05 vs. 2.33). As the research protocol was discontinued for these participants, they were excluded from study analyses. Excluding these participants, as well as participants with ADH1B*2, ALDH1A*2 or ALDH1A*3 alleles, resulted in a sample size of 91 for study analyses.
Table 1 also presents genotype frequencies for the final sample used in study analyses. For analytic purposes, ADH1B and ADH1C gene status was coded as two levels, comparing subjects with at least one copy of either ADH1B*3 or ADH1C*2 to those with no copies (homozygous for either ADH1B*1 or ADH1C*1). Analyses of ADH1B were conducted within subjects homozygous for ADH1C*1 (ADH1B*3(-)/ADH1C*1 compared to ADH1B*3(+)/ADH1C*1), while analyses of ADH1C were conducted within those homozygous for ADH1B*1.
Table 2 presents gender, weight, mean peak BrAC and self-reported past month quantity and frequency of alcohol consumption for participants included in study analyses separately by ADH1B and ADH1C allele status. There were no significant differences in these variables for the alleles of either ADH1B or ADH1C.
Table 2.
Gender, Weight, BrAC, and Alcohol Consumption by Gene Status.
| Final Sample | ADH1B | ADH1C | |||
|---|---|---|---|---|---|
|
| |||||
| n = 91 | ADH1B*1 (n = 60) | ADH1B*3 (n = 31) | ADH1C*1 (n = 67) | ADH1C*2 (n = 24) | |
| Female (%) | 57% | 58% | 55% | 55% | 63% |
| Weight (pounds) | 172.0 (42.26) | 175.6 (45.63) | 165.2 (34.48) | 174.0 (45.57) | 166.5 (31.41) |
| Peak BrAC (mg%) | .077 (.01) | .076 (.01) | .078 (.01) | .078 (.01) | .074 (.01) |
| Alcohol Frequency (days/month) | 6.98 (7.47) | 6.9 (6.56) | 7.2 (9.10) | 6.9 (7.71) | 7.3 (6.92) |
| Alcohol Quantity (drinks/occasion) | 2.92 (1.49) | 3.0 (1.53) | 2.7 (1.42) | 3.0 (1.54) | 2.7 (1.36) |
Note: Values are percents, means, or standard deviations (in parentheses).
ADH1B and Alcohol Response
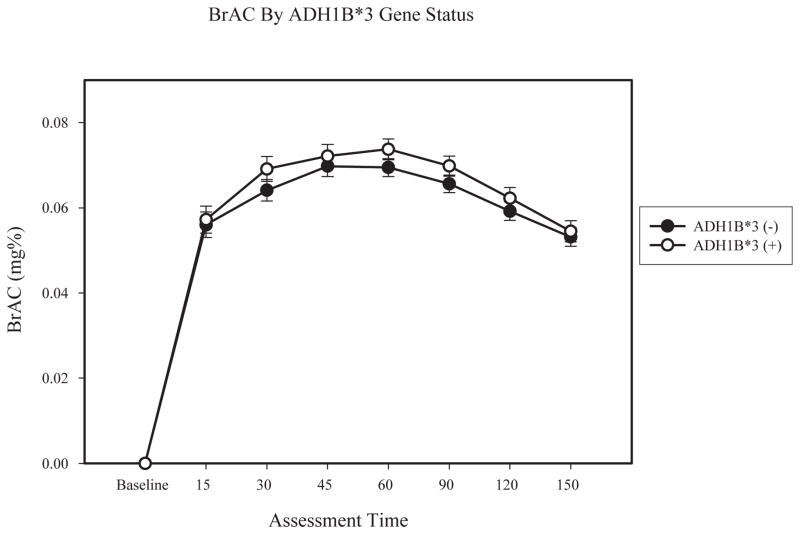
We first tested whether changes in BrAC varied as a function of ADH1B gene status. A 7 (time) × 2 (gender) × 2 (ADH1B gene status) mixed factorial analysis of variance (ANOVA) was conducted. Baseline BrAC was not included, as it was a constant. Results indicated a main effect of time (F (6, 62) = 41.91, p < .01; partial η2 = .41) and gender on BrAC (F (1, 62) = 4.46, p < .05; partial η2 = .07). Both the main effect of ADH1B gene status (F (1, 62) = 1.07, p = .31; partial η2 = .02) and the time x ADH1B interaction (F (6, 62) = 0.51, p = .80; partial η2 = .01) were not significant. Follow-up ANOVAs indicated that BrAC did not differ by ADH1B gene status at any time point (see Figure 1).
Figure 1.
Lines represent BrAC separately by ADH1B gene status, within participants homozygous for ADH1C*1. Error bars are based on the standard error of the mean.
A series of 8 (time) × 2 (gender) × 2 (ADH1B gene status) mixed factorial ANOVAs were then conducted to examine change in alcohol response measures (sedation, stimulation, and pulse rate) by ADH1B gene status. For these analyses, BrAC at the 60 minute time point was included as a covariate, as this time point represented the average peak BrAC for most participants.
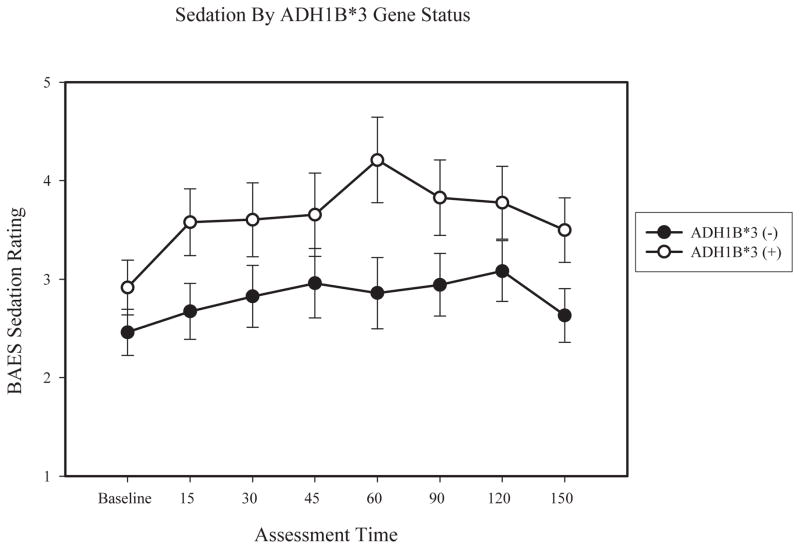
Results for sedation indicated a main effect of ADH1B gene status (F (1, 62) = 4.23, p < .05; partial η2 = .07), gender (F (1, 62) = 5.90, p < .05; partial η2 = .09). There was also a significant time x gender interaction (F (7, 62) = 3.59, p < .01; partial η2 = .06), with females reporting greater increases in sedation. Follow-up analysis of covariance (ANCOVA) was then used to compare the groups differentiated by allele status at each time point. BrAC for each time point was included as a covariate, except for at baseline, where it was a constant. Results indicated no difference in self-reported sedation across ADH1B groups at baseline (F (1, 66) = 1.10, p = .30; partial η2 = .02). ADH1B*3 participants reported significantly higher sedation at the 15 minute (F (1, 66) = 4.84, p < .05; partial η2 = .07), 60 minute (F (1, 66) = 4.65; p < .05; partial η2 = .07), and 150 minute (F (1, 66) = 5.16; p < .05; partial η2 = .08) time points, with a marginally significant difference at the 90 minute (F (1, 66) = 3.55; p = .06; partial η2 = .05) time point. ADH1B groups did not significantly differ at the 30 minute (F (1, 66) = 2.86, p = .10; partial η2 = .04), 45 minute (F (1, 66) = 1.26, p = .27; partial η2 = .02) and 120 minute (F (1, 66) = 1.83, p = .18; partial η2 = .03) time points. Figure 2 presents estimated means of BAES sedation separately by ADH1B allele status.
Figure 2.
Lines represent means of self-reported sedation from the BAES separately by ADH1B gene status, within participants homozygous for ADH1C*1, controlling for BrAC at 60 minutes. Error bars are based on the standard error of the mean.
Results did not indicate significant ADH1B main effects or interactions for self-reported stimulation. For stimulation, results indicated a time x gender interaction (F (7, 62) = 2.50, p < .05; partial η2 = .04), with males reporting a sharper increase in stimulation than females. No other main effects or interactions were significant.
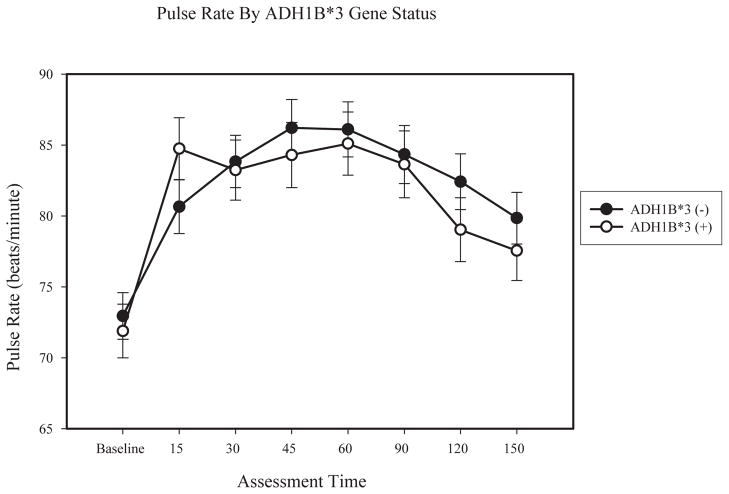
For pulse rate, there was a significant main effect of time (F (7, 62) = 2.18, p < .05; partial η2 = .04), and gender (F (1, 62) = 13.07, p < .01; partial η2 = .18), as well as a time x ADH1B interaction (F (7, 62) = 2.16, p < .05; partial η2 = .04). Examination of means indicated that males exhibited higher pulse rate than females. Although ANCOVA results did not indicate significant differences across ADH1B groups at any time point, as shown in Figure 3, participants with ADH1B*3 alleles experienced a sharper increase in pulse rate from the baseline to 15 minute assessments, and presents estimated means of pulse rate separately by ADH1B gene status.
Figure 3.
Lines represent means of pulse rate separately by ADH1B gene status, within participants homozygous for ADH1C*1, controlling for BrAC at 60 minutes. Error bars are based on the standard error of the mean.
ADH1C and Alcohol Response
A 7 (time) × 2 (gender) × 2 (ADH1B gene status) mixed factorial analysis of variance (ANOVA) was conducted to test whether BrAC varied as a function of ADH1C gene status. Baseline BrAC was not included, as it was a constant. Results indicated a main effect of time (F (6, 55) = 39.94, p < .01; partial η2 = .43) on BrAC, but no significant main effect of ADH1C (F (1, 55) = 0.04, p = .84; partial η2 = .001) or time x ADH1C interaction (F (6, 55) = 0.64, p = .70; partial η2 = .01).
A parallel series of 8 (time) × 2 (gender) × 2 (ADH1C gene status) mixed factor ANOVAs were then conducted to examine change in alcohol response measures by ADH1C gene status. No significant main effects were observed for ADH1C gene status on sedation (F (1, 55) = 1.78, p = .19; partial η2 = .03), stimulation (F (1, 55) = 0.95, p = .34; partial η2 = .02), or pulse rate (F (1, 55) = 1.15, p = .29; partial η2 = .02). Interactions involving ADH1C were also not significant.
Discussion
The ADH1B*3 allele, observed in up to one-third of participants in African American samples, has been found to be associated with reduced risk for alcohol dependence (Edenberg et al., 2006; Luo et al., 2006; Wall et al., 2003). One hypothesized mechanism for this is that the faster elimination of alcohol in carriers of this allele leads to higher levels of acetaldehyde and a stronger response to alcohol. Results of the present study provide the first test of subjective and physiological indicators of alcohol response as a function of ADH1B*3 alleles.
Despite similar recent alcohol use and post-consumption BrAC levels, individuals in this study with at least one ADH1B*3 allele reported experiencing greater sedation following a moderate dose of alcohol. This difference was most pronounced at the 60 minute assessment, when most participants were at their peak BrAC. A time x gene status interaction effect indicated that subjects with ADH1B*3 alleles also experienced a sharper increase in pulse rate immediately after consumption compared to those homozygous for ADH1B*1. An increase in pulse rate is consistently associated with alcohol-induced flushing and ADH/ALDH polymorphisms (Peng et al., 1999; Wall et al., 1992). The sharp elevation in pulse rate, followed by a rapid return, is consistent with the short-term acetaldehyde “burst” hypothesized to result from faster alcohol conversion (Quintanilla et al., 2007). The pattern of results is similar to studies comparing ADH1B*1 and ADH1B*2 in an Asian sample (Cook et al., 2005) as well as a mixed Caucasian and African American sample (Duranceaux et al., 2006). The isoenzymes encoded by ADH1B*2 and ADH1B*3 are fairly similar in their kinetic constants and should produce faster alcohol elimination rates compared to ADH1B*1 (Edenberg, 2007).
No significant differences were observed between those with two ADH1C*1 alleles and those with at least one ADH1C*2 allele. Findings have been inconsistent regarding whether ADH1C polymorphisms influence alcohol dependence risk over and above the effect of ADH1B alleles (Luo et al., 2006; Osier et al., 1999). Some studies have identified modest differences between ADH1C*1 and ADH1C*2 in alcohol response (Duranceaux et al., 2006) and alcohol metabolism (Lee et al., 2006) after accounting for ADH1B status.
As noted, ALDH1A1 polymorphisms are also found to occur at low frequencies in African Americans. The association of ALDH1A1 variants with risk for alcohol dependence is unclear (Ehlers et al., 2004; Moore et al., 2007; Spence et al., 2003). Although ALDH1A1 is associated with alcohol metabolism, it is also unclear whether variations in the ALDH1A1 promoter region contribute to differences in alcohol metabolism or alcohol response. In the present study, the frequencies of ALDH1A1 variants (ALDH1A1*2 and ALDH1A1*3) were too low to allow for separate analysis, so participants with these variants were excluded from the analyses, so as not to confound their possible effects on alcohol metabolism with the effects of the tested alleles for ADH1B and ADH1C.
While accounting for ADH1C is most relevant to testing the effect of ADH1B*3, there is also evidence for the influence of other ADH genes on alcohol dependence risk, including ADH4 (Edenberg et al., 2006), ADH5, and ADH7 (Luo et al., 2006). Molecular genetic studies of alcohol dependence risk are increasingly incorporating a range of ADH genes to test their joint and unique effects (Edenberg, 2007). An important direction for future studies of genetic influences on behavioral and psychological factors, such as alcohol response, is to examine a broader range of ADH genes in order to examine the distinct contribution of each and test for potential gene-gene interactions.
While ADH1B*3 is thought to result in faster metabolism of alcohol and higher levels of acetaldehyde, participants in the present study with ADH1B*3 alleles did not differ in BrAC from those with ADH1B*1. This is consistent with prior studies using oral alcohol administration (Taylor et al., 2008). Studies using intravenous alcohol administration and a BrAC clamping method are better able to detect differences in alcohol elimination rate as a function of ADH polymorphisms (Neumark et al., 2004). In addition, more research is needed to determine if ADH1B*3 results in faster production of acetaldehyde.
Another limitation of the current study is that alcohol response was tested for a single dose level across all participants. There is some evidence that the kinetic properties of ADH enzymes in those with ADH1B*3 have greater activity at higher alcohol concentrations (Lee et al., 2006). Effect size results for the current study are relatively modest, with partial η2 values ranging from .04-.08 for sedation main effects. It may be that greater differences in alcohol response would be observed at higher alcohol doses. The study also lacked a placebo condition, which prevents testing of potential expectancy effects. However, both the participants and experimenter were blind to the participants’ genetic status, which makes expectancy effects an unlikely explanation for observed ADH1B*3 differences.
Although previous studies have observed differences in drinking behavior in those with at least one ADH1B*3 allele (Ehlers et al., 2001), no such differences were observed in the current study. This may be the result of eligibility requirements for the study. Participants were only enrolled in the alcohol challenge portion of the study if they reported recent drinking at quantities at least equal to the amount of alcohol administered in the study. The similarity in recent drinking behavior between those with ADH1B*1 and ADH1B*3 alleles reduces the likelihood that ADH1B differences in alcohol response are due to differences in recent drinking history or alcohol tolerance.
Convergent evidence has suggested that the reduced risk for alcohol dependence seen in African Americans with ADH1B*3 alleles may be in part due to differences in alcohol metabolism (Scott & Taylor, 2007). Results of this study provide the first test of behavioral and physiological differences in those with ADH1B*3 alleles following alcohol consumption. Further research is required to more fully understand the role that ADH1B*3 and response to alcohol play in determining alcohol use topography and alcohol dependence risk in African Americans.
Acknowledgments
Supported by grants (K02 AA00269; R21 AA015218; T32 AA13526; F31AA017571) from the National Institute of Alcohol Abuse and Alcoholism.
We would like to acknowledge Amy Doebber for her technical assistance and Bruce Bartholow for his comments on the manuscript.
Footnotes
As the influence of ALDH1A variants on alcohol response is unclear, we chose to conduct analyses excluding participants with ALDH1A*2 and ALDH1A*3 (n = 11). The main findings of the study do not change if these participants are included in analyses. The main effect of ADH1B on sedation is slightly stronger (F (1, 73) = 5.86, p < .05; partial η2 = .09) and the time x pulse rate interaction remains significant (F (7, 73) = 2.08, p < .05; partial η2 = .04).
Contributor Information
Denis M. McCarthy, University of Missouri
Sarah L. Pedersen, University of Missouri
Elizabeth A. Lobos, Washington University, School of Medicine
Richard D. Todd, Washington University, School of Medicine
Tamara L. Wall, University of California, San Diego
References
- Bachman JG, Wallace JM, O’Malley PM, Johnston LD, Kurth CL, Neighbors HW. Racial/ethnic differences in smoking, drinking, and illicit drug use among American high school seniors, 1976–89. Am J Public Health. 1991;81:372–377. doi: 10.2105/ajph.81.3.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosron WF, Li TK. Catalytic properties of human liver alcohol dehydronase isoenzymes. Enzyme. 1987;37:19–28. doi: 10.1159/000469238. [DOI] [PubMed] [Google Scholar]
- Breslau J, Aguilar-Gaxiola S, Kendler KS, Su M, Williams D, Kessler RC. Specifying race-ethnic differences in risk for psychiatric disorder in a USA national sample. Psychol Med. 2006;36:57–68. doi: 10.1017/S0033291705006161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucholz KK, Cadoret R, Cloninger CR, Dinwiddle SH, Hesselbrock VM, Nurnberger JI. A new semi-structured psychiatric interview for use with genetic linkage studies: A report on the reliability of the SSAGA. J Stud Alcohol. 1994;55:149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- Conrod PJ, Peterson JB, Pihl RO. Reliability and validity of alcohol-induced heart rate increase as a measure of sensitivity to the stimulant properties of alcohol. Psychopharmacology. 2001;157:20–30. doi: 10.1007/s002130100741. [DOI] [PubMed] [Google Scholar]
- Cook TAR, Luczak SE, Shea SH, Ehlers CL, Carr LG, Wall TL. Association of ALDH2 and ADH1B genotypes with response to alcohol in Asian Americans. J Stud Alcohol. 2005;66:196–204. doi: 10.15288/jsa.2005.66.196. [DOI] [PubMed] [Google Scholar]
- Duranceaux NC, Schuckit MA, Eng MY, Robinson SK, Carr LG, Wall TL. Associations of variations in alcohol dehydrogenase genes with the level of response to alcohol in non-Asians. Alcohol Clin Exp Res. 2006;30:1470–1478. doi: 10.1111/j.1530-0277.2006.00178.x. [DOI] [PubMed] [Google Scholar]
- Earleywine M, Erblich J. A confirmed factor structure for the biphasic effects of alcohol scale. Exp Clin Psychopharmacol. 1996;4:107–113. [Google Scholar]
- Edenberg HJ. The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Research and Health. 2007;30:5–13. [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Xuei X, Chen HJ, Tian H, Wetherill LF, Dick DM, et al. Association of alcohol dehydrogenase genes with alcohol dependence: a comprehensive analysis. Hum Mol Genet. 2006;15:1539–1549. doi: 10.1093/hmg/ddl073. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Gilder DA, Harris L, Carr L. Association of the ADH2*3 allele with a negative family history of alcoholism in African American young adults. Alcohol Clin Exp Res. 2001;25:1773–1777. [PubMed] [Google Scholar]
- Ehlers CL, Spence JP, Wall TL, Gilder DA, Carr LG. Association of ALDH1 promoter polymorphisms with alcohol-related phenotypes in southwest California Indians. Alcohol Clin Exp Res. 2004;28:1481–1486. doi: 10.1097/01.alc.0000141821.06062.20. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Montane-Jaime K, Moore S, Shafe S, Joseph R, Carr LG. Association of the ADHIB*3 allele with alcohol-related phenotypes in Trinidad. Alcohol Clin Exp Res. 2007;31:216–20. doi: 10.1111/j.1530-0277.2006.00298.x. [DOI] [PubMed] [Google Scholar]
- Goedde HW, Agarwal DP, Fritze G, Meier-Tackmann D, Singh S, Beckman G, et al. Distribution of ADH2 and ALDH2 genotypes in different populations. Hum Genet. 1992;88:344–346. doi: 10.1007/BF00197271. [DOI] [PubMed] [Google Scholar]
- Heath AC, Madden PAF, Bucholz KK, Dinwiddie SH, Slutske WS, Bierut LJ, et al. Genetic differences in alcohol sensitivity and the inheritance of alcoholism risk. Psychol Med. 1999;29:1069–1081. doi: 10.1017/s0033291799008909. [DOI] [PubMed] [Google Scholar]
- Higuchi S, Matsushita S, Muramatsu T, Murayama M, Hayashida M. Alcohol and aldehyde dehydrogenase genotypes and drinking behavior in Japanese. Alcohol Clin Exp Res. 1996;20:493–497. doi: 10.1111/j.1530-0277.1996.tb01080.x. [DOI] [PubMed] [Google Scholar]
- Lee SL, Höög JO, Yin SJ. Functionality of allelic variations in human alcohol dehydrogenase gene family: Assessment of a functional window for protection against alcoholism. Pharmacogenetics. 2004;14:725–732. doi: 10.1097/00008571-200411000-00003. [DOI] [PubMed] [Google Scholar]
- Lee SL, Chau GY, Yao CT, Wu CW, Yin SJ. Functional assessment of human alcohol dehydrogenase family in ethanol metabolism: Significance of first-pass metabolism. Alcohol Clin Exp Res. 2006;30:1132–1142. doi: 10.1111/j.1530-0277.2006.00139.x. [DOI] [PubMed] [Google Scholar]
- Li TK. Pharmacogenetics of responses to alcohol and genes that influence alcohol drinking. J Stud Alcohol. 2000;61:5–12. doi: 10.15288/jsa.2000.61.5. [DOI] [PubMed] [Google Scholar]
- Li TK, Yin S, Crabb DW, O’Connor S, Ramchandi VA. Genetic and Environmental Influences on Alcohol Metabolism in Humans. Alcohol Clin Exp Res. 2001;25:136–144. [PubMed] [Google Scholar]
- Luczak SE, Glatt SJ, Wall TL. Meta-analyses of ALDH2 and ADH1B with alcohol dependence in Asians. Psychol Bull. 2006;132:607–621. doi: 10.1037/0033-2909.132.4.607. [DOI] [PubMed] [Google Scholar]
- Luo X, Kranzler HR, Zuo L, Wang S, Schork NJ, Gelernter J. Diplotype trend regression analysis of the ADH gene cluster and the ALDH2 gene: Multiple significant associations with alcohol dependence. Am J Hum Genet. 2006;78:973–987. doi: 10.1086/504113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin CS, Earleywine M, Musty R, Perrine M, Swift R. Development and validation of the biphasic effects of alcohol scale. Alcohol Clin Exp Res. 1993;17:140–146. doi: 10.1111/j.1530-0277.1993.tb00739.x. [DOI] [PubMed] [Google Scholar]
- McCarver DG, Thomasson HR, Martier SS, Sokol RJ, Li TK. Alcohol dehydrogenasse-2*3 allele protects against alcohol-related birth defects among African Americans. J Pharmacol Exp Ther. 1997;283:1095–1101. [PubMed] [Google Scholar]
- Moore S, Montane-Jaime K, Shafe S, Joseph R, Crooks H, Carr LC, Ehlers CL. Association of ALDH1 promoter polymorphisms with alcohol-related phenotypes in Trinidad and Tobago. Journal of Studies on Alcohol and Drugs. 2007;68:192–196. doi: 10.15288/jsad.2007.68.192. [DOI] [PubMed] [Google Scholar]
- National Advisory Council on Alcohol Abuse and Alcoholism. Recommended council guidelines on ethyl alcohol administration in human experimentation. Maryland: National Institute on Alcohol Abuse and Alcoholism; 2005. [Google Scholar]
- Neumark YD, Friedlander Y, Durst R, Leitersdorf E, Jaffe D, Ramchandani VA, O’Connor S, Carr LG, Li TK. Alcohol dehydrogenase polymorphisms influence alcohol-elimination rates in a male Jewish population. Alcohol Clin Exp Res. 2004;28:10–14. doi: 10.1097/01.ALC.0000108667.79219.4D. [DOI] [PubMed] [Google Scholar]
- O’Malley PM, Johnston LD. Epidemiology of alcohol and other drug use among American college students. J Stud Alcohol Suppl. 2002;14:23–39. doi: 10.15288/jsas.2002.s14.23. [DOI] [PubMed] [Google Scholar]
- Osier M, Pakstis AJ, Kidd JR, Lee JF, Yin SJ, Ko HC, et al. Linkage disequilibrium at the ADH2 and ADH3 loci and risk for alcoholism. Am J Hum Genet. 1999;64:1147–1157. doi: 10.1086/302317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osier MV, Pakstis Aj, Soodyall H, Comas D, Goldman D, Odusni A, et al. A global perspective on genetic variation at the ADH genes reveals unusual patterns of linkage disequilibrium and diversity. Am J Hum Genet. 2002;71:84–99. doi: 10.1086/341290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng GS, Wang MF, Chen CY, Luu SY, Chau HC, Li TK, Yin SJ. Involvement of acetaldehyde for full protection against alcoholism by homozygosity of the variant allele of mitochondrial aldehyde dehydrogenase gene in Asians. Pharmacogenetics. 1999;9:463–476. [PubMed] [Google Scholar]
- Peterson RJ, Goldman D, Long JC. Effects of worldwide population subdivision on ALDH2 linkage disequilibrium. Genome Res. 1999;9:844–52. doi: 10.1101/gr.9.9.844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintanilla ME, Tampier L, Sapag A, Gerdtzen Z, Israel Y. Sex differences, alcohol dehydrogenase, acetaldehyde burst, and aversion to ethanol in the rat: a systems perspective. Am J Physiol - Endocrinology and Metabolism. 2007;293:E531–7. doi: 10.1152/ajpendo.00187.2007. [DOI] [PubMed] [Google Scholar]
- Rodriguez S, Gaunt TR, Day INM. Hardy-Weinberg Equilibrium testing of biological ascertainment for mendelian randomization studies. American Journal of Epidemiology. 2009;169:505–514. doi: 10.1093/aje/kwn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL. The clinical course of alcohol dependence associated with a low level of response to alcohol. Addiction. 2001;96:903–910. doi: 10.1046/j.1360-0443.2001.96690311.x. [DOI] [PubMed] [Google Scholar]
- Scott DM, Taylor RE. Health-related effects of genetic variations of alcohol-metabolizing enzymes in African Americans. Alcohol Research and Health. 2007;30:18–21. [PMC free article] [PubMed] [Google Scholar]
- Shen YC, Fan JH, Edenberg HJ, Li TK, Cui YH, Wang YF, et al. Polymorphisms of ADH and ALDH genes among four ethnic groups in China and effects upon the risk for alcoholism. Alcohol Clin Exp Res. 1997;21:1272–1277. [PubMed] [Google Scholar]
- Sher KJ, Walitzer KS. Individual differences in the stress-response-dampening effect on alcohol: a dose-response study. J Abnorm Psychol. 1986;95:159–67. doi: 10.1037//0021-843x.95.2.159. [DOI] [PubMed] [Google Scholar]
- Smith GT, McCarthy DM, Goldman MS. Self-reported drinking and alcohol-related problems among early adolescents: Dimensionality and validity over 24 months. J Stud Alcohol. 1995;56:383–394. doi: 10.15288/jsa.1995.56.383. [DOI] [PubMed] [Google Scholar]
- Spence JP, Liang T, Eriksson CJP, Taylor RE, Wall TL, Ehlers CL, Carr LG. Evaluation of aldehyde dehydrogenase 1 promotor polymorphisms identified in human populations. Alcohol Clin Exp Res. 2003;27:1389–1394. doi: 10.1097/01.ALC.0000087086.50089.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Substance Abuse and Mental Health Services Administration. Results from the 2002 National Survey on Drug Use and Health: National Findings. 2003 Retrieved July 30, 2007 from http://www.niaaa.nih.gov/Resources/DatabaseResources/QuickFacts/AlcoholConsumption/dkpat2.htm.
- Taylor RE, Raysor BR, Kwagyan J, Ramchandani VA, Kalu N, Powell-Davis M, Ferguson CL, Carr L, Scott DM. Alterations in ethyl alcohol pharmacokinetics during oral consumption of malt liquor beverages in African Americans. Alcohol Clin Exp Res. 2008;32:2074–2080. doi: 10.1111/j.1530-0277.2008.00795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomasson HR, Beard JD, Li TK. ADH2 gene polymorphisms are determinants of alcohol pharmacokinetics. Alcohol Clin Exp Res. 1995;19:1495–1499. doi: 10.1111/j.1530-0277.1995.tb01013.x. [DOI] [PubMed] [Google Scholar]
- Wall TL. Genetic associations of alcohol and aldehyde dehydrogenase with alcohol dependence and their mechanisms of action. Ther Drug Monit. 2005;27:700–703. doi: 10.1097/01.ftd.0000179840.78762.33. [DOI] [PubMed] [Google Scholar]
- Wall TL, Carr LG, Ehlers CL. Protective association of genetic variation in alcohol dehydrogenase with alcohol dependence in Native American mission Indians. Am J Psychiatry. 2003;160:41–46. doi: 10.1176/appi.ajp.160.1.41. [DOI] [PubMed] [Google Scholar]
- Wall TL, Garcia-Andrade C, Thomasson HR, Carr LG, Ehlers CL. Alcohol dehydrogenase polymorphisms in Native Americans: Identification of the ADH2*3 allele. Alcohol Alcohol. 1997;32:129–132. doi: 10.1093/oxfordjournals.alcalc.a008246. [DOI] [PubMed] [Google Scholar]
- Wall TL, Thomasson HR, Ehlers CL. Investigator-observed alcohol-induced flushing but not self-report of flushing is a valid predictor of ALDH2 genotype. J Stud Alcohol. 1996;57:267–272. doi: 10.15288/jsa.1996.57.267. [DOI] [PubMed] [Google Scholar]
- Wall TL, Thomasson HR, Schuckit MA, Ehlers CL. Subjective feelings of alcohol intoxication in Asians with genetic variations of ALDH2 alleles. Alcohol Clin Exp Res. 1992;16:991–995. doi: 10.1111/j.1530-0277.1992.tb01907.x. [DOI] [PubMed] [Google Scholar]
- Wallace JM, Bachman JG, O’Malley PM, Schulenberg JE, Cooper SM, Johnston LD. Gender and ethnic differences in smoking, drinking and illicit drug use among American 8th, 10th and 12th grade students, 1976–2000. Addiction. 2003;98:225–234. doi: 10.1046/j.1360-0443.2003.00282.x. [DOI] [PubMed] [Google Scholar]
- Warheit GJ, Vega WA, Khoury EL, Gil AA, Elfenbein PH. A comparative analysis of cigarette, alcohol, and illicit drug use among an ethically diverse sample of Hispanic, African American, and non-Hispanic White adolescents. Journal of Drug Issues. 1996;26:901–922. [Google Scholar]
- Whitfield JB. Alcohol dehydrogenase and alcohol dependence: Variation in genotype-associated risk between populations. Am J Hum Genet. 2002;71:1247–1250. doi: 10.1086/344287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield JB. Meta-analysis of the effects of alcohol dehydrogenase genotype on alcohol dependence and alcoholic liver disease. Alcohol Alcohol. 1997;32:613–619. doi: 10.1093/oxfordjournals.alcalc.a008303. [DOI] [PubMed] [Google Scholar]