Abstract
Next-generation sequencing has lead to an unparalleled pace of Mendelian disease gene discovery in recent years. To address the challenges of analysis and sharing of large datasets, we had previously introduced the collaborative web-based GEM.app software (Gonzalez et al., 2013). Here we are presenting the results of using GEM.app over nearly three years and introducing the next generation of this platform. Firstly, GEM.app has been renamed to GENESIS since it is now part of ‘The Genesis Project’ (501c3), a non-for-profit foundation that is committed to providing the best technology to enable research scientists and to connecting patients and clinicians to genomic information. Secondly, GENESIS (GEM.app) has grown to nearly 600 registered users from 44 countries, who have collectively achieved 62 gene identifications or published studies that have expanded phenotype/genotype correlations. Our concept of user driven data sharing and matchmaking is now the main cause for gene discoveries within GENESIS. In many of these findings, researchers from across the globe have been connected, which gave rise to the genetic evidence needed to successfully pinpoint specific gene mutations that explained patients’ disease. Here we present an overview of the various novel insights that have been made possible through the data sharing capabilities of GENESIS/GEM.app.
Keywords: Next-generation sequencing, data sharing, exome, Mendelian disease, Matchmaker Exchange
Introduction
The rate of gene discoveries for Mendelian disease has increased substantially over the past 6 years. This has been made possible by the introduction of affordable whole exome sequencing and the development of effective genome-wide variant filtering strategies for identifying a small subset of potentially causative variants in a given patient. In the coming year we will likely experience a shift from whole exome sequencing to whole genome sequencing for many research applications.
The amount of genomic data will grow substantially with the development of many government driven, academic/hospital, non-for-profit, and commercial genomic databases. A number of for-profit companies, such as Google (https://cloud.google.com/genomics/), 23AndMe (https://www.23andme.com), and IBM (http://researcher.watson.ibm.com/researcher/) are actively establishing new genomic databases. Meanwhile, the NIH has established dbGaP (Mailman et al., 2007) as a relatively passive and classic approach (upload/download model). RD-Connect (Thompson et al., 2014) in Europe is planning a well funded and interactive data repository, whose mission will be similar to dbGaP. As more of these initiatives develop, it is unlikely that they will ever merge to one centralized ‘super-database’. Instead, the abilities of these repositories to exchange specific information and thus integrate will become a key element. This is a key aim of the Matchmaker Exchange.
The advance in next generation sequencing analysis needs to be paired with a harmonized capture of phenotypes and phenotypic elements across many different diseases. Phenotyping is a key element in understanding the complex structure of phenotype/genotype relationships. Key examples of the renewed interest in phenotyping include the Human Phenotype Ontology (HPO) (Kohler et al., 2014) and the phenotypic data elements initiative by the NIH (Thompson et al., 2014). Creating standardized machine-readable phenotype datasets in combination with genomic data and the data sharing efforts via our existing software platforms (GENESIS/GEM.app) will ensure the discovery of phenotypic overlaps, very rare disease gene variants, and global matchmaking based on patient phenotype/genotype data. The GENESIS/GEM.app endeavor believes that this will become a crowd-sourced community effort. These efforts will likely reshape clinical classifications of diseases in many fields.
In many Mendelian diseases, a portion of patients still cannot be genetically diagnosed; thus, identifying the genes that account for these cases will be very informative in our understanding of diseases and phenotypes. In addition, this will allow for the development of affordable and effective diagnostic tests needed for ‘precision medicine’. Mendelian disease genes usually represent strong genetic factors; therefore they will often be the first line of personalized medicine implementations. Furthermore, much of the key biological and mechanistic knowledge is based on initial gene discoveries. Initial drug targets based on Mendelian genes have begun to evolve into successful new therapies. With the sequencing and analysis technologies available today, efficient methods and crowd-sourcing efforts will enhance the pace to discovery and ultimately lead to better health care.
We have developed the GENESIS/GEM.app software to encourage scientists to perform genetic analysis and matchmaking. This has in fact led to self-organizing, investigator driven networks involving several hundred registered scientists. Large consortia and smaller labs have realized that limited numbers of families are not sufficient to resolve all families, thus they have volunteered data to this networking effort. The motivation has been that investigators are able to freely determine their level of engagement in sharing their own data and ultimately that matchmaking events in GENESIS/GEM.app have led to a higher success rate and more publications. This in conjunction with efficient and secure data exchange protocols between databases will be the basis of a comprehensive global network of genetic knowledge.
Description of the GENESIS system
The GENESIS/GEM.app software is a cloud-based system that allows users to directly process and analyze next-generation sequencing data through a user-friendly graphical user interface (GUI). The first objectives of the platform are: 1) to assist scientists/clinicians in transferring and processing genomic data, 2) to produce accurate, high quality, and reproducible results, 3) to provide a highly available and scalable analytical framework for analyzing variant data, and 4) to provide tools for user-driven data-sharing and collaboration. Hereby, GENESIS/GEM.app enables users of varying computational experience to iteratively test many different filtering strategies in a matter of seconds and to browse very large sets of full exomes and genomes in real-time.
Bioinformatics pipeline
The GENESIS/GEM.app platform can receive data via a client-side application or on hard drives. In order to alleviate the data transfer process, we have developed the GENESIS client that automatically transfers raw data from any client directly into our bioinformatics pipeline. The common file formats are accepted, including FASTQ, BAM, or VCF format. In cases where raw sequence reads are submitted (FASTQ or BAM files), we have developed a scalable cloud-based pipeline that adheres to the best practices pipeline suggested by the Broad Institute based on BWA-MEM alignment (0.7.12) and GATK joint genotyping (3.3.0) (https://www.broadinstitute.org/gatk/guide/best-practices). Our pipeline currently processes whole exomes in under 4 hours and whole genomes in under 16 hours, which then are automatically uploaded into the GENESIS/GEM.app platform. Each pipeline run delivers a number of quality metrics that can be viewed within GENESIS/GEM.app. These include: 1) read depth, 2) percentage of read duplicates, 3) metrics on sequence read quality, and 4) metrics on sequence GC content.
GENESIS/GEM.app Knowledgebase
Users have full access to over 140 sources of variant annotations ranging from predictions on evolutionary constraint to allele frequencies in public databases. These annotations have been organized and structured in order for users to be able to easily apply them when filtering their genomic data. Due to the enormous possible combinations of filters, we have provided three predefined filters for ease of use: 1) Relaxed – filter for evolutionary neutral variants with minor allele frequency < 2%, 2) Moderate – filter for variants under slight purifying selection and minor allele frequency < 0.5%, and 3) Strict – filter for variants under purifying selection, minor allele frequency < 0.1%, and predicted to have a functional impact on protein. Users are also able to save their own filter settings for later use. Furthermore, the GENESIS/GEM.app user interface provides direct links to various online resources including PubMed, ClinVar, UCSC genome browser, ENSEMBL genome browser, String-DB, etc.
GENESIS/GEM.app Analysis Queries
GENESIS/GEM.app supports two main query types: Mendelian and case-control queries. In Mendelian queries, users can select a particular mode of inheritance to analyze, which will group related individuals into families and filter for variants that adhere to the Mendelian inheritance pattern selected. In addition, users can look for enrichment of certain variants or variants in certain genes in a group of unrelated individuals. This type of query is particularly useful when searching for overlapping candidate variants or genes in several families sharing phenotypic features. In case-control queries, users can select to filter cases and/or controls based on heterozygous and/or homozygous allele counts. This query is meant to be a basic first step to prepare data for statistics packages and custom R scripts for further analysis.
Real-time data analyses
The GENESIS/GEM.app software uses a combination of a massively parallel processing (MPP) query engine and a columnar binary storage format to allow real-time queries of single families to thousands of whole genomes. The unique customized data architecture of our database can reliably scale and provide real time access to over 30 billion genotypes currently housed in GENESIS/GEM.app. Depending on the complexity of the query, the average query response time is under 5 seconds. Even the most complex queries are generally completed within 45 seconds. The total response time of a query depends on the query time plus data transfer over the internet and is almost independent of the device the query was launched from.
Phenotype representation in GENESIS
The rarity of many Mendelian phenotypes on the one hand and their extreme genetic heterogeneity on the other hand necessitate assembly of large cohorts of MD families with similar phenotypes to enable successful identification of Mendelian disease genes. To standardize phenotypic information derived from multiple cohorts and across numerous disease groups, we have selected the Human Phenotype Ontology (HPO) (Kohler et al., 2014) to represent phenotypic features, modes of inheritance and age at onset in a standardized way. The HPO – far more than just a naming convention for phenotypic features - conceptualizes the phenotypic domain by defining types, properties and relationships of phenotypic elements. In addition we allow to enter diagnosis terms as defined by the International Classification of Disease (ICD) and others. Use of the HPO enables us to develop combined phenotype- genotype analysis algorithms that have enormous potential to accelerate identification of Mendelian disease genes.
Description of the user community and data in GENESIS/GEM.app
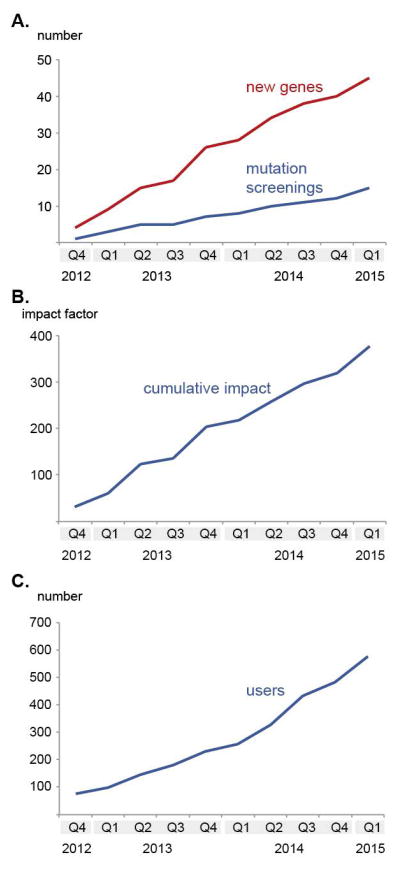
GENESIS/GEM.app has currently 582 registered users with an average of 50 new users per quarter (Figure 1C). The user community is spread worldwide over 5 continents (North America, South America, Europe, Asia, Australia) and 44 countries (Figure 2A) with particularly strong participation from North America and Europe. The vast majority of users are affiliated with academic institutions or academic hospitals (95%), and only about 5% of users are associated with non-academic entities, including commercial genetic testing labs and for-profit organizations providing related services. Users that contribute data to GENESIS/GEM.app are organized in labs around a PI; by default all users associated with this PI have access to his/her data. 72 such data contributors are registered in GENESIS/GEM.app and have collectively contributed 5,200 exomes and 800 genomes, ranging from small-scale contributors uploading just single exomes at a time to large-scale uploaders of more than 1.000 exomes (Figure 2B).
Figure 1.
Basic characteristics of GENESIS/GEM.app. A) Discovery of novel genes for Mendelian diseases and mutation screening studies. B) The cumulative publication impact score of published papers using GENESIS/GEM.app. C) The cumulative growth of the user base until to 3/2015.
Figure 2.
World maps of the current distribution of data contributors and overall users of GENESIS/GEM.app. A) Location of all registered and approved users of GENESIS/GEM.app until 3/2015. B) Color and size of circles indicate number of available data sets.
GENESIS/GEM.app currently has a strong focus on Mendelian diseases although data and query structure also support studies on complex traits. Several large disease-specific consortia and institutions (EuroEpinomics – epilepsy; Inherited Neuropathies Consortium RDCRC; Hertie Institute for Clinical Brain Research – neurodegenerative diseases; NeurOmics – neuromuscular and neurodegenerative diseases) are contributing data to GENESIS/GEM.app, leading to a strong representation of neurological phenotypes (Figure 3). Other strong phenotypic groups include autism, mild cognitive impairment, Alzheimer’s disease, dilated cardiomyopathies, syndromic and non-syndromic forms of deafness and many others.
Figure 3.
Major phenotype distribution of data sets in GENESIS/GEM.app. Size of circles corresponds to relative contribution to the 5,200 whole exomes and 800 genomes. AD - Alzheimer Disease, MCI - Mild Cognitive Impairment, ALS - Amyotrophic lateral sclerosis, park - Parkinson Disease, mito - mitochondrial disease, HSP - Hereditary spastic paraplegia, ocular - ocular disorders.
Empowering the user
Unlike an electropherogram from a Sanger sequencing experiment, the output of a next generation sequencing run is not directly human-readable. Multiple steps involving bioinformatics expertise as well as high-performance computing are necessary to reduce the complexity of the data to a degree that a clinician or geneticist can interpret the results. This reduction of complexity often comes at the expense of an enormous loss of information. In many cases, a sequencing run is reduced to a spreadsheet with a handful of variants, the output of the bioinformatics pipeline that is transferred to the clinician or geneticist. This approach leaves a huge resource untapped: the expert knowledge of a clinician or geneticist ideally suited to interpret variants in the context of a specific phenotype. GENESIS/GEM.app puts the clinician and geneticist back in control of their data.
Full access to pre-processed data
GENESIS/GEM.app aligns the raw sequence reads and calls variants for our users and uploads genotype calls so that users can define queries and analyze this data with multiple annotations (described above). This full, richly annotated dataset is then available to the user to be evaluated in entirety or filtered flexibly. A user may often want to start analysis by applying one of the predefined query filters in GEM.app (e.g. filter for rare and conserved variants that follow an autosomal recessive inheritance - homozygous or compound heterozygous - pattern in exomes of a recessive family), or to modify these settings in the course of the analysis. Therefore GEM.app gives the user the flexibility to explore the data guided by his/her scientific creativity and the requirements of a specific project. The system provides tools to interpret the data and may make suggestions regarding analysis, a feature GENESIS/GEM.app will greatly extend in the future, but the user remains in control of filtering and data display.
Graphical user interface
The GEM.app user connects to data through a simple graphical user interface that runs in any browser and can therefore be accessed from almost any device with web capabilities, including computers, tablets or smartphones. Via this interface the user can manage their samples, manage access rights for their data, upload new data, analyze data, and start collaborations with other GEM.app users, all without writing a single line of code. Users can customize the appearance of ‘their GEM.app’ account (e.g. by selecting the information to be displayed on the start page or annotation to include in the default output).
Free for academic users
The true cost of genomic data processing is underappreciated in academia, where local institutional computing resources are commonly available for very low or no cost. With the switch to WGS and the general adoption of cloud computing as a computing device, this will change, as most university computer clusters are under-equipped for the required standardized large-scale analysis. The cost for WGS raw data analysis can be substantial ($40–100 per genome from FASTQ to VCF). Thus, this will become a routine part of budgeting genome projects. However, the cost for analysis, reanalysis, daily availability of the entire data set, is much more difficult to organize for an individual study or a research institution. GENESIS/GEM.app will provide this latter part free of charge, regardless of sample size. The hope of The Genesis Foundation is that such a resource would encourage investigators to organize them in such a way that they are more efficient and more collaborative in identifying genetic causes of disease.
Genomic matchmaking
In most genetically heterogeneous Mendelian disorders, there are a proportion of patients lacking a genetic diagnosis. The current hypothesis is that extremely rare or private damaging variants are the cause for these cases. Large sample sizes will be necessary to identify these rare novel disease alleles. For example, to identify a gene that is responsible for 2% of recessive hereditary spastic paraplegia cases, 500 samples are required to achieve ~80% power (Zhi and Chen, 2012). Initial discovery publications may contain only a few small families or, in special cases, even individual patients. Crowd-sourcing efforts between many laboratories around the world can effectively collect larger sample sizes of even the rarest phenotypes. While this is a routine strategy for geneticists working in this field for many years, the new tools we have developed offer standardized methodology and often reduce this effort to a few mouse clicks.
We have been pioneering the first of its kind data-sharing platform that allows investigators to effectively and securely delegate access to genomic datasets in real time with anyone in the world. GENESIS/GEM.app users are in full control and can decide with whom to share specific datasets. By providing a sophisticated analytical platform, users can share data without the need for moving sizable quantities of data (i.e. downloading/uploading of large files). This allows collaborations to occur instantly and enables collaborators to simultaneously analyze datasets. It is now known as ‘matchmaking’ when the GENESIS/GEM.app platform connects two or more scientists in search for evidence for a novel candidate gene. These matchmaking events are now happening on a weekly basis. The discovery and publication of 37 novel Mendelian disease genes since the first launch of GEM.app (31 since the first web-version of GEM.app was launched in 10/2012) and numerous publications re-defining genotype-phenotype correlations and phenotypic spectra of Mendelian disease genes, collectively measuring >484 impact points (374 since 10/2012) are an impressive documentation of the success of the matchmaking strategy (Figure 1 A–B).
GENESIS/GEM.app contains powerful ‘social’ tools to connect scientists from the same or entirely different fields. For instance, users can invite their colleagues to join their virtual lab to work together on lab-specific datasets. In cases where multiple investigators/labs want to collaborate, they can easily create collaborative networks where several parties contribute datasets and manage access to this network. This allows unlimited numbers of users to compile and access a shared data resource, effectively increasing their sample sizes in a collaborative fashion. Thus far, the system has executed ~80,000 queries without degrading performance. Further, we are currently implementing the APIs developed within the Matchmaker Exchange group to interact with all participating databases. The standardized and safe communication of multiple heterogeneous database resources that collect phenotype and genotype information will greatly complement and support the work done within GENESIS/GEM.app.
Real live matchmaking – discovery of the PNPLA6 gene
Various tools use phenotype- or genotype-based matchmaking strategies. GENESIS/GEM.app supports both and we will use the discovery of the PNPLA6 gene, a real-live example of matchmaking on the GENESIS/GEM.app platform, to demonstrate the power of combining both approaches. Boucher-Neuhäuser-Syndrome (BNS) and Gordon-Holmes Syndrome (GHS) are two clinically defined separate disease entities that share the clinical features of ataxia and hypogonadism. In a small Italian family with BNS exome sequencing of two siblings failed to provide a genetic diagnosis (ARCA-05 family in Synofzik et al, 2014). Using phenotypic matchmaking, GENESIS/GEM.app was therefore queried for additional families with phenotypes including ataxia and hypogonadism. Two additional families, one of Brazilian and one of French origin, were identified among the > 2,000 recessive families with neurological phenotypes available in the database at that time. After each of the three involved PIs gave their consent to a joint data analysis, exome data was queried for recessive mutations affecting the same gene across all three families. Only one gene – PNPLA6 – was mutated in all three families. Phenotypic matchmaking therefore efficiently led to identification of PNPLA6 gene as the gene responsible for both Gordon-Holmes Syndrome and Boucher-Neuhäuser-Syndrome (Synofzik et al, 2014). Surprisingly, PNPLA6 had been previously published to cause SPG39, a pure motor neuron disease involving both upper and lower motor neurons (Rainier et al, 2008). We therefore suspected that PNPLA6 might cause a broader phenotypic spectrum. Using a genotype-based matchmaking approach we queried GENESIS/GEM.app for recessive mutations in PNPLA6 in families with a broad phenotypic spectrum and promptly identified two additional PNPLA6 families, one diagnosed with spastic ataxia and one with Hereditary Spastic Paraplegia complicated by mild motor neuropathy. Matchmaking via the GENESIS/GEM.app platform successfully connected five PIs from three continents and allowed rapid discovery of a new disease gene and delineation of the phenotypic spectrum of PNPLA6-associated disease over the course of just a few weeks.
The Genesis Project
As the academic GEM.app platform has grown beyond what a typical academic lab could responsibly maintain, a new solution in a private-public space was needed. As data is being accumulated from many sources, and countries, an appropriate legal structure is becoming a key issue and universities are often not prepared to take on this role for their research faculty. Further, as such data aggregation efforts carry multiple interests, the correct terms and conditions, distribution of potential intellectual property, and guardianship of expensive data archives are better served in a dedicated organization. Our solution is the non-for-profit foundation called ‘The Genesis Project” (GENESIS). The goal of GENESIS is to provide a platform that enables scientists to collaborate, share, and analyze data in a superior way at the lowest cost possible. As a non-for-profit, GENESIS is able to partner with foundations, scientific-consortia, universities, and individual researchers alike. Besides adopting GEM.app, GENESIS is partnering with MSeqDR (Falk et al., 2015), large consortia (Inherited Neuropathy Consortium, CReATe ALS consortium, EuroEPINOMICS, etc), has thus co-sponsored research projects (Mayo Clinic), and is partnering with patient Advocacy Groups (PAG). In addition, GENESIS has the opportunity to bridge the gap between basic science and development of commercial applications.
Integration with disease-specific community resources
GENESIS is partnering with a community effort for mitochondrial diseases called the Mitochondrial Sequencing Data repository (MSeqDR). MSeqDR has been established by the global mitochondrial disease community (https://mseqdr.org) to enable common genomic data deposition, curation, annotation, and effective mining through a centralized suite of custom and publicly accessible bioinformatics tools (Falk et al., 2015). MSeqDR also collates and enables visualization of all disease, gene, and variant-level data on known and likely mitochondrial disease genes, and links to public resources to enable users to efficiently interrogate all known information on the 37 mtDNA genes and already more than 250 nuclear genes that are currently recognized to cause human mitochondrial diseases (Koopman et al, 2012). We anticipate that as more users deposit genomic data into the common MSeqDR resource, either at the individual exome level with patient consent or by clinical and research laboratories sharing their aggregated data from all variants present within their cohorts, that the speed and accuracy of connecting cases and diagnosing suspected mitochondrial disease patients, who currently lack genetic etiologies, will further improve. All data and data analysis tools hosted within MSeqDR are shared with GENESIS, including a common login for users that works across both sites.
Conclusion
The excess of rare variation in the human genome has led to a high locus and allelic heterogeneity for many rare Mendelian diseases, and likely will also inform so-called common diseases. Only sufficiently high-quality sequencing based studies will resolve the puzzle of pathogenic rare variation. But those studies will present the community with an unprecedented amount of data. Extracting knowledge from these resources is the true challenge that must be our key objective. This will take a great number of investigators from different backgrounds, including geneticists, bioinformaticians, biologists, and clinician scientists. The tools to browse through large datasets at relative ease are now being developed and improved, and GENESIS/GEM.app is an example of the possibilities and the achievable results. Equally important is the development of standards in data analysis and data capture to permit the next phase, which is to enable such databases to talk to each other in a secure and efficient manner. This will ultimately give investigators access to a global network of genetic resources to answer specific questions derived from potentially very removed geographic areas. The matchmaker exchange is one of the first serious attempts to achieve this goal and we expect GENESIS/GEM.app to fully support the matchmaker API in the very near future.
We have learned in the past two years that crowdsourcing of genotype/phenotype data and analysis will lead to a remarkable pace of novel disease gene discovery. This may or may not require strict consortia rules, but is left to investigators to identify the best organizational structure for maximal competiveness of a particular study. The current tremendous pace of discovery will likely continue as new opportunities lie in the non-coding regions of the genome, deeper phenotyping, and increasingly large data sets available for analysis.
In summary, we have been pioneering the first of its kind data-sharing platform that allows investigators to effectively and securely delegate access to genomic datasets in real time with anyone in the world. GENESIS/GEM.app users are in full control and can decide with whom to share specific datasets. By providing a sophisticated analytical platform, users can share data without the need for moving data. This ‘marketplace’ driven approach has led to an impressive number of ‘genetic matchmaking’ events on GENESIS/GEM.app platform and soon in the context of the Matchmaker Exchange.
Acknowledgments
Funding:
This tool is used and supported by the following grants: NIH R01NS075764, 5R01NS072248, and U54NS065712 to S.Z., and by the CMT Association, and the Muscular Dystrophy Association. It was further supported by the European Union within the 7th European Community Framework Program through funding for the E-Rare Network NEUROLIPID (01GM1408B to RS), and a Marie Curie International Outgoing Fellowship (grant PIOF-GA-2012-326681 to R.S.). Support was also received from “The United Mitochondrial Disease Foundation” and the “National Institutes of Health (U54-NS078059 - North American Mitochondrial Disease Consortium, pilot award #NAMDC7407 to MJF and XG; and U41-HG006834).
We are thankful to the donors to The Genesis Project for keeping this resource free to the academic community and thus supporting these studies. Drs. Züchner, Schüle, and Falk are members of scientific advisory board for The Genesis Project Inc.
References
- Falk MJ, Shen L, Gonzalez M, Leipzig J, Lott MT, Stassen AP, Diroma MA, Navarro-Gomez D, Yeske P, Bai R, Boles RG, Brilhante V, Ralph D, et al. Mitochondrial disease sequence data resource (MSeqDR): A global grass-roots consortium to facilitate deposition, curation, annotation, and integrated analysis of genomic data for the mitochondrial disease clinical and research communities. Mol Genet Metab. 2015;114:388–396. doi: 10.1016/j.ymgme.2014.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez MA, Lebrigio RF, Van Booven D, Ulloa RH, Powell E, Speziani F, Tekin M, Schule R, Zuchner S. GEnomes management application (GEM.app): A new software tool for large-scale collaborative genome analysis. Hum Mutat. 2013;34:842–846. doi: 10.1002/humu.22305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler S, Doelken SC, Mungall CJ, Bauer S, Firth HV, Bailleul-Forestier I, Black GC, Brown DL, Brudno M, Campbell J, FitzPatrick DR, Eppig JT, et al. The human phenotype ontology project: Linking molecular biology and disease through phenotype data. Nucleic Acids Res. 2014;42:D966–74. doi: 10.1093/nar/gkt1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, Hao L, Kiang A, Paschall J, Phan L, Popova N, Pretel S, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet. 2007;39:1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson R, Johnston L, Taruscio D, Monaco L, Beroud C, Gut IG, Hansson MG, ‘t Hoen PB, Patrinos GP, Dawkins H, Ensini M, Zatloukal K, et al. RD-connect: An integrated platform connecting databases, registries, biobanks and clinical bioinformatics for rare disease research. J Gen Intern Med. 2014;29(Suppl 3):S780–7. doi: 10.1007/s11606-014-2908-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi D, Chen R. Statistical guidance for experimental design and data analysis of mutation detection in rare monogenic mendelian diseases by exome sequencing. PLoS One. 2012;7:e31358. doi: 10.1371/journal.pone.0031358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman WJ, Willems PH, Smeitink JA. Monogenic mitochondrial disorders. N Engl J Med. 2012;366(12):1132–41. doi: 10.1056/NEJMra1012478. [DOI] [PubMed] [Google Scholar]
- Synofzik MA, Gonzalez MA, Lourenco CM, Coutelier M, Haack TB, Rebelo A, Hannequin D, Strom TM, Prokisch H, Kernstock C, Durr A, Schöls L, et al. PNPLA6 mutations cause Boucher-Neuhäuser and Gordon Holmes syndromes as part of a broad neurodegenerative spectrum. Brain. 2014;137:69–77. doi: 10.1093/brain/awt326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainier S, Bui M, Mark E, Thomas D, Tokarz D, Ming L, Delaney C, Richardson RJ, Albers JW, Matsunami N, Stevens J, Coon H, et al. Neuropathy Target Esterase Gene Mutations Cause Motor Neuron Disease. Am J Hum Genet. 2008;82:780–785. doi: 10.1016/j.ajhg.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]