Summary
Background
MST1 and MST2 are the mammalian Ste20-related protein kinases most closely related to Drosophila Hippo, a major regulator of cell proliferation and survival during development. Overexpression of MST1 or MST2 in mammalian cells is proapototic; however, little is known concerning the physiologic regulation of the endogenous MST1/MST2 kinases, their role in mammalian cell proliferation, or the identity of the MST1/MST2 substrates critical to proliferative regulation.
Results
We show that MST1 and MST2 activity increases during mitosis, especially in nocodazole-arrested mitotic cells, where these kinases exhibit both an increase in both abundance and activation. MST1 and MST2 also can be activated nonphysiologically by okadaic acid or H2O2. The MOBKL1A and MOBKL1B polypeptides, homologs of the Drosophila MATS polypeptide, are identified as preferred MST1/MST2 substrates in vitro and are phosphorylated in cells in an MST1/MST2-dependent manner in mitosis and in response to okadaic acid or H2O2. MST1/MST2-catalyzed MOBKL1A/MOBKL1B phosphorylation alters the ability of MOBKL1A/MOBKL1B to bind and regulate downstream targets such as the NDR-family protein kinases. Thus, MOBKL1A/MOBKL1B phosphorylation in cells promotes MOBKL1A/MOBKL1B binding to the LATS1 kinase and enables H2O2-stimulated LATS1 activation loop phosphorylation. Most importantly, replacement of endogenous MOBKL1A/MOBKL1B by a nonphosphorylatable mutant is sufficient to accelerate cell proliferation substantially by speeding progression through G1/S as well as mitotic exit.
Conclusions
These results establish that MST1 and MST2 are activated in mitosis and catalyze the mitotic phosphorylation of MOBKL1A/MOBKL1B. MOBKL1A/MOBKL1B phosphorylation, in turn, is sufficient to inhibit proliferation through actions at several points in the cell cycle.
Introduction
MST1 (stk4) and MST2 (stk3) are class II GC kinases [1], initially identified by their Ste20-related catalytic domain [2] and independently as KRS1 and KRS2, protein kinases activated late after transformation by v-src [3]. The regulation and physiologic functions of these two kinases in mammalian cells are not completely understood. Early studies showed that overexpression of MST1 or MST2 in mammalian cells promotes apoptosis; in addition, apoptosis, once initiated by a variety of stimuli, results in the activation of MST1/MST2, and a caspase-catalyzed cleavage generates a highly active catalytic fragment [4–6]. MST1 also can be activated in cells by okadaic acid, H2O2, or heat shock at 55°C. A more physiologic activation of MST1 is reflected by the requirement for a Nore1B/RAPL-MST1 complex in the rap1-induced increase in integrin avidity that occurs in response to TCR or chemokine stimulation of murine T cells [7], although the MST1 substrates responsible for this action are as yet unknown.
MST1 and MST2 are the closest mammalian orthologs of the Drosophila Hippo protein kinase (for reviews, see [8–10]), and human MST2 (but not MST1) can complement Hippo deficiency in the fly [11]. Hippo deficiency in the developing eye results in massive overgrowth due to an accelerated rate of proliferation and a failure of developmental apoptosis. The phenotype of Hippo deficiency is very similar to that seen with loss of function (LOF) of Warts/LATS, another protein kinase. Warts is orthologous to the murine LATS1 kinase, and biallelic deletion of the murine LATS1 gene results in soft tissue sarcomas and ovarian tumors [12]. The noncatalytic protein Salvador/Shar-pei, whose deficiency results in a phenotype resembling a weak LOF of both Hippo and Warts/LATS, binds to both kinases, and is believed to facilitate Hippo-catalyzed phosphorylation and activation of Warts/LATS. In turn, Warts/LATS phosphorylates and inhibits the transcriptional regulator Yorkie, whose overexpression phenocopies the LOF of Hippo and Warts/LATS [13]. LOF of the Drosophila MATS gene yields a phenotype closely resembling those of Hippo and Warts/LATS [14]; MATS is homologous to the S. cerevisae polypeptides Mob1 and Mob2, which act during mitotic exit to facilitate the ability of Cdc15, a Hippo/MST1-2 homolog, to phosphorylate and activate Dbf2, a Warts/LATS-like protein kinase (see [15, 16] for review). Wei et al. found that MATS is a Hippo substrate that serves a similar role in the Hippo activation of Warts/LATS [17]. In turn, recent evidence indicates that mammalian MST2 can phosphorylate and activate LATS1 [18] and that the LATS1 and LATS2 kinases also may regulate mitotic exit [19, 20]. Other candidate MST substrates are histone H2B, which is phosphorylated at Ser14 during apoptosis, possibly by a caspase-cleaved catalytic fragment of MST1 [21]; and the FoxO1 and 3 polypeptides, whose MST1-catalyzed phosphorylation interferes with negative regulation of FoxO by Akt [22].
Thus, kinases orthologous with MST1 and MST2 appear to contribute to a variety of cellular programs, including mitotic progression, integrin affinity, apoptosis, and tumor suppression. Herein we identify the MOBKL1A/MOBKL1B polypeptides as major physiologic substrates of MST1/MST2 and characterize the effects of this phosphorylation on MOBKL1A/MOBKL1B binding to NDR-family kinases. Importantly, we demonstrate that the MST1/MST2-catalyzed phosphorylation of MOBKL1A/MOBKL1B in intact cells is sufficient to substantially retard cell-cycle progression.
Results
Screening for MST1 Substrates In Vitro
Extracts of murine L1210 preB-cell leukemia cells (ATCC CCL219) were fractionated on anion and cation exchange columns, and fractions were briefly phosphorylated in vitro [23] with recombinant MST1, which had been preactivated in vitro by incubation with Mg + ATP [24]. The most prominent MST1-phosphorylated band was a polypeptide of approximately 25 kDa recovered in the eluate of the cation exchange column (Figure 1A). Gel filtration of the pooled fractions containing this putative substrate resulted in a large portion emerging just behind the void volume, Mr > 500 kDa, with the remainder eluting at a molecular weight of approximately 30 kDa. The latter peak was pooled and subjected to SDS-PAGE, and the polypeptides migrating between Mr 20–40 kDa were digested in situ with trypsin; the tryptic peptides were analyzed by ESI-MS-MS. The largest number of peptides identified corresponded to peroxyredoxin1; however, a substantial number of peptides corresponded to MOBKL1A and MOBKL1B, mammalian homologs of the yeast Mob and Drosophila MATS polypeptides. The mouse and human genomes each contain two genes encoding MATS-related polypeptides that are identical between the two species and that differ from each other at only 8/216 amino acids (96.3% identity); the human polypeptide described by NP_775739 is MOBKL1A (encoded on Chr 4q13.3), and the human polypeptide described by NP_060691 is MOBKL1B (encoded on Chr 2p13.1); the tryptic peptides we retrieved did not distinguish between MOBKL1A and MOBKL1B; MATS is almost 90% identical to both.
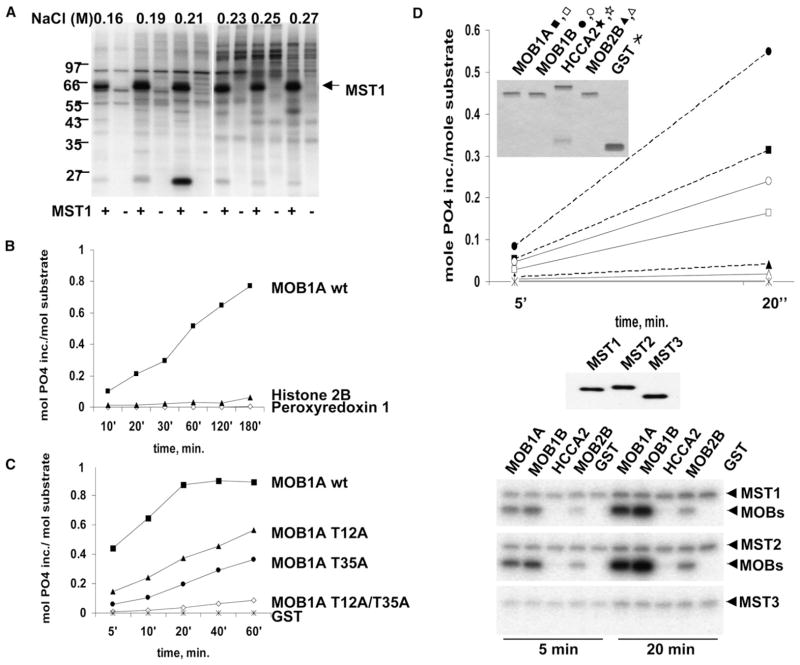
Figure 1. Identification and Characterization of a Substrate for the MST1 and MST2 Kinases.
(A) Shown is the detection of an MST1 substrate in vitro. L1210 cell extract was fractionated by NaCl gradient elution. Fractions were subjected to phosphorylation ± MST1 as described in the Supplemental Experimental Procedures.
(B) Time course of MST1-catalyzed 32P incorporation into MOBKL1A (■), histone H2B (▲), and Peroxiredoxin-1 (Prx-1) (◇).
(C) The effect of mutation of MOBKL1A Thr12 and Thr35 on MOBKL1A phosphorylation by MST1.
(D) Different MOB family members as substrates for MST1/MST2/MST3. Substrates were phosphorylated by preactivated MST1 (solid lines), MST2 (dashed lines), and MST3 as in (B) for the time points indicated. The insert shows a Coomassie blue stain reflecting the relative amount of the substrates in the reaction. The open symbols are MST1, and the closed symbols are MST2. The panel below the graph is an anti-FLAG blot reflecting the relative amount of each kinase in the reaction. The autoradiograph shows the relative amount of 32P incorporation into the kinases and the substrates.
MOBKL1A and MOBKL1B Are Selectively Phosphorylated by MST1 and MST2 In Vitro
We examined comparable concentrations of recombinant peroxyredoxin1, MOBKL1A, and histone H2B as potential in vitro substrates for the MST1 kinase. As seen in Figure 1B, MOBKL1A is phosphorylated at a rate 10-fold (or more) faster than is H2B and it rapidly reaches nearly 1 mol PO4 incorporation/molMOBKL1A polypeptide (Figure 1B), whereas peroxyredoxin 1 is not phosphorylated. MOBKL1A phosphorylated by MST1 to approximately 0.3 mol PO4/mol was subjected to tryptic digestion, and phosphopeptides were identified and sequenced by MS-MS; two tryptic phosphopeptides were retrieved and found to be phosphorylated at Thr12 and Thr35. These MOBKL1A threonine residues were converted to alanine singly and together; mutation of each threonine substantially reduced the extent of 32P incorporation into MOBKL1A, and MOBKL1A (Thr12Ala/Thr35Ala) was phosphorylated at <5% the rate and extent as wild-type MOBKL1A (Figure 1C). We also examined the ability of recombinant preactivated FLAG-MST1, -MST2, and -MST3, each at comparable polypeptide concentrations, to phosphorylate mammalian recombinant MOBKL1A, MOBKL1B, MOBKL2B, and MOB2/HCCA2 (Figure S1 available online), all at identical concentrations (Figure 1D). MOBKL1B was phosphorylated at the highest rate, approximately 2-fold faster than MOBKL1A and 9- to 14-fold faster than MOBKL2B; MOB2/HCCA2 was not phosphorylated at all. The MST1, MST2, and MST3 polypeptides each were preactivated by incubation in vitro with Mg + ATP and added at identical polypeptide concentrations; under these conditions, MST2 catalyzed phosphorylation at approximately twice the rate of MST1, whereas MST3, although capable of autophosphorylation to an extent approximately half that of MST1 and MST2, does not phosphorylate any of these Mob-related polypeptides. Thus MOBKL1A and MOBKL1B are selective substrates of the MST1 and MST2 kinases.
MOBKL1 Binds Activated MST2 Preferentially, and MOBKL1 Phosphorylation Inhibits the Binding of MST2
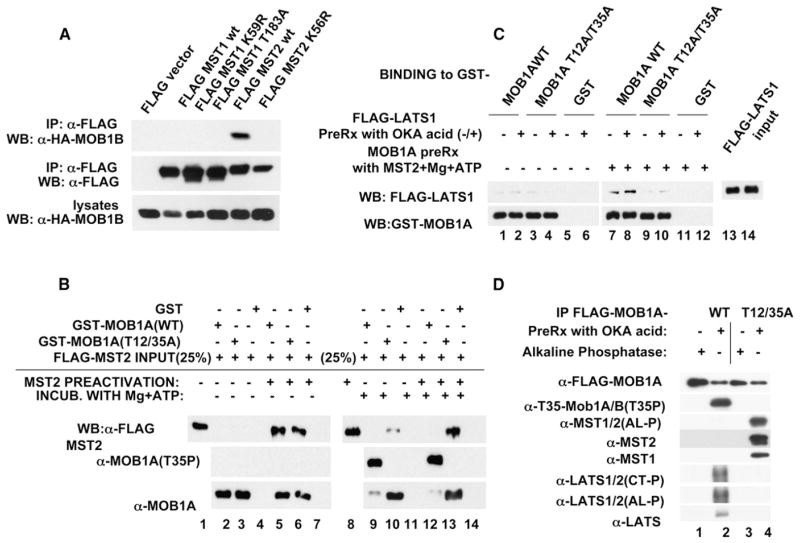
Overexpressed recombinant MOBKL1A coprecipitates endogenous MST1, MST2, LATS1, LATS2 (Figure S2A), and NDR1 (identified by MS/MS, data not shown) from HEK293T cells. Recombinant MOBKL1B binds coexpressed MST2 much better than MST1; however, the inactive MST2 mutant K56R binds MOBKL1B much more poorly than wild-type MST2 (Figure 2A); this suggests that activated MST2 may bind MOBKL1B much better than the inactive kinase. We showed previously [24] that when transiently expressed in HEK293T cells, MST1 and MST2 are retrieved predominantly in an unactivated state but can be fully activated by pretreatement of cells with okadaic acid or in vitro by autophosphorylation. We therefore compared the ability of MST2 to bind in vitro to prokaryotic recombinant MOBKL1A polypeptides, with or without in vitro autophosphorylation/preactivation of MST2 (Figure 2B, lanes 2–7). Autophosphorylated, preactivated MST2 binds MOBKL1A wild-type and MOBKL1A (Thr12/35Ala) comparably (Figure 2B, compare lanes 5 and 6) and much better than unactivated MST2 (Figure 2B compare lanes 2 and 3 to lanes 5 and 6). If Mg++ and ATP are included in the incubation of MST2 with the MOBKL1A wild-type and Thr12/Thr35 polypeptides, then concomitant with the phosphorylation of wild-type MOBKL1A, its ability to bind MST2 is greatly diminished (Figure 2B, compare lane 5 to lane 12), whereas the ability of MST2 to bind to MOBKL1A (Thr12/35Ala) is unaffected by the presence of Mg++ and ATP (Figure 2B, compare lane 6 to lane 13). The results indicate that MST2 (and probably MST1) must be activated to bind MOBKL1A. Moreover, once phosphorylated by MST2, the ability of MOBKL1A to bind MST2 is greatly diminished.
Figure 2. Characterization of MOBKL1A/MOBKL1B Binding to MST1/MST2 and LATS1.
(A) MOBKL1B binds native MST2 in preference to kinase-dead MST2 or native MST1. HA-MOBKL1B was cotrasfected in HEK293T cells with the Flag-tagged MST1 and MST2 variants indicated.
(B) MST2 autophosphorylation enhances, whereas MST2-catalyzed MOBKL1A phosphorylation inhibits, the MST-MOBKL1A interaction in vitro. Immobilized GST (lanes 4, 7, 11, and 14), GST-MOBKL1A wild-type (lanes 3, 6, 10, and 13), and the GST-MOBKL1A (Thr12/35Ala) (lanes 2, 5, 9, and 12) were incubated with purified MST2, nonpreactivated (lanes 2–4 and 9–11), or preactivated (lanes 5–7 and 12–14), with (lanes 9–14) or without (lanes 2–7) ATP. Note that phosphorylation of MOBKL1A (lanes 9 and 12) reduces its reactivity with the anti-MOBKL1A/MOBKL1B antibody (lowest panel).
(C) Characterization of LATS1 binding to unphosphorylated and phosphorylated MOBKL1A in vitro is shown. Immobilized GST, GST-MOB1A wild-type, or GST-MOBKL1A (Thr12/35Ala) were preincubated with preactivated MST2, with (lanes 7–12) or without (lanes 1–6) MgCl2 and ATP. After washing, okadaic-acid-treated (lanes 2, 4, 6, 8, 10, and 12) or untreated (1, 3, 5, 7, 9, and 11) HEK293T cell lysates containing Flag-LATS1 were added for pull-down assay.
(D) Characterization of the effect of okadaic acid on the ability of recombinant MOBKL1A wild-type or MOBKL1A (Thr12/35Ala) to bind endogenous MST1/MST2 and LATS1 during transient expression is shown. HEK293T cells expressing FLAG-tagged MOBKL1A wild-type (lanes 1 and 2) or MOBKL1A (Thr12/T35Ala) (lanes 3 and 4) were treated with okadaic acid (OA) (lanes 2 and 4) or not (lanes 1 and 3). Aliquots of the cell extracts containing 2 mg protein were incubated with either okadaic acid (lanes 2 and 4) or alkaline phosphatase (lanes 1 and 3) at room temperature for 30 min. and then subjected to anti-Flag immunoprecipitation.
MOBKL1 Phosphorylation Enhances Its Binding of LATS1 and NDR1
The NDR kinases, such as the mammalian LATS1/LATS2 and NDR1/NDR2 are AGC-family kinases that are activated by phosphorylation at two regulatory sites, one in a hydrophobic motif located carboxy-terminal to the canonical catalytic domain, and a second in the activation loop of the catalytic domain (see [25] for review). An upstream kinase is required for phosphorylation of the carboxy-terminal site, whereas most evidence indicates that the activation loop phosphorylation is an autophosphorylation [26–28]. In Drosophila, the Hippo kinase was shown to phosphorylate and activate the Warts/LATS kinase, and one of the six Drosophila Mob homologs (i.e., MATS) was shown to bind Warts/LATS and potentiate its activation; moreover loss of MATS yielded an overgrowth phenotype similar to that seen with loss of Hippo or Warts/LATS [14]. MOBKL1A/MOBKL1B can bind to LATS1/LATS2 (Figure 2C and Figures S2A and S2B) and to NDR1 (Figure S2B), and Bischel et al. [29] reported that okadaic acid treatment prior to extraction of Mob1 greatly increased the ability of recombinant Mob1 to bind NDR1 in vitro. Moreover, Mob1 binds to an amino-terminal segment of NDR1 and is accompanied by kinase activation, presumably due to release from an intramolecular autoinhibition. We find that MST2-catalyzed phosphorylation of MOBKL1A strongly enhances the binding of LATS1 (Figure 2C). Mutation of Thr12 to alanine modestly diminishes LATS1 binding, whereas mutation of Thr35 has a much stronger inhibitory effect (Figure S2B), and conversion of both threonines to alanines nearly eliminates LATS1 binding (Figure 2C). MST2-phosphorylated MOBKL1A binds LATS1 isolated from control or okadaic-acid-treated cells to a similar extent (Figure 2C). Mutation of Thr12/Thr35 to Glu does not mimic the effect of MOBKL1A/MOBKL1B phosphorylation but, rather, alters MOBKL1A binding to MST2 and LATS1 in a manner similar to the MOBKL1A Thr12/35Ala mutations (data not shown). MST2-catalyzed MOBKL1A phosphorylation also enhances the binding in vitro of NDR1 (Figure S2B). The operation of these effects in the intact cell is illustrated in Figure 2D; thus whereas okadaic acid pretreatment enhances the ability of recombinant wild-type MOBKL1A to bind endogenous LATS1, okadaic acid pretreatment does not enable the binding of LATS1 to MOBKL1A (Thr12/35Ala); with regard to MST1/MST2, okadaic acid pretreatment enhances the binding of endogenous MST1 and MST2 to MOBKL1A (Thr12/35Ala) much more than to wild-type MOBKL1A, presumably reflecting the ability of okadaic acid to promote MST1/MST2 activation, together with the inability of the mutant MOBKL1A to be phosphorylated, which would inhibit its ability to bind MST1/MST2.
MST1/MST2 Mediates MOBKL1A/MOBKL1B and LATS1 (Thr1079) Phosphorylation in Response to Okadaic Acid and H2O2 in Intact Cells
Little is known concerning the physiologic regulation of MST1 or MST2. Nevertheless, MST1/MST2 can be activated by incubation of cells with okadaic acid or by subjecting cells to certain severe stressors such as H2O2 [3, 24]. Under these conditions, the phosphorylation of MST1 and MST2 in their activation loop at Thr183 and Thr180, respectively, is increased (Figures 3A and 3B, fourth row from top) and MOBKL1A/B phosphorylation at both Thr12 and Thr35 is markedly increased (Figures 3A and 3B, top two rows). To probe the role of MST1/MST2 in these phosphorylations, we selected stable U2OS transformants that overexpress either a wild-type or a kinase-dead mutant of Flag-MST2 in a tetracycline-responsive manner. The recombinant inactive Flag-MST2, through its carboxy-terminal SARAH domain, is capable of dimerizing with endogenous full-length MST1 and MST2 [24]. Inasmuch as MST1/MST2 activation requires transphosphorylation by the dimer partner [24], the heterodimers of MST2 (K56R) with endogenous MST1 or MST2 are inactive. Induction of MST2 (K56R) markedly inhibits the ability of okadaic acid to promote MOBKL1A/MOBKL1B phosphorylation (Figure 3A, top two rows) and blocks the okadaic-acid-induced increase in LATS1 carboxy-terminal phosphorylation at Thr1079 (Figure 3A, fourth row from bottom). Notably, okadaic-acid-stimulated phosphorylation of the LATS1 activation loop at Ser909, thought to be an autophosphorylation, is not affected by overexpression of MST2 (K56R) (Figure 3A, third row from bottom). Validation of the specificity of the MOBKL1A/MOBKL1B and LATS1/LATS2 phosphospecific antibodies is provided in Figure S3.
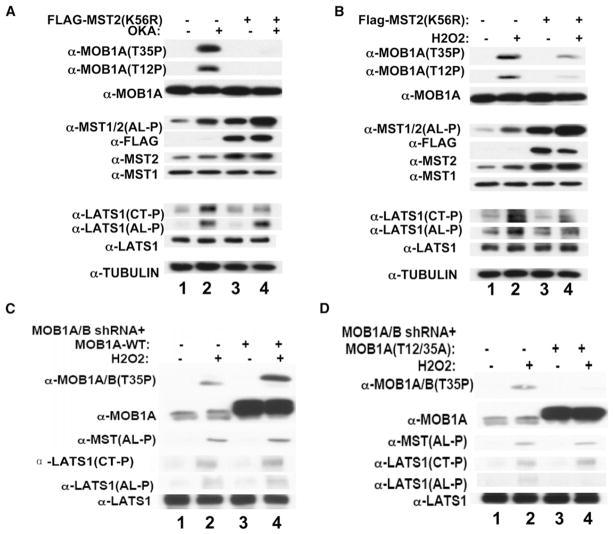
Figure 3. The Role of MST1/MST2 in MOBKL1A/MOBKL1B and LATS1 Phosphorylation in Response to Okadaic Acid and H2O2.
(A) Overexpression of a kinase-dead MST2 inhibits okadaic-acid-induced MOBKL1A/MOBKL1B and LATS1/LATS2 (Thr1079), but not LATS1/LATS2 (Ser909) phosphorylation. U2OS cells stably expressing a tetracycline (Tet) inducible Flag-MST2 (K56R) were treated with doxycycline (lanes 3 and 4) or carrier (lanes 1 and 2) and 12 hr later with okadaic acid (1 μM) (lanes 2 and 4) or carrier (lanes 1 and 3) for 30 min prior to harvest.
(B) Overexpression of a kinase-dead MST2 inhibits H2O2-induced MOBKL1A/MOBKL1B and phosphorylation of both LATS1/LATS2 (Thr1079) and LATS1 (Ser909). The experiment was carried out exactly as in (A), except the stimulation was with H2O2 (2.5 mM), which was added for 20 min.
(C) Replacement of endogenous MOBKL1A/MOBKL1B with recombinant MOBKL1A wild-type does not alter H2O2 stimulated LATS1/LATS2 phosphorylation. U2OS cells carry cassettes of shRNA against 3′ UTR of MOBKL1A, shRNA directed against the coding region of MOBKL1B, and cDNA encoding Flag-MOBKL1A. The first cassette is constitutively expressed, whereas the latter two are tetracycline inducible. Five days prior to stimulation, doxycycline (5 μg/ml) (lanes 3 and 4) or carrier (lanes 1 and 2) was added. Cells were treated with H2O2 as in (B) (lanes 2 and 4). Note that the recombinant Flag-MOBKL1A runs slightly slower than the endogenous MOBKLIA/MOBKL1B.
(D) Elimination of endogenous MOBKL1A/MOBKL1B phosphorylation selectively eliminates H2O2-stimulated phosphorylation of LATS1/LATS2 (Ser909), but not of LATS1/LATS2 (Thr1079). The experiment was carried out exactly as in (C), except that the cDNA sequences employed encodes MOBKL1A (Thr12/35Ala).
H2O2 is a strong activator of endogenous MST1/MST2 and also results in robust MOBKL1A/MOBKL1B phosphorylation; induction of MST2 (K56R) markedly inhibits H2O2-induced MOBKL1A/MOBKL1B phosphorylation (Figure 3B, top two rows). The MST2 (K56R) also inhibits the H2O2-induced phosphorylation of the LATS1 carboxy-terminal site and results in a partial inhibition of LATS1 activation loop phosphorylation. Overexpression of MST2 (K56R) does not alter the H2O2-induced phosphorylation of endogenous ERK or Jnk (Figure S4A). Tetracycline-induced overexpression of wild-type MST2 causes a pronounced increase in H2O2-stimulated LATS1 (Thr1079) phosphorylation (Figure S4B, third row from bottom) and a modest increase in the phosphorylation of endogenous MOBKL1A/MOBKL1B and in LATS1 (Ser909) phosphorylation. Together, these data provide strong evidence that MST1/MST2 are the kinases responsible for the okadaic-acid- and H2O2-stimulated phosphorylation of MOBKL1A/MOBKL1B and LATS1 (Thr1079).
MST1/MST2-Stimulated MOBKL1A/MOBKL1B Phosphorylation Is Required for H2O2-Stimulated LATS1 (Ser909) Phosphorylation
In the presence of okadaic acid, LATS1 (Ser909) activation loop phosphorylation persists in the presence of MST2 (K56R) despite inhibition of LATS1 (Thr1079) and MOBKL1A/MOBKL1B phosphorylation (Figure 3A), consistent with the view that LATS1 (Ser909) phosphorylation is catalyzed by an intramolecular autophosphorylation rather than by MST1/MST2. Nevertheless, the observation that overexpressed MST2 (K56R) inhibits H2O2-stimulated LATS1 (Ser909) phosphorylation (Figure 3B) impelled us to examine further the significance of MOBKL1A/MOBKL1B phosphorylation for LATS1 phosphorylation. For this purpose, we selected U2OS cell lines that expressed, in a tetracycline-inducible manner, shRNAs directed at endogenous MOBKL1A and MOBKL1B, as well as the MOBKL1A wild-type (Figure 3C) or MOBKL1A (Thr12/35Ala) (Figure 3D) polypeptides. Tetracycline induction virtually eliminated expression of the endogenous MOBKL1A/MOBKL1B polypeptides while greatly increasing the abundance of the recombinant MOBKL1A polypeptides, which exhibit a slightly slower electrophoretic mobility (Figures 3C and 3D, second row from top). The recombinant wild-type MOBKL1A polypeptide, like the endogenous, exhibits H2O2-stimulated phosphorylation (Figure 3C, top row, lanes 3 and 4), whereas no MOBKL1A phosphorylation is detectable in the cells expressing recombinant MOBKL1A (Thr12/35Ala) (Figure 3D, top row, lanes 3 and 4), indicating that the endogenous MOBKL1A/MOBKL1B had been greatly reduced and replaced by recombinant MOBKL1A (Thr12/35Ala). Whereas tetracycline-induced replacement of endogenous MOBKL1A/MOBKL1B by recombinant overexpressed wild-type MOBKL1A does not significantly affect H2O2-stimulated phosphorylation of LATS1 (Ser909) (Figure 3C, fifth row from top, compare lanes 3 and 4 to lanes 1 and 2), replacement of endogenous MOBKL1A/MOBKL1B by recombinant overexpressed MOBKL1A (Thr12/35Ala) inhibits H2O2-stimulated phosphorylation of LATS1 (Ser909) (Figure 3D, fifth row from top, compare lanes 3 and 4 to lanes 1 and 2). In contrast, replacement of endogenous MOBKL1A/MOBKL1B by either recombinant MOBKL1A wild-type or MOBKL1A (Thr12/35Ala) does not alter H2O2-stimulated MST1/MST2 activation loop phosphorylation, (Figures 3C and 3D, third row from top), H2O2-stimulated LATS1/LATS2 (Thr1079) phosphorylation (Figures 3C and 3D, fourth row from top), or LATS1 polypeptide levels (Figures 3C and 3D, bottom row). These results indicate that phosphorylated MOBKL1A promotes LATS1 (Ser909) activation loop autophosphorylation in response to H2O2.
MST1 and MST2 Are Activated in Mitosis and Catalyze the Mitotic Phosphorylation of MOBKL1A/MOBKL1B and the LATS1 (Thr1079)
Although the yeast kinases most closely related to MST1/MSTT2, Cdc15p in S. cerevisiae and cdc7 and Sid1p in S. pombe, are known cell-cycle components necessary for mitotic exit and cytokinesis, respectively, little or no information is available concerning the state of MST1, MST2, or mammalian MOBKL1A/MOBKL1B in mitosis. We therefore characterized the abundance and activity of the MST1/MST2 kinases in mitosis in U2OS cells. Rapidly growing, unsynchronized U2OS cells were extracted directly or after mitotic arrest, obtained by shake-off after incubation overnight with nocodazole; in addition, nocodazole-arrested mitotic cells were washed free of the agent and replated, and extracts were prepared at intervals thereafter (Figure 4A). As expected, cyclin B abundance is substantially increased in the nocodazole-arrested cells and declines rapidly after nocodazole washout (Figure 4A, third row from bottom); reciprocally, the robust cdk1(Tyr15) phosphorylation seen in unsynchronized cells is undetectable in the nocodazole-arrested cells (Figure 4A, second row from bottom). Nocodazole arrest also is accompanied by an increase in the abundance of both MST1 and MST2 in comparison to the levels detected in freely cycling cells (Figure 4A, top two rows, and Figure 4B); moreover, the abundance of MST1 and MST2 decline rapidly after nocodazole washout, although levels somewhat greater than those seen in cycling cells are still evident at 1 hr after washout (Figure 4A). Activation of the MST1 and MST2 kinase activity in vitro requires phosphorylation on the activation loop at Thr180 and Thr183, respectively [24]. MST1/MST2 phosphorylation at these sites is increased in the nocodazole-arrested cells; however, it is not possible to judge reliably whether this is due entirely to the increase in MST1/MST2 polypeptide abundance or whether MST1 and/or MST2 also is activated. Activation can be obtained in vitro by incubation of immunoprecipitated MST1/MST2 with relatively “high” levels of Mg – ATP (approximate EC50 ATP~50 μM) and can be monitored quantitatively by brief assay under conditions that minimize further autoactivation during assay (e.g., 2 min at 10 μM ATP) [24]. Immunoprecipitates of MST1 and MST2 therefore were prepared from cycling and nocodazole-arrested cells, and the fractional activation of MST1 and MST2 was quantitated by measuring kinase activity under nonactivating conditions with and without Mg + ATP preactivation in vitro, as described previously [24]. The absolute values from three experiments are summarized in Figure 4B, and the fractional activation is described in the legend. The MST1 kinase immunoprecipitated from cycling cells exhibits approximately 10% maximal activation, whereas this increases to approximately 40% in the nocodazole-arrested cells (Figure 4B, left; see figure legend). MST2 exhibits ~26% activation in cycling cells, which increases to ~45% in mitotic cells (Figure 4B, right; see figure legend). The higher “basal” activation state of MST2, as compared with MST1, was observed previously with transiently expressed recombinant polypeptides [24]. The increase in MST1/MST2 abundance in mitotic cells (estimated at ~3 fold), together with the increased fractional activation of the MST1/MST2 kinases, indicates that the available activity of both isoforms increases approximately 6- to 12-fold during mitosis. Nocodazole arrest of U2OS cells also is accompanied by an increased MOBKL1A/MOBKL1B phosphorylation at Thr12 and Thr35, LATS1 (Thr1079), and LATS1 (Ser909), which decline during washout roughly in parallel to the decrease in MST1/MST2 (Thr183/Thr180) phosphorylation (Figure 4C).
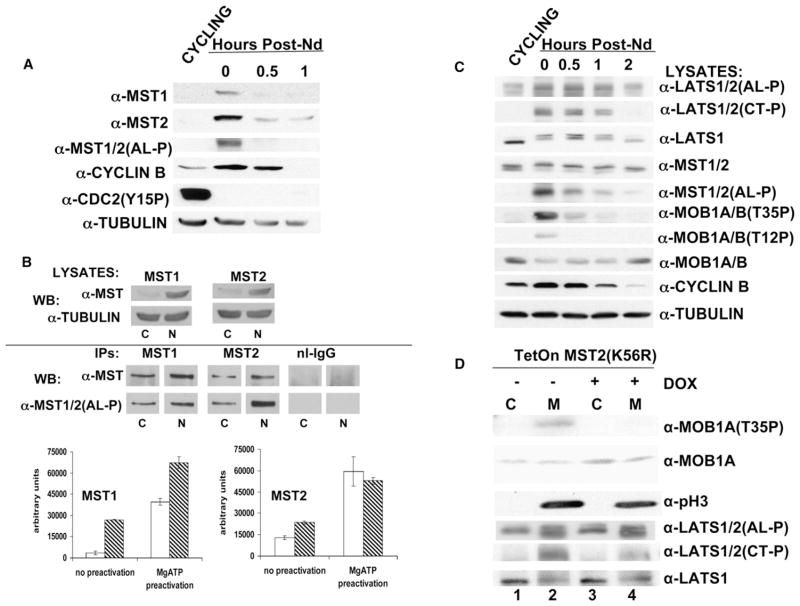
Figure 4. MST1/MST2 Activity, MOBKL1A/MOBKL1B, and LATS1 Phosphorylation in Nocodazole-Arrested and Unimpeded Mitotic U2OS Cells.
(A) The MST1 and MST2 kinases exhibit increased abundance in nocodazole-arrested mitotic cells. U2OS cells either were exponentially growing (cycling) or were arrested in metaphase by nocodazole (0 hr) and then harvested 0.5 and 1 hr after washing.
(B) The MST1 and MST2 kinases show increased fractional activation in nocodazole-arrested cells. Top: Extracts from cycling (marked as “C”) or nocodazole (“N”)-arrested mitotic U2OS were analyzed by immunoblot for MST1 and MST2. Bottom: Immunoprecipitates of MST1 and MST2 were prepared from the extracts of cycling and nocodazole-arrested U2OS cells shown in the top panels. The bar graphs show the MST activities from three independent MBP kinase assays. The fractional activation (% maximal) in the cycling and nocodazole-arrested cells (n = 3) were as follows. MST1: cycling, 10.3 ± 1.2; mitotic, 40.8 ± 6.4 (p < 0.01); MST2, cycling 26.4 ± 6.7; mitotic, 45.6 ± 1.3 (p < 0.03).
(C) Characterization of the effect of nocodazole-induced mitotic arrest on the phosphorylation of MOBKL1A/MOBKL1B and LATS1/LATS2 is shown. U2OS cells were treated as in (A).
(D) Overexpression of kinase-dead MST2 in unimpeded mitotic cells eliminates mitotic MOBKL1A/MOBKL1B phosphorylation, modestly reduces LATS1 (Thr1079P), and does not alter LATS (Ser909P). U2OS cells carrying a tetracycline-inducible kinase-dead MST2 (K56R) were treated with doxycycline (1 μg/ml) (lanes 3 and 4) or carrier (lanes 1 and 2) 12 hr prior to a mitotic shake-off in the absence of nocodazole. Histone H3 (Ser10) phosphorylation marks mitosis.
Nocodazole arrests cells at metaphase by disruption of the spindle structure and, thus, activates the spindle checkpoint; to determine whether MST1/MST2 activation occurs in the course of a normal mitosis, we collected cells easily shaken off plates of freely cycling cultures. The presence of robust histone H3 (Ser10) phosphorylation in these cells, as compared to freely cycling, adherent cells, provided an indicator that a substantial fraction of the cells collected by shake-off were in mitosis (Figure 4D, third row from top). These untreated mitotic cells also show enhanced MOBKL1A/MOBKL1B (Thr35) phosphorylation (top row) and LATS1 (Thr1079) phosphorylation (Figure 4D, fifth row from top), as compared to the nonmitotic cells. Notably, however, the intensity of MOBKL1A/MOBKL1B phosphorylation in untreated mitotic cells was less than 10% that seen in nocodazole-arrested cells, and an increased phosphorylation of the LATS1/LATS2 activation loop (Figure 4D, fourth row from top) was not observed, in contrast to nocodazole-arrested cells (Figure 4C, top row), suggesting that nocodazole, presumably by activation of the spindle checkpoint, contributes substantially to extent of MST1/MST2 activation beyond that occurring in an unimpeded mitosis.
Tetracycline induction of MST2 (K56R) prior to shake-off of unimpeded mitotic cells eliminated mitotic phosphorylation of endogenous MOBKL1A/MOBKL1B and modestly reduced LATS1/LATS2 (Thr1079) phosphorylation in the nonarrested mitotic cells (Figure 4D, fifth row from top, compare lanes 3 and 4 to lanes 1 and 2) but did not alter the LATS1/LATS2 (Ser909P) immunoreactivity (Figure 4D, fourth row from top, compare lanes 3 and 4 to lanes 1 and 2) These results indicate that MST1 and/or MST2 is activated during a normal mitosis and is responsible for the mitotic phosphorylation of MOBKL1A/MOBKL1B. The inability of MST2 (K56R) to fully inhibit LATS1 (Thr1079) phosphorylation may indicate that phosphorylation of LATS1 (1079) requires lesser amounts of active MST1/MST2 than does phosphorylation of MOBKL1A/MOBKL1B or that another kinase is responsible, in part, for the mitotic phosphorylation of LATS1/LATS2 (Thr1079). The failure to observe an alteration in LATS1 (Ser909) phosphorylation in an unimpeded mitosis is qualified somewhat by the broadening of the LATS1/LATS2 band in mitosis; however, the LATS1/LATS2 (Thr1079P) immunoreactivity, although broadened, is unmistakably increased in M (Figure 4D, fifth row from top). The inability of overexpressed MST2 (K56R) to inhibit LATS1/LATS2 (Ser909) phosphorylation in M, whereas it effectively suppresses this phosphorylation in response to H2O2, is a further indication that the LATS1/LATS2 regulation in M is likely to involve kinases other than MST1/MST2.
In U2OS cells, stable expression of lentiviral-encoded shRNA against MST2 also can inhibit nocodazole-induced MOBKL1A/MOBKL1B and LATS1/LATS2 phosphorylation, but it is observable only in clones in which the suppression of MST2 expression exceeds 90% (Figure S5), a level of inhibition well beyond that achieved in unselected mixtures of cells infected with the lentiviral-encoded shRNA or by using RNAi oligonucleotides.
Elimination of MOBKL1A/MOBKL1B Phosphorylation Stimulates Cell Proliferation
The ability to replace endogenous MOBKL1A/MOBKL1B with recombinant wild-type or mutant MOBKL1A (Figures 3C and 3D) enabled an examination of the effects of MOBKL1A/MOBKL1B phosphorylation on cell proliferation. Replacement of endogenous MOBKL1A/MOBKL1B with recombinant wild-type MOBKL1A (Figure 5A, left) had no discernable affect on proliferative rate (Figure 5B, left), whereas replacement with MOBKL1A (Thr12/35Ala) (Figure 5A, right) increased proliferative rate by 2- to 3-fold (Figure 5B, right). Despite the enhanced proliferative rate seen with MOBKL1A/MOBKL1B(Thr12/35Ala), the distribution of the cells in various phases of the cell cycle was not significantly altered by replacement of endogenous MOBKL1A/MOBKL1B with either wild-type MOBKL1A or MOBKL1A (Thr12/35Ala) (Figure 6A) in the absence or presence of doxycycline. This suggested that the stimulatory effect on proliferation of replacing endogenous MOBKL1A/MOBKL1B with a nonphosphorylatable MOBKL1A may be due to actions exerted at several sites in the cell cycle. We estimated the effect of MOBKL1A/MOBKL1B replacement on the rate of progression through G1/S by examining the rate at which cells accumulate in G2/M after addition of nocodazole (Figure 6B). Twenty-four hours after addition of nocodazole, the fraction of cells in G2/M had increased from 24.6% to 46.4% (a 21.8% increment) in one cell line and from 28.3% to 48.4% in a second cell line (a 20.1% increment). Doxycycline-induced replacement of endogenous MOBKL1A/MOBKL1B by wild-type MOBKL1A resulted in a slightly lesser increase in progression to G2/M (24.9% to 43.4%, an 18.5% increment). In contrast, replacement of endogenous MOBKL1A/MOBKL1B by nonphosphorylatable MOBKL1A greatly increased the fraction of cells that had progressed to G2/M over the same interval, from 21.0% to 58.1% (a 37.1% increment). This nearly 2-fold increase in the rate of passage through G1/S into G2/M seen when MOBKL1A (Thr12/35Ala) is overexpressed, as compared with wild-type MOBKL1A, is likely to underlie a substantial component of the increased rate of proliferation (Figure 5B) that results from replacement of endogenous MOBKL1A/MOBKL1B with the nonphosphorylatable MOBKL1A mutant. We also examined whether MOBKL1A/MOBKL1B phosphorylation affects the rate of exit from mitosis into G1 (Figures 6C and 6D). Elimination of endogenous MOBKL1A/MOBKL1B and its replacement with overexpressed, wild-type MOBKL1A causes a modest slowing in the rate of entry from M into G1 (Figure 6C), whereas replacement with MOBKL1A (Thr12/35Ala) results in an acceleration of mitotic exit of a comparable extent (Figure 6D). These data indicate that MOBKL1A/MOBKL1B phosphorylation by MST1 and/or MST2 delays mitotic exit.
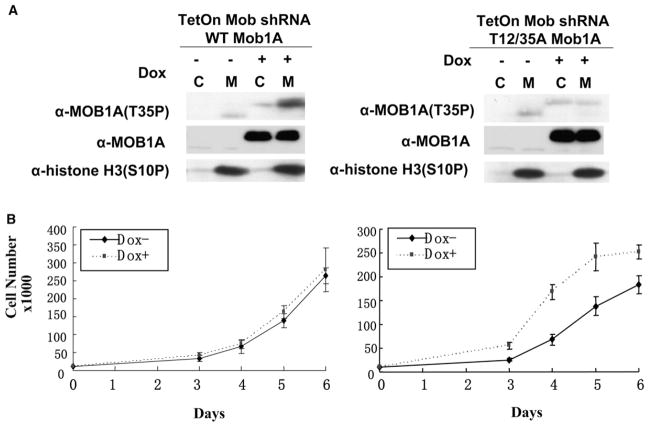
Figure 5. Elimination of MST1/MST2-Catalyzed MOBKL1A/MOBKL1B Phosphorylation Accelerates U2OS Cell Proliferation.
(A) Doxycycline-induced replacement of endogenous MOBKL1A/MOBKL1B with recombinant MOBKL1A wild-type or MOBKL1A (Thr12/35Ala) is shown. U2OS cells described in Figures 3C and 3D were treated with doxycycline for 5 days and then replated at an appropriate density. Unimpeded mitotic cells were collected as in Figure 4D. Note in the experiment illustrated on the left that the mitotic phosphorylation of the endogenous MOBKL1A/MOBKL1B polypeptides is greatly reduced after doxycycline induction, whereas the doxycycline-induced recombinant, overexpressed wild-type MOBKL1A is heavily phosphorylated in mitosis. In the experiment illustrated on the right, endogenous MOBKL1A/MOBKL1B polypeptides and phosphorylation also are abolished. The Thr35P immunoreactivity of the MOBKL1A (Thr12/35Ala) is indistinguishable in the cycling and shaken-off cells; this signal probably reflects the very minor nonphosphospecific immunoreactivity of this antibody, uncovered by the marked overexpression of recombinant MOBKL1A (Thr12/35Ala).
(B) Elimination of MST1/MST2-catalyzed MOBKL1A/MOBKL1B phosphorylation accelerates U2OS cell proliferation. The doxycycline induction and the replating was carried out exactly as described in (A). The cell counts at each day after replating are shown. The error bars represent the SEM. On the right, the differences between Dox− and Dox+ on days 3–6 are all significant at p < 0.01.
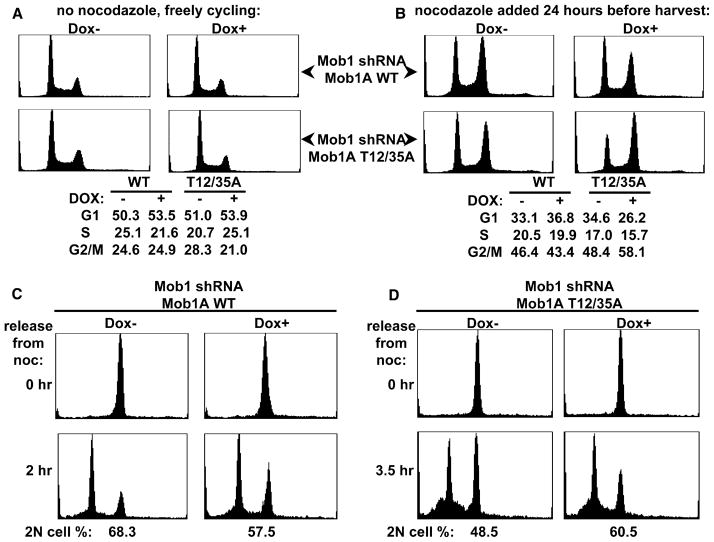
Figure 6. Elimination of MOBKL1A Phosphorylation Accelerates Progression through G1/S and Mitotic Exit.
(A and B) Elimination of MOBKL1A phosphorylation accelerates progression through G1/S. The U2OS cells were treated with doxycycline and replated exactly as described in Figure 5A. (A) Three days after replating, aliquots of the cells were subjected to DNA content analysis by FACS. Note that when growing exponentially in the absence of nocodazole, neither cell line shows significant effects of doxycycline on cell-cycle distribution. (B) A parallel set of cells were treated with nocodazole, harvested 24 hr later, and analyzed for DNA content by FACS. Note that replacement with MOBKL1A (Thr12/35Ala) is accompanied by fewer cells in G1 and more in G2/M.
(C and D) Elimination of MOBKL1A phosphorylation accelerates exit from mitosis into G1. Doxycycline, replating, and nocodazole treatments were performed as in (A) and (B). Cells arrested in mitosis (4N) were then collected by shake-off. A portion of mitotic cells were washed free of nocodazole and replated. The cells expressing MOBKL1A wild-type (C) were harvested 2 hr after replating, whereas the cells expressing MOBKL1A (Thr12/35Ala) (D) were harvested 3.5 hr after replating. Cells are analyzed by FACS (lower FACS scans) The fraction of replated cells exhibiting 2N DNA content is shown below the FACS scans and reflects the fraction of cells exiting mitosis into G1. Note that replacement of endogenous MOBKL1A/MOBKL1B with recombinant MOBKL1A wild-type diminishes the fraction of cells exiting M, whereas replacement with MOBKL1A (Thr12/35Ala) increases the fraction of cells exiting M into G1.
Discussion
MOBKL1A/MOBKL1B Are Physiologic Substrates of the MST1 and MST2 Kinases
The present data identify MOBKL1A and MOBKL1B as avid and physiologic substrates of the MST1 and/or MST2 kinases. MST2 activation enables its binding to MOBKL1A/MOBKL1B, and MST2-catalyzed MOBKL1A/MOKL1B phosphorylation inhibits MST2 binding while enhancing MOBKL1A/MOBKL1B binding of LATS1 (and NDR1); consequently it is likely that MOBKL1A/MOBKL1B binding to MST2 and LATS1 is sequential. Moreover, the ability of overexpressed MST2 (K56R) to strongly inhibit MOBKL1A/MOBKL1B phosphorylation in response to okadaic acid and H2O2, treatments known to activate a wide variety of protein kinases, indicates that MST1/MST2 are the dominant—perhaps the only—kinases responsible for MOBKL1A/MOBKL1B phosphorylation in vivo. MST2 (K56R) also will bind through its SARAH domain to the RASSF1-6 polypeptides and to WW45; we have not examined the role of these polypeptides in MOBKL1A/MOBKL1B phosphorylation; however, the ability of MST2-directed shRNA, when accompanied by >90% suppression of MST2 polypeptide expression, to inhibit nocodazole-induced MOBKL1A/MOBKL1B phosphorylation (Figure S5) provides further support for the primary role of MST1/MST2.
MST1/MST2 and MOBKL1A/MOBKL1B Regulate LATS1/LATS2 by Multiple Mechanisms
Previous genetic and biochemical evidence has shown that MST2 and its orthologs are required for the phosphorylation and activation of LATS1 and orthologs ([25]; reviewed in [8–10]). The ability of MST2 (K56R) overexpression to inhibit LATS1 (Thr1079) phosphorylation in response to okadaic acid (Figure 4A) provides strong evidence in support of the earlier suggestion that MST1 and/or MST2 is/are the kinase(s) responsible for this phosphorylation in vivo. By contrast, LATS1 (909) phosphorylation in response to okadaic acid is unaffected by MST2 (K56R) overexpression, a finding compatible with the view that the latter is an intramolecular autophosphorylation. In response to H2O2, however, the phosphorylation of both LATS1 (Thr1079) and LATS1 (Ser909) is strongly suppressed by MST2 (K56R) (Figure 3B), indicating that with this stimulus, MST1/MST2, probably through the generation of phospho-MOBKL1A/MOBKL1B (Figures 3C and 3D), also is crucial for LATS1 (Ser909) phosphorylation. Thus, MST1/MST2 promotes LATS1 multisite phosphorylation through two parallel, independent mechanisms: by direct phosphorylation at Thr1079 and by provision of phospho-MOBKL1A, which promotes LATS1 autophosphorylation at Ser909 in vivo. In addition, others have shown that MOBKL1A/MOBKL1B polypeptides may determine the subcellular localization of LATS1-related kinases [27, 30–32]. The importance of MOBKL1A/MOBKL1B phosphorylation for MOBKL1A cellular localization is not yet known. Phosphorylated MOBKL1A also binds NDR1 in vitro (Figure S2B) and, during transient expression, however, previous work in S. cerevisae and mammalian cells suggests that members of the MOBKL2A/MOBKL1B/MOBKL1C polypeptides are more likely to be the physiologic partners of the NDR1/NDR2-kinase subfamily [25]. Stegert, et al. [33] reported that MST3 can phosphorylate the NDR1 carboxy-terminal hydrophobic site; however, we were unable to detect MST3-catalyzed MOBKL1 or MOBKL2 phosphorylation in vitro (Figure 1D); thus more work on the phosphorylation of the MOBKL2 polypeptides and their relation to NDR1/NDR2 activation will be required.
MOBKL1A/MOBKL1B Phosphorylation Regulates Cell Proliferation
The present data establish that MST1 and MST2, when activated in mitosis, phosphorylate MOBKL1A/MOBKL1B and appear to be the kinases exclusively responsible for mitotic MOBKL1A/MOBKL1B phosphorylation. Moreover elimination of endogenous MOBKL1A/MOBKL1B, together with overexpression of a nonphosphorylatable MOBKL1A polypeptide, exerts a stimulatory effect on cell proliferation by promoting progression through several phases of the cell cycle. Although eliminating MOBKL1A/MOBKL1B phosphorylation accelerates cell proliferation by 2- to 3-fold (Figure 5B), cells lacking phosphorylatable MOBKL1A/MOBKL1B are unaltered in their distribution within the phases of the cell cycle, as compared with cells expressing endogenous MOBKL1A/MOBKL1B or overexpressed recombinant MOBKL1A (Figure 6A). An increase in proliferative rate without a change in cell-cycle distribution also was observed with Drosophila cells bearing Hippo LOF mutations [11, 34]. This property suggests that passage through more than one phase of the cell cycle is likely to be enhanced by the elimination of MOBKL1A/MOBKL1B phosphorylation, and the present data support this view. Thus progression of cells through G1/S into G2/M is accelerated by elimination of MOBKL1A phosphorylation (Figure 6B); however, cells lacking MOBKL1A phosphorylation do not accumulate in M, probably because exit from mitosis also is accelerated (Figures 6C and 6D). The mechanism underlying these actions of MOBKL1A/MOBKL1B phosphorylation remains to be determined. Although the effect of MOBKL1 phosphorylation on G1/S progression is consistent with the phenotype seen with Hippo LOF, the activity of MST1 and MST2 and the levels of MOBKL1A/MOBKL1B phosphorylation in G1/S are very low as compared with those seen in M, and the P-MOBKL1A/MOBKL1B target relevant to G1/S progression is unknown. Whereas overexpression of wild-type MOBKL1A, which undergoes a robust mitotic phosphorylation (Figure 6A, left), substantially delays mitotic exit (Figure 6C), it does not significantly alter proliferative rate (Figure 5B, left) and has a minimal effect on progression through G1/S (Figure 6B). Although it is possible that the minimal levels of MST1/MST2 activity and MOBKL1 phosphorylation in G1/S exert a nearly maximal inhibitory effect on progression through this phase, it is more likely that as with Hippo, active MST1/MST2 and P-MOBKL1 generated during G2/M exert a more sustained transcriptional effect that is responsible for the acceleration of G1/S. As for the role of MOBKL1A/MOBKL1B in mitotic exit, although it is plausible to infer that the inhibitory effect of MOBKL1A phosphorylation is mediated by changes in the activity of LATS1, the evidence on this point is not yet conclusive. Whereas overexpression of MST(K56R) is sufficient to suppress H2O2-stimulated LATS1/LATS2 phosphorylation at Thr1079 and Ser909 (Figure 3B), it does not fully suppress the mitotic phosphorylation of LATS1/LATS2 (Thr1079) or appreciably alter the mitotic phosphorylation of LATS1/LATS2 (Ser909), raising the possibility that LATS1/LATS2 activity in mitosis is controlled by inputs other than or in addition to MST1/MST2. Moreover, these results raise the possibility that P-MOBKL1 targets other than LATS1/LATS2 relevant to the control of cell proliferation and mitotic exit remain to be identified.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grant DK17776 and by institutional funds. We thank Lillian Chow, Ph.D., for purification of the LATS1 antibodies and Yumi Aoyama, M.D., for generation of the U2OS clones expressing MST2 shRNA.
Footnotes
Supplemental Data
Supplemental Experimental Procedures and five figures are available at http://www.current-biology.com/cgi/content/full/18/5/311/DC1/.
References
- 1.Dan I, Watanabe NM, Kusumi A. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 2001;11:220–230. doi: 10.1016/s0962-8924(01)01980-8. [DOI] [PubMed] [Google Scholar]
- 2.Creasy CL, Chernoff J. Cloning and characterization of a human protein kinase with homology to Ste20. J Biol Chem. 1995;270:21695–21700. doi: 10.1074/jbc.270.37.21695. [DOI] [PubMed] [Google Scholar]
- 3.Taylor LK, Wang WCR, Erikson RL. Newly identified stress-responsive protein kinases, Krs-1 and Krs-2. Proc Natl Acad Sci USA. 1996;93:10099–10104. doi: 10.1073/pnas.93.19.10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graves JD, Gotoh Y, Draves KE, Ambrose D, Han DK, Wright M, Chernoff J, Clark EA, Krebs EG. Caspase-mediated activation and induction of apoptosis by the mammalian Ste20-like MST1. EMBO J. 1998;17:2224–2234. doi: 10.1093/emboj/17.8.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee KK, Murakawa M, Nishida E, Tsubuki S, Kawashima SI, Safkamaki K, Yonehara S. Proteolytic activation of MST/Krs, STE20-related protein kinase by caspase during apoptosis. Oncogene. 1998;16:3029–3037. doi: 10.1038/sj.onc.1201840. [DOI] [PubMed] [Google Scholar]
- 6.Reszka AA, Halasy-Nagy JM, Masarachia PJ, Gideon RA. Bisphosphonates act directly on the osteoclast to induce caspase cleavage of Mst1 kinase during apoptosis. A link between inhibition of the mevalonate pathway and regulation of an apoptosis-promoting kinase. J Biol Chem. 1999;274:34967–34973. doi: 10.1074/jbc.274.49.34967. [DOI] [PubMed] [Google Scholar]
- 7.Katagiri K, Imamura M, Kinashi T. Spatiotemporal regulation of the kinase Mst1 by binding protein RAPL is critical for lymphocyte polarity and adhesion. Nat Immunol. 2006;7:919–928. doi: 10.1038/ni1374. [DOI] [PubMed] [Google Scholar]
- 8.Harvey K, Tapon N. The Salvador-Warts-Hippo pathway - an emerging tumour-suppressor network. Nat Rev Cancer. 2007;7:182–191. doi: 10.1038/nrc2070. [DOI] [PubMed] [Google Scholar]
- 9.Pan D. Hippo signaling in organ size control. Genes Dev. 2007;21:886–897. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- 10.Saucedo LJ, Edgar BA. Filling out the Hippo pathway. Nat Rev Mol Cell Biol. 2007;8:613–621. doi: 10.1038/nrm2221. [DOI] [PubMed] [Google Scholar]
- 11.Wu S, Huang J, Dong J, Pan D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- 12.St John MA, Tao W, Fei X, Fukumoto R, Carcangiu ML, Brownstein DG, Parlow AF, McGrath J, Xu T. Mice deficient of Lats1 develop soft-tissue sarcomas, ovarian tumours and pituitary dysfunction. Nat Genet. 1999;21:182–186. doi: 10.1038/5965. [DOI] [PubMed] [Google Scholar]
- 13.Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 14.Lai ZC, Wei X, Shimizu T, Ramos E, Rohrbaugh M, Nikolaidis N, Ho LL, Li Y. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell. 2005;120:675–685. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 15.Simanis V. Events at the end of mitosis in the budding and fission yeasts. J Cell Sci. 2003;116:4263–4275. doi: 10.1242/jcs.00807. [DOI] [PubMed] [Google Scholar]
- 16.Bardin AJ, Amon A. Men and sin: what’s the difference? Nat Rev Mol Cell Biol. 2001;2:815–826. doi: 10.1038/35099020. [DOI] [PubMed] [Google Scholar]
- 17.Wei X, Shimizu T, Lai ZC. Mob as tumor suppressor is activated by Hippo kinase for growth inhibition in Drosophila. EMBO J. 2007;26:1772–1781. doi: 10.1038/sj.emboj.7601630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan EH, Nousiainen M, Chalamalasetty RB, Schäfer A, Nigg EA, Silljé HH. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene. 2005;24:2076–2086. doi: 10.1038/sj.onc.1208445. [DOI] [PubMed] [Google Scholar]
- 19.Bothos J, Tuttle RL, Ottey M, Luca FC, Halazonetis TD. Human LATS1 is a mitotic exit network kinase. Cancer Res. 2005;65:6568–6575. doi: 10.1158/0008-5472.CAN-05-0862. [DOI] [PubMed] [Google Scholar]
- 20.Yabuta N, Okada N, Ito A, Hosomi T, Nishihara S, Sasayama Y, Fujimori A, Okuzaki D, Zhao H, Ikawa M, et al. Lats2 is an essential mitotic regulator required for the coordination of cell division. J Biol Chem. 2007;282:19259–19271. doi: 10.1074/jbc.M608562200. [DOI] [PubMed] [Google Scholar]
- 21.Cheung WL, Cheung WL, Ajiro K, Samejima K, Kloc M, Cheung P, Mizzen CA, Beeser A, Etkin LD, Chernoff J, et al. Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase l. Cell. 2003;113:507–517. doi: 10.1016/s0092-8674(03)00355-6. [DOI] [PubMed] [Google Scholar]
- 22.Lehtinen MK, Yuan Z, Boag PR, Yang Y, Villen J, Becker EB, DiBacco S, de la Iglesia N, Gygi S, Blackwell TK, Bonni A. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. 2006;125:987–1001. doi: 10.1016/j.cell.2006.03.046. [DOI] [PubMed] [Google Scholar]
- 23.Knebel A, Morrice N, Cohen P. A novel method to identify protein kinase substrates: eEF2 kinase is phosphorylated and inhibited by SAPK4/p38delta. EMBO J. 2001;20:4360–4369. doi: 10.1093/emboj/20.16.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Praskova M, Khoklatchev A, Ortiz-Vega S, Avruch J. Regulation of the MST1 kinase by autophosphorylation, by the growth inhibitory proteins, RASSF1 and NORE1 and by Ras. Biochem J. 2004;381:453–462. doi: 10.1042/BJ20040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hergovich A, Stegert MR, Schmitz D, Hemmings BA. NDR kinases regulate essential cell processes from yeast to humans. Nat Rev Mol Cell Biol. 2006;7:253–264. doi: 10.1038/nrm1891. [DOI] [PubMed] [Google Scholar]
- 26.Mah AS, Jang J, Deshaies RJ. Protein kinase Cdc15 activates the Dbf2-Mob1 kinase complex. Proc Natl Acad Sci USA. 2001;98:7325–7330. doi: 10.1073/pnas.141098998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hou MC, Salek J, McCollum D. Mob1p interacts with the Sid2p kinase and is required for cytokinesis in fission yeast. Curr Biol. 2000;10:619–622. doi: 10.1016/s0960-9822(00)00492-9. [DOI] [PubMed] [Google Scholar]
- 28.Hou MC, Wiley DJ, Verde F, McCollum D. Mob2p interacts with the protein kinase Orb6p to promote coordination of cell polarity with cell cycle progression. J Cell Sci. 2003;116:125–135. doi: 10.1242/jcs.00206. [DOI] [PubMed] [Google Scholar]
- 29.Bichsel SJ, Tamaskovic R, Stegert MR, Hemmings BA. Mechanism of activation of NDR (nuclear Dbf2-related) protein kinase by the hMOB1 protein. J Biol Chem. 2004;279:35228–35235. doi: 10.1074/jbc.M404542200. [DOI] [PubMed] [Google Scholar]
- 30.Luca FC, Mody M, Kurischko C, Roof DM, Giddings TH, Winey M. Saccharomyces cerevisiae Mob1p is required for cytokinesis and mitotic exit. Mol Cell Biol. 2001;21:6972–6983. doi: 10.1128/MCB.21.20.6972-6983.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hou MC, Guertin DA, McCollum D. Initiation of cytokinesis is controlled through multiple modes of regulation of the Sid2p-Mob1p kinase complex. Mol Cell Biol. 2004;24:3262–3276. doi: 10.1128/MCB.24.8.3262-3276.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hergovich A, Schmitz D, Hemmings BA. The human tumour suppressor LATS1 is activated by human MOB1 at the membrane. Biochem Biophys Res Commun. 2006;345:50–58. doi: 10.1016/j.bbrc.2006.03.244. [DOI] [PubMed] [Google Scholar]
- 33.Stegert MR, Hergovich A, Tamaskovic R, Bichsel SJ, Hemmings BA. Regulation of NDR protein kinase by hydrophobic motif phosphorylation mediated by the mammalian Ste20-like kinase MST3. Mol Cell Biol. 2005;25:11019–11029. doi: 10.1128/MCB.25.24.11019-11029.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.