Abstract
Non‐coding RNAs play a key role in organizing the nucleus into functional subcompartments. By combining fluorescence microscopy and RNA deep‐sequencing‐based analysis, we found that RNA polymerase II transcripts originating from intronic Alu elements (alu RNAs) were enriched in the nucleolus. Antisense‐oligo‐mediated depletion of alu RNAs or drug‐induced inhibition of RNA polymerase II activity disrupted nucleolar structure and impaired RNA polymerase I‐dependent transcription of rRNA genes. In contrast, overexpression of a prototypic alu RNA sequence increased both nucleolus size and levels of pre‐rRNA, suggesting a functional link between alu RNA, nucleolus integrity and pre‐rRNA synthesis. Furthermore, we show that alu RNAs interact with nucleolin and target ectopic genomic loci to the nucleolus. Our study suggests an alu RNA‐based mechanism that links RNA polymerase I and II activities and modulates nucleolar structure and rRNA production.
Keywords: Alu repeat‐containing RNA, nucleolus structure and function, RNA‐dependent phase separation
Subject Categories: Chromatin, Epigenetics, Genomics & Functional Genomics; Transcription; RNA Biology
Introduction
Non‐coding RNAs (ncRNAs) regulate a diverse set of nuclear activities and shape nuclear organization as an architectural factor (Caudron‐Herger & Rippe, 2012; Batista & Chang, 2013; Mercer & Mattick, 2013; Bergmann & Spector, 2014). They are involved in the formation of nuclear bodies (Mao et al, 2011; Shevtsov & Dundr, 2011), establish active or repressive chromatin states (Wong et al, 2007; Deng et al, 2009; Caudron‐Herger et al, 2011; Yang et al, 2011) and regulate gene expression (Yao et al, 2010; Mercer & Mattick, 2013; Yang et al, 2013; Bergmann & Spector, 2014).
The nucleolus is an exemplary case for the complex network between structure and function, demonstrating how RNA‐dependent spatial organization affects transcriptional activity (Pederson, 1998; Carmo‐Fonseca et al, 2000; Olson et al, 2000; Boisvert et al, 2007; McKeown & Shaw, 2009). The nucleolus is highly sensitive to cellular stress, and environmental cues dynamically regulate its structure and activity (Rubbi & Milner, 2003; Olson, 2004; Sirri et al, 2008; Boulon et al, 2010). An important role for RNA in this process can be inferred from experiments that perturb transcription. Although RNA polymerase I (Pol I) transcribes nucleolar rRNA genes (rDNAs), inhibition of RNA polymerase II (Pol II) by α‐amanitin, 5,6‐dichloro‐1‐beta‐D‐ribofuranosylbenzimidazole (DRB) or roscovitine (Chafin et al, 1995; Sirri et al, 2002) leads to disintegration of nucleoli (Granick, 1975; Scheer et al, 1984; Chafin et al, 1995; Haaf & Ward, 1996; Sirri et al, 2002; Burger et al, 2010). Inhibition of Pol II transcription is accompanied by down‐regulation of rRNA synthesis, indicating that yet‐to‐be‐identified Pol II transcripts may regulate nucleolar organization and Pol I transcriptional activity (David‐Pfeuty et al, 2001).
Transcripts from the intergenic spacer that separates individual rRNA genes have been linked to epigenetic regulation of Pol I transcription (Mayer et al, 2006), to sequestering proteins in the nucleolus upon cellular stress (Audas et al, 2012) and to remodeling of the nucleolus (Jacob et al, 2013). A recent study identified Pol II transcripts in antisense orientation to the pre‐rRNA coding region, which recruit the histone methyltransferase SUV4‐20H2 to rRNA genes to induce histone H4 lysine 20 trimethylation and chromatin compaction in growth‐arrested cells (Bierhoff et al, 2014). In addition, novel functional transcripts originating from the intergenic spacer of rDNA repeats and synthesized by Pol I have been implicated in the nucleolar stress response to maintain cellular homeostasis (Audas et al, 2012; Jacob et al, 2012, 2013). Finally, the association of the NAD+‐dependent deacetylase SIRT7 to nascent pre‐rRNA stabilizes the interaction of SIRT7 with Pol I, being a prerequisite for SIRT7‐dependent transcription activation (Chen et al, 2013). However, a detailed analysis of the role of regulatory nucleolar transcripts for the structure and activity of the nucleolus is lacking.
Here, we combined high‐resolution fluorescence microscopy and RNA deep sequencing to address this issue. We found that Pol II transcripts originating from intronic Alu elements (aluRNAs) play an important role in the assembly and function of the nucleolus. Alu repeats are the most abundant repetitive elements of primates constituting more than 10% of the human genome (Batzer & Deininger, 2002). Alu transcripts are synthesized by both RNA polymerase III (Pol III) and II. “Free Alu RNAs”, that is, RNAs transcribed from Alu elements that are not embedded within genes, as well as the related 7SL RNA, are produced by Pol III using the internal promoter of Alu elements. In contrast, “embedded Alu RNAs” are localized within introns, transcribed by Pol II and spliced out from pre‐mRNA during mRNA maturation (Deininger, 2011). Alu transcripts have been shown to regulate gene expression post‐transcriptionally, being involved in alternative splicing (Singer et al, 2004), RNA editing (Mattick & Mehler, 2008) and translation efficiency (Capshew et al, 2012; Fitzpatrick & Huang, 2012), but have not been linked to the functional organization of nuclear subcompartments. In the present study, we show that Pol II‐dependent aluRNAs regulate nucleolar structure and rRNA synthesis via interaction with nucleolin (NCL), a major structural and multifunctional nucleolar protein with pivotal functions in ribosome biogenesis (Ginisty et al, 1999). Our results uncover a novel mechanism that links Pol I transcription levels with global Pol II activity via nuclear aluRNA production and suggest an aluRNA‐dependent mechanism that preserves nucleolar structure and function.
Results
Functional nucleolus structure requires Pol II transcription
To address the response of nucleolar structure to the inhibition of transcription, we treated HeLa cells for 5 h with drugs that specifically inhibit each of the three polymerases (Fig 1A). Notably, a Pol II‐specific structural phenotype was observed after inhibition of Pol II transcription by α‐amanitin (Weinmann & Roeder, 1974) or DRB (Fig 1A and Appendix Fig S1A). Nucleoli were disrupted into small domains, and rDNA was scattered throughout the nucleus. These observations suggest that active Pol II transcription is needed for the structural integrity of the nucleolus, although this nuclear compartment has very little apparent Pol II activity (Roeder & Rutter, 1970; Sirri et al, 2008). Drug‐mediated perturbations of nucleolar structure correlated with a clearly impaired nucleolar function as revealed by strong reduction in pre‐rRNA synthesis (Appendix Fig S1B and C) (David‐Pfeuty et al, 2001). Moreover, NCL, Pol I and the Pol I‐specific transcription factor UBF became dispersed throughout the nucle‐oplasm after α‐amanitin treatment (Fig 1A and Appendix Fig S1D and E). In the following, we refer to this phenotype as nucleolar dispersion or dispersed nucleoli, which are characterized by the dissociation of nucleoli into small nucleolar domains and the loss of co‐localization of the nucleolar components. These changes of nucleolar structure after Pol II inhibition were clearly different from those induced by treatment of cells with low doses of actinomycin D (AMD), which blocks Pol I transcription elongation and causes nucleolar segregation (Fig 1A), that is, condensation of rDNA in nucleolar caps (Reynolds et al, 1964). Moreover, efficient inhibition of Pol III transcription by ML‐60218 (Wu et al, 2003; Di Ruscio et al, 2013) did not result in nucleolar dispersion (Fig 1A and Appendix Fig S1F). Likewise, treatment with cycloheximide for 5 h did not affect nucleolar structure, demonstrating that disruption of nucleoli after α‐amanitin treatment was not due to abrogation of protein synthesis (Appendix Fig S1G). Thus, the absence of a nucleolar dispersion phenotype after specific inhibition of Pol I, III or protein synthesis demonstrates that Pol II transcription is needed to maintain the structure of the nucleolus and the efficient production of pre‐rRNA by Pol I.
Figure 1. Pol II transcription is essential for nucleolar structure and function.
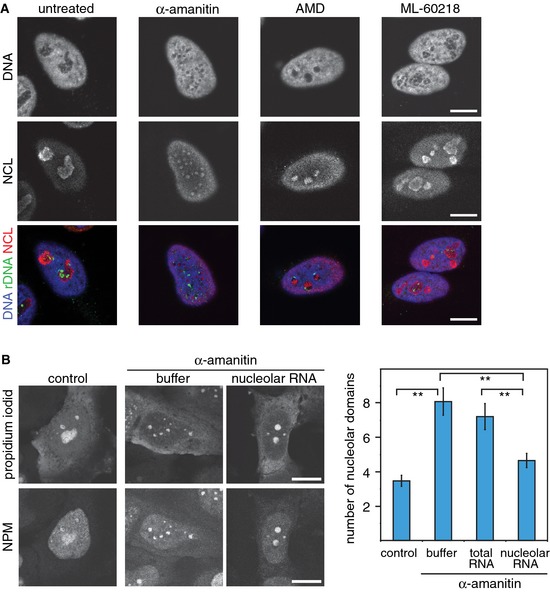
- Confocal laser scanning fluorescence microscopy (CLSM) images showing DNA staining (blue in the merged images), rDNA FISH (green) and NCL immunofluorescence (red) in untreated HeLa cells or in cells treated with α‐amanitin (50 μg/ml), AMD (50 ng/ml) or the Pol III inhibitor ML‐60218 (200 μM) for 5 h.
- CLSM images of propidium iodide‐stained RNA after microinjection of buffer or nucleolar RNA into α‐amanitin‐treated HeLa cells. Nucleoli were visualized by immunofluorescence of nucleophosmin (NPM). The graph represents the average number of nucleolar domains (± 95% CI) based on the analysis of 90, 87, 80 and 86 cells, respectively. **P‐value < 0.01, t‐test.
Data information: Scale bars, 10 μm.
See also Appendix Figs S1 and S2.
Nucleolar RNA partially rescues α‐amanitin‐mediated disruption of nucleoli
Our results obtained after α‐amanitin treatment do not distinguish whether Pol II transcription per se or Pol II transcripts are required for maintaining nucleolar structure. If Pol II transcripts stabilize nucleoli, Pol II transcripts should be present in nucleoli. To test this, we isolated RNA from purified nucleoli that were devoid of substantial contaminations by cytoplasmic or nucleoplasmic RNA (Appendix Fig S2). Small nucleolar RNAs (snoRNA) displayed strong enrichment in the nucleolar RNA fraction (Richard et al, 2003). In contrast MALAT1, a highly abundant ncRNA that localizes to nuclear speckles (Hutchinson et al, 2007), was absent as compared to total RNA and the nucleoplasmic fraction. Consistent with a small fraction of Cajal bodies being associated with nucleoli (Raska et al, 1990), Cajal body‐specific RNAs (scaRNAs) were present in the nucleoplasmic and nucleolar fractions.
Next, we tested the capability of the purified nucleolar RNA to rescue the disrupted nucleolar phenotype observed upon the inhibition of Pol II transcription. As reported above, α‐amanitin treatment induced dispersion of nucleoli into smaller nucleolar domains. This process was quantified by a more than two‐fold increase in their number from n = 3.3 ± 0.3 to n = 8.1 ± 1.0 with an average size of 1.1 ± 0.1 μm2 (Fig 1B). Microinjection of RNA from purified nucleoli but not from total RNA was capable of rescuing nucleolar perturbation caused by α‐amanitin treatment. As shown in Fig 1B, larger and fewer nucleolar domains (n = 4.7 ± 0.4 with an average size of 2.3 ± 0.3 μm2) were observed after microinjection of nucleolar RNA into α‐amanitin‐treated HeLa cells. This indicates that the nucleolar RNA fraction contains RNA transcripts that counteract the α‐amanitin‐mediated segregation of active nucleolus organizer regions.
Alu element‐containing Pol II transcripts are enriched in the nucleolus
To identify the RNA transcripts that stabilize nucleolar structure, we looked for transcripts that were specifically enriched in the nucleolar RNA fraction. We performed RNA‐seq and a comparative bioinformatic analysis of data sets obtained from nucleolar and nucleoplasmic RNA fractions as well as total RNA. Nucleolar RNA was markedly enriched in reads mapping to “intronic‐only” sequences, that is, sequences not associated with exonic parts of the corresponding primary transcripts (Appendix Table S1). The nucleolar “intronic‐only” sequences were enriched in Alu repeat elements as compared to the total RNA or nucleoplasmic RNA fractions. We here refer to these Alu repeat‐containing transcripts that are overrepresented in nucleolar RNA as aluRNAs. Generally, intron‐encoded aluRNAs clustered into transcripts covering either the whole Alu sequence or only a truncated left (aluRNAL) or right (aluRNAR) arm in either forward or reverse orientation within the primary transcript (Fig 2A). In the total RNA fraction, most aluRNAs were found in long primary transcripts (> 2,000 nt) (Appendix Table S2). In the nucleolar RNA fraction, however, isolated aluRNA sequences lacking flanking sequences were markedly enriched and, on an average, about 100‐fold more abundant than in total RNA or nucleoplasmic RNA (Fig 2B and C, Appendix Tables S2 and S3). The most abundant nucleolar aluRNAs were about 250 nt in length and corresponded to a forward aluRNAR sequence that lacked the box A and box B element (Fig 2A and B, Appendix Figs S3 and S4). The level of most nucleolar aluRNAs was decreased after α‐amanitin treatment, whereas the inhibition of Pol III transcription did not affect the abundance of nucleolar aluRNAs (Fig 2D), confirming that Pol II synthesizes nucleolar aluRNAs.
Figure 2. Nucleolar RNA is enriched in specific intron‐encoded aluRNAs.
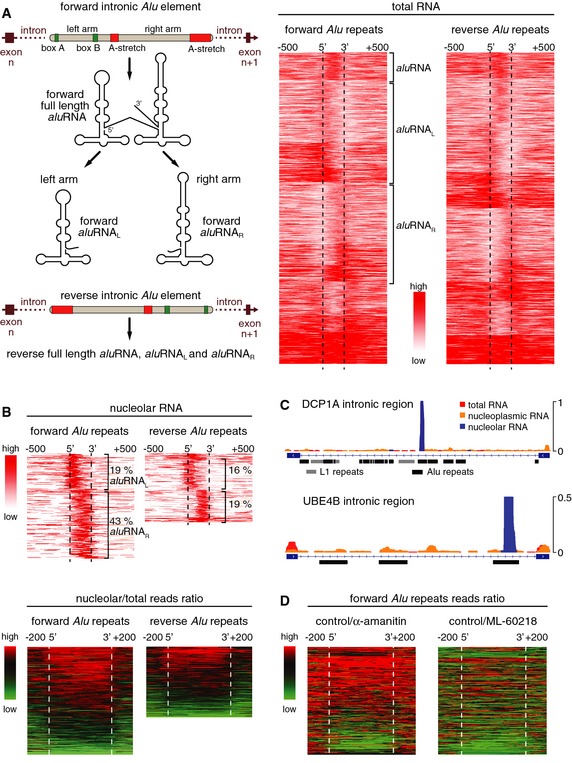
- Left: Scheme illustrating the origin of forward or reverse intronic aluRNA and processing into left arm aluRNAs (aluRNAL) and right arm aluRNAs (aluRNAR). Right: Heatmaps of read density of forward and reverse intronic Alu RNA species present in the total RNA sample.
- Top: Heatmaps of read density of intronic Alu repeats in the nucleolar RNA fraction. For each aluRNA variant, the percentage over all transcribed intronic Alu repeats is indicated. Bottom: Ratio of nucleolar versus total RNA from higher (red) to lower (green) read density.
- View of nucleolus‐enriched Alu elements exemplified at two genomic loci. Mapping of normalized reads from total (red), nucleoplasmic (orange) and nucleolar (blue) RNA to intron #5 of DCP1A and intron #22 of UBE4B.
- Heatmaps of total RNA read density ratios between untreated (control) and α‐amanitin‐ or ML‐60218‐treated HeLa cells.
To corroborate that aluRNA sequences were enriched in nucleoli, we conducted RNA FISH experiments with probes that hybridize to nucleolar aluRNA in forward or reverse orientation. The results confirmed that both types of aluRNAs accumulated in the nucleoli (Fig 3A and Appendix Fig S5A–C). Furthermore, a Northern blot analysis of total RNA revealed the presence of aluRNA‐containing long primary transcripts (< 2,000 nt) in addition to transcripts of about 300 nt and 90 nt (Appendix Fig S5D). This finding is consistent with the aluRNA length distribution of nucleolar aluRNA as inferred from the RNA‐seq data (Appendix Fig S4). Thus, we conclude that Pol II‐dependent aluRNAs are largely overrepresented in nucleoli.
Figure 3. aluRNA is localized to nucleoli and partially rescues α‐amanitin‐induced nucleolar dispersion.
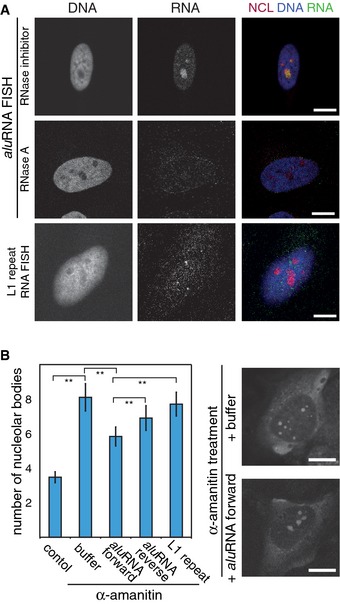
- CLSM images showing nucleolar co‐localization of NCL (immunofluorescence) with aluRNA (RNA FISH) but not with L1‐repeat RNA. Cells were pre‐treated in situ with RNase A or an RNase inhibitor. Nuclei were counterstained with DAPI. The signal intensity of nucleolar aluRNA was two‐fold higher compared to nucleoplasmic signal (n = 92, P‐value < 0.05, t‐test).
- Graphs representing the average number of nucleolar bodies after microinjection of in vitro transcribed RNA into HeLa cells that were pre‐treated with α‐amanitin (50 μg/ml) for 5 h or left untreated (control) (± 95% CI. **P‐value < 0.01, n = 90, 87, 86, 83 or 86 cells, respectively). Representative CLSM images of propidium iodide‐stained RNA are shown on the right side.
Data information: Scale bars, 10 μm.
See also Appendix Fig S5, Appendix Table S4.
Changes in aluRNA levels affect nucleolar structure and rRNA synthesis
Next, we tested whether ectopic aluRNA was capable of rescuing α‐amanitin‐induced perturbation of nucleolar organization. Indeed, microinjection of in vitro transcribed aluRNAs triggered reassembly of nucleolar bodies after α‐amanitin inhibition of Pol II (Fig 3B). The forward aluRNA variant was more efficient in rescuing nucleolar dispersion than the reverse aluRNA or an L1‐repeat RNA transcript (Fig 3B and Appendix Table S4).
To further investigate the functional relevance of aluRNAs for nucleolar structure and function, we depleted aluRNA with different specific antisense oligonucleotides (ASOs) (Ideue et al, 2009) corresponding to 20 nucleotides of the nucleolar aluRNA consensus sequence (Appendix Fig S3 and Appendix Table S4). The ASOs, which were transfected into the cell via transfection reagent, hybridized to the target RNAs that were subsequently cleaved by endogenous RNase H. Knockdown of aluRNA, as measured from RNA‐seq analysis (Appendix Fig S6A and B), preserved the level of housekeeping genes (Appendix Fig S6C) (Eisenberg & Levanon, 2013) but produced a phenotype resembling that observed after α‐amanitin treatment: dispersion of nucleoli and the nucleolar marker protein NPM (Fig 4A and Appendix Fig S1D and E), reduction in aluRNA FISH signal (Fig 4B and Appendix Fig S6D) and repression of pre‐rRNA synthesis (Fig 4C and D and Appendix Fig S1B and C). In addition, α‐amanitin treatment induced a particular strong reduction in intronic Alu repeat‐containing transcripts (Appendix Fig S6E), which were included in the sequences targeted by the aluRNA ASOs. Furthermore, ASO‐mediated knockdown of aluRNA triggered the dispersion of nucleolar marker proteins such as NCL, NPM and Pol I (Fig 4A and B and Appendix Fig S7A). This phenotype was not observed in cells treated with ASOs targeting the highly abundant L1‐repeat or 7SL RNA families (Appendix Table S4 and Appendix Fig S7B and C), for which cells showed a phenotype similar to the control ASO‐treated cells. Notably, dispersion of nucleoli upon depletion of aluRNA was not restricted to HeLa cells but also observed in human keratinocytes or fibroblasts (Appendix Fig S7B and C). We conclude from these results that a reduction of aluRNA levels induced strong perturbations in nucleolar structure and rRNA synthesis.
Figure 4. Knockdown or overexpression of aluRNA impact size of nucleoli and rDNA transcription.
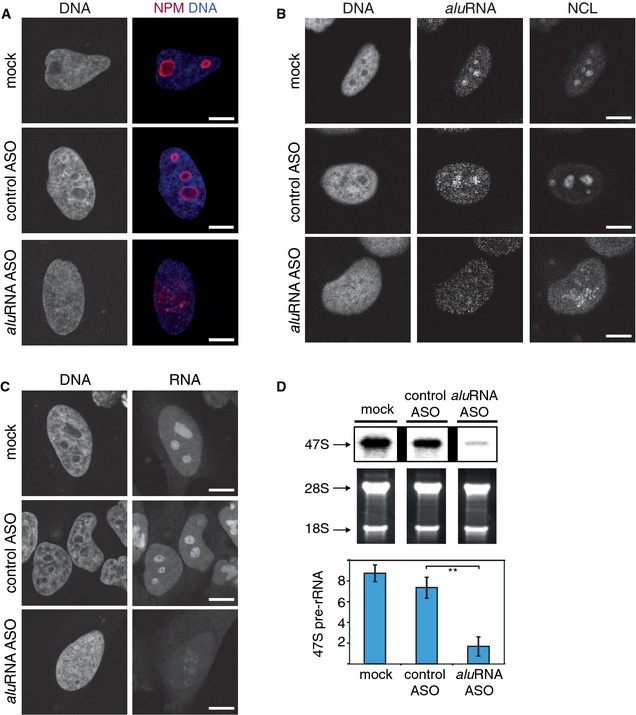
- CLSM images showing NPM (immunofluorescence, red) in HeLa cells transfected with control ASO, aluRNA ASO, or treated with transfection reagent only (mock). Nuclei were counterstained with DAPI (blue). A fraction of 80 ± 8% of the cells treated with aluRNA ASO (n = 143) showed irregular nucleoli or dispersed nucleoli (53% of the cells). Only 26 ± 9% of the cells treated with control ASO presented abnormal nucleoli, with none of them showing a dispersion of nucleolar domains (n = 123), P‐value < 0.01, t‐test.
- CLSM images showing NCL localization (immunofluorescence), aluRNA (RNA FISH) and DNA (DAPI). Transfection of HeLa cells was done as in (A).
- HeLa cells were transfected as in (A), and nascent RNAs were visualized by pulse labeling with ethynyl uridine (EU) for 30 min. Nuclei were counterstained with DAPI (DNA).
- Levels of 47S pre‐rRNA in mock‐, control ASO‐ and aluRNA ASO‐treated HeLa cells. Top: Northern blot. Center: Agarose gel electrophoretic analysis of RNA. Bottom panel: Quantification from RT–qPCR levels of 47S pre‐rRNA normalized to 18S rRNA. Error bars represent the standard deviation (n = 6). **P‐value < 0.01, t‐test.
Data information: Scale bars, 10 μm. See also Appendix Figs S1C, S6, S7 and S8, Appendix Table S4.
Source data are available online for this figure.
Time course analysis by immunofluorescence microscopy revealed that dispersion of nucleoli after aluRNA knockdown occurred in two steps. First, the size of nucleoli increased within 1–3 h after α‐amanitin treatment or 6–10 h after knockdown of aluRNA (Appendix Fig S8A and B). When nucleoli reached a critical size, they dispersed into smaller particles. Swelling of nucleoli and formation of numerous small ectopic NPM‐containing particles were also observed in cells treated with LNA antisense “blocker” probes that were designed to hybridize to aluRNAs without inducing their degradation (Appendix Fig S8C). A control LNA blocker directed against L1‐repeat transcripts did not disturb nucleolar structures (Appendix Fig S8D).
To examine whether overexpression of aluRNA would increase the size of nucleoli, we transfected cells with a plasmid that simultaneously expresses GFP from a Pol II CMV promoter and RNAs from a Pol III U6 promoter. As shown in Fig 5A, the size of nucleoli increased by 30% upon overexpression of forward aluRNA, as quantified by 3D measurements of imaging stacks. In contrast, overexpression of an L1‐repeat transcript had no significant effect. This finding was confirmed by tracing single living cells over time using an RFP‐tagged NCL as nucleolar marker. Clearly, overexpression of the forward aluRNA sequence resulted in an enlargement of nucleoli within a single cell over a time period of 10 h (Fig 5B). In addition, RNA pulse labeling of nascent RNA under the same conditions revealed an increase in nucleolar activity by 27% in the presence of ectopic aluRNA, but not of L1‐repeat RNA, based on measuring the nucleolar RNA fluorescence signal intensity (Fig 5C and D). Thus, both the loss‐of‐function and the gain‐of‐function experiments showed a consistent correlation between aluRNA levels, nucleoli size and rRNA production.
Figure 5. aluRNA overexpression induces an increase in nucleolar volume and activity.
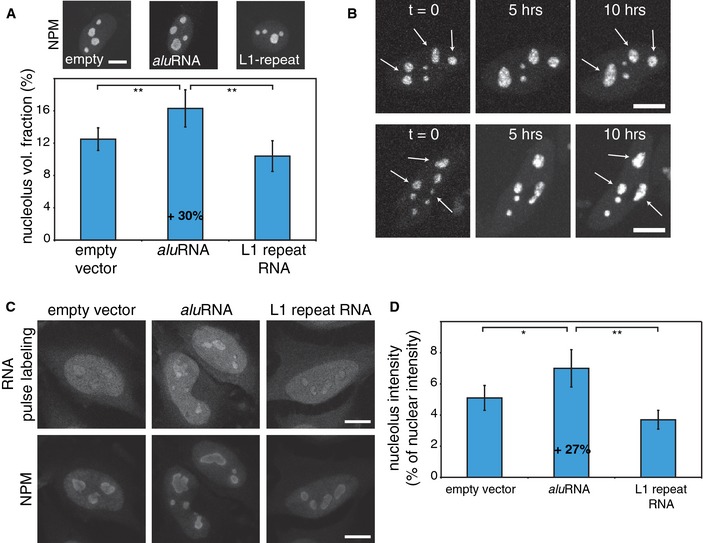
- HeLa cells were transfected with an empty vector or plasmids expressing aluRNA or L1‐repeat RNA and GFP from a separate promoter. Transfected cells were identified by GFP fluorescence, and nucleoli volumes were evaluated based on the immunofluorescence signal of the nucleolar marker NPM. The graphs show that the volume of nucleoli is increased by 30% in the presence of ectopic aluRNA (± 95% CI, n = 20). **P‐value < 0.01, t‐test. Scale bar, 10 μm.
- CLSM live‐cell imaging of RFP‐NCL in cells transfected with a plasmid expressing aluRNA and imaged for 10 h. The arrows indicate nucleolar domains displaying time‐dependent increase in the nucleolar size or fusion of two domains into one. Scale bars, 10 μm.
- HeLa cells were transfected as in (A) and incubated for 24 h. Nascent RNA was pulse‐labeled for 30 min, and nucleoli were visualized by immunofluorescence of NPM. Scale bars, 10 μm.
- Graphs represent the intensity of nucleolar fluorescence signals as percentage of the intensity of nuclear signals and reveal a 27% increase in the presence of ectopic aluRNA (± 95% CI, n = 30). **P‐value < 0.01, *P‐value = 0.01, t‐test.
B1‐containing RNAs maintain nucleolar structure in mouse cells
We observed that the structure of nucleoli was also disrupted in mouse NIH 3T3 cells upon Pol II inhibition in a manner very similar to what we found in human cells (Fig 6A) (Caudron‐Herger et al, 2011). Accordingly, we investigated whether this phenotype could be assigned to a specific type of RNA transcripts in this organism. Alu elements are primate specific (Liu et al, 2009), but the closely related B1 repeats are present in the mouse genome (Nishihara et al, 2002). We hypothesized that transcripts from B1 elements could represent the functional equivalent of human aluRNAs. This assumption was validated by our observation that treatment with ASOs against B1‐containing RNA transcripts (Appendix Table S4) induced a significant 30% reduction of B1‐containing RNA levels (P‐value = 0.04, n = 3) and nucleolar dispersion in NIH 3T3 cells (Fig 6B), similar to that revealed in human cell lines (Fig 4A and Appendix Fig S7B and C). As expected, the human aluRNA ASO had no effect on nucleolar structure in the mouse fibroblast cell line demonstrating specificity. Thus, B1‐containing RNA transcripts are important to maintain nucleolar structure in mouse cells.
Figure 6. Nucleoli disruption in mouse cells by depletion of B1‐containing RNAs.

- CLSM images of UBF immunofluorescence in mouse NIH 3T3 cells treated for 5 h with α‐amanitin to inhibit Pol II transcription show the dispersion of nucleoli, as previously also imaged via RNA staining (Caudron‐Herger et al, 2011).
- CLSM images showing the localization of NPM (immunofluorescence, red) in mouse NIH 3T3 cells treated with ASO as indicated and counterstained with DAPI (blue).
aluRNA interacts with NCL and targets genomic loci to the nucleolus
To further investigate the link between aluRNA and nucleolar organization, we conducted a set of experiments with forward aluRNA fused to MS2 stem‐loop hairpins (MS2‐aluRNA). First, we observed that ectopically expressed MS2‐aluRNA was enriched in nucleoli (Fig 7A) as compared to a control RNA sequence. This supports our previous conclusion of aluRNA being enriched in the nucleolus that was based on the analysis of endogenous aluRNAs by deep sequencing (Fig 2C) and by RNA FISH (Fig 3A). Next, we made use of a U2OS cell line (F4IIB8) containing a stably integrated lacO array at a single genomic locus (Jegou et al, 2009). We used a lac repressor (LacI) protein construct fused to the MS2 stem‐loops binding protein (Shevtsov & Dundr, 2011) to recruit MS2‐RNAs to the lacO array (Fig 7B). Notably, we observed that MS2‐aluRNAR induced recruitment of NCL to the lacO array (Fig 7B). Furthermore, tethering of both MS2‐aluRNA and MS2‐aluRNAR variants to these sites significantly increased the number of lacO arrays localizing in nucleoli (Fig 7C). These observations suggest that aluRNA interacts with NCL and can target genomic loci to nucleoli.
Figure 7. Ectopically expressed aluRNA accumulates in the nucleolus together with associated genomic loci.

- CLSM images of MS2‐tagged aluRNA or MS2‐tagged control RNA (a fragment from the transcript of the CDV3 gene). MS2‐loop‐containing RNAs were visualized by RNA FISH using MS2‐loop‐specific FISH probes (green); nucleoli were visualized by immunofluorescence of NCL (red).
- The top panel depicts the experimental approach used to tether MS2‐loop‐containing RNAs to lacO arrays via a GFP‐tagged LacI‐MS2 coat fusion protein. The lacO arrays were stably integrated in the genome of U2OS cells. In the bottom panel, CLSM images of U2OS cells are shown that were transfected with MS2‐aluRNAR. They reveal the localization of LacI‐GFP‐MS2 and NCL (immunofluorescence, red). The arrow indicates the lacO array, which is associated with a nucleolar domain.
- MS2‐loop‐containing forward aluRNA or aluRNAL or aluRNAR were recruited to the single stably integrated lacO array in the U2OS F4IIB8 cell line. The propensity for nucleolar localization was evaluated as the average number of lacO arrays with tethered RNA detected in nucleoli (± 95% CI) and is plotted in the bar chart. Calculations are based on analysis of more than 100 cells. **P‐value < 0.01, t‐test, from the analysis of two independent biological replicates. As controls, transcripts of the MS2 loops only, the MS2‐CDV3, MS2‐RepA, MS2‐STARD7 and MS2‐CORO1C RNAs were used. The inset shows a CLSM image with MS2‐GFP‐LacI (green) and NPM (red, immunofluorescence) after recruitment of MS2‐aluRNA.
Nucleolin and nucleophosmin interact with aluRNA and target genomic loci to the nucleolus
To elucidate the mechanism by which aluRNA targets genomic loci to the nucleolus, we investigated the interaction of aluRNAs with specific nucleolar proteins. We analyzed the association of aluRNA with NCL and NPM, abundant nucleolar proteins that interact with RNA and are important for the structure of nucleoli (Ugrinova et al, 2007; Amin et al, 2008) using a U2OS cell line (F6B2) that contains three lacO array integrations at different chromosomes (Jegou et al, 2009). To visualize recruitment of GFP‐tagged proteins to the lacO arrays, a previously described fusion construct of LacI with a GFP‐binding protein was used (Chung et al, 2011) (Fig 8A). Analysis of the localization of aluRNA by RNA FISH revealed that aluRNA was enriched at lacO arrays that were associated with GFP‐tagged NCL or NPM (Fig 8A and Appendix Fig S9A). Interestingly, GFP‐tagged fibrillarin, involved in the site‐specific 2′‐O‐methylation of ribose (Reichow et al, 2007), was also recruiting aluRNA at the lacO arrays (Appendix Fig S9A). In contrast, only background levels of aluRNA were found at lacO arrays associated with GFP or the RNA‐binding domain of TIP5, a subunit of the nucleolar remodeling complex NoRC (Fig 8A) (Mayer et al, 2006). Moreover, recruitment of NCL to lacO arrays was accompanied by enrichment of UBF and NPM (Appendix Fig S9B), confirming previously reported interactions between NCL, UBF and NPM (Li et al, 1996; Hisaoka et al, 2010).
Figure 8. NCL and NPM interact with aluRNA and target genomic loci to the nucleolus.
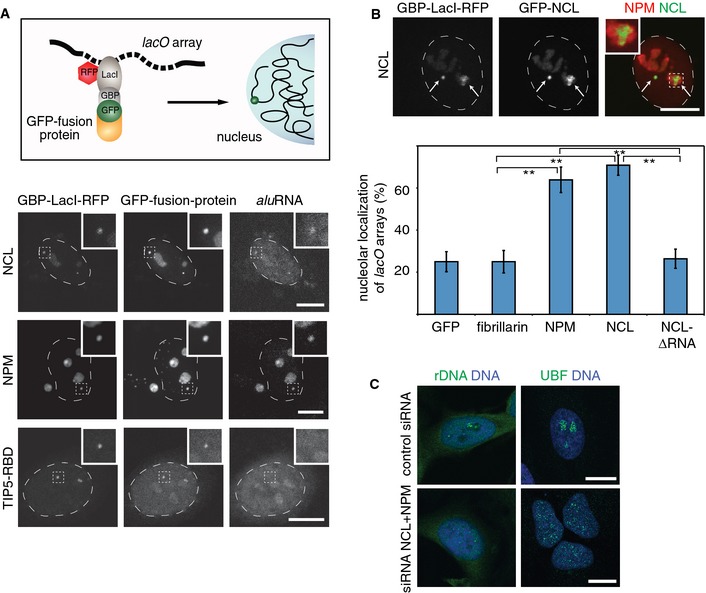
- Top: Scheme illustrating the experimental approach used to tether GFP‐tagged LacI fusion proteins via a GFP‐binding protein (GBP) to lacO arrays. The lacO arrays are stably integrated in the genome of U2OS cells. Bottom: CLSM images showing the localization of GBP‐LacI‐RFP, GFP‐NCL, GFP‐NPM or GFP‐TIP5‐RBD fusion proteins relative to aluRNA that was visualized by RNA FISH. The insets contain an enlarged image of one of the lacO loci.
- CLSM images showing GBP‐LacI‐RFP and GFP‐NCL (green) recruited to lacO arrays as indicated by arrows. Immunofluorescence of endogenous NPM (red) marks nucleoli. The inset shows decondensation of lacO arrays in the nucleolus. The graph at the bottom depicts the average number of lacO loci tethered to nucleoli (± 95% CI) by the indicated proteins (n = 72). NCL‐∆RNA is an NCL deletion mutant lacking the RBD and GAR domains required for RNA binding. **P‐value < 0.01, t‐test.
- CLSM images of UBF (immunofluorescence, green) or rDNA (DNA FISH, green) and DNA (DAPI, blue) after knockdown of NCL and NPM by siRNA in HeLa cells.
Data information: Scale bars, 10 μm.
See also Appendix Fig S9.
Next, we evaluated the localization of the lacO arrays upon tethering of tagged NCL, NPM or fibrillarin. We found that NCL or NPM targeted 60–70% of the megabase‐long lacO arrays to nucleoli (Fig 8B and Appendix Fig S9C). In contrast, GFP or GFP‐tagged fibrillarin failed to promote the translocation of the lacO arrays to nucleoli (Fig 8B and Appendix Fig S9C). Moreover, deletion of the RNA‐binding domains and the C‐terminal GAR domain of nucleolin (NCL‐∆RNA) abolished its nucleolar enrichment, interaction with NPM and, importantly, nucleolar targeting of the lacO arrays (Fig 8B and Appendix Fig S9D).
The interaction of NCL with aluRNA variants was further confirmed by pull‐down assays using nuclear extracts and biotinylated aluRNA transcripts immobilized to streptavidin beads (Appendix Fig S9E). Even when raising the salt concentration up to 450 mM, a stable NCL–aluRNA interaction was found in the pull‐down (Appendix Fig S9F). However, under the same experimental conditions and even at lower salt concentrations, no interaction between NPM and aluRNA was observed (Appendix Fig S9G).
In support of our conclusion that aluRNA interacts with NCL and may indirectly involve NPM to maintain the structure of nucleoli, simultaneous siRNA‐mediated knockdown of NCL and NPM yielded a phenotype very similar to that observed after α‐amanitin treatment or aluRNA knockdown. Notably, dispersion of nucleoli and redistribution of rDNA throughout the nucleoplasm were more pronounced in cells depleted of both, NPM and NCL, compared to cells depleted of one of the two proteins only (Fig 8C and Appendix Fig S9H and I). Together, these results demonstrate that tethering of NCL, NPM and aluRNA to chromatin is sufficient to target large genomic regions to the nucleolus and strongly suggest that the interaction of NCL with aluRNA, and co‐recruitment of NPM, is important to build up a functional nucleolus.
Discussion
In previous studies, it has been shown that number, size and morphology of nucleoli are closely related to rRNA production and the activity of chromatin modifiers (Laferte et al, 2006; Boisvert et al, 2007; Hernandez‐Verdun et al, 2010; Pontvianne et al, 2013). Through its dynamic structural properties, the nucleolus can act as a cellular stress sensor (Rubbi & Milner, 2003; Olson, 2004; Sirri et al, 2008; Boulon et al, 2010). Here, we were able to link a fraction of intronic Alu element‐containing Pol II transcripts termed aluRNA with an essential role for maintaining nucleolar structure and function. These RNAs are part of the large fraction of about 50% of the total pool of genomic Alu repeats that reside in introns (Deininger, 2011). Loss of aluRNAs due to Pol II inhibition or ASO knockdown led to dispersion of nucleoli into smaller domains and reduction in rDNA transcription (Figs 1A and 4 and Appendix Fig S1B and C). In contrast, microinjection of in vitro transcribed aluRNA promoted nucleolar (re)assembly (Fig 3B). These activities were most pronounced for the forward aluRNA‐type sequence, which was highly enriched in the nucleolus as shown by RNA‐seq (Fig 2) and RNA FISH (Fig 3A). Based on the RNA‐seq (Appendix Fig S4) and Northern blot analysis (Appendix Fig S5D), we conclude that relatively short Alu element‐containing RNAs of 100–300 nt in size are stably present in the cell. This finding is consistent with a previous study that identified a class of intron‐encoded Pol II Alu‐related transcripts of 100–200 nt in length termed AluACA RNAs in the nucleus (Jady et al, 2012). We found no evidence that the expression of aluRNAs was also related to Pol III transcription although Alu elements contain an internal Pol III promoter in their left arm (Dieci et al, 2007). Consistently, Pol III inhibition was neither associated with nucleolus dispersion nor with the reduction in pre‐rRNA synthesis (Fig 1A and Appendix Fig S1B and C). It is noted that the internal Pol III promoter, which differs from the original sequence found in the 7SL RNA, is relatively weak and subject to additional silencing mechanisms (Li & Schmid, 2001; Hasler & Strub, 2006). Accordingly, Alu repeat expression in general is dependent on promoters located in their flanking regions (Roy et al, 2000). We conclude that maintenance of nucleolar structure is related to Pol II activity by spliced out aluRNAs from Pol II transcripts. In turn, this affects rRNA synthesis since Pol I transcriptional activity is intrinsically linked to nucleolar structure and suggests that the amount of Pol II‐produced aluRNA modulates Pol I transcription levels.
In agreement with a role of aluRNA in promoting nucleolar domain assembly, we observed that overexpression of aluRNA induced an increase in nucleolus size as well as an enhanced nucleolar transcriptional activity (Fig 5). Accumulation of Alu repeat‐containing RNA transcripts beyond a critical level can also become cytotoxic for the cell as reported previously for retinal pigmented epithelium (Kaneko et al, 2011). This phenotype was linked to a deficit in DICER, a component of the RNA‐induced silencing complex (Kaneko et al, 2011). Interestingly, a significant increase in nucleolar size was observed in cells depleted of DICER (Liang & Crooke, 2011). These previous studies suggest that Alu repeat‐containing RNA levels are also subject to post‐transcriptional regulation. Furthermore, they corroborate our own observation that overexpression of aluRNA induces the formation of larger nucleoli.
Our analysis suggests that aluRNA exerts its nucleolar maintenance activity via interaction with nucleolar proteins. Using the lacO array recruiting system (Figs 7 and 8), we detected binding of aluRNA to NCL and NPM in living cells. Our additional pull‐down experiments confirmed an efficient interaction of NCL with aluRNA but also with other RNAs (Appendix Fig S9E) in addition to the previously reported NCL–rRNA interaction (Tajrishi et al, 2011). It is noted that the concentration of a given NCL–RNA complex will be proportional to the product of the association constant and respective RNA binding site concentration. Given that aluRNAs are highly abundant, this would favor their NCL binding over that of other RNA species to some extent. Nevertheless, it appears likely that the ~100‐fold enrichment of aluRNA in the nucleus cannot be explained solely by binding to soluble NCL in the nucleoplasm but involves additional interactions with other nucleolar proteins, RNAs or DNA components within the nucleolus. Furthermore, our data suggest that a possibly indirect NPM–aluRNA recruitment occurs in the cell as detected in the lacO experiments. It might contribute to the aluRNA enrichment directly in the nucleolus, where NPM can additionally interact with UBF and rRNA (Hisaoka et al, 2010).
In support of aluRNA promoting nucleolar assembly, recruitment of aluRNA and/or NCL and NPM to lacO arrays led to a significantly increased nucleolar localization of these arrays. This raises the question how intron‐encoded nucleolar aluRNA exerts such an activity. A possible mechanism is related to the physico‐chemical properties of RNA–protein interactions (Weber & Brangwynne, 2012). According to the liquid‐drop model of the nucleolus proposed previously, unmixing of nucleolar components from the surrounding nucleoplasm as observed here for NCL/NPM and aluRNA is due to their interaction properties (Brangwynne et al, 2011). Notably, both NCL and NPM contain disordered and low‐complexity sequences (Emmott & Hiscox, 2009; Hisaoka et al, 2014). These sequences form protein domains that are able to drive RNA‐dependent intracellular phase‐separation processes in the cell through conformational changes upon RNA binding (Han et al, 2012; Hyman & Simons, 2012; Weber & Brangwynne, 2012). Such a behavior has been reported for NPM (Hisaoka et al, 2010, 2014). It is consistent with our findings of an emulsion‐like dispersion of the nucleolus observed after Pol II inhibition or aluRNA depletion (Appendix Fig S8A and B) as well as a RNA‐dependent nucleolar targeting activity of NCL (Fig 8B). In such a scenario, aluRNA would serve as “glue” or scaffold to orchestrate nucleolus formation via promoting the self‐organization of NCL/NPM‐containing “droplets” into domains that efficiently associate with rDNA and support Pol I transcriptional activity within the nucleolus (Fig 9). This could serve to increase the local concentration of reactants and the efficiency of rRNA synthesis (Boisvert et al, 2007; Weber & Brangwynne, 2012).
Figure 9. Model of aluRNA‐driven maintenance of the nucleolus.
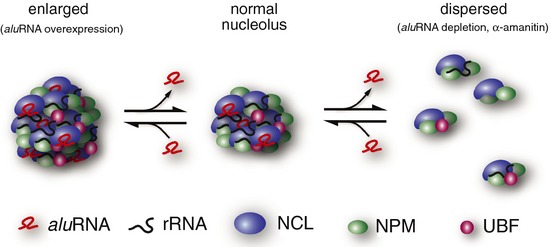
The knockdown of aluRNA or treatment with α‐amanitin induces the segregation of nucleolar compartments into smaller domains (dispersion process) as observed (Figs 1A and 4A, and Appendix Fig S2). With regard to rRNA production, those “droplets,” which may vary in composition, are less efficient but some rRNAs are still produced albeit at low levels (Appendix Fig S1B and C and Fig 4C and D). Overexpression of aluRNA promotes assembly into larger nucleolar subcompartments, with enhanced Pol I activity (see Fig 5). This model is in excellent agreement with the previously reported liquid‐like model of the nucleolus (Brangwynne et al, 2011; Weber & Brangwynne, 2012) and suggests a RNA binding‐driven liquid‐like transition between dispersed nucleolar domains and intact and fully functional nucleoli.
NCL, NPM and fibrillarin are constituents of the prenucleolar bodies that associate with the nucleolar organizer regions at early G1 phase (Dousset et al, 2000). Their enrichment at rDNA loci could originate from a direct interaction of NCL with rDNA (Olson et al, 1983). At the same time, UBF has been identified as a key player in nucleating the assembly of nucleoli (Grob et al, 2014). It suggests that the aluRNA‐directed fusion into larger domains could occur via the interaction of NCL and/or NPM with UBF bound to nucleolar organizer regions. Consistent with this view and in accord with the observation that NCL co‐localizes with UBF and interacts with NPM (Li et al, 1996), we show that recruitment of NCL to lacO arrays was accompanied by the enrichment of both UBF and NPM at these arrays (Appendix Fig S8B). Thus, the assembly of the nucleolus requires multiple coordinated macromolecular interactions including: (i) DNA–protein interactions such as binding of UBF to rDNA and possibly direct interactions of NCL with rDNA (Olson et al, 1983; Cong et al, 2012); (ii) protein–protein interactions as, for example, interaction of NCL with UBF and NPM, or NPM with UBF (Hisaoka et al, 2010); and (iii) RNA–protein interactions that involve the association of aluRNA with NCL and other nucleolar components as suggested here.
The nucleolus is a nuclear subcompartment that is found in almost all eukaryotic cells (Lamaye et al, 2011), while Alu elements are primate specific (Liu et al, 2009). This raises the question whether the mechanism revealed here in studying human cells can also operate in other eukaryotic organisms. In favor of this possibility, we found that nucleoli in mouse fibroblast cells showed a similar disruption of their structure upon Pol II inhibition (Fig 6A) and observed that ASO targeting of Alu‐related B1 repeat‐containing RNAs induced nucleolar disassembly (Fig 6B). This strongly suggests that transcripts from B1 elements could represent the functional equivalent of human aluRNAs. It is noted that Alu and B1 repeats have a common ancestor in evolution, namely the 7SL RNA (Nishihara et al, 2002). This RNA is a crucial component of the signal recognition particle and is present in all eukaryotic cells (Nagai et al, 2003). One could speculate that RNAs derived from the 7SL RNA family share a common function for nucleolus structure throughout eukaryotic organisms.
Maintaining large numbers of Alu repeats in the human genome imposes a significant risk to genome stability (Callinan & Batzer, 2006; Belancio et al, 2010). Furthermore, intronic Alu elements may also compromise correct mRNA production. As reported recently, the cell has developed a dedicated mechanism to prevent misguided splicing that would lead to exonization of intronic Alu repeats (Zarnack et al, 2013). The important function of aluRNA to maintain a functional nucleoli structure revealed in our study could represent a selection pressure to keep Alu sequences in introns.
Materials and Methods
Cell culture and purification of nucleoli
HeLa and HeLa S3 cells were grown at 37°C/5% CO2 in RPMI 1640 or DMEM containing 1 g/l glucose, respectively, and supplemented with 10% FCS, 2 mM L‐glutamine and 1% penicillin/streptomycin. U2OS and NIH 3T3 cells were cultured under the same conditions in DMEM containing 1 g/l or 4.5 g/l glucose, respectively. The U2OS cell clones F6B2 (stable insertion of three lacO arrays) and F4IIB8 (stable insertion of one lacO array) and transfection of cells with expression plasmids for lacO array tagging and protein recruitment were conducted as described previously (Jegou et al, 2009; Chung et al, 2011). The human keratinocyte and fibroblast cells were kindly provided by Aubry Miller and Nikolas Gunkel (German Cancer Research Center, Germany).
Nucleoli were isolated from HeLa S3 cells as described (Busch et al, 1963; Sullivan et al, 2001), and RNA was purified from unfractionated cells (total RNA) or nucleoli by TRIzol (Caudron‐Herger et al, 2011). Pre‐rRNA and aluRNAs from total RNA were assayed by Northern blotting using a radiolabeled 5′‐ETS antisense riboprobe (from +150 to +1) or a radiolabeled antisense Alu probe (Appendix Table S4). Alternatively, pre‐rRNA levels were quantified by RT–qPCR as reported before (Hoppe et al, 2009). Quantification of rRNA amounts depends on synthesis and degradation rates. 47S has a very short half‐life time in the range of minutes (Popov et al, 2013) as compared to 18S and 28S that are stable for days (Defoiche et al, 2009). Therefore, 47S steady‐state measurements can be correlated with the synthesis rate of the precursor (before the cleavage of the 5′‐ETS), as done here.
Drug treatments
RNA polymerases were inhibited by culturing cells for 5 h in medium supplemented with the following inhibitors: actinomycin D (AMD, 50 ng/ml) for Pol I, α‐amanitin (50 μg/ml) or 5,6‐dichloro‐1‐beta‐D‐ribofuranosylbenzimidazole (DRB, 50 μg/ml) for Pol II, and ML‐60218 (2‐chloro‐N‐[3‐(5‐chloro‐3‐methylbenzo[b]thien‐2‐yl)‐1‐methyl‐1H‐pyrazol‐5‐yl]‐benzenesulfonamide) for Pol III. Translation was inhibited by cycloheximide treatment (50 μg/ml) for 5 h.
Microinjection
Microinjection of cells grown in Nunc Lab‐Tek chamber slides in F‐15 medium (Thermo Scientific) was performed with a computer‐assisted system (AIS2, CellBiology Trading). A volume of about 50 femtoliters was injected using a needle with a tip diameter of about 300 nm, 150 hPA pressure and 0.5 s injection time. Ten microliters of injection mix contained 2 μl propidium iodide (PI, 1 mg/ml, Thermo Fisher Scientific) and 1 μg of RNA in PBS. Following injection, cells were cultured under standard conditions for 30 min and processed for confocal microscopy image acquisition.
Immunofluorescence and RNA labeling
Cells grown on coverslips were fixed 24 h after transfection with 4% paraformaldehyde/PBS. For immunostaining, cells were permeabilized for 5 min with ice‐cold 0.5% (v/v) Triton X‐100/PBS. After three washes, samples were incubated for at least 15 min with 10% goat serum in PBS followed by addition of antibodies specific to NCL (sc‐13057), NPM (sc‐56622), UBF (sc‐13125), SC35 (BD Pharmingen, 556363) or Pol I (Percipalle et al, 2006) for 1 h at room temperature (NCL, Pol I) or overnight at 4°C (NPM, UBF). After washing, cells were incubated for 25 min with the appropriate secondary antibodies conjugated to Alexa Fluor 488, 568 or 633 (Thermo Fisher Scientific, Molecular Probes) and for another 5 min with 4′,6‐diamidino‐2‐phenylindoledihydrochloride (DAPI). The coverslips were mounted with Mowiol. For pulse labeling of RNA, cells were incubated for 30 min with 1 mM 5‐ethynyl uridine (EU) and fixed with 4% paraformaldehyde/PBS. EU‐labeled transcripts were detected using Alexa Fluor 488 azide (Click‐it RNA imaging Kit, Thermo Fisher Scientific).
DNA and RNA FISH
For DNA FISH, cells grown on coverslips were fixed with 4% paraformaldehyde/PBS, washed in PBS and permeabilized with 0.5% (v/v) Triton X‐100/PBS. After serial washes with ethanol (70, 80 and 100%) and air‐drying, DNA was denatured at 80°C for 5 min and hybridized overnight at 42°C to a biotin‐labeled human rDNA probe (from nucleotides 18063 to 30486, GenBank Accession No.: U13369.1) in hybridization buffer (50% formamide, 2× SSC and 10% dextran sulfate). The plasmid pHr4 (Mais et al, 2005), a kind gift from Brian McStay (NUI Galway, Ireland), was used to generate about 2,500‐bp‐long body‐labeled fragments by nick translation. Slides were washed consecutively with hybridization buffer, 2× SSC, 0.2× SSC containing 0.1% (v/v) Tween‐20 at 55°C, 2× SSC containing 0.05% (v/v) Tween‐20 and finally PBS.
For RNA FISH, cells grown on coverslips were permeabilized in CSK buffer (100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 10 mM PIPES, 0.5% (v/v) Triton X‐100) containing 10 mM vanadyl ribonucleoside complex (VRC) or 50 μg/ml RNase A. Cells were fixed with 4% paraformaldehyde/PBS, dehydrated by sequential washes with ethanol (70, 85 and 100%) and air‐dried. The RNA was hybridized overnight in hybridization buffer at 37°C with 2.5 μl of digoxigenin‐labeled aluRNA or control RNA probes (each 50 ng/μl). Slides were washed consecutively with hybridization buffer, 0.2× SSC containing 0.1% (v/v) Tween‐20 at 40°C, 2× SSC, and PBS. Digoxigenin‐labeled RNA was detected with anti‐digoxigenin antibody (Roche, clone 1.71.256). The MS2 RNA stem‐loops were visualized using a 5′‐Atto‐565‐labeled antisense probe that targets the MS2 loop sequence (Chaumeil et al, 2002). Sequences of the FISH probes are listed in Appendix Table S4.
Plasmids and in vitro transcription
GFP‐tagged proteins were generated by cloning the corresponding cDNAs into pEGFP‐C1 (Thermo Fisher Scientific). The pEGFP‐NCL and pEGFP‐fibrillarin vectors were obtained from Addgene (#28176 and #26673, respectively). pTagRFP‐NCL was produced by cloning the NCL into KpnI and BamHI sites of the pTagRFP plasmid (Evrogen). pEGFP‐NPM was kindly provided by Mitsuru Okuwaki (University of Tsukuba, Japan). GFP‐TIP5‐RBD and GBP‐LacI‐mRFP plasmids have been described (Jegou et al, 2009; Zillner et al, 2013). The GFP‐NCL‐∆RNA deletion mutant missing the RBD and GAR domains was generated by cloning of the corresponding cDNA PCR fragment into the KpnI and BamHI sites of the pEGFP‐C1 plasmid. MS2 coat protein fused to GFP‐LacI was cloned by introducing the MS2 protein into the NheI and AgeI sites of pSV2‐GFP‐LacI (Jegou et al, 2009). The MS2‐aluRNA and the corresponding control RNA sequences were synthesized as DNA (Integrated DNA Technologies, Inc) and were cloned into a pcDNA3 plasmid containing 18 repeats of the MS2 stem‐loop as described (Schmidt et al, 2011). The MS2‐aluRNA was cloned using the AgeI and NotI restriction sites, and the control sequences were cloned into the NotI site. The sequences are listed in Appendix Table S4. The aluRNA and L1‐repeat sequences were introduced into the pBS/U6 plasmid expressing either GFP or TagRFP using two BbsI restriction sites (Grimm et al, 2006). The plasmid was kindly provided by Dirk Grimm (University of Heidelberg, Germany).
DNA templates for in vitro transcription of aluRNA and L1‐repeat (Appendix Table S4) were amplified using genomic DNA from HeLa cells and primers specific to T7 promoter sequences. In vitro transcription was performed using the RNA polymerase T7 High Yield RNA Synthesis Kit (NEB) according to the manufacturer's instructions.
Confocal fluorescence microscopy and image analysis
Imaging was done with a Leica TCS SP5 confocal laser scanning microscope (CLSM) equipped with a HCX PL APO lambda blue 63×/1.4 NA oil immersion objective (Leica Microsystems CMS GmbH, Mannheim, Germany). A near UV diode, diode‐pumped solid‐state, argon and helium–neon lasers were used for DAPI (λ = 405 nm), Alexa 488 or GFP (λ = 488 nm), Alexa 568 or Atto‐565 or TagRFP (λ = 561 nm) and Alexa 633 (λ = 633 nm) excitation. For multi‐color analysis, sequential image acquisition was applied and emission detection ranges were adjusted to minimize crosstalk between the different signals. The detection pinhole had a diameter corresponding to one airy disk.
After microinjection, random pictures of microinjected cells were taken and analyzed using ImageJ (Schneider et al, 2012). The images were segmented via thresholding, and the function “Analyze Particles” was used to automatically count the number and size of particles in each picture. For nucleoli volume estimation, confocal z‐stacks were acquired, each slice was segmented via thresholding, and the function “Analyze Particles” was used to automatically calculate the nucleolar area in each image. Summing and multiplying each area by the z‐step provided an estimation of the nucleolar volume. A similar procedure was repeated to evaluate the nuclear volume based on the DAPI staining of the DNA. For live‐cells image acquisitions of cells transfected with pBS_U6_aluRNA and RFP‐NCL using X‐tremeGENE (Roche Life Science), z‐stacks of cells were acquired every 30 min. The microscope was equipped with an incubation chamber allowing normal growth conditions (5% CO2 and 37°C). To determine the fluorescence intensity of nuclei and nucleoli, each slice was segmented via thresholding and the function “Analyze Particles” was used to automatically calculate the intensities. To determine the position of the lacO arrays, the fluorescence signal of GBP‐LacI‐RFP and a nucleolar marker (labeled with Alexa 633, Thermo Fisher Scientific) were overlaid. P‐values were calculated according to Student's t‐test.
RNA and protein knockdown
For RNA knockdown, cells were transfected with Lipofectamine 2000 according to the manufacturer's instruction (Thermo Fisher Scientific) using 40 nM of antisense oligonucleotides (ASOs), that is aluRNA ASO or control ASO (see Appendix Table S4 for sequences) 5′‐labeled with Cy3. Unless otherwise indicated (see Appendix Fig S8), cells were fixed 14 h after transfection for 10 min in 4% paraformaldehyde/PBS and processed for immunofluorescence. Custom LNA blocker probes (Exiqon) were designed to functionally inactivate target RNAs. The sequences are listed in Appendix Table S4. For siRNA‐mediated knockdown of NCL and NPM, 1.5 × 105 cells were transfected with Lipofectamine 2000 and siRNAs against NCL (Dharmacon L‐003854‐00‐0005) or NPM (Dharmacon L‐015737‐00‐0005). ON‐TARGETplus Non‐targeting Pool (Dharmacon, D‐001810‐10‐50) was used as control siRNA. Forty‐eight hours after transfection, cells were processed for immunofluorescence microscopy and rDNA FISH or for Western blot analysis using antibodies against NCL (sc‐13057), NPM (sc53175) and histone H3 (ab1791, Abcam).
Protein pull‐down with biotinylated RNA transcript
Cell nuclei were isolated from HeLa S3 to prepare a nuclear extract using nuclear lysis buffer (200 mM NaCl; 20 mM Tris–HCl, pH 8.0; 0.2% Tween‐20; 1 mM EDTA; 1 mM EGTA). Biotinylated RNA transcripts were in vitro transcribed in the presence of biotin‐16‐UTP (Sigma‐Aldrich). Five micrograms of labeled RNA were incubated with 50 μl of streptavidin‐coated magnetic beads (Thermo Fisher Scientific) for 30 min at room temperature in 100 mM NaCl solution. Beads were washed in washing buffer (5 mM Tris–HCl, pH 7.5; 0.5 mM EDTA; 0.5 M NaCl; 0.05% Tween‐20) and further incubated for 3 h in 70 μg or 35 μg nuclear extract in a final volume of 100 μl. After 3 steps of washing in nuclear lysis buffer, the beads were resuspended in 50 μl of nuclear lysis buffer and bound proteins were eluted by adding 3 μl of RNase A (10 mg/ml; Thermo Fisher Scientific) for 30 min at 4°C. Beads were retrieved on a magnet, and eluted proteins were analyzed by Western blotting.
RNA sequencing
Total RNA, nucleoplasmic RNA or nucleolar RNA were isolated from HeLa cells or purified nucleoli, respectively, using the RNeasy Mini Kit (Qiagen). High‐throughput RNA sequencing was done with two biological replicates. Ribosomal RNA was removed using the Ribo‐Zero rRNA Removal Kit (Epicentre/Illumina) according to the manufacturer's protocol. For the first data set, RNAs were fragmented with the RNA fragmentation reagent kit from Ambion (Thermo Fisher Scientific, AM8740). For Illumina sequencing, libraries were generated according to the standard protocol for mRNA (Illumina) comprising first‐ and second‐strand cDNA synthesis, end repair, addition of a single A base and adapter ligation. PCR products of about 200 bp were excised from a 2% E‐Gel Size Select (Thermo Fisher Scientific). After determining the concentration and quality on a Qubit fluorometer (Thermo Fisher Scientific) and Bioanalyzer system (Agilent Technologies), 36‐nt‐long sequencing reactions were performed on the Illumina GAIIx platform (Deep Sequencing Core Facility of the Cell Networks cluster of excellence, University of Heidelberg, Germany). For the second data set, strand‐specific libraries were prepared using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (#E7420L, New England BioLabs Inc.). Sequencing of 100‐base‐pair reads was performed on the Illumina HiSeq 2000 platform (Genomics and Proteomics Core Facility of the DKFZ, Heidelberg, Germany).
RNA‐seq data analysis
RNA‐seq reactions were performed with 36‐nt and 100‐nt read length and yielded the number of total and mapped reads given in Appendix Table S5. Reads were quality‐controlled and aligned with Bowtie (Langmead et al, 2009) on the GRCh37/hg19 (2009) assembly version of the human genome reporting unique hits and allowing up to two mismatches. Within the 100‐nt read length, we estimated that 90% of the sequences mapping to Alu elements were unambiguously mapped, given that this read length allows precise identification of the majority of the Alu elements (Umylny et al, 2007). For a selected set of genes (see Appendix Fig S2), comparison of various samples was done by normalizing expression levels to the total number of mapped reads and calculating the relative amounts. Read clusters showing the expression values normalized to the total number of mapped reads were calculated using Cufflinks (Trapnell et al, 2010). For annotation, the overlaps with genomic regions were evaluated based on Genomatix software suite (Genomatix, Munich, Germany). Clusters annotated as “exon–intron overlapping” comprise clusters overlapping exonic and intronic regions. After annotation, the clusters were compared to a primary transcripts (PT) database and re‐grouped into PT when both exonic and intronic clusters corresponded to the same PT, or spliced transcripts when only exons were found.
A list of human Alu repeats was produced using the RepeatMasker track in the Table Browser (www.genome.ucsc.edu) as well as a list of tRNAs. The average and normalized tRNA level was calculated for various samples (Appendix Fig S1) using the function “intersect” of the genome arithmetic suite bedtools (Quinlan & Hall, 2010). The same function was applied to list clusters overlapping with Alu repeats. The normalized expression (NE) values were used to classify the clusters and determine the highest enrichment in nucleoli or in the total RNA samples (see Appendix Table S2). For the graphical representation of the RNA‐seq data, coverage files were produced with the Integrative Genomics Viewer toolbox and uploaded in the Integrative Genomics Viewer (Robinson et al, 2011). Multiple sequences alignment was performed using the alignment tool MultAlin (http://multalin.toulouse.inra.fr) (Corpet, 1988). Heatmaps were produced using the programs seqMINER (Ye et al, 2011) and ngs.plot (Shen et al, 2014).
Data availability
RNA‐seq data generated can be accessed via the EBI Array Express archive under the accession number E‐MTAB‐3460.
Author contributions
MC‐H and KR conceived the project, coordinated the study and designed the experimental plan with help from IG, RV and AN. MC‐H, TP, JS and RV conducted experiments and/or data analysis, and AN provided reagents. MC‐H, KR, IG and RV wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Review Process File
Source Data for Figure 4
Acknowledgements
We are grateful to Joël Beaudouin, Ulrike Engel (Nikon Imaging Center, Heidelberg), David Ibberson (CellNetworks Deep Sequencing Core Facility, Heidelberg), Benjamin Schwalm and Claudio Flores for their help and advice; Gernot Längst, Sven Diederichs and Argyris Papantonis for helpful discussions; and Dirk Grimm, Nikolas Gunkel, Brian McStay, Aubry Miller and Mitsuru Okuwaki for providing reagents. This work was funded by a DKFZ young investigator grant to MC‐H, the German Federal Ministry of Education and Research in the EU FP7 ERA‐NET Plus program (grant 0315712A to KR) and the DFG (grants GR475/22‐1 and SFB1036 to IG and grant SFB960 to AN).
The EMBO Journal (2015) 34: 2758–2774
See also: M Carmo‐Fonseca (November 2015)
References
- Amin MA, Matsunaga S, Uchiyama S, Fukui K (2008) Depletion of nucleophosmin leads to distortion of nucleolar and nuclear structures in HeLa cells. Biochem J 415: 345–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audas TE, Jacob MD, Lee S (2012) Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol Cell 45: 147–157 [DOI] [PubMed] [Google Scholar]
- Batista PJ, Chang HY (2013) Long noncoding RNAs: cellular address codes in development and disease. Cell 152: 1298–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL (2002) Alu repeats and human genomic diversity. Nat Rev Genet 3: 370–379 [DOI] [PubMed] [Google Scholar]
- Belancio VP, Roy‐Engel AM, Deininger PL (2010) All y'all need to know ‘bout retroelements in cancer. Semin Cancer Biol 20: 200–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann JH, Spector DL (2014) Long non‐coding RNAs: modulators of nuclear structure and function. Curr Opin Cell Biol 26: 10–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierhoff H, Dammert MA, Brocks D, Dambacher S, Schotta G, Grummt I (2014) Quiescence‐induced LncRNAs trigger H4K20 trimethylation and transcriptional silencing. Mol Cell 54: 675–682 [DOI] [PubMed] [Google Scholar]
- Boisvert F‐M, van Koningsbruggen S, Navascués J, Lamond AI (2007) The multifunctional nucleolus. Nat Rev Mol Cell Biol 8: 574–585 [DOI] [PubMed] [Google Scholar]
- Boulon S, Westman BJ, Hutten S, Boisvert F‐M, Lamond AI (2010) The nucleolus under stress. Mol Cell 40: 216–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP, Mitchison TJ, Hyman AA (2011) Active liquid‐like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci USA 108: 4334–4339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger K, Mühl B, Harasim T, Rohrmoser M, Malamoussi A, Orban M, Kellner M, Gruber‐Eber A, Kremmer E, Hölzel M, Eick D (2010) Chemotherapeutic drugs inhibit ribosome biogenesis at various levels. J Biol Chem 285: 12416–12425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch H, Muramatsu M, Adams H, Steele WJ, Liau MC, Smetana K (1963) Isolation of nucleoli. Exp Cell Res 24(Suppl. 9): 150–163 [PubMed] [Google Scholar]
- Callinan PA, Batzer MA (2006) Retrotransposable elements and human disease. Genome Dyn 1: 104–115 [DOI] [PubMed] [Google Scholar]
- Capshew CR, Dusenbury KL, Hundley HA (2012) Inverted Alu dsRNA structures do not affect localization but can alter translation efficiency of human mRNAs independent of RNA editing. Nucleic Acids Res 40: 8637–8645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo‐Fonseca M, Mendes‐Soares L, Campos I (2000) To be or not to be in the nucleolus. Nat Cell Biol 2: E107–E112 [DOI] [PubMed] [Google Scholar]
- Caudron‐Herger M, Muller‐Ott K, Mallm JP, Marth C, Schmidt U, Fejes‐Toth K, Rippe K (2011) Coding RNAs with a non‐coding function: maintenance of open chromatin structure. Nucleus 2: 410–424 [DOI] [PubMed] [Google Scholar]
- Caudron‐Herger M, Rippe K (2012) Nuclear architecture by RNA. Curr Opin Genet Dev 22: 179–187 [DOI] [PubMed] [Google Scholar]
- Chafin DR, Guo H, Price DH (1995) Action of alpha‐amanitin during pyrophosphorolysis and elongation by RNA polymerase II. J Biol Chem 270: 19114–19119 [DOI] [PubMed] [Google Scholar]
- Chaumeil J, Okamoto I, Guggiari M, Heard E (2002) Integrated kinetics of X chromosome inactivation in differentiating embryonic stem cells. Cytogenet Genome Res 99: 75–84 [DOI] [PubMed] [Google Scholar]
- Chen S, Seiler J, Santiago‐Reichelt M, Felbel K, Grummt I, Voit R (2013) Repression of RNA polymerase I upon stress is caused by inhibition of RNA‐dependent deacetylation of PAF53 by SIRT7. Mol Cell 52: 303–313 [DOI] [PubMed] [Google Scholar]
- Chung I, Leonhardt H, Rippe K (2011) De novo assembly of a PML nuclear subcompartment occurs through multiple pathways and induces telomere elongation. J Cell Sci 124: 3603–3618 [DOI] [PubMed] [Google Scholar]
- Cong R, Das S, Ugrinova I, Kumar S, Mongelard F, Wong J, Bouvet P (2012) Interaction of nucleolin with ribosomal RNA genes and its role in RNA polymerase I transcription. Nucleic Acids Res 40: 9441–9454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16: 10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David‐Pfeuty T, Nouvian‐Dooghe Y, Sirri V, Roussel P, Hernandez‐Verdun D (2001) Common and reversible regulation of wild‐type p53 function and of ribosomal biogenesis by protein kinases in human cells. Oncogene 20: 5951–5963 [DOI] [PubMed] [Google Scholar]
- Defoiche J, Zhang Y, Lagneaux L, Pettengell R, Hegedus A, Willems L, Macallan DC (2009) Measurement of ribosomal RNA turnover in vivo by use of deuterium‐labeled glucose. Clin Chem 55: 1824–1833 [DOI] [PubMed] [Google Scholar]
- Deininger P (2011) Alu elements: know the SINEs. Genome Biol 12: 236–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM (2009) TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol Cell 35: 403–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J, Figueroa ME, De Figueiredo Pontes LL, Alberich‐Jorda M, Zhang P, Wu M, D'Alo F, Melnick A, Leone G, Ebralidze KK, Pradhan S, Rinn JL, Tenen DG (2013) DNMT1‐interacting RNAs block gene‐specific DNA methylation. Nature 503: 371–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A (2007) The expanding RNA polymerase III transcriptome. Trends Gen 23: 614–622 [DOI] [PubMed] [Google Scholar]
- Dousset T, Wang C, Verheggen C, Chen D, Hernandez‐Verdun D, Huang S (2000) Initiation of nucleolar assembly is independent of RNA polymerase I transcription. Mol Biol Cell 11: 2705–2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg E, Levanon EY (2013) Human housekeeping genes, revisited. Trends Genet 29: 569–574 [DOI] [PubMed] [Google Scholar]
- Emmott E, Hiscox JA (2009) Nucleolar targeting: the hub of the matter. EMBO Rep 10: 231–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick T, Huang S (2012) 3′‐UTR‐located inverted Alu repeats facilitate mRNA translational repression and stress granule accumulation. Nucleus 3: 359–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginisty H, Sicard H, Roger B, Bouvet P (1999) Structure and functions of nucleolin. J Cell Sci 112: 761–772 [DOI] [PubMed] [Google Scholar]
- Granick D (1975) Nucleolar necklaces in chick embryo fibroblast cells. II. Microscope observations of the effect of adenosine analogues on nucleolar necklace formation. J Cell Biol 65: 418–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA (2006) Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 441: 537–541 [DOI] [PubMed] [Google Scholar]
- Grob A, Colleran C, McStay B (2014) Construction of synthetic nucleoli in human cells reveals how a major functional nuclear domain is formed and propagated through cell division. Genes Dev 28: 220–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaf T, Ward DC (1996) Inhibition of RNA polymerase II transcription causes chromatin decondensation, loss of nucleolar structure, and dispersion of chromosomal domains. Exp Cell Res 224: 163–173 [DOI] [PubMed] [Google Scholar]
- Han TW, Kato M, Xie S, Wu LC, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G, McKnight SL (2012) Cell‐free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 149: 768–779 [DOI] [PubMed] [Google Scholar]
- Hasler J, Strub K (2006) Alu elements as regulators of gene expression. Nucleic Acids Res 34: 5491–5497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez‐Verdun D, Roussel P, Thiry M, Sirri V, Lafontaine DL (2010) The nucleolus: structure/function relationship in RNA metabolism. Wiley Interdiscip Rev RNA 1: 415–431 [DOI] [PubMed] [Google Scholar]
- Hisaoka M, Ueshima S, Murano K, Nagata K, Okuwaki M (2010) Regulation of nucleolar chromatin by B23/nucleophosmin jointly depends upon its RNA binding activity and transcription factor UBF. Mol Cell Biol 30: 4952–4964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisaoka M, Nagata K, Okuwaki M (2014) Intrinsically disordered regions of nucleophosmin/B23 regulate its RNA binding activity through their inter‐ and intra‐molecular association. Nucleic Acids Res 42: 1180–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe S, Bierhoff H, Cado I, Weber A, Tiebe M, Grummt I, Voit R (2009) AMP‐activated protein kinase adapts rRNA synthesis to cellular energy supply. Proc Natl Acad Sci USA 106: 17781–17786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A (2007) A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genom 8: 39–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, Simons K (2012) Cell biology. Beyond oil and water–phase transitions in cells. Science 337: 1047–1049 [DOI] [PubMed] [Google Scholar]
- Ideue T, Hino K, Kitao S, Yokoi T, Hirose T (2009) Efficient oligonucleotide‐mediated degradation of nuclear noncoding RNAs in mammalian cultured cells. RNA 15: 1578–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob MD, Audas TE, Mullineux S‐T, Lee S (2012) Where no RNA polymerase has gone before: novel functional transcripts derived from the ribosomal intergenic spacer. Nucleus 3: 315–319 [DOI] [PubMed] [Google Scholar]
- Jacob MD, Audas TE, Uniacke J, Trinkle‐Mulcahy L, Lee S (2013) Environmental cues induce a long noncoding RNA‐dependent remodeling of the nucleolus. Mol Biol Cell 24: 2943–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jady BE, Ketele A, Kiss T (2012) Human intron‐encoded Alu RNAs are processed and packaged into Wdr79‐associated nucleoplasmic box H/ACA RNPs. Genes Dev 26: 1897–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jegou T, Chung I, Heuvelmann G, Wachsmuth M, Görisch SM, Greulich‐Bode K, Boukamp P, Lichter P, Rippe K (2009) Dynamics of telomeres and promyelocytic leukemia nuclear bodies in a telomerase negative human cell line. Mol Biol Cell 20: 2070–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko H, Dridi S, Tarallo V, Gelfand BD, Fowler BJ, Cho WG, Kleinman ME, Ponicsan SL, Hauswirth WW, Chiodo VA, Kariko K, Yoo JW, Lee DK, Hadziahmetovic M, Song Y, Misra S, Chaudhuri G, Buaas FW, Braun RE, Hinton DR et al (2011) DICER1 deficit induces Alu RNA toxicity in age‐related macular degeneration. Nature 471: 325–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laferte A, Favry E, Sentenac A, Riva M, Carles C, Chedin S (2006) The transcriptional activity of RNA polymerase I is a key determinant for the level of all ribosome components. Genes Dev 20: 2030–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamaye F, Galliot S, Alibardi L, Lafontaine DLJ, Thiry M (2011) Nucleolar structure across evolution: the transition between bi‐ and tri‐compartmentalized nucleoli lies within the class Reptilia. J Struct Biol 174: 352–359 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory‐efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YP, Busch RK, Valdez BC, Busch H (1996) C23 interacts with B23, a putative nucleolar‐localization‐signal‐binding protein. Eur J Biochem 237: 153–158 [DOI] [PubMed] [Google Scholar]
- Li TH, Schmid CW (2001) Differential stress induction of individual Alu loci: implications for transcription and retrotransposition. Gene 276: 135–141 [DOI] [PubMed] [Google Scholar]
- Liang XH, Crooke ST (2011) Depletion of key protein components of the RISC pathway impairs pre‐ribosomal RNA processing. Nucleic Acids Res 39: 4875–4889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GE, Alkan C, Jiang L, Zhao S, Eichler EE (2009) Comparative analysis of Alu repeats in primate genomes. Genome Res 19: 876–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mais C, Wright JE, José‐Luis P, Raggett SL, McStay B (2005) UBF‐binding site arrays form pseudo‐NORs and sequester the RNA polymerase I transcription machinery. Genes Dev 19: 50–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao YS, Sunwoo H, Zhang B, Spector DL (2011) Direct visualization of the co‐transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol 13: 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS, Mehler MF (2008) RNA editing, DNA recoding and the evolution of human cognition. Trends Neurosci 31: 227–233 [DOI] [PubMed] [Google Scholar]
- Mayer C, Schmitz K‐M, Li J, Grummt I, Santoro R (2006) Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol Cell 22: 351–361 [DOI] [PubMed] [Google Scholar]
- McKeown PC, Shaw PJ (2009) Chromatin: linking structure and function in the nucleolus. Chromosoma 118: 11–23 [DOI] [PubMed] [Google Scholar]
- Mercer TR, Mattick JS (2013) Understanding the regulatory and transcriptional complexity of the genome through structure. Genome Res 23: 1081–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai K, Oubridge C, Kuglstatter A, Menichelli E, Isel C, Jovine L (2003) Structure, function and evolution of the signal recognition particle. EMBO J 22: 3479–3485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara H, Terai Y, Okada N (2002) Characterization of novel Alu‐ and tRNA‐related SINEs from the tree shrew and evolutionary implications of their origins. Mol Biol Evol 19: 1964–1972 [DOI] [PubMed] [Google Scholar]
- Olson MO, Rivers ZM, Thompson BA, Kao WY, Case ST (1983) Interaction of nucleolar phosphoprotein C23 with cloned segments of rat ribosomal deoxyribonucleic acid. Biochemistry 22: 3345–3351 [DOI] [PubMed] [Google Scholar]
- Olson MO, Dundr M, Szebeni A (2000) The nucleolus: an old factory with unexpected capabilities. Trends Cell Biol 10: 189–196 [DOI] [PubMed] [Google Scholar]
- Olson MO (2004) Sensing cellular stress: another new function for the nucleolus? Sci STKE 224: pe10 [DOI] [PubMed] [Google Scholar]
- Pederson T (1998) The plurifunctional nucleolus. Nucleic Acids Res 26: 3871–3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percipalle P, Fomproix N, Cavellan E, Voit R, Reimer G, Kruger T, Thyberg J, Scheer U, Grummt I, Farrants AK (2006) The chromatin remodelling complex WSTF‐SNF2 h interacts with nuclear myosin 1 and has a role in RNA polymerase I transcription. EMBO Rep 7: 525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F, Blevins T, Chandrasekhara C, Mozgova I, Hassel C, Pontes OM, Tucker S, Mokros P, Muchova V, Fajkus J, Pikaard CS (2013) Subnuclear partitioning of rRNA genes between the nucleolus and nucleoplasm reflects alternative epiallelic states. Genes Dev 27: 1545–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov A, Smirnov E, Kovacik L, Raska O, Hagen G, Stixova L, Raska I (2013) Duration of the first steps of the human rRNA processing. Nucleus 4: 134–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raska I, Ochs RL, Andrade LE, Chan EK, Burlingame R, Peebles C, Gruol D, Tan EM (1990) Association between the nucleolus and the coiled body. J Struct Biol 104: 120–127 [DOI] [PubMed] [Google Scholar]
- Reichow SL, Hamma T, Ferré‐D'Amaré AR, Varani G (2007) The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res 35: 1452–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds RC, Montgomery PO, Hughes B (1964) Nucleolar “caps” produced by actinomycin D. Cancer Res 24: 1269–1277 [PubMed] [Google Scholar]
- Richard P, Darzacq X, Bertrand E, Jady BE, Verheggen C, Kiss T (2003) A common sequence motif determines the Cajal body‐specific localization of box H/ACA scaRNAs. EMBO J 22: 4283–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer. Nat Biotechnol 29: 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder RG, Rutter WJ (1970) Specific nucleolar and nucleoplasmic RNA polymerases. Proc Natl Acad Sci USA 65: 675–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy AM, West NC, Rao A, Adhikari P, Aleman C, Barnes AP, Deininger PL (2000) Upstream flanking sequences and transcription of SINEs. J Mol Biol 302: 17–25 [DOI] [PubMed] [Google Scholar]
- Rubbi CP, Milner J (2003) Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J 22: 6068–6077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer U, Hügle B, Hazan R, Rose KM (1984) Drug‐induced dispersal of transcribed rRNA genes and transcriptional products: immunolocalization and silver staining of different nucleolar components in rat cells treated with 5,6‐dichloro‐beta‐D‐ribofuranosylbenzimidazole. J Cell Biol 99: 672–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt U, Basyuk E, Robert MC, Yoshida M, Villemin JP, Auboeuf D, Aitken S, Bertrand E (2011) Real‐time imaging of cotranscriptional splicing reveals a kinetic model that reduces noise: implications for alternative splicing regulation. J Cell Biol 193: 819–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Shao N, Liu X, Nestler E (2014) ngs.plot: quick mining and visualization of next‐generation sequencing data by integrating genomic databases. BMC Genom 15: 284–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevtsov SP, Dundr M (2011) Nucleation of nuclear bodies by RNA. Nat Cell Biol 13: 167–173 [DOI] [PubMed] [Google Scholar]
- Singer SS, Mannel DN, Hehlgans T, Brosius J, Schmitz J (2004) From “junk” to gene: curriculum vitae of a primate receptor isoform gene. J Mol Biol 341: 883–886 [DOI] [PubMed] [Google Scholar]
- Sirri V, Hernandez‐Verdun D, Roussel P (2002) Cyclin‐dependent kinases govern formation and maintenance of the nucleolus. J Cell Biol 156: 969–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirri V, Urcuqui‐Inchima S, Roussel P, Hernandez‐Verdun D (2008) Nucleolus: the fascinating nuclear body. Histochem Cell Biol 129: 13–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan GJ, Bridger JM, Cuthbert AP, Newbold RF, Bickmore WA, McStay B (2001) Human acrocentric chromosomes with transcriptionally silent nucleolar organizer regions associate with nucleoli. EMBO J 20: 2867–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajrishi MM, Tuteja R, Tuteja N (2011) Nucleolin: the most abundant multifunctional phosphoprotein of nucleolus. Commun Integr Biol 4: 267–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA‐Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugrinova I, Monier K, Ivaldi C, Thiry M, Storck S, Mongelard F, Bouvet P (2007) Inactivation of nucleolin leads to nucleolar disruption, cell cycle arrest and defects in centrosome duplication. BMC Mol Biol 8: 66–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umylny B, Presting G, Efird JT, Klimovitsky BI, Ward WS (2007) Most human Alu and murine B1 repeats are unique. J Cell Biochem 102: 110–121 [DOI] [PubMed] [Google Scholar]
- Weber SC, Brangwynne CP (2012) Getting RNA and protein in phase. Cell 149: 1188–1191 [DOI] [PubMed] [Google Scholar]
- Weinmann R, Roeder RG (1974) Role of DNA‐dependent RNA polymerase 3 in the transcription of the tRNA and 5S RNA genes. Proc Natl Acad Sci USA 71: 1790–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LH, Brettingham‐Moore KH, Chan L, Quach JM, Anderson MA, Northrop EL, Hannan R, Saffery R, Shaw ML, Williams E, Choo KHA (2007) Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res 17: 1146–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Pan J, Thoroddsen V, Wysong DR, Blackman RK, Bulawa CE, Gould AE, Ocain TD, Dick LR, Errada P, Dorr PK, Parkinson T, Wood T, Kornitzer D, Weissman Z, Willis IM, McGovern K (2003) Novel small‐molecule inhibitors of RNA polymerase III. Eukaryot Cell 2: 256–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG (2011) ncRNA‐ and Pc2 methylation‐dependent gene relocation between nuclear structures mediates gene activation programs. Cell 147: 773–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Lin C, Jin C, Yang JC, Tanasa B, Li W, Merkurjev D, Ohgi KA, Meng D, Zhang J, Evans CP, Rosenfeld MG (2013) lncRNA‐dependent mechanisms of androgen‐receptor‐regulated gene activation programs. Nature 500: 598–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H, Brick K, Evrard Y, Xiao T, Camerini‐Otero RD, Felsenfeld G (2010) Mediation of CTCF transcriptional insulation by DEAD‐box RNA‐binding protein p68 and steroid receptor RNA activator SRA. Genes Dev 24: 2543–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye T, Krebs AR, Choukrallah MA, Keime C, Plewniak F, Davidson I, Tora L (2011) seqMINER: an integrated ChIP‐seq data interpretation platform. Nucleic Acids Res 39: e35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnack K, Konig J, Tajnik M, Martincorena I, Eustermann S, Stevant I, Reyes A, Anders S, Luscombe NM, Ule J (2013) Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell 152: 453–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillner K, Filarsky M, Rachow K, Weinberger M, Längst G, Németh A (2013) Large‐scale organization of ribosomal DNA chromatin is regulated by Tip5. Nucleic Acids Res 41: 5251–5262 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Review Process File
Source Data for Figure 4
Data Availability Statement
RNA‐seq data generated can be accessed via the EBI Array Express archive under the accession number E‐MTAB‐3460.


