Abstract
The biochemical and biophysical properties of the extracellular matrix (ECM) dictate tissue-specific cell behaviour. The molecules that are associated with the ECM of each tissue, including collagens, proteoglycans, laminins and fibronectin, and the manner in which they are assembled determine the structure and the organization of the resultant ECM. The product is a specific ECM signature that is comprised of unique compositional and topographical features that both reflect and facilitate the functional requirements of the tissue.
The extracellular matrix (ECM) has important roles in regulating the development, function and homeostasis of all eukaryotic cells1–3. In addition to providing physical support for cells, the ECM actively participates in the establishment, separation and maintenance of differentiated tissues and organs by regulating the abundance of growth factors and receptors, the level of hydration and the pH of the local environment1,4. These diverse functions are achieved through its complex chemical composition and organization. Although it is primarily composed of water, proteins and polysaccharides, the ECM shows exquisite tissue specificity as a result of the unique ECM compositions and topographies that are generated through a dynamic biochemical and biophysical interplay between the various cells in each tissue and the evolving microenvironment4. The mature ECM can also undergo dynamic remodelling in response to environmental stimuli, such as applied force or injury, which enables the tissue to maintain homeostasis and to respond to physiological challenges and stresses, including disease2,5–8.
Understanding cellular differentiation, tissue morphogenesis and physiological remodelling requires an understanding of the ECM components that are produced by cells, as well as the assembly of those macromolecules into a functional three-dimensional structure2,3. The macromolecules that constitute the ECM have evolved structural and chemical properties that are particularly suited to their specific biological functions in their respective tissues. Small, modular subunits form homopolymers and heteropolymers that become supramolecular assemblies with highly specialized organizations1–3. These assemblies contain binding domains for growth factors and chemokines, establish complex adhesion surfaces and form diffusion barriers between different cellular layers; thus, ECM molecules can function both as support for cells and as active participants in signalling to control cell behaviour9,10. In all cases, each class of ECM molecule has evolved the ability to interact with another class to produce unique physical and signalling properties that support tissue structure, growth and function1. Growing insight into the biological importance of the ECM in development, pathophysiology and the normal function of tissues throughout the body has confirmed the importance of elucidating the mechanisms of ECM assembly.
This Review provides a general overview of our current understanding of the ‘structural principles’ that underlie the assembly of the ECM at several levels, highlighting key molecular components and their organization into an interlocking network. We also consider the biochemical and mechanical cues that affect ECM assembly and how its organization directly relates to its tissue-specific functions, including as a structural support, as a barrier between different tissues and as a source of both chemical and physical cues. We highlight unanswered questions that require further investigation and the techniques and model systems that may facilitate this research.
Key molecular players in the ECM
Our understanding of the composition, structure and function of the ECM is continuously evolving, aided by the discovery of novel ECM molecules, the mapping of internal sites within ECM proteins that are crucial for self-assembly and for interactions with other ECM components, the characterization of the proteases and the protease inhibitors that are responsible for ECM degradation and turnover, and the identification of novel receptors and signalling mechanisms that mediate cellular responses to the ECM1–3. The architecture of the ECM is highly organized as a result of the innate properties of its constituent molecules and their interactions, as well as the activities of resident cells. The interactions between cells and the environment that are created by the ECM in turn have important roles in directing processes in development, homeostasis and pathogenesis. In this Review, we define the ECM as all secreted molecules that are immobilized outside a cell, including growth factors, cytokines and cell adhesion molecules, although we primarily focus on the macromolecules that are mainly responsible for tissue-type specific extracellular architecture.
The protein and non-protein constituents of the ECM vary not only in terms of their functional roles but also in terms of their structure. The polypeptide chains of ECM macromolecules often consist of many individual domains, and homologous domains between macromolecules can have sequence and structural differences that impart different functions. Individual ECM macromolecules rarely exist in isolation but instead often integrate into supramolecular structures containing different molecular species that differ in both their identity and their relative abundance. The ECM is primarily composed of two main classes of macromolecules: fibrous proteins (including collagens and elastin) and glycoproteins (including fibronectin, proteoglycans (PGs) and laminin)1.
Fibrillar collagens
Collagens are major proteins of the ECM and are arguably the most dominant. Decades of research have uncovered 28 different collagen types, and each type is comprised of homotrimers and heterotrimers that are formed by three polypeptide chains (known as α-chains) (FIG. 1; TABLE 1). More than 40 distinct α-chains have been identified in humans as well as multiple other proteins containing collagen-like domains11,12. The structural hallmark of all collagens is their triple helix — a tight right-handed helix of three α-chains, each of which contains one or more regions characterized by the repeating amino acid motif Gly-X-Y, where X and Y can be any amino acid13 (FIG. 1). Some collagen molecules assemble as homotrimers, whereas others assemble as heterotrimers that are comprised of two or three distinguishable α-chain types. The large family of collagens and proteins with collagenous domains has revealed that the collagen triple helix is a basic motif that can be adopted by proteins that have different functions. The rod-like domain has the potential to engage in various modes of self-association and the capacity to bind to cell surface receptors, other proteins, glycosaminoglycans (GAGs) and nucleic acids. A detailed description of the structural organizations of the different types of collagens can be found in REF. 14. For the purposes of this Review, we specifically focus on the assembly of fibrillar collagen, which comprises multiple complex intracellular and extracellular post-translational processes from the translational product to a fibrillar structure that is capable of withstanding tensile forces. The unique mechanical properties of fibrillar collagen are mainly controlled by its structure, which shows the important relationship between three-dimensional protein structure and the function of the resultant ECM6.
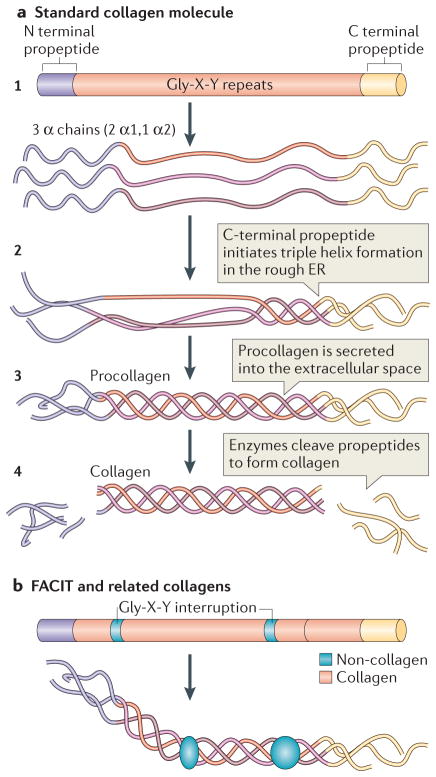
Figure 1. Collagen structure.
a | The standard fibrillar collagen molecule is characterized by amino- and carboxy-terminal propeptide sequences, which flank a series of Gly-X-Y repeats (where X and Y represent any amino acids but are frequently proline and hydroxyproline) (step 1). These form the central triple helical structure of procollagen and collagen. Three α-chains (the illustration shows two α1-chains and one α2-chain, which is representative of type I collagen) are intracellularly assembled into the triple helix following initiation of this process by the C-terminal domain (step 2). Procollagen is secreted by cells into the extracellular space (step 3) and converted into collagen by the removal of the N- and C-propeptides via metalloproteinase enzymes (step 4).
b | Fibril-associated collagens with interrupted triple helices (FACIT) and related collagens have a different structure to standard fibrillar collagen; they contain non-collagenous regions — that is, non-triple helical sequences. These lead to kinks in the resulting macromolecular structure that straighten under small strains. Figure part a is modified, with permission, from REF 135 © 2012 Fan et al.; licensee BioMed Central Ltd, and from REF. 136, Klug, William S.; Cummings, Michael R., Concepts of Genetics, 5th Edition, © 1997. Reprinted by permission of Pearson Education, Inc., Upper Saddle River, NJ. Figure part b: this figure was originally published in Biochem. J. Jäälinoja, J., Ylöstalo, J., Beckett, W., Hulmes, D. J. S. & Ala-Kokko, L., Trimerization of collagen IX alpha-chains does not require the presence of the COL1 and NC1 domains. Biochem. J. 2008; 409: 545–554 © the Biochemical Society.
Table 1.
Primary types of collagens*
| Classes | Type | Genes encoding proteins typical of collagen composition | Description |
|---|---|---|---|
| Fibrillar | I | COL1A1 and COL1A2 | Quarter-stagger packed fibrils that are primarily found in fibrous stromal matrices, such as skin, bone, tendons and ligaments |
| II | COL2A1 | ||
| III | COL3A1 | ||
| V | COL5A1, COL5A2 and COL5A3 | ||
| XI | COL11A1, COL11A2 and COL11A3 | ||
| XXIV | COL24A1 | ||
| XXVII | COL27A1 | ||
| FACIT | IX | COL9A1, COL9A2 and COL9A3 | Fibril-associated molecular bridges that are associated with type I (XII, XVI, XIX, XXI) and type II (IX, XVI, XIX) collagen fibrils |
| XII | COL12A1 | ||
| XIV | COL14A1 | ||
| XVI | COL16A1 | ||
| XIX | COL19A1 | ||
| XX | COL20A1 | ||
| XXI | COL21A1 | ||
| XXII | COL22A1 | ||
| Basement membrane | IV | COL4A1, COL4A2, COL4A3, COL4A4, COL4A5 and COL4A6 | Network structure comprised of laminins and basement membrane proteins |
| Long chain | VII | COL7A13 | Anchoring fibrils that are associated with the basement membrane |
| Filamentous | VI | COL6A1, COL6A2, COL6A3, COL6A5 and COL6A6 | Beaded microfibrils |
| Short chain | VIII | COL8A1 | Hexagonal lattice structure |
| X | COL10A1 | These collagens both regulate, and are regulated by, hypertrophy in cartilage | |
| Multiplexins | XV | COL15A1 | Multiple triple helix domains with interruptions containing chondroitin sulphate and heparin sulphate glycosaminoglycans |
| XVIII | COL18A1 | ||
| MACIT (transmembrane domain) | XIII | COL13A1 | Cell surface molecules with extracellular, membrane-spanning and intracellular domains |
| XVII | COL17A1 | ||
| XXIII | COL23A1 |
Collagen types are classified on the basis of domain structure homology and suprastructural assembly.
FACIT, fibril-associated collagens with interrupted triple helices; MACIT, membrane-associated collagens with interrupted triple helices.
Four distinct stages define collagen fibril assembly after the transcription and translation of the procollagen α-chains: import into the rough endoplasmic reticulum, where the α-chains are modified so that they can form triple-helical procollagen; modification of procollagen in the Golgi apparatus and its packaging into secretory vesicles; cleavage of the procollagen to form the collagen molecule in the extracellular space; and lysyl oxidase-catalysed crosslinking between collagen molecules to stabilize the supramolecular collagen structure6,11,12,15,16 (FIGS 1,2a). All fibrillar collagens are initially synthesized as precursor molecules that contain large amino- and carboxy-terminal propeptides and a signal recognition sequence, which promotes targeting to the rough endoplasmic reticulum, where post-translational modifications take place, leading to the assembly of procollagen molecules17. Within the endoplasmic reticulum, particular lysine and proline residues within the propeptides are hydroxylated by lysyl and prolyl hydroxylase enzymes to promote the stability and glycosylation of the triple helices following formation17. At the same time, a limited number of lysine hydroxyl groups are glycosylated. These proline and hydroxyproline amino acids can comprise 20% of the molecule, contributing to triple helix stabilization. Although the exact mechanism that controls the stabilization of the triple helix has generated much controversy, the current consensus suggests that increased stabilization is partly due to its high content of amino acids, increased interchain hydrogen bonds through hydration networks and electrostatic interactions between lysine residues13,18,19. Following lysyl and prolyl hydroxylation and O-linked glycosylation, α-chains associate to form procollagen, and triple helix trimerization is initiated by the association of the C-propeptide domains through both specific chain recognition and the formation of a stable nucleus, which favours triple-helical assembly15,20–23. Recent work elucidating the crystal structure of the C-propeptide domain from procollagen III suggests that there is a structural mechanism by which α-chains are selectively recognized, with the chain recognition sequence (CRS) of one chain interacting with the CRS of a neighbouring chain during intracellular trimerization of procollagen to ensure correct homotrimeric and heterotrimeric chain stoichiometry20. In addition, protein disulphide isomerase catalyses the formation and the rearrangement of intramolecular and intermolecular disulphide bonds24. Many other proteins influence and guide triple helix formation during procollagen assembly, including prolyl 4-hydroxylase, 78 kDa glucose-regulated protein (GRP78) and GRP94 (REF. 15). Importantly, the tight packing of the three α-chains near the common axis places steric constraints on every third amino acid position, with only the smallest amino acid, glycine, accommodated without chain distortion; this generates the Gly-X-Y repeating motif and imparts further stability to the triple helix13. Detailed reviews discussing the collagen triple helix structure and assembly can be found in REFS 13,15,25,26.
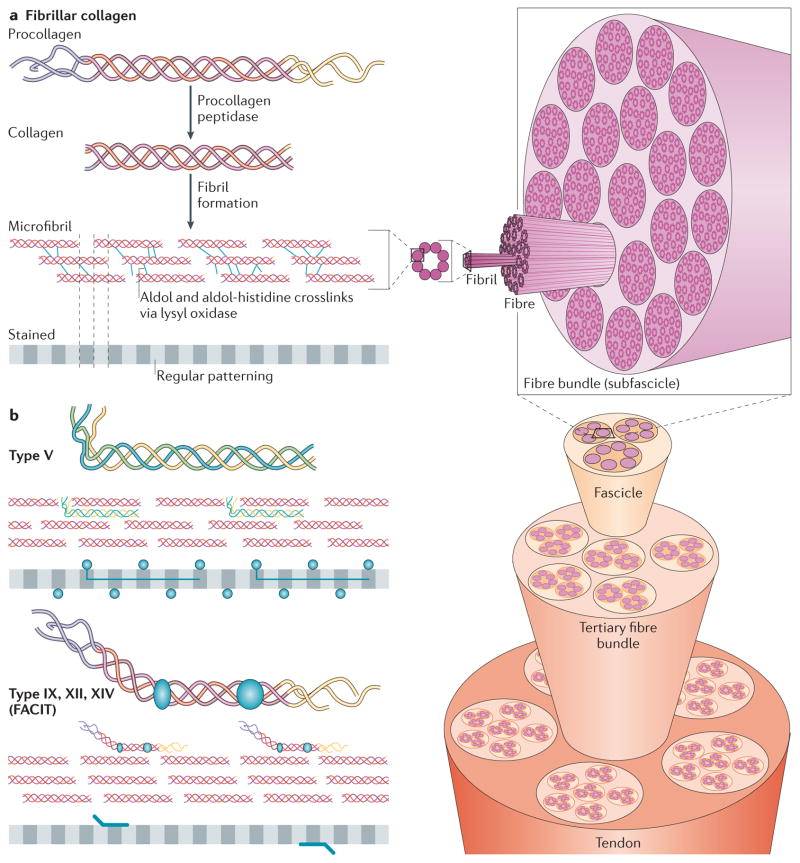
Figure 2. Fibrillar collagen assembly.
a | After procollagen is secreted into the extracellular space, collagen type-specific metalloproteinase enzymes remove the amino- and carboxy-propeptides. Collagen is assembled into cross-striated microfibrils that occur in the extracellular matrix of connective tissues. This can be observed via electron microscopy as regularly spaced bands, as represented by the grey, striped bar. The position of non-fibrillar collagens in this structure is shown in similar diagrams in part b. Short microfibrils merge into mature fibrils through longitudinal and axial growth. To form mature fibres, lysyl oxidase catalyses the formation of intramolecular and intermolecular covalent crosslinks between collagen molecules. Fibres are bundled together within a connective tissue and stabilized via interactions with fibril-associated collagens with interrupted triple helices (FACIT) collagens and small leucine-rich repeat proteoglycans (SLRPs) into linear structures capable of transmitting tensile forces. Specifically in the case of tendons, fibres ranging in size from 1 μm to 20 μm are bundled into 15 400 μm diameter triangular subfascicles, subfascicles are bundled into 150 1000 μm fascicles and fascicles are grouped into tertiary bundles ranging from 1000 μm to 3000 μm. Finally, multiple tertiary bundles are enclosed by the epitenon connective tissue, which contains the vascular, lymphatic and nerve sources for the tendon tissue.
b | In addition to fibrillar collagen, other collagen subtypes, such as type V and FACIT collagens, are incorporated into the fibril structure. Type V collagen is inserted between strands of the microfibril, and FACIT collagens cling to the surface of the microfibril and work with SLRPs to stabilize higher order structures. Figure part a is modified, with permission, from REF. 135 © 2012 Fan et al.; licensee BioMed Central Ltd, from Klug, William S.; Cummings, Michael R., Concepts of Genetics, 5th Edition, © 1997 (REF. 136). Reprinted by permission of Pearson Education, Inc., Upper Saddle River, NJ, and from REF. 138, reprinted from Trends Biotechnol., 26, Liu, Y., Ramanath, H. S. & Wang, D.-A., Tendon tissue engineering using scaffold enhancing strategies, 201–209, Copyright (2008), with permission from Elsevier. Figure part b: this figure was originally published in Biochem. J. Jäälinoja, J., Ylöstalo, J., Beckett, W., Hulmes, D. J. S. & Ala-Kokko, L., Trimerization of collagen IX alpha-chains does not require the presence of the COL1 and NC1 domains. Biochem. J. 2008; 409: 545–554 © the Biochemical Society.
In the second step, after assembly in the endoplasmic reticulum, procollagen is packaged in the Golgi apparatus for export into the extracellular matrix. For the third step of collagen fibril synthesis, evidence suggests that procollagen processing, exocytosis and initial fibril assembly are closely associated with membrane-enclosed compartments15. Early models proposed that cells exert considerable spatial control over this pericellular space, with procollagen excreted by exocytosis and modified within deep, narrow recesses between cellular protrusions close to the cell body27,28. Although procollagen processing was originally viewed as a solely extracellular event (with the second and third steps of collagen fibril synthesis being distinct and mutually exclusive), later work provided evidence that procollagen processing and collagen fibril assembly are initiated in compartments between the Golgi apparatus and the plasma membrane that are targeted to plasma membrane extrusions (known as fibripositors) that project from the cell surface. Recent electron microscopy studies confirmed the presence of collagen fibrils in cell surface fibripositors29–32. Following or during secretion of procollagen into the ECM, proteolytic processing of the large procollagen NH2- and COOH-propeptide domains leads to the production of mature collagen molecules that then self-assemble into fibrils. Enzymatic removal of the propeptide domains is carried out by collagen type-specific metalloproteinases from the a disintegrin and metalloproteinase with thrombospondin motif (ADAMTS) and bone morphogenetic protein 1 (BMP1) and tolloid-like families, as well as by furin-like proprotein convertases16,33–36 (FIG. 1). If the C-propeptide remains attached, procollagen solubility in the extracellular space remains high, which inhibits premature fibril assembly; by contrast, the persistence of the N-propeptide influences fibril shape and diameter, possibly by increasing the surface area to volume ratio, without affecting fibril formation16,37. In collagens I, II and III, the N-propeptides are completely removed, leaving short N-propeptides15. More detailed information on the roles and processing of the C- and N-propeptides can be found in REF. 16.
After cleavage of procollagen into collagen, initial fibril formation events occur at the cell surface38. Collagen organization in fibrils has been intensely studied by electron microscopy and X-ray diffraction39–44. Collagen self-assembles to form microfibrils with a quarter stagger axial D periodicity of approximately 67 nm to create the characteristic ‘striation’ that was recognized in initial electron microscopy observations of collagen-containing tissues16,45 (FIG. 2a). Intermediate-sized microfibrils are thought to form via a multistage process that includes the nucleation, organization and unilateral elongation of short primary fibrils. Current models suggest that microfibril assembly is nucleated and modulated by the inclusion of collagens V and/or XI, which, as discussed above, retain portions of their N-terminal propeptide to co-assemble into fibrils with types I, II and III (FIGS 1,2b). The N-terminal propeptides of collagens V and/or XI are thought to project outwards through the gap between adjacent collagen molecules to interact with the fibril surface, which limits lateral growth via steric hindrance and charge interactions38 (FIG. 2b). Although collagen V is a quantitatively minor component relative to collagen I and comprises less than 5% of the total collagen content in most tissues, it is considered to be a dominant regulator of fibrillogenesis, as deletion of collagen V in the mouse is associated with a lack of fibril assembly38.
After collagen microfibrils have been assembled, short microfibrils may merge or grow into fibrils, increasing both longitudinally and axially; these transitions are regulated by various molecules38,46 (FIG. 2). For example, the large body of evidence gained using in vitro techniques and in vivo knockout transgenic experiments shows that small leucine-rich repeat proteoglycans (SLRPs) with collagen-binding properties, including decorin, biglycan, fibromodulin and lumican, can affect fibril growth rate, size, morphology and content15,47,48. Most heterotypic collagen fibrils are composed of a mixture of fibrillar collagens, with the FACIT (fibril-associated collagens with interrupted triple helices) collagens integrating into the fibrils, selectively altering the surface properties of the collagen fibril and its interactions with additional ECM molecules such as the SLRPs37,49. Throughout the process of fibril assembly and growth, interactions among the assembling fibrils, accessory macromolecules and cell adhesion receptors actively organize the maturing fibril.
To form mature collagen fibres, further modifications are made during the assembly of collagen fibril aggregates. In the final step of collagen biosynthesis, covalent crosslinks are introduced into the supramolecular assembly to provide stability and enhanced mechanical properties50–52. Extracellular lysyl oxidases catalyse and facilitate the oxidative deamination of targeted peptidyl lysine residues, which results in the formation of aldols or β-ketoamines by reacting with aldehydes or amino groups on lysines between collagen monomers to form intramolecular and intermolecular covalent crosslinks49,51–53. The N- and C-terminals of neighbouring collagen monomers interact with each other and are covalently crosslinked by lysyl oxidase, both within and between microfibres52–54. Overall, interactions between the fibrillar and the FACIT collagens, as well as among collagens, proteoglycans and crosslinking enzymes, can function as key regulators of fibrillar collagen organization, resulting in immense tissue-, stage- and age-specific differences in both fibril composition and structure. These variations are finely tuned to fulfil bespoke biochemical, structural and biomechanical roles25. Importantly, the assembly of collagen fibrils is mainly driven by the unique chemical and physical interactions of the interacting components. This cell-driven self-assembly results in a multi-hierarchical collagen structure, which provides binding sites for other proteins and cells and is crucial for the mechanical integrity of the ECM55.
Proteoglycans
In contrast to the predominantly fibrillar structure of collagens, proteoglycans form the basis of higher order ECM structures around cells. The primary biological function of proteoglycans derives from the biochemical and hydrodynamic characteristics of the GAG components of the molecules, which bind water to provide hydration and compressive resistance. As such, proteoglycans are found in abundance in cartilage and neural ECM56,57. Proteoglycans are characterized by a core protein that is covalently linked to GAGs, which are long, negatively charged, linear chains of disaccharide repeats. They are classified into subtypes on the basis of the structure of these GAG carbohydrate chains, as well as on the distribution and the density of these chains along the core protein58,59. Major GAGs include heparin sulphate, chondroitin sulphate, dermatan sulphate, hyaluronan and keratin sulphate58,60–62 (the structures of heparin sulphate and chondroitin sulphate are shown in FIG. 3a).
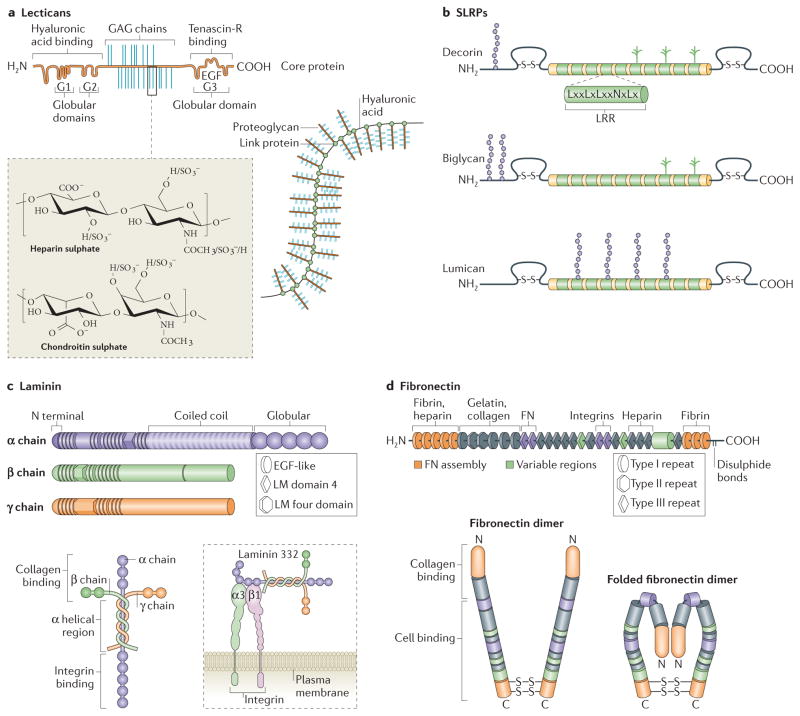
Figure 3. Non-collagenous molecules of the ECM.
Lecticans (aggrecan is shown; part a) and small leucine-rich repeat proteoglycans (SLRPs; decorin, biglycan and lumican are shown; part b) are major proteoglycans (PGs). The SLRPs are characterized by differences in their amino terminus structure and highly conserved leucine rich repeats (LRR) in the core molecule. Lecticans have a core protein with binding domains for glycosaminoglycan (GAG) chains flanked by globular domains that interact with hyaluronic acid (at the N terminus) and tenascin R (at the carboxy terminus). Common GAGs include chondroitin sulphate and heparan sulphate, the chemical structures of which are depicted. The assembled bottle-brush-like aggrecan link protein hyaluronic acid structure is shown. Laminins are formed by the incorporation of α, β and γ chains into a cruciform, Y-shaped or rod-like structure (part c). These chains are characterized by different domains, as shown. The domain structures depicted represent only one isoform for each chain type, but major differences between the basic composition of the chains (such as the lack of globular regions in β and γ chains) are also present in the other isoforms. Laminins interact with cell surface receptors, such as integrins, primarily through globular domains in the chain. Fibronectin domain structure and the domains to which extracellular matrix (ECM) molecules and cell surface receptors bind are indicated (part d). The fibronectin molecule forms a dimer through disulphide bonds on its C terminus. The folded fibronectin molecule forms via ionic interactions between type III domains of neighbouring molecules and is deformed by mechanical force to reveal cryptic binding sites for other fibronectin molecules and cell surface receptors when interacting with cells. Figure part a: this image originally appeared in Diamond Light Source Ltd’s Annual Report 2010 (p72–73) and has been reproduced with permission from Diamond Light Source Ltd, the University of Liverpool, T. R. Rudd, M. A. Skidmore, S. E. Guimond, R. Xu, R. Hussain, D. G. Fernig, G. Siligardi and E. A. Yates http://www.diamond.ac.uk/Science/Research/Highlights/study2.html. Figure part b is modified from REF. 139, Nature Publishing Group. Figure part c is modified, with permission, from Yurchenco, P. D. Basement membranes: cell scaffoldings and signaling platforms. Cold Spring Harb. Perspect. Biol. 3, a004911 (2011), Cold Spring Harbour Laboratory Press, and REF. 141, Nature Publishing Group. Figure part d is modified, with permission, from Schwarzbauer, J. E. & DeSimone, D. W. Fibronectins, their fibrillogenesis, and in vivo functions. Cold Spring Harb. Perspect. Biol. 3, a005041 (2011), Cold Spring Harbour Laboratory Press, and REF. 82, modified with permission from Annual Review of Cell Dev. Biol., Volume 26 by Annual Reviews, http://www.annualreviews.org. EGF, epidermal growth factor; LM domain, laminin domain.
Heparan sulphate proteoglycans (HSPGs) are a major part of the basement membrane, which is a type of ECM structure (see below). Their GAG chains are comprised of highly sulphated chains of hexuronic acid and -glucosamine disaccharide repeats. HSPGs can be cell surface-bound, such as the syndecans and CD44v3, glycophosphatidylinositol (GPI)-linked, such as glypicans, or secreted ECM proteins, such as agrin, collagen XVIII and perlecan63. Variety in HSPGs is introduced through splice isoforms that control GAG chain lengths, the spacing of GAG chain attachments along the protein core and the extent of sulphation and epimerization of the sugars within the GAG chains62,63. Resulting proteoglycans show different binding affinities for molecules within the ECM. For example, fibroblast growth factors (FGFs) are particularly sensitive to the spacing and sulphation of GAG chains in their interactions with HSPGs64. HSPGs have high negative charges that enable them to easily bind to other proteins, including growth factors such as FGF, chemokines, other ECM proteins, such as laminin and fibronectin, and cell surface receptors63,65,66. As such, they have been implicated in regulating many cellular processes, including cell growth and migration63,67,68.
Chondroitin sulphate proteoglycans (CSPGs) can be found in cartilage and neural ECM. CSPGs are functionalized with sulphated polysaccharides with repeating disaccharides of glucuronic acid and N-acetylgalactosamine68. Lecticans (also known as hyalectins) are the most common CSPGs in the neural ECM68,69 (FIG. 3a). This family consists of four members: aggrecan, neurocan, veriscan and brevican61,69. The core protein is highly homologous across lecticans, which are characterized by globular domains at both the N and the C termini. Lecticans contain binding domains for hyaluronic acid (which is a common glycosaminoglycan in cartilage and the nervous system), lectins and growth factors56,69,70. As with HSPGs, alternative splicing and different densities of GAG chains on the core protein lead to considerable variations in the properties of these proteins56.
In addition to being dominant components of the ECM, proteoglycans can also function as accessory proteins in tissues rich in other matrix proteins. For example, the SLRPs are a family of proteoglycans that have been implicated in fibrillar collagen assembly48. This family includes well-known members such as decorin, biglycan and lumican, with new members still being discovered and characterized. All SLRPs have two regions — a variable N-terminal domain that contains sulphated tyrosine or acidic amino acid residues and a conserved carboxy terminus that contains the leucine-rich repeats (LRRs) for which the family was named48,71 (FIG. 3b). GAG chains can then be added to these core proteins48. The LRRs contain collagen-binding regions and have been well characterized as a result of this property; the function of the N-terminal domains is less extensively understood48. Elucidation of the crystal structure of decorin suggests that these N-terminal domains function as a capping mechanism, whereby cysteine residues within this domain form disulphide bonds that link back to the C terminus via leucines in the consensus sequence72.
Five classes of SLRPs have so far been established, and each class is distinguished by their homology on the protein and genomic levels, by the presence of N-terminal cysteine-rich clusters and by chromosomal placement. Classes I–III are so-called ‘canonical’ classes, which are distinguished by the relative abundance of data describing their function and localization within the body, whereas classes IV–V have only been recently described and there are many unanswered questions about their function. A full listing and description of SLRPs is beyond the scope of this Review, but such information can be found in REFS 73,74. The Class I SLRPs decorin and biglycan have been extensively studied and participate in collagen fibrillogenesis57,75. Not only do these proteins facilitate the formation of collagen fibrils but they also regulate fibril width and growth through mechanisms that have not been fully elucidated74. In addition to their roles as structural proteins in ECM assembly, SLRPs are considered to be ‘active’ members of the ECM and to participate in cell signalling. Binding sites for growth factors, cytokines and ECM proteins other than collagen have recently been found on SLRPs71.
Connectors in the ECM
Collagen, proteoglycans, and hyaluronic acid represent major structural components within the ECM. They provide much of the supportive framework within which other ECM components and cells interact. Additional ECM components, such as laminin or fibronectin, function as bridges between structural ECM molecules to reinforce this network, as well as to connect the ECM to cells and to soluble molecules within the extracellular space. These connectors tend to be multidomain glycoproteins that contain binding motifs for other ECM proteins, growth factors and cell surface receptors1. In this Review, we focus on laminin and fibronectin as examples and point to others in TABLE 2 for further reference.
Table 2.
Non-collagenous ECM proteins
| Protein | Location | Description | Refs |
|---|---|---|---|
| Agrin | Skeletal muscles | Large PG that is crucial in the development of the neuromuscular junction; contains laminin G domains, Kazal-type serine protease inhibitor domains and epidermal growth factor domains | 142–144 |
| Integrin-binding sialoprotein (also known as bone sialoprotein) | Bone | Phosphorylated and sulphated glycoprotein that is crucial for the structure of mineralized matrix; binds to calcium and hydroxyapatite and mediates cell attachment through an RGD sequence that recognizes the vitronectin receptor | 145–147 |
| Matrilins | Cartilage and connective tissues | Non-collagenous, disulphide-bonded proteins that are involved in the formation of filamentous networks; contains von Willebrand factor A domains | 148–151 |
| Elastin | Arteries, connective tissue, lungs, skin and bladder | One of two components of elastic fibres that provide structural integrity along with reversible extensibility and deformability; forms covalently crosslinked polymers | 152–155 |
| Fibrinogen | Blood | Soluble glycoprotein, which, after cleavage by thrombin, is converted to fibrin monomer molecules that self-associate to form insoluble, homopolymeric ‘clots’ | 153,156, 157 |
| Fibrillin | Connective tissues and cardiovascular tissues | Predominant component of microfibrils, which forms a sheath around elastin in the formation of elastic fibres | 158,159 |
| Fibulins | Basement membranes, blood and arteries | Calcium-binding glycoprotein that is found in fibrillar matrices and blood | 153,160, 161 |
| Netrins | Basement membranes | Laminin-related protein that is involved in axon guidance and the development of the vasculature, lung, pancreas, muscle and the mammary gland | 162,163 |
| SPARC (also known as osteonectin) | Bone | Cysteine-rich, acidic, calcium-binding glycoprotein that is required for collagen calcification, mineralization initiation and mineral crystal formation | 164,165 |
| Secreted phosphoprotein 1 (also known as osteopontin) | Bone and kidneys | Negatively-charged glycoprotein that is involved in the attachment of cells to mineralized bone matrix | 146,166, 167 |
| Reelin | Brain | Large glycoprotein that is involved in guiding neuron and glial cell positioning in the developing brain | 168,169 |
| Tenascins | Connective tissues | Glycoproteins that are involved in mediating inflammatory and fibrotic processes | 119,153, 170,171 |
| Vitronectin | Blood | Adhesive glycoprotein that is involved in blood coagulation and wound healing | 172,173 |
ECM, extracellular matrix; PG, proteoglycan.
Laminins
Laminins are a family of large, mosaic glycoproteins composed of globular, laminin-type epidermal growth factor (EGF)-like repeats and α-helical domains (FIG. 3c). They are primarily located in the basal lamina and some mesenchymal compartments. Interactions between modular domains within the laminin molecule and other proteins enable laminins to mediate interactions between cells via cell surface receptors (such as integrins and dystroglycan) and other components of the ECM (such as nidogens (also known as entactins), perlecans and collagens)76 (FIG. 3c).
Laminins consist of α, β and γ chains that combine via the triple-helical coiled-coil domain in the centre of each chain to form structures with a cruciform (cross-shaped), Y-shaped (three arms) or rod-shaped (single arm) structure76 (FIG. 3c). Different chain isoforms combine to generate distinct assembled heterotrimers76. These laminins are described by their combined chain numbers; for example, α1β1γ1 is known as laminin 111. This nomenclature was preceded by single number assignments when there were fewer known isoforms. The original designations remain preferred in some settings so, for example, it is possible to find references to laminin 5 instead of α3Aβ3γ2. For a useful table that cross-references these different systems, see REF. 77. So far, five α-chains (α1–α5), four β-chains (β1–β4) and three γ-chains (γ1–γ3) are known, and they combine to form 16 different laminin heterotrimers77,78 Although laminin 111 has been shown to self-assemble into aggregates, laminins primarily function as bridges between other molecules79,80.
Fibronectin
Many of the ECM proteins described above interact with cells through crucial connections with the multidomain protein fibronectin to regulate cell adhesion, migration and differentiation81. Fibronectin is secreted as a large ECM glycoprotein that assembles via cell-mediated processes into fibrillar structures around cells82. Each fibronectin subunit consists of three modules of repeating units, each of which has distinct structures: type I, type II and type III structures81,82 (FIG. 3d). These modules contain binding motifs that are important in facilitating the interaction of fibronectin with cell surface receptors, such as integrins, collagen and gelatin, and intramolecular units that enable self-assembly of the molecule82,83. Intramolecular disulphide bonds between each type I and type II module stabilize the folded tertiary structure of the fibronectin subunit, and fibronectin dimers form via antiparallel disulphide bonds at the C terminus82.
Fibronectin fibril assembly involves interactions between its RGD and synergy sequences with corresponding binding sites within cell surface receptors such as integrins81,82,84. Initial binding between fibronectin dimers and integrins leads to the activation and clustering of these receptors82. Integrin clustering is thought to promote further fibronectin–fibronectin intermolecular interactions, and tethering of fibronectin molecules to the cell surface enables cell-mediated contractility to exert force on the fibronectin molecule and to change its conformation81. This change in conformation exposes cryptic binding sites within the fibronectin molecule and also enables it to take a form that is more conducive to self-assembly into fibrils. The maturation and the movement of fibrillar adhesions within the cell is thought to guide the formation of fibronectin fibrils82,85. Generation of force along these adhesions further controls the assembly of the fibronectin network and the maturation of multiple clusters of integrin-based adhesions brings nascent fibronectin bundles together to form a larger structure82,85.
In addition to the key molecular players we have described in this Review, the ECM is composed of many other classes of macromolecules that we have not included owing to space constraints. For brief descriptions of their chemical composition and where in the body they may be found, as well as excellent references that may enrich understanding of these important components, see TABLE 2.
Organizing the ECM
The ECM is not simply a collection of proteins. Different tissues have unique and specialized ECM components and ECM organization, which enables each ECM to carry out tissue-specific roles, including structural support, the transmission of forces and macromolecular filtration1,86. Three ECM landscapes, spanning diverse structures and functions, are discussed in this section, with a focus on how each supramolecular ECM is fabricated and organized. With a specialized axial and longitudinal organization, the first ECM assembly example consists of the fibrillar collagen-rich matrix that comprises mechanical load-bearing tendons. To provide an example of a non-fibrillar but protein-rich ECM, the basal lamina is discussed as the second example. With a much more amorphous, GAG-rich structure, the final example discussed is the perineuronal net (PNN), which is found in brain ECM. The unique structure–function relationships exhibited by these ECMs are highlighted. This section describes the ECMs of two specific tissues (tendon and brain) as well as the basal lamina, which is found in many tissues.
Fibrillar collagen-rich tendons
Fibrillar ECM that consists of substantial amounts of collagens I, II and III, such as the ECM found in tendons, ligaments and cartilage, has distinct structural and biomechanical properties that facilitate the effective transmission and absorption of cyclical tensile forces while avoiding injury87–89. These properties arise from the specialized axial and longitudinal structural organization of the collagen fibre hierarchy, which provides a scaffold for cell and macromolecular attachment89,90. The tendon is primarily a uniaxial force-transmitting connective tissue consisting of bundles of parallel fibrillar ECM and resident tendon fibroblasts (known as tenoblasts) that connect bone to muscle. Both the structure and the function of this fibrillar ECM provide biochemical and mechanical signals that cooperate in the integrated regulation of tenoblast proliferation, survival, differentiation and migration, which ultimately feedforward to maintain physiological ECM synthesis and assembly91.
The ECM that constitutes the tendon consists of collagen (65–80% of its dry mass, primarily collagen I) and elastin (1–2% of its dry mass) embedded in a well-hydrated, proteoglycan-rich matrix synthesized by tenoblasts90. In the mature tendon, bundles of aligned collagen fibrils are organized into a functionally continuous fibrous ECM, which enables them to guide motion and to transmit forces89,92. Throughout tendon development, multiple independent steps of collagen fibrillogenesis and ECM assembly take place and eventually contribute to a functionally mature tissue that has a complex ECM hierarchical structure.
In the first of three distinct steps in tendon collagen fibrillogenesis, initial collagen fibril assembly occurs near the surface of tendon fibroblasts, with collagen incorporating into microfibrils, which are the lowest order filamentous structures (approximately 4 nm in diameter), having an axial D periodicity16,88–90,93 (FIG. 2a). Through spatially regulated protein secretion, tenoblasts help to guide this initial assembly step; evidence suggests that fibripositors of tenoblasts contiguously present these microfibrils from one cell to another along the tendon axis through side-to-side and end-to-end tenoblast alignment88,94,95. Procollagen processing, homotypic and heterotypic collagen interactions and other matrix macromolecules influence the nucleation and the growth of these microfibril intermediates (see above). Importantly, these micrfibrils remain semi-flexible, which enables the repositioning of these structures within the tendon tissue throughout tissue maturation and force transmission88.
In the second step, longitudinal fibril growth occurs with microfibril intermediates fusing tip-to-tip as a result of lateral associations between adjacent tapered ends, and thus incorporating into continuous, mechanically functional fibrils87,96,97 (FIG. 2a). Intermediate microfibrils are present in two structural forms — unipolar fibrils that have C and N termini and N,N-bipolar fibrils, in which the molecules have their N termini pointed towards one of the tips15. End-to-end fusion specifically requires the C terminus of unipolar fibrils97. SLRPs often coat the body of microfibril intermediates, which promotes the fusion of the proteoglycan-free ends and inhibits premature side-to-side fusions97.
The third step involves the axial growth of the microfibrils via incorporation of, and interactions with, other microfibrils, as well as with microdomains of extracellular macromolecules such as FACIT collagens and proteoglycans (FIG. 2b). These collagen-interacting molecules regulate and restrict this axial growth and limit fibril diameter, which provides resident tendon fibroblasts with a mechanism for regulating this ECM assembly, resulting in fibril structures ranging in size from 10 nm to 500nm88,89 (FIG. 2a).
Fibres ranging in size from 1 μm to 20 μm form the next level of structure, with multiple fibrils grouped together by an endotenon connective tissue containing nerves, as well as blood and lymphatic vessels89. Fibres are bundled into 15–400 μm diameter triangular subfascicles, at which point they are compressed by surrounding structures and stabilized via interactions with FACIT collagens and SLRPs89,90. Subfascicles are bundled into 150–1000 μm fascicles, which are further bundled into tertiary bundles ranging from 1000 μm to 3000 μm89,90. Finally, multiple tertiary bundles are enclosed by the epitenon connective tissue, which, similar to the endotenon, contains the vascular, lymphatic and nerve sources for the tendon tissue89,90 (FIG. 2a).
Throughout the tissue, non-collagenous matrix, which consists of glycoproteins, proteoglycans, GAGs and other small molecules, surrounds and stabilizes collagen fibrils and fibres. The negatively charged groups of the sulphated proteoglycans and GAGs bind water, which improves the mechanical properties of the tissue under both shear and compressive stresses90. The stability and mechanical properties of this tissue are further strengthened and stabilized by collagen crosslinking, which is carried out by enzymes such as lysyl oxidase, as well as by non-enzymatic glycation89. Taken together, this hierarchical fibrillar structure results in tendon tissue that is capable of transmitting forces from muscle to bone without sustaining injury91.
Although collagen fibrillogenesis has been intensively studied for multiple tissues, many outstanding questions and areas of active research remain. Further elucidation of the biochemical mechanisms that underlie collagen fibril assembly will enable the development of rational therapies and tissue-engineering strategies for tissue repair and/or for the regeneration of adult tissues with mechanical properties that are similar to those that are observed in normal development and function. In addition, through the continued investigation and clarification of the structures and functions of the various domains along the collagen fibril, along with how these domains affect supramolecular assembly, novel insights into the roles of collagen at a tissue level in fibrotic diseases may highlight new targets to limit or to inhibit fibrosis. Improvements in high-resolution imaging, the establishment of in vivo genetic knockout (and overexpression) models and the introduction of novel in vitro self-assembling ECMs add to the available range of tools that investigators can use to continue elucidating the underlying mechanisms responsible for the different levels of supramolecular collagen assembly15.
Basal lamina
The basal lamina is a set of cell-adherent, organized layers of ECM molecules that form the supporting structure for epithelial, endothelial, muscle and fat cells, the central nervous system and the Schwann cells of peripheral nerves98–100. These ‘sheet-like’ layers appear early in development, segregating tissues, functioning as macromolecular filters and providing sites for cell adhesion86,101,102. The basal lamina shields tissues from disruptive biochemical and biophysical stresses and mediates communication among cells in the tissue and between cells and their external environment101. Although ‘basal lamina’ and ‘basement membrane’ are often used interchangeably in some tissues, such as skeletal muscle, the basement membrane is comprised of two distinct structures: the basal lamina and an external, fibrillar reticular lamina, which is directly linked to the cell membrane by the basal lamina103.
The basal lamina predominately assembles at or near cell surfaces104. The resulting ECM architecture consists of interconnected polymers of collagen IV and VII (FIG. 4a,b), laminins bound to nidogen glycoproteins and HSPGs99. Laminin deposits on the surface of cells self-assemble into sheets through terminal domain interactions that are organized by cell surface molecules, such as integrins and dystroglycan, and by binding to nidogen through a single globular domain102,105 (FIG. 4c). Collagen IV networks formed through interactions between its N-and C-terminal domains (FIG. 4a) then provide a three-dimensional scaffold for the association of laminin–nidogen complexes into less ordered aggregates, which interact with the collagen IV structure through binding sites on the nidogen molecule102,106. These collagen IV networks are thought to stabilize the assembling basal lamina through covalent crosslinks101. In addition, sulphated proteoglycans, such as agrin, perlecan and collagen XVIII, can incorporate into the lamina, which provides areas of negative charge and tethers growth factors107. These layers then recruit other basal lamina proteins, many of which have binding sites for one another, which leads to the formation of a mature basal lamina. Collagen VII can further stabilize this structure, in addition to connecting the basal lamina to the reticular lamina to form the basement membrane, via interactions with collagen IV (FIG. 4b,c). The inclusion of tissue-specific isoforms of laminin and collagen IV with different proteoglycan species and accessory proteins provides specific properties to the basal lamina in different locations. The basal lamina seems to be predominantly attached through laminins to cell surface receptors and sulphated glycoproteins104. These interactions are responsible for the activation of intracellular signalling, such as that involved in establishing cell polarity.
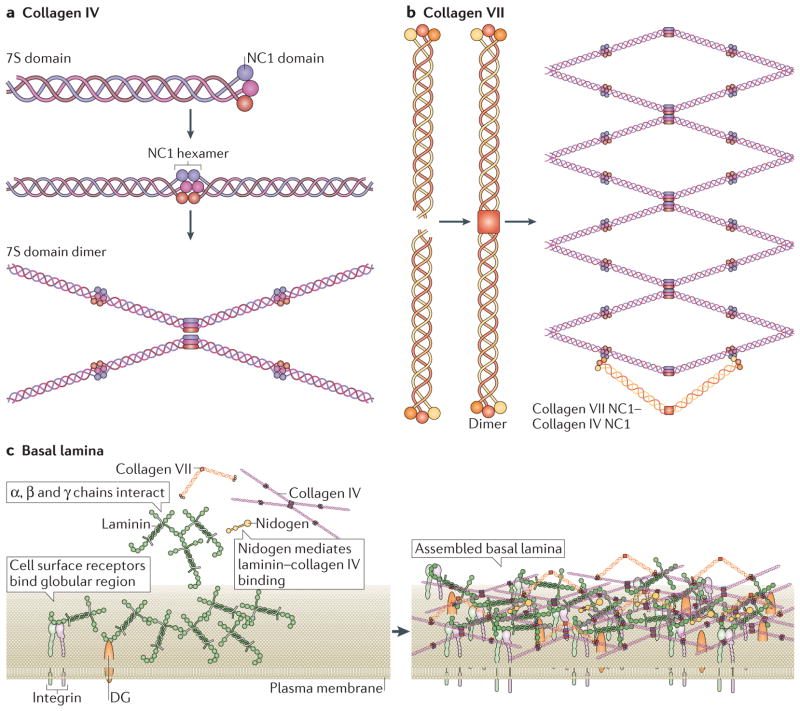
Figure 4. Basal lamina assembly.
Two major collagens participate in the assembly of the basal lamina: collagen IV and collagen VII.
a | Collagen IV is characterized by an amino-terminal 7S domain and a carboxy-terminal NC1 domain these are globular domains that facilitate the formation of hexamers and dimers, respectively. These then form highly ordered lattice structures, as shown.
b | Collagen VII forms long dimers that then bend and connect with the globular regions of collagen IV lattices.
c | Components of the basal lamina, and how they interact, are shown on the left-hand side. Within the basal lamina, laminins form a sheet on the cell surface by recruiting soluble laminin. This sheet is reinforced via interactions among chains of different laminin molecules and by binding between laminin globular regions and cell surface receptors (for example, integrins and dystroglycan). Collagen IV binds to laminins via nidogens and forms a scaffold on top of the laminins. This scaffold is further secured by collagen VII, which, in its unbent form (sometimes termed anchoring fibrils), can link the basal lamina to the reticular lamina to form the basement membrane. Assembled basal lamina is shown on the right-hand side. Figure parts a and b are modified from REF. 100, Nature Publishing Group. Figure part c is modified from REF. 100, Nature Publishing Group, with permission, from Yurchenco, P. D. Basement membranes: cell scaffoldings and signaling platforms. Cold Spring Harb. Perspect. Biol. 3, a004911 (2011), Cold Spring Harbor Laboratory Press and from REF. 141, Nature Publishing Group. DG, dystroglycan.
The structure and composition of the basal lamina directly contributes to its functions within tissues: to anchor the epithelium to the dermis, to guide development and differentiation and to function as a mechanical barrier to cells and macromolecules. The basement membrane is deposited early in development (soon after implantation) and helps to establish polarity in the epiblast108. Expression of ECM components within the basal lamina is tightly controlled in order to regulate intracellular signalling via activation of different receptors for these proteins and, in turn, to regulate cell fate determination at different stages of development109–111. The scaffold structure of the basal lamina, along with its secure connections to the dermis, means it provides excellent support for the epithelium. It is highly crosslinked and functions as an efficient barrier to both cellular and molecular traffic between distinct layers of tissue. However, it is also thin enough to permit transit in certain conditions, such as when immune cells infiltrate during inflammation112. Many of its components contain binding domains that allow the basement membrane to bind and to retain growth factors and chemokines, which have crucial roles in directing differentiation and guiding tissue repair and regeneration103,113,114.
Although much has been elucidated about the basal lamina, our understanding of its functions and interactions with cells is by no means complete. There remain interesting unanswered questions regarding the regulatory processes in place for the production of various components of the basal lamina, how the basal lamina imposes cell polarity and, through this, influences cell behaviour, the many roles of the proteoglycans in directing cell fate, and more. Research to address these questions uses a combination of cell biology, novel approaches in synthetic chemistry and glycobiology, and cutting-edge imaging techniques to visualize large-scale ECM structures.
Proteoglycan-rich matrices of the brain
Proteoglycan-rich ECM can be found in cartilage and in the central nervous system and lacks the distinctive superstructural organization that characterizes ECM surrounding other tissues. Instead, it is characterized by amorphous aggregates of HSPGs and CSPGs, crosslinked by other small proteoglycans. However, that is not to say that these ECMs lack organization altogether. In this section, we focus on the brain ECM as an example.
ECM in the brain contains fibrous components such as collagens, fibronectins and other molecules discussed above. However, these proteins represent a small percentage of the brain ECM, which predominantly consists of hyaluronic acid, proteoglycans and GAGs. Thus, it is a useful tissue in which to highlight proteoglycan-rich ECM properties and functions. For a full discussion of neural ECM and its various components, see REFS 56,115,116.
Early in development, the neural ECM is produced and secreted by both neurons and glial cells117. The composition and organization of neural ECM is highly dynamic throughout development and varies in different parts of the adult brain56,59,118. It is characterized by amorphous protein aggregates and is similar in organization to the proteoglycan component of cartilage. In both developing and adult brains, hyaluronic acid, which is a large, linear polysaccharide, functions as a central organizing molecule around which the other components of the ECM aggregate61,117. Glycoproteins within the tenascin family have an oligomeric structure that facilitates this assembly, and proteoglycans with hyaluronic acid-binding domains on their N termini and glycoprotein-binding domains on their C termini connect these proteins69,70,119. In contrast to the highly regulated structures in collagen-rich matrices or in the basal lamina, neural ECM aggregates form a latticed network that varies in its composition and organization throughout development120–122. Depending on the stage of development, this network can be mostly made up of neurocans, versicans, and tenascin C, as is found in the first few weeks of life, or brevican and aggrecan, as is the case in the adult brain122–124. These different proteoglycan profiles correspond with a ‘loose’ network that is made up of larger proteoglycans in early development, which is replaced and tightened with smaller proteoglycans as the brain develops69,122. These loose networks provide space for the developing brain to grow and to extend the neural network; tight networks in later life restrict growth and reinforce existing connections.
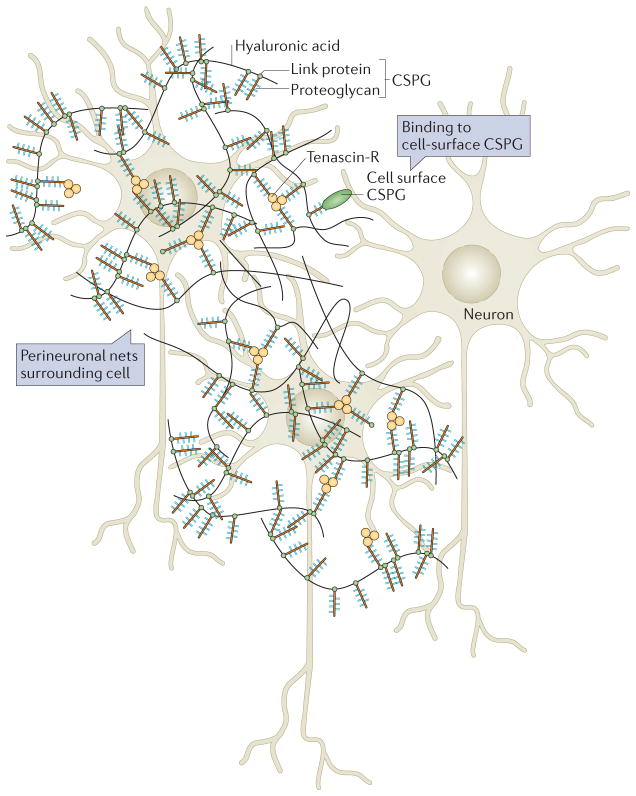
An exception to the mainly unorganized nature of the brain ECM are PNNs, so-called because they form a regular structure around the cell bodies of neurons, dendrites and the initial segments of axons122. The formation of PNNs is initiated by the production of hyaluronan by hyaluronan synthases on the inner surface of the plasma membrane125. Linear unsulphated hyaluronic acid chains deposited on the cell surface bind to extracellular CSPGs; these interactions are reinforced by link proteins (which are poorly characterized glycoproteins known to bind to both hyaluronic acid and proteoglycans) and tenascin R125 (FIG. 5). In this network, the N terminus of CSPG binds to hyaluronic acid and the C terminus of CSPG binds to tenascin R, forming trimeric crosslinkers that set up a stoichiometric relationship that determines the organization of the PNN125.
Figure 5. Perineuronal nets.
Perineuronal nets (PNNs) surround cells of the brain and act as inhibitors of both growth and migration. PNNs are lattices of hyaluronic acid, proteoglycans and tenascin molecules. The basic structure of a PNN is depicted here. Hyaluronic acid chains coated with proteoglycans are linked by trimeric tenascin R molecules to form a fairly organized structure around neurons. The chondroitin sulphate proteoglycans (CSPGs) within the net bind to cell surface CSPGs and can thus control cell behaviour. Figure modified from REF. 115, Nature Publishing Group.
The brain ECM, as with all other ECMs, provides structural support for cells within the organ. It also actively participates in the function of the brain; for example, the formation of PNNs is perfectly timed with the termination of plasticity in the developing brain, termed the ‘critical period’ (REFS 125,126). Conversely, delay of PNN formation or degradation of the PNN is associated with prolonged, or a return of, plasticity127. Furthermore, the degradation of CSPGs within the PNN is associated with a restoration of neural plasticity124,127. The density and ‘tightness’ of the ECM around neurons can inhibit their growth at the end of development69. Deposition of proteoglycans provides an adhesive surface for growth factors and then cells during axon regeneration after injury or in neurite growth125,128–130. The differential expression of proteoglycans has been implicated in various neurological conditions, including Alzheimer’s disease and epilepsy65. In particular, it has been suggested that increased secretion of CSPGs after injury inhibits regeneration and repair, which may exacerbate an initially small injury131,132. The manner in which ECM deposition and degeneration is related to neural pathogenesis is an active area of research. As researchers unravel the complexities that are responsible for the context-dependent effects of ECM alteration in the nervous system, they are also making progress towards understanding how to mitigate long-term effects of damage to the neural ECM.
Conclusions and future perspectives
The ECM has important roles in regulating cell and tissue development and function1,86. The ECM acts with specificity in different tissues of the body by taking advantage of the wide variety of ECM proteins that exist, as well as unique structures formed by interactions among these ECM components4. This diversity results in highly distinctive ECMs that can be directly linked to the particular roles they have in their respective tissues. As such, the production and assembly of the ECM is an important aspect of maintaining cell and tissue homeostasis. These processes follow different temporal and spatial rules in various tissues, with load-bearing tissues such as tendons exhibiting highly ordered, fibrillar structures and the continually evolving brain showing a less organized, GAG-rich ECM28,61,117. Therefore, disruption of the relative abundance of ECM proteins or their interactions with one another has important consequences for the behaviour and the fate of cells within that tissue5,133,134. In order to study how perturbations to ECM remodelling influence pathological conditions such as cancer and cardiovascular disease, it is necessary to understand its ground state. Researchers have made great progress in characterizing the many components of the ECM and the manners in which these components interact with one another. Although this remains an ongoing area of investigation, the next step is to more thoroughly describe the temporal and spatial mechanisms by which ECM production and assembly are regulated in cells and tissues. Genetic modifications offer a way to perturb these factors in a controlled manner, and advances in in vivo imaging techniques will facilitate observations of the resulting effects. Not only would this understanding benefit tissue engineers seeking to build organs in vitro, but pinpointing such details may also identify more nuanced approaches to therapy in cancer, inflammatory diseases and more.
Acknowledgments
The authors apologize to all colleagues whose work cannot be cited owing to space limitations. This work was supported by DOD Breast Cancer Research Program (BCRP) grant W81XWH-07-1-0538 (to J.K.M.), US National Science Foundation (NSF) Graduate Research Fellowship (to G.O.), DOD BCRP grants W81XWH-05-1-0330 and W81XWH-13-1-0216 (to V.M.W.), US National Institutes of Health National Cancer Institute (NCI) grants R01 CA138818, U54 CA143836, R01 CA085492 and U01 ES019458 (to V.M.W.), and Susan G. Komen grant KG110560PP (to V.M.W.).
Glossary
- Topographies
The three-dimensional qualities of surfaces or structures, including contours and relief. In the context of the ECM this includes features such as peaks and valleys, changes in roughness and geometric features.
- Morphogenesis
The process of cell movement during embryonic development that controls the size, the shape and the patterning of tissues and organs.
- Proteoglycans
(PGs). Glycoproteins that consist of a core protein to which one or more glycosaminoglycan chain is attached.
- Glycosaminoglycans
(GAGs). Long, linear, charged polysaccharides that comprise repeating pairs of sugars, of which one is an amino sugar.
- Fibrillar collagen
Polymerized, supramolecular collagen that has been organized into fibrils; collagen types I, II and III form fibrils.
- Tensile forces
The forces required to exert a certain amount of tension (which is a type of normal stress) on a one-dimensional object. In this context, a pulling force on a cable tending to cause extension of the cable.
- Procollagen
A trimeric collagen precursor molecule containing large amino- and carboxy-terminal propeptide domains.
- Lysyl oxidase
An enzyme that participates in the formation of collagen polymers by facilitating oxidative deamination of peptidyl lysine residues on collagen monomers.
- Pericellular space
The space surrounding a cell.
- Fibripositors
(Also known as fibropositors). Plasma membrane protrusions projecting from the cell surface, where procollagen can potentially be processed, assembled and organized by individual cells.
- Fibrillogenesis
The development or formation of fibrils, used to refer to collagen- or fibronectin-rich structures.
- Small leucine-rich repeat proteoglycans
(SLRPs). A family of proteoglycans that share a common leucine-rich-repeat motif in their conserved carboxy termini; they have been strongly implicated in modulating fibrillar collagen assembly.
- FACIT
(Fibril-associated collagens with interrupted triple helices). A type of collagen that does not form fibrils by itself but that is associated with the surface of fibrillar collagen.
- Basement membrane
A thin, complex extracellular matrix that separates endothelial and epithelial cells from their subjacent connective tissues. It is composed of various collagens, proteoglycans and adhesive glycoproteins.
- Syndecans
One of the two major families of heparin sulphate proteoglycans.
- Glypicans
One of the two major families of heparin sulphate proteoglycans.
- Epimerization
The formation of stereoisomers.
- Integrins
A large family of heterodimeric transmembrane proteins, which are present in the plasma membrane as heterodimers of α- and β-subunits and function as receptors for cell adhesion molecules.
- Nidogens
(Also known as entactins). Glycoproteins and key parts of the basement membrane.
- Fibrillar adhesions
Cell–matrix connections that are typified by their location near the centre of the cell and their higher length to width ratio compared with focal adhesions at the periphery of cells.
- Fascicles
Bundles of fibres.
- Reticular lamina
A collagen-rich layer that is often found below the basal lamina that connects the combined basement membrane to the connective tissue.
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Mecham RP. Overview of extracellular matrix. Curr Protoc Cell Biol. 2012;10(Unit 10.1) doi: 10.1002/0471143030.cb1001s00. [DOI] [PubMed] [Google Scholar]
- 2.Lu P, Takai K, Weaver VM, Werb Z. Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harb Perspect Biol. 2011;3:a005058. doi: 10.1101/cshperspect.a005058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rozario T, DeSimone DW. The extracellular matrix in development and morphogenesis: a dynamic view. Dev Biol. 2010;341:126–140. doi: 10.1016/j.ydbio.2009.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Naba A, et al. The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Mol Cell Proteomics. 2012;11:M111.014647. doi: 10.1074/mcp.M111.014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jakeman LB, Williams KE, Brautigam B. In the presence of danger: the extracellular matrix defensive response to central nervous system injury. Neural Regen Res. 2014;9:377–384. doi: 10.4103/1673-5374.128238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fratzl P, et al. Fibrillar structure and mechanical properties of collagen. J Struct Biol. 1998;122:119–122. doi: 10.1006/jsbi.1998.3966. [DOI] [PubMed] [Google Scholar]
- 7.Bonnans C, Chou J, Werb Z. Remodeling the extracellular matrix in development and diseases. Nature Rev Cell Mol Biol. 2014 doi: 10.1038/nrm3904. http://dx.doi.org/10.1038/nrm3904. [DOI] [PMC free article] [PubMed]
- 8.Humphrey JD, Dufresne ER, Schwartz AM. Mechanotransduction and extracellular matrix homeostasis. Nature Rev Cell Mol Biol. 2014 doi: 10.1038/nrm3896. http://dx.doi.org/10.1038/nrm3896. [DOI] [PMC free article] [PubMed]
- 9.Rowe RG, Weiss SJ. Breaching the basement membrane: who, when and how? Trends Cell Biol. 2008;18:560–574. doi: 10.1016/j.tcb.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 10.Schaefer L, Schaefer RM. Proteoglycans: from structural compounds to signaling molecules. Cell Tissue Res. 2010;339:237–246. doi: 10.1007/s00441-009-0821-y. This paper shows the biological structure and functions of the primarily extracellular SLRPs and highlights SLRP-associated genetic diseases and signalling events. [DOI] [PubMed] [Google Scholar]
- 11.Myllyharju J, Kivirikko KI. Collagens, modifying enzymes and their mutations in humans, flies and worms. Trends Genet. 2004;20:33–43. doi: 10.1016/j.tig.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 12.Ricard-Blum S, Ruggiero F. The collagen superfamily: from the extracellular matrix to the cell membrane. Pathol Biol (Paris) 2005;53:430–442. doi: 10.1016/j.patbio.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 13.Brodsky B, Persikov AV. Molecular structure of the collagen triple helix. Adv Protein Chem. 2005;70:301–339. doi: 10.1016/S0065-3233(05)70009-7. [DOI] [PubMed] [Google Scholar]
- 14.Ricard-Blum S. The collagen family. Cold Spring Harb Perspect Biol. 2011;3:a004978. doi: 10.1101/cshperspect.a004978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hulmes DJS. In: Collagen Struct Mech. Fratzl P, editor. Springer; 2008. pp. 15–47. This is a comprehensive review of intracellular and extracellular collagen biosynthesis and collagen fibril assembly. [Google Scholar]
- 16.Hulmes DJS. Building collagen molecules, fibrils, and suprafibrillar structures. J Struct Biol. 2002;137:2–10. doi: 10.1006/jsbi.2002.4450. [DOI] [PubMed] [Google Scholar]
- 17.Myllyharju J. Intracellular post-translational modifications of collagens. Top Curr Chem. 2005;247:115–147. [Google Scholar]
- 18.Bella J, Eaton M, Brodsky B, Berman HM. Crystal and molecular structure of a collagen-like peptide at 1.9 A resolution. Science. 1994;266:75–81. doi: 10.1126/science.7695699. [DOI] [PubMed] [Google Scholar]
- 19.Persikov AV, Ramshaw JAM, Kirkpatrick A, Brodsky B. Electrostatic interactions involving lysine make major contributions to collagen triple-helix stability. Biochemistry. 2005;44:1414–1422. doi: 10.1021/bi048216r. [DOI] [PubMed] [Google Scholar]
- 20.Bourhis J-M, et al. Structural basis of fibrillar collagen trimerization and related genetic disorders. Nature Struct Mol Biol. 2012;19:1031–1036. doi: 10.1038/nsmb.2389. This paper elucidates the crystal structure of the procollagen C propeptide to reveal an exquisite structural mechanism of α-chain selectivity during intracellular trimerization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McLaughlin SH, Bulleid NJ. Molecular recognition in procollagen chain assembly. Matrix Biol. 1998;16:369–377. doi: 10.1016/s0945-053x(98)90010-5. [DOI] [PubMed] [Google Scholar]
- 22.Lees JF, Tasab M, Bulleid NJ. Identification of the molecular recognition sequence which determines the type-specific assembly of procollagen. EMBO J. 1997;16:908–916. doi: 10.1093/emboj/16.5.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boudko SP, Engel J, Bächinger HP. The crucial role of trimerization domains in collagen folding. Int J Biochem Cell Biol. 2012;44:21–32. doi: 10.1016/j.biocel.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 24.Ellgaard L, Ruddock LW. The human protein disulphide isomerase family: substrate interactions and functional properties. EMBO Rep. 2005;6:28–32. doi: 10.1038/sj.embor.7400311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wess TJ. Collagen fibril form and function. Adv Protein Chem. 2005;70:341–374. doi: 10.1016/S0065-3233(05)70010-3. [DOI] [PubMed] [Google Scholar]
- 26.Shoulders MD, Raines RT. Collagen structure and stability. Annu Rev Biochem. 2009;78:929–958. doi: 10.1146/annurev.biochem.77.032207.120833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trelstad RL, Hayashi K. Tendon collagen fibrillogenesis: intracellular subassemblies and cell surface changes associated with fibril growth. Dev Biol. 1979;71:228–242. doi: 10.1016/0012-1606(79)90166-0. [DOI] [PubMed] [Google Scholar]
- 28.Birk DE, Trelstad RL. Extracellular compartments in tendon morphogenesis: collagen fibril, bundle, and macroaggregate formation. J Cell Biol. 1986;103:231–240. doi: 10.1083/jcb.103.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Birk DE, Zycband EI, Winkelmann DA, Trelstad RL. Collagen fibrillogenesis in situ: fibril segments are intermediates in matrix assembly. Proc Natl Acad Sci USA. 1989;86:4549–4553. doi: 10.1073/pnas.86.12.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Canty EG, et al. Coalignment of plasma membrane channels and protrusions (fibripositors) specifies the parallelism of tendon. J Cell Biol. 2004;165:553–563. doi: 10.1083/jcb.200312071. This paper shows how embryonic fibroblasts produce an ECM that is rich in elongated, parallel, collagen fibrils, through the initiation of collagen fibrillogenesis by targeting collagen fibril-containing Golgi-to-plasma membrane carriers (GPCs) to plasma membrane protrusions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Starborg T, et al. Using transmission electron microscopy and 3View to determine collagen fibril size and three-dimensional organization. Nature Protoc. 2013;8:1433–1448. doi: 10.1038/nprot.2013.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalson NS, et al. Nonmuscle myosin II powered transport of newly formed collagen fibrils at the plasma membrane. Proc Natl Acad Sci USA. 2013;110:E4743–E4752. doi: 10.1073/pnas.1314348110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Porter S, Clark IM, Kevorkian L, Edwards DR. The ADAMTS metalloproteinases. Biochem J. 2005;386:15–27. doi: 10.1042/BJ20040424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hopkins DR, Keles S, Greenspan DS. The bone morphogenetic protein 1/tolloid-like metalloproteinases. Matrix Biol. 2007;26:508–523. doi: 10.1016/j.matbio.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seidah NG, Prat A. Precursor convertases in the secretory pathway, cytosol and extracellular milieu. Essays Biochem. 2002;38:79–94. doi: 10.1042/bse0380079. [DOI] [PubMed] [Google Scholar]
- 36.Pappano WN, Steiglitz BM, Scott IC, Keene DR, Greenspan DS. Use of BMP1/TLL1 doubly homozygous null mice and proteomics to identify and validate in vivo substrates of bone morphogenetic protein 1/tolloid-like metalloproteinases. Mol Cell Biol. 2003;23:4428–4438. doi: 10.1128/MCB.23.13.4428-4438.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kadler KE, Holmes DF, Trotter JA, Chapman JA. Collagen fibril formation. Biochem J. 1996;316:1–11. doi: 10.1042/bj3160001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wenstrup RJ, et al. Type V collagen controls the initiation of collagen fibril assembly. J Biol Chem. 2004;279:53331–53337. doi: 10.1074/jbc.M409622200. [DOI] [PubMed] [Google Scholar]
- 39.Chapman JA. The staining pattern of collagen fibrils. I. An analysis of electron micrographs. Connect Tissue Res. 1974;2:137–150. doi: 10.3109/03008207409152099. [DOI] [PubMed] [Google Scholar]
- 40.Olsen BR. Electron microscope studies on collagen. I. Native collagen fibrils. Z Zellforsch Mikrosk Anat. 1963;59:184–198. doi: 10.1007/BF00320444. [DOI] [PubMed] [Google Scholar]
- 41.Petruska JA, Hodge AJ. A subunit model for the tropocollagen macromolecule. Proc Natl Acad Sci USA. 1964;51:871–876. doi: 10.1073/pnas.51.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hulmes DJ, Miller A. Quasi-hexagonal molecular packing in collagen fibrils. Nature. 1979;282:878–880. doi: 10.1038/282878a0. [DOI] [PubMed] [Google Scholar]
- 43.Hulmes DJ, Wess TJ, Prockop DJ, Fratzl P. Radial packing, order, and disorder in collagen fibrils. Biophys J. 1995;68:1661–1670. doi: 10.1016/S0006-3495(95)80391-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orgel JPRO, Irving TC, Miller A, Wess TJ. Microfibrillar structure of type I collagen in situ. Proc Natl Acad Sci USA. 2006;103:9001–9005. doi: 10.1073/pnas.0502718103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmitt FO, Hall CE, Jakus MA. Electron microscope investigations of the structure of collagen. J Cell Comp Physiol. 1942;20:11–33. [Google Scholar]
- 46.Bruckner P. Suprastructures of extracellular matrices: paradigms of functions controlled by aggregates rather than molecules. Cell Tissue Res. 2010;339:7–18. doi: 10.1007/s00441-009-0864-0. [DOI] [PubMed] [Google Scholar]
- 47.Ameye L, Young MF. Mice deficient in small leucine-rich proteoglycans: novel in vivo models for osteoporosis, osteoarthritis, Ehlers–Danlos syndrome, muscular dystrophy, and corneal diseases. Glycobiology. 2002;12:107R–116R. doi: 10.1093/glycob/cwf065. [DOI] [PubMed] [Google Scholar]
- 48.Kalamajski S, Oldberg A. The role of small leucine-rich proteoglycans in collagen fibrillogenesis. Matrix Biol. 2010;29:248–253. doi: 10.1016/j.matbio.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 49.Molnar J, et al. Structural and functional diversity of lysyl oxidase and the LOX-like proteins. Biochim Biophys Acta. 2003;1647:220–224. doi: 10.1016/s1570-9639(03)00053-0. [DOI] [PubMed] [Google Scholar]
- 50.Eyre DR, Weis MA, Wu JJ. Advances in collagen cross-link analysis. Methods. 2008;45:65–74. doi: 10.1016/j.ymeth.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eyre DR, Glimcher MJ. Collagen cross-linking. Isolation of cross-linked peptides from collagen of chicken bone. Biochem J. 1973;135:393–403. doi: 10.1042/bj1350393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lucero HA, Kagan HM. Lysyl oxidase: an oxidative enzyme and effector of cell function. Cell Mol Life Sci. 2006;63:2304–2316. doi: 10.1007/s00018-006-6149-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rucker RB, Murray J. Cross-linking amino acids in collagen and elastin. Am J Clin Nutr. 1978;31:1221–1236. doi: 10.1093/ajcn/31.7.1221. [DOI] [PubMed] [Google Scholar]
- 54.Orgel JP, Wess TJ, Miller A. The in situ conformation and axial location of the intermolecular cross-linked non-helical telopeptides of type I collagen. Structure. 2000;8:137–142. doi: 10.1016/s0969-2126(00)00089-7. [DOI] [PubMed] [Google Scholar]
- 55.O’Leary LER, Fallas JA, Bakota EL, Kang MK, Hartgerink JD. Multi-hierarchical self-assembly of a collagen mimetic peptide from triple helix to nanofibre and hydrogel. Nature Chem. 2011;3:821–828. doi: 10.1038/nchem.1123. [DOI] [PubMed] [Google Scholar]
- 56.Bandtlow CE, Zimmermann DR. Proteoglycans in the developing brain: new conceptual insights for old proteins. Physiol Rev. 2000;80:1267–1290. doi: 10.1152/physrev.2000.80.4.1267. [DOI] [PubMed] [Google Scholar]
- 57.Knudson CB, Knudson W. Cartilage proteoglycans. Semin Cell Dev Biol. 2001;12:69–78. doi: 10.1006/scdb.2000.0243. [DOI] [PubMed] [Google Scholar]
- 58.Cui H, Freeman C, Jacobson GA, Small DH. Proteoglycans in the central nervous system: role in development, neural repair, and Alzheimer’s disease. IUBMB Life. 2013;65:108–120. doi: 10.1002/iub.1118. [DOI] [PubMed] [Google Scholar]
- 59.Schwartz NB, Domowicz M. Proteoglycans in brain development. Glycoconj J. 2004;21:329–341. doi: 10.1023/B:GLYC.0000046278.34016.36. [DOI] [PubMed] [Google Scholar]
- 60.Zhang L, editor. Glycosaminoglycans in Development, Health and Disease. Academic; 2010. [DOI] [PubMed] [Google Scholar]
- 61.Deepa SS, et al. Composition of perineuronal net extracellular matrix in rat brain: a different disaccharide composition for the net-associated proteoglycans. J Biol Chem. 2006;281:17789–17800. doi: 10.1074/jbc.M600544200. [DOI] [PubMed] [Google Scholar]
- 62.Simon Davis DA, Parish CR. Heparan sulfate: a ubiquitous glycosaminoglycan with multiple roles in immunity. Front Immunol. 2013;4:470. doi: 10.3389/fimmu.2013.00470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bishop JR, Schuksz M, Esko JD. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature. 2007;446:1030–1037. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- 64.Esko JD, Kimata K, Lindahl U. Essentials of Glycobiology. 2. Ch 16. Cold Spring Harbor; 2009. [Google Scholar]
- 65.Wade A, McKinney A, Phillips JJ. Matrix regulators in neural stem cell functions. Biochim Biophys Acta. 2014 doi: 10.1016/j.bbagen.2014.01.017. http://dx.doi.org/10.1016/j.bbagen.2014.01.017. [DOI] [PMC free article] [PubMed]
- 66.Johnson KG, et al. Axonal heparan sulfate proteoglycans regulate the distribution and efficiency of the repellent slit during midline axon guidance. Curr Biol. 2004;14:499–504. doi: 10.1016/j.cub.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 67.Häcker U, Nybakken K, Perrimon N. Heparan sulphate proteoglycans: the sweet side of development. Nature Rev Mol Cell Biol. 2005;6:530–541. doi: 10.1038/nrm1681. [DOI] [PubMed] [Google Scholar]
- 68.Maeda N, Ishii M, Nishimura K, Kamimura K. Functions of chondroitin sulfate and heparan sulfate in the developing brain. Neurochem Res. 2011;36:1228–1240. doi: 10.1007/s11064-010-0324-y. [DOI] [PubMed] [Google Scholar]
- 69.Yamaguchi Y. Lecticans: organizers of the brain extracellular matrix. Cell Mol Life Sci. 2000;57:276–289. doi: 10.1007/PL00000690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lundell A, et al. Structural basis for interactions between tenascins and lectican C-type lectin domains: evidence for a crosslinking role for tenascins. Structure. 2004;12:1495–1506. doi: 10.1016/j.str.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 71.Dellett M, Hu W, Papadaki V, Ohnuma S. Small leucine rich proteoglycan family regulates multiple signalling pathways in neural development and maintenance. Dev Growth Differ. 2012;54:327–340. doi: 10.1111/j.1440-169X.2012.01339.x. [DOI] [PubMed] [Google Scholar]
- 72.McEwan PA, Scott PG, Bishop PN, Bella J. Structural correlations in the family of small leucine-rich repeat proteins and proteoglycans. J Struct Biol. 2006;155:294–305. doi: 10.1016/j.jsb.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 73.Schaefer L, Iozzo RV. Biological functions of the small leucine-rich proteoglycans: from genetics to signal transduction. J Biol Chem. 2008;283:21305–21309. doi: 10.1074/jbc.R800020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen S, Birk DE. The regulatory roles of small leucine-rich proteoglycans in extracellular matrix assembly. FEBS J. 2013;280:2120–2137. doi: 10.1111/febs.12136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Oohira A, Matsui F, Tokita Y, Yamauchi S, Aono S. Molecular interactions of neural chondroitin sulfate proteoglycans in the brain development. Arch Biochem Biophys. 2000;374:24–34. doi: 10.1006/abbi.1999.1598. [DOI] [PubMed] [Google Scholar]
- 76.Timpl R, Brown JC. The laminins. Matrix Biol. 1994;14:275–281. doi: 10.1016/0945-053x(94)90192-9. [DOI] [PubMed] [Google Scholar]
- 77.Aumailley M, et al. A simplified laminin nomenclature. Matrix Biol. 2005;24:326–332. doi: 10.1016/j.matbio.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 78.Miner JH, Yurchenco PD. Laminin functions in tissue morphogenesis. Annu Rev Cell Dev Biol. 2004;20:255–284. doi: 10.1146/annurev.cellbio.20.010403.094555. This paper discusses how different laminin proteins affect invertebrate and vertebrate tissue morphogenesis through the induction and the maintenance of cell polarity, the establishment of barriers between tissue compartments and the protection of adherent cells from anoikis. [DOI] [PubMed] [Google Scholar]
- 79.Mercurio AM, Shaw LM. Laminin binding proteins. Bioessays. 1991;13:469–473. doi: 10.1002/bies.950130907. [DOI] [PubMed] [Google Scholar]
- 80.Yurchenco PD, Smirnov S, Mathus T. Analysis of basement membrane self-assembly and cellular interactions with native and recombinant glycoproteins. Methods Cell Biol. 2002;69:111–144. doi: 10.1016/s0091-679x(02)69010-7. [DOI] [PubMed] [Google Scholar]
- 81.Schwarzbauer JE, DeSimone DW. Fibronectins, their fibrillogenesis, and in vivo functions. Cold Spring Harb Perspect Biol. 2011;3:a005041. doi: 10.1101/cshperspect.a005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Singh P, Carraher C, Schwarzbauer JE. Assembly of fibronectin extracellular matrix. Annu Rev Cell Dev Biol. 2010;26:397–419. doi: 10.1146/annurev-cellbio-100109-104020. This is an excellent review highlighting the major steps, molecular interactions and cellular mechanisms involved in assembling fibronectin into a fibrillar matrix. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hynes RO. In: Guidebook to the Extracellular Matrix, Anchor and Adhesion Proteins. 2. Kreis T, Vale R, editors. Oxford University Press; 1999. pp. 422–425. [Google Scholar]
- 84.García AJ, Schwarzbauer JE, Boettiger D. Distinct activation states of α5β1 integrin show differential binding to RGD and synergy domains of fibronectin. Biochemistry. 2002;41:9063–9069. doi: 10.1021/bi025752f. [DOI] [PubMed] [Google Scholar]
- 85.Ilić D, et al. FAK promotes organization of fibronectin matrix and fibrillar adhesions. J Cell Sci. 2004;117:177–187. doi: 10.1242/jcs.00845. [DOI] [PubMed] [Google Scholar]
- 86.Hay ED. Extracellular matrix alters epithelial differentiation. Curr Opin Cell Biol. 1993;5:1029–1035. doi: 10.1016/0955-0674(93)90088-8. [DOI] [PubMed] [Google Scholar]
- 87.Birk DE, Zycband EI, Woodruff S, Winkelmann DA, Trelstad RL. Collagen fibrillogenesis in situ: fibril segments become long fibrils as the developing tendon matures. Dev Dyn. 1997;208:291–298. doi: 10.1002/(SICI)1097-0177(199703)208:3<291::AID-AJA1>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 88.Zhang G, et al. Development of tendon structure and function: regulation of collagen fibrillogenesis. J Musculoskelet Neuronal Interact. 2005;5:5–21. [PubMed] [Google Scholar]
- 89.Wang JHC. Mechanobiology of tendon. J Biomech. 2006;39:1563–1582. doi: 10.1016/j.jbiomech.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 90.Kannus P. Structure of the tendon connective tissue. Scand J Med Sci Sports. 2000;10:312–320. doi: 10.1034/j.1600-0838.2000.010006312.x. [DOI] [PubMed] [Google Scholar]
- 91.Tresoldi I, et al. Tendon’s ultrastructure. Muscles Ligaments Tendons J. 2013;3:2–6. doi: 10.11138/mltj/2013.3.1.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mienaltowski MJ, Birk DE. Structure, physiology, and biochemistry of collagens. Adv Exp Med Biol. 2014;802:5–29. doi: 10.1007/978-94-007-7893-1_2. [DOI] [PubMed] [Google Scholar]
- 93.Craig AS, Birtles MJ, Conway JF, Parry DA. An estimate of the mean length of collagen fibrils in rat tail-tendon as a function of age. Connect Tissue Res. 1989;19:51–62. doi: 10.3109/03008208909016814. [DOI] [PubMed] [Google Scholar]
- 94.Tozer S, Duprez D. Tendon and ligament: development, repair and disease. Birth Defects Res C Embryo Today. 2005;75:226–236. doi: 10.1002/bdrc.20049. [DOI] [PubMed] [Google Scholar]
- 95.Richardson SH, et al. Tendon development requires regulation of cell condensation and cell shape via cadherin-11-mediated cell-cell junctions. Mol Cell Biol. 2007;27:6218–6228. doi: 10.1128/MCB.00261-07. This paper describes how the precise regulation of cell shape via cell–cell junctions and the coaxial alignment of plasma membrane channels in longitudinally stacked cells drives tendon formation in the developing limb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sasaki N, Odajima S. Elongation mechanism of collagen fibrils and force-strain relations of tendon at each level of structural hierarchy. J Biomech. 1996;29:1131–1136. doi: 10.1016/0021-9290(96)00024-3. [DOI] [PubMed] [Google Scholar]
- 97.Graham HK, Holmes DF, Watson RB, Kadler KE. Identification of collagen fibril fusion during vertebrate tendon morphogenesis. The process relies on unipolar fibrils and is regulated by collagen-proteoglycan interaction. J Mol Biol. 2000;295:891–902. doi: 10.1006/jmbi.1999.3384. [DOI] [PubMed] [Google Scholar]
- 98.Paulsson M. Basement membrane proteins: structure, assembly, and cellular interactions. Crit Rev Biochem Mol Biol. 1992;27:93–127. doi: 10.3109/10409239209082560. [DOI] [PubMed] [Google Scholar]
- 99.Martin Rupert GRT. Laminin and other basement membrane components. Annu Rev Cell Biol. 1987;3:57–85. doi: 10.1146/annurev.cb.03.110187.000421. [DOI] [PubMed] [Google Scholar]
- 100.Kalluri R. Basement membranes: structure, assembly and role in tumour angiogenesis. Nature Rev Cancer. 2003;3:422–433. doi: 10.1038/nrc1094. This is an excellent review highlighting the constituents, structure and assembly of the basement membrane, with emphasis on angiogenesis and vascular basement membranes. [DOI] [PubMed] [Google Scholar]
- 101.Yurchenco PD. Basement membranes: cell scaffoldings and signaling platforms. Cold Spring Harb Perspect Biol. 2011;3:a004911. doi: 10.1101/cshperspect.a004911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yurchenco PD, Schittny JC. Molecular architecture of basement membranes. FASEB J. 1990;4:1577–1590. doi: 10.1096/fasebj.4.6.2180767. [DOI] [PubMed] [Google Scholar]
- 103.Sanes JR. The basement membrane/basal lamina of skeletal muscle. J Biol Chem. 2003;278:12601–12604. doi: 10.1074/jbc.R200027200. [DOI] [PubMed] [Google Scholar]
- 104.Yurchenco PD, Patton BL. Developmental and pathogenic mechanisms of basement membrane assembly. Curr Pharm Des. 2009;15:1277–1294. doi: 10.2174/138161209787846766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Takagi J, Yang Y, Liu JH, Wang JH, Springer TA. Complex between nidogen and laminin fragments reveals a paradigmatic β-propeller interface. Nature. 2003;424:969–974. doi: 10.1038/nature01873. [DOI] [PubMed] [Google Scholar]
- 106.Pöschl E, et al. Collagen IV is essential for basement membrane stability but dispensable for initiation of its assembly during early development. Development. 2004;131:1619–1628. doi: 10.1242/dev.01037. [DOI] [PubMed] [Google Scholar]
- 107.Iozzo RV, Zoeller JJ, Nyström A. Basement membrane proteoglycans: modulators Par Excellence of cancer growth and angiogenesis. Mol Cells. 2009;27:503–513. doi: 10.1007/s10059-009-0069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Costello I, Biondi CA, Taylor JM, Bikoff EK, Robertson EJ. Smad4-dependent pathways control basement membrane deposition and endodermal cell migration at early stages of mouse development. BMC Dev Biol. 2009;9:54. doi: 10.1186/1471-213X-9-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wiradjaja F, DiTommaso T, Smyth I. Basement membranes in development and disease. Birth Defects Res C Embryo Today. 2010;90:8–31. doi: 10.1002/bdrc.20172. [DOI] [PubMed] [Google Scholar]
- 110.Paralkar VM, Vukicevic S, Reddi AH. Transforming growth factor-β type 1 binds to collagen IV of basement membrane matrix: Implications for development. Dev Biol. 1991;143:303–308. doi: 10.1016/0012-1606(91)90081-d. [DOI] [PubMed] [Google Scholar]
- 111.Streuli CH. Control of mammary epithelial differentiation: basement membrane induces tissue-specific gene expression in the absence of cell–cell interaction and morphological polarity. J Cell Biol. 1991;115:1383–1395. doi: 10.1083/jcb.115.5.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Camelo A, Dunmore R, Sleeman MA, Clarke DL. The epithelium in idiopathic pulmonary fibrosis: breaking the barrier. Front Pharmacol. 2014;4:173. doi: 10.3389/fphar.2013.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Torricelli AAM, Singh V, Santhiago MR, Wilson SE. The corneal epithelial basement membrane: structure, function, and disease. Invest Ophthalmol Vis Sci. 2013;54:6390–6400. doi: 10.1167/iovs.13-12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Brownell AG, Slavkin HC. Role of basal lamina in tissue interactions. Ren Physiol. 1980;3:193–204. doi: 10.1159/000172761. [DOI] [PubMed] [Google Scholar]
- 115.Lau LW, Cua R, Keough MB, Haylock-Jacobs S, Yong VW. Pathophysiology of the brain extracellular matrix: a new target for remyelination. Nature Rev Neurosci. 2013;14:722–729. doi: 10.1038/nrn3550. This paper shows the organization of the brain ECM, emphasizing the pathophysiological roles of CSPGs after neural injury. [DOI] [PubMed] [Google Scholar]
- 116.Plantman S. Proregenerative properties of ECM molecules. Biomed Res Int. 2013;2013:981695. doi: 10.1155/2013/981695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ruoslahti E. Brain extracellular matrix. Glycobiology. 1996;6:489–492. doi: 10.1093/glycob/6.5.489. [DOI] [PubMed] [Google Scholar]
- 118.Barros CS, Franco SJ, Müller U. Extracellular matrix: functions in the nervous system. Cold Spring Harb Perspect Biol. 2011;3:a005108. doi: 10.1101/cshperspect.a005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chiquet-Ehrismann R, Tucker RP. Tenascins and the importance of adhesion modulation. Cold Spring Harb Perspect Biol. 2011;3:a004960. doi: 10.1101/cshperspect.a004960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dityatev A. Remodeling of extracellular matrix and epileptogenesis. Epilepsia. 2010;51(Suppl 3):61–65. doi: 10.1111/j.1528-1167.2010.02612.x. [DOI] [PubMed] [Google Scholar]
- 121.Lajtha A, Banik N. Handbook of Neurochemistry and Molecular Neurobiology: Neural Protein Metabolism and Function. Vol. 3. Springer; 2007. [Google Scholar]
- 122.Howell MD, Gottschall PE. Lectican proteoglycans, their cleaving metalloproteinases, and plasticity in the central nervous system extracellular microenvironment. Neuroscience. 2012;217:6–18. doi: 10.1016/j.neuroscience.2012.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Carulli D, Rhodes KE, Fawcett JW. Upregulation of aggrecan, link protein 1, and hyaluronan synthases during formation of perineuronal nets in the rat cerebellum. J Comp Neurol. 2007;501:83–94. doi: 10.1002/cne.21231. [DOI] [PubMed] [Google Scholar]
- 124.Frischknecht R, Gundelfinger ED. The brain’s extracellular matrix and its role in synaptic plasticity. Adv Exp Med Biol. 2012;970:153–171. doi: 10.1007/978-3-7091-0932-8_7. [DOI] [PubMed] [Google Scholar]
- 125.Kwok JCF, Dick G, Wang D, Fawcett JW. Extracellular matrix and perineuronal nets in CNS repair. Dev Neurobiol. 2011;71:1073–1089. doi: 10.1002/dneu.20974. [DOI] [PubMed] [Google Scholar]
- 126.Wang D, Fawcett J. The perineuronal net and the control of CNS plasticity. Cell Tissue Res. 2012;349:147–160. doi: 10.1007/s00441-012-1375-y. [DOI] [PubMed] [Google Scholar]
- 127.Romberg C, et al. Depletion of perineuronal nets enhances recognition memory and long-term depression in the perirhinal cortex. J Neurosci. 2013;33:7057–7065. doi: 10.1523/JNEUROSCI.6267-11.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Grimpe B, Silver J. The extracellular matrix in axon regeneration. Prog Brain Res. 2002;137:333–349. doi: 10.1016/s0079-6123(02)37025-0. [DOI] [PubMed] [Google Scholar]
- 129.Fitch MT, Silver J. Glial cell extracellular matrix: boundaries for axon growth in development and regeneration. Cell Tissue Res. 1997;290:379–384. doi: 10.1007/s004410050944. [DOI] [PubMed] [Google Scholar]
- 130.Ishii M, Maeda N. Oversulfated chondroitin sulfate plays critical roles in the neuronal migration in the cerebral cortex. J Biol Chem. 2008;283:32610–32620. doi: 10.1074/jbc.M806331200. [DOI] [PubMed] [Google Scholar]
- 131.Sharma K, Selzer ME, Li S. Scar-mediated inhibition and CSPG receptors in the CNS. Exp Neurol. 2012;237:370–378. doi: 10.1016/j.expneurol.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bartus K, James ND, Bosch KD, Bradbury EJ. Chondroitin sulphate proteoglycans: key modulators of spinal cord and brain plasticity. Exp Neurol. 2012;235:5–17. doi: 10.1016/j.expneurol.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 133.Li AH, Liu PP, Villarreal FJ, Garcia RA. Dynamic changes in myocardial matrix and relevance to disease: translational perspectives. Circ Res. 2014;114:916–927. doi: 10.1161/CIRCRESAHA.114.302819. [DOI] [PubMed] [Google Scholar]
- 134.Page-McCaw A, Ewald AJ, Werb Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nature Rev Mol Cell Biol. 2007;8:221–233. doi: 10.1038/nrm2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fan D, Takawale A, Lee J, Kassiri Z. Cardiac fibroblasts, fibrosis and extracellular matrix remodeling in heart disease. Fibrogen Tissue Repair. 2012;5:15. doi: 10.1186/1755-1536-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nickla H, Klug WS, Cummings MR. Concepts of Genetics. Prentice Hall; 1997. [Google Scholar]
- 137.Jäälinoja J, Ylöstalo J, Beckett W, Hulmes DJS, Ala-Kokko L. Trimerization of collagen IX α-chains does not require the presence of the COL1 and NC1 domains. Biochem J. 2008;409:545–554. doi: 10.1042/BJ20070984. [DOI] [PubMed] [Google Scholar]
- 138.Liu Y, Ramanath HS, Wang DA. Tendon tissue engineering using scaffold enhancing strategies. Trends Biotechnol. 2008;26:201–209. doi: 10.1016/j.tibtech.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 139.Edwards IJ. Proteoglycans in prostate cancer. Nature Rev Urol. 2012;9:196–206. doi: 10.1038/nrurol.2012.19. [DOI] [PubMed] [Google Scholar]
- 140.Rudd TR, et al. A key biological signalling system promises new routes to intervention in medically important processes. Diamond Science. 2014 online http://www.diamond.ac.uk/Science/Research/Highlights/study2.html.
- 141.Marinkovich MP. Tumour microenvironment: laminin 332 in squamous-cell carcinoma. Nature Rev Cancer. 2007;7:370–380. doi: 10.1038/nrc2089. [DOI] [PubMed] [Google Scholar]
- 142.Bezakova G, Ruegg MA. New insights into the roles of agrin. Nature Rev Mol Cell Biol. 2003;4:295–308. doi: 10.1038/nrm1074. [DOI] [PubMed] [Google Scholar]
- 143.Meier T, Wallace BG. Formation of the neuromuscular junction: molecules and mechanisms. Bioessays. 1998;20:819–829. doi: 10.1002/(SICI)1521-1878(199810)20:10<819::AID-BIES7>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 144.McMahan UJ, et al. Agrin isoforms and their role in synaptogenesis. Curr Opin Cell Biol. 1992;4:869–874. doi: 10.1016/0955-0674(92)90113-q. [DOI] [PubMed] [Google Scholar]
- 145.Fisher LW, Torchia DA, Fohr B, Young MF, Fedarko NS. Flexible structures of SIBLING proteins, bone sialoprotein, and osteopontin. Biochem Biophys Res Commun. 2001;280:460–465. doi: 10.1006/bbrc.2000.4146. [DOI] [PubMed] [Google Scholar]
- 146.Bellahcène A, Castronovo V, Ogbureke KUE, Fisher LW, Fedarko NS. Small integrin-binding ligand N-linked glycoproteins (SIBLINGs): multifunctional proteins in cancer. Nature Rev Cancer. 2008;8:212–226. doi: 10.1038/nrc2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ogata Y. Bone sialoprotein and its transcriptional regulatory mechanism. J Periodontal Res. 2008;43:127–135. doi: 10.1111/j.1600-0765.2007.01014.x. [DOI] [PubMed] [Google Scholar]
- 148.Deák F, Wagener R, Kiss I, Paulsson M. The matrilins: a novel family of oligomeric extracellular matrix proteins. Matrix Biol. 1999;18:55–64. doi: 10.1016/s0945-053x(98)00006-7. [DOI] [PubMed] [Google Scholar]
- 149.Klatt AR, Becker AKA, Neacsu CD, Paulsson M, Wagener R. The matrilins: modulators of extracellular matrix assembly. Int J Biochem Cell Biol. 2011;43:320–330. doi: 10.1016/j.biocel.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 150.Wagener R, et al. The matrilins — adaptor proteins in the extracellular matrix. FEBS Lett. 2005;579:3323–3329. doi: 10.1016/j.febslet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 151.Chen Q, Johnson DM, Haudenschild DR, Goetinck PF. Cartilage matrix protein: expression patterns in chicken, mouse, and human. Ann NY Acad Sci. 1996;785:238–240. doi: 10.1111/j.1749-6632.1996.tb56271.x. [DOI] [PubMed] [Google Scholar]
- 152.Mithieux SM, Weiss AS. Elastin. Adv Protein Chem. 2005;70:437–461. doi: 10.1016/S0065-3233(05)70013-9. [DOI] [PubMed] [Google Scholar]
- 153.Halper J, Kjaer M. Basic components of connective tissues and extracellular matrix: elastin, fibrillin, fibulins, fibrinogen, fibronectin, laminin, tenascins and thrombospondins. Adv Exp Med Biol. 2014;802:31–47. doi: 10.1007/978-94-007-7893-1_3. [DOI] [PubMed] [Google Scholar]
- 154.Pepe A, Bochicchio B, Tamburro AM. Supramolecular organization of elastin and elastin-related nanostructured biopolymers. Nanomed. 2007;2:203–218. doi: 10.2217/17435889.2.2.203. [DOI] [PubMed] [Google Scholar]
- 155.Wagenseil JE, Mecham RP. New insights into elastic fiber assembly. Birth Defects Res C Embryo Today. 2007;81:229–240. doi: 10.1002/bdrc.20111. [DOI] [PubMed] [Google Scholar]
- 156.Weisel JW. Fibrinogen and fibrin. Adv Protein Chem. 2005;70:247–299. doi: 10.1016/S0065-3233(05)70008-5. [DOI] [PubMed] [Google Scholar]
- 157.Mosesson MW, Siebenlist KR, Meh DA. The structure and biological features of fibrinogen and fibrin. Ann NY Acad Sci. 2001;936:11–30. doi: 10.1111/j.1749-6632.2001.tb03491.x. [DOI] [PubMed] [Google Scholar]
- 158.Ramirez F, Sakai LY, Dietz HC, Rifkin DB. Fibrillin microfibrils: multipurpose extracellular networks in organismal physiology. Physiol Genom. 2004;19:151–154. doi: 10.1152/physiolgenomics.00092.2004. [DOI] [PubMed] [Google Scholar]
- 159.Kielty CM, et al. Fibrillin: from microfibril assembly to biomechanical function. Phil Trans R Soc. 2002;357:207–217. doi: 10.1098/rstb.2001.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Timpl R, Sasaki T, Kostka G, Chu ML. Fibulins: a versatile family of extracellular matrix proteins. Nature Rev Mol Cell Biol. 2003;4:479–489. doi: 10.1038/nrm1130. [DOI] [PubMed] [Google Scholar]
- 161.De Vega S, Iwamoto T, Yamada Y. Fibulins: multiple roles in matrix structures and tissue functions. Cell Mol Life Sci. 2009;66:1890–1902. doi: 10.1007/s00018-009-8632-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cirulli V, Yebra M. Netrins: beyond the brain. Nature Rev Mol Cell Biol. 2007;8:296–306. doi: 10.1038/nrm2142. [DOI] [PubMed] [Google Scholar]
- 163.Lai Wing Sun K, Correia JP, Kennedy TE. Netrins: versatile extracellular cues with diverse functions. Development. 2011;138:2153–2169. doi: 10.1242/dev.044529. [DOI] [PubMed] [Google Scholar]
- 164.Bradshaw AD. Diverse biological functions of the SPARC family of proteins. Int J Biochem Cell Biol. 2012;44:480–488. doi: 10.1016/j.biocel.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Brekken RA, Sage EH. SPARC, a matricellular protein: at the crossroads of cell-matrix communication. Matrix Biol. 2001;19:816–827. doi: 10.1016/s0945-053x(00)00133-5. [DOI] [PubMed] [Google Scholar]
- 166.Giachelli CM, Steitz S. Osteopontin: a versatile regulator of inflammation and biomineralization. Matrix Biol. 2000;19:615–622. doi: 10.1016/s0945-053x(00)00108-6. [DOI] [PubMed] [Google Scholar]
- 167.Hunter GK. Role of osteopontin in modulation of hydroxyapatite formation. Calcif Tissue Int. 2013;93:348–354. doi: 10.1007/s00223-013-9698-6. [DOI] [PubMed] [Google Scholar]
- 168.Fatemi SH. Reelin glycoprotein: structure, biology and roles in health and disease. Mol Psychiatry. 2005;10:251–257. doi: 10.1038/sj.mp.4001613. [DOI] [PubMed] [Google Scholar]
- 169.Magdaleno SM, Curran T. Brain development: integrins and the Reelin pathway. Curr Biol. 2001;11:R1032–R1035. doi: 10.1016/s0960-9822(01)00618-2. [DOI] [PubMed] [Google Scholar]
- 170.Vincent TL, Woolfson DN, Adams JC. Prediction and analysis of higher-order coiled-coils: insights from proteins of the extracellular matrix, tenascins and thrombospondins. Int J Biochem Cell Biol. 2013;45:2392–2401. doi: 10.1016/j.biocel.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 171.Tucker RP, Chiquet-Ehrismann R. The regulation of tenascin expression by tissue microenvironments. Biochim Biophys Acta. 2009;1793:888–892. doi: 10.1016/j.bbamcr.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 172.Leavesley DI, et al. Vitronectin — master controller or micromanager? IUBMB Life. 2013;65:807–818. doi: 10.1002/iub.1203. [DOI] [PubMed] [Google Scholar]
- 173.Preissner KT, Reuning U. Vitronectin in vascular context: facets of a multitalented matricellular protein. Semin Thromb Hemost. 2011;37:408–424. doi: 10.1055/s-0031-1276590. [DOI] [PubMed] [Google Scholar]