Summary
Incursions of new pathogenic viruses into humans from animal reservoirs are occurring with alarming frequency. The molecular underpinnings of immune recognition, host responses, and pathogenesis in this setting are poorly understood. We studied pandemic influenza viruses to determine the mechanism by which increasing glycosylation during evolution of surface proteins facilitates diminished pathogenicity in adapted viruses. ER stress during infection with poorly glycosylated pandemic strains activated the unfolded protein response, leading to inflammation, acute lung injury, and mortality. Seasonal strains or viruses engineered to mimic adapted viruses displaying excess glycans on the hemagglutinin did not cause ER stress, allowing preservation of the lungs and survival. We propose that ER stress resulting from recognition of non-adapted viruses is utilized to discriminate “non-self” at the level of protein processing and to activate immune responses, with unintended consequences on pathogenesis. Understanding this mechanism should improve strategies for treating acute lung injury from zoonotic viral infections.
Graphical Abstract
Highlights
-
•
ER stress pathways can mediate immune recognition of zoonotic viruses
-
•
Glycosylation status of viral proteins regulates activation of ER stress
-
•
Acute lung injury from pandemic influenza viruses is dependent on this activation
-
•
Adaptation through glycan addition mediates immune escape of seasonal IAV
Hrincius et al. show that the recognition of poorly glycosylated viral proteins of pandemic influenza by an ER stress pathway is utilized as an intracellular sensor of non-self at the level of protein processing and results in acute lung injury.
Introduction
Societal changes, characterized by increased global traffic and a breakdown in historical barriers to transit, have fundamentally altered the natural history of viral infections, allowing sporadic spread of infectious agents around the world within days. Some of these outbreaks, exemplified by the severe acute respiratory syndrome virus (SARS), highly pathogenic avian influenza viruses (e.g., H5N1 subtype strains), and the Middle Eastern respiratory syndrome coronavirus (MERS-CoV), result in high morbidity and mortality and cause significant disruptions to society due to public fears of infection and the necessary governmental responses (Enserink, 2014, van den Brand et al., 2014). Interestingly, not all viral infections result in pathology. Many viruses have adapted well enough to humans that they can undergo robust replication cycles with little to no evidence of immune recognition and response or at least no immune responses that are damaging to the host. As an example, Sendai virus, a close relative of the human respiratory pathogen parainfluenza virus-1, infects humans in an analogous manner to related respiratory viruses but causes no disease (Faísca and Desmecht, 2007). The interplay of virus and host factors that dictate pathology in these various infection scenarios is very poorly understood.
Influenza A viruses (IAVs) represent an interesting and relevant model to study this phenomenon. All IAVs are zoonoses, emerging periodically from animal reservoirs and sometimes establishing endemicity in humans (Webster et al., 1992). When these initial incursions spread worldwide, they are termed pandemics and are often characterized by severe morbidity and high mortality as exemplified by the 1918 H1N1 pandemic, which killed more than 50 million persons. However, over several decades of circulation, these strains adapt and generally become less pathogenic. The changes to the virus that result in reduced morbidity and the corresponding host pathways and processes that are involved are only now being explicated. Although several mechanisms are likely responsible, the tendency of seasonal influenza strains to gain excess glycosylation on the surface protein hemagglutinin (HA) has been identified as a key factor (El Moussi et al., 2014, Kim and Park, 2012, Medina et al., 2013, Sun et al., 2013, Vigerust et al., 2007, Zhang et al., 2013). Mechanistically, gain of glycosylation sites on the HA globular head has been linked to increased collagenous lectin (collectin) binding, resulting in virus neutralization (Tate et al., 2014, Vigerust et al., 2007). Physical alteration of HA-receptor-binding sites, thereby affecting receptor binding specificity and avidity and potentially altering virus tropism (Jayaraman et al., 2012, Wang et al., 2009), has also been studied. However, the observation of differential inflammatory potential based on glycosylation differences with downstream effects on pathogenesis has not been explored.
The HA protein, a 200-kDa homotrimer with an ectodomain composed of a globular head and a stalk region (Wilson et al., 1981) mediates attachment of the viral particle to the host cell and escape from the endosome (Luo, 2012). Newly produced HA is sorted into the protein secretory pathway through translocation into the ER. During transport to the plasma membrane, the HA undergoes posttranslational modifications in the ER and Golgi apparatus, where glycans are added to specific sites for N-linked glycosylation. Some of these glycans, primarily in the stalk region, are needed for proper protein folding and transport (Daniels et al., 2003, Roberts et al., 1993). Others, on the globular head, have no known function other than a proposed role in facilitation of immune escape from antibodies (Job et al., 2013, Tate et al., 2014). In general, glycosylation has important roles in protein folding and quality control with the ER integrating protein quality control mechanisms, such as cellular responses to the accumulation of non-native or misfolded proteins (the ER stress or unfolded protein response [UPR]) and their potential elimination (Roth et al., 2010). Alterations and/or disturbances in protein glycosylation have been connected to human disease (Gregersen et al., 2006, Roth et al., 2010).
Because previous studies have linked HA glycosylation to IAV pathogenicity, we wanted to further explore the interplay between the HA protein and host pathways to better understand fundamental relationships in virus recognition, induction of immune responses, and subsequent pathology. Our findings revealed a mechanism for self/non-self-discrimination by which poorly glycosylated virus proteins, such as those from pandemic IAV strains, are recognized as non-self, mediating virus recognition, airway inflammatory responses, and pathogenicity. Viruses that have adapted to humans through selection for excess glycosylation sites or viruses artificially constructed to carry excess glycans are not recognized by this innate sensing mechanism and are able to replicate without pathologic consequences.
Results
The Pandemic HA Drives Pathogenesis
To understand the differences in the pathobiology of pandemic and seasonal IAVs, we investigated immune responses and pathology using relevant, naturally occurring strains. Infection of mice with the most recent 2009 pandemic H1N1 IAV resulted in much-stronger lung immune responses and weight loss than infection with recent seasonal H1N1 and H3N2 IAVs (Figures 1A and 1B). The HA protein of this pandemic IAV is characterized by poor glycosylation of the globular head, as are all known pandemic strains. After previous introductions of pandemic IAVs including the 1918 H1N1 and 1968 H3N2 strains into humans, these IAV viruses subsequently acquired additional glycosylation sites on the HA globular head during adaptation to humans (Vigerust et al., 2007). To understand the impact of these glycosylation changes on IAV pathogenicity and adaptation to humans, we utilized viruses carrying either 1968 pandemic (HK; poorly glycosylated) or 1999 seasonal (Pan; heavily glycosylated) H3N2 HA/NA proteins in various combinations. These “6:2” IAVs created by the eight-plasmid reverse genetics system carry the six internal gene segments from the IAV laboratory strain PR8 and the appropriate gene segments to express the pandemic (HK) or seasonal (Pan) surface proteins. Inflammatory responses and pathology could be clearly linked to pandemic HA expression (Figures 1C and 1D). To avoid potential confounding caused by differences in virus accumulation of the different 6:2 viruses (Figure S1), we next tested whether we could seasonalize a pandemic IAV by addition of the corresponding glycosylation sites. Isogenic viruses representing the 1968 pandemic H3N2 IAV (WT) and two viruses carrying two (+II) or four (+IV) additional glycosylation sites on the globular head of HA were constructed as described (Vigerust et al., 2007). The stepwise addition of glycans to the HA in the absence of other changes resulted in correspondingly reduced airway inflammation and pathology upon infection with the different IAV (Figures 1E and 1F). Further analyses demonstrated reduced morbidity on the level of body weight loss (Figures 1G and 1H) and mortality (Figure 1I) associated with increased glycosylation. Infection of a second mouse line revealed comparable results (Figures S2A and S2B), broadening the phenotype. Finally, the addition of at least two additional glycans to the pandemic HA protein was not associated with reduced viral growth (Figure 1J), excluding a virus propagation disadvantage as the primary mechanism of reduced pathogenicity in these experiments. The addition of further two glycans (+IV virus) did result in attenuated growth (Figure 1J), likely due to collectin neutralization (Reading et al., 2007, Vigerust et al., 2007), which led us to exclude this virus from later molecular mechanistic studies so as not to confound the results.
Figure 1.
Lack of HA Globular Head Glycosylation Triggers Lung Inflammation and Pathogenicity
Groups of (A and B) ten and (C–J) five BALB/c mice were infected intranasally (A and B) with 104 PFU of pandemic (p) 2009 H1N1 HH04, seasonal (s) H3N2 Pan/99, or seasonal (s) H1N1 Mem/01, (C and D) with 104 PFU of viruses carrying different combinations of pandemic (HK) and seasonal (Pan) HA/NA surface proteins on a six-gene-segment PR8 backbone (HK/HK 6:2, Pan/Pan 6:2, HK/Pan 6:2, and Pan/HK 6:2) and (E–J) with (E, F, G, and J) 5 × 105 and (H and I) 106 PFU of isogenic viruses differing in the number of N-linked glycosylation sites on the HA globular head (HK WT, HK +II, and HK +IV). (A–F) PBS-inoculated mice served as control. (A, C, and E) Lung mRNA levels of Il6 and Ccl2 3 days post infection are depicted as the fold change of levels of the PBS-inoculated mice, and (B, D, and F) body weight (%; compared to starting weight) is shown. (G and H) Body weight changes over 15 days are depicted, and (I) Kaplan-Meier survival curve analysis was performed. (J) Virus lung titers on d2, d3, d5, and d7 post-infection are depicted as PFU/ml. (A–H) Data are represented as mean with SD, (J) each dot represents an individual mouse, and bars show mean. An asterisk (∗) indicates statistically significant differences (p < 0.05) between (A and B) HH04 pH1N1 and all other groups, (C and D) HK/HK 6:2 and HK/PAN 6:2 and all other groups, (E–I) HK WT and all other groups, and (J) HK +II and HK +IV to HK WT. See also Figures S1 and S2.
Poor Glycosylation Is Associated with Inflammation and Pathology
After the initial finding of increased pro-inflammatory cytokine expression in lungs infected with viruses expressing HAs from pandemic IAVs, we assessed the impact of HA glycosylation on airway inflammation. Investigations of responses to isogenic IAVs only differing in the amount of glycosylation sites on the HA demonstrated a dampening of inflammation with the stepwise addition of glycans. Assessment of pro-inflammatory cytokine/chemokine expression on the mRNA level (Figure 2 A) as well as the protein level (Figure 2B) revealed a strong inflammatory response upon WT virus infection but stepwise reduced inflammation upon infection with the +II and +IV virus variants. Infection with the WT virus also resulted in infiltration of immune cells into airways compared to lower cell counts upon +II and +IV virus infection (Figure 2C). An analysis of specific cell types demonstrated that depletion of resident airway macrophages, as shown earlier by our group (Ghoneim et al., 2013), and accumulation of inflammatory cells, including exudative macrophages and neutrophils, was strongest upon WT infection with a lesser or no impact during +II and +IV virus infections (Figure 2C). Analysis of the ratio of resident macrophages, exudative macrophages, and neutrophils in BALF showed an almost complete shift from resident macrophages in naive mice to infiltrating inflammatory cells in WT virus infection, a shift seen to lesser extent in +II and only to a marginal extent in +IV virus infections (Figure 2D). Taken together, these data clearly demonstrate an impact of HA glycosylation on airway inflammation. Along with these differences, we detected an HA glycosylation-dependent increase in lung pathology using pathology markers such as total BALF protein (Figure S3A), BALF LDH levels (Figure S3B), and lung edema (Figures S3C and S3D). Histology analyses confirmed gross differences in lung pathology upon WT, +II, and +IV virus infection (Figure S3E), with marked denudation of bronchioles following only the WT virus infection (Figure S3F). Finally, analyses of markers of lung physiology revealed adverse effects on lung homeostasis and functionality characterized by generally increased airway resistance and decreased lung compliance upon infection with the WT virus, findings that were reduced upon infection with the higher glycosylated IAV viruses (Figure S4) and are consistent with the observation of decreased inflammatory responses and decreased lung injury. Although the findings of diminished pathogenicity for the +IV virus may be partially due to differences in viral titer, alternate mechanisms must be responsible for the differences in WT and +II viruses because these variants to not differ in replication or viral lung load.
Figure 2.
Addition of Glycosylation Sites to the Pandemic HA Abolishes Lung Inflammation upon IAV Infection
Five (A, C, and D) and four to five (B) BALB/c mice per group were infected with 5 × 105 PFU of isogenic viruses carrying differentially glycosylated HA globular heads (HK WT, HK +II, and HK +IV). PBS-inoculated mice served as control. (A) Total lung mRNA levels of Ifnb1, Il6, Ccl2, Tnf, and Cxcl1 2 days post-infection are shown as n-fold of the PBS-inoculated mice, and (B) total lung protein levels for IL6, CCL2, and TNF in pg/ml 3 and 5 days after infection are depicted. (C and D) Five days after infection, BALF was collected and cellular composition was analyzed. (C) Total cell counts and counts for specific cell types (resident macrophages, exudative macrophages, and neutrophils) are depicted as cells in BALF/mouse. (D) Relative proportions of cell types within the BALF cell composition are shown as parts of whole. (A–C) Data are represented as mean with SD, and (D) mean values for each experimental group were used. An asterisk (∗) indicates statistically significant differences (p < 0.05) between HK WT and all other groups. In (C) resident macrophages, an asterisk (∗) indicates statistically significant differences (p < 0.05) between HK WT and PBS and +IV. See also Figures S3 and S4.
Global Impact of HA Glycosylation on the Lung Transcriptome
In order to understand the mechanisms underlying the association between HA glycosylation and inflammation upon IAV infections, we assessed the whole lung transcriptome. Microarray analysis revealed clear differences between the experimental groups. The WT-virus-infected lungs displayed a different gene expression profile by principal-component analysis (PCA) compared to lungs from naive mice and from +II- and +IV-virus-infected mice (Figure 3 A). The expression profile of +II-virus-infected lungs was closer to but remained distinct from the WT-virus-infected lungs, whereas the gene expression profile of the +IV-infected lungs was comparable to but distinct from naive mouse lungs (Figure 3A). Heatmap analyses revealed a candidate group of genes that were upregulated in WT-virus-infected lungs compared to naive mice with a lesser extent of upregulation in +II and an even weaker signal in +IV-virus-infected lungs (Figure 3B). Focusing on this group of more than 1,000 genes with the strongest associations with glycosylation demonstrated that they were enriched in genes involved in inflammatory responses (Figure 3C). Of interest, genes linked to ER stress and the UPR were highly correlated with glycosylation status, prompting further investigation (Figure 3D).
Figure 3.
Global Lung Transcriptome upon IAV Infection Is Altered due to HA Globular Head Glycosylation
Five BALB/c mice per group were infected intranasally with 5 × 105 PFU of isogenic viruses carrying differentially glycosylated HA globular heads (HK WT, HK +II, and HK +IV). PBS-inoculated mice served as controls. Two days after infection, total lung RNA was used for analysis of the global lung transcriptome.
(A) Principal-component analysis (PCA) visualization of IAV-infected and PBS control mice is depicted. This visualization captures 40.3% of the variability across samples in the top three components.
(B) The Z transformed complete data set shows color-coded expression (heatmap) clustered by gene and displayed by experimental class.
(C) Data from a select group (bar indication in B) of genes showing the highest upregulation in HK WT infection was tested for GO enrichment using DAVID. Redundant categories were removed, and the p value of enrichment was −log10 transformed to create a score for visualization in the bar chart.
(D) A select group of HK WT-induced genes (ER stress genes) are visualized in this heatmap.
ER Stress Mediates Inflammatory Responses to Viral Infections
To further probe the molecular mechanism of increased inflammation upon pandemic IAV infections, we next assessed the potential involvement of ER stress and the UPR. Isolating the infection process to lung epithelial cells, which are the main cell type productively infected by IAV, and only focusing on viruses having comparable growth characteristics (WT and +II virus) to avoid confounding, profound differences in the inflammatory response were evident upon infection (Figure 4 A). Infection with the WT virus resulted in higher induction of IFNB1, IL6, and CCL2 mRNA compared to +II virus infection. This result was not affected by reduced accumulation of viral material, a concern that could confound the data, because the +II virus replicated comparably to the WT virus in this system (Figures 4B and S5A) and showed generally comparable to slightly better growth in relevant cell lines (Figures S5B and S5C). Expanding the number of genes investigated, a qPCR-based analysis revealed strong upregulation of multiple inflammatory genes in WT virus compared to +II-virus-infected lung epithelial cells (Figure 4C). Additional experiments in primary normal human bronchial epithelial cells (NHBEs) confirmed the finding of more-pronounced inflammatory responses upon WT virus infection in a second relevant human cell model (Figure 4D) in the absence of differences in replication (Figure 4E). We next assessed the contribution of different ER stress pathways to the inflammatory response upon inflammation and the involvement in altered immune reactions upon infection with IAV in the setting of differentially glycosylated HA proteins. Analyzing IRE1a phosphorylation, we demonstrated activation in WT-virus-infected cells (Figure 4F) with lesser activation in +II-virus-infected cells; this was consistent with subsequent induction of the mitogen-activated protein (MAP) kinase JNK pathway (Figure 4F), which has been linked to ER stress (Darling and Cook, 2014) and is involved in inflammatory responses to IAV infection (Ehrhardt et al., 2010). Examining the activation of PERK and ATF6 as additional pathways triggered upon ER stress, we could illustrate PERK phosphorylation in WT compared to +II infections and found that this was virus MOI dependent (Figure 4G). ATF6 activation cleavage was if any minimal, arguing that the contribution of ATF6 in IAV-induced, ER-stress-mediated inflammation seems to be minimal or none (Figure 4G). Analyzing expression of several UPR-dependent genes revealed at most weak upregulation of some genes upon WT infection without a general pattern of induction (Figure 4H) at the point in time of strong induction of inflammatory responses (Figure 4I). These data do not exclude a specific contribution of particular UPR genes but do suggest that activation of the ER stress kinases IRE1a and PERK and subsequent initiation of inflammatory pathways is potentially the more critical event. Probing the hypothesis through use of the regulatory compound phenylbutyric acid (PBA) to inhibit ER stress, we could illustrate the interplay of this pathway with induction of inflammatory responses upon infection with pandemic IAV (Figure 5 A). Activation of the MAP kinase JNK and p38 pathways was stronger upon WT virus infection compared to +II virus infection, and inhibition of ER stress resulted in a reduction of WT-virus-induced activation of these pathways along with diminished IRE1a activation (Figure 5A). Cytokine/chemokine analyses of inflammatory responses upon pandemic IAV infection supported this interpretation with decreased induction of inflammatory genes after inhibition (Figure 5B), data confirmed with a second molecular chaperone (TUDCA) also decreasing ER stress (Figure 5C). To broaden the relevance of the explicated mechanism of ER stress induction on inflammatory responses related to glycosylation status, we utilized different respiratory syncytial viruses (RSVs) propagated either in HEp2 or Vero cells, because the major glycosylated surface protein of RSV is truncated when propagated in Vero cells, resulting in a significant loss of glycosylation (Derscheid et al., 2013, Kwilas et al., 2009, Rixon et al., 2002). Analyzing the inflammatory response upon infection with these two versions of RSV revealed differences in expression of pro-inflammatory cytokines and differential induction of DDIT3 expression (Figure 5D) along with comparable virus growth (Figure S5D), showing that the glycosylation status of the RSV surface protein also affects the induction of cellular immune responses.
Figure 4.
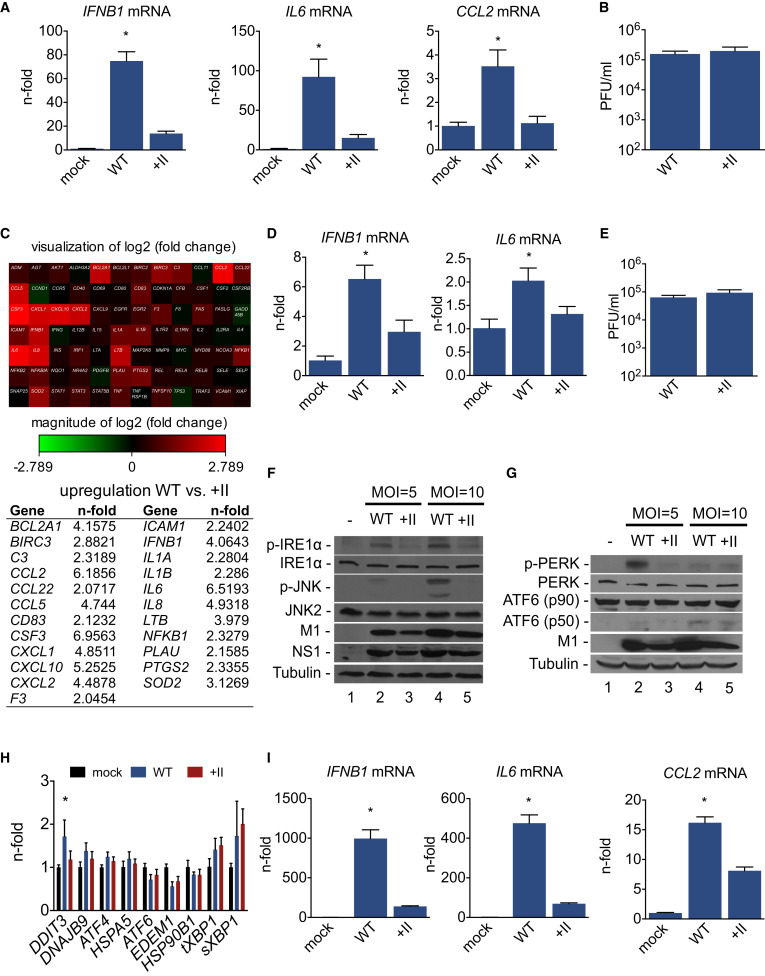
ER-Stress-Mediated Sensing of Pandemic HA as Mechanism of Immune Recognition
A549 lung epithelial cells (A–C and F–I) or primary NHBE cells (D and E) were infected with isogenic viruses carrying differentially glycosylated HA globular heads (HK WT and HK +II) or (A, D, and F–I) mock infected. The MOI used was (A, B, F, and G) MOI = 5 and (C–I) MOI = 10. (A, D, and I) Cellular mRNA levels of (A, D, and I) IFNB1, (A, D, and I) IL6, and (A and I) CCL2 are shown as n-fold of the mock infected cells (A and D) 24 hr and (I) 12 hr after infection, and (B and E) virus titers are depicted in PFU/ml. (H) Cellular mRNA level of the depicted ER stress genes 12 hr after infection are shown as n-fold of the mock infected cells. (C) qPCR (RT2 Profiler)-based analysis of inflammatory gene expression of HK WT- and HK +II-infected cells is shown. (C, upper panel) Heatmap visualization of fold increase of inflammatory genes after HK WT infection compared to HK +II infection and (C, lower panel) list of genes upregulated in HK WT compared to HK +II infection are depicted. (F and G) Cells were harvested 12 hr after infection for western blot analysis. Detection of the indicated proteins was done with specific antibodies. (A, B, D, E, H, and I) Data are represented as mean with SD. (A, B, D, E, and I) Representative data of three independent experiments each consisting of three biological samples are shown. (F and G) Representative blots of three independent experiments are shown and (H) mean values from three independent experiments, each consisting of three biological samples, are shown. (C) cDNA of three different biological samples were pooled and analyzed. An asterisk (∗) indicates statistically significant differences (p < 0.05) between HK WT and all other groups. See also Figure S5.
Figure 5.
ER Stress as a Central Regulator for Inflammatory Responses upon Viral Infections
A549 lung epithelial cells were infected with (A–C) isogenic IAV carrying differentially glycosylated HA globular heads (HK WT and HK +II; MOI = 10) and (D) two RSV A2 virus variants differing in surface protein glycosylation (RSV HEp2 and RSV Vero; MOI = 5). Mock infected cells served as control. After infection, cells were treated with (A and B) 10 mM PBA or (C) 500 μg/ml TUDCA. (A) Cells were harvested 12 hr after infection for western blot analysis, and indicated proteins were detected with specific antibodies. (B–D) Cellular mRNA levels of (B–D) IFNB1, IL6, CCL2, and (D) DDIT3 are shown as n-fold of the mock-infected cells (B and C) 12 hr and (D) 24 hr after infection. (B–D) Data are represented as mean with SD. (A) Representative blots of three independent experiments; (B and C) representative data of three independent experiments, each consisting of three biological samples; and (D) representative data of two independent experiments, each consisting of four biological samples, are shown. An asterisk (∗) indicates statistically significant differences (p < 0.05) between (B and C) HK WT and all other groups and (D) RSV HEp2 to all other groups. See also Figure S5.
ER Stress Is Responsible for the Pathogenicity of Pandemic IAV
Having clearly established the link between HA glycosylation, ER stress activation, and inflammatory responses in epithelial cells and the lungs, we next sought to demonstrate that these over-exuberant inflammatory responses are the basis for pandemic IAV pathogenicity. Therefore, we manipulated the inflammatory response using several approaches and analyzed the outcome on IAV pathogenesis. First, artificially stimulating inflammatory responses using exogenous lipoteichoic acid (LTA) in +II-virus-infected lungs resulted in elevated pathogenicity to the level of WT virus infection (Figure 6 A), showing that the dampened inflammation upon +II virus infection is the basis for reduced morbidity and mortality in the model. In addition, reduction of inflammatory responses upon WT virus infection rescued infected animals, either using CCL2-deficient mice (Figure 6B), which have alterations in inflammatory responses such as less infiltration of inflammatory cells (Conrady et al., 2013, Dessing et al., 2007, Frangogiannis et al., 2007, van Zoelen et al., 2011), or using imatinib (Figure 6C) as a broadly active anti-inflammatory drug (Wolf et al., 2005). Both approaches helped WT-infected animals to survive, whereas the control animals died (Figures 6B and 6C). In further support of the concept of ER stress as the source of this pathological inflammation, treatment of mice with a chemical chaperone that decreases ER stress by promoting protein folding rescued WT-infected animals (Figure 6D). To confirm our key findings with a second, recent pandemic IAV, the 2009 H1N1pdm virus utilized in the initial comparisons to seasonal strains (Figures 1A and 1B), we constructed an otherwise isogenic +II version and repeated select experiments. Reduced immune responses upon ER stress inhibition were seen in a comparison of untreated WT virus to treatment with PBA (Figures 7A and 7B). Addition of two glycans to the globular head of the HA resulted in reduced inflammatory responses in line with reduced ER stress induction on the level of Ddit3 mRNA expression (Figure 7C), as well as decreased morbidity and mortality (Figures 7D and 7E). Taken together, our data illuminate the interplay of poorly glycosylated HA proteins, zoonotically introduced into humans during pandemic outbreaks, with induction of inflammation through an ER stress pathway, resulting in pathology. It can be speculated that these pandemic viruses adapted to the human host by selection of variants with increased glycosylation because avoidance of ER-stress-mediated immune recognition and the resulting massive airway inflammation and pathology benefitted the virus.
Figure 6.
ER-Stress-Mediated Inflammation Triggers Pandemic IAV Pathogenicity
(A) Five BALB/c mice per group were intranasally infected with 106 PFU of isogenic viruses carrying differentially glycosylated HA globular heads (HK WT and HK +II) or PBS for control. At d2 post-infection, different groups were either treated with 100 μg LTA or PBS intranasally. (B) Ten C57BL/6 WT and eleven Ccl2−/− mice per group were infected intranasally with 105 PFU HK WT virus. (C and D) Five BALB/c mice per group were infected intranasally with 106 PFU HK WT virus. (C) Animals were treated i.p. with 25 or 50 mg/kg/day imatinib and (D) 200 mg/kg/day PBA or solvent control. Survival was monitored for 15 days, and Kaplan-Meier survival curve analysis was performed. An asterisk (∗) indicates statistically significant differences (p < 0.05) between (A) HK +II PBS d2 and HK +II LTA d2 treated mice, (B) C57BL/6 WT and Ccl2−/− mice, and (C and D) between HK WT-infected mice and all treatment groups.
Figure 7.
Addition of Glycosylation Sites to the 2009 Pandemic Virus HA Abrogates Lung Inflammation and Pathogenicity
(A and B) A549 lung epithelial cells were infected with the pandemic 2009 H1N1 (HH04) virus (MOI = 5) and treated with 10 mM PBA or left untreated. Mock infected cells served as control, and cells were harvested 12 hr after infection. (A) Western blot analysis was conducted, and detection of the indicated proteins was done with specific antibodies. (B) IL6 mRNA level is shown as n-fold of the mock infected cells.
(C–E) Five BALB/c mice per group were infected intranasally with (C) 103 PFU or (D and E) 104 of isogenic viruses carrying different glycosylated HA globular heads (HH04 and HH04 HA +II), (C) with PBS-infected animals serving as control. (C) Total lung mRNA levels of Ifnb1, Il6, Ccl2, and Ddit3 3 days after infection are shown as n-fold of the PBS-inoculated mice. (D) Body weight changes over the course of infection (15 days) are depicted, and (E) Kaplan-Meier survival curve analysis was performed. (B–D) Data are represented as mean with SD. (A) Representative blots of three independent experiments and (B) representative data of three independent experiments, each consisting of three biological samples, are shown. An asterisk (∗) indicates statistically significant differences (p < 0.05) between HH04 and the other groups and (C) between HH04 and PBS for Ddit3 mRNA.
Discussion
The versatile interplay of invading pathogens and host immune responses is critical for determining both the severity and the outcome of infections. Pathogen-host co-evolution has resulted in manifold receptors for sensing pathogens (Mulhern et al., 2009). Although recognition of viral RNA by sensors such as RIG-I is thought to be one of the most important mechanisms of sensing, IAVs have developed countermeasures for these systems and the complexity of the interactions suggests that other pathways must be operable. In our current study, we addressed the crucial involvement of ER stress pathways such as the UPR in inflammatory responses and pathogenicity upon viral infections including both IAV and RSV. Innate sensing occurred at the level of protein processing, recognizing the nascent HA proteins from pandemic strains as “non-self” as a result of poor surface glycosylation. Almost all identified innate sensors of viruses rely on recognition of differences in nucleic acid composition to trigger immune responses. Interestingly, the pathology in this model stemmed from these immune responses, not direct damage from the virus(es). We propose that this pathway operates as an intracellular innate sensor, distinguishing “self” from non-self by recognizing differentially glycosylated proteins as foreign and triggering immune responses. In support of our data, evidence for a regulatory involvement of ER stress in microbial infections was recently shown (Muralidharan and Mandrekar, 2013) and an interplay of ER stress and viral infections was suggested for human pathogens such as HBV (Liu et al., 2013), HCV (Chan, 2014), and influenza virus (Hassan et al., 2012, Roberson et al., 2012). More recently, a potential direct contribution of ER stress to recognition of viral infections was speculated upon (Smith, 2014). Finally, a general contribution of ER stress to lung inflammation as well as acute lung injury in human patients was recently described (Kim et al., 2013). Here, we link these observations and provide the mechanism that ties them together—proteins that are differentially glycosylated relative to other, perhaps more “natural” proteins trigger ER stress, and the activation of signaling events downstream of the UPR results in inflammation and pathology.
The natural host niche of IAV is the gut of wild birds and waterfowl. All previous pandemic viruses, having left this environment and crossed species barriers to establish endemic infections in the respiratory tract of humans, have been poorly glycosylated. Such viruses would be predicted to be recognized through this mechanism, promoting virulence and morbidity. Indeed, pandemic viruses including the 2009 H1N1 strain are typically more virulent than related seasonal strains (Reed et al., 2014). Virulence of the 1918 pandemic strain has been linked clearly to the HA and to its glycosylation status, adding further support for the importance of this mechanism (Sun et al., 2013, Tumpey et al., 2004). Over decades of circulation, negative selection for this trait that allows immune recognition would favor accumulation of glycans on the HA surface to avoid triggering ER stress, a pattern that has been recapitulated in nature with both the H1N1 and H3N2 lineages. Glycosylation variants of the 2009 H1N1 pandemic strain are already being detected in nature, suggesting that adaptation to avoid this sensing is already taking place (Mullick et al., 2011) and emphasizing the need to evaluate the consequences on IAV pathogenicity for different virus isolates (Medina et al., 2013, Zhang et al., 2013). More broadly, it is likely that this ability to discriminate non-self at the level of protein folding and processing has utility in maintaining cellular homeostasis in the face of viral invasion and provides an alternative to nucleic-acid-based innate sensors, which can be circumvented by a variety of strategies. However, our data demonstrate that this may also have untoward consequences, because pathology can result from the unrestrained inflammation. As societal and climate change drive further emergence of pathogens new to humans from animal reservoirs, a deeper understanding of virus-host interactions at a molecular level is needed to prevent or mitigate disease.
Experimental Procedures
Cells, Viruses, Chemicals, and Infection Conditions
Madin-Darby canine kidney (MDCK) cells were cultured in MEM, whereas the human lung epithelial cell line (A549) and the human embryonic kidney cells (HEK293T) were cultivated in DMEM. NHBEs (Lonza) were cultured according to the manufacturer’s protocol using BEGM bullet kit (Lonza). After refreezing and counting upon initial cell expansion, single vials were thawed and directly seeded for experiments. Cell culture media were supplemented with 10% heat-inactivated fetal bovine serum. The seasonal IAV isolates A/Memphis/7/2001 (H1N1; Mem/01) and A/Panama/2007/1999 (H3N2; Pan/99) were taken from the strain collection at the St. Jude Children’s Research Hospital. All recombinant IAVs were created as described earlier (Vigerust et al., 2007). The 2009 pandemic IAV A/Hamburg/04/2009 (H1N1; HH04) and the isogenic mutant virus A/Hamburg/04/2009 HA +II were rescued from the reverse genetics plasmid system kindly provided by Dr. Thorsten Wolff (Robert Koch Institute). Two additional glycosylation sites found in seasonal H1N1 IAV before 2009 were introduced into the 2009 pandemic HA to create the A/Hamburg/04/2009 HA +II virus (142NHT and 177NLS; HH04 HA +II) for comparison to HH04 WT (142NHE and 177KLS). Of note, the reverse genetic plasmid for A/Hamburg/04/2009 HA (protein accession number ACR10223.1) carries the mutation D144E. The different viruses consisting of six internal gene segments of A/Puerto Rico/8/34 (H1N1) and different combinations of seasonal (A/Panama/2007/99 H3N2; Pan) and pandemic (A/Hong Kong/1/68 H3N2; HK) HA and NA segments (referred to as 6:2 viruses) were created as described earlier (Vigerust et al., 2007) as were the isogenic viruses consisting of PR8 internal genes and A/Hong Kong/1/68 H3N2 (HK) HA and NA segments either wild-type HA (HK WT) or carrying two (HK +II) or four additional (HK +IV) glycosylation sites (Vigerust et al., 2007). The presence of the desired mutation(s) in each virus was confirmed by sequencing. All viruses were grown on MDCK cells. The infection procedure of cells was conducted as described earlier (Hrincius et al., 2010). The human RSVs A2 for alteration in glycosylation studies were grown either in Vero cells or HEp2 cells for alteration in surface proteins as described earlier (Kwilas et al., 2009). The chemical chaperones PBA (Sigma-Aldrich; sodium salt) and TUDCA (Sigma-Aldrich; sodium salt), the chemical inhibitor imatinib mesylate (STI571; Selleckchem), and the bacterial cell wall polymer LTA (InvivoGen) were dissolved in water and used as indicated.
Standard Plaque Titration
Analysis of the number of infectious particles (plaque titers) was conducted by collecting samples (tissue culture supernatants and lung homogenates) at the indicated points in time post-infection (p.i.) and assessed in standard plaque titration as described earlier in Hrincius et al. (2010).
Mouse Infections
All mouse studies were performed in compliance with animal welfare regulations under an animal protocol approved by the Animal Care and Use Committee of St. Jude Children’s Research Hospital. Seven to eight-week-old female BALB/c mice or C57BL/6 WT and Ccl2 −/− (B6.129S4-Ccl2 tm1Rol /J mice; Jackson Laboratory) were used for the mouse studies. The animals were lightly anesthetized with 2.5% inhaled isoflurane (Baxter) and afterward infected intranasally with the indicated viruses and plaque-forming units (PFU) in a final volume of 50 μl. Mice were monitored daily for weight loss, disease symptoms, and mortality. In accordance with protection of animal welfare restrictions, mice were sacrificed after weight loss of 30% and considered to have died that specific day.
Viral Lung Titers and Cytokine/Chemokine Protein Analysis
For determination of virus titers and cytokine/chemokine protein levels, mouse lungs were collected in PBS, homogenized using a FastPrep-24 homogenizer with Lysing Matrix D (both from MP Biomedical), and adjusted with PBS to obtain a 10% tissue homogenate. The samples were cleared of debris, and supernatants were used for virus titer determination in standard plaque titration and for measuring the concentration of interleukin 6 (IL6), tumor necrosis factor (TNF), and chemokine (C-C motif) ligand 2 (CCL2) using a custom multiplex cytokine/chemokine assay (Millipore) analyzed with a Luminex xPOTENT 3.1 system (Bio-Rad) following manufacturer’s protocol.
Total RNA Isolation and Quantitative Real-Time PCR
Total RNA from cells was isolated using the RNeasy Mini Kit (QIAGEN) according to the manufacturer’s protocol. For analysis of cytokine/chemokine levels, mouse lungs were collected in Trizol and RNA was isolated using Trizol method (Chomczynski and Sacchi, 1987) and PureLink RNA Mini Kit (Ambion). For reverse transcription of mRNA, 1 or 2 μg total RNA and 0.5 μg oligo(dT) primer were used. The reverse transcription was done using M-MLV Reverse Transcriptase (Promega) according to the manufacturer’s protocol. For qPCR reaction, QuantiTect SYBR Green PCR Kit (QIAGEN) was used according to manufacturer’s recommendation. Relative mRNA concentrations were determined after 40 cycles of amplification with Applied Biosystems 7300 Rea-Time PCR System by using the 2−ΔΔCT method (Livak and Schmittgen, 2001). The housekeeping gene GAPDH served as internal standard. The following primers were used: QuantiTect primer (QIAGEN); Mm_Gapdh_3_SG; Mm_Il6_1_SG; Mm_Ccl2_1_SG; Mm_Ifnb1_1_SG; Mm_Tnf_1_SG; Mm_Cxcl1_1_SG; Mm_Ddit3_1_SG; HS_GAPDH_2_SG; HS_IFNB1_1_SG; HS_IL6_1_SG; HS_CCL2_1_SG; HS_DNAJB9_1_SG; HS_ATF4_1_SG; HS_ATF6_1_SG; HS_TRA1_1_SG (HSP90B1); and primer for human DDIT3: fwd: 5′AGAACCAGGAAACGGAAACAGA and rev: 5′TCTCCTTCATGCGCTGCTTT, human HSPA5: fwd: 5′TGTTCAACCAATTATCAGCAAACTC and rev: 5′TTCTGCTGTATCCTCTTCACCAGT, human EDEM1: fwd: 5′CAAGTGTGGGTACGCCACG and rev: 5′AAAGAAGCTCTCCATCCGGTC, human total XBP1 (tXBP1): fwd: 5′TGGCCGGGTCTGCTGAGTCCG and rev: 5′ATCCATGGGGAGATGTTCTGG, and human spliced XBP1 (sXBP1): fwd: 5′CTGAGTCCGAATCAGGTGCAG and rev: 5′ATCCATGGGGAGATGTTCTGG. ER stress genes primer sequences were taken from Oslowski and Urano (2011). For quantification of influenza M1 mRNA, fwd: CTTCTAACCGAGGTCGAAACGTA and rev: GGATTGGTCTTGTCTTTAGCCA (Di Trani et al., 2006) primer were used. For quantification of RSV NS1 gene, real-time RT-PCR was performed as previously published (Boukhvalova et al., 2010). Total RNA was reverse transcribed to cDNA with NS1 antigenomic primer (5′-CAATGAACTAGGATATCAAGAC-3′) or with oligo(dT) primers for the house-keeping gene HPRT1 using SuperScript III first-strand synthesis system (Life Technology). Real-time PCR was performed with NS1 primers (5′-CACAACAATGCCAGTGCTACAA-3′ and 5′-TTAGACCATTAGGTTGAGAGCAATGT-3′) using Power SYBR Green PCR Master Mix (Life Technology) on LightCycler 480 II (Roche). The relative amount of RSV NS1 was normalized to HPRT1 (primers: 5′-GGCTCCGTTATGGCGACCCG-3′ and 5′-CGAGCAAGACGTTCAGTCCTGTCC-3′) using 2−ΔΔCT method.
The quantitative real-time PCR-based cytokine/chemokine analysis (RT2 Profiler PCR Array; human NFκB signaling targets [PAHS-225Z]; QIAGEN) was conducted according to manufacturer’s protocol. Data analysis was conducted by RT2 Profiler PCR Array Data Analysis software on QIAGEN webpage.
Western Blot Analysis and Antibodies
Western blot analysis was conducted as described earlier (Hrincius et al., 2012). In brief, cells were lysed and cleared from debris, and Bradford method was used for protein concentration measurement. Finally, cell lysates were used for protein expression analysis using western blot technique. Activation of ER stress was analyzed by phosphorylation of IRE1α (anti-IRE1α [pS724] rabbit polyclonal antibody [pAb]; Novus Biologicals) and PERK (anti-PERK [pT980] [16F8] rabbit monoclonal antibody [mAb]; Cell Signaling Technology) and ATF6 cleavage (anti-ATF6 [70B1413.1] mouse mAb; Novus Biologicals). Analysis of MAPK activation was conducted by analysis of JNK phosphorylation (anti-JNK [pThr183/pTyr185] [81E11] rabbit mAb; Cell Signaling Technology) and p38 phosphorylation (anti-p38 [pThr180/pTyr182] [D3F9] rabbit mAb; Cell Signaling Technology). Expression of influenza M1 and NS1 protein was detected using a specific anti-M1 (GA2B) mouse mAb (Abcam) and specific anti-NS1 (23-1) mouse mAb (developed at the IMV). Detection of JNK2 (anti-JNK2 [56G8] rabbit mAb; Cell Signaling Technology), IRE1α (anti-IRE1α [14C10] rabbit mAb; Cell Signaling Technology), PERK (anti-PERK mouse pAb; ROCKLAND), and tubulin (anti-β-tubulin [9F3] rabbit mAb; Cell Signaling Technology) was used for control. Protein bands were finally visualized using a standard enhanced chemiluminescence (ECL) reaction.
Microarray
For microarray analysis of gene expression in mouse lungs, total lung RNA was isolated as described. RNA quality was confirmed by analysis on the Agilient 2100 Bioanalyzer. Total RNA (100 ng) was processed in the Hartwell Center microarray core according to the Affymetrix GeneChip Whole Transcript Labeling protocol. 5.5 μg of biotinylated cDNA was fragmented and hybridized to Affymetrix GeneChip Mouse Gene 2.0 ST arrays and then stained and scanned according to the manufacturer’s instructions. Data were RMA summarized (Irizarry et al., 2003) and visualized by PCA. Next, each probe set was statistically tested using ANOVA, and the resultant p values were adjusted for multiple testing by the Bonferroni and Storey q value methods (Storey and Tibshirani, 2003) using Partek Genomics Suite 6.6. Hierarchical clustering and heatmap visualization of Z score normalized data for complete data or select genes were performed using Spotfire Decision Site. Gene Ontology (GO) enrichment analysis was performed by the DAVID Bioinformatics Resources 6.7 software developed and provided by the National Institute of Allergy and Infectious Diseases (NIAID) (Huang et al., 2009). GO bar charts were scored and visualized with STATA/MP 11.2.
BALF Collection, Flow Cytometry, and Pathologic Marker
For analysis of infiltration of inflammatory cells into mouse lungs and markers of lung injury, BALF was collected at the indicated point in time as described earlier (Hrincius et al., 2012). The harvested BALF cells were purified, counted, and FACS analyzed as described previously (Hrincius et al., 2012). In brief, FACS analyses were conducted to obtain counts for resident macrophages (F4/80+, Gr1(low), CD11c(high), and MHC-II(low)), exudative macrophages (F4/80+, Gr1(high), CD11c(int), and MHC-II(low)), and neutrophils (Gr1+, F4/80−, and CD11c−). The remaining cell-free BALF was used to investigate markers of lung injury. Total protein concentration measurement via the Bradford assay (Bio-Rad) and LDH measurement via an LDH assay (Sigma-Aldrich) were executed according to the manufacturer’s protocol.
Histopathology and Edema
For detailed pathological investigation of mice lungs, animals were euthanized and lungs were fixed via intratracheal infusion followed by immersion in 10% neutral buffered formalin solution. Afterward, tissues samples were embedded in paraffin, automatically sectioned, and finally stained with H&E. A review was conducted by a veterinary pathologist blinded to the purpose of the study and to group composition to obtain gross pathology evaluation and pathology scores. For lung edema investigation, total lungs were collected and wet lung weight was determined as marker for inflammation and pathology in lung. Afterward, lungs were dried at 70°C for 48 hr and again weighed to obtain dry lung weight. Finally, the wet-dry lung weight ratio was calculated as marker for lung edema.
Lung Physiology
For analysis of lung physiology upon influenza virus infections, animals were anesthetized with 1 mg/ml xylazine (Vedco) and 7 mg/ml Euthasol (Virbac) prior to surgical intubation and attachment to the flexiVent FX1 small animal ventilator (SCIREQ). Using the “mouse inhaled dose response” script, baseline and responses to 25 mg/ml acetyl-β-methacholine (Sigma-Aldrich) were gathered. Software-generated values for the respiratory system resistance (Rrs), compliance (Crs), and elastance (Ers) and Newtonian resistance (Rn), tissue damping (G), and tissue elastance (H) were recorded. The mean for each was calculated for each animal prior to obtaining the mean and SD of the mean for the group.
Statistical Analyses
Comparison of survival curves from groups of mice over a period of 15 days was conducted using log rank (chi-square) test for statistical analysis of Kaplan-Meier survival data. ANOVA with Tukey test for multiple comparisons was used for all multiple comparisons of data (cytokine/chemokine level, single-day weight loss, in vivo viral titer, cell counts, lung pathology marker, pathology scores, lung physiology data, etc.). Student’s t test (with Welch’s correction for samples with unequal variance) was used for comparison of viral titer and viral RNA in vitro, and Student’s t test analysis with correction for multiple tests (Holm-Sidak method) and two-way ANOVA for repeated measurements with Tukey test for multiple comparisons was conducted for weight loss data over a period of 15 days. A p value of <0.05 was considered significant for these comparisons. Prism6 (GraphPad) software was used for all statistical analyses.
Author Contributions
E.R.H., S.L., and J.A.M. designed the project and experiments, analyzed data, and set up the manuscript. Experiments were conducted by E.R.H. with support from S.G., D.F. (microarray data), P.V. (lung pathology), and A.E.S. (lung physiology). D.Y. and S.A.C. (RSV HEp2 and RSV Vero) were involved in experimental procedures and/or data analysis.
Acknowledgments
We thank the St. Jude Hartwell Center, Animal Resources Center (ARC), and Veterinary Pathology core for support. This work was supported by the American Lebanese Associated Charities (ALSAC), grants to author S.A.C. (NIEHS [R01ES015050 and P42ES013648] and NIAID [R01AI090059]), and by a research fellowship from the German Research Foundation (DFG) to E.R.H. (HR 90/1-1). Original data are archived at the St. Jude Children’s Research Hospital and University of Tennessee Health Science Center.
Published: June 4, 2015
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information includes five figures and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2015.05.012.
Accession Numbers
Raw and summarized microarray have been deposited to the NCBI GEO and are available under accession numbers GEO:GSE65075.
Supplemental Information
References
- Boukhvalova M.S., Yim K.C., Prince G.A., Blanco J.C. Methods for monitoring dynamics of pulmonary RSV replication by viral culture and by real-time reverse transcription-PCR in vivo: Detection of abortive viral replication. Curr. Protoc. Cell Biol. 2010;Chapter 26:6. doi: 10.1002/0471143030.cb2606s46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S.W. Unfolded protein response in hepatitis C virus infection. Front Microbiol. 2014;5:233. doi: 10.3389/fmicb.2014.00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Conrady C.D., Zheng M., Mandal N.A., van Rooijen N., Carr D.J. IFN-α-driven CCL2 production recruits inflammatory monocytes to infection site in mice. Mucosal Immunol. 2013;6:45–55. doi: 10.1038/mi.2012.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels R., Kurowski B., Johnson A.E., Hebert D.N. N-linked glycans direct the cotranslational folding pathway of influenza hemagglutinin. Mol. Cell. 2003;11:79–90. doi: 10.1016/s1097-2765(02)00821-3. [DOI] [PubMed] [Google Scholar]
- Darling N.J., Cook S.J. The role of MAPK signalling pathways in the response to endoplasmic reticulum stress. Biochim. Biophys. Acta. 2014;1843:2150–2163. doi: 10.1016/j.bbamcr.2014.01.009. [DOI] [PubMed] [Google Scholar]
- Derscheid R.J., van Geelen A., McGill J.L., Gallup J.M., Cihlar T., Sacco R.E., Ackermann M.R. Human respiratory syncytial virus Memphis 37 grown in HEp-2 cells causes more severe disease in lambs than virus grown in Vero cells. Viruses. 2013;5:2881–2897. doi: 10.3390/v5112881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessing M.C., van der Sluijs K.F., Florquin S., van der Poll T. Monocyte chemoattractant protein 1 contributes to an adequate immune response in influenza pneumonia. Clin. Immunol. 2007;125:328–336. doi: 10.1016/j.clim.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Di Trani L., Bedini B., Donatelli I., Campitelli L., Chiappini B., De Marco M.A., Delogu M., Buonavoglia C., Vaccari G. A sensitive one-step real-time PCR for detection of avian influenza viruses using a MGB probe and an internal positive control. BMC Infect. Dis. 2006;6:87. doi: 10.1186/1471-2334-6-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt C., Seyer R., Hrincius E.R., Eierhoff T., Wolff T., Ludwig S. Interplay between influenza A virus and the innate immune signaling. Microbes Infect. 2010;12:81–87. doi: 10.1016/j.micinf.2009.09.007. [DOI] [PubMed] [Google Scholar]
- El Moussi A., Ben Hadj Kacem M.A., Slim A. Loss and gain of N-linked glycosylation sites in globular head and stem of HA found in A/H3N2 flu fatal and severe cases during 2013 Tunisia flu seasonal survey. Virus Genes. 2014;48:189–192. doi: 10.1007/s11262-013-0993-0. [DOI] [PubMed] [Google Scholar]
- Enserink M. Mission to MERS. Science. 2014;344:1218–1220. doi: 10.1126/science.344.6189.1218. [DOI] [PubMed] [Google Scholar]
- Faísca P., Desmecht D. Sendai virus, the mouse parainfluenza type 1: a longstanding pathogen that remains up-to-date. Res. Vet. Sci. 2007;82:115–125. doi: 10.1016/j.rvsc.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Frangogiannis N.G., Dewald O., Xia Y., Ren G., Haudek S., Leucker T., Kraemer D., Taffet G., Rollins B.J., Entman M.L. Critical role of monocyte chemoattractant protein-1/CC chemokine ligand 2 in the pathogenesis of ischemic cardiomyopathy. Circulation. 2007;115:584–592. doi: 10.1161/CIRCULATIONAHA.106.646091. [DOI] [PubMed] [Google Scholar]
- Ghoneim H.E., Thomas P.G., McCullers J.A. Depletion of alveolar macrophages during influenza infection facilitates bacterial superinfections. J. Immunol. 2013;191:1250–1259. doi: 10.4049/jimmunol.1300014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregersen N., Bross P., Vang S., Christensen J.H. Protein misfolding and human disease. Annu. Rev. Genomics Hum. Genet. 2006;7:103–124. doi: 10.1146/annurev.genom.7.080505.115737. [DOI] [PubMed] [Google Scholar]
- Hassan I.H., Zhang M.S., Powers L.S., Shao J.Q., Baltrusaitis J., Rutkowski D.T., Legge K., Monick M.M. Influenza A viral replication is blocked by inhibition of the inositol-requiring enzyme 1 (IRE1) stress pathway. J. Biol. Chem. 2012;287:4679–4689. doi: 10.1074/jbc.M111.284695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrincius E.R., Wixler V., Wolff T., Wagner R., Ludwig S., Ehrhardt C. CRK adaptor protein expression is required for efficient replication of avian influenza A viruses and controls JNK-mediated apoptotic responses. Cell. Microbiol. 2010;12:831–843. doi: 10.1111/j.1462-5822.2010.01436.x. [DOI] [PubMed] [Google Scholar]
- Hrincius E.R., Hennecke A.K., Gensler L., Nordhoff C., Anhlan D., Vogel P., McCullers J.A., Ludwig S., Ehrhardt C. A single point mutation (Y89F) within the non-structural protein 1 of influenza A viruses limits epithelial cell tropism and virulence in mice. Am. J. Pathol. 2012;180:2361–2374. doi: 10.1016/j.ajpath.2012.02.029. [DOI] [PubMed] [Google Scholar]
- Huang W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Jayaraman A., Koh X., Li J., Raman R., Viswanathan K., Shriver Z., Sasisekharan R. Glycosylation at Asn91 of H1N1 haemagglutinin affects binding to glycan receptors. Biochem. J. 2012;444:429–435. doi: 10.1042/BJ20112101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Job E.R., Deng Y.M., Barfod K.K., Tate M.D., Caldwell N., Reddiex S., Maurer-Stroh S., Brooks A.G., Reading P.C. Addition of glycosylation to influenza A virus hemagglutinin modulates antibody-mediated recognition of H1N1 2009 pandemic viruses. J. Immunol. 2013;190:2169–2177. doi: 10.4049/jimmunol.1202433. [DOI] [PubMed] [Google Scholar]
- Kim J.I., Park M.S. N-linked glycosylation in the hemagglutinin of influenza A viruses. Yonsei Med. J. 2012;53:886–893. doi: 10.3349/ymj.2012.53.5.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.J., Jeong J.S., Kim S.R., Park S.Y., Chae H.J., Lee Y.C. Inhibition of endoplasmic reticulum stress alleviates lipopolysaccharide-induced lung inflammation through modulation of NF-κB/HIF-1α signaling pathway. Sci Rep. 2013;3:1142. doi: 10.1038/srep01142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwilas S., Liesman R.M., Zhang L., Walsh E., Pickles R.J., Peeples M.E. Respiratory syncytial virus grown in Vero cells contains a truncated attachment protein that alters its infectivity and dependence on glycosaminoglycans. J. Virol. 2009;83:10710–10718. doi: 10.1128/JVI.00986-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Cao Y., Wang T., Xiang G., Lu J., Zhang J., Hou P. The N-glycosylation modification of LHBs (large surface proteins of HBV) effects on endoplasmic reticulum stress, cell proliferation and its secretion. Hepat. Mon. 2013;13:e12280. doi: 10.5812/hepatmon.12280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luo M. Influenza virus entry. Adv. Exp. Med. Biol. 2012;726:201–221. doi: 10.1007/978-1-4614-0980-9_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina R.A., Stertz S., Manicassamy B., Zimmermann P., Sun X., Albrecht R.A., Uusi-Kerttula H., Zagordi O., Belshe R.B., Frey S.E. Glycosylations in the globular head of the hemagglutinin protein modulate the virulence and antigenic properties of the H1N1 influenza viruses. Sci. Transl. Med. 2013;5:87ra70. doi: 10.1126/scitranslmed.3005996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulhern O., Harrington B., Bowie A.G. Modulation of innate immune signalling pathways by viral proteins. Adv. Exp. Med. Biol. 2009;666:49–63. doi: 10.1007/978-1-4419-1601-3_4. [DOI] [PubMed] [Google Scholar]
- Mullick J., Cherian S.S., Potdar V.A., Chadha M.S., Mishra A.C. Evolutionary dynamics of the influenza A pandemic (H1N1) 2009 virus with emphasis on Indian isolates: evidence for adaptive evolution in the HA gene. Infect. Genet. Evol. 2011;11:997–1005. doi: 10.1016/j.meegid.2011.03.015. [DOI] [PubMed] [Google Scholar]
- Muralidharan S., Mandrekar P. Cellular stress response and innate immune signaling: integrating pathways in host defense and inflammation. J. Leukoc. Biol. 2013;94:1167–1184. doi: 10.1189/jlb.0313153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oslowski C.M., Urano F. Measuring ER stress and the unfolded protein response using mammalian tissue culture system. Methods Enzymol. 2011;490:71–92. doi: 10.1016/B978-0-12-385114-7.00004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reading P.C., Tate M.D., Pickett D.L., Brooks A.G. Glycosylation as a target for recognition of influenza viruses by the innate immune system. Adv. Exp. Med. Biol. 2007;598:279–292. doi: 10.1007/978-0-387-71767-8_20. [DOI] [PubMed] [Google Scholar]
- Reed C., Chaves S.S., Perez A., D’Mello T., Daily Kirley P., Aragon D., Meek J.I., Farley M.M., Ryan P., Lynfield R. Complications among adults hospitalized with influenza: a comparison of seasonal influenza and the 2009 H1N1 pandemic. Clin. Infect. Dis. 2014;59:166–174. doi: 10.1093/cid/ciu285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rixon H.W., Brown C., Brown G., Sugrue R.J. Multiple glycosylated forms of the respiratory syncytial virus fusion protein are expressed in virus-infected cells. J. Gen. Virol. 2002;83:61–66. doi: 10.1099/0022-1317-83-1-61. [DOI] [PubMed] [Google Scholar]
- Roberson E.C., Tully J.E., Guala A.S., Reiss J.N., Godburn K.E., Pociask D.A., Alcorn J.F., Riches D.W., Dienz O., Janssen-Heininger Y.M., Anathy V. Influenza induces endoplasmic reticulum stress, caspase-12-dependent apoptosis, and c-Jun N-terminal kinase-mediated transforming growth factor-β release in lung epithelial cells. Am. J. Respir. Cell Mol. Biol. 2012;46:573–581. doi: 10.1165/rcmb.2010-0460OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts P.C., Garten W., Klenk H.D. Role of conserved glycosylation sites in maturation and transport of influenza A virus hemagglutinin. J. Virol. 1993;67:3048–3060. doi: 10.1128/jvi.67.6.3048-3060.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth J., Zuber C., Park S., Jang I., Lee Y., Kysela K.G., Le Fourn V., Santimaria R., Guhl B., Cho J.W. Protein N-glycosylation, protein folding, and protein quality control. Mol. Cells. 2010;30:497–506. doi: 10.1007/s10059-010-0159-z. [DOI] [PubMed] [Google Scholar]
- Smith J.A. A new paradigm: innate immune sensing of viruses via the unfolded protein response. Front Microbiol. 2014;5:222. doi: 10.3389/fmicb.2014.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Jayaraman A., Maniprasad P., Raman R., Houser K.V., Pappas C., Zeng H., Sasisekharan R., Katz J.M., Tumpey T.M. N-linked glycosylation of the hemagglutinin protein influences virulence and antigenicity of the 1918 pandemic and seasonal H1N1 influenza A viruses. J. Virol. 2013;87:8756–8766. doi: 10.1128/JVI.00593-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate M.D., Job E.R., Deng Y.M., Gunalan V., Maurer-Stroh S., Reading P.C. Playing hide and seek: how glycosylation of the influenza virus hemagglutinin can modulate the immune response to infection. Viruses. 2014;6:1294–1316. doi: 10.3390/v6031294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumpey T.M., García-Sastre A., Taubenberger J.K., Palese P., Swayne D.E., Basler C.F. Pathogenicity and immunogenicity of influenza viruses with genes from the 1918 pandemic virus. Proc. Natl. Acad. Sci. USA. 2004;101:3166–3171. doi: 10.1073/pnas.0308391100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brand J.M., Haagmans B.L., van Riel D., Osterhaus A.D., Kuiken T. The pathology and pathogenesis of experimental severe acute respiratory syndrome and influenza in animal models. J. Comp. Pathol. 2014;151:83–112. doi: 10.1016/j.jcpa.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zoelen M.A., Verstege M.I., Draing C., de Beer R., van’t Veer C., Florquin S., Bresser P., van der Zee J.S., te Velde A.A., von Aulock S., van der Poll T. Endogenous MCP-1 promotes lung inflammation induced by LPS and LTA. Mol. Immunol. 2011;48:1468–1476. doi: 10.1016/j.molimm.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Vigerust D.J., Ulett K.B., Boyd K.L., Madsen J., Hawgood S., McCullers J.A. N-linked glycosylation attenuates H3N2 influenza viruses. J. Virol. 2007;81:8593–8600. doi: 10.1128/JVI.00769-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.C., Chen J.R., Tseng Y.C., Hsu C.H., Hung Y.F., Chen S.W., Chen C.M., Khoo K.H., Cheng T.J., Cheng Y.S. Glycans on influenza hemagglutinin affect receptor binding and immune response. Proc. Natl. Acad. Sci. USA. 2009;106:18137–18142. doi: 10.1073/pnas.0909696106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster R.G., Bean W.J., Gorman O.T., Chambers T.M., Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol. Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson I.A., Skehel J.J., Wiley D.C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- Wolf A.M., Wolf D., Rumpold H., Ludwiczek S., Enrich B., Gastl G., Weiss G., Tilg H. The kinase inhibitor imatinib mesylate inhibits TNF-alpha production in vitro and prevents TNF-dependent acute hepatic inflammation. Proc. Natl. Acad. Sci. USA. 2005;102:13622–13627. doi: 10.1073/pnas.0501758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhu J., Li Y., Bradley K.C., Cao J., Chen H., Jin M., Zhou H. Glycosylation on hemagglutinin affects the virulence and pathogenicity of pandemic H1N1/2009 influenza A virus in mice. PLoS ONE. 2013;8:e61397. doi: 10.1371/journal.pone.0061397. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


