Abstract
Gingipain proteases are important virulence factors from the periodontal pathogen Porphyromonas gingivalis and are the target of many in vitro studies. Due to their close biochemical properties, purification of individual gingipains is difficult and requires multiple chromatographic steps. In this study, we demonstrate that insertion of a hexahistidine affinity tag upstream of a C-terminal outer membrane translocation signal in RgpB gingipain leads to the secretion of a soluble, mature form of RgpB bearing the affinity tag which can easily be purified by nickel-chelating affinity chromatography. The final product obtained in high yielding and high purity is biochemically indistinguishable from the native RgpB enzyme.
Keywords: cysteine protease, periodontitis, posttranslational modification, proteolytic processing, protein secretion, virulence factor
RgpB belongs to a group of cysteine endoproteases, the gingipains, that are unique to the periodontal pathogen Porphyromonas gingivalis. Being an asaccharolytic, anaerobic Gram-negative bacterium adapted to reside within the crevice between the teeth and the gums in humans, the organism is highly dependent on its proteases to obtain proteinaceous nutrients for growth. The gingipains account for the majority (85%) of the general proteolytic activity detectable in this organism and have also been implicated as important virulence factors in many studies (Potempa et al., 1997). Together, they have been reported to be essential for many stages of the host infection process including: bacterial attachment to host cells; degradation of intercellular adhesion molecules for tissue invasion; promoting inflammation and bleeding through activation and degradation of factors of the complement, kinin and coagulation cascades, and dysregulation of the host’s immune response through degradation of cytokines and their receptors. Furthermore, they partake in the acquisition of iron and haem through degradation of haemoglobin (Guo et al., 2010; Imamura, 2003; O’Brien-Simpson et al., 2009).
There are three members to the gingipain family: RgpA and RgpB hydrolyse Arg-Xaa while Kgp cleaves Lys-Xaa peptide bonds (Veillard et al., 2012). Gingipains are encoded by three genes: rgpA, rgpB and kgp at separate loci in the P. gingivalis genome. While RgpB is a single chain protease, RgpA and Kgp are assembled into multifunctional and non-covalent complexes, composed of the catalytic domain and several hemagglutinin/adhesin domains (Sztukowska et al., 2012). All three enzymes undergo complex post-translational processing and modification steps before being secreted outside the cell to mediate various functions. The newly translated products are translocated through the inner membrane via signal peptide targeting to the Sec machinery. Subsequently, the N-terminal pro-domains, acting as catalytic inhibitors, are removed through a 2-step process (de Diego et al., 2013; Mikolajczyk et al., 2003; Veillard et al., 2013) and the maturing enzymes are targeted by their C-terminal domains (CTD) to a newly described Type 9 Secretion System (T9SS) for translocation through the outer membrane (Nguyen et al., 2007; Sato et al., 2009; Shoji et al., 2011). As part of the maturation process, the CTDs are cleaved off and glycan attachment to the peptide backbone serves to affix the proteases to the outer leaflet of the outer membrane (Glew et al., 2012; Paramonov et al., 2005).
Being important virulence factors for P. gingivalis, the gingipains have been the target of many in vitro and in vivo studies. Towards this aim, producing sufficient amounts of pure enzyme for study is critical. Purification of the native forms of the gingipains has been difficult as in most strains, the proteases are attached to the cell surface by glycan modification during maturation and purification inevitably requires the use of detergents with variable results of the purified product, including co-purification of lipopolysaccharide from the outer membrane. Fortunately, one laboratory strain, HG66, has spontaneously mutated in vitro to release all gingipains into the growth medium and researchers in the gingipain research community have relied on this mutant as a source of soluble gingipains for purification (Eichinger et al., 1999; Shoji et al., 2014). However, purification of soluble gingipains is not a simple process and includes concentrating the protein component of the media fraction by acetone precipitation followed by gel filtration and arginine-affinity chromatography. For RgpB, additional ion exchange and arginine-affinity chromatography steps are required to remove the co-purifying hemin pigment contamination (Potempa and Nguyen, 2007).
To circumvent the complex purification protocol of native gingipains, many laboratories around the world have also tried to produce gingipains recombinantly with affinity tags for ease of purification but all have met with little success. RgpB has been reported to be functionally expressed in a yeast host but the yield was only 10–20 μg/L of culture (Mikolajczyk et al., 2003). The probable reason is that gingipains are probably too toxic to be expressed in heterologous hosts due to the low specificity of substrate recognition. In this manuscript, we present work detailing the development and purification of recombinant, affinity-tagged soluble RgpB expressed within P. gingivalis itself and carried out a comprehensive characterisation of its behaviour to compare it to that of the wild-type RgpB.
Affinity tag chromatography is a powerful technique widely used to purify recombinant proteins expressed in homo- or heterologous hosts. The most commonly used approach is the insertion of an affinity motif at one extremity of the recombinant protein for accessibility to affinity purification matrix. This strategy was used to tag RgpB with 6×His residues for expression within P. gingivalis itself to ensure proper folding and maturation of the protein. Because both the N-terminal inhibitory pro-fragment and CTD domains of RgpB are cleaved off during its maturation, we were able to generate a C-terminally-tagged version of mature RgpB by inserting 6×His just N-terminal to the CTD cleavage site (at position 662) by site-directed mutagenesis to create the mutant 662i6H (Zhou et al., 2013). To prevent the highly homologous rgpA gene from interfering with genetic manipulation of rgpB, creation of the tagged RgpB was carried out in an RgpA-deficient strain (RgpA-C) (Nguyen et al., 2007). Fortuitously, the presence of these additional 6×His residues at the cleavage junction also interferes with the glycan modification step of RgpB maturation and the protein is secreted into the culture medium.
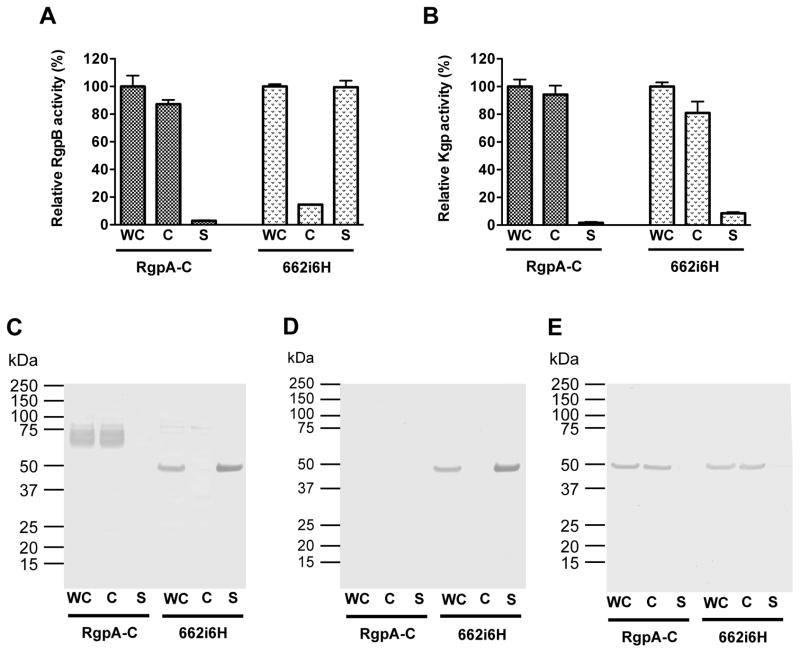
Phenotypically, in contrast to the parental RgpA-C strain whereby RgpB activity is cell-associated, in mutant 662i6H, the majority of RgpB is released into the culture medium (Figure 1A). By Western blot using anti-RgpB, wild-type RgpB from the parental RgpA-C strain migrates as a diffuse band between 70 and 90 kDa representing highly glycosylated forms of the protein whereas mature RgpB from 662i6H migrates at the predicted mature molecular mass of 48 kDa as a sharp band, presumably, in a non-glycosylated form (Figure 1C). Importantly, RgpB from 662i6H is recognized by anti-6×His antibody, indicating that the tag is present as designed and could be used for affinity chromatography (Figure 1D). As expected, Kgp cellular distribution remains unaffected in the 662i6H mutant and the majority was cell-bound as indicated by activity assays and anti-Kgpcat Western-blot (Figures 1B and 1E). Together, these results indicate that the spent culture medium of the 662i6H mutant is a good source of soluble, mature and active 6×His-tagged RgpB for downstream affinity purification.
Figure 1.
Enzymatic activity and Western-blot analysis of RgpB and Kgp in P. gingivalis parental RgpA-null (RgpA-C) strain or the mutant 662i6H. P. gingivalis mutant 662i6H expressing His-tagged RgpB was created as previously described (Zhou et al., 2013). Bacteria were cultivated in enriched tryptic soy broth medium (eTSB) in anaerobic conditions to early stationary phase and cultures were adjusted to the same ODA600 = 1.5 to be designated as “whole culture” (WC). Cells were separated from the spent media fraction by centrifugation (5,000 × g, 20 min) and the cell-free media was designated as “supernatant” (S). Cells were washed and resuspended in PBS to the original volume to obtain the washed “cells” (C) fractions. For each fraction, Rgp (A) and Kgp (B) amidolytic activities were assessed by hydrolysis of the chromogenic substrates benzoyl-l-arginine-pnitroanilide (BAPNA) and acetyl-l-lysine p-nitroanilide (Ac-Lys-pNA) at 37 °C in Assay buffer (200 mM Tris-HCl, 100 mM NaCl, 5 mM CaCl2, pH 7.6 supplemented with fresh 10 mM L-cysteine). For Western-blot analysis, 30 μl of each fraction were resolved on 4–12% Bis-Tris SDS-PAGE gels and electro-transferred onto nitrocellulose membrane. After blocking with skim milk, membranes were probed with rabbit pAb anti-Rgp (C), rabbit pAb anti-6His tag (GenScript, Atlanta, GA, USA) (D) and mouse mAb anti-Kgp (E). After incubation with the corresponding secondary antibodies: anti-rabbit HRP-conjugated (Sigma) or anti-mouse AP-conjugated (Sigma); specific bands were visualized using TMB Membrane Peroxidase Substrate (KPL, Gaithersburg, MD) or AP conjugate Substrate Kit (BioRad, Hercules, CA, USA).
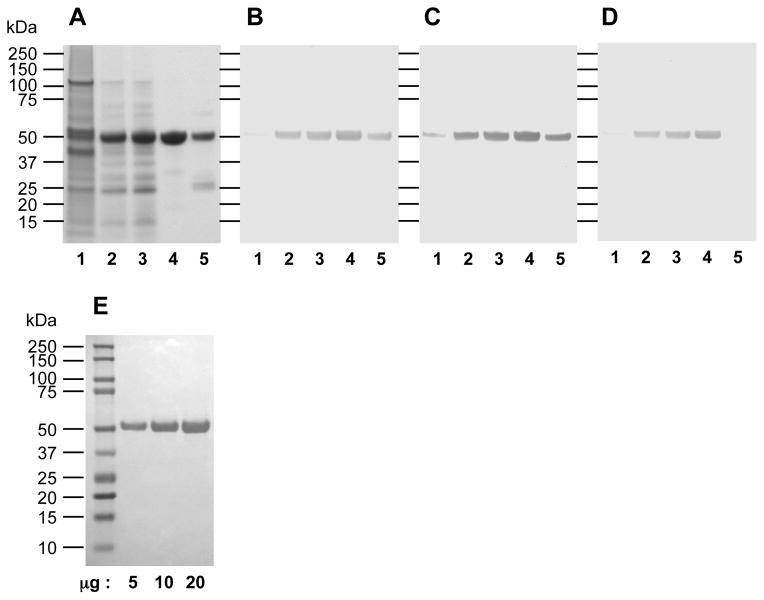
To affinity purify RgpB from mutant 662i6H, bacteria were grown in eTSB medium in anaerobic conditions to early stationary phase and the culture medium was clarified by centrifugation. Similar to the process developed for purification of wild-type gingipains from strain HG66, purification of RgpB-6His started with concentration of the protease from the clarified medium by acetone precipitation. This step of protein recovery from a large volume presents two major advantages over other techniques such as ammonium sulfate precipitation. Firstly, a large amount of hemin and hemin-derived pigments was removed in the process and secondly, it allows a high recovery of active gingipains. The obtained precipitate was dissolved in 4,4′-dithiopyridine disulphide buffer to allow the oxidizing reagent to reversibly modify free thiol residues to inactivate gingipain activity. Gingipain activity could subsequently be regenerated in the presence of a reducing agent (eg. with dithiothreitol or L-cysteine). Such treatment protects gingipains from auto-degradation when highly concentrated and probably protects the catalytic cysteine from more drastic and irreversible oxidation during the purification procedure as observed for other cysteine proteases (Godat et al., 2008). The remaining hemin pigment contamination was removed by adsorption onto DE-52 ion exchange matrix. This ion-exchange matrix is the preferred choice due to its extremely high affinity and binding capacity for hemin and hemin derived-pigments. RgpB-6His does not bind the matrix and is found in the flow-through fraction along with the majority of other proteins (Figure 2A, lane 3). However, this step eliminates the residual hemin-derived pigments as indicated by the substantial increase in the specific activity of the sample (Table 1). While large variabilities in ODA280 were observed after acetone precipitation from various purification runs (ODA280 range: 94.5–777), the yield is more consistent after the DE-52 purification step (ODA280 range: 7.9–18). Thus, the combination of these two steps is necessary to allow for concentration of soluble protein in the culture medium as well as elimination of the majority of hemin-derived pigments.
Figure 2.
SDS-PAGE and Western-blot analysis of RgpB-6His purification steps as described in Table 1. Lanes: 1, initial culture supernatant; 2, after acetone precipitation; 3, after ion exchange chromatography on DE-52 matrix; 4, eluted fraction from Ni-Sepharose affinity chromatography; 5, flow-through fraction from Ni-Sepharose chromatography. 10 μg of each sample were electrophoresed on a 4–12% BisTris SDS-PAGE gel and stained with Simple Blue Safe Stain (Invitrogen) (A). Alternatively, 1 μg of proteins were electrophoresed under the same condition and electro-blotted onto nitrocellulose membrane. After blocking with skim milk, membranes were probed with rabbit pAb antiRgp (B), or mouse mAb anti-6His detecting exclusively hexa-histidine tags (Roche, Indianapolis, IN, USA) (C).
Table 1.
Purification of RgpB-6His from 3 representative purifications from 1 liter of culture
| Purification Stepa | Protein | Activity | Specific activity | Purification | Yield |
|---|---|---|---|---|---|
|
| |||||
| mg | Units b | Units/ODA280 | Fold | % | |
| Culture fluid (range) | 2353 (2033–2611) | 6980 (6000–8439) | 3.0 (2.48–3.2) | - | - |
| Acetone precipitation | 406.5 (94.5–777) | 3895 (3640–4195) | 24.8 (5.4–38.5) | 6.0 (1.7–11.9) | 56 (43–65) |
| DE-52 | 12.2 (7.9–18.0) | 2317 (1065–4730) | 217.6 (54–441) | 69.3 (22–137) | 33 (16–56) |
| Ni-Sepharose/elution | 2.4 (1.1–3.1) | 1634 (1041–2755) | 737.3 (359–992) | 259.6 (112–400) | 22 (16–33) |
| Ni-Sepharose/flow through | 5 (3.5–6.61) | 215 (74–450) | 37.8 (21–68) | 12.5 (6.6–21.1) | 3 (1–5) |
1L of early stationary phase cultures were clarified by centrifugation (18,000 × g, 30 min) and soluble proteins in the medium were precipitated by the slow addition (50 ml/min) of 1.5 L of chilled acetone (−20°C) with vigorous stirring on dry ice. After 1 hour, the precipitant was pelleted (10,000 × g, 20 min) and re-dissolved for 1 hour on ice into 150 mL 20 mM BisTris buffer, 150 mM NaCl, pH 6.5, supplemented with 1.5 mM dithiodipyridine and 0.02% [w:v] NaN3. The sample was dialysed at 4°C against ion exchange buffer (50 mM BisTris buffer, 5 mM CaCl2, pH 6.5 with 0.02% NaN3), then clarified by ultracentrifugation at 4°C for 1 hour at 100,000 × g. The resulting product was loaded onto 50 g of pre-equilibrated DE-52 Whatman anion-exchange resin (GE Healthcare, Pittsburgh, PA, USA) at a flow rate of 1 mL/min. The column was washed with 200 mL of the same buffer and a gradient of NaCl (0 to 500 mM in 400 mL) was applied to elute bound proteins. Fractions were assayed for the RgpB activity using L-BAPNA as substrate. The fractions of flow through containing the majority of RgpB were pooled and dialysed against Ni-Sepharose binding buffer (20 mM sodium phosphate buffer, 500 mM NaCl, 20 mM imidazole, pH 7.4 with 0.02% NaN3). The retentate obtained was applied to a 10 mL pre-equilibrated Ni2+-Sepharose 6 Fast Flow matrix (GE Healthcare, Pittsburgh, PA) and bound proteins were eluted with binding buffer supplemented with 500 mM imidazole. Two peaks containing protease activity were separated in this manner corresponding to the affinity-bound and unbound proteins. Fractions of those two peaks were combined separately and dialysed at 4 °C against 20 mM BisTris 150 mM NaCl, 5 mM CaCl2, pH 6.8 with 0.02% NaN3. Protein concentration of the final samples was determined by BCA Assay (Sigma) and the active fraction of RgpB-6His was determined by titration against D-Phe-Phe-Arg-chloromethylketone (FFR-CMK) as published previously (Potempa and Nguyen, 2007).
Amidase activity using L-BAPNA as substrate; 1 unit = A405nm of 1.00/ml/min at 37°C
The concentrated protein sample was subsequently applied onto nickel-chelating Sepharose matrix to affinity purify RgpB-6His. A majority of the RgpB activity was found to be retained on the matrix as expected and eluted into the elution fraction but a small quantity of RgpB (<3%; Table 1) did not bind and was lost in the flow-through fraction. Western-blot analysis of bound and un-bound RgpB forms separated on the affinity column clearly shows that the latter did not interact with the anti-6×His antibody (Figure 2C, lane 5). This C-terminal heterogeneity could be explained by the action of endogenous carboxy-peptidase(s) of P. gingivalis during the secretion and maturation processes or during the purification procedure. The affinity purified RgpB-6His product is highly purified (>95%) as confirmed by SDS-PAGE (Figure 2A) as well as gel filtration analysis (data not shown). Importantly, no contaminating Kgp proteolytic activity could be detected using the classical chromogenic substrate Ac-Lys-pNA or with the more sensitive fluorogenic tosyl-Gly-Pro-Lys-AMC substrate (data not shown). To test the quality of the purified RgpB-6His, the active RgpB fraction was determined using the titrating inhibitor FFR-CMK as compared to the theoretical concentration based on RgpB-6His molecular mass and protein concentration obtained from BCA assay. Based on this titration, the final RgpB-6His is 88% active (mean of 3 purifications; range: 82–92%), thus, confirming high purity of the enzyme and indicates that the majority is not irreversibly inactivated during the purification process.
In comparison to conventionally purified wild-type RgpB (the complete and detailed protocol is described in (Potempa and Nguyen, 2007)), RgpB-6His purification has significantly higher yield than that of native RgpB (22% versus 13%, respectively) and in greater quantity from 1 liter of original culture (2.4 mg versus 0.5 mg, respectively) (Potempa et al., 1998). Further, RgpB-6His obtained after Ni-Sepharose affinity constitutes the final purification product being highly purified, whereas wild-type RgpB purification requires additional steps to eliminate cross-contamination by the other gingipains. Indeed, the gel filtration chromatography step of the native protocol fails to eliminate contaminating RgpA and Kgp activities (Potempa and Nguyen, 2007). Additional chromatographic steps, such as Arg-sepharose affinity chromatography, high-resolution ion exchange chromatography or chromatofocusing are usually required to further purify wild-type RgpB (Potempa et al., 1998). While each of the supplementary purification steps increased the purity of RgpB, it also reduced the yield of recovery. Kgp contamination in the wild-type RgpB preparation could be inhibited with a Lys-specific protease inhibitor Ac-Leu-Val-Lys-aldehyde (Ac-LVK-ald), but no easy solution exists for RgpA contamination because of the identical enzymatic specificities between RgpA and RgpB. The same applies to the detection and quantification of the contaminant since no synthetic substrate is able to discriminate RgpA from RgpB activity.
In this regard, purification of RgpB-6His from 662i6H presents great advantages. First, it is expressed in an RgpA-deficient strain, eliminating the risk of undetectable contamination with this enzyme. Second, the majority of the expressed Kgp is glycosylated and remained bound to the bacterial membrane, thus, reducing greatly the amount of contaminating Kgp in the initial cell-free culture fluid of 662i6H as compared to the HG66 strain. Finally, high specificity of the hexahistidine-Ni-Sepharose matrix interaction ensures the elimination of the remaining residual Kgp. Altogether, this approach reduces the number of chromatographic steps needed to obtain pure RgpB, thus increasing significantly the final yield of purification. Interestingly, the specific activity of RgpB-6His was higher than that of the native RgpB. This difference in specific activity was directly proportional to the difference in the content of the active enzyme determined by the active site titration.
Protein tagging with hexahistidine tag is a powerful method to facilitate the purification of recombinant proteins. However, a potential downside of this approach is that the insertion of this highly polar motif can lead to structural changes and results in alteration to the catalytic properties of the enzyme. To discount this possibility, we proceeded to compare the enzymatic properties of the recombinant RgpB-6His to wild-type RgpB.
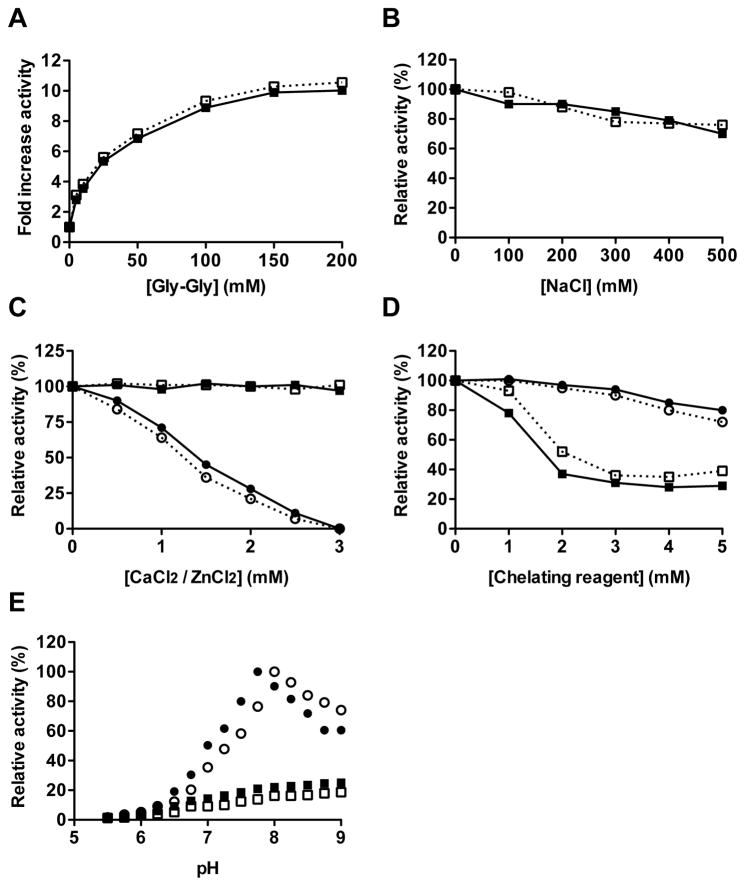
One singular property of RgpB is the stimulation of its amidolytic activity by amino-acids and dipeptides, particularly glycyl-glycine (Chen et al., 1991). In this regard, the activity of RgpB-6His is stimulated up to 10-fold in presence of 200 mM Gly-Gly – a value comparable to RgpB (Supplementary Figure 1A). Moreover, this dipeptide is also known to influence the pH optimum of RgpB. Without the dipeptide, both forms of RgpB shows a very broad spectrum of activity from pH 6 to an optimum pH 9. However, in presence of 50 mM Gly-Gly, the pH optimum for RgpB and RgpB-6His activity are 7.75 and 8, respectively (Supplementary Figure 1E). At higher pH values, the activity decreased rapidly probably due to an enhanced inactivation rate (Potempa et al., 1998). Ionic strength is not known to have a strong effect on RgpB proteolytic activity. Accordingly, the RgpB activity is stable with increasing concentrations of salt, losing only 15% of its activity at 500 mM NaCl (Supplementary Figure 1B). The crystallographic structure of wild-type RgpB has shown that the enzyme possesses two binding sites for the divalent Ca2+ cations (Eichinger et al., 1999). Accordingly, the native and recombinant RgpB are both sensitive to the general chelating reagent EDTA with 60% of inhibition at 2 mM, but show no susceptibility to the more zinc-specific 1,10-orthophenanthroline (Supplementary Figure 1D). In the same manner, Ca2+ has no effect on both enzymes, whereas increasing concentrations of ZnCl2 lead to their inactivation (Supplementary Figure 1C). ZnCl2, up to 10 mM concentration, neither affected pH of the assay buffer nor caused precipitation of RgpB-6xHis. The observed inhibition most likely results from chelating of Zn2+ by the catalytic cysteine residue of RgpB.. Altogether, these results indicate that the addition of the hexahistidine tag has no effect on RgpB biochemical properties.
To confirm this observation, we determined the kinetic constant of the hydrolysis of the most commonly used synthetic substrates by non-linear regression using Graphpad Prism software version 5 (GraphPad Software, La Jolla, USA). Given its low specificity, native RgpB is expected to hydrolyse every synthetic substrate with the arginine residue at position P1. The Km values for synthetic substrates are in the range of 10 to 20 μM, except for substrates with a P2 proline residue that results in a higher Km (up to 100 μM). Similarly, the kcat values are also within a narrow range (from 5 to 50 s−1) and are independent of the sequence of the substrate (Potempa et al., 1998). Comparatively, recombinant RgpB-6His exhibits similar kinetic properties: the Km values obtained with the chromogenic substrate benzoyl-l-arginine-p-nitroanilide (L-BAPNA: 9.14 μM), the fluorogenic substrates acetyl-L-arginine 7-amino-4-methylcoumarin (Ac-Arg-AMC: 9.24 μM) and N-benzyloxycarbonyl-phenylalanine-arginine 7-amino-4-methylcoumarin (Z-FR-AMC: 14.66 μM) were found in the range previously reported. The same observation can be made with regard to the kcat with values of 8.32 s−1 (L-BAPNA), 7.88 s−1 (Ac-Arg-AMC) and 3.61 s−1 (Z-FR-AMC).
Altogether, these results indicate that the hexahistidine tag present at the C-terminus of the mature, recombinant RgpB has no effect on the biochemical behaviour of the enzyme and does not modify its catalytic properties. In this regard, RgpB-6His could be a useful alternate source to replace wild-type RgpB for in vitro experimentation.
Historically, the isolation of the P. gingivalis HG66 strain eased the purification of gingipains by providing a reliable source of soluble enzymes. However, due to their close biochemical properties, the individual isolation of each of the three gingipains is laborious and difficult, requiring multiple chromatography steps. In this study, we demonstrate that the insertion of a hexahistidine tag upstream of the CTD cleavage site in RgpB leads to the secretion of a soluble, mature form of RgpB bearing a C-terminal hexahistidine tag to facilitate its purification by affinity chromatography. The resulting His-tagged RgpB of high yield and purity was observed to possess the same biochemical and catalytic properties as wild-type RgpB.
The CTD domain is not limited to gingipains but is found in other virulence proteins from P. gingivalis and other periodontal pathogens belonging to the Bacteriodetes phylum (Nguyen et al., 2007). Similar to CTD in RgpB, it acts as a targeting signal for the translocation of these proteins through the T9SS and is cleaved off during the process (Veith et al., 2013). There is evidence that the processing mechanism and membrane attachment is conserved for all CTD-bearing proteins (Veith et al., 2014). Further, genetic manipulation to insert hexahistidine residues into the CTD cleavage site in a carboxypeptidase Cpg70 has been reported to result in the secretion of Cpg70 into the extracellular medium in a manner similar to RgpB (Zhou et al., 2013). Thus, the approach developed in this work could be useful for the purification and characterisation of other virulence factors secreted by P. gingivalis and in other closely related periodontal pathogens such as Prevotella intermedia and Tannerella forsythia.
Supplementary Material
Figure 3.
Comparison of activity of RgpB-6His and native RgpB in various conditions. The amidolytic activities of Rgp-6His (open symbols) and native RgpB (filled symbols) were determined at 37°C using the chromogenic substrate L-BAPNA (λ = 405 nm; n = 3) in activity assay buffer (200 mM Tris, 10 mM L-cysteine, pH 7.6) supplemented with increasing concentration of: the dipeptide Gly-Gly (A); NaCl (B); divalent cations Ca2+ (square) or Zn2+ (circle) (C); and the chelating reagents EDTA (square) and 1,10 orthophenanthroline (circle) (D). Alternatively, the effect of the pH on the amidolytic activity was determined in 0.1 M Tris, 0.05 M MES and 0.05 M acetic acid buffer adjusted from pH 5 to 9 with (circle) or without (square) 50 mM Gly-Gly (E).
Acknowledgments
This study was supported by grants from National Science Center, Poland (2012/04/A/NZ1/00051), US NIH (DE 09761 and DE 022597), the European Commission (FP7-PEOPLE-2011-ITN-290246 “RAPID” and FP7-HEALTH-F3-2012-306029 “TRIGGER”), and Polish Ministry of Science and Higher Education (UMO-2795/7.PR/13/2017/2). MK obtained a doctoral scholarship (2013/08/T/NZ1/00315) from National Science Center, Poland. The Faculty of Biochemistry, Biophysics and Biotechnology is a partner of the Leading National Research Center (KNOW) supported by the Ministry of Science and Higher Education.
References
- Chen ZX, Potempa J, Polanowski A, Renvert S, Wikstrom M, Travis J. Stimulation of proteinase and amidase activities in Porphyromonas (Bacteroides) gingivalis by amino acids and dipeptides. Infect Immun. 1991;59:2846–2850. doi: 10.1128/iai.59.8.2846-2850.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu J, March PE, Lee R, Tillett D. Site-directed, Ligase-Independent Mutagenesis (SLIM): a single-tube methodology approaching 100% efficiency in 4 h. Nucleic Acids Res. 2004;32:e174. doi: 10.1093/nar/gnh172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Diego I, Veillard FT, Guevara T, Potempa B, Sztukowska M, Potempa J, Gomis-Ruth FX. Porphyromonas gingivalis virulence factor gingipain RgpB shows a unique zymogenic mechanism for cysteine peptidases. J Biol Chem. 2013;288:14287–14296. doi: 10.1074/jbc.M112.444927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichinger A, Beisel HG, Jacob U, Huber R, Medrano FJ, Banbula A, Potempa J, Travis J, Bode W. Crystal structure of gingipain R: an Arg-specific bacterial cysteine proteinase with a caspase-like fold. EMBO J. 1999;18:5453–5462. doi: 10.1093/emboj/18.20.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glew MD, Veith PD, Peng B, Chen YY, Gorasia DG, Yang Q, Slakeski N, Chen D, Moore C, Crawford S, Reynolds EC. PG0026 is the C-terminal signal peptidase of a novel secretion system of Porphyromonas gingivalis. J Biol Chem. 2012;287:24605–24617. doi: 10.1074/jbc.M112.369223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godat E, Herve-Grvepinet V, Veillard F, Lecaille F, Belghazi M, Bromme D, Lalmanach G. Regulation of cathepsin K activity by hydrogen peroxide. Biol Chem. 2008;389:1123–1126. doi: 10.1515/BC.2008.109. [DOI] [PubMed] [Google Scholar]
- Guo Y, Nguyen KA, Potempa J. Dichotomy of gingipains action as virulence factors: from cleaving substrates with the precision of a surgeon’s knife to a meat chopper-like brutal degradation of proteins. Periodontol 2000. 2010;54:15–44. doi: 10.1111/j.1600-0757.2010.00377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura T. The role of gingipains in the pathogenesis of periodontal disease. J Periodontol. 2003;74:111–118. doi: 10.1902/jop.2003.74.1.111. [DOI] [PubMed] [Google Scholar]
- Mikolajczyk J, Boatright KM, Stennicke HR, Nazif T, Potempa J, Bogyo M, Salvesen GS. Sequential autolytic processing activates the zymogen of Arg-gingipain. J Biol Chem. 2003;278:10458–10464. doi: 10.1074/jbc.M210564200. [DOI] [PubMed] [Google Scholar]
- Nguyen KA, Travis J, Potempa J. Does the importance of the C-terminal residues in the maturation of RgpB from Porphyromonas gingivalis reveal a novel mechanism for protein export in a subgroup of Gram-Negative bacteria? J Bacteriol. 2007;189:833–843. doi: 10.1128/JB.01530-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien-Simpson NM, Pathirana RD, Walker GD, Reynolds EC. Porphyromonas gingivalis RgpA-Kgp proteinase-adhesin complexes penetrate gingival tissue and induce proinflammatory cytokines or apoptosis in a concentration-dependent manner. Infect Immun. 2009;77:1246–1261. doi: 10.1128/IAI.01038-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paramonov N, Rangarajan M, Hashim A, Gallagher A, Aduse-Opoku J, Slaney JM, Hounsell E, Curtis MA. Structural analysis of a novel anionic polysaccharide from Porphyromonas gingivalis strain W50 related to Arg-gingipain glycans. Mol Microbiol. 2005;58:847–863. doi: 10.1111/j.1365-2958.2005.04871.x. [DOI] [PubMed] [Google Scholar]
- Potempa J, Mikolajczyk-Pawlinska J, Brassell D, Nelson D, Thogersen IB, Enghild JJ, Travis J. Comparative properties of two cysteine proteinases (gingipains R), the products of two related but individual genes of Porphyromonas gingivalis. J Biol Chem. 1998;273:21648–21657. doi: 10.1074/jbc.273.34.21648. [DOI] [PubMed] [Google Scholar]
- Potempa J, Nguyen KA. Purification and characterization of gingipains. Curr Protoc Protein Sci. 2007;Chapter 21(Unit 21):20. doi: 10.1002/0471140864.ps2120s49. [DOI] [PubMed] [Google Scholar]
- Potempa J, Pike R, Travis J. Titration and mapping of the active site of cysteine proteinases from Porphyromonas gingivalis (gingipains) using peptidyl chloromethanes. Biol Chem. 1997;378:223–230. doi: 10.1515/bchm.1997.378.3-4.223. [DOI] [PubMed] [Google Scholar]
- Sato K, Kido N, Murakami Y, Hoover CI, Nakayama K, Yoshimura F. Lipopolysaccharide biosynthesis-related genes are required for colony pigmentation of Porphyromonas gingivalis. Microbiology. 2009;155:1282–1293. doi: 10.1099/mic.0.025163-0. [DOI] [PubMed] [Google Scholar]
- Shoji M, Sato K, Yukitake H, Kondo Y, Narita Y, Kadowaki T, Naito M, Nakayama K. Por secretion system-dependent secretion and glycosylation of Porphyromonas gingivalis hemin-binding protein 35. PLoS One. 2011;6:e21372. doi: 10.1371/journal.pone.0021372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji M, Sato K, Yukitake H, Naito M, Nakayama K. Involvement of the Wbp pathway in the biosynthesis of Porphyromonas gingivalis lipopolysaccharide with anionic polysaccharide. Sci Rep. 2014;4:5056. doi: 10.1038/srep05056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztukowska M, Veillard F, Potempa B, Bogyo M, Enghild JJ, Thogersen IB, Nguyen KA, Potempa J. Disruption of gingipain oligomerization into non-covalent cell-surface attached complexes. Biol Chem. 2012;393:971–977. doi: 10.1515/hsz-2012-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veillard F, Potempa B, Poreba M, Drag M, Potempa J. Gingipain aminopeptidase activities in Porphyromonas gingivalis. Biol Chem. 2012;393:1471–1476. doi: 10.1515/hsz-2012-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veillard F, Sztukowska M, Mizgalska D, Ksiazek M, Houston J, Potempa B, Enghild JJ, Thogersen IB, Gomis-Ruth FX, Nguyen KA, Potempa J. Inhibition of gingipains by their profragments as the mechanism protecting Porphyromonas gingivalis against premature activation of secreted proteases. Biochim Biophys Acta. 2013;1830:4218–4228. doi: 10.1016/j.bbagen.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veith PD, Chen YY, Gorasia DG, Chen D, Glew MD, O’Brien-Simpson NM, Cecil JD, Holden JA, Reynolds EC. Porphyromonas gingivalis outer membrane vesicles exclusively contain outer membrane and periplasmic proteins and carry a cargo enriched with virulence factors. J Proteome Res. 2014;13:2420–2432. doi: 10.1021/pr401227e. [DOI] [PubMed] [Google Scholar]
- Veith PD, Nor Muhammad NA, Dashper SG, Likic VA, Gorasia DG, Chen D, Byrne SJ, Catmull DV, Reynolds EC. Protein substrates of a novel secretion system are numerous in the Bacteroidetes phylum and have in common a cleavable C-terminal secretion signal, extensive post-translational modification, and cell-surface attachment. J Proteome Res. 2013;12:4449–4461. doi: 10.1021/pr400487b. [DOI] [PubMed] [Google Scholar]
- Zhou XY, Gao JL, Hunter N, Potempa J, Nguyen KA. Sequence-independent processing site of the C-terminal domain (CTD) influences maturation of the RgpB protease from Porphyromonas gingivalis. Mol Microbiol. 2013;89:903–917. doi: 10.1111/mmi.12319. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.