Abstract
RNA binding proteins are capable of regulating translation initiation by a variety of mechanisms. Although the vast majority of these regulatory mechanisms involve translational repression, one example of translational activation has been characterized in detail. The RNA recognition targets of these regulatory proteins exhibit a wide range in structural complexity, with some proteins recognizing complex pseudoknot structures and others binding to simple RNA hairpins and/or short repeated single-stranded sequences. In some instances the bound protein directly competes with ribosome binding, and in other instances the bound protein promotes formation of an RNA structure that inhibits ribosome binding. Examples also exist in which the bound protein traps the ribosome in a complex that is incapable of initiating translation.
Keywords: repression, activation, autoregulation, protein-RNA interaction, RNA structure
INTRODUCTION
Bacteria and their viruses are capable of rapid alterations in gene expression in response to changing environmental and physiological conditions. Gene expression in bacteria is regulated at many levels, including transcription, translation, and mRNA stability. Translation initiation in bacteria begins with the formation of a binary complex consisting of the 30S ribosomal subunit and either mRNA or fmet-tRNAfMet. The binary complex is converted to a preternary complex following binding of the other RNA molecule; however, the two RNA molecules in the preternary complex do not interact with one another. Conversion of the preternary complex to the translation-competent ternary complex involves proper interaction of mRNA with fmet-tRNAfMet (32). The Shine-Dalgarno (SD) sequence in mRNA base-pairs with the anti-SD sequence in 16S rRNA within the 30S subunit and thereby correctly positions the initiation codon in the ribosome. Efficient translation often leads to increased mRNA stability, as translating ribosomes can protect the mRNA from nucleolytic attack.
Studies on the regulation of protein synthesis have shown that RNA structural features present in the 5′-untranslated leaders can influence translation initiation in bacteria (32, 47, 77). A variety of translational control mechanisms that involve regulated access of ribosomes to mRNA in bacteria and bacteriophages have been identified. It is well documented that RNA secondary structures can sequester the SD sequence and prevent ribosome binding. Protein-independent mechanisms in which the kinetics of RNA folding plays a crucial role in regulating the rate of SD-sequestering hairpin formation have been characterized (64). Protein-dependent formation (14), stabilization (33, 42), or destabilization (27, 72) of SD sequestering hairpins controls translation initiation of a variety of genes. Several examples exist in which an RNA binding protein can directly compete with ribosome binding. Most of these cases involve a structured RNA recognition target (8, 10, 33, 42, 45), although there are examples in which a sequence-specific RNA binding protein can compete with ribosome binding (6, 19).
This review describes some of the best-characterized examples in which RNA binding proteins specifically control translation initiation of their cognate mRNA targets. In most cases, the protein either directly or indirectly competes with 30S ribosomal subunit binding to the mRNA; however, two examples have been identified in which protein-mRNA interaction entraps the 30S subunit in an inactive complex (17, 48). Virtually all known examples of protein-dependent translational control are repression mechanisms; however, one bacterial translation activation mechanism has been described in detail (26).
TRANSLATIONAL REPRESSION: FEEDBACK REGULATION
Translation of most Escherichia coli ribosomal protein genes is regulated by autoregulatory feedback mechanisms in which one gene in the operon encodes a ribosomal protein (r-protein) that can bind either to rRNA during ribosome biogenesis or to its mRNA and repress translation (31, 77). The alternative r-protein binding sites are similar in structure and hence constitute interesting examples of molecular mimicry. During rapid growth the r-protein binds to its target in newly synthesized rRNA; however, under unfavorable growth conditions rRNA synthesis is reduced such that free r-protein instead binds to its mRNA target. The bound r-protein typically inhibits translation of one cistron in the mRNA, while the downstream cistrons are often repressed via translational coupling, a process in which translation of a downstream cistron is at least partially dependent on translation of the cistron just upstream. In several cases repression occurs by a competition mechanism in which the r-protein prevents 30S ribosomal subunit binding. In contrast, two entrapment mechanisms in which the regulatory r-protein functions by trapping the 30S subunit in an inactive complex with the mRNA have been identified (31, 77). Feedback regulation is not restricted to r-protein genes. An E. coli tRNA synthetase enzyme that can either charge its cognate tRNAs or bind to its own mRNA and repress translation has been identified (44).
E. coli L20
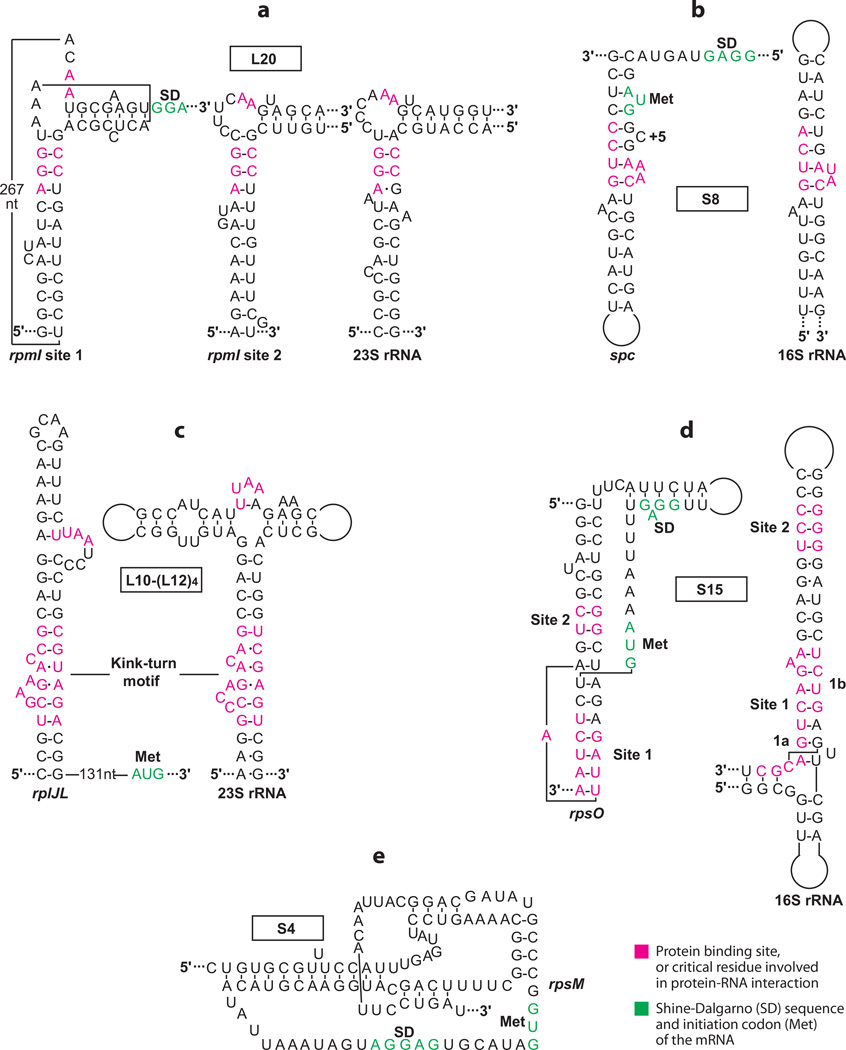
The L35 operon of E. coli (rpmI-rplT) encodes r-proteins L35 and L20 (31, 77). The translational repressor protein L20, encoded by rplT, binds to its leader transcript and competes with 30S ribosomal subunit binding (25). Bound L20 directly represses L35 synthesis, while translation of its own gene is inhibited via translational coupling. The regulatory region of this operon is particularly long (~450 nt) and includes several stem-loop structures, as well as a long-range pseudoknot. This pseudo-knot forms between the loop of one hairpin and a single-stranded segment just upstream from the rpmI SD sequence (10). L20 is capable of binding to one of two sites in the leader transcript: the pseudoknot structure (binding site 1) and a bulged stem-loop (binding site 2) positioned a few bases upstream from the rpmI SD sequence. Both binding sites are similar to the L20 binding site in 23S rRNA (Figure 1a) (23). Although both binding sites appear to be important for autogenous control, L20 is unable to bind to both sites simultaneously (1). Because the stem-loop of binding site 2 is capable of forming prior to the pseudoknot containing binding site 1, a translation control model in which L20 first binds to binding site 2 has been proposed. Once the pseudoknot structure forms, a repressing complex forms between L20 and the mRNA, although it is not known which of the two binding sites would be occupied by L20 (25). A structure of the repressing complex would likely answer this question.
Figure 1.
Feedback regulation of E. coli ribosomal protein operons. Structures of the mRNA (a–e) and rRNA (a–d ) binding sites for each protein are shown. (a) L20 binding sites in the rpmI leader and 23S rRNA transcripts (23). (b) S8 binding sites in the spc and 16S rRNA transcripts (39). (c) L10-(L12)4 binding sites in the rplJ leader and 23S rRNA transcripts (29). (d ) S15 binding sites in the rpsO and 16S rRNA transcripts (17). (e) Pseudoknot involved in S4 binding to the rpsM transcript (48).
E. coli S8
The spc operon of E. coli (rplNXE-rpsNH-rplFR-rpsE-rpmD-rplO-secY-rpmJ) encodes 11 r-proteins and SecY, a component of the secretion apparatus (31, 77). The translation re-pressor protein S8, encoded by rpsH, binds to the rplE translation initiation region and represses L5 synthesis by competing with 30S ribosomal subunit binding. Repression is extended to several of the downstream cistrons via translational coupling. The S8 binding site in rplE is included within a bulged RNA hairpin that begins just upstream of the initiation codon (8). Although the structure of this hairpin is similar to that of the S8 binding site in 16S rRNA (Figure 1b), the affinity of S8 for its rRNA target is about fivefold higher (21). Two extra bulged nucleotides within the mRNA target (+2 and +5, Figure 1b) reduce the affinity of S8 for its mRNA target (71). Crystal structures of Methanococcus jannaschii S8 bound to a 16S rRNA fragment (59), as well as E. coli S8 bound to a synthetic mRNA target, have been reported (39). The two structures are highly similar despite the sequence differences of the two RNAs. S8 binds to one face of the synthetic mRNA target, the internal loop and the terminal loop; however, the spc operon mRNA does not include a terminal loop equivalent to the synthetic mRNA target (39). Thus, it appears that the most relevant interaction from the crystal structure is the packing of an antiparallel β-sheet in the C-terminal domain of S8 against the minor grove of the internal loop, as side chains within this β-sheet make the only two base-specific hydrogen bond contacts in the entire complex. The two extra bulged nucleotides that reduce affinity of S8 for spc mRNA had no effect on the conformation of the RNA that interacts with S8 (39). Although the structural basis for reduced affinity of S8 for it smRNA target remains unresolved, the S8-mRNA structure is consistent with a model in which bound S8 would compete with 30S subunit binding.
E. coli L10-(L12)4
The L10 operon of E. coli (rplJL) encodes r-proteins L10 and L12 (31, 77). The translation repressor of the L10 operon consists of a complex of one L10 subunit and four L12 subunits. The L10-(L12)4 complex binds to the rplJL leader ~150 bases upstream from the rplJ initiation codon and directly represses translation of the first cistron. However, the mechanism by which the bound complex represses translation at such a considerable distance has not been identified. A recent modeling and RNA mutagenesis study identified key features of the binding targets in 23S rRNA and mRNA (29). The minimal mRNA recognition structure, which is conserved among several bacterial species, includes a kink-turn motif and a UUAA bulge loop sequence (Figure 1c) (11, 29). Both of these elements are also part of the 23S rRNA binding target, although the structural context of the UUAA sequence differs; the recognized loop in rRNA is a U-turn motif (Figure 1c). Nevertheless, the different structural contexts of the loops apparently evolved so that they contribute similarly to the L10-(L12)4 binding affinity (29). While the repressor complex binds to both RNAs with comparable affinity, cooperative interaction of L11 and the L10-(L12)4 complex with the rRNA target leads to increased affinity of the L10-(L12)4 complex for 23S rRNA, thereby providing an explanation for how ribosomes affectively compete with mRNA for L10-(L12)4 binding (29). The location of putative L10-(L12)4 mRNA binding sites in a variety of bacterial species suggests that similar autoregulatory mechanisms might control expression of their respective operons; however, L10-(L12)4-mediated regulation has only been reported for E. coli (29).
E. coli S4
The α operon of E. coli (rpsMKD-rpoD-rplQ) encodes four ribosomal subunits and the α-subunit of RNA polymerase. Ribosomal subunit S4, encoded by rpsD, binds to the mRNA leader and directly inhibits translation of rpsM (31, 77). The binding site for S4 consists of a nested pseudoknot structure that includes the SD sequence and translation initiation region; the pseudoknot extends from about −75 to +35 nt relative to the rpsM initiation codon (Figure 1e) (48). Results from RNA toeprint and electrophoretic mobility experiments indicated that the leader RNA is capable of undergoing a conformational switch between active and inactive states of the pseudoknot (48, 56). Although 30S ribosomal subunits are capable of binding to both conformations, only the active conformation is proficient for translation (57). S4 binds only the inactive mRNA conformation and traps 30S ribosomal subunits in a dead-end complex that is unable to bind initiator tRNA (56). This repression mechanism is unusual in that S4 functions as an allosteric effector that drives the equilibrium between two conformation states of the RNA toward the inactive state. Moreover, S4 acts at a second irreversible step leading to entrapment of the 30S subunit in an inactive complex with mRNA (48). As structural information of S4 interaction with its mRNA target is not available, the critical contacts of this r-protein with the pseudoknot structure have not been firmly established. However, the finding that both S4 and 30S subunits can simultaneously bind to the mRNA provides strong evidence that the S4 binding site does not include the rpsD SD sequence and initiation codon. A similar translation repression mechanism may be responsible for controlling expression of the α operon of Bacillus subtilis, although details of the S4-mediated translation repression mechanism were not investigated in this organism (22).
E. coli S15
The S15 operon of E. coli (rpsO-pnp) encodes r-protein S15 and PNPase, a 3′ to 5′ exoribonuclease (13, 31, 77). The translation repressor protein S15 binds to a region that contains the rpsO translation initiation region. As in the case of S4-mediated translational repression, S15 binds to a pseudoknot structure in the mRNA and blocks translation via an entrapment mechanism. Crystal structures of the Thermus thermophilus 30S ribosomal subunits (49, 68) allowed modeling of E. coli S15 with 16S rRNA (52). S15 recognizes two sites in 16S rRNA that are close to one another (Figure 1d). Site 1 is subdivided into subsites 1a and 1b. Subsite 1a is part of a three-way helical junction, and subsite 1b is formed by one of the stems. Site 2 consists of adjacent GU and GC base pairs and is located near subsite 1b on the same stem (17). The pseudoknot of rpsO mRNA consists of two stacked helices that are bridged by a single adenine, and the SD sequence and AUG initiation codon are contained within a large connecting loop that is not part of the S15 binding site (Figure 1d). The S15 binding sites in 16S rRNA and rpsO mRNA share the GU and GC base pairs of site 2 but additional sequence and/or structural conservation is not readily apparent. Footprinting and mutagenesis studies of both S15 and its binding site in rpsO mRNA identified important contacts involved in S15 binding (17, 53). The GC base pair of the GU/GC motif in both mRNA and rRNA is recognized by His41 and Asp48 of S15. Although the details of how S15 recognizes the GU pair are not firmly established, recognition of the GU/GC motif in mRNA and rRNA is similar (17). A likely interaction unique to rpsO mRNA is between Arg57 and the single A residue that bridges the two stacked helices of the pseudo-knot. As mentioned above, binding of S15 to rpsO mRNA does not prevent 30S subunit binding but instead traps the 30S subunit in a nonproductive complex. A recent cryo-electron microscopy study identified the S15-rpsO mRNA docking site on the ribosome. Once S15 dissociates from the complex, the mRNA unfolds such that productive start codon–initiator tRNA interactions can take place (36).
E. coli ThrRS
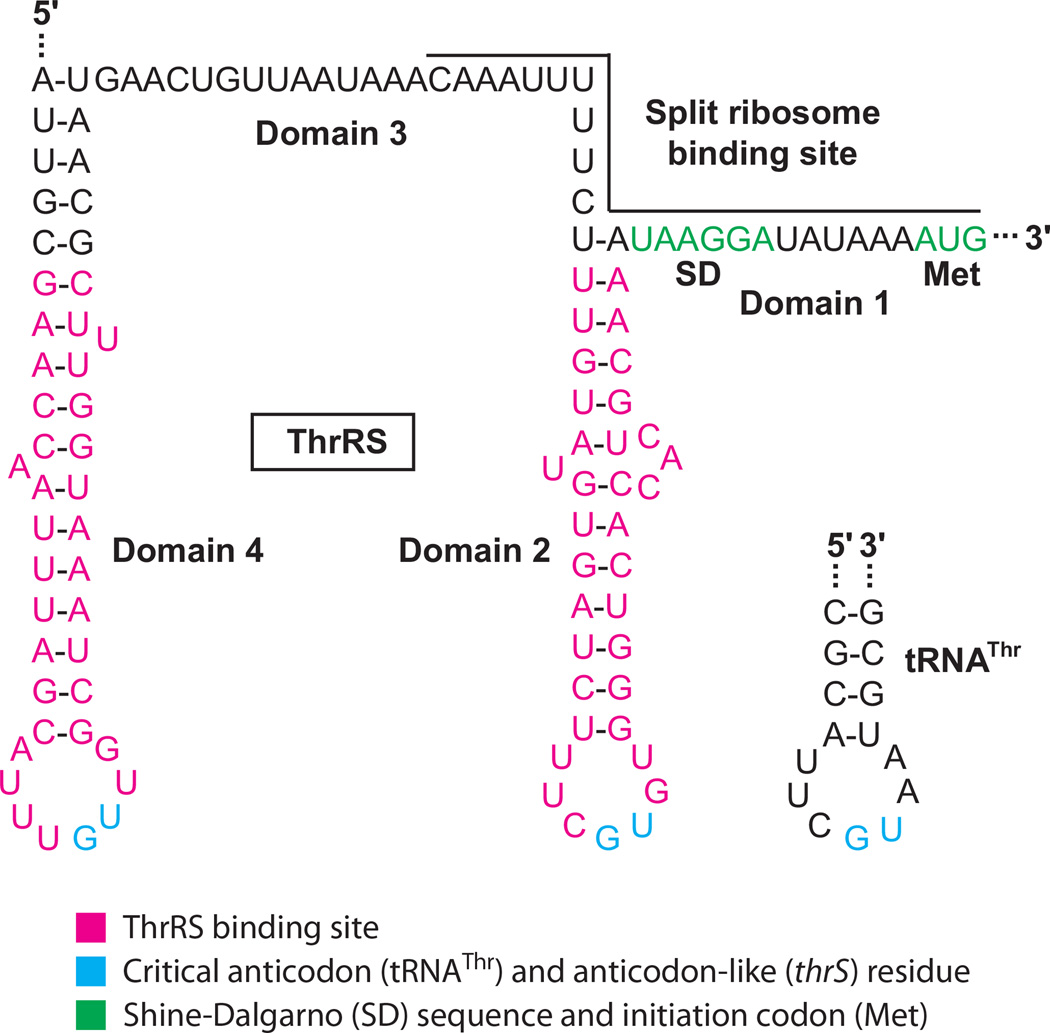
Threonyl-tRNA synthetase (ThrRS), the enzyme responsible for charging tRNAThr, represses translation of its own gene (thrS) by competing with ribosome binding (44). The thrS translation initiation and autoregulatory regions consist of four domains (Figure 2). Domain 1 contains the initiation codon and the SD sequence, domains 2 and 4 consist of RNA hairpins, and domain 3 is a single-stranded segment separating the two hairpins. thrS is unusual in that it contains a split ribosome binding site consisting of domain 1 and part of domain 3 (45). ThrRS functions as a homodimer that can bind to two tRNAThr molecules for aminoacylation. Alternatively, ThrRS can bind to the domain 2 and 4 hairpins within its own leader transcript. Binding of these two alternative substrates provides an exquisite example of molecular mimicry, as both of the thrS leader hairpins closely resemble the an-ticodon loop of the tRNA substrate (Figure 2). The first base of the anticodon-like loops in thrS mRNA can be changed without adverse affects on regulation, whereas changing the second or third position eliminates repression (43). This pattern of mRNA recognition mirrors that of the tRNAThr isoacceptors for which the first position of the anticodon can be C, G, or U, whereas the second and third positions must be G and U, respectively (44). Crystal structures of ThrRS complexed with two tRNAThr molecules (46) or two domain 2 binding sites (60) indicate that recognition of the anticodon and anticodon-like loops involves interaction with the same amino acids within the C-terminal domain of the protein. In addition, the backbone of the tRNA anticodon stem and the stem of domain 2 bind in a similar fashion to the catalytic domain of ThrRS. Biochemical and mutagenesis studies indicate that domain 4 is recognized in the same way as domain 2 (43, 44). Although the binding sites for ThrRS and 30S ribosomal subunits do not strictly overlap, bound ThrRS competes with ribosome binding (44). A cocrystal structure of the T. thermophilus ribosome bound to a fragment of thrS mRNA provides an explanation of how ThrRS could block ribosome binding. Molecular modeling indicates that a steric clash between the N-terminal domain of ThrRS and the 30S subunit would prevent simultaneous binding of ThrRS and the ribosome to the same thrS mRNA molecule (30). In support of this model, deletion of the N-terminal domain of ThrRS prevents translational regulation in vivo. Furthermore, the truncated form of the enzyme was unable to compete with ribosome binding, despite its ability to bind to its mRNA target in vitro (7).
Figure 2.
Feedback regulation of Escherichia coli thrS. In the absence of bound ThrRS, ribosomes can bind to the split ribosome binding site (domains 1 and 3) and initiate translation of thrS. Dimeric ThrRS can bind to two tRNAThr molecules for aminoacylation. Alternatively, ThrRS can bind to two sites in thrS (domains 2 and 4), which inhibits ribosome binding and represses translation. The ThrRS binding sites in thrS, as well as thrS Shine-Dalgarno (SD) sequence and initiation codon, are shown. The anticodon loop of tRNAThr is shown for comparison (44). The critical anticodon (tRNAThr) and anticodon-like (thrS) residues are shown.
As is the case with r-proteins of E. coli, the synthesis of most aminoacyl-tRNA synthetases is dependent on growth rate (44). If growth rate increases, the synthesis of tRNAThr increases, and ThrRS binds to its tRNA substrate rather than to its mRNA. As a consequence, synthesis of ThrRS increases. However, when growth rate slows the synthesis of tRNAThr decreases such that free ThrRS can bind to its own mRNA and repress translation.
TRANSLATIONAL REPRESSION: STRUCTURED AUTOREGULATORY BINDING SITES
Autogenous translational repression mechanisms in which the RNA binding target includes the proteins own SD sequence have been identified. These binding sites are often called translational operators. The binding site in these systems typically contains an RNA structure with critical single-stranded residues. In some instances additional protein contacts are made with downstream single-stranded sequences. In many ways these mechanisms are reminiscent of feedback regulation, with the important distinction that the proteins do not have an alternative rRNA or tRNA binding site.
RNA Phage Coat Proteins
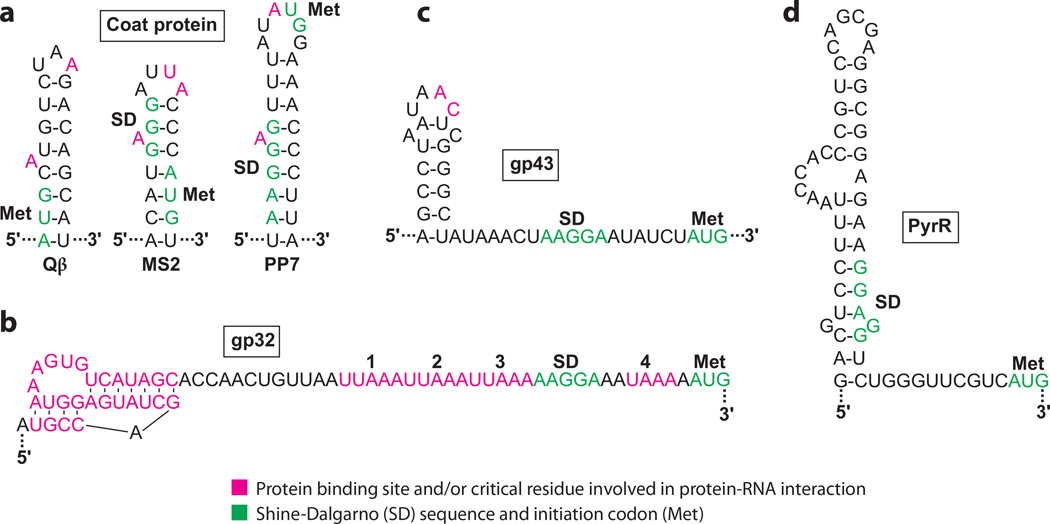
Qβ, MS2, and PP7 are RNA phages whose coat proteins serve as structural proteins of the phage capsid. Alternatively, these proteins bind to replicase mRNA and repress translation by competing with ribosome binding, thereby switching from viral replication to virion assembly (33, 42, 69). Each binding site forms a hairpin that sequesters the cognate SD sequence and/or initiation codon. Although the RNA structures are similar, the specificity of recognition by the cognate proteins involves the length of the stem, the size and sequence of the loop, and the loop’s position on the stem (Figure 3a). Furthermore, the importance of the bulged A residue varies; it is essential in MS2 but not in Qβ (28, 69). The coat proteins bind to RNA as dimers and the amino acid residues important for binding are located on a large β-sheet. The crystal structure of the MS2 protein bound to its RNA target indicates that the protein contacts seven phosphate groups on the 5′ side of the hairpin, A and U residues of the loop, and the bulged A residue (63). SELEX experiments with the PP7 coat protein identified a preferred binding site containing a 4-bp lower stem, the bulged A residue, a 4-bp upper stem, and a conserved six-base loop at the apex of the structure (33). The PP7 coat protein-RNA cocrystal structure indicates that the first three purines in the loop form a purine stack that continues the base stacking of the helical stem (9). The A residues in the bulge and initiation codon are recognized by nearly identical interactions made by symmetric pockets on the protein dimer. This arrangement contrasts with the MS2 coat protein-RNA structure in which both of these pockets are contained within the same subunit of the dimer (9).
Figure 3.
Autogenous translational repression of mRNAs containing structured binding sites. The bound regulatory protein competes with ribosome binding in each case. Protein binding sites and/or critical residues involved in protein-RNA interaction are highlighted. Shine-Dalgarno (SD) sequences and initiation codons (Met) of the mRNAs are shown. (a) Binding sites for the coat proteins of bacteriophages Qβ, MS2, and PP7 (33). (b) Binding site for bacteriophage T4 gp32 (5). (c) Binding site for bacteriophage T4 gp43 (40). (d) Binding site for Mycobacterium smegmatis PyrR. The distance between the putative SD sequence and the AUG codon (13 nt) is unusually long (18).
Bacteriophage T4 gp32
The bacteriophage T4 ssDNA binding protein (gp32) is a zinc-finger protein that functions in phage DNA replication, repair, and recombination. This protein also binds to its own mRNA and represses its translation (37, 55). gp32 first binds to an RNA pseudoknot upstream of its SD sequence, followed by cooperative binding of gp32 to the downstream sequence, which includes four UUAAA/UAAA repeats between the pseudoknot and the initiation codon (Figure 3b) (5, 37, 55). Both the pseudoknot and the unstructured repeat region are essential for autoregulation (37). Although the binding of gp32 to ssDNA is not sequence specific, it has a higher affinity for ssDNA than for its mRNA target. The crystal structure of gp32 complexed with ssDNA indicates that the phosphate backbone contacts an electropositive cleft of the protein (54). As a structure with RNA is not available, it is not known how the protein interacts with RNA; however, it is unlikely that the same protein cleft could bind specifically to the pseudoknot and the downstream repeats.
Bacteriophage T4 gp43
The T4 DNA polymerase, gp43, functions in phage DNA replication and as an RNA binding protein responsible for repressing its own translation (2). The gp43 mRNA target includes a hairpin and a single-stranded tail, which includes the SD sequence (Figure 3c) (40). Thus, bound protein competes with ribosome binding. Formation of the hairpin and the length of the tail (sequence-independent) contribute to the specificity and the stability of gp43-RNA interaction. Specificity determinants in the hairpin include the A-C dinucleotide in the hairpin loop and the restricted positioning of purine and pyrimidine residues in the stem (40). It is of historical interest to note that SELEX was first developed for gp43-RNA interaction studies; the RNA variants selected had an affinity similar to that of the natural target (61).
M. smegmatis PyrR
PyrR of B. subtilis regulates expression of the pyrimidine biosynthetic (pyr) operon by a well-characterized transcription attenuation mechanism. When activated by uridine nucleotides, PyrR binds to three positions in the polycistronic transcript, including the leader region upstream of pyrR, between pyrR and pyrP, and between pyrP and pyrB (62). In each case, bound PyrR stabilizes an anti-antiterminator structure called the binding loop. Thus, bound PyrR promotes transcription termination in three regions of the pyr transcript when sufficient levels of uridine nucleotides are present in the cell. Regulation of pyr gene expression in M. smegmatis by exogenous uracil occurs by a translational repression mechanism rather than by transcription attenuation. In this case, the PyrR binding loop apparently functions as an SD-sequestering hairpin (Figure 3d) (18). Bound PyrR stabilizes this structure and competes with 30S ribosomal subunit binding (62).
TRANSLATIONAL REPRESSION: SINGLE-STRANDED BINDING SITES
B. subtilis TRAP
The B. subtilis trp RNA binding attenuation protein (TRAP) plays a central role in controlling tryptophan synthesis and transport by sensing the concentration of tryptophan in the cell. TRAP regulates tryptophan metabolism by participating in transcription attenuation and translational repression mechanisms (19). In the transcription attenuation mechanism of the trpEDCFBA biosynthetic operon, tryptophan-activated TRAP binds to the nascent transcript and promotes formation of an intrinsic terminator by blocking formation of an overlapping antiterminator structure. In the absence of TRAP binding, the antiterminator structure forms and the operon is expressed. Tryptophan-activated TRAP also represses translation of trpE, trpG (tryptophan synthesis), trpP (tryptophan transport), and ycbK (putative efflux protein). In the case of trpE, TRAP binding promotes formation of a trpE SD-sequestering hairpin (14). In contrast, TRAP regulates translation of trpG, trpP, and ycbK directly by binding to RNA segments that overlap their cognate SD sequences and/or translation initiation regions (15, 74, 75).
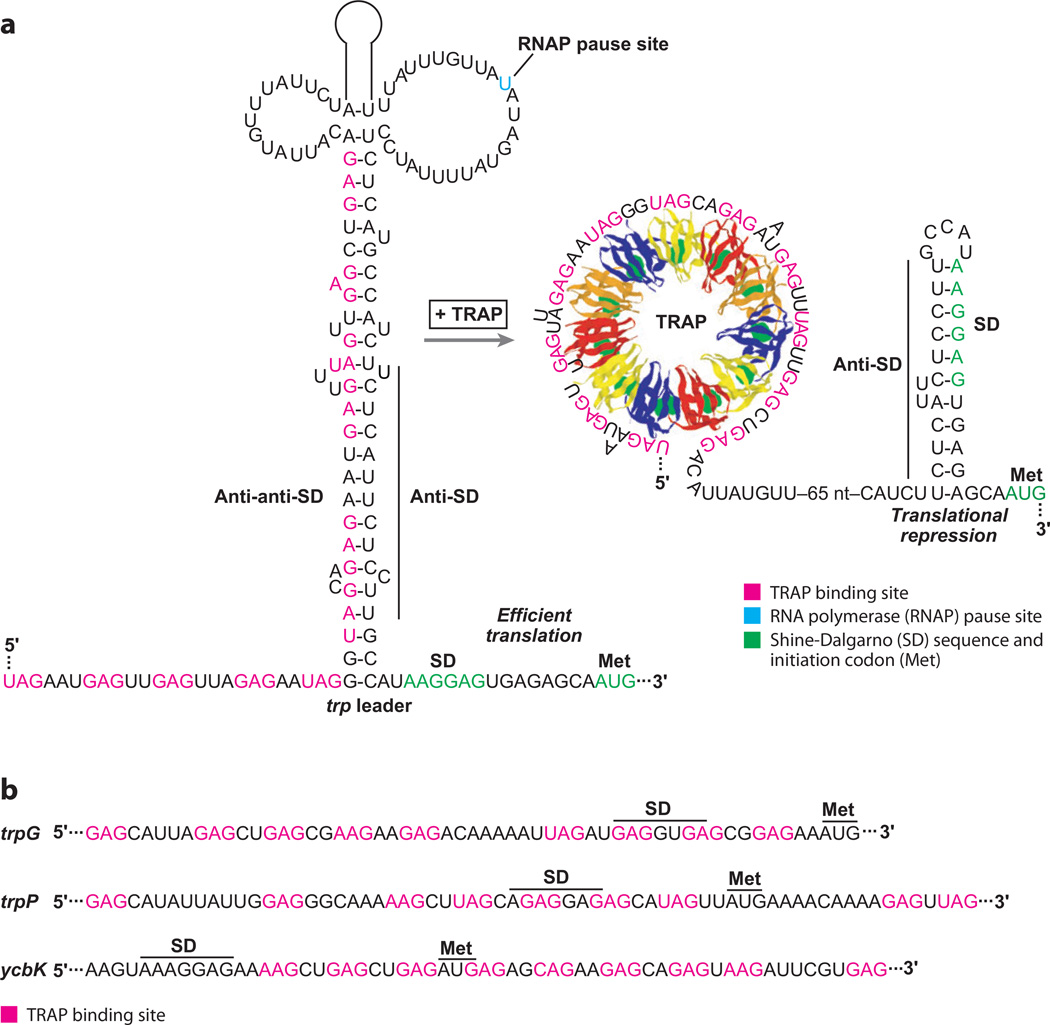
TRAP is a sequence-specific single-stranded RNA binding protein that binds to trinucleotide repeats (GAG > UAG > AAG >CAG), which are separated by two to three nonconserved spacer nucleotides (Figure 4). The TRAP binding sites in the trp leader, trpG, trpP, and ycbK transcripts contain 9–11 repeats. TRAP consists of 11 identical subunits arranged in a ring, with tryptophan binding between adjacent subunits via a network of hydrogen bonds and stacking interactions (19). Eleven repeated KKR motifs on the perimeter of TRAP are crucial for the interaction of tryptophan-activated TRAP with RNA (76). The crystal structure of a B. stearothermophilus TRAP-RNA complex revealed that the RNA wraps around the outside of the protein ring (3). The phosphodiester backbone is on the outside of the RNA ring, with the bases pointing in toward the protein. The KKR motifs form hydrogen bonds with the NAG repeats; Lys37 hydrogen bonds to the A residue and both Lys56 and Arg58 hydrogen bond with the third G of each repeat.
Figure 4.
Translational repression of Bacillus subtilis tryptophan metabolism genes. (a) Model of the trpE translational repression mechanism. RNA polymerase pausing during transcription provides additional time for binding of tryptophan-activated trp RNA binding attenuation protein (TRAP). In the absence of bound TRAP (limiting tryptophan), the RNA adopts a structure such that the trpE Shine-Dalgarno (SD) sequence is single stranded and available for ribosome binding. In the presence of excess tryptophan, TRAP binding promotes formation of the trpE SD-sequestering hairpin. (b) TRAP binding sites in the trpG, trpP, and ycbK transcripts. In each case, bound TRAP competes with ribosome binding and represses translation. The SD sequence and initiation codon (Met) for each mRNA is marked.
The TRAP binding site in the trp leader consists of 11 repeats. In the trpE translational repression mechanism, TRAP binding to these repeats promotes formation of the trpE SD-sequestering hairpin, which inhibits TrpE synthesis by preventing ribosome binding (Figure 4a) (14). Translation of trpD is also controlled by the trpE SD-sequestering hairpin via translational coupling (19). Remarkably, the TRAP binding site ends ~100 nt upstream from the trpE SD sequence. In the absence of bound TRAP, an alternative RNA structure forms between a portion of the TRAP binding site (anti-anti-SD sequence) and the anti-SD sequence such that the SD sequence is single-stranded and available for ribosome binding. The general transcription elongation factors NusA and NusG stimulate RNA polymerase pausing during transcription of the trp leader, thereby providing additional time for TRAP to bind and promote formation of the trpE SD-sequestering hairpin (73).
A third TRAP-dependent regulatory mechanism is responsible for controlling translation initiation of trpG, trpP, and ycbK in which bound TRAP directly blocks 30S ribosomal subunit binding (15, 74, 75). The TRAP binding site in trpG includes nine triplet repeats that overlap its SD sequence (Figure 4b). The TRAP binding site in the trpP transcript also contains nine triplet repeats; however, in this case the TRAP binding site overlaps the trpP SD sequence and extends into the trpP coding sequence. In the case of ycbK, all nine triplet repeats are downstream from the SD sequence and extend further into the ycbK coding sequence. Thus, the unusual mechanism of TRAP-RNA interaction in which multiple triplet repeats extend over a long linear distance allows the protein to specifically bind to a subset of translation initiation regions and repress translation.
TRAP is found in a few other closely related gram-positive organisms. In each case, a putative TRAP binding site is positioned in the trp operon leader, suggesting that TRAP is capable of controlling expression of these operons by attenuation and/or translational repression mechanisms (24). While experimental support for transcription attenuation exists for some of these organisms, translational repression by an SD-sequestering hairpin has not been demonstrated for any organism other than B. subtilis (19).
Bacteriophage T4 RegA
T4 RegA is a single-stranded RNA binding protein that represses translation of several T4 genes, including its own mRNA, by competing with ribosome binding to the cognate translation initiation region. Although the mRNA targets are AU rich, a consensus binding site has not been identified (66). While RegA exists as a dimer when free in solution, this protein binds RNA as a monomer (41). RegA’s RNA binding domain consists of a surface pocket formed by residues on two loops and an N-terminal α-helix (20). T4 RegA is 78% identical to that from bacteriophage RB69. SELEX studies identified high-affinity RNA targets for both RegA proteins: AAAAUUGUUAUGUAA for T4 and UAA repeat sequence UAAUAAUAAUAAUAAUA for RB69 (6, 12). Both RegA proteins exhibit a hierarchy of affinities for their cognate RNAs: gene 44 > gene 45 > regA (51). Thus, the relative affinities of RegA for target sequences explain its capacity to regulate translation of a variety of genes prior to repressing its own synthesis.
TRANSLATIONAL REPRESSION: MULTIPLE BINDING SITES
E. coli CsrA
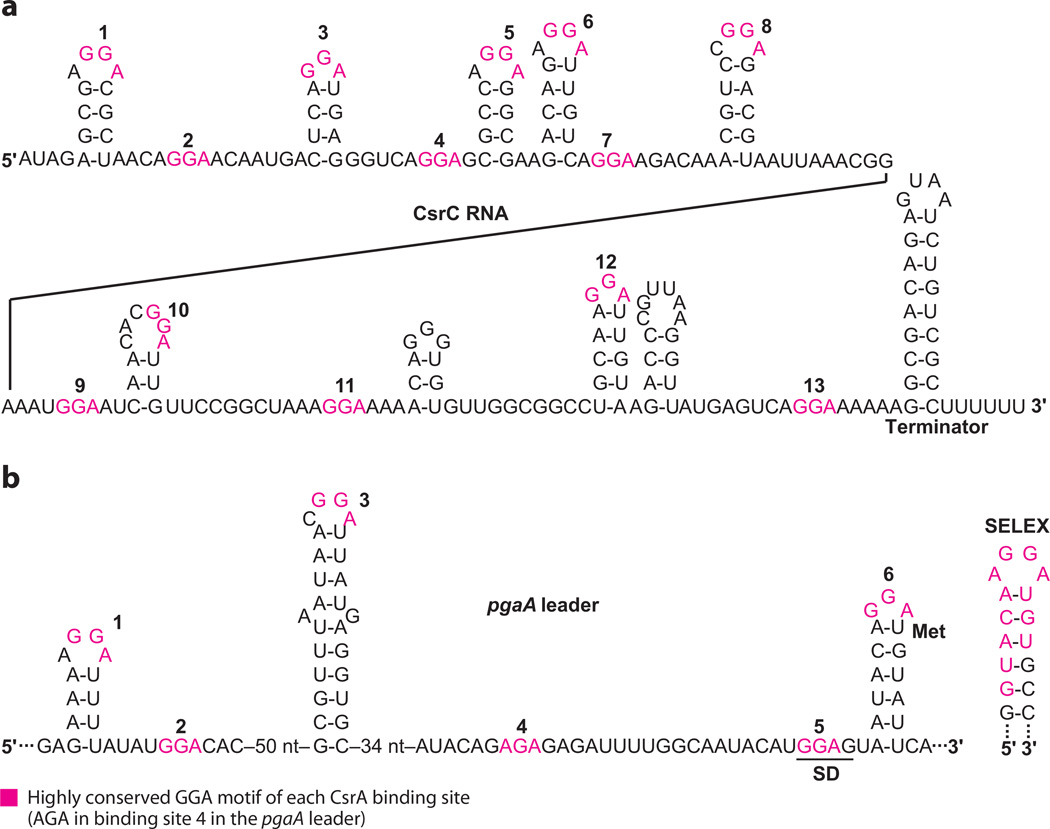
The E. coli carbon storage regulator (CsrA) protein is the key component of a global regulatory system that both represses translation of genes that are induced upon the approach to stationary-phase growth and activates genes that are expressed during exponential-phase growth. csrA is broadly distributed among eubacteria, and it regulates virulence factors, quorum sensing, motility, carbon metabolism, peptide uptake, and biofilm development in various species (4). CsrA-mediated repression typically involves binding of this protein to multiple sites, one of which overlaps the cognate SD sequence. Thus, bound CsrA competes with 30S ribosomal subunit binding. In contrast, the mechanism(s) of CsrA-mediated activation has not been elucidated. CsrA activity is controlled by its interaction with two noncoding sRNAs (CsrB and CsrC) that contain multiple binding sites that enable them to sequester and antagonize CsrA (35, 67). In this example of molecular mimicry, the CsrA binding sites of CsrB and CsrC resemble those of mRNA target molecules (Figure 5). The homeostatic Csr circuitry is controlled by two negative feedback loops: (a) CsrA somehow activates signaling by a two-component signal transduction system (BarA-UvrY) that activates transcription of the sRNA antagonists, CsrB/C. (b) CsrA represses expression of the CsrD protein, which along with RNase E mediates specific turnover of CsrB/C (58).
Figure 5.
CsrA binding sites in CsrC and pgaA leader transcripts. (a) CsrC contains 13 putative CsrA binding sites (numbered ) and functions as a CsrA antagonist. Because several of the binding sites have slight overlaps, only the highly conserved GGA motifs are colored to improve clarity. (b) The pgaA leader transcript contains six CsrA binding sites (numbered), three of which are present in RNA hairpins. Bound CsrA competes with ribosome binding and represses translation. The pgaA Shine-Dalgarno (SD) sequence and initiation codon (Met) are shown. A high-affinity SELEX-derived RNA target is shown for comparison. Note that this motif is AGA in binding site 4.
CsrA binds to single-stranded sequences present in unstructured RNA or in the loops of short hairpins. Binding sites in mRNAs, CsrB/C, and SELEX-derived ligands contain an almost universal GGA sequence bracketed by additional conserved residues (Figure 5). Substitution analysis of a SELEX-derived target confirmed that the GGA motif is critical for high-affinity binding and that secondary structure of the stem-loop was beneficial but not essential for binding (16). Structural studies of three CsrA orthologs have shown that this protein forms a homodimer composed of five interdigitated β-strands of each polypeptide and protruding C-terminal α-helices. Critical amino acids that contact bound RNA are located on the β-1 and β-5 strands of opposing polypeptides, which lie together and in parallel on each side of the protein, and on the base of the α-helix. Thus, an identical RNA binding surface is found on each side of this symmetrical protein dimer. The critical GG residues of RNA come in contact with R44 and V42, the amino acid residues most important for RNA binding (38, 50).
CsrA target transcripts contain between 1 (hfq) and ~20 (CsrB) binding sites. Typically, at least two CsrA binding sites are present in the mRNA leader of a repressed gene, one of which invariably overlaps the SD sequence. Cooperative binding is often observed for transcripts containing multiple binding sites. CsrA targets resemble SD sequences, thus mutation of the SD sequence to promote CsrA binding is relatively facile and likely facilitated the evolutionary expansion of the Csr regulon. Six CsrA binding sites are present in the pgaA mRNA leader, two of which overlap the SD sequence and initiation codon (Figure 5b) (65). For pgaA and other mRNA leaders containing a CsrA binding hairpin close to the SD sequence, it is likely that one RNA binding surface of CsrA first binds with high affinity to the stem-loop and the second surface bridges to the SD sequence, with the formation of a repression loop. Translational repression by CsrA is usually associated with decreased mRNA stability. The mechanism for altered mRNA stability may be passively related to the inhibition of translation and/or directly associated with nucleolytic cleavage and turnover due to the presence of bound CsrA. Finally, the CsrA-CsrB ribonucle-oprotein complex is globular in form and contains ~1 CsrA dimer per two binding sites in CsrB (35), suggesting that CsrA dimers might tether nonadjacent binding sites in the complex.
TRANSLATIONAL ACTIVATION
Bacteriophage Mu Com
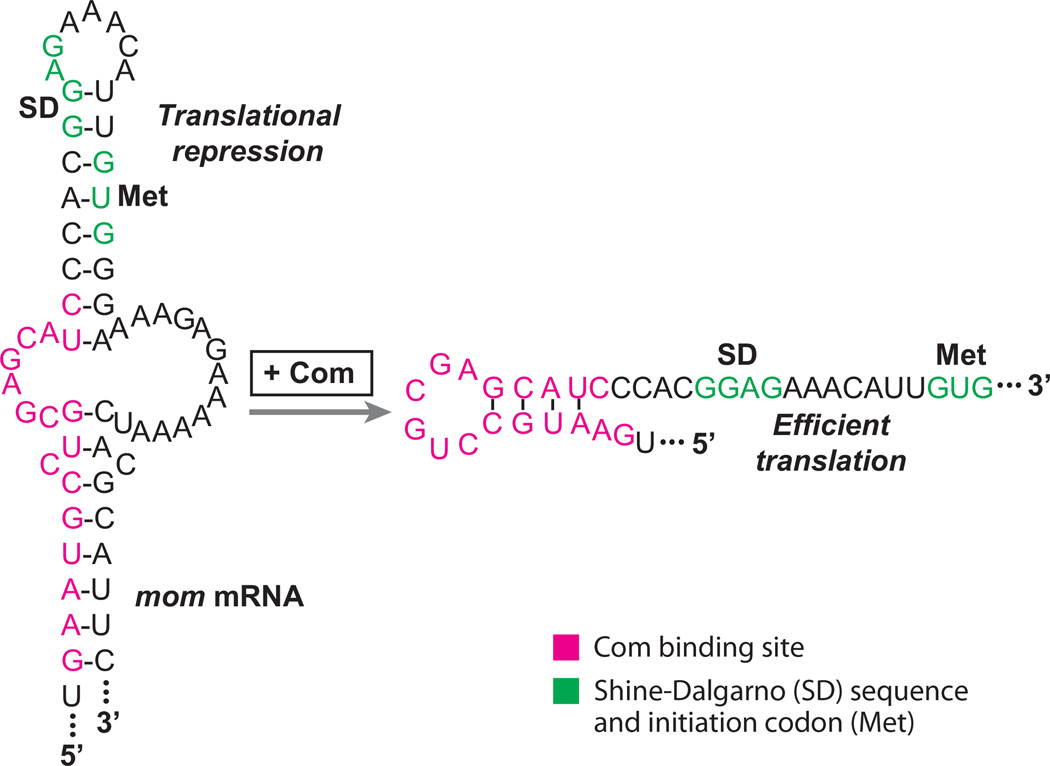
Although there are numerous examples of translational repression mechanisms, examples of translational activation are rare. Investigation of the com-mom operon of bacteriophage Mu has provided one example of an RNA binding protein-mediated translational activation mechanism. mom encodes a DNA modification enzyme that protects Mu DNA from a variety of restriction enzymes (26). Translation of mom is activated by Com (control of mom), a zinc-finger RNA binding protein. In the absence of Com, the intercistronic region of the com-mom transcript adopts a repressive secondary structure that sequesters the mom initiation codon and a portion of the mom SD sequence (Figure 6). Com binding destabilizes the inhibitory structure such that Mom synthesis can proceed (27, 72). Results from mutagenesis studies indicate that the Com binding site includes both secondary and primary structural features (70). Although the precise structure of the RNA target bound by Com has not been firmly established, Com can bind to a 19-nt sequence containing the hairpin, as shown in Figure 6 (34).
Figure 6.
Translational activation model of the bacteriophage Mu mom transcript. In the absence of bound Com protein, the RNA adopts an RNA hairpin that sequesters the mom Shine-Dalgarno (SD) sequence and initiation codon (Met). Bound Com stabilizes an alternative RNA hairpin such that the mom SD sequence and initiation codon are single-stranded and available for ribosome binding (26).
SUMMARY POINTS.
RNA binding proteins can repress translation initiation by a wide variety of mechanisms, including directly competing with 30S ribosomal subunits, promoting formation of an SD-sequestering hairpin, and entrapping 30S subunits in an inactive complex on mRNA.
Autogenous repression of translation is particularly common in E. coli r-protein operons and in certain bacteriophages.
Translational activation by RNA binding proteins is rare.
The size of the regulon for an RNA binding protein can range from one to several genes.
The complexity of RNA recognition targets ranges from complex pseudoknot structures to single-stranded triplet repeats, and the number of separate binding sites in target transcripts ranges from 1 to ~20.
ACKNOWLEDGMENTS
This work was supported by grants GM52840 (PB), GM59969 (TR and PB), and GM66794 (TR) from the National Institutes of Health.
Glossary
- SD
Shine-Dalgarno
- Secondary structure
intramolecular base-pairing interactions within an RNA molecule
- SD-sequestering hairpin
an inhibitory RNA secondary structure that contains the SD sequence of a gene
- Pseudoknot
an RNA secondary structure in which part of one stem is intercalated between two halves of another stem
- SELEX
systematic evolution of ligands by exponential enrichment
- TRAP
trp RNA binding attenuation protein
- Csr
carbon storage regulator
- Com
control of mom
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Contributor Information
Paul Babitzke, Email: pxb28@psu.edu.
Carol S. Baker, Email: csb3@psu.edu.
Tony Romeo, Email: tromeo@ufl.edu.
LITERATURE CITED
- 1.Allemand F, Haentjens J, Chiaruttini C, Royer C, Springer M. Escherichia coli ribosomal protein L20 binds as a single monomer to its own mRNA bearing two potential binding sites. Nucleic Acids Res. 2007;35:3016–3031. doi: 10.1093/nar/gkm197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrake M, Guild N, Hsu T, Gold L, Tuerk C, Karam J. DNA polymerase of bacteriophage T4 is an autogenous translational repressor. Proc. Natl. Acad. Sci. USA. 1988;85:7942–7946. doi: 10.1073/pnas.85.21.7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antson AA, Dodson EJ, Dodson G, Greaves RB, Chen X-P, Gollnick P. Structure of the trp RNA-binding attenuation protein, TRAP, bound to RNA. Nature. 1999;401:235–242. doi: 10.1038/45730. [DOI] [PubMed] [Google Scholar]
- 4.Babitzke P, Romeo T. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr. Opin. Microbiol. 2007;10:156–163. doi: 10.1016/j.mib.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Borjac-Natour JM, Petrov VM, Karam JD. Divergence of the mRNA targets for the Ssb proteins of bacteriophages T4 and RB69. Virology J. 2004;1:4. doi: 10.1186/1743-422X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown D, Brown J, Kang C, Gold L, Allen P. Single-stranded RNA recognition by the bacteriophage T4 translational repressor, regA. J. Biol. Chem. 1997;272:14969–14974. doi: 10.1074/jbc.272.23.14969. [DOI] [PubMed] [Google Scholar]
- 7.Caillet J, Nogueira T, Masquida B, Winter F, Graffe M, et al. The modular structure of Escherichia coli threonyl-tRNA synthetase as both an enzyme and a regulator of gene expression. Mol. Microbiol. 2003;47:961–974. doi: 10.1046/j.1365-2958.2003.03364.x. [DOI] [PubMed] [Google Scholar]
- 8.Cerretti DP, Mattheakis LC, Kearney KR, Vu L, Nomura M. Translational regulation of the spc operon in Escherichia coli. Identification and structural analysis of the target site for S8 repressor protein. J. Mol. Biol. 1998;204:309–329. doi: 10.1016/0022-2836(88)90578-5. [DOI] [PubMed] [Google Scholar]
- 9. Chao JA, Patskovsky Y, Almo SC, Singer RH. Structural basis for the coevolution of a viral RNA-protein complex. Nat. Struct. Mol. Biol. 2008;15:103–105. doi: 10.1038/nsmb1327. Provides the structural basis for bacteriophage MS2 and PP7 coat protein interaction with their mRNA targets.
- 10. Chiaruttini C, Milet M, Springer M. A long-range RNA-RNA interaction forms a pseudo-knot required for translational control of the IF3-L35-L20 ribosomal protein operon in Escherichia coli. EMBO J. 1996;15:4402–4413. Demonstrates that a long-range interaction results in the formation of a pseudoknot in the ~450 nt untranslated rpmI-rplT leader RNA.
- 11.Climie SC, Friesen JD. In vivo and in vitro structural analysis of the rplJ mRNA leader of Escherichia coli. Protection by bound L10-L7/L12. J. Biol. Chem. 1988;263:15166–15175. [PubMed] [Google Scholar]
- 12.Dean TR, Allen SV, Miller ES. In vitro selection of phage RB69 RegA RNA binding sites yields UAA repeats. Virology. 2005;336:26–36. doi: 10.1016/j.virol.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Deutscher MP. Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res. 2006;34:659–666. doi: 10.1093/nar/gkj472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du H, Babitzke P. trp RNA-binding attenuation protein-mediated long distance RNA refolding regulates translation of trpE in Bacillus subtilis. J. Biol. Chem. 1998;273:20494–20503. doi: 10.1074/jbc.273.32.20494. [DOI] [PubMed] [Google Scholar]
- 15.Du H, Tarpey R, Babitzke P. The trp RNA-binding attenuation protein regulates TrpG synthesis by binding to the trpG ribosome binding site of Bacillus subtilis. J. Bacteriol. 1997;179:2582–2586. doi: 10.1128/jb.179.8.2582-2586.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubey AK, Baker CS, Romeo T, Babitzke P. RNA sequence and secondary structure participate in high-affinity CsrA-RNA interaction. RNA. 2005;11:1579–1587. doi: 10.1261/rna.2990205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehresmann C, Ehresmann B, Ennifar E, Dumas P, Garber M, et al. Molecular mimicry in transla-tional regulation: the case of ribosomal protein S15. RNA Biol. 2004;1:66–73. [PubMed] [Google Scholar]
- 18.Fields CJ, Switzer RL. Regulation of pyr gene expression in Mycobacterium smegmatis by PyrR-dependent translational repression. J. Bacteriol. 2007;189:6236–6245. doi: 10.1128/JB.00803-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gollnick P, Babitzke P, Antson A, Yanofsky C. Complexity in regulation of tryptophan biosynthesis in Bacillus subtilis. Annu. Rev. Genet. 2005;39:47–68. doi: 10.1146/annurev.genet.39.073003.093745. [DOI] [PubMed] [Google Scholar]
- 20.Gordon J, Sengupta TK, Phillips CA, O’Malley SM, Williams KR, Spicer EK. Identification of the RNA binding domain of T4 RegA protein by structure-based mutagenesis. J. Biol. Chem. 1999;274:32265–32273. doi: 10.1074/jbc.274.45.32265. [DOI] [PubMed] [Google Scholar]
- 21.Gregory RJ, Cahill PBF, Thurlow DL, Zimmermann RA. Interaction of Escherichia coli ribosomal protein S8 with its binding sites in ribosomal RNA and messenger RNA. J. Mol. Biol. 1988;204:295–307. doi: 10.1016/0022-2836(88)90577-3. [DOI] [PubMed] [Google Scholar]
- 22.Grundy FJ, Henkin TM. The rpsD gene, encoding ribosomal protein S4, is autogenously regulated in Bacillus subtilis. J. Bacteriol. 1991;173:4595–4602. doi: 10.1128/jb.173.15.4595-4602.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guillier M, Allemand F, Dardel F, Royer CA, Springer M, Chiaruttini C. Double molecular mimicry in Escherichia coli: Binding of ribosomal protein L20 to its two sites in mRNA is similar to its binding to 23S rRNA. Mol. Microbiol. 2005;56:1441–1456. doi: 10.1111/j.1365-2958.2005.04644.x. [DOI] [PubMed] [Google Scholar]
- 24.Gutiérrez-Preciado A, Yyanofsky C, Merino E. Comparison of tryptophan biosynthetic operon regulation in different Gram-positive bacterial species. Trends Genet. 2007;23:422–426. doi: 10.1016/j.tig.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 25.Haentjens-Sitri J, Allemand F, Springer M, Chiaruttini C. A competition mechanism regulates the translation of the Escherichia coli operon encoding ribosomal proteins L35 and L20. J. Mol. Biol. 2008;375:612–625. doi: 10.1016/j.jmb.2007.10.058. [DOI] [PubMed] [Google Scholar]
- 26.Hattman S. Unusual transcriptional and translational regulation of the bacteriophage Mu mom operon. Pharmacol. Ther. 1999;84:367–388. doi: 10.1016/s0163-7258(99)00042-x. [DOI] [PubMed] [Google Scholar]
- 27. Hattman S, Newman L, Murthy HMK, Nagaraja V. Com, the phage Mu mom translational activator, is a zinc-binding protein that binds specifically to its cognate mRNA. Proc. Natl. Acad. Sci. USA. 1991;88:10027–10031. doi: 10.1073/pnas.88.22.10027. Along with Reference 72, this pape demonstrates that Com activates translation of the Mu mom gene by destabilizing the mom SD sequestering hairpin.
- 28.Helgstrand C, Grahn E, Moss T, Stonehouse NJ, Tars K, et al. Investigating the structural basis of purine specificity in the structures of MS2 coat protein RNA translational operator hairpins. Nucleic Acids Res. 2002;30:2678–2685. doi: 10.1093/nar/gkf371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iben JR, Draper DE. Specific interactionsof the L10(L12)4 ribosomal protein complex with mRNA, rRNA, and L11. Biochemistry. 2008;47:2721–2731. doi: 10.1021/bi701838y. [DOI] [PubMed] [Google Scholar]
- 30.Jenner L, Romby P, Rees B, Schulze-Briese C, Springer M, et al. Translational operator of mRNA on the ribosome: how repressor proteins exclude ribosome binding. Science. 2005;308:120–123. doi: 10.1126/science.1105639. [DOI] [PubMed] [Google Scholar]
- 31.Keener J, Nomura M. Regulation of ribosome synthesis. In: Neidhardt FC, Curtis R III, Ingraham JL, Lin ECC, Low KB, et al., editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. 2nd ed. Washington, DC: Am. Soc. Microbiol; 1996. pp. 1417–1431. [Google Scholar]
- 32.Laursen BS, Sorensen HP, Mortensen KK, Sperling-Petersen HU. Initiation of protein synthesis in bacteria. Microbiol. Mol. Biol. Rev. 2005;69:101–123. doi: 10.1128/MMBR.69.1.101-123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim F, Peabody DS. RNA recognition site of PP7 coat protein. Nucleic Acids Res. 2002;30:4138–4144. doi: 10.1093/nar/gkf552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lima S, Hildenbrand J, Korostelev A, Hattman S, Li SH. Crystal structure of an RNA helix recognized by a zinc-finger protein: an 18-bp duplex at 1.6 Å resolution. RNA. 2002;8:924–932. doi: 10.1017/s1355838202028893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu MY, Gui G, Wei B, Preston JF, 3rd, Oakford L, et al. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J. Biol. Chem. 1997;272:17502–17510. doi: 10.1074/jbc.272.28.17502. This paper provides the first demonstration that a regulatory sRNA can sequester an RNA binding protein.
- 36.Marzi S, Myasnikov AG, Serganov A, Ehresmann C, Romby P, et al. Structured mRNAs regulate translation initiation by binding to the platform of the ribosome. Cell. 2007;130:1019–1031. doi: 10.1016/j.cell.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 37.McPheeters DS, Stormo GD, Gold L. Autogenous regulatory site on the bacteriophage T4 gene 32 messenger RNA. J. Mol. Biol. 1988;201:517–535. doi: 10.1016/0022-2836(88)90634-1. [DOI] [PubMed] [Google Scholar]
- 38.Mercante J, Suzuki K, Cheng X, Babitzke P, Romeo T. Comprehensive alanine-scanning muta-genesis of Escherichia coli CsrA defines two subdomains of critical functional importance. J. Biol. Chem. 2006;281:31832–31842. doi: 10.1074/jbc.M606057200. [DOI] [PubMed] [Google Scholar]
- 39.Merianos HJ, Wang J, Moore PB. The structure of a ribosomal protein S8/spc operon mRNA complex. RNA. 2004;10:954–964. doi: 10.1261/rna.7030704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pavlov AR, Karam JD. Nucleotide-sequence-specific and non-specific interactions of T4 DNA polymerase with its own mRNA. Nucleic Acids Res. 2000;28:4657–4664. doi: 10.1093/nar/28.23.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phillips CA, Gordon J, Spicer EK. Bacteriophage T4 regA protein binds RNA as a monomer, overcoming dimer interactions. Nucleic Acids Res. 1996;24:4319–4326. doi: 10.1093/nar/24.21.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Romaniuk PJ, Lowary P, Wu HN, Stormo G, Uhlenbeck OC. RNA binding site of R17 coat protein. Biochemistry. 1987;26:1563–1568. doi: 10.1021/bi00380a011. [DOI] [PubMed] [Google Scholar]
- 43.Romby P, Caillet J, Ebel C, Sacerdot C, Graffe M, et al. The expression of E. coli threonyl-tRNA synthetase is regulated at the translational level by symmetrical operator-repressor interactions. EMBO J. 1996;15:5976–5987. [PMC free article] [PubMed] [Google Scholar]
- 44.Romby P, Springer M. Bacterial translational control at atomic resolution. Trends Genet. 2003;19:155–161. doi: 10.1016/S0168-9525(03)00020-9. [DOI] [PubMed] [Google Scholar]
- 45. Sacerdot C, Caillet J, Graffe M, Eyermann F, Ehresmann B, et al. The Escherichia coli threonyl-tRNA synthetase gene contains a split ribosomal binding site interrupted by a hairpin structure that is essential for autoregulation. Mol. Microbiol. 1998;29:1077–1090. doi: 10.1046/j.1365-2958.1998.00995.x. Shows that thrS contains an unusual split ribosome binding site, which is interrupted by a hairpin comprising one of the ThrRS binding sites.
- 46.Sankaranarayanan R, Dock-Bregeon A-C, Romby P, Caillet J, Springer M, et al. The structure of threonyl-tRNA synthetase-tRNAThr complex enlightens its repressor activity and reveals an essential zinc ion in the active site. Cell. 1999;97:371–381. doi: 10.1016/s0092-8674(00)80746-1. [DOI] [PubMed] [Google Scholar]
- 47.Schlax PJ, Worhunsky DJ. Translational repression mechanisms in prokaryotes. Mol. Microbiol. 2003;48:1157–1169. doi: 10.1046/j.1365-2958.2003.03517.x. [DOI] [PubMed] [Google Scholar]
- 48. Schlax PJ, Xavier KA, Gluick TC, Draper DE. Translational repression of the Escherichia coli α operon mRNA: importance of an mRNA conformational switch and a ternary entrapment complex. J. Biol. Chem. 2001;276:38494–38501. doi: 10.1074/jbc.M106934200. Shows that S4 represses translation of rpsM by binding to a nested pseudoknot in the rpsM transcript.
- 49.Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, et al. Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell. 2000;102:615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 50.Schubert M, Lapouge K, Duss O, Oberstrass FC, Jelesarov I, et al. Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA. Nat. Struct. Mol. Biol. 2007;14:807–813. doi: 10.1038/nsmb1285. [DOI] [PubMed] [Google Scholar]
- 51.Sengupta TK, Gordon J, Spicer EK. RegA proteins from phage T4 and RB69 have conserved helix-loop groove RNA binding motifs but different RNA binding specificities. Nucleic Acids Res. 2001;29:1175–1184. doi: 10.1093/nar/29.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Serganov A, Bénard L, Portier C, Ennifar E, Garber M, Ehresmann B, Ehresmann C. Role of conserved nucleotides in building the 16S rRNA binding site for ribosomal protein S15. J. Mol. Biol. 2001;305:785–803. doi: 10.1006/jmbi.2000.4354. [DOI] [PubMed] [Google Scholar]
- 53.Serganov A, Ennifar E, Portier C, Ehresmann B, Ehresmann C. Do mRNA and rRNA binding sites of E. coli ribosomal protein S15 share common structural determinants? J. Mol. Biol. 2002;320:963–978. doi: 10.1016/s0022-2836(02)00553-3. [DOI] [PubMed] [Google Scholar]
- 54.Shamoo Y, Friedman AM, Parsons MR, Konigsberg WH, Steitz TA. Crystal structure of a replication fork single-stranded DNA binding protein (T4 gp32) complexed to DNA. Nature. 1995;376:362–366. doi: 10.1038/376362a0. [DOI] [PubMed] [Google Scholar]
- 55.Shamoo Y, Tam A, Konigsberg WH, Williams KR. Translational repression by the bacteriophage T4 gene 32 protein involves specific recognition of an RNA pseudoknot structure. J. Mol. Biol. 1993;232:89–104. doi: 10.1006/jmbi.1993.1372. [DOI] [PubMed] [Google Scholar]
- 56.Spedding G, Draper DE. Allosteric mechanism for translational repression in the Escherichia coli α operon. Proc. Natl. Acad. Sci. USA. 1993;90:4399–4403. doi: 10.1073/pnas.90.10.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spedding G, Gluick TC, Draper DE. Ribosome initiation complex formation with the pseudoknotted a operon messenger RNA. J. Mol. Biol. 1993;229:609–622. doi: 10.1006/jmbi.1993.1067. [DOI] [PubMed] [Google Scholar]
- 58.Suzuki K, Babitzke P, Kushner SR, Romeo T. Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes. Dev. 2006;20:2605–2617. doi: 10.1101/gad.1461606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tishchenko S, Nikulin A, Fomenkova N, Nevskaya N, Nikonov O, et al. Detailed analysis of RNA-protein interactions within the ribosomal protein S8-rRNA complex from the archaeon Methanococcus jannaschii. J. Mol. Biol. 2001;311:311–324. doi: 10.1006/jmbi.2001.4877. [DOI] [PubMed] [Google Scholar]
- 60.Torres-Larios A, Dock-Bregeon A-C, Romby P, Rees B, Sankaranarayanan R, et al. Structural basis of translational control by Escherichia coli threonyl tRNA synthetase. Nat. Struct. Biol. 2002;9:343–347. doi: 10.1038/nsb789. [DOI] [PubMed] [Google Scholar]
- 61.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 62.Turnbough CL, Jr, Switzer RL. Regulation of pyrimidine biosynthetic gene expression in bacteria: repression without repressors. Microbiol. Mol. Biol. Rev. 2008;72:266–300. doi: 10.1128/MMBR.00001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Valegård K, Murray JB, Stonehouse NJ, van den Worm S, Stockley PG, Liljas L. The three-dimensional structures of two complexes between recombinant MS2 capsids and RNA operator fragments reveal sequence-specific protein-RNA interactions. J. Mol. Biol. 1997;270:724–738. doi: 10.1006/jmbi.1997.1144. [DOI] [PubMed] [Google Scholar]
- 64.van Meerten D, Girard G, van Duin J. Translational control by delayed RNA folding: identification of the kinetic trap. RNA. 2001;7:483–494. doi: 10.1017/s1355838201001984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang X, Dubey AK, Suzuki K, Baker CS, Babitzke P, Romeo T. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol. Microbiol. 2005;56:1648–1663. doi: 10.1111/j.1365-2958.2005.04648.x. [DOI] [PubMed] [Google Scholar]
- 66.Webster KR, Spicer EK. Characterization of bacteriophage T4 regA protein-nucleic acid interactions. J. Biol. Chem. 1990;265:19007–19014. [PubMed] [Google Scholar]
- 67.Weilbacher T, Suzuki K, Dubey AK, Wang X, Gudapaty S, et al. A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol. Microbiol. 2003;48:657–670. doi: 10.1046/j.1365-2958.2003.03459.x. [DOI] [PubMed] [Google Scholar]
- 68.Wimberly BT, Brodersen DE, Clemons WM, Jr, Morgan-Warren RJ, Carter AP, et al. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 69.Witherell GW, Uhlenbeck OC. Specific RNA binding by Qβ coat protein. Biochemistry. 1989;28:71–76. doi: 10.1021/bi00427a011. [DOI] [PubMed] [Google Scholar]
- 70.Witkowski RT, Hattman S, Newman L, Clark K, Tierney DL, et al. The zinc coordination site of the bacteriophage Mu translational activator protein, Com. J. Mol. Biol. 1995;247:753–764. doi: 10.1006/jmbi.1995.0178. [DOI] [PubMed] [Google Scholar]
- 71.Wu H, Jiang L, Zimmermann RA. The binding site for ribosomal protein S8 in 16S rRNA and spc mRNA from Escherichia coli: minimum structural requirements and the effects of single bulged bases on S8-RNA interaction. Nucleic Acids Res. 1994;22:1687–1695. doi: 10.1093/nar/22.9.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wulczyn FG, Kahmann R. Translational stimulation: RNA sequence and structural requirements for binding of Com protein. Cell. 1991;65:259–269. doi: 10.1016/0092-8674(91)90160-z. [DOI] [PubMed] [Google Scholar]
- 73. Yakhnin AV, Yakhnin H, Babitzke P. Function of the Bacillus subtilis transcription elongation factor NusG in hairpin-dependent RNA polymerase pausing in the trp leader. Proc. Natl. Acad. Sci. USA. 2008;105:16131–16136. doi: 10.1073/pnas.0808842105. Demonstrates that NusA- and NusG-stimulated RNA polymerase pausing participates in the trpE translational repression mechanism by providing additional time for TRAP binding.
- 74.Yakhnin H, Yakhnin AV, Babitzke P. The trp RNA-binding attenuation protein (TRAP) of Bacillus subtilis regulates translation initiation of ycbK, a gene encoding a putative efflux protein, by blocking ribosome binding. Mol. Microbiol. 2006;61:1252–1266. doi: 10.1111/j.1365-2958.2006.05278.x. [DOI] [PubMed] [Google Scholar]
- 75.Yakhnin H, Zhang H, Yakhnin AV, Babitzke P. The trp RNA-binding attenuation protein of Bacillus subtilis regulates translation of the tryptophan transport gene, trpP (yhaG), by blocking ribosome binding. J. Bacteriol. 2004;186:278–286. doi: 10.1128/JB.186.2.278-286.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang M, Chen X-P, Militello K, Hoffman R, Fernandez B, et al. Alanine-scanning mutagenesis of Bacillus subtilis trp RNA-binding attenuation protein (TRAP) reveals residues involved in tryptophan binding and RNA binding. J. Mol. Biol. 1997;270:696–710. doi: 10.1006/jmbi.1997.1149. [DOI] [PubMed] [Google Scholar]
- 77.Zengel JM, Lindahl L. Diverse mechanisms for regulating ribosomal protein synthesis in Escherichia coli. Prog. Nucleic Acid Res. Mol. Biol. 1994;47:331–370. doi: 10.1016/s0079-6603(08)60256-1. [DOI] [PubMed] [Google Scholar]