Abstract
Genetically encoded photosensitizers, proteins that produce reactive oxygen species when illuminated with visible light, are increasingly used as optogenetic tools. Their applications range from ablation of specific cell populations to precise optical inactivation of cellular proteins. Here, we report an orange mutant of red fluorescent protein KillerRed that becomes toxic when illuminated with blue or green light. This new protein, KillerOrange, carries a tryptophan-based chromophore that is novel for photosensitizers. We show that KillerOrange can be used simultaneously and independently from KillerRed in both bacterial and mammalian cells offering chromatic orthogonality for light-activated toxicity.
Introduction
Fluorescent proteins of GFP family are widely used in molecular and cellular biology as passive reporters of the cellular structures and processes [1]. The chromophore of a fluorescent protein is buried deeply inside a protein beta-barrel and shielded from the surrounding solvent. This is believed to explain a much lower level of phototoxicity of a typical fluorescent protein comparing to small organic dyes [2,3].
In one documented case, however, the structure of beta-barrel maintains a path through the protein that reaches the chromophore and is accessible by the water and oxygen molecules [4,5]. This unusual example belongs to the red fluorescent protein KillerRed developed from the jellyfish chromoprotein anm2CP by directed evolution [6]. When illuminated with green or orange light, KillerRed produces reactive oxygen species and demonstrates three orders of magnitude higher level of phototoxicity than other fluorescent proteins [6].
It was shown that the reactive oxygen species formation in KillerRed proceeds through the Type I (radical-based), and not the Type II (singlet oxygen formation) mechanism [7]. The mechanism of KillerRed action possibly involves unusual reductive electron transfer to the singlet excited protein chromophore forming the radical dianion intermediate [8]. The latter subsequently transfers an electron to the oxygen molecule forming the superoxide anion radical.
Depending on the subcellular localization and the dose of light irradiation, reactive oxygen species produced by KillerRed can lead to a variety of outcomes ranging from inactivation of a fusion partner protein [6], DNA damage and cell division arrest [9,10] to cell death through necrosis or apoptosis [6,11].
KillerRed is thus used as an optogenetic tool in cell biology to inactivate particular proteins by light [6,12–14] and to study local intracellular oxidative stress [15,16], and in developmental biology and neuroscience to ablate a specific cell population [17–21]. Besides that, KillerRed has been used as a photosensitizer for tumor treatment in model systems [22–25]
KillerRed was the first genetically encoded photosensitizer developed in 2006 [6]. Since then a few other proteins that generate reactive oxygen species have been reported: SuperNova [26], the spectrally similar monomeric version of KillerRed, and miniSOG and Pp2FbFP L30M–flavoproteins that generate singlet oxygen upon blue light irradiation mainly via Type II mechanism [27,28]. Together they cover the spectral range from 430 to 590 nm that can be used for reactive oxygen species production.
In the current work we aimed to further expand the toolkit of available phototoxic proteins by developing a KillerRed mutant of distinct color. Our goal was to develop a protein that can be excited independently of KillerRed, so the concurrent biomolecular processes or cell populations fates can be regulated in an orthogonal and non-interfering fashion. Such fine tuning leading to high selectivity is rapidly developing in a number of chemical and biochemical systems [29,30].
Methods
Plasmid construction and mutagenesis
Synthetic DNA oligonucleotides for mutagenesis were purchased from Evrogen. PCRs were carried out using PTC-100 thermal cycler (MJ Research). Purification of PCR products and products of digestion was performed by gel electrophoresis and extraction using the Cleanup Standard Kit (Evrogen). Site-directed mutagenesis was performed by self-assembly cloning [31].
Random mutagenesis was performed by error-prone PCR [32] using an in house analogue of the Diversify PCR Random Mutagenesis kit (Clontech, 50x mix: 0.2 мМ dGTP, 0.2 мМ dATP, 1 мМ dCTP, 1 мМ dTTP). The average mutation rate, which is a function of buffer composition and number of PCR cycles, was selected to result in approximately 2–3 amino acid mutations per gene sequence.
For bacterial expression, a fluorescent protein gene was cloned into the pQE-30 vector (Qiagen) using EcoRI and HindIII restriction sites (in random mutagenesis experiments) or by self-assembly cloning [33]. Restriction endonucleases were purchased from Sibenzyme. We selected mutants by visual screening of Escherichia coli colonies expressing mutant proteins using Olympus SZX12 fluorescence stereo zoom microscope.
The plasmid for the expression of mitochondria-targeted KillerOrange was constructed by inserting KillerOrange coding sequence into the pKillerRed-dMito vector (Evrogen) instead of KillerRed sequence (using AgeI and NotI restriction sites).
Purification and characterization proteins in vitro
Fluorescent proteins were expressed in E. coli XL1 Blue strain (Invitrogen) in the LB medium, centrifuged, sonicated in PBS (pH 7.4), then purified using Talon metal-affinity resin (Clontech). Cary 100 UV/VIS Spectrophotometer and Varian Cary Eclipse Fluorescence spectrophotometer were used to measure absorption and excitation-emission spectra. Quantum yield was determined by direct comparison with TagRFP [34].
Extinction coefficient was determined as follows. Synthetic CFP chromophore in basic dimethylformamide possesses double-peaked absorption curve with two close maxima at 460 and 473 nm and extinction coefficient 46000 M−1cm−1 at both wavelengths [35]. This value was used to calculate the concentration of mature protein in a sample of alkali-denatured KillerOrange (these conditions are believed to result in the reduction of acylimin bond leading to the formation of CFP chromophore from KillerOrange’s mature chromophore). The extinction coefficients of the protein were then obtained spectrophotometrically using the known concentration.
To measure the pH dependence of spectra, we used buffers in the pH range from 3 to 11 (citrate buffers in the pH range 3.0–4.5, phosphate buffers in the pH range 5.0–7.5, and borate buffers in the pH range 8.0–11.0). An aliquot of purified protein was diluted in the corresponding buffer solution. Spectra were measured after about 15 min of incubation in buffer.
Phototoxicity test in E. coli
To compare the phototoxicity in bacteria, we mixed a suspension of E. coli XL1 Blue cells (PBS pH 7.4, PanEco) transformed with EGFP (non-phototoxic control) with the suspension of cells transformed with the phototoxic protein. The E. coli cells used for the experiment were incubated overnight at 4°C to increase the fraction of the mature protein. We used genes cloned in pQE-30 vector under the leaky promoter and did not use induction for expression.
We diluted suspension to the optical density of 0.05 and aliquoted it into transparent PCR tubes, 40 mkl of the suspension per tube. We kept one aliquot in the darkness while others were subjected to illumination with light. All aliquots were then spread over the Petri dishes, cultivated for 12–16 hours at 37°C, washed out and analyzed with fluorescence-activated cell sorter (FACS Aria III, BD Biosciences).
For the experiment with EGFP-KillerOrange mixture we illuminated the sample for 60 seconds with custom LED illuminator (assembly of 7 Luxeon Star LXML-PB01-0040 Rebel LEDs, Lumileds, 470/20 nm, spectral half-width of 20 nm), with the light intensity of about 1 W/cm2 at the sample.
For the experiment with EGFP-KillerOrange-KillerRed mixture we illuminated samples with one of LEDs of X-Cite XLED1 illuminator (Lumen Dynamics Group Inc.) We used LEDs named “BDX” (450–495 nm), “BGX” (505–545 nm) and “GYX” (540–600 nm), their full spectra are shown at S3 Fig. The power of all LEDs was set to be about 80 mW/cm2 at the sample.
Phototoxicity test in mammalian cells
The phototoxicity test in mammalian cells followed the same principle as the test in bacteria. We grew HEK 293 cells in DMEM (Paneco, Russia) on 6-cm Petri dishes until about 70% confluency. We then transiently transfected them using FuGene HD (Roche) with one of the following plasmids: pKillerRed-dMito (Evrogen), pKillerOrange-dMito (for this construct we replaced KillerRed coding sequence with KillerOrange in pKillerRed-dMito) or pEGFP-N1 (Clontech). We then imaged cells using a Zeiss LSM-710 confocal inverted microscope (Carl Zeiss, Germany) to confirm correct protein localization.
24 hours after transfection we washed cells off from the dish, centrifuged, resuspended in PBS pH7.4 (Paneco, Russia) and mixed in pairs (KillerOrange with EGFP, KillerRed with EGFP) in polystyrene optical cuvettes (Sarstedt, Germany). We then splitted suspensions in two halves, illuminated first one with custom LEDs while keeping the second in the darkness for the same amount of time. For blue light illumination we used assembly of 7 Luxeon Star LXML-PR01-0500 Rebel LEDs, Lumileds (dominant wavelength is 447 nm, spectral half-width is 20 nm), for orange illumination we used 7 Luxeon Star 7 LXML-PL01-0040 Rebel LEDs, Lumileds (dominant wavelength is 590 nm, spectral half-width is 20 nm). The light power at the sample was set to be about 60 mW for 447 nm light and about 20 mW for 590 nm light.
After illumination we transferred the cells back to Petri dishes, incubated cells for 24 hours, washed off again, and analyzed cell fluorescence with flow cytometer (Beckman Coulter, USA).
TCSPC measurements
We measured fluorescence lifetime using an Edinburgh Instruments time-correlated single photon counting (TCSPC) system. Fluorescence was excited with a 467 nm picosecond excitation pulses diode laser (10 MHz, Picoquant) and measured by a detection system consisted of a high speed MicroChannel Plate PhotoMultiplier Tube (MCP-PMT, Hamamatsu R3809U-50) and TCSPC electronics. The time resolution of the system was 30 ps after deconvolution with an IRF signal.
Results
KillerRed possesses a DsRed-like chromophore (Fig 1a) [4,5]. Proteins carrying this type of chromophore were shown to pass through the green- or blue-emitting states before their chromophore matures into the red-emitting structure [2]. In order to develop a non-red mutant we first performed a round of random mutagenesis on KillerRed gene expecting to find a substitution that disrupts acylimine group formation during the chromophore maturation. However, visual screening of about 100,000 colonies did not yield any green- or blue-fluorescent mutants.
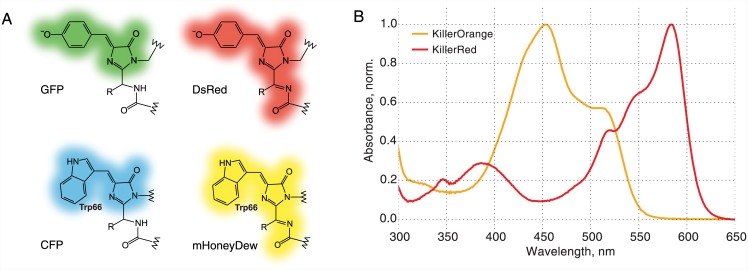
Fig 1. Structures of discussed chromophore types, absorption spectra of KillerRed and KillerOrange.
(A) Structures of GFP, CFP, DsRed and mHoneyDew chromophores. (B) Normalized absorption spectra of KillerRed and KillerOrange.
We thus changed the strategy and made three mutants of KillerRed with substitutions of the chromophore’s tyrosine (Tyr66) by other aromatic amino acids. Y66H and Y66F substitutions made KillerRed completely non-fluorescent, and we failed to find compensatory substitutions during the following round of random mutagenesis. The mutant with Y66W substitution was weakly fluorescent in orange, and we managed to enhance its brightness in three rounds of random mutagenesis. The final mutant carrying 7 substitutions (KillerRed G3C Y66W D113S N145S F177L Y221H E236Q, see the full protein sequence as S1 Text) was a bright orange fluorescent protein with excitation maximum at 512 nm and emission maximum at 555 nm. We named it KillerOrange.
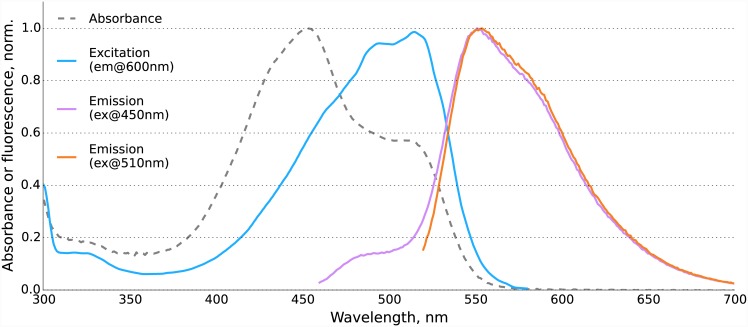
Absorbance spectrum of KillerOrange demonstrated two peaks at 455 and 514 nm (Figs 1b and 2) likely corresponding to an immature CFP-like chromophore and a mature mHoneyDew-like tryptophan-based acylimine-containing chromophore [36], respectively (Fig 1a). Both forms seemed to possess the double-peak signature characteristic for neutral tryptophan-based chromophores [37]. We noticed that the ratio of the two peaks was influenced by the expression conditions, for example, expression on Petri dishes could result in a more complete maturation of the chromophore as compared with the expression in a liquid medium. The extinction coefficients of KillerOrange expressed in liquid medium were 41200 M-1·cm-1 at 455 nm and 22600 M-1·cm-1 at 514 nm. Despite the presence of two forms, the emission of the protein was almost purely orange (Fig 2) with the maximum at 555 nm (Stokes shift of over 40 nm) and quantum yield of 0.42. The immature form contributes little to the emitted light despite being dominant on the absorption spectrum due to its low quantum yield and/or efficient Förster resonance energy transfer within a dimer.
Fig 2. Absorption, excitation and emission spectra of KillerOrange.
The fluorescence decay of KillerOrange was polyexponential, with the average fluorescence lifetime of 1.5 ns at pH 5.5, 1.9 ns at pH 7.4, and 2.6 ns at pH 9.5 (S1 Fig). The increase in fluorescence lifetime at alkaline conditions was accompanied by an increase in fluorescence intensity, as can be seen from the pH-dependence of KillerOrange spectra (S2 Fig), indicating the rise of quantum yield over pH.
To assess the phototoxicity of KillerOrange under various light conditions we designed an assay for Escherichia coli based on fluorescence-activated cell sorting. Briefly, we mixed E. coli cells expressing EGFP with E. coli cells expressing KillerOrange, and illuminated half of the suspension, while keeping another half in darkness. We then plated each aliquot on Petri dishes, grew overnight, washed the bacteria off from the plates, and analyzed the KillerOrange/EGFP ratio in the suspension using fluorescence-activated cell sorting. This approach assays only the relative phototoxicity of a protein, but is more precise and less time-consuming than the approach used earlier [38].
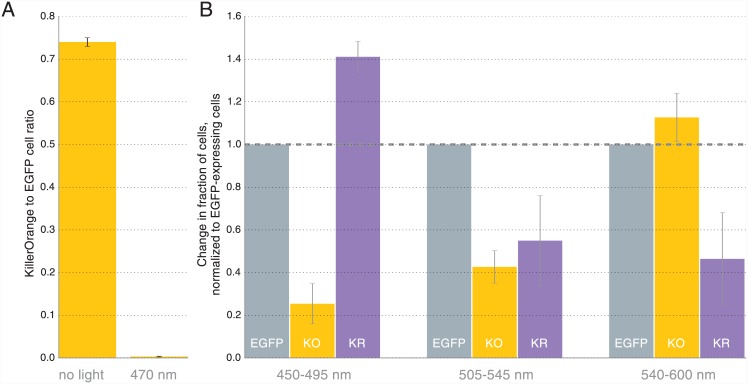
Using a very intense blue light (470/20 nm, 1 W/cm2) we found that KillerOrange-expressing cells almost disappeared (222-fold decrease) from the population as a result of just 60 seconds of illumination (Fig 3a) confirming the high level of phototoxicity of KillerOrange.
Fig 3. Phototoxicity of KillerOrange and KillerRed in E. coli.
(A) Blue light (470/20) nm illumination results in selective elimination (222-fold) of KillerOrange-expressing cells from the mixture with EGFP-expressing cells. Error bars represent SD, N = 3. (B) KillerOrange (KO) and KillerRed (KR) toxicity under light illumination at different wavelengths. Error bars represent SD, N = 4.
To test if KillerOrange and KillerRed can be used independently to kill selected cell populations with light of different wavelengths we have performed similar experiment with three proteins. We mixed E. coli cells expressing KillerOrange with the cells expressing EGFP and cells expressing KillerRed. Using another light source and milder intensities, we illuminated aliquots of cell suspension with 450–495, 505–545 and 540–600 nm light (LED spectra are shown in S3 Fig). Illumination with 540–600 nm light resulted in the selective removal of KillerRed-expressing cells, while the illumination with 450–495 nm light killed the majority of KillerOrange-expressing cells (Fig 3b, S4 Fig). Interestingly, 505–545 nm light illumination was almost equally efficient in killing both KillerOrange and KillerRed cells. Thus, by combining different light sources one can achieve precise control over cell populations expressing KillerOrange and KillerRed.
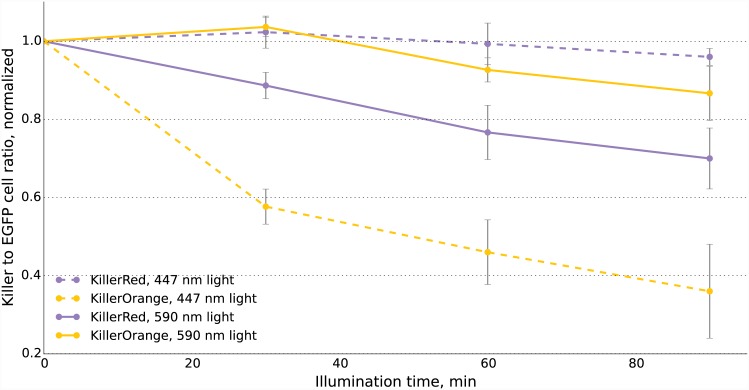
We also tested whether it is possible to use KillerOrange and KillerRed to independently ablate populations of mammalian cells. We mixed cells expressing the phototoxic protein (KillerRed or KillerOrange) with cells expressing EGFP, illuminated mixture with blue (447/20 nm) or orange (590/20 nm) light, cultivated cells for two days and then analyzed their fluorescence. As expected, we found a strong decrease in the fraction of KillerRed-expressing cells in the population that passed through the orange light irradiation (Fig 4). In contrast, KillerOrange-expressing cells were substantially removed from the population of cells that was illuminated with the blue light. Taken together, these results show that KillerOrange is phototoxic to mammalian cells and that KillerRed and KillerOrange can be used simultaneously to independently control fates of two cell populations.
Fig 4. Phototoxicity of KillerOrange and KillerRed in mammalian cells.
KillerOrange, KillerRed and EGFP-expressing cells were mixed and illuminated with 477 nm or 590 nm light. Y-axis depicts changes in fractions of KillerOrange (yellow lines) or KillerRed-expressing (red lines) cells in the population. Cell fractions were normalized to the fractions in non-illuminated sample. Error bars represent SD, N = 3.
Discussion
Here we report the development of a new genetically encoded photosensitizer KillerOrange—an orange mutant of the red fluorescent protein KillerRed. KillerOrange is phototoxic upon blue and green light illumination and can be used simultaneously with KillerRed to independently control the fate of cell populations by tuning the light source wavelength.
KillerOrange possesses two spectral forms and absorbs light efficiently in the range from ~420 nm to ~530 nm. Based on the substantial ablation of KillerOrange-expressing cells in both 450–495 nm and 505–545 nm light-illuminated samples (Fig 3) we think that both spectral forms are phototoxic.
The blue-shifted spectra of KillerOrange makes it potentially more suitable for two-photon microscopy than the parental KillerRed. Also, the large Stokes shift of over 40 nm would make it possible to spectrally separate signals of KillerOrange from cyan and green fluorescent proteins when the proteins are being excited simultaneously by blue light. One can thus use green and cyan indicators to observe the effects of phototoxicity in real time without the need to change the excitation light.
With its broad absorption spectrum KillerOrange has a substantial overlap in absorbance with another genetically encoded photosensitizer—miniSOG. With both proteins being different in many aspects we believe that the choice between them should be based on particular experimental conditions. KillerOrange is brighter and possesses strongly red-shifted emission. Like all other fluorescent proteins, KillerOrange chromophore maturation does not depend on any cofactors except for molecular oxygen (which is in any case required for reactive oxygen species production). As for miniSOG, its phototoxicity is dependent on the presence of a cofactor, flavin mononucleotide, which abundance can vary between cell compartments and cell types and can depend on the physiological state of a cell. MiniSOG was successfully used for cell ablation in C. elegans [39] and for chromophore assisted light inactivation [40] but, for unknown reasons, was shown to be inefficient when applied to mouse tumor xenografts in vivo [41]. Also, miniSOG and KillerRed differ in mechanism of reactive species production. At the moment the mechanism of KillerOrange phototoxicity is unknown, but we believe it to be the Type I, similarly to the parental KillerRed. Thus, KillerOrange and miniSOG are possibly not equivalent, and should be chosen with regards to the experimental system.
We believe that the expansion of the palette of available genetically encoded photosensitizers will open up new possibilities. KillerOrange-KillerRed pair can potentially be used to independently ablate two cell populations during development, or to cut off two distinct neural circuits in the brain. This pair also promise the orthogonal optical control of the propagation of signaling cascades either by chromophore-assisted light inactivation of the participating proteins or by triggering cascades with hydrogen peroxide produced by KillerRed and likely KillerOrange.
Due to same-channel excitation compatibility with the majority of green fluorescent protein-based sensors KillerOrange can be used in systems where immediate registration of the toxic effects is required, such as nervous system or fast intracellular processes. Combination of various photosensitizers in one cassette, or KillerOrange-KillerRed tandem fusions, may enhance phototoxicity under white light irradiation and may be useful both as a research tool in biology and as a potential anti-tumor agent in photodynamic therapy.
Supporting Information
Fluorescence was excited at 467 nm and detected at 565 nm.
(EPS)
Absorption (A), fluorescence excitation (C) and emission (B, D) spectra of KillerOrange at various pH (pH values are indicated in the legend). Note the correspondence between the pH-dependent change of emission intensity (B, D) and the average fluorescence lifetimes (S1 Fig) measured at similar conditions.
(EPS)
Absorption spectra of KillerOrange and KillerRed and emission spectra of LEDs used for phototoxicity experiments with KillerOrange, KillerRed and EGFP in bacteria. Blue-filled area represents normalized spectrum of “BDX” LED, green-filled—spectrum of “BGX” LED and orange-filled—spectrum of “GYX” LED.
(EPS)
Illumination of mixtures KillerOrange, KillerRed and EGFP-expressing E.coli cells with 450–495 nm (A), 505–545 nm (B) and 540–600 nm (C) light. Error bars represent SD, N = 4.
(EPS)
(DOCX)
Acknowledgments
We would like to thank J.T. Ripper (Department of Surgery, Whitechapel District Institute, London, UK) for fruitful discussions and technical assistance. The work was supported by Russian Science Foundation (14-25-00129). KSS was supported by OPTEC grant for young scientists (21/2014/51-Msk) and Science and Innovation Youth Contest Participant grant (UMNIK, 3616ГУ2/2014). Experiments were partially carried out using the equipment provided by the IBCH сore facility (CKP IBCH). KMS acknowledges generous support from the National Science Foundation (CHE-1213047).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The work was supported by Russian Science Foundation (14-25-00129). KSS was supported by OPTEC grant for young scientists (21/2014/51-Msk) and Science and Innovation Youth Contest Participant grant (UMNIK, 3616ГУ2/2014). Experiments were partially carried out using the equipment provided by the IBCH сore facility (CKP IBCH). KMS acknowledges generous support from the National Science Foundation (CHE-1213047).
References
- 1. Mishin AS, Belousov VV, Solntsev KM, Lukyanov KA. Novel uses of fluorescent proteins. Curr Opin Chem Biol. 2015;27: 1–9. 10.1016/j.cbpa.2015.05.002 [DOI] [PubMed] [Google Scholar]
- 2. Chudakov DM, Matz MV, Lukyanov S, Lukyanov KA. Fluorescent proteins and their applications in imaging living cells and tissues. Physiol Rev. 2010;90: 1103–1163. 10.1152/physrev.00038.2009 [DOI] [PubMed] [Google Scholar]
- 3. Lukyanov KA, Serebrovskaya EO, Lukyanov S, Chudakov DM. Fluorescent proteins as light-inducible photochemical partners. Photochem Photobiol Sci. 2010;9: 1301–1306. 10.1039/c0pp00114g [DOI] [PubMed] [Google Scholar]
- 4. Pletnev S, Gurskaya NG, Pletneva NV, Lukyanov KA, Chudakov DM, Martynov VI, et al. Structural basis for phototoxicity of the genetically encoded photosensitizer KillerRed. J Biol Chem. 2009;284: 32028–32039. 10.1074/jbc.M109.054973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carpentier P, Violot S, Blanchoin L, Bourgeois D. Structural basis for the phototoxicity of the fluorescent protein KillerRed. FEBS Lett. 2009;583: 2839–2842. 10.1016/j.febslet.2009.07.041 [DOI] [PubMed] [Google Scholar]
- 6. Bulina ME, Chudakov DM, Britanova OV, Yanushevich YG, Staroverov DB, Chepurnykh TV, et al. A genetically encoded photosensitizer. Nat Biotechnol. 2006;24: 95–99. [DOI] [PubMed] [Google Scholar]
- 7. Vegh RB, Solntsev KM, Kuimova MK, Cho S, Liang Y, Loo BLW, et al. Reactive oxygen species in photochemistry of the red fluorescent protein “Killer Red.” Chem Commun. 2011;47: 4887–4889. [DOI] [PubMed] [Google Scholar]
- 8. Vegh RB, Bravaya KB, Bloch DA, Bommarius AS, Tolbert LM, Verkhovsky M, et al. Chromophore photoreduction in red fluorescent proteins is responsible for bleaching and phototoxicity. J Phys Chem B. 2014;118: 4527–4534. 10.1021/jp500919a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Serebrovskaya EO, Gorodnicheva TV, Ermakova GV, Solovieva EA, Sharonov GV, Zagaynova EV, et al. Light-induced blockage of cell division with a chromatin-targeted phototoxic fluorescent protein. Biochem J. 2011;435: 65–71. 10.1042/BJ20101217 [DOI] [PubMed] [Google Scholar]
- 10. Lan L, Nakajima S, Wei L, Sun L, Hsieh C-L, Sobol RW, et al. Novel method for site-specific induction of oxidative DNA damage reveals differences in recruitment of repair proteins to heterochromatin and euchromatin. Nucleic Acids Res. 2014;42: 2330–2345. 10.1093/nar/gkt1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Serebrovskaya EO, Ryumina AP, Boulina ME, Shirmanova MV, Zagaynova EV, Bogdanova EA, et al. Phototoxic effects of lysosome-associated genetically encoded photosensitizer KillerRed. J Biomed Opt. 2014;19: 071403 10.1117/1.JBO.19.7.071403 [DOI] [PubMed] [Google Scholar]
- 12. Jarvela T, Linstedt AD. Isoform-specific tethering links the Golgi ribbon to maintain compartmentalization. Mol Biol Cell. 2014;25: 133–144. 10.1091/mbc.E13-07-0395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Destaing O, Planus E, Bouvard D, Oddou C, Badowski C, Bossy V, et al. β1A integrin is a master regulator of invadosome organization and function. Mol Biol Cell. 2010;21: 4108–4119. 10.1091/mbc.E10-07-0580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Matsunaga S, Takata H, Morimoto A, Hayashihara K, Higashi T, Akatsuchi K, et al. RBMX: a regulator for maintenance and centromeric protection of sister chromatid cohesion. Cell Rep. 2012;1: 299–308. 10.1016/j.celrep.2012.02.005 [DOI] [PubMed] [Google Scholar]
- 15. Ivashchenko O, Van Veldhoven PP, Brees C, Ho Y-S, Terlecky SR, Fransen M. Intraperoxisomal redox balance in mammalian cells: oxidative stress and interorganellar cross-talk. Mol Biol Cell. 2011;22: 1440–1451. 10.1091/mbc.E10-11-0919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shibuya T, Tsujimoto Y. Deleterious effects of mitochondrial ROS generated by KillerRed photodynamic action in human cell lines and C. elegans. J Photochem Photobiol B. 2012;117: 1–12. 10.1016/j.jphotobiol.2012.08.005 [DOI] [PubMed] [Google Scholar]
- 17. Byrne LC, Khalid F, Lee T, Zin EA, Greenberg KP, Visel M, et al. AAV-mediated, optogenetic ablation of Müller Glia leads to structural and functional changes in the mouse retina. PLoS One. 2013;8: e76075 10.1371/journal.pone.0076075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Williams DC, Bejjani RE, Ramirez PM, Coakley S, Kim SA, Lee H, et al. Rapid and permanent neuronal inactivation in vivo via subcellular generation of reactive oxygen with the use of KillerRed. Cell Rep. 2013;5: 553–563. 10.1016/j.celrep.2013.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kobayashi J, Shidara H, Morisawa Y, Kawakami M, Tanahashi Y, Hotta K, et al. A method for selective ablation of neurons in C. elegans using the phototoxic fluorescent protein, KillerRed. Neurosci Lett. 2013;548: 261–264. 10.1016/j.neulet.2013.05.053 [DOI] [PubMed] [Google Scholar]
- 20. Del Bene F, Wyart C, Robles E, Tran A, Looger L, Scott EK, et al. Filtering of visual information in the tectum by an identified neural circuit. Science. 2010;330: 669–673. 10.1126/science.1192949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jewhurst K, Levin M, McLaughlin KA. Optogenetic Control of Apoptosis in Targeted Tissues of Xenopus laevis Embryos. J Cell Death. 2014;7: 25–31. 10.4137/JCD.S18368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liao Z-X, Li Y-C, Lu H-M, Sung H-W. A genetically-encoded KillerRed protein as an intrinsically generated photosensitizer for photodynamic therapy. Biomaterials. 2014;35: 500–508. 10.1016/j.biomaterials.2013.09.075 [DOI] [PubMed] [Google Scholar]
- 23. Shirmanova MV, Serebrovskaya EO, Lukyanov KA, Snopova LB, Sirotkina MA, Prodanetz NN, et al. Phototoxic effects of fluorescent protein KillerRed on tumor cells in mice. J Biophotonics. 2013;6: 283–290. 10.1002/jbio.201200056 [DOI] [PubMed] [Google Scholar]
- 24. Kuznetsova DS, Shirmanova MV, Dudenkova VV, Subochev PV, Turchin IV, Zagaynova EV, et al. Photobleaching and phototoxicity of KillerRed in tumor spheroids induced by continuous wave and pulsed laser illumination. J Biophotonics. 2015;9999. 10.1002/jbio.201400130 [DOI] [PubMed] [Google Scholar]
- 25. Yan L, Kanada M, Zhang J, Okazaki S, Terakawa S. Photodynamic Treatment of Tumor with Bacteria Expressing KillerRed. PLoS One. 2015;10: e0131518 10.1371/journal.pone.0131518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Takemoto K, Matsuda T, Sakai N, Fu D, Noda M, Uchiyama S, et al. SuperNova, a monomeric photosensitizing fluorescent protein for chromophore-assisted light inactivation. Sci Rep. 2013;3: 2629 10.1038/srep02629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shu X, Lev-Ram V, Deerinck TJ, Qi Y, Ramko EB, Davidson MW, et al. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 2011;9: e1001041 10.1371/journal.pbio.1001041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Torra J, Burgos-Caminal A, Endres S, Wingen M, Drepper T, Gensch T, et al. Singlet oxygen photosensitisation by the fluorescent protein Pp2FbFP L30M, a novel derivative of Pseudomonas putida flavin-binding Pp2FbFP. Photochem Photobiol Sci. 2015;14: 280–287. 10.1039/c4pp00338a [DOI] [PubMed] [Google Scholar]
- 29. Levin E, Mavila S, Eivgi O, Tzur E, Lemcoff NG. Regioselective Chromatic Orthogonality with Light-Activated Metathesis Catalysts. Angew Chem Int Ed Engl. WILEY-VCH Verlag; 2015; 10.1002/ange.201500740 [DOI] [PubMed] [Google Scholar]
- 30. Hansen MJ, Velema WA, Lerch MM, Szymanski W, Feringa BL. Wavelength-selective cleavage of photoprotecting groups: strategies and applications in dynamic systems. Chem Soc Rev. The Royal Society of Chemistry; 2015;44: 3358–3377. [DOI] [PubMed] [Google Scholar]
- 31. Matsumoto A, Itoh TQ. Self-assembly cloning: a rapid construction method for recombinant molecules from multiple fragments. Biotechniques. 2011;51: 55–56. 10.2144/000113705 [DOI] [PubMed] [Google Scholar]
- 32. Vartanian JP, Henry M, Wain-Hobson S. Hypermutagenic PCR involving all four transitions and a sizeable proportion of transversions. Nucleic Acids Res. 1996;24: 2627–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Matsumoto A, Itoh TQ. Self-assembly cloning: a rapid construction method for recombinant molecules from multiple fragments. Biotechniques. 2011;51: 55–56. 10.2144/000113705 [DOI] [PubMed] [Google Scholar]
- 34. Merzlyak EM, Goedhart J, Shcherbo D, Bulina ME, Shcheglov AS, Fradkov AF, et al. Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat Methods. 2007;4: 555–557. [DOI] [PubMed] [Google Scholar]
- 35. Sarkisyan KS, Yampolsky IV, Solntsev KM, Lukyanov SA, Lukyanov KA, Mishin AS. Tryptophan-based chromophore in fluorescent proteins can be anionic. Sci Rep. 2012;2: 608 10.1038/srep00608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ai HW, Shaner NC, Cheng Z, Tsien RY, Campbell RE. Exploration of new chromophore structures leads to the identification of improved blue fluorescent proteins. Biochemistry. 2007;46: 5904–5910. [DOI] [PubMed] [Google Scholar]
- 37. Lelimousin M, Noirclerc-Savoye M, Lazareno-Saez C, Paetzold B, Le Vot S, Chazal R, et al. Intrinsic dynamics in ECFP and Cerulean control fluorescence quantum yield. Biochemistry. 2009;48: 10038–10046. 10.1021/bi901093w [DOI] [PubMed] [Google Scholar]
- 38. Bulina ME, Chudakov DM, Britanova OV, Yanushevich YG, Staroverov DB, Chepurnykh TV, et al. A genetically encoded photosensitizer. Nat Biotechnol. 2006;24: 95–99. [DOI] [PubMed] [Google Scholar]
- 39. Qi YB, Garren EJ, Shu X, Tsien RY, Jin Y. Photo-inducible cell ablation in Caenorhabditis elegans using the genetically encoded singlet oxygen generating protein miniSOG. Proc Natl Acad Sci U S A. 2012;109: 7499–7504. 10.1073/pnas.1204096109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lin JY, Sann SB, Zhou K, Nabavi S, Proulx CD, Malinow R, et al. Optogenetic inhibition of synaptic release with chromophore-assisted light inactivation (CALI). Neuron. 2013;79: 241–253. 10.1016/j.neuron.2013.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ryumina AP, Serebrovskaya EO, Shirmanova MV, Snopova LB, Kuznetsova MM, Turchin IV, et al. Flavoprotein miniSOG as a genetically encoded photosensitizer for cancer cells. Biochim Biophys Acta. 2013;1830: 5059–5067. 10.1016/j.bbagen.2013.07.015 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fluorescence was excited at 467 nm and detected at 565 nm.
(EPS)
Absorption (A), fluorescence excitation (C) and emission (B, D) spectra of KillerOrange at various pH (pH values are indicated in the legend). Note the correspondence between the pH-dependent change of emission intensity (B, D) and the average fluorescence lifetimes (S1 Fig) measured at similar conditions.
(EPS)
Absorption spectra of KillerOrange and KillerRed and emission spectra of LEDs used for phototoxicity experiments with KillerOrange, KillerRed and EGFP in bacteria. Blue-filled area represents normalized spectrum of “BDX” LED, green-filled—spectrum of “BGX” LED and orange-filled—spectrum of “GYX” LED.
(EPS)
Illumination of mixtures KillerOrange, KillerRed and EGFP-expressing E.coli cells with 450–495 nm (A), 505–545 nm (B) and 540–600 nm (C) light. Error bars represent SD, N = 4.
(EPS)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.