Abstract
Among frequent events in prostate cancer are loss of the tumor-suppressor phosphatase and tensin homologue (PTEN) and overexpression of Prostate-Specific G-protein-coupled Receptor (PSGR), but the potential tumorigenic synergy between these lesions is unknown. Here, we report a new mouse model (PSGR- PtenΔ/Δ) combining prostate-specific loss of Pten with probasin promoter-driven PSGR overexpression. By 12 months PSGR-PtenΔ/Δ mice developed invasive prostate tumors featuring Akt activation and extensive inflammatory cell infiltration. PSGR-PtenΔ/Δ tumors exhibited E-cadherin loss and increased stromal androgen receptor (AR) expression. PSGR overexpression increased LnCaP proliferation, while PSGR shRNA knockdown inhibited proliferation and migration. In conclusion, we demonstrate that PSGR overexpression synergizes with loss of PTEN to accelerate prostate cancer development, and present a novel bigenic mouse model that mimics the human condition, where both PSGR overexpression and loss of PTEN occur concordantly in the majority of advanced prostate cancers, yielding an environment more relevant to studying human prostate cancer.
Keywords: PSGR, PTEN, AR, prostate cancer, stromal/epithelial interactions
Introduction
Prostate cancer continues to be a major and growing health problem today (1). The likelihood of developing prostate cancer is as high as 1 in 6 men, and of these, 1 in 30 will die as a direct consequence of metastatic disease (2). Although a number of genetic and lifestyle factors have been implicated in the development of prostate cancer, along with mediating conditions such as chronic inflammation, how specific genetic lesions interact and contribute to cancer progression remains an area of active discovery.
Phosphatase and tensin homologue (PTEN) is the most frequently deleted tumor-suppressor gene in prostate cancer and its loss is predictive of poor prognosis (3-5). PTEN inactivation or loss is also implicated in prostate cancer progression to a castration-resistant phenotype (6-8). Down-regulation of PTEN function induces a concomitant activation of AKT, which can regulate cell survival and growth pathways, including nuclear factor (NF)-κB-dependent inflammation (6), as well as mechanistic target of Rapamycin, glycogen synthase kinase-3β, and Raf (9). Although PTEN loss has been reported to cooperate with ERG transactivation (10) or p53 loss (11) in prostate cancer development, at present, our understanding of the oncogenic events that cooperate with PTEN loss during tumor progression remains limited.
The prostate-specific G-protein coupled receptor (PSGR) is exclusively expressed in the human prostate and is overexpressed in prostate tumors, with PSGR levels correlating with tumor progression (12-15). PSGR expression is controlled in part by interleukin-6 signaling (16), suggesting potential PSGR upregulation in response to inflammatory stimuli. Recent findings in our lab show that transgenic PSGR expression induces a chronic inflammatory reaction in the prostate through AKT-mediated NF-κB activation, resulting in low grade Prostatic Intraepithelial Neoplasia (PIN) (17). However, PSGR overexpression by itself was unable to cause complete carcinoma formation, suggesting the contribution of additional lesions during prostate carcinogenesis.
Here, we sought to determine how down-regulation of PTEN synergizes with over-expression of PSGR, a genetic combination frequently seen in human prostate cancer (6, 12), to enhance prostate cancer development. We generated a novel mouse model encompassing a prostate-specific combination of these genetic alterations and examined the effects on cell proliferation, migration, and tumor progression. We observed accelerated tumorigenesis in double mutant mice accompanied by AKT-NFκB driven inflammation, loss of E-cadherin suggestive of epithelial to mesenchymal-like transition, and paracrine regulation of stromal AR. Our model represents a new approach for analyzing combinatorial effects of lesions frequently observed in human cancers.
Results
PSGR over-expression cooperates with Pten loss to facilitate prostate cancer progression
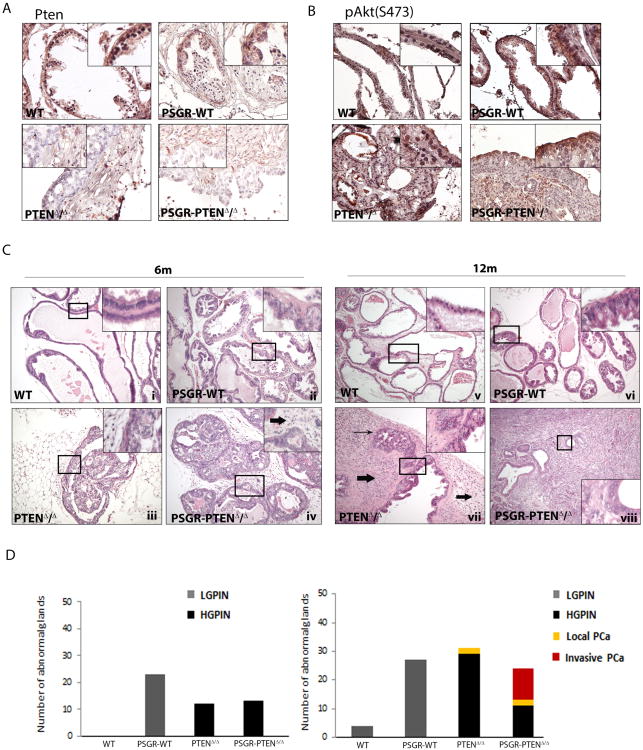
PSGR expression increases upon transition from benign prostate tissue to primary prostate cancer, as we have previously shown (12). High expression levels are maintained in advanced prostate cancer, suggesting a possible role in this disease stage. Genomic data set analysis (Oncomine (Compendia Bioscience, Ann Arbor, MI, USA – http://www.oncomine.org))(18, 19)) was performed on tumor datasets available online. Four data sets out of ten showed upregulation (expression change ≥ 1.5 fold) of PSGR, or OR51E2, in 89% and 50%, 76% and 70% of tumors, respectively (Supplementary Figure 1A). These microarray results are in accord with two previous reports using in situ hybridization and quantitative PCR analysis that found PSGR overexpression in 76% and 64% of prostate cancers (12, 20)). PTEN inactivation is common in advanced prostate cancer,(7) suggesting that co-occurrence and potential synergism between PSGR over-expression and PTEN loss is highly probable. In the Oncomine™ (Compendia Bioscience) database, we found that PSGR was frequently over-expressed but PTEN expression was not significantly changed in prostate tumor tissue (P<0.001; Supplementary Figure 1B); by TCGA analysis PTEN was mutated or deleted in 21% (103 out of 487 tumors) of prostate cancers, but no significant correlation in expression between PSGR and PTEN genes could be found (data not shown). However, the limitations of tumor mRNA microarrays, including a smaller dynamic range and less reliable quantitation than quantitative PCR, and lack of information on protein levels, led us to adopt an alternative approach. To further investigate the possibility of synergy between PSGR overexpression and PTEN loss in prostate tumor development, we created a bigenic mouse model over-expressing human PSGR with a conditional Pten knockout, both driven by the ARR2PB promoter. Immunohistochemistry showed downregulation of Pten expression in epithelial cells (Figure 1A), with consequent elevation of phosphoAKT (S473) upon PSGR over-expression and complete loss of Pten (Figure 1B). Tumor initiation and progression of Pten deleted mice was much slower than originally described for the conditional Pten knockout model (21). Analysis of mouse tissues at 6 and 12 months of age revealed striking differences in tumor progression. Of note, at 6 months of age, PSGR transgenics (PSGR-Ptenf/f-Cre- or PSGR-PtenWT) showed an increased amount of LGPIN lesions, characterized by cell layer conglomeration and a tufting growth pattern, but no nuclear enlargement or prominent nucleoli (Figure 1C-ii). Mice with only a complete deletion of Pten (Pten f/f-Cre+ or PtenΔ/Δ) showed PIN lesions of a cribiform pattern, with cells beginning to grow towards the center of the lumen and a small increase in stromal cells (Figure 1C-iii). Pten knockouts with an added PSGR transgene (PSGR-Ptenf/f-Cre+ or PSGR-PtenΔ/Δ), however, show glands with cribiform PIN that have already reached the center of the lumen and a higher extent of cell infiltration in the stroma (Figure 1C – iv). No carcinoma was detected at this time-point. At 12 months of age, the phenotype is significantly different. PSGR-PtenWT prostates continued to show LGPIN lesions, but no progression to carcinoma, as previously described (Figure 1C-vi), whereas PtenΔ/Δ mice displayed an important inflammatory cell infiltration with carcinoma in situ lesions growing adjacent to HGPIN glands (Figure 1C – vii). PSGR-PtenΔ/Δ mice, however, already showed invasive adenocarcinomas with tumor masses disrupting the normal prostatic architecture, including the basal membranes and smooth muscle cells normally surrounding the glands (Figure 1C – viii). PSGR over-expressing mice were analyzed with conditional loss of one Pten allele (Ptenf/w-Cre+ (PTENΔ/+) vs. PSGR-Ptenf/w-Cre+ (PSGR-PTENΔ/+), and while they did show glands with LGPIN, no significant differences were found between these genotypes at either time point (data not shown).
Figure 1. Presence of PSGR cooperates with Pten deletion.
A Immunohistochemistry showing intensity of Pten expression in12 month old prostate sections of each genotype analyzed. B. Immunohistochemistry showing activation of AKT (phosphoS473) in genotypes analyzed. C. Mice harboring a complete Pten deletion together with PSGR overexpression (PSGR-PTENΔ/Δ) show more aggressive progression both at six months (iv) and twelve months (viii) compared with PSGR over-expression alone (PSGR-WT (ii and vi)) or complete Pten deletion alone (PTENΔ/Δ (iii and vii)) Thick arrows indicate infiltrating inflammatory cells; long arrows indicate carcinoma in situ. D. Representative bar graph showing morphological differences between each genotype at 6 (left) and 12 (right) months of age (n=4 mice/age/genotype, 10 analyzed fields per mouse). All images shown were taken using 20× objectives (40× for inserts) and are representative of N = 4 mice per genotype and time point.
PSGR combined with Pten loss increases prostate cell proliferation
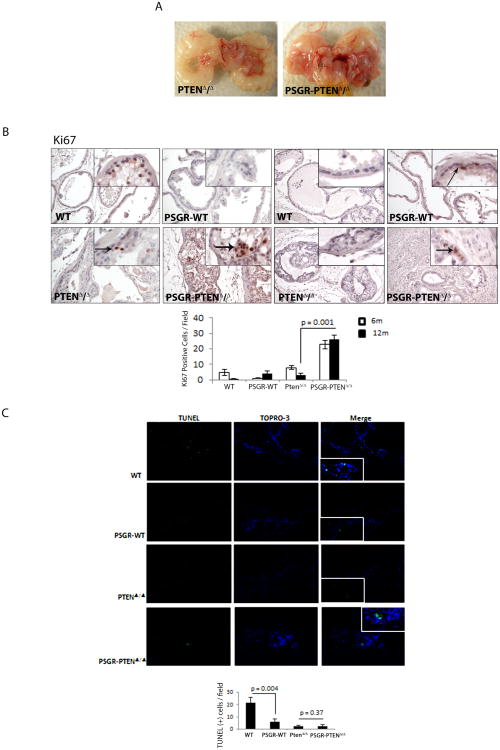
Macroscopically, loss of Pten alone produced an altered phenotype in the prostate, but identification of individual lobes was still possible, whereas addition of PSGR frequently caused deformities in prostatic structure and occlusion of the adjacent seminal vesicles (Figure 2A). In an effort to understand the underlying mechanisms of this phenotype, we performed Ki67 staining to measure differences in proliferation. As shown in Figure 2B, over-expression of PSGR alone was enough to induce a small increase in proliferation levels in 12-month-old mice, but when combined with a complete loss of Pten expression, the effect on proliferation was greater than in Pten knockouts alone. Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) analysis for apoptosis showed that PSGR over-expression or Pten deletion alone were enough to significantly reduce apoptosis, but these genetic alterations do not appear to act synergistically to decrease cell death (Figure 2C).
Figure 2. PSGR overexpression induces proliferation in a Pten deleted background.
A Representative macroscopic image of male genitourinary tract showing differences between 12 month old Pten deleted mice (PTENΔ/Δ) and Pten deleted mice overexpressing PSGR (PSGR-PTENΔ/Δ) B. Ki67 staining showing increased proliferation in PSGR overexpressing Pten-deleted mice, both at 6 months (left) and 12 months (right). Bar graph represents average Ki67 positive cells/field, ± standard error (n=4 per genotype per age). p = 0.001, t-test. Arrows indicate location of positive staining cells. C. TUNEL assay showing differences in cell apoptosis between different genotypes. Bar graph represents average TUNEL positive cells/field, ± standard error (minimum 10 fields analyzed per section; n=4 per genotype) p = 0.004 and p = 0.37, t-test.
PSGR in conjunction with loss of Pten induces an invasive phenotype in the prostate
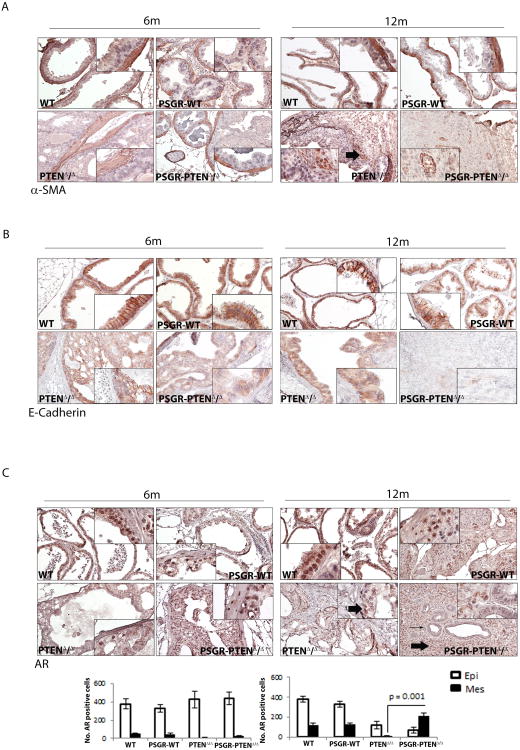
Analysis of smooth muscle actin showed that smooth muscle cells begin to disappear in PtenΔ/Δ mice at 12 months of age, with a fragmented layer of cells surrounding the glands. PSGR-PtenΔ/Δ mice, however, showed no actual glandular structure and had a complete loss of smooth muscle cells, including in areas surrounding the tumor mass. These mice did show positive smooth muscle cells in intra-tumoral areas, which marked neoangiogenic formations surrounded by cancer cells (Figure 3A). Because of this increased tumorigenicity, we analyzed the samples for both E-cadherin and androgen receptor (AR). E-cadherin expression (Figure 3B) was decreased both by the over-expression of PSGR (PSGR-PtenWT at 12 months) and by the complete loss of Pten, with a synergistic effect observed in PSGR-PtenΔ/Δ mice at 12 months of age, which had an almost complete loss of E-cadherin expression. AR was also affected by this genetic combination, as both epithelial and stromal AR was lost in PtenΔ/Δ mice, but stromal AR expression was regained in PSGR-PtenΔ/Δ mice (Figure 3C), suggesting that PSGR may increase stromal AR expression upon complete loss of Pten and contribute to the elevated cell proliferation observed in these mice (Figure 2B).
Figure 3. Characterization of PSGR-Pten mice.
A. Smooth muscle actin staining shows breakage of smooth muscle barrier surrounding prostate glands in PTENΔ/Δ mice at 12 months of age. Overexpression of PSGR with Pten deletion (PSGR-PTENΔ/Δ) shows disintegration of smooth muscle gland barrier, with emergence of new intra-tumoral blood vessel structures. Arrows indicate positive stained cells. B. E-cadherin staining shows down-regulation of E-cadherin in PTENΔ/Δ prostates at each time point. Downregulation of E-cadherin in PSGR-WT prostates is seen at 12 months of age and there is a synergistic decrease in E-cadherin expression in PSGR-PTENΔ/Δ at both 6 months and 12 months of age. C. Androgen receptor (AR) expression is down-regulated in PTENΔ/Δ in both epithelial and stromal cells at both time points. PSGR-PTENΔ/Δ mice show re-emergence of AR in stromal cells. Bar graph represents average number of AR-positive cells per compartment ± standard error (n=4 mice per genotype), p = 0.001, t-test. Thick arrows indicate location of positive staining cells; long arrow indicates gland with negative AR staining. 6m, 6 months; 12m, 12 months.
Upon Pten loss, PSGR potentiates inflammatory cell infiltration in the stroma
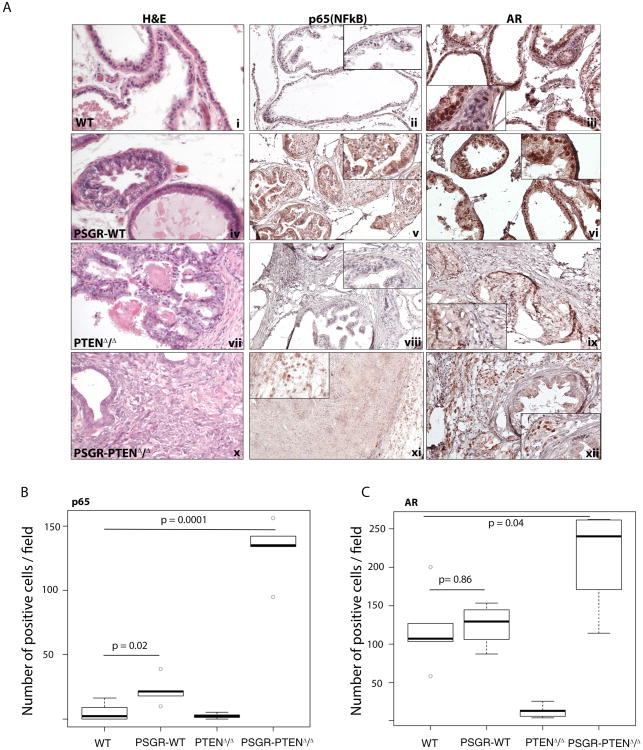
Because PSGR is capable of inducing a chronic inflammatory reaction in the prostate, we hypothesized that this plays a role in AR expression differences upon Pten deletion. In the presence of Pten, p65 NF-κB expression is increased by PSGR, but its localization is both nuclear and cytoplasmic (Figures 4A-v and 4B wild type (WT) vs PSGR-WT). AR expression is not significantly changed between PtenWT and PSGR-PtenWT mice (Figure 4A-iii and vi Figure 4C WT vs PSGR-WT). However, in the absence of epithelial Pten (PtenΔ/Δ), no p65 expression can be detected (Figure 4A-xiii, Figure 4B PTENΔ/Δ), and both epithelial and stromal AR are significantly decreased (Figure 4A-ix, Figure 4C PTEN Δ/Δ). When PSGR is over-expressed (PSGR-PtenΔ/Δ), stromal p65 nuclear localization increased (Figure 4A xi, Figure 4B PSGR-PTEN Δ/Δ) accompanied by a significant up-regulation of stromal AR (Figure 4A.xii, Figure 4C PSGR-PTEN Δ/Δ), suggesting that in a Pten-deleted context, PSGR may regulate stromal AR expression through stromal NF-κB. Quantitations are shown in Figures 4B and C. To further characterize this immune cell infiltration, we performed IHC for CD3 as well as macrophage markers. We observed an increase in infiltrating macrophages, and synergy between PSGR overexpression and Pten loss on increasing numbers of infiltrating CD3+ T-cells (Supplementary Figure 2A). Therefore, PSGR over-expression together with Pten loss enhances immune cell infiltration in prostate tumors.
Figure 4. Androgen receptor (AR) expression pattern is correlated with p65 (NF-κB) expression pattern.
A. Staining shows expression pattern of p65 (NF-κB) and AR in prostate sections of 12 month old mice. PSGR-WT mice show both cytoplasmic and nuclear localization of p65 and AR (v and vi). Nuclear localization is correlated with protein activity. PSGR-PTENΔ/Δ mice show increased levels of nuclear p65 and AR in the stromal compartment of the prostate (xi and xii). B. Box plots represent the average number of positive cells per field for each genotype. Only nuclear p65 and AR staining was considered as positive (n=4mice/group), Comparison of p65 staining between WT and PSGR over-expressing mice is significantly increased (p-value = 0.02), whereas concomitant deletion of PTEN and PSGR over-expression further increases p65 nuclear staining (p-value = 0.0001). C. For AR, there is no significant difference in stromal AR staining (p-value = 0.86) between WT and PSGR over-expressing mice, and deletion of PTEN alone is enough to induce a significant loss of stromal AR (p-value = 0.009). Double mutant mice, however, show a statistically significant increase in stromal AR staining (p-value = 0.04), suggesting deletion of PTEN is necessary for PSGR to induce expression of stromal AR.
PSGR over-expression affects cell oncogenic properties both directly and indirectly
We next investigated specific oncogenic effects of PSGR using human prostate cancer cell lines. We previously showed PSGR expression in LnCAP and MDA PCa 2a cells. The prostate cancer cell line LnCAP has an inactivating mutation in PTEN (22), therefore, we selected it to generate cells stably expressing either PSGR (LnCaP-PSGR) or shRNA targeting PSGR (LnCaP-KD) by lentiviral transduction. PSGR levels were measured by real-time PCR and found to be increased approximately 500-fold (data not shown) in the over-expressing cell lines and decreased by approximately 90% in the KD cells (Supplementary Figure 3A). We observed an increase in proliferation in LnCAP-PSGR (Figure 5A) cells, but no significant changes in cell migration (Figure 5B), whereas LnCAP-KD (Figure 5C) cells showed decreased cell proliferation compared to control, and a stronger negative effect on cell migration (Figures 5D-E). PSGR over-expression did not affect cell invasion or colony formation (Figures 5F-G). Using PSGR over-expressing cells, we analyzed the expression of AR and AR-dependent genes, including PSA and NKX3.1. As with our findings in the mouse model (Figure 3C), there was a down-regulation of epithelial AR in PSGR over-expressing cells. AR function decreased as well, as evidenced by a decrease in PSA and NKX3.1 expression (Figure 6A). Because PSGR is normally expressed in prostate epithelial cells, we tested whether PSGR can regulate stromal AR expression through paracrine mechanisms, using an in vitro two-dimensional co-culture system. We cultured stromal WPMY-1 cells together with LnCAP-Ctrl or LnCAP-PSGR cells in the presence or absence of Raw264.7 cells, a mouse macrophage cell line, using a CoStar Transwell co-culture system. PSGR overexpression in tumor epithelial cells (i.e. LnCaP-PSGR) resulted in 5-fold higher PSGR levels in co-cultured stromal (WPMY-1) cells but did not affect WPMY-1 AR expression (Figures 6B-C). Addition of macrophages had no effect on PSGR expression in WPMY-1 cells; however, co-culture with both macrophages and LnCAP-PSGR cells profoundly increased PSGR expression in WPMY-1 cells, suggesting the existence of a paracrine mechanism between the PSGR-expressing epithelial and stromal cells that is aided by activated macrophages (Figure 6C). Importantly, AR expression increased (∼5-fold) in stromal cells exposed to PSGR-activated macrophages. This suggests that PSGR activation of macrophages in the microenvironment could play a role in regulating AR expression in stromal cells, possibly through NF-κB.
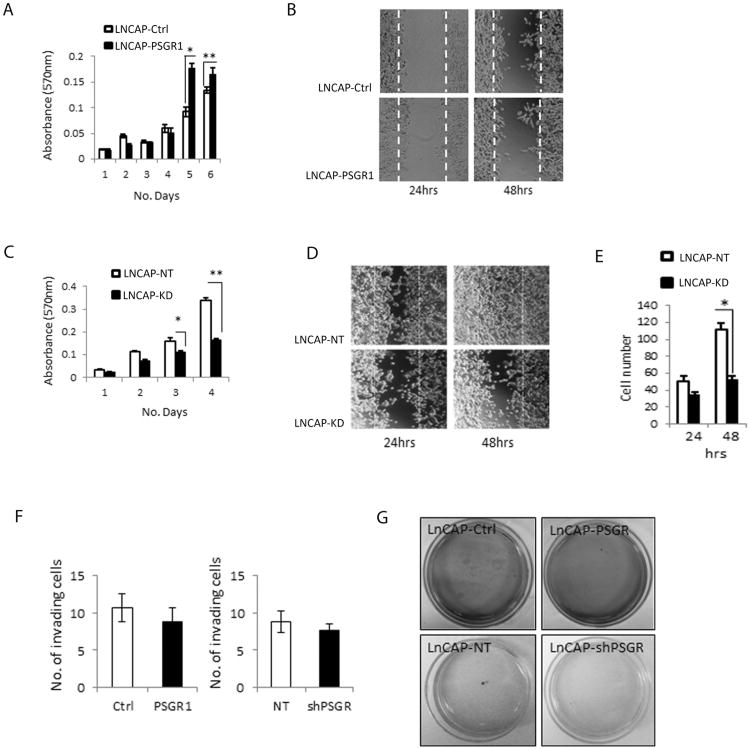
Figure 5. PSGR overexpression affects LnCaP oncogenic properties.
A PSGR over-expression in a PTEN inactivated cell line (LnCAP) induced cell proliferation (* p < 0.01, ** p < 0.05, t-test) n=6 per day. B. PSGR over-expression did not have significant effects on cell migration compared to controls (n=3) at 24 or 48 hours after scratch assay. C. PSGR knockdown decreased cell proliferation in LnCAP cells compared with non-targeted controls (* p < 0.05, ** p < 0.01, t-test) n=6 per day. D. PSGR knockdown decreased cell migration compared to non-targeted controls. E. Bar graph represents average number of migrating cells in LnCAP-KD versus LnCAP-NT (non-targeted controls) ± standard error (* p < 0.05, t-test). F. PSGR over-expression or knockdown did not significantly affect cell invasion. Bar graphs represent average number of invading cells ± standard error. G. PSGR over-expression or knockdown had no significant effect on LnCAP colony formation.
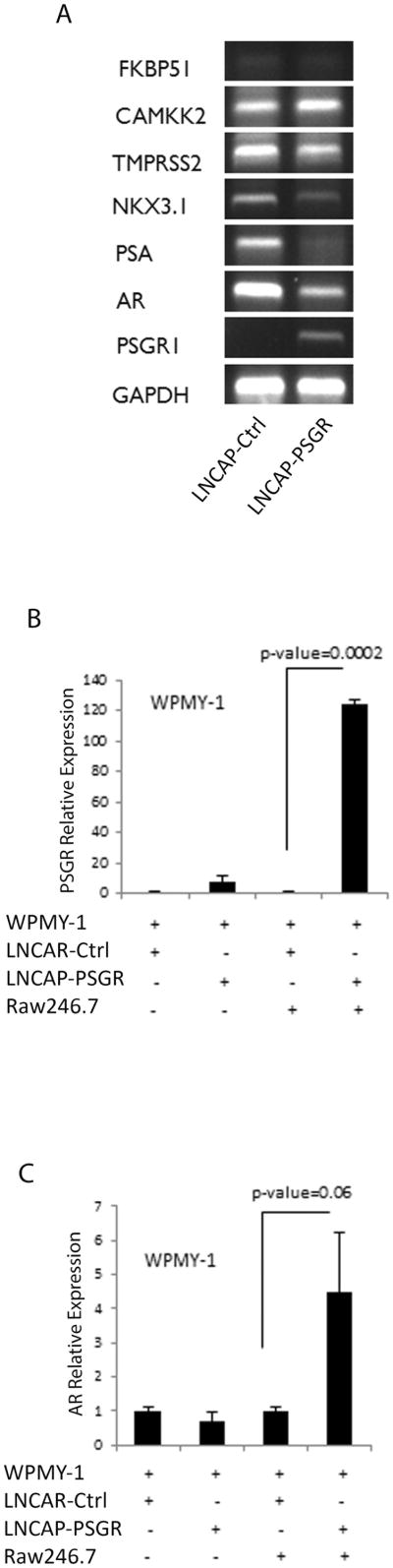
Figure 6. PSGR can regulate stromal androgen receptor (AR) in vitro in the presence of inflammatory cells.
A. Reverse transcription-PCR of AR signaling-related gene products shows decreased AR and target gene mRNAs (NKX3.1, PSA) following PSGR overexpression. N = 2. B. Co-culture experiments using the stromal cell line WPMY-1 with indicated additional cell lines show increased WPMY-1 PSGR mRNA expression in the presence of both PSGR-overexpressing LnCaP cells and the Raw264.7 macrophage cell line (p < 0.0002, t-test). Bar graph represents mean ± standard error of n=3 experiments, each done in triplicate. C. Co-culture with PSGR overexpressing LnCaP cells induced WPMY-1 cell AR mRNA expression only in the presence of Raw264.7 macrophages (p<0.06). Bar graph represents mean ± standard error of n=3 different experiments, each in triplicate.
Discussion
PSGR is overexpressed in ∼60-75% of tumor samples (12, 20). Prostate cancers frequently have other concomitant genetic mutations, such as PTEN, which is deleted in 70% of advanced prostate cancers (6). Inactivation of PTEN leads to further AKT activation, which alone is sufficient to establish prostate cancer (21). NF-κB activation plays a very important role in prostate cancer progression, and high levels of NF-κB activity are a predictor of poor outcome for patients with early forms of prostate cancer (23). Furthermore, NF-κB is also up-regulated in subsets of castration-resistant prostate cancer patients, highlighting its potential as a therapeutic target in both early and advanced prostate cancer (24). We sought to discover whether augmented AKT activity from Pten deletion would further increase NF-κB activity in PSGR over-expressing conditions and facilitate prostate cancer progression.
Using our bigenic mouse model (PSGR over-expression with conditional Pten deletion) we found that, indeed, PSGR-PtenΔ/Δ mice had faster tumor progression and developed larger tumors capable of compromising the prostatic architecture. Several major mechanisms were involved in the phenotype of these mice. First, there was a small increase in proliferation of PSGR over-expressing mice which was significantly enhanced in double mutant PSGR-PtenΔ/Δ mice, suggesting a potential synergistic mechanism. This was also seen in vitro, where over-expression of PSGR induced cell proliferation and PSGR knockdown had the opposite effect. Second, in accord with previous reports (25), PSGR regulated cell migration, as PSGR knockdown in LnCAP cells showed a significant decrease in migration. Curiously, in our mouse model, the loss of E-cadherin was significantly greater in the double mutant mice (PSGR-PtenΔ/Δ mice) when compared to either PSGR over-expressing or Pten deleted single mutant mice, suggesting a potential for increased invasion in PSGR-PtenΔ/Δ mice in vivo.
AR signaling is vital to prostate function. Stromal AR expression is required for prostate development [5]. Androgen binding and activation of AR within the stromal compartment sends proliferative cues to the epithelium, whereas activation of the AR in the epithelium drives cell differentiation and expression of secretory proteins (26). In addition to its role during prostate development, the androgen receptor plays a central role in prostate cancer (27). The development of human prostate cancer occurs in the presence of an AR positive stroma (28) and AR expression levels are directly related to advanced stages of prostate cancer (28). However, low expression of stromal AR in either tumor stroma or normal stroma was also correlated with shorter cancer-specific survival and more rapid progression to castration-resistant prostate cancer (29), suggesting the clinical significance of stromal AR is far from understood. Little is known about the role of androgen receptor in early or non-metastatic prostate cancer. It is believed that the key molecular event driving change in prostate cancer involves a transition from AR induced luminal cell differentiation to luminal cell proliferation (30), but the exact mechanisms are not yet understood. Our data show that PSGR over-expression decreases epithelial AR expression both in vitro and in vivo. Concomitantly, in vivo stromal AR expression is significantly increased, which could enable increased epithelial cell proliferation. Induction of cell proliferation could be further enhanced by activation of concomitant pathways. Our in vitro data shows increased cell proliferation in PSGR over-expressing cells despite down-regulation of AR, suggesting a reduced AR-dependent control on luminal cell proliferation. PSGR over-expression can itself activate NF-κB (17), which can also induce prostate epithelial cell proliferation (31).
We observed increased stromal AR expression concomitant to PSGR over-expression and Pten deletion, suggesting that up-regulation of NF-κB activity as a consequence of PSGR over-expression may regulate AR expression levels in the stroma. Re-emergence of AR expression is a hallmark of castration resistant prostate cancer (6). As stromal AR drives epithelial cell proliferation (28), in the case of PSGR-PtenΔ/Δ mice it may represent a potential mechanism for progression to advanced prostate cancer. The role of stromal AR in advanced prostate cancer is still controversial (32-34), although our work suggests that in these genetic conditions, stromal AR may act as an enhancer of tumor progression.
Inflammation plays a prominent role in prostate cancer development (1, 35, 36), and we have previously reported an inflammatory response in mice transgenically expressing PSGR, with accompanying elevation in NF-κB (17). The relationship between AR and NF-κB signaling is complex; while AR is a downstream target gene of NF-κB, the consequences of NF-κB activation on AR expression are believed to be determined by tissue-specific expression of cofactors. An inverse correlation between NF-κB activity and AR status has been reported in prostate epithelial cells (31). The opposite is also true, where NF-κB was shown to transcriptionally modulate AR expression in prostate epithelial cells (37). The underlying mechanisms behind these signaling differences remains to be determined. Here, we found that PSGR over-expression moderately increased inflammatory cell infiltration. When Pten was completely deleted, but PSGR levels were unchanged (PtenΔ/Δ), NF-κB levels were undetectable, either in the epithelia or the stroma. PSGR-PtenΔ/Δ mice, however, had increased NF-κB positive stromal inflammatory cell infiltration, predominantly of CD3+ T cells (Supplementary Figure 2), which was correlated with stromal AR expression. We confirmed the role of stromal inflammatory cells on stromal AR expression using a co-culture system where LnCAP-Ctrl or LnCAP-PSGR cells were grown with stromal WPMY-1 cells in the presence or absence of Raw264.7 macrophages. Only in the presence of both inflammatory cells and PSGR-overexpressing cancer cells was there a significant (five-fold) increase of stromal AR, accompanied by a further increase in PSGR expression, suggesting an intercellular crosstalk. If replicated in vivo, a requirement for both PSGR-overexpresing prostate epithelium and paracrine factors from infiltrating inflammatory cells would suggest a more complex, indirect relationship between NF-κB elevation and stromal AR regulation than the direct regulatory models cited above. Further experiments are needed to understand the potential relationship between PSGR-NFκB-AR and whether this is a targetable new pathway that would potentially limit the re-emergence of AR in advanced prostate cancer, thus inhibiting progression to castration resistant prostate cancer.
Our findings also hint at a cooperative role between elevated PSGR expression and PTEN loss in human prostate cancer development. We have previously reported that PSGR expression remains at normal levels in benign prostate hyperplasia tissue but is frequently elevated in PIN lesions and remains elevated in the majority of primary and metastatic tumors (12). In contrast, PTEN expression has been reported to be elevated in PIN lesions, with heterogeneous expression in primary prostate carcinomas where frequency of PTEN loss correlated with increasing tumor stage and grade (21, 38-41). From these observations we speculate that PSGR overexpression may be an earlier event in human prostate cancer progression, occurring during the benign prostate hyperplasia-PIN transition and instigating an inflammatory response that places pressure on the PIN cells to select for PTEN loss. However, this speculation remains to be tested experimentally.
In conclusion, we present a novel mouse model of prostate cancer development. By combining a prostate-specific loss of Pten with transgenic overexpression of PSGR, we generated tumors with high penetrance that exhibit a robust inflammatory response and re-emergence of stromal AR expression. Furthermore, our work shows how these two common human prostate cancer lesions result in hyperactivation of NF-κB and drive further tumor progression through several possible mechanisms, potentially explaining the poorer prognosis observed in human patients with hyperactivation of these pathways. Finally, our data support further research investigating PSGR as a potential therapeutic target in prostate cancer.
Materials and Methods
Generation of PSGR/PTEN mouse line
All animal experiments were performed in accordance with an animal use protocol approved by the IBT Institutional Animal Care and Use Committee. Mouse lines were created by crossing PSGR transgenic FVB mice with floxed Pten-Cre transgenic FVB mice (The Jackson Laboratory) to produce PSGR/Ptenf/w-Cre+ mice (PSGR-PtenΔ/+) under an ARR2PB promoter. PSGR-PteΔ/+ mice were then intercrossed to obtain six different genotypes: Ptenf/f-Cre- (WT), PSGR-Ptenf/f-Cre- (PSGR-PtenWT), Ptenf/w-Cre+ (PtenΔ/+), PSGR-Ptenf/w-Cre+ (PSGR-PtenΔ/+), Ptenf/f-Cre+ (PtenΔ/Δ), PSGR-Ptenf/f-Cre+ (PSGR-PtenΔ/Δ). Male animals were analyzed at 6 and 12 months of age. Littermates were compared when possible; when not possible, controls were assigned on the basis of similar age and genetic background. Sample sizes were selected based on our experience of inter-mouse variability in the PSGR transgenic line. Exclusion criteria included non-experimental morbidity (i.e. severe fight wounds or sickness other than cancer).
Tissue handling and histology
Prostate tissue was extracted from the surrounding genitourinary system under the microscope and fixed in zinc formalin for 7 hours (Richard-Allan Scientific). Samples were then dehydrated in graded ethanols (70, 85, 95 and 100%), embedded in paraffin, and cut into 5 μm sections.
Immunohistochemistry
Samples were processed and stained according to the manufacturer's instructions (Vectastain ABC Kit). Antibodies include: PTEN (Cell Signaling Technology (CST #9559 - 1:100)), pAKT(S473) #4060 (CST 1:200), Ki67 (1:500, Novus Biochemicals #NB110-89719), α-SMA (Sigma-Aldrich #A7607, 1:1000), E-cadherin (CST #3195 1:200), AR (Santa Cruz Biotechnology (SCBT) #N-20, 1:400), p65 (CST #6956 1:200). TUNEL assay performed using DeadEnd Fluorimetric TUNEL Assay (Promega, #G3250). Photomicrographs were taken using a Nikon Eclipse 80i microscope and acquired using NIS-Elements BR 3.2. Investigators were not blinded during immunohistochemical analysis.
Real-time PCR
A total of 3μg of RNA was used to generate cDNA for PCR reactions. PSGR and AR expression levels were detected using the following primers: rt-OR51E2-F1: (5′-TTCCTGCAACTTCACACATGCCAC-3′), rt-OR51E2-R1: (5′-TGGATGTGGATAAGGCCA GGTCAA-3′) and AR-F: (5′-GGATAGCTACTCCGGACCTTAC-3′), AR-R: 5′-GGGTGG AAAGTAATAGTCAATGGG-3′).
Cell lines
LnCaP cells were obtained from American Type Culture Collection (Manassas, VA). LnCAP stable cell lines (LnCAP-Ctrl, LnCAP-PSGR, LnCAP-NT and LnCAP-shPSGR1.1) were grown in RPMI 1640 media (Hyclone) with 10% FBS (Atlanta Biologicals). For over-expression, hPSGR was cloned into a pLVX-zsGreen plasmid (Clontech) and transfected into 293FT (Invitrogen) cells together with packaging plasmids pMD2.G and psPAX2 (Addgene). Media were harvested 24 hours later and then used to infect LnCAP cells. For silencing PSGR, hPSGR specific short hairpin RNA sequences were cloned into a pLL3.7 plasmid (Addgene) and the same transfection/infection procedure was used as for over-expression. Two different sequences were used: #1 F: (TGGCCTGTCTTACCCAGATGTTttcaagagaAACATCTGGGTAAGACAGGCCTTTTTTC); R:(TCGAGAAAAAAGGCCTGTCT TACCCAGATGTTtctcttgaaAACATCTGGGTAAGACAGGCCA); and #2 F: (TGTTGCCCATA AGCACATCAGTttcaagagaACTGATGTGCTTATGGGCAACTTTTTTC); R: (TCGAGAAAAAATTGCCCATAAGCACATCAGTtctcttgaaACTGATGTGCTTATGGGCAA CA). WPMY-1 prostate stromal cells were purchased from ATCC. Raw 264.7 cells were a gift from the Dekai Zhang lab. WPMY-1 and Raw264 cells were grown in DMEM media (Hyclone) with 10% FBS. Cells were co-cultured in Costar Transwell permeable chambers with 0.4 μm pores (24 and 96 wells) (Corning Incorporated).
Cell transformation assays
Cell proliferation was measured by MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide) analysis (Sigma). A total of 1000 cells per well were seeded in 96 wells and measured for optical density every day for six days. For cell migration, 5000 cells were seeded in 6-well plates (in triplicate) and grown until full confluence. Cells were then treated with 40 μM Mitomycin C (EMD Millipore, Billerica, MA) for 2 hours and scratched. Migration was analyzed after 24 and 48 hours. Cell invasion was performed using a Boyden invasion chamber, seeding cells (2000 cells per well) with low-nutrient Matrigel (BD Biosciences) to stimulate cell invasion. Colony formation was performed using agar plates (Noble Agar, Sigma-Aldrich) with cells seeded at a density of 5000 cells per plate, and left to grow for 30 days. Cell media was changed every 48 hours.
Statistical Analysis
Data analysis is shown as mean with standard error. Statistical differences between two groups were compared using two-tailed Welch's t-test due to unequal variances, as indicated by Fisher's F-test analysis of intra-group variability. P-values and correlation data were obtained using R statistical package software. A probability of p < 0.05 was considered statistically significant.
Supplementary Material
Figure S1. PSGR over-expression in human prostate cancers. A Boxplots showing mRNA expression data of PSGR in normal versus prostate cancer microarray datasets in Oncomine. Grasso et al, 5.098-fold change (p = 6.69E-11); LaPointe et al, 2.689-fold change (p = 1.21E-6); Liu et al, 2.225-fold change (p = 2.28E-4); and Vanaja et al, 3.151-fold change (p = 1.62E-4) Cutoff = 1.5-fold change. B. Oncomine database analysis from indicated prostate cDNA microarrays shows prostate tumor tissues overexpressing PSGR; no significant change in PTEN mRNA levels was detected in these datasets. Red lines show basal expression levels in adjacent normal tissue as control. C. Genotyping results shown for all analyzed genotypes of PSGR-PTEN mice.
Figure S2. PSGR-PTENΔ/Δ mice show increased inflammatory cell infiltration into the stroma. A. CD3 positive T cells were significantly increased in PSGR-PTENΔ/Δ mice at 12 months of age compared to other time intervals (p < 0.05). Arrows indicate positive staining cells. Bar graph represents average positive number of cells ± standard error (n = 4). B. Macrophages infiltrate prostate tissues upon complete Pten deletion. The presence of PSGR over-expression further increased the number of infiltrating macrophages (p < 0.05).
Figure S3. PSGR knockdown efficiency in LnCAP cells A Bar graph represents lentiviral PSGR knockdown efficiency in LnCAP cells as determined by real-time PCR.
Acknowledgments
We thank Dr. Michael Ittmann (Baylor College of Medicine) for his insights and suggestions. We thank Dr. Fen Wang (Texas A&M Health Science Center) for the gift of the PTENfl/fl Cre mice and for valuable discussions and suggestions, and Dr. Dekai Zhang (Texas A&M Health Science Center) for the gift of the RAW264.7 cells. This work was partially supported by grants from the State Key Development Programs of China (2012CB910400), DOD Prostate Cancer Research Program (PCRP, W81XWH-10-1-0612, W81XWH-10-10147), and NIH (5R01CA134731).
Footnotes
The authors disclose no potential conflicts of interest.
Author Contributions: Melissa Rodriguez: Experimental design, data collection, data analysis, manuscript writing, Stefan Siwko: Experimental design, data analysis, manuscript writing, Li Zeng: Data collection and analysis, Jinsong Li: Data collection and analysis, Zhengfang Yi: Data collection and analysis, Mingyao Liu: Overall direction, experimental design and analysis, final approval of completed manuscript
Financial Conflict of Interest: The authors declare no conflicts of interest.
References
- 1.Giles GG. Epidemiological investigation of prostate cancer. Methods in molecular medicine. 2003;81:1–19. doi: 10.1385/1-59259-372-0:1. [DOI] [PubMed] [Google Scholar]
- 2.Nelson WG, De Marzo AM, Isaacs WB. Prostate cancer. The New England journal of medicine. 2003;349(4):366–81. doi: 10.1056/NEJMra021562. [DOI] [PubMed] [Google Scholar]
- 3.Yoshimoto M, Cunha IW, Coudry RA, Fonseca FP, Torres CH, Soares FA, et al. FISH analysis of 107 prostate cancers shows that PTEN genomic deletion is associated with poor clinical outcome. British journal of cancer. 2007;97(5):678–85. doi: 10.1038/sj.bjc.6603924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoshimoto M, Joshua AM, Cunha IW, Coudry RA, Fonseca FP, Ludkovski O, et al. Absence of TMPRSS2:ERG fusions and PTEN losses in prostate cancer is associated with a favorable outcome. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2008;21(12):1451–60. doi: 10.1038/modpathol.2008.96. [DOI] [PubMed] [Google Scholar]
- 5.Cairns P, Okami K, Halachmi S, Halachmi N, Esteller M, Herman JG, et al. Frequent inactivation of PTEN/MMAC1 in primary prostate cancer. Cancer research. 1997;57(22):4997–5000. [PubMed] [Google Scholar]
- 6.Shen MM, Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes & development. 2010;24(18):1967–2000. doi: 10.1101/gad.1965810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen MM, Abate-Shen C. Pten inactivation and the emergence of androgen-independent prostate cancer. Cancer research. 2007;67(14):6535–8. doi: 10.1158/0008-5472.CAN-07-1271. [DOI] [PubMed] [Google Scholar]
- 8.Trotman LC, Niki M, Dotan ZA, Koutcher JA, Di Cristofano A, Xiao A, et al. Pten dose dictates cancer progression in the prostate. PLoS biology. 2003;1(3):E59. doi: 10.1371/journal.pbio.0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee JT, Lehmann BD, Terrian DM, Chappell WH, Stivala F, Libra M, et al. Targeting prostate cancer based on signal transduction and cell cycle pathways. Cell cycle. 2008;7(12):1745–62. doi: 10.4161/cc.7.12.6166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carver BS, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nature genetics. 2009;41(5):619–24. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436(7051):725–30. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weng J, Wang J, Cai Y, Stafford LJ, Mitchell D, Ittmann M, et al. Increased expression of prostate-specific G-protein-coupled receptor in human prostate intraepithelial neoplasia and prostate cancers. International journal of cancer Journal international du cancer. 2005;113(5):811–8. doi: 10.1002/ijc.20635. [DOI] [PubMed] [Google Scholar]
- 13.Xia C, Ma W, Wang F, Hua S, Liu M. Identification of a prostate-specific G-protein coupled receptor in prostate cancer. Oncogene. 2001;20(41):5903–7. doi: 10.1038/sj.onc.1204803. [DOI] [PubMed] [Google Scholar]
- 14.Xu LL, Stackhouse BG, Florence K, Zhang W, Shanmugam N, Sesterhenn IA, et al. PSGR, a novel prostate-specific gene with homology to a G protein-coupled receptor, is overexpressed in prostate cancer. Cancer research. 2000;60(23):6568–72. [PubMed] [Google Scholar]
- 15.Xu LL, Sun C, Petrovics G, Makarem M, Furusato B, Zhang W, et al. Quantitative expression profile of PSGR in prostate cancer. Prostate cancer and prostatic diseases. 2006;9(1):56–61. doi: 10.1038/sj.pcan.4500836. [DOI] [PubMed] [Google Scholar]
- 16.Weng J, Ma W, Mitchell D, Zhang J, Liu M. Regulation of human prostate-specific G-protein coupled receptor, PSGR, by two distinct promoters and growth factors. Journal of cellular biochemistry. 2005;96(5):1034–48. doi: 10.1002/jcb.20600. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez M, Luo W, Weng J, Zeng L, Yi Z, Siwko S, et al. PSGR promotes prostatic intraepithelial neoplasia and prostate cancer xenograft growth through NF-kappaB. Oncogenesis. 2014;3:e114. doi: 10.1038/oncsis.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2(5):401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Weng J, Cai Y, Penland R, Liu M, Ittmann M. The prostate-specific G-protein coupled receptors PSGR and PSGR2 are prostate cancer biomarkers that are complementary to alpha-methylacyl-CoA racemase. The Prostate. 2006;66(8):847–57. doi: 10.1002/pros.20389. [DOI] [PubMed] [Google Scholar]
- 21.Wang S, Gao J, Lei Q, Rozengurt N, Pritchard C, Jiao J, et al. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer cell. 2003;4(3):209–21. doi: 10.1016/s1535-6108(03)00215-0. [DOI] [PubMed] [Google Scholar]
- 22.Russell PJ, Kingsley EA. Human prostate cancer cell lines. Methods in molecular medicine. 2003;81:21–39. doi: 10.1385/1-59259-372-0:21. [DOI] [PubMed] [Google Scholar]
- 23.MacKenzie L, McCall P, Hatziieremia S, Catlow J, Adams C, McArdle P, et al. Nuclear factor kappaB predicts poor outcome in patients with hormone-naive prostate cancer with high nuclear androgen receptor. Human pathology. 2012;43(9):1491–500. doi: 10.1016/j.humpath.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 24.McCall P, Bennett L, Ahmad I, Mackenzie LM, Forbes IW, Leung HY, et al. NFkappaB signalling is upregulated in a subset of castrate-resistant prostate cancer patients and correlates with disease progression. British journal of cancer. 2012;107(9):1554–63. doi: 10.1038/bjc.2012.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanz G, Leray I, Dewaele A, Sobilo J, Lerondel S, Bouet S, et al. Promotion of cancer cell invasiveness and metastasis emergence caused by olfactory receptor stimulation. PloS one. 2014;9(1):e85110. doi: 10.1371/journal.pone.0085110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niu Y, Altuwaijri S, Yeh S, Lai KP, Yu S, Chuang KH, et al. Targeting the stromal androgen receptor in primary prostate tumors at earlier stages. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(34):12188–93. doi: 10.1073/pnas.0804701105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roy AK, Lavrovsky Y, Song CS, Chen S, Jung MH, Velu NK, et al. Regulation of androgen action. Vitamins and hormones. 1999;55:309–52. doi: 10.1016/s0083-6729(08)60938-3. [DOI] [PubMed] [Google Scholar]
- 28.Cano P, Godoy A, Escamilla R, Dhir R, Onate SA. Stromal-epithelial cell interactions and androgen receptor-coregulator recruitment is altered in the tissue microenvironment of prostate cancer. Cancer research. 2007;67(2):511–9. doi: 10.1158/0008-5472.CAN-06-1478. [DOI] [PubMed] [Google Scholar]
- 29.Wikstrom P, Marusic J, Stattin P, Bergh A. Low stroma androgen receptor level in normal and tumor prostate tissue is related to poor outcome in prostate cancer patients. The Prostate. 2009;69(8):799–809. doi: 10.1002/pros.20927. [DOI] [PubMed] [Google Scholar]
- 30.Zhou Y, Bolton EC, Jones JO. Androgens and androgen receptor signaling in prostate tumorigenesis. Journal of molecular endocrinology. 2015;54(1):R15–R29. doi: 10.1530/JME-14-0203. [DOI] [PubMed] [Google Scholar]
- 31.Suh J, Rabson AB. NF-kappaB activation in human prostate cancer: important mediator or epiphenomenon? Journal of cellular biochemistry. 2004;91(1):100–17. doi: 10.1002/jcb.10729. [DOI] [PubMed] [Google Scholar]
- 32.Cunha GR, Hayward SW, Wang YZ, Ricke WA. Role of the stromal microenvironment in carcinogenesis of the prostate. International journal of cancer Journal international du cancer. 2003;107(1):1–10. doi: 10.1002/ijc.11335. [DOI] [PubMed] [Google Scholar]
- 33.Niu Y, Altuwaijri S, Lai KP, Wu CT, Ricke WA, Messing EM, et al. Androgen receptor is a tumor suppressor and proliferator in prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(34):12182–7. doi: 10.1073/pnas.0804700105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lai KP, Yamashita S, Huang CK, Yeh S, Chang C. Loss of stromal androgen receptor leads to suppressed prostate tumourigenesis via modulation of pro-inflammatory cytokines/chemokines. EMBO molecular medicine. 2012;4(8):791–807. doi: 10.1002/emmm.201101140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gurel B, Lucia MS, Thompson IM, Jr, Goodman PJ, Tangen CM, Kristal AR, et al. Chronic inflammation in benign prostate tissue is associated with high-grade prostate cancer in the placebo arm of the prostate cancer prevention trial. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2014;23(5):847–56. doi: 10.1158/1055-9965.EPI-13-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwon OJ, Zhang L, Ittmann MM, Xin L. Prostatic inflammation enhances basal-to-luminal differentiation and accelerates initiation of prostate cancer with a basal cell origin. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(5):E592–600. doi: 10.1073/pnas.1318157111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang L, Altuwaijri S, Deng F, Chen L, Lal P, Bhanot UK, et al. NF-kappaB regulates androgen receptor expression and prostate cancer growth. The American journal of pathology. 2009;175(2):489–99. doi: 10.2353/ajpath.2009.080727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fenic I, Franke F, Failing K, Steger K, Woenckhaus J. Expression of PTEN in malignant and non-malignant human prostate tissues: comparison with p27 protein expression. The Journal of pathology. 2004;203(1):559–66. doi: 10.1002/path.1545. [DOI] [PubMed] [Google Scholar]
- 39.Koksal IT, Dirice E, Yasar D, Sanlioglu AD, Ciftcioglu A, Gulkesen KH, et al. The assessment of PTEN tumor suppressor gene in combination with Gleason scoring and serum PSA to evaluate progression of prostate carcinoma. Urologic oncology. 2004;22(4):307–12. doi: 10.1016/j.urolonc.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 40.McMenamin ME, Soung P, Perera S, Kaplan I, Loda M, Sellers WR. Loss of PTEN expression in paraffin-embedded primary prostate cancer correlates with high Gleason score and advanced stage. Cancer research. 1999;59(17):4291–6. [PubMed] [Google Scholar]
- 41.Dahia PL. PTEN, a unique tumor suppressor gene. Endocrine-related cancer. 2000;7(2):115–29. doi: 10.1677/erc.0.0070115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. PSGR over-expression in human prostate cancers. A Boxplots showing mRNA expression data of PSGR in normal versus prostate cancer microarray datasets in Oncomine. Grasso et al, 5.098-fold change (p = 6.69E-11); LaPointe et al, 2.689-fold change (p = 1.21E-6); Liu et al, 2.225-fold change (p = 2.28E-4); and Vanaja et al, 3.151-fold change (p = 1.62E-4) Cutoff = 1.5-fold change. B. Oncomine database analysis from indicated prostate cDNA microarrays shows prostate tumor tissues overexpressing PSGR; no significant change in PTEN mRNA levels was detected in these datasets. Red lines show basal expression levels in adjacent normal tissue as control. C. Genotyping results shown for all analyzed genotypes of PSGR-PTEN mice.
Figure S2. PSGR-PTENΔ/Δ mice show increased inflammatory cell infiltration into the stroma. A. CD3 positive T cells were significantly increased in PSGR-PTENΔ/Δ mice at 12 months of age compared to other time intervals (p < 0.05). Arrows indicate positive staining cells. Bar graph represents average positive number of cells ± standard error (n = 4). B. Macrophages infiltrate prostate tissues upon complete Pten deletion. The presence of PSGR over-expression further increased the number of infiltrating macrophages (p < 0.05).
Figure S3. PSGR knockdown efficiency in LnCAP cells A Bar graph represents lentiviral PSGR knockdown efficiency in LnCAP cells as determined by real-time PCR.