Abstract
Computational prediction of nucleic acid binding sites in proteins are necessary to disentangle functional mechanisms in most biological processes and to explore the binding mechanisms. Several strategies have been proposed, but the state-of-the-art approaches display a great diversity in i) the definition of nucleic acid binding sites; ii) the training and test datasets; iii) the algorithmic methods for the prediction strategies; iv) the performance measures and v) the distribution and availability of the prediction programs. Here we report a large-scale assessment of 19 web servers and 3 stand-alone programs on 41 datasets including more than 5000 proteins derived from 3D structures of protein-nucleic acid complexes. Well-defined binary assessment criteria (specificity, sensitivity, precision, accuracy…) are applied. We found that i) the tools have been greatly improved over the years; ii) some of the approaches suffer from theoretical defects and there is still room for sorting out the essential mechanisms of binding; iii) RNA binding and DNA binding appear to follow similar driving forces and iv) dataset bias may exist in some methods.
Author Summary
Nucleic acid binding sites in proteins are functionally important in a majority of biological processes. Computationally predicting these binding sites can help the biological community in understanding the nucleic acid binding proteins in the very first step. The emergence of nucleic acid binding site prediction programs and web servers during the last decade shows a great diversity in various aspects. Besides, some binding site prediction related questions, such as i) can RNA binding sites be distinguished from DNA binding sites? ii) can RNA and DNA binding sites be predicted in the same model?, have not been fully answered. Here, we benchmarked 19 web servers and 3 stand-alone programs on 41 previously reported data sets and analyzed the prediction results in different aspects to show a more complete view of how well these programs can perform and how they can be used. We hope to demonstrate some key points for unbiased comparison for further development of similar prediction programs.
Introduction
Protein-nucleic acid (RNA/DNA) bindings play crucial roles in most biological processes[1] and the detection of the functional sites/regions in proteins is an important step for structurally understanding the molecular mechanism of the biological processes. Compared with the vast number of protein-nucleic acid interactions in bio-systems (Supplementary Note 1 in S1 Text), the experimental determination of binding sites is always difficult, demanding and not always readily feasible. Hence, computational prediction of nucleic acid binding sites has been an established field in computational and molecular biology over the past two decades.
The prediction approaches are diverse in many aspects, which results in controversies over technical details and renders difficult to make totally fair comparisons[2]. Further, previous reviews[3–6] only assessed at small scale the available datasets. Currently, RNA- and DNA-binding residue predictions are always treated as different problems, or trained with different data sets within the same model[7–11]. However, whether RNA- and DNA-binding proteins (Supplementary Note 2 in S1 Text) exploit different driving forces is not established and it is known that some proteins do bind both types of nucleic acids. Very recently, Yan and coworkers noticed also that prediction programs are unable to distinguish between DNA and RNA binding proteins and concluded that one should compare RNA- and DNA-binding site predictors together[6]. The definition of a nucleic acid binding residue is not standardized with definitions ranging from distance cutoffs[8,9,12–15] to the enumeration of non-covalent contacts[16–19](Supplementary Note 3 in S1 Text). This leads to ambiguities goal in the problem and variations in prediction accuracy. Besides, tens of training and test sets of variable sizes have now been curated by developers during the development of computational approaches. How to avoid bias in a dataset is nontrivial. Further, the assessment criteria are still arguable, e.g. whether all residues from different proteins should be compared together (Supplementary Note 4 in S1 Text). In addition, programs differ in their approaches (Supplementary Note 5 in S1 Text) making a fair assessment difficult. Finally, the distribution and ease-of-use of the programs greatly determine their help to the users in the biological community.
In this report, we present a large-scale assessment of 19 currently available web servers and 3 stand-alone prediction programs in nucleic acid binding site prediction, which is 24 predictors in total, on 41 different datasets derived from structures of protein-nucleic acid complexes in the PDB, including more than 5000 proteins. We use a hierarchical definition of binding sites and various assessment criteria for reference. We analyze differences i) between RNA binding site prediction and DNA binding sites predictions; ii) between binary prediction and continuous scores; iii) between sequence-based prediction and structure-based ones; and finally iv) between original and updated programs. The large-scale analysis should be helpful to developers and users.
Results
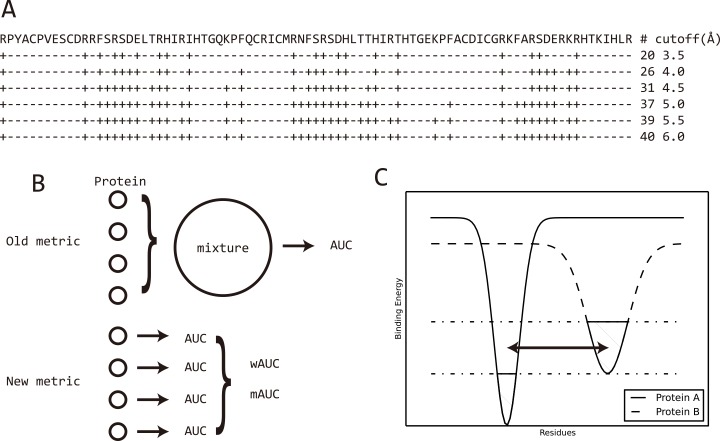
The prediction of nucleic acid binding sites is usually determined by three main factors: the definition of a binding site, the assessment criteria and the datasets. Currently, there is no universal definition of a binding site and a minimum distance cutoff between interacting residues is most frequently applied. However, different distance cutoffs lead to accuracy variations while a single cutoff biases certain prediction programs. In Fig 1A is displayed a real-world case where a distance cutoff of 6.0Å leads to a two times higher number of binding sites than that obtained with a cutoff of 3.5Å. S1 Table and S12 Fig show that this difference is a general distribution rather than a rare case. And, in S2 Table, the data show that with a cutoff of 3.5Å used for prediction and 6Å for the definition of the binding sites, the final specificity is 100% but the sensitivity is as low as 51–62% (high false negative rate) with a total accuracy (ACC) decrease down to ~90% (details are discussed in Supplementary Note 6 in S1 Text). To fully capture the accuracy variance resulting from a distance cutoff, a hierarchical definition with distance cutoffs ranging from 3.5 to 6Å using 0.5Å as step was used and the distributions of the accuracies were plotted. Besides, the prediction accuracy highly depends on the dataset used for testing. Normally, the largest the training set the better the prediction model, thus making it difficult to compare different programs. However, a good prediction program should show stable accuracy on all the datasets, a feature that cannot be achieved by biased predictions even with a large training set (Supplementary Note 7 in S1 Text). We used 41 datasets to accentuate the possibility of biased prediction by the programs. Finally, different criteria are measured to show different aspects of the programs. The webservers assessed include BindN[8], BindN+[9], RNABindR[20], RNABindRPlus[21], DBS-Pred[12], DBS-PSSM[13], KYG[14], PRBR[22], PPRInt[23], DNABINDPROT[24], ProteDNA[25], DISPLAR[10], DR_bind1[26], aaRNA[27], RBscore[28], RBRDetector[29], DNABind[30], xypan[31] and RNAProSite (lilab.ecust.edu.cn/NABind/), while the programs are Predict_RBP[17], PRNA[32] and RBRIdent[33]. Previously reported prediction approaches have been summarized in Table 1. Slow programs DR_bind1 and RBRDetector were only tested on part of the datasets and the results are provided in the supplementary information. metaDBSite[34] shows same result as BindN and was not tested explicitly.
Fig 1. Binding site definition and assessment metrics can result in accuracy variation.
A) Binding site definition of Zif268 protein based on different distance cutoffs. ‘+’ marks the binding sites while ‘-’ marks non-binding sites. With a distance cutoff of 6.0Å, 40 residues are defined as binding sites, which is twice that obtained with a cutoff of 3.5Å; B) Two metrics to measure prediction accuracy in terms of AUC. Old metric mix all the residues from all the proteins together for comparison, then measure AUC on the mixed data. Metric in this work measures AUC for each protein and average the AUC values considering protein length. C) A scheme to illustrate the irrelevant comparison between binding sites of a protein and the non-binding sites on another protein. As protein A and protein B may have different size of nucleic acid binding region and binding affinity they are possible to have different energy funnels. The dashed region shows the binding region of the two proteins. Binary assessment, which mixes all residues together, will certainly include comparison between non-binding sites of protein A and binding sites of protein B, shown by the double arrowed line.
Table 1. Summary of the existing approaches in nucleic acid binding site prediction.
| RNA | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sequence-based | Brief/Features | ||||||||||||||||
| Year | Name | Server | Program | Website | Binding Site Definition | PSSM | RP | ASA | HP | SS | EC | Q | SA | Training | window | Dataset | reference |
| 2006 | BindN | ✓ | http://bioinfo.ggc.org/bindn/ | 3.5Å | ✓ | ✓ | SVM | 11 | R107(PRINR25),D62(PDNA-62) | [8] | |||||||
| 2007 | RNABindR | ✓ | http://einstein.cs.iastate.edu/RNABindR/ | 5Å | ✓ | NB | 25 | R147 | [20] | ||||||||
| 2008 | PPRInt | ✓ | http://www.imtech.res.in/raghava/pprint/ | 6Å | ✓ | ✓ | SVM | 17 | R107 from BindN, R86 from PPRInt | [23] | |||||||
| 2008 | RNAproB | 6Å/3.5Å | ✓ | SVM | 25 | R107 from BindN, R86 from PPRInt, R109 from RNABindR | [35] | ||||||||||
| 2008 | PRINTR | ✓ | http://210.42.106.80/printr/ | ENTANGLE | ✓ | SVM | 15 | R109 from RNABindR | [16] | ||||||||
| 2008 | RISP | ✓ | http://grc.seu.edu.cn/RISP | 3.5Å | ✓ | SVM | 7 | R147 from RNABindR, R71(PRNA-71,NA) | [36] | ||||||||
| 2009 | PiRaNhA | ✓ | http://www.bioinformatics.sussex.ac.uk/PIRANHA | 3.9A | ✓ | ✓ | ✓ | ✓ | SVM | 23 | R81 from PPRint,R42(NA) | [37] | |||||
| 2010 | BindN+ | ✓ | http://bioinfo.ggc.org/bindn+/ | 3.5Å | ✓ | ✓ | ✓ | SVM | 11 | R107(PRINR25),D62(PDNA-62) | [9] | ||||||
| 2010 | NAPS | ✓ | http://proteomics.bioengr.uic.edu/NAPS/ | 4.5Å | ✓ | ✓ | DT | 7 | R109 from RNABindR, D84 from Pro-dna, D274 from DISIS, D62(PDNA-62) | [7] | |||||||
| 2010 | PRBR | ✓ | http://www.cbi.seu.edu.cn/PRBR/optional.php | 3.5Å | ✓ | ✓ | RF | 11 | R180 (RBP-180) | [22] | |||||||
| 2011 | Sungwook | H-bond | ✓ | ✓ | ✓ | SVM | 9 | R3149(PRI3149),R727(PRI727),R267(PRI267) | [38] | ||||||||
| 2011 | SRCPred | ✓ | http://tardis.nibio.go.jp/netasa/srcpred/ | 3.5Å | ✓ | NN | 5 | R160(PRNA160) | [39] | ||||||||
| 2011 | Predict_RBP | ✓ | cic.scu.edu.cn/bioinformatics/Predict_RBP.rar | ENTANGLE | ✓ | ✓ | ✓ | SVM | 15 | R107 from BindN, R86 from PPRInt, R109 from RNABindR | [17] | ||||||
| 2011 | meta2 | ✓ | https://genesilico.pl/meta2 | 3.5Å | Meta-server | R44, R38 from OPRA, R180 from PRBR, R111 from RNABindR, R81 from PiRaNha, R86 from KYG | [4] | ||||||||||
| 2012 | Qian-Zhong | 6Å/3.5Å/ENTANGLE | ✓ | ✓ | SVM | 25 | R107 from BindN, R86 from PPRInt, R109 from RNABindR | [40]. | |||||||||
| 2014 | RNABindRPlus | ✓ | http://einstein.cs.iastate.edu/RNABindRPlus/ | 5Å | ✓ | SVM | 21 | R28,R44,R111,R198 | [21] | ||||||||
| 2015 | RBRIdent | ✓ | http://166.111.152.91/RBRIdent | ENTANGLE | ✓ | ✓ | ✓ | RF | 9 | R281 | [41] | ||||||
| Structure-based | |||||||||||||||||
| 2006 | KYG | ✓ | http://cib.cf.ocha.ac.jp/KYG/ | 7Å | ✓ | ✓ | Function | R86 | [14] | ||||||||
| 2008 | RsiteDB | ✓ | http://bioinfo3d.cs.tau.ac.il/RsiteDB/ | 7Å | ✓ | Clustering | [15] | ||||||||||
| 2008 | DR_bind1 | ✓ | http://drbind.limlab.ibms.sinica.edutw/ | HBPLUS | ✓ | ✓ | ✓ | Function | D56 from Susan 2003, D69, R81 | [42] | |||||||
| 2009 | PRIP | ✓ | http://www.qfab.org/PRIP | 5Å | ✓ | ✓ | SVM | 19 | R147(R144) and R109 from RNABindR | [43] | |||||||
| 2010 | OPRA | By contact | 4Å | statistical potentials | Function | R316,R38 | [44] | ||||||||||
| 2011 | DRNA | ✓ | 4.5Å | ✓ | Function | R250(RB250),R212(RB212),RBD292(NA) | [45] | ||||||||||
| 2010 | Struct-NB | 5Å | ✓ | NB | R147 from RNABindR | [46] | |||||||||||
| 2010 | PRNA | ✓ | http://doc.aporc.org/wiki/PRNA | ENTANGLE | ✓ | ✓ | ✓ | ✓ | RF | 5 | R205 | [32] | |||||
| 2014 | aaRNA | ✓ | http://sysimm.ifrec.osaka-u.ac.jp/aarna/ | 3.5Å | ✓ | ✓ | ✓ | ✓ | NN | 11 | R67,R141,R205 | [27] | |||||
| 2014 | RBRDetector | ✓ | http://ibi.hzau.edu.cn/rbrdetector | 4.5Å, 10%rASA | ✓ | SVM | 11 | R264, R75 | [29] | ||||||||
| 2014 | Xiaoyong | ENTANGLE | ✓ | ✓ | ✓ | ✓ | RF | 5 | R205(PRNA) | [31] | |||||||
| 2015 | RBscore | ✓ | http://ahsoka.u-strasbg.fr/rbscore/ | 3.5-6Å | ✓ | ✓ | ✓ | ✓ | R130,R116 | [28] | |||||||
| 2015 | RNAProSite | ✓ | lilab.ecust.edu.cn/NABind/ | ||||||||||||||
| DNA | |||||||||||||||||
| Sequence-based | |||||||||||||||||
| 2004 | DBS-Pred | ✓ | http://www.abren.net/dbs-pred/ | 3.5Å | ✓ | ✓ | ✓ | NN | 3 | D62(PDNA-62), NRTF-915 | [12] | ||||||
| 2005 | DBS-PSSM | ✓ | http://www.abren.net/dbs-pssm/ | 3.5Å | ✓ | NN | 5 | D62(PDNA-62), PDNA-RDN(NA), PDNA-NR90(NA) | [13] | ||||||||
| 2006 | DNABindR | ΔASA>1 | ✓ | ✓ | ✓ | ✓ | ✓ | NB | 9 | D171 | [47] | ||||||
| 2007 | DISIS | ✓ | http://cubic.bioc.columbia.edu/services/disis | 6Å | ✓ | ✓ | ✓ | SVM | 9 | D274 | [48] | ||||||
| 2007 | DP-Bind | ✓ | http://lcg.rit.albany.edu/dp-bind/ | 3.5Å | ✓ | kernel regression | D62(PDNA-62) | [49] | |||||||||
| 2009 | ProteDNA | ✓ | http://protedna.csbb.ntu.edu.tw/ | 4.5Å | ✓ | SVM, SSEA | 11 | D253 | [50] | ||||||||
| 2009 | DbindR | ✓ | http://www.cbi.seu.edu.cn/DBindR/DBindR.htm | 3.5Å | ✓ | ✓ | RF | 11 | D374 | [51] | |||||||
| 2009 | SDCPred | ✓ | http://sdcpred.netasa.org/ | 3.5Å | ✓ | ✓ | NN | 5 | D159(PDNA159) | [52] | |||||||
| 2014 | Byungkyu | H-bond | ✓ | SVM | 9 | D143 | [53] | ||||||||||
| Structure-based | |||||||||||||||||
| 1999 | Hidetoshi | 3.5Å | Function | D52 | [54] | ||||||||||||
| 2003 | Susan | ΔASA>1 | ✓ | Patch analysis | D56 | [55] | |||||||||||
| 2005 | DBS-kernel | 4.5Å | ✓ | ✓ | SVM | D83(NA) | [56] | ||||||||||
| 2005 | Pro-dna | ✓ | bioinformatics.bioengr.uic.edu/pro-dna/ | 4.5Å | ✓ | ✓ | ✓ | SVM | D99 (D96,D50) | [57] | |||||||
| 2005 | PreDs | ✓ | http://pre-s.protein.osaka-u.ac.jp/~preds/ | 3.0Å | ✓ | Function | D63 | [58]. | |||||||||
| 2007 | DISPLAR | ✓ | http://pipe.scs.fsu.edu/displar.html | 5Å | ✓ | NN | 15 | D428 | [10] | ||||||||
| 2007 | DR_bind1 | ✓ | http://drbind.limlab.ibms.sinica.edu.tw/ | HBPLUS | Energy based | Function | D56 from Susan 2003, D69, R81 | [26] | |||||||||
| 2008 | DBD-Hunter | ✓ | http://cssb.biology.gatech.edu/skolnick/webservice/DBD-Hunter/ | 4.5Å | ✓ | Function | D179(DB179) | [59] | |||||||||
| 2010 | DNABINDPROT | ✓ | http://www.prc.boun.edu.tr/appserv/prc/dnabindprot/ | NUCPLOT | ✓ | ✓ | GNM | 3 | D54 | [24] | |||||||
| 2011 | metaDBSite | ✓ | http://projects.biotec.tu-dresden.de/metadbsite/ | 3.5Å | Meta-server | NA | D316(PDNA-316),D232(PDNA-232) | [34] | |||||||||
| 2011 | Xiong | 4.5Å, 10%rASA | ✓ | ✓ | SVM | 11 | D206 | [60] | |||||||||
| 2012 | Sucharita | ΔASA>0.1 | ✓ | SVM | D130(NA) | [61] | |||||||||||
| 2013 | Duo-Duo | 4.0Å | ✓ | SVM | 11 | D62(PDNA-62) | [62]. | ||||||||||
| 2013 | PreDNA | ✓ | http://202.207.14.178/predna/ | 3.5Å | ✓ | ✓ | ✓ | SVM | 11 | D62(PDNA-62), D224 | [63] | ||||||
| 2013 | DNABind | ✓ | http://mleg.cse.sc.edu/DNABind/ | 4.5Å, 10%rASA | ✓ | ✓ | SVM, template | 11 | D206 | [30] | |||||||
| 2014 | Bi-Qing | 6Å | ✓ | ✓ | ✓ | SVM | 9 | D90 | [64] | ||||||||
PSSM: position specific scoring matrix derived from sequence alignment
RP: residue propensity
ASA: accessible surface area
HP: hydrophobicity
SS: secondary structure
EC: conservation entropy
Q: electrostatic/pKa
SA: structural alignment
Overall prediction performance of accuracy and stability
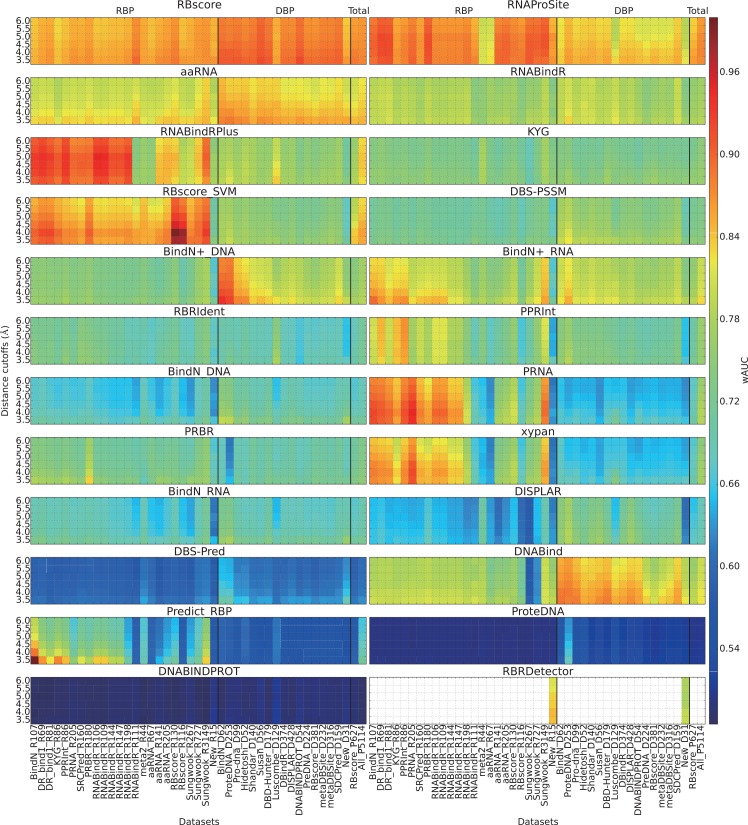
As demonstrated in Fig 1B and 1C, the standard way to measure the area under the receiver operating characteristic (ROC) curve (AUC) would include irrelevant comparisons between binding sites on one protein and non-binding sites on another protein. We assessed the nucleic acid binding site prediction ability by calculating the AUC for each protein and average AUCs on a dataset (wAUC or mAUC, Supplementary Note 4 in S1 Text). Fig 2 demonstrates a general assessment result in terms of wAUC (mean AUC considering the protein length), while mAUC (mean AUC) and tAUC (total AUC that compare all proteins together) are found in S1 Fig and S2 Fig. Although fluctuations are found, the general distributions of wAUC and mAUC are similar. BindN+, RNABindRPlus, aaRNA, RNAProSite and RBscore rank at the top while DNABind works well on DNA binding protein (DBP). Obviously, dataset bias is a very severe problem for some predictors and require special attention.
Fig 2. General accuracy distribution based on wAUC.
wAUC as assessment criterion of all programs on all datasets with hierarchical definition of binding sites from 3.5 to 6Å. wAUC is the weighted arithmetic mean of AUC and is considered as the criterion to assess the prediction accuracy of the predictors. It is plotted as rainbow colors from highest accuracy of red to lowest accuracy of blue. Each grid in the plot show the wAUC of a predictor (subtitle) on a certain data set (x-axis) assessed according to the binding sites defined by a certain distance cutoff (y-axis). For each subplot, DBP data sets and RBP data sets are separated by bold line. The last two data sets are mixed with DBP and RBP. RBscore_P627 is a non-redundant data set by removing cases of sequence identity >25%. All_P5114 is a mixture of all data sets.
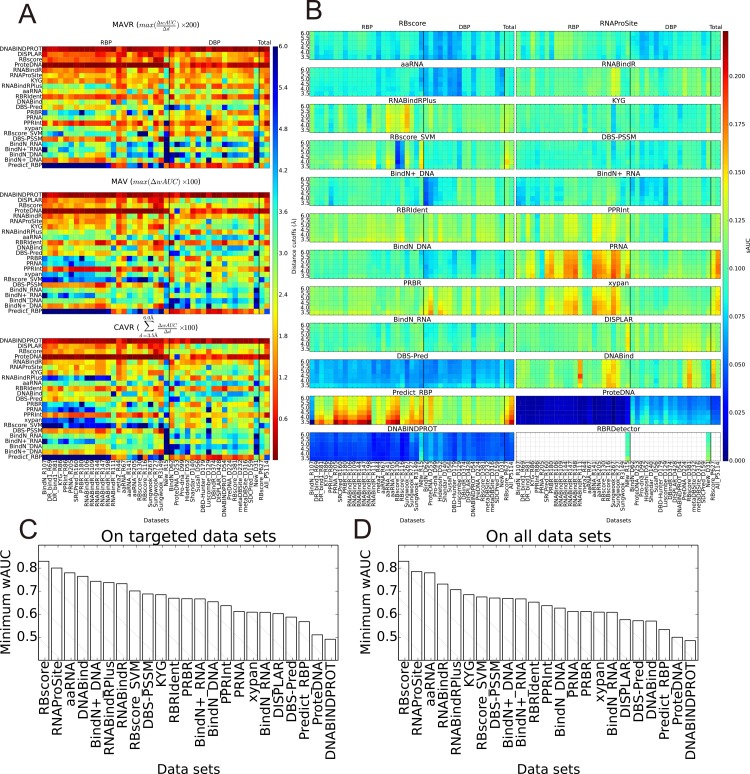
A successful prediction program should demonstrate stable predictive ability on all the criteria of the assessment. Three criteria (MAVR, MAV and CAVR, described in Methods), stand for distance cutoff dependent accuracy variances (wAUC) were used to assess the stability. It can be deduced from Fig 3A that the prediction KYG, RNAProSite, RNABindR, ProteDNA, RBscore, DISPLAR and DNABINDPROT are less dependent on distance cutoff (mAUC and tAUC based results could be found in S3 Fig and S4 Fig). Generally, programs tend to favor the distance cutoff used during training (Supplementary Note 11 in S1 Text). In terms of data set, standard deviations of AUC (sAUC) were measured as criteria to assess the stability and is shown in Fig 3B. The accuracies of DBS-PSSM, RBscore, aaRNA, DBS-Pred, ProteDNA and DNABINDPROT are more stable when varying the assessment data sets. Considering both prediction accuracy and stability, the ‘barrel effect’ applies to the predictions with dataset bias and a program can best be assessed by its minimum accuracy value in all the datasets. In Fig 3C, RNA binding site predictors are assessed on RBP while DNA binding predictors on DBP, the minimum wAUC measured by all distance cutoffs is taken as their prediction ability. Fig 3D shows the minimum wAUC applied on all the datasets. A more complete list can be found in Table 2.
Fig 3. Accuracy variations resulted from binding site definition and data set bias.
A) Accuracy variation resulted from distance cutoff based definition of binding sites. Three metrics, MAVR, MAV and CAVR (see methods for details), were used to describe the distance cutoff resulted accuracy variation. The higher the values are, the less stable the predictor is, indicating a less reliable predictor; B) Standard deviation of AUC (sAUC) on all data sets. Similarly, the higher values of sAUC indicate an unstable predictor that cannot guarantee stable accuracy or it is more likely to have data set bias; C) Minimum wAUC of all programs on their targeted data sets: DNA binding site prediction programs are tested on DBP and minimum wAUC are plotted, while the same for RNA binding site prediction programs on RBP. As prediction accuracy of a predictor can vary with data set of assessment and distance cutoff to define binding sites, minimum wAUC demonstrates the bottom line of accuracy that a predictor can guarantee. D) Minimum wAUC of all programs on all data sets.
Table 2. Minimum performance of all programs.
| DBP datasets | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sequence-based | ||||||||||||
| Programs | Distance Cutoff | Dataset | wAUC | mAUC | sAUC | tAUC | SEN | SPC | PPV | ACC | F1 | MCC |
| DBS-Pred | 6 | RBscore_D381 | - | - | - | - | 0.418 | 0.781 | 0.214 | 0.736 | 0.283 | 0.154 |
| ProteDNA | 6 | RBscore_D381 | - | - | - | - | 0.029 | 0.999 | 0.799 | 0.878 | 0.055 | 0.137 |
| BindN+_DNA | 6 | New_D31 | 0.743 | 0.780 | 0.091 | 0.776 | 0.421 | 0.908 | 0.358 | 0.856 | 0.387 | 0.307 |
| RNABindR | 6 | New_D31 | 0.731 | 0.753 | 0.116 | 0.779 | 0.648 | 0.759 | 0.246 | 0.747 | 0.356 | 0.280 |
| RNABindRPlus | 6 | New_D31 | 0.707 | 0.732 | 0.117 | 0.759 | 0.229 | 0.956 | 0.390 | 0.878 | 0.289 | 0.236 |
| DBS-PSSM | 5.5 | New_D31 | 0.688 | 0.728 | 0.111 | 0.748 | 0.541 | 0.825 | 0.265 | 0.795 | 0.356 | 0.273 |
| RBscore_SVM | 6 | New_D31 | 0.684 | 0.703 | 0.104 | 0.737 | 0.000 | 1.000 | 0.000 | 0.892 | 0.000 | 0.000 |
| BindN_DNA | 6 | Luscombe_D129 | 0.654 | 0.678 | 0.093 | 0.680 | 0.341 | 0.856 | 0.282 | 0.783 | 0.308 | 0.182 |
| RBRIdent | 4.5 | New_D31 | 0.652 | 0.683 | 0.104 | 0.655 | 0.105 | 0.953 | 0.167 | 0.883 | 0.129 | 0.071 |
| PPRInt | 4 | New_D31 | 0.652 | 0.691 | 0.105 | 0.690 | 0.450 | 0.799 | 0.148 | 0.774 | 0.222 | 0.155 |
| xypan | 6 | DNABINDPROT_D54 | 0.635 | 0.628 | 0.109 | 0.641 | 0.092 | 0.982 | 0.469 | 0.850 | 0.153 | 0.156 |
| PRNA | 6 | New_D31 | 0.626 | 0.653 | 0.110 | 0.646 | 0.336 | 0.866 | 0.234 | 0.809 | 0.276 | 0.173 |
| PRBR | 6 | ProteDNA_D253 | 0.612 | 0.587 | 0.139 | 0.602 | 0.317 | 0.824 | 0.362 | 0.702 | 0.338 | 0.148 |
| Predict_RBP | 4.5 | New_D31 | 0.534 | 0.554 | 0.130 | 0.556 | 0.000 | 1.000 | 0.000 | 0.917 | 0.000 | 0.000 |
| Structure-based | ||||||||||||
| DISPLAR | 6 | New_D31 | - | - | - | - | 0.359 | 0.939 | 0.416 | 0.876 | 0.386 | 0.318 |
| DNABINDPROT | 5.5 | BindN_D62 | - | - | - | - | 0.075 | 0.929 | 0.233 | 0.737 | 0.113 | 0.005 |
| RBscore | 6 | New_D31 | 0.837 | 0.839 | 0.089 | 0.843 | 0.438 | 0.918 | 0.395 | 0.866 | 0.415 | 0.341 |
| aaRNA | 6 | New_D31 | 0.804 | 0.813 | 0.060 | 0.834 | 0.552 | 0.891 | 0.381 | 0.854 | 0.451 | 0.378 |
| RNAProSite | 6 | Luscombe_D129 | 0.785 | 0.803 | 0.081 | 0.790 | 0.760 | 0.696 | 0.290 | 0.705 | 0.420 | 0.329 |
| DNABind | 6 | RBscore_D381 | 0.765 | 0.798 | 0.152 | 0.774 | 0.614 | 0.898 | 0.492 | 0.859 | 0.546 | 0.467 |
| KYG | 6 | DNABINDPROT_D54 | 0.707 | 0.703 | 0.074 | 0.712 | 0.444 | 0.802 | 0.281 | 0.749 | 0.344 | 0.206 |
| RBP datasets | ||||||||||||
| Sequence-based | ||||||||||||
| DBS-Pred | 6 | aaRNA_R141 | - | - | - | - | 0.359 | 0.790 | 0.190 | 0.738 | 0.248 | 0.116 |
| ProteDNA | 4.5 | Sungwook_R267 | - | - | - | - | 0.001 | 0.999 | 0.140 | 0.888 | 0.002 | 0.003 |
| RNABindRPlus | 6 | RNABindR_R111 | 0.738 | 0.725 | 0.113 | 0.720 | 0.321 | 0.914 | 0.316 | 0.848 | 0.319 | 0.233 |
| RNABindR | 6 | New_R15 | 0.733 | 0.756 | 0.093 | 0.737 | 0.661 | 0.681 | 0.279 | 0.678 | 0.393 | 0.258 |
| RBscore_SVM | 6 | New_R15 | 0.687 | 0.697 | 0.135 | 0.697 | 0.046 | 0.987 | 0.396 | 0.839 | 0.083 | 0.090 |
| DBS-PSSM | 5.5 | New_R15 | 0.670 | 0.695 | 0.089 | 0.672 | 0.473 | 0.780 | 0.277 | 0.734 | 0.349 | 0.207 |
| RBRIdent | 4 | New_R15 | 0.670 | 0.699 | 0.127 | 0.671 | 0.169 | 0.952 | 0.313 | 0.863 | 0.219 | 0.160 |
| PRBR | 6 | New_R15 | 0.667 | 0.680 | 0.112 | 0.664 | 0.305 | 0.877 | 0.317 | 0.787 | 0.311 | 0.185 |
| BindN+_RNA | 6 | New_R15 | 0.667 | 0.687 | 0.089 | 0.672 | 0.378 | 0.835 | 0.300 | 0.763 | 0.334 | 0.194 |
| PPRInt | 5.5 | New_R15 | 0.637 | 0.658 | 0.131 | 0.648 | 0.356 | 0.814 | 0.255 | 0.745 | 0.297 | 0.150 |
| PRNA | 6 | New_R15 | 0.612 | 0.629 | 0.141 | 0.624 | 0.295 | 0.865 | 0.290 | 0.775 | 0.293 | 0.159 |
| xypan | 6 | New_R15 | 0.609 | 0.631 | 0.106 | 0.620 | 0.115 | 0.981 | 0.534 | 0.845 | 0.189 | 0.193 |
| BindN_RNA | 4 | New_R15 | 0.608 | 0.642 | 0.121 | 0.626 | 0.324 | 0.842 | 0.209 | 0.783 | 0.254 | 0.139 |
| Predict_RBP | 6 | New_R15 | 0.568 | 0.581 | 0.155 | 0.576 | 0.022 | 1.000 | 1.000 | 0.846 | 0.043 | 0.136 |
| Structure-based | ||||||||||||
| DISPLAR | 6 | Sungwook_R267 | - | - | - | - | 0.214 | 0.956 | 0.433 | 0.856 | 0.287 | 0.234 |
| DNABINDPROT | 5 | Sungwook_R3149 | - | - | - | - | 0.038 | 0.948 | 0.239 | 0.677 | 0.066 | -0.029 |
| DR_bind1 | 4.5 | meta2_R44 | - | - | - | - | 0.285 | 0.942 | 0.655 | 0.758 | 0.397 | 0.311 |
| RBscore | 6 | KYG_R86 | 0.830 | 0.845 | 0.095 | 0.862 | 0.502 | 0.936 | 0.697 | 0.837 | 0.584 | 0.496 |
| RNAProSite | 6 | meta2_R44 | 0.801 | 0.805 | 0.101 | 0.798 | 0.675 | 0.770 | 0.573 | 0.740 | 0.620 | 0.427 |
| aaRNA | 5.5 | New_R15 | 0.780 | 0.785 | 0.070 | 0.777 | 0.539 | 0.854 | 0.396 | 0.806 | 0.457 | 0.348 |
| KYG | 6 | Sungwook_R267 | 0.685 | 0.690 | 0.073 | 0.685 | 0.362 | 0.816 | 0.235 | 0.755 | 0.285 | 0.150 |
| DNABind | 4.5 | Sungwook_R267 | 0.570 | 0.608 | 0.149 | 0.600 | 0.260 | 0.827 | 0.169 | 0.759 | 0.205 | 0.073 |
-: binary predictors and AUC not applicable.
Detailed comparisons
Some programs tested here are only designed to predict RNA binding sites while others DNA binding sites. However, when tested together, we find that some of the programs show predictive ability on both types of proteins. For instance, RNABindR, KYG, aaRNA, RNAProSite and RBscore are developed on RBP and never trained on DBP, but they also show predictive ability on DBP. In terms of sequence-based methods, RNABindR even demonstrates higher prediction accuracy than most DNA-binding site predictors. aaRNA and RBscore show even higher accuracies for DBP than for RBP. The RNA binding site prediction mode of BindN+ also shows prediction ability on DBP, but its DBP prediction mode has a much lower accuracy on RBP.
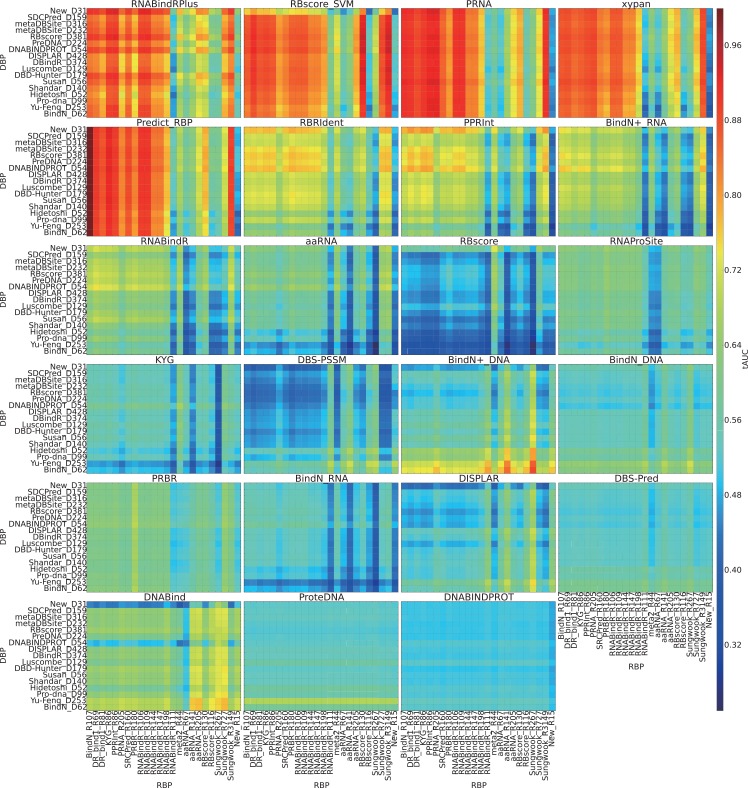
Together with the problem of binding site prediction, it is very interesting to find out whether there is a program that can discriminate RNA binding residues from DNA binding residues. This discrimination require three assumptions: i) residues from different proteins can be compared; ii) RNA and DNA binding is driven by different driving forces; iii) such driving forces have been explored by current programs. Previous work from Yan et al.[6] analysed the cross-prediction between RNA- and DNA-binding residue predictors and concluded that they are unable to properly separate DNA- from RNA-binding residues. We performed a more explicit large-scale test and assessed this discrimination by mixing DNA binding residues of a data set with RNA binding residues of another data set. According to Fig 4, we find several machine learning based approaches display a discriminative ability for RNA binding residues, including PRNA, Predict_RBP, RNABindRPlus and RBscore_SVM. However, this cannot guarantee predictive ability, since all of the programs have AUC <0.5 on some data sets, which means the programs favor the wrong type of residue. Therefore, we come to the same conclusion than reached by Yan et al. [6], i.e. that none of the current existing predictors can properly distinguish DNA-binding residues from RNA binding ones. Further, we find that the programs PRNA, Predict_RBP, RNABindRPlus and RBscore_SVM have similar distribution for this test while their wAUC distributions on Fig 2 are also similar. This result implies that these methods have similar prediction accuracies and similar preferences on datasets.
Fig 4. Discrimination between RNA binding sites and DNA binding sites.
For DNA binding site prediction programs, DNA binding sites (3.5Å as distance cutoff) from a data set are taken as positive while RNA binding sites from another data set are taken as negative, the AUC values of these discriminations are plotted with different colors. RNA binding site prediction programs take the opposite. Each grid on the heat map show the assessment result in terms of AUC on such a DNA binding sites vs. RNA binding sites data set. i.e. The grid BindN_R107(x-axis)-Susan_D56(y-axis) of program PRNA is to use all the RNA binding sites of BindN_R107 data set as positive while using all DNA binding sites of Susan_D56 data set as negative and measure the AUC value of program PRNA.
Comparing the updated webservers with the old ones, we find obvious improvement over the years. BindN+ shows consistent improvement over BindN, when programs integrating an homologous search approach, RNABindRPlus, DNABind and RBRDetector show accuracy increase on some of the datasets. New upcoming webservers, aaRNA, RNAProSite and RBscore, show the top ranking performances. Still, xypan and RBRIdent, both improved versions of PRNA, do not display enough effectiveness in this large-scale test.
The AUC is a criterion of assessment that presents a bias towards the score-based predictions rather than binary predictions (binding or not-binding). However, some programs, such as DISPLAR, include a predefined prediction process and give only binary results. On the contrary, in order to obtain binary predictions, some score-based programs only include arbitrary cutoffs, which are not favored by the binary assessment criteria. In a binary comparison, we have to mix up all the proteins in assessment and balance between specificity and sensitivity. Traditional binary assessment criteria are used, including specificity (S5 Fig), sensitivity (S6 Fig), precision (S7 Fig), accuracy (S8 Fig), F1 score (S9 Fig) and Matthew correlation coefficiency (S10 Fig). A general summary of minimum performance is listed in Table 2.
Comparing structure-based predictors with sequence-based ones in Table 2 and Fig 2, we can easily find out that some structure-based predictors, aaRNA, RBscore and RNAProSite, are consistently better in performance regardless of the nucleic acid type. This implies that sequence-based approaches that attempt to incorporate predicted structure features such as solvent accessibility and electrostatics cannot capture the real structural features that govern nucleic acid binding. Many sequence-based predictors do not guarantee an AUC above 0.7, which can hardly be considered as meaningful predictions, since an AUC ~0.5 is equivalent random guess.
We find that aaRNA shows similar level of wAUC on dataset meta_R44 (0.82) and Sungwook_R267(0.83), but its sensitivity on meta_R44 (0.8) is much higher than on Sungwook_R267(0.52) while specificity shows the opposite, 0.73 vs. 0.89. Similar cases could also be found in many other programs. In fact, these programs show stable prediction accuracies on both of the sets, but the binary defined sensitivity is a trade-off of specificity and determined by a pre-set cutoff. DNABINDPROT and DISPLAR show top rank specificities and accuracies, which was not described by the AUC distribution. However, when regard to sensitivity, DNABINDPROT ranks at the bottom and DISPLAR is lower than median. This implies that these programs sacrifice sensitivity to gain specificity and accuracy, leading to low true positive rates. This demonstrates that proteins in different environments can have different affinities to nucleic acids and vary in the size of their binding interface. Thus, the use of fixed cutoffs to define binary prediction may misinterpret the binding reality (Supplementary Note 4 in S1 Text).
Can the predictions represent a binding funnel around the binding interface?
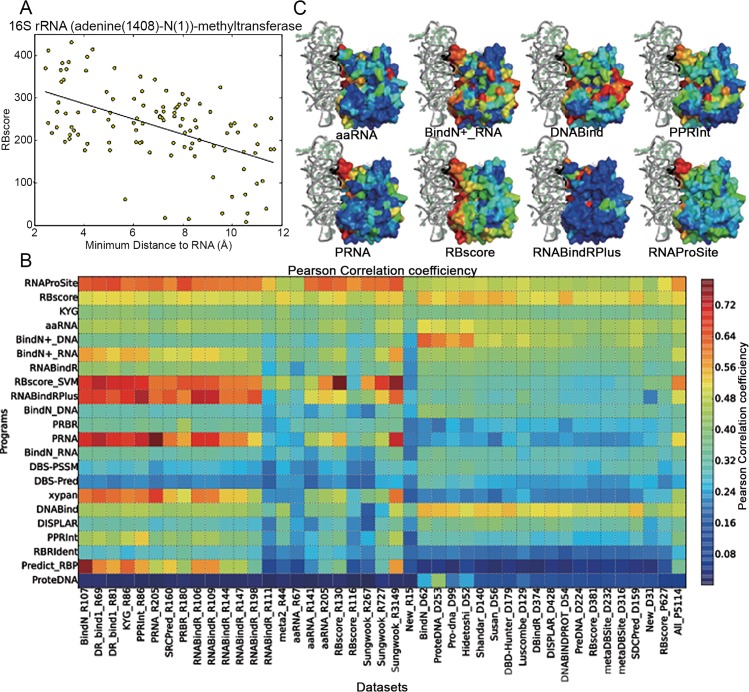
When we color the protein surface residues with the prediction scores as hierarchical colors, we find that the prediction score of some non-binding residues near the binding region are lower than that of the binding residues but higher than other non-binding ones, Fig 5A. The residues around a protein surface are more likely to form a binding funnel than abruptly change from binding region to non-binding ones, which leads to the conclusion that the use of only binary definition and single distance cutoff in the assessment is not feasible. As the residues can gradually change from binding to non-binding region, there could be a correlation between the distance from a residue to the core binding region and the predicted binding score. In Fig 5B, the distance to the core binding region is partly represented by the distance to the RNA ligand, and we do find that such a correlation exists in residues around the binding region (within 12Å from the RNA ligands). Although this region maybe smaller for small proteins and the correlation is not necessarily linear, we can roughly measure the correlation by the Pearson correlation coefficient. Fig 5C illustrates this distribution for some programs. We find that the programs of higher accuracies also display higher Pearson correlation coefficients. This assessment could be further optimized if the distance to the core binding region is well defined.
Fig 5. Correlation between prediction score and nucleic acid binding funnel on protein surface.
A) Relationship between the minimum distance from a residue around the binding interface (within 12Å) to its RNA ligand (x-axis) and the RBscore of the residue (y-axis). Generally, the RBscore drops when the distance to RNA ligand increases. B) Pearson correlation coefficient, color-coded in rainbow colors, between minimum distance from a residue around the binding interface (within 12Å) to its RNA ligand and the prediction score. The higher Pearson correlation coefficient a predictor has, the more likely its prediction score can display the energy funnel of nucleic acid binding. C) Examples of prediction scores plotted on protein (16S rRNA (adenine(1408)-N(1))-methyltransferase) surfaces as rainbow color, higher binding score region are shown in red.
Tests on newly solved structures
Another way to show the predictive ability is to test the newly solve complex structures, since most of the predictors were developed on datasets before 2013. We, thus, collected the protein-nucleic acid complexes solved after 2014, 31 DBP and 15 RBP, as test sets. Because these cases are non-homologous with all the training and test sets of all the programs, they can be taken as an independent test set. This test is similar to the blind tests of CASP[65], RNA-Puzzles[66,67] and CAFA[68]. If a prediction program really has predictive ability rather than the ability of interpolation, it should show high accuracy on these new data. The homologous search approaches of some programs, such as RNABindRPlus and DNABind, could be excluded by this assessment. According to Fig 2, we find that some of the programs show much lower accuracies on these new data than expected according to previous performance (Supplementary Note 7 in S1 Text). But, overall, the results on the newly published data sets are similar to the performance on some ‘difficult sets’ such as meta_R44 and RBscore_R117. The values presented in Table 2 indicate also that many machine learning based methods show their minimum performance in these two new datasets. This highlights the data set bias problem of the machine learning approaches that can only demonstrate predictive ability in terms of interpolation. For this reason, a fair comparison indeed require for blind tests independent datasets that do not have any relation to the known datasets.
Discussion
Rather than achieving a high level of accuracy on existing datasets, both an increase in knowledge and understanding of the driving forces and mechanisms for protein-nucleic acids binding are required in order to improve the accuracy on all datasets. Although most of the programs have been validated with curated datasets, we find that some of the programs do not consistently show high predictive ability on all the datasets. That is to say, these programs are poor at recapitulating the main factors key to protein-nucleic acid binding.
According to results on all the programs, some RNA binding site prediction programs (RBscore, RNAProSite, aaRNA and BindN+) show predictive ability in DNA binding site prediction, but hardly any DNA binding site prediction program demonstrates high level of accuracy in RNA binding site prediction. Thus, the key features of RNA binding residues and DNA binding residues are similar and can be better captured by RBP-based datasets than by DBP-based datasets. Besides, we find the most stable and accurate programs are aaRNA, RNAProSite and RBscore, all programs that take the advantage of protein structure, while other sequence-based programs are less stable in terms of both data set and distance cutoff, implying that important structural features cannot be fully captured by sequence-based programs making them less accurate.
The previous assessments of the predictive ability were mainly focused on two approaches, comparisons between previously reported results[4] and with small tests[6] (Supplementary Note 8 in S1 Text). However, as shown by this large-scale test, datasets can be biased and tests on one or a few test sets cannot testify the bottom line of predictive ability of a program. Also, comparisons with previously reported results are indirect, stressing the importance of large-scale tests and comparisons. This work has systematically benchmarked most currently existing programs in various aspects to complement the loophole of previous assessments. Further, we provide and regularly maintain all the test sets in this work on our web site allowing benchmarking of novel methods on all these data sets. Thus, new programs can be directly compared with all the existing programs and merits of the programs can be demonstrated in a straightforward manner.
Finally, we notice also that some existing binding site prediction approaches contain theoretical drawbacks: 1. For predictions leading to a binary classification, the mixing together of all proteins is arguable (Supplementary Note 4 in S1 Text); 2. The use of non-orthogonal but redundant features in prediction renders loose the relationships between feature and prediction (Supplementary Note 10 in S1 Text); 3. Slide-window approaches in sequence-based methods do not consider the real spatial environment of the residues (Supplementary Note 9 in S1 Text). Therefore, there is still room for the search for better alternatives.
In order to be useful to the research community, it is very important to make the prediction programs and web servers available and user-friendly. Some current programs require special computational skills, some are slow in efficiency and some require special formatted input, stressing the importance of ease-of-use and robustness of a prediction web server.
Methods
Datasets
All of the data sets were built based on protein-nucleic acid co-crystal structures extracted from the PDB. 23 RNA binding protein datasets and 16 DNA binding protein datasets were collected from previous studies and listed in Table 3. Some unreasonable cases were excluded from the assessment datasets: 1) the presence of a DBP in a RBP set (PDB ID 1a1v); 2) superseded PDB structures; 3) peptides shorter than 20 residues; 4) weak and uncertain nucleic acid binding proteins including those with less than three binding residues; 5) PDB chains containing only Cα atoms; 6) proteins constituted by two separate short peptides. Other two data sets of RBP and DBP after 2014 have been curated. Sequence similarity of 25% have been used as cutoff to remove redundancy from other data sets by PISCES[69]. These two data sets include 15RBP and 31 DBP respectively. All the data sets could be downloaded on website: http://ahsoka.u-strasbg.fr/nbench/.
Table 3. Summary of datasets used in tests.
| Name | Protein# | After screening | Reference | Seq id | Str id | resolution |
|---|---|---|---|---|---|---|
| BindN_R107 | 107 | 95 | [8] | 25 | NA | 3.5 |
| DR_bind1_R69 | 69 | 69 | [42] | NA | CATH | 3 |
| DR_bind1_R81 | 81 | 79 | [42] | NA | CATH | 3 |
| KYG_R86 | 86 | 85 | [14] | 50 | NA | NA |
| PPRInt_R86 | 86 | 83 | [23] | 70 | NA | 3 |
| PRNA_R205 | 205 | 189 | [19] | 25 | NA | 3 |
| SRCPred_R160 | 160 | 124 | [39] | 25 | NA | NA |
| PRBR_R180 | 180 | 142 | [22] | 25 | NA | 3.5 |
| RNABindR_R106 | 106 | 100 | [20,21] | NA | NA | NA |
| RNABindR_R109 | 109 | 100 | [20,21] | 30 | NA | 3.5 |
| RNABindR_R144 | 144 | 137 | [20,21] | NA | NA | NA |
| RNABindR_R147 | 147 | 138 | [20,21] | 30 | NA | 3.5 |
| RNABindR_R198 | 198 | 187 | [20,21] | 30 | NA | 3.5 |
| RNABindR_R111 | 111 | 101 | [20,21] | 30 | NA | 3.5 |
| meta2_R44 | 44 | 44 | [4] | 40 | NA | NA |
| aaRNA_R67 | 67 | 67 | [27] | 30 | NA | NA |
| aaRNA_R141 | 141 | 136 | [27] | 25 | NA | 3 |
| aaRNA_R205 | 205 | 200 | [27] | 25 | NA | 3 |
| RBscore_R130 | 130 | 130 | [28] | 25 | TMscore<0.7 | 3.5 |
| RBscore_R116 | 117 | 116 | [28] | 25 | TMscore<0.7 | 3.5 |
| Sungwook_R267 | 267 | 178 | [38] | 60 | NA | 3 |
| Sungwook_R727 | 727 | 574 | [38] | NA | NA | 3 |
| Sungwook_R3149 | 3149 | 2632 | [38] | NA | NA | 3 |
| New_R15 | 15 | 15 | 25 | NA | 5 | |
| BindN_D62 | 62 | 66 | [8] | 25 | NA | NA |
| ProteDNA_D253 | 253 | 253 | [70] | 20 | NA | 3.5 |
| Pro-dna_D99 | 99 | 188 | [56] | 20 | NA | 3 |
| Hidetoshi_D52 | 52 | 49 | [54] | NA | NA | 3.2 |
| Shandar_D140 | 140 | 138 | [12] | 25 | NA | 2.5 |
| Susan_D56 | 56 | 54 | [55] | NA | CATH | 3 |
| DBD-Hunter_D179 | 179 | 177 | [59] | 35 | NA | 3 |
| Luscombe_D129 | 129 | 182 | [71] | NA | NA | 3 |
| DBindR_D374 | 374 | 329 | [51] | 25 | NA | 3.5 |
| DISPLAR_D428 | 428 | 390 | [10] | 50 | NA | NA |
| DNABINDPROT_D54 | 54 | 50 | [24] | NA | NA | NA |
| PreDNA_D224 | 224 | 216 | [63] | 25 | NA | 3 |
| RBscore_D381 | 381 | 381 | [28] | 25 | NA | 3.5 |
| metaDBSite_D232 | 232 | 225 | [34] | 30 | NA | 3 |
| metaDBSite_D316 | 316 | 308 | [34] | 30 | NA | 3 |
| SDCPred_D159 | 159 | 158 | [52] | 25 | NA | 2.5 |
| New_D31 | 31 | 31 | 25 | NA | 5 | |
| RBscore_P627 | 628 | 627 | [28] | NA | NA | 3.5 |
| All_P5114 | 5058 | 5058 | NA | NA | NA |
NA: not applicable
Definition of binding sites
The minimum distance from any atom of a protein residue to any nucleic acid atom defines the distance from a protein residue and the nucleic acid. Nucleic acid binding sites are defined when such distances to the nucleic acid are shorter than certain thresholds. The range between 3.5 to 6Å, with 0.5Å step, was used as hierarchical thresholds to define binding residues in test sets. Besides, a nucleic acid binding residue always requires accessible surface area change (ΔASA>0Å2) upon complex formation with the nucleic acid. Accessible surface area is measured by NACCESS[72] with default parameters.
Assessment criteria
Receiver Operating Characteristic (ROC) curve together with Area Under Curve (AUC) is always used as criterion for accuracy[73]. We define the accuracy of a set of proteins by averaging accuracies of all proteins. We suggest the weighted arithmetic mean of AUC (wAUC) and mean of AUC (mAUC) as two criteria of accuracy for a set of proteins:
| (1) |
| (2) |
For a protein i, AUC(i) is its AUC value and len(i) is length of the protein, while N is the number of proteins in a dataset. We call the AUC that compare all the residues in a dataset together as total AUC (tAUC) and use it as a reference for comparison.
We define standard deviation of AUC of a data set as sAUC to show the accuracy stability varying the data sets:
| (3) |
Other binary criteria include specificity, sensitivity, precision, accuracy, F1 score and Matthews correlation coefficient:
| (4) |
| (5) |
| (6) |
| (7) |
| (8) |
| (9) |
TP is true positive, TN is true negative, FP is false positive and FN is false negative. P is total positive and N is total negative.
MAVR, maximum accuracy variation rate, is defined as:
| (10) |
Δd is the difference between two distance cutoffs used to define binding sites. ΔwAUC is the resulted accuracy variance defined by wAUC, this can also be replaced by mAUC or tAUC.
| (11) |
MAV is the maximum accuracy variation and CAVR is the cumulated accuracy variation rate:
| (12) |
MAVR, MAV and CAVR are criteria to assess the distance cutoff dependent variation in accuracy.
The correlation coefficient between prediction score and minimum distance to nucleic acid is defined by the Pearson correlation coefficient:
| (13) |
min(d) is the minimum distance from a residue to any nucleic acid atom, s is the prediction score, cov is the covariation and σx is the standard deviation.
Use of prediction programs
Li lab and Xiaoyong Pan provided RNAProSite and xypan prediction results respectively, with default parameter. aaRNA was used by inputting the structure file with default parameters. RNABindR and RNABindRPlus were predicted with default parameters, while removing 95% sequence identity in RNABindRPlus. KYG was predicted with command line using “method_type = 8” option. RBscore_SVM was based on the training set of R246. DBS-PSSM, DBS-Pred, RBRIdent, PPRInt, PRBR, DNABind, RBRDetector and ProteDNA were used with default parameter. The DISPLAR program was provided by Sanbo Qin and was run with default parameters. BindN and BindN+ were used by default parameters, while suffix “_RNA” and “_DNA” are RNA mode and DNA mode respectively. PRNA and Predict_RBP were trained on PRNA_R205 and BindN_R107 respectively, without cross-validation and applied with default parameters. DNABINDPROT was predicted with option “Fast 1” and DR_bind1 was predicted in the RNA mode. N-terminal and C-terminal residues not predicted by PRBR are taken as non-binding sites and assigned 0 as prediction score. For binary predictions of DNABINDPROT, DISPLAR, DBS-Pred and DBS-PSSM, positive sites are assigned a prediction score of 1, while negatives are assigned 0.
Availability
All data in this assessment are available on NBench web site http://ahsoka.u-strasbg.fr/nbench/.
Supporting Information
(DOCX)
(XLSX)
Binding sites are defined by 3.5Å and 6Å defined sites are taken as prediction, accuracies are assessed and vice versa.
(XLSX)
(XLSX)
(EPS)
(EPS)
(EPS)
(EPS)
(EPS)
(EPS)
(EPS)
(EPS)
(EPS)
(EPS)
(EPS)
Distributions on other data sets are available as zip file on NBench website.
(EPS)
Case of poly(A)-binding protein (PDB id: 1cvj chain A). Residue F102 is mutated into His, Glu and Asp. Variations around F102 is easy to comprehend but two other regions close in space are also related. RBscore consider the spatial neighbors, and show difference in region 127–129 and region 172–179. BindN+ only show certain difference in the region 172–179. PPRInt and RNABindRPlus hardly show difference in prediction. As demonstrated by deep mutational scanning, the single point mutation to Glu and Asp obviously make difference in the binding and should also affect other residue neighbors. With a sequence based slide-window approach it is difficult to capture such structural differences.
(EPS)
Approach 1 is to input directly the residue information into the learning machine, approach 2 is first to map the residue information to a function and input the resulted feature vector into the learning machine. If we take the mapping step together with the learning machine as another learning machine, approach 2 is not different from approach 1.
(EPS)
The plot is based on test on data set ‘DR_bind1_R69’. X-axis show the distance cutoff used to define nucleic acid binding sites, while Y-axis show the resulted wAUC value. We find the wAUC values vary for different distance cutoffs, and the distributions for different programs are different.
(EPS)
Acknowledgments
We gratefully thank Pascal Auffinger for helpful discussions on the manuscript, Drena Dobbs and Rasna R. Walia for valuable discussions on machine learning methods and tests of RNABindR and RNABindRPlus, Shandar Ahmad for beneficial discussions on assessment criteria and tests of DBS-PSSM, DBS-Pred, SRCPred and SDCPred, Carmay Lim, Yao-Chi Chen and Jon Wright for the small scaled test of DR_bind1, aaRNA developers for scripts in running aaRNA in batch mode, Piero Fariselli for helpful discussion of machine learning theories and techniques, Sanbo Qin for program package of DISPLAR, Xiaoyang Pan for providing prediction results of his method, G.P.S.Raghava and Manish Kumar for technical details in PPRInt, Haipeng Gong and Xiong Dapeng for discussion of RBRIdent, Kei Yura, Yaping Fang, Zhiping Liu for technical details in KYG, Predict_RBP and PRNA respectively.
Data Availability
All data in this assessment are available on NBench web site http://ahsoka.u-strasbg.fr/nbench/.
Funding Statement
This work is supported by the French Government grant ANR-10-BINF-02-02 "BACNET ". The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Gromiha MM, Nagarajan R (2013) Chapter Three—Computational Approaches for Predicting the Binding Sites and Understanding the Recognition Mechanism of Protein–DNA Complexes In: Rossen D, editor. Advances in Protein Chemistry and Structural Biology: Academic Press; pp. 65–99. [DOI] [PubMed] [Google Scholar]
- 2.(2015) The difficulty of a fair comparison. Nat Meth 12: 273–273. [DOI] [PubMed] [Google Scholar]
- 3. Zhao HY, Yang YD, Zhou YQ (2013) Prediction of RNA binding proteins comes of age from low resolution to high resolution. Mol Biosyst 9: 2417–2425. 10.1039/c3mb70167k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Puton T, Kozlowski L, Tuszynska I, Rother K, Bujnicki JM (2012) Computational methods for prediction of protein-RNA interactions. J Struct Biol 179: 261–268. 10.1016/j.jsb.2011.10.001 [DOI] [PubMed] [Google Scholar]
- 5. Cirillo D, Agostini F, Tartaglia GG (2013) Predictions of protein-RNA interactions. Wires Comput Mol Sci 3: 161–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yan J, Friedrich S, Kurgan L (2015) A comprehensive comparative review of sequence-based predictors of DNA- and RNA-binding residues. Briefings in bioinformatics 10.1093/bib/bbv023 [DOI] [PubMed] [Google Scholar]
- 7. Carson MB, Langlois R, Lu H (2010) NAPS: a residue-level nucleic acid-binding prediction server. Nucleic Acids Research 38: W431–W435. 10.1093/nar/gkq361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang LJ, Brown SJ (2006) BindN: a web-based tool for efficient prediction of DNA and RNA binding sites in amino acid sequences. Nucleic Acids Research 34: W243–W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang LJ, Huang CY, Yang MQ, Yang JY (2010) BindN plus for accurate prediction of DNA and RNA-binding residues from protein sequence features. Bmc Syst Biol 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tjong H, Zhou HX (2007) DISPLAR: an accurate method for predicting DNA-binding sites on protein surfaces. Nucleic Acids Research 35: 1465–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Peng Z, Kurgan L (2015) High-throughput prediction of RNA, DNA and protein binding regions mediated by intrinsic disorder. Nucleic Acids Res 10.1093/nar/gkv585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ahmad S, Gromiha MM, Sarai A (2004) Analysis and prediction of DNA-binding proteins and their binding residues based on composition, sequence and structural information. Bioinformatics 20: 477–486. [DOI] [PubMed] [Google Scholar]
- 13. Ahmad S, Sarai A (2005) PSSM-based prediction of DNA binding sites in proteins. Bmc Bioinformatics 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim OTP, Yura K, Go N (2006) Amino acid residue doublet propensity in the protein-RNA interface and its application to RNA interface prediction. Nucleic Acids Research 34: 6450–6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shulman-Peleg A, Shatsky M, Nussinov R, Wolfson HJ (2008) Prediction of interacting single-stranded RNA bases by protein-binding patterns. J Mol Biol 379: 299–316. 10.1016/j.jmb.2008.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang Y, Xue Z, Shen G, Xu J (2008) PRINTR: Prediction of RNA binding sites in proteins using SVM and profiles. Amino Acids 35: 295–302. 10.1007/s00726-007-0634-9 [DOI] [PubMed] [Google Scholar]
- 17. Wang CC, Fang YP, Xiao JM, Li ML (2011) Identification of RNA-binding sites in proteins by integrating various sequence information. Amino Acids 40: 239–248. 10.1007/s00726-010-0639-7 [DOI] [PubMed] [Google Scholar]
- 18. Allers J, Shamoo Y (2001) Structure-based analysis of Protein-RNA interactions using the program ENTANGLE. J Mol Biol 311: 75–86. [DOI] [PubMed] [Google Scholar]
- 19. Freddolino PL, Harrison CB, Liu Y, Schulten K (2010) Nat Phys 6: 751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Terribilini M, Sander JD, Lee JH, Zaback P, Jernigan RL, et al. (2007) RNABindR: a server for analyzing and predicting RNA-binding sites in proteins. Nucleic Acids Research 35: W578–W584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Walia RR, Xue LC, Wilkins K, El-Manzalawy Y, Dobbs D, et al. (2014) RNABindRPlus: A Predictor that Combines Machine Learning and Sequence Homology-Based Methods to Improve the Reliability of Predicted RNA-Binding Residues in Proteins. Plos One 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ma X, Guo J, Wu JS, Liu HD, Yu JF, et al. (2011) Prediction of RNA-binding residues in proteins from primary sequence using an enriched random forest model with a novel hybrid feature. Proteins 79: 1230–1239. 10.1002/prot.22958 [DOI] [PubMed] [Google Scholar]
- 23. Kumar M, Gromiha AM, Raghava GPS (2008) Prediction of RNA binding sites in a protein using SVM and PSSM profile. Proteins 71: 189–194. [DOI] [PubMed] [Google Scholar]
- 24. Ozbek P, Soner S, Erman B, Haliloglu T (2010) DNABINDPROT: fluctuation-based predictor of DNA-binding residues within a network of interacting residues. Nucleic Acids Res 38: W417–W423. 10.1093/nar/gkq396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chu WY, Huang YF, Huang CC, Cheng YS, Huang CK, et al. (2009) ProteDNA: a sequence-based predictor of sequence-specific DNA-binding residues in transcription factors. Nucleic Acids Res 37: W396–401. 10.1093/nar/gkp449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen YC, Wu CY, Lim C (2007) Predicting DNA-binding amino acid residues from electrostatic stabilization upon mutation to Asp/Glu and evolutionary conservation. Proteins 67: 671–680. [DOI] [PubMed] [Google Scholar]
- 27. Li S, Yamashita K, Amada KM, Standley DM (2014) Quantifying sequence and structural features of protein–RNA interactions. Nucleic Acids Res 10.1093/nar/gku681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miao Z, Westhof E (2015) Prediction of nucleic acid binding probability in proteins: a neighboring residue network based score. Nucleic Acids Res 10.1093/nar/gkv446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang XX, Deng ZL, Liu R (2014) RBRDetector: improved prediction of binding residues on RNA-binding protein structures using complementary feature- and template-based strategies. Proteins 82: 2455–2471. 10.1002/prot.24610 [DOI] [PubMed] [Google Scholar]
- 30. Liu R, Hu J (2013) DNABind: a hybrid algorithm for structure-based prediction of DNA-binding residues by combining machine learning- and template-based approaches. Proteins 81: 1885–1899. 10.1002/prot.24330 [DOI] [PubMed] [Google Scholar]
- 31. Pan X, Zhu L, Fan Y-X, Yan J (2014) Predicting protein–RNA interaction amino acids using random forest based on submodularity subset selection. Computational Biology and Chemistry 53, Part B: 324–330. [DOI] [PubMed] [Google Scholar]
- 32. Liu ZP, Wu LY, Wang Y, Zhang XS, Chen LN (2010) Prediction of protein-RNA binding sites by a random forest method with combined features. Bioinformatics 26: 1616–1622. 10.1093/bioinformatics/btq253 [DOI] [PubMed] [Google Scholar]
- 33. Xiong D, Zeng J, Gong H (2015) RBRIdent: An algorithm for improved identification of RNA-binding residues in proteins from primary sequences. Proteins 10.1002/prot.24806 [DOI] [PubMed] [Google Scholar]
- 34. Si JN, Zhang ZM, Lin BY, Schroeder M, Huang BD (2011) MetaDBSite: a meta approach to improve protein DNA-binding sites prediction. Bmc Syst Biol 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cheng CW, Su ECY, Hwang JK, Sung TY, Hsu WL (2008) Predicting RNA-binding sites of proteins using support vector machines and evolutionary information. Bmc Bioinformatics 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tong J, Jiang P, Lu ZH (2008) RISP: A web-based server for prediction of RNA-binding sites in proteins. Comput Meth Prog Bio 90: 148–153. [DOI] [PubMed] [Google Scholar]
- 37. Murakami Y, Spriggs RV, Nakamura H, Jones S (2010) PiRaNhA: a server for the computational prediction of RNA-binding residues in protein sequences. Nucleic Acids Research 38: W412–W416. 10.1093/nar/gkq474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Choi S, Han K (2011) Prediction of RNA-binding amino acids from protein and RNA sequences. Bmc Bioinformatics 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fernandez M, Kumagai Y, Standley DM, Sarai A, Mizuguchi K, et al. (2011) Prediction of dinucleotide-specific RNA-binding sites in proteins. Bmc Bioinformatics 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li T, Li QZ (2012) Annotating the protein-RNA interaction sites in proteins using evolutionary information and protein backbone structure. J Theor Biol 312: 55–64. 10.1016/j.jtbi.2012.07.020 [DOI] [PubMed] [Google Scholar]
- 41. Xiong D, Zeng J, Gong H (2015) RBRIdent: An algorithm for improved identification of RNA-binding residues in proteins from primary sequences. Proteins 83: 1068–1077. 10.1002/prot.24806 [DOI] [PubMed] [Google Scholar]
- 42. Chen YC, Lim C (2008) Predicting RNA-binding sites from the protein structure based on electrostatics, evolution and geometry. Nucleic Acids Res 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Maetschke SR, Yuan Z (2009) Exploiting structural and topological information to improve prediction of RNA-protein binding sites. Bmc Bioinformatics 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Perez-Cano L, Fernandez-Recio J (2010) Optimal Protein-RNA Area, OPRA: A propensity-based method to identify RNA-binding sites on proteins. Proteins 78: 25–35. 10.1002/prot.22527 [DOI] [PubMed] [Google Scholar]
- 45. Zhao HY, Yang YD, Zhou YQ (2011) Structure-based prediction of RNA-binding domains and RNA-binding sites and application to structural genomics targets. Nucleic Acids Research 39: 3017–3025. 10.1093/nar/gkq1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Towfic F, Caragea C, Gemperline DC, Dobbs D, Honavar V (2010) Struct-NB: predicting protein-RNA binding sites using structural features. Int J Data Min Bioin 4: 21–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yan CH, Terribilini M, Wu FH, Jernigan RL, Dobbs D, et al. (2006) Predicting DNA-binding sites of proteins from amino acid sequence. Bmc Bioinformatics 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ofran Y, Mysore V, Rost B (2007) Prediction of DNA-binding residues from sequence. Bioinformatics 23: I347–I353. [DOI] [PubMed] [Google Scholar]
- 49. Hwang S, Gou ZK, Kuznetsov IB (2007) DP-Bind: a Web server for sequence-based prediction of DNA-binding residues in DNA-binding proteins. Bioinformatics 23: 634–636. [DOI] [PubMed] [Google Scholar]
- 50. Chu WY, Huang YF, Huang CC, Cheng YS, Huang CK, et al. (2009) ProteDNA: a sequence-based predictor of sequence-specific DNA-binding residues in transcription factors. Nucleic Acids Res 37: W396–W401. 10.1093/nar/gkp449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wu JS, Liu HD, Duan XY, Ding Y, Wu HT, et al. (2009) Prediction of DNA-binding residues in proteins from amino acid sequences using a random forest model with a hybrid feature. Bioinformatics 25: 30–35. 10.1093/bioinformatics/btn583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Andrabi M, Mizuguchi K, Sarai A, Ahmad S (2009) Prediction of mono- and di-nucleotide-specific DNA-binding sites in proteins using neural networks. Bmc Struct Biol 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Park B, Im J, Tuvshinjargal N, Lee W, Han K (2014) Sequence-based prediction of protein-binding sites in DNA: Comparative study of two SVM models. Comput Meth Prog Bio 117: 158–167. [DOI] [PubMed] [Google Scholar]
- 54. Kono H, Sarai A (1999) Structure-based prediction of DNA target sites by regulatory proteins. Proteins-Structure Function And Genetics 35: 114–131. [PubMed] [Google Scholar]
- 55. Jones S, Shanahan HP, Berman HM, Thornton JM (2003) Using electrostatic potentials to predict DNA-binding sites on DNA-binding proteins. Nucleic Acids Res 31: 7189–7198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bhardwaj N, Langlois RE, Zhao GJ, Lu H (2005) Kernel-based machine learning protocol for predicting DNA-binding proteins. Nucleic Acids Research 33: 6486–6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bhardwaj N, Lu H (2007) Residue-level prediction of DNA-binding sites and its application on DNA-binding protein predictions. Febs Lett 581: 1058–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tsuchiya Y, Kinoshita K, Nakamura H (2005) PreDs: a server for predicting dsDNA-binding site on protein molecular surfaces. Bioinformatics 21: 1721–1723. [DOI] [PubMed] [Google Scholar]
- 59. Gao M, Skolnick J (2008) DBD-Hunter: a knowledge-based method for the prediction of DNA-protein interactions. Nucleic Acids Research 36: 3978–3992. 10.1093/nar/gkn332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Xiong Y, Liu J, Wei DQ (2011) An accurate feature-based method for identifying DNA-binding residues on protein surfaces. Proteins 79: 509–517. 10.1002/prot.22898 [DOI] [PubMed] [Google Scholar]
- 61. Dey S, Pal A, Guharoy M, Sonavane S, Chakrabarti P (2012) Characterization and prediction of the binding site in DNA-binding proteins: improvement of accuracy by combining residue composition, evolutionary conservation and structural parameters. Nucleic Acids Research 40: 7150–7161. 10.1093/nar/gks405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wang DD, Li TH, Sun JM, Li DP, Xiong WW, et al. (2013) Shape string: A new feature for prediction of DNA-binding residues. Biochimie 95: 354–358. 10.1016/j.biochi.2012.10.006 [DOI] [PubMed] [Google Scholar]
- 63. Li T, Li QZ, Liu S, Fan GL, Zuo YC, et al. (2013) PreDNA: accurate prediction of DNA-binding sites in proteins by integrating sequence and geometric structure information. Bioinformatics 29: 678–685. 10.1093/bioinformatics/btt029 [DOI] [PubMed] [Google Scholar]
- 64. Li BQ, Feng KY, Ding J, Cai YD (2014) Predicting DNA-binding sites of proteins based on sequential and 3D structural information. Mol Genet Genomics 289: 489–499. 10.1007/s00438-014-0812-x [DOI] [PubMed] [Google Scholar]
- 65. Moult J, Fidelis K, Kryshtafovych A, Schwede T, Tramontano A (2014) Critical assessment of methods of protein structure prediction (CASP)—round x. Proteins 82 Suppl 2: 1–6. 10.1002/prot.24452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Miao Z, Adamiak RW, Blanchet MF, Boniecki M, Bujnicki JM, et al. (2015) RNA-Puzzles Round II: assessment of RNA structure prediction programs applied to three large RNA structures. Rna 21: 1066–1084. 10.1261/rna.049502.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cruz JA, Blanchet MF, Boniecki M, Bujnicki JM, Chen SJ, et al. (2012) RNA-Puzzles: a CASP-like evaluation of RNA three-dimensional structure prediction. Rna 18: 610–625. 10.1261/rna.031054.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Radivojac P, Clark WT, Oron TR, Schnoes AM, Wittkop T, et al. (2013) A large-scale evaluation of computational protein function prediction. Nature methods 10: 221–227. 10.1038/nmeth.2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wang GL, Dunbrack RL (2003) PISCES: a protein sequence culling server. Bioinformatics 19: 1589–1591. [DOI] [PubMed] [Google Scholar]
- 70. Huang Y-F, Huang C-C, Liu Y-C, Oyang Y-J, Huang C-K (2009) DNA-binding residues and binding mode prediction with binding-mechanism concerned models. Bmc Genomics 10 Suppl 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Luscombe NM, Laskowski RA, Thornton JM (2001) Amino acid–base interactions: a three-dimensional analysis of protein–DNA interactions at an atomic level. Nucleic Acids Res 29: 2860–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mcdonald IK, Thornton JM (1994) Satisfying Hydrogen-Bonding Potential In Proteins. J Mol Biol 238: 777–793. [DOI] [PubMed] [Google Scholar]
- 73. Bradley AP (1997) The use of the area under the roc curve in the evaluation of machine learning algorithms. Pattern Recogn 30: 1145–1159. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(XLSX)
Binding sites are defined by 3.5Å and 6Å defined sites are taken as prediction, accuracies are assessed and vice versa.
(XLSX)
(XLSX)
(EPS)
(EPS)
(EPS)
(EPS)
(EPS)
(EPS)
(EPS)
(EPS)
(EPS)
(EPS)
(EPS)
Distributions on other data sets are available as zip file on NBench website.
(EPS)
Case of poly(A)-binding protein (PDB id: 1cvj chain A). Residue F102 is mutated into His, Glu and Asp. Variations around F102 is easy to comprehend but two other regions close in space are also related. RBscore consider the spatial neighbors, and show difference in region 127–129 and region 172–179. BindN+ only show certain difference in the region 172–179. PPRInt and RNABindRPlus hardly show difference in prediction. As demonstrated by deep mutational scanning, the single point mutation to Glu and Asp obviously make difference in the binding and should also affect other residue neighbors. With a sequence based slide-window approach it is difficult to capture such structural differences.
(EPS)
Approach 1 is to input directly the residue information into the learning machine, approach 2 is first to map the residue information to a function and input the resulted feature vector into the learning machine. If we take the mapping step together with the learning machine as another learning machine, approach 2 is not different from approach 1.
(EPS)
The plot is based on test on data set ‘DR_bind1_R69’. X-axis show the distance cutoff used to define nucleic acid binding sites, while Y-axis show the resulted wAUC value. We find the wAUC values vary for different distance cutoffs, and the distributions for different programs are different.
(EPS)
Data Availability Statement
All data in this assessment are available on NBench web site http://ahsoka.u-strasbg.fr/nbench/.