Abstract
Phytohormone auxin is a master regulator in plant growth and development. Regulation of cellular auxin level plays a central role in plant development. Auxin polar transport system modulates an auxin gradient that determines plant developmental process in response to environmental conditions and developmental programs. Photolabile caged auxins allow optical control of artificial auxin gradients at cellular resolution. Especially, two-photon uncaging system achieves high spatiotemporal control of photolysis reaction at two-photon cross-section. However, the development of caged versions of auxin has been limited by the instability of the caged auxins to higher plant metabolic activities. Here, we describe the synthesis and application of highly stable caged auxins, 4-methoxy-7-nitroindolinyl (MNI)-caged auxins. Natural auxin, indole 3-acetic acid, and two synthetic auxins, 1-NAA and 2,4-D were caged by MNI caging group. MNI-caged auxins showed a high stability in planta and a rapid release the original auxin when photolyzed. We demonstrated that optical control of auxin-responsive gene expression and auxin-related physiological responses by using MNI-caged auxins. We anticipate that MNI-caged auxins will be an effective tool for high-resolution control of endogenous auxin level.
Keywords: Plant hormone, Auxin, Caged auxin, Two-photon uncaging
The plant hormone auxin plays a crucial role in almost every aspects of plant development including embryogenesis, cell elongation, vascular tissue differentiation, tropic responses to light and gravity, and lateral branching of shoots and roots.1,2 The cellular auxin levels are modulated by auxin biosynthesis, auxin polar transport, and auxin catabolism. Indole-3-acetic acid (IAA), the major naturally occurring auxin, is biosynthesized from tryptophan in the indole 3-pyruvate pathway and is polar transported to generate auxin concentration gradients in plant tissues.1–3 Auxin polar streams function in the establishment of the embryonic development, lateral development, and vascular patterning. Additionally, asymmetric redistribution of auxin in response to environmental and developmental cues drives the asymmetric tropic growth that allows plants to maintain positioning against the gravity vector and light. Therefore, spatiotemporal information of auxin levels would be crucial for dissecting the underlying mechanisms of the physiological roles of auxin gradients.3,4
Molecular biology and genetic studies have unraveled that the AUX1 family of auxin influx symporters, PIN family of auxin efflux carrier proteins, and ATP-binding cassette group B (ABCB) auxin transporters, coordinately modulate auxin transport.4 The cellular level and localization of auxin transport proteins on plasma membrane was thought to determine the direction and rate of auxin movement to generate asymmetric auxin gradient in plant tissue. Furthermore recent studies demonstrated that locally synthesized auxin also participates in the formation of auxin gradients, and cellular auxin levels are preciously maintained by auxin inactivating enzymes, such as GH3, IAA-amino acid conjugating enzymes and auxin oxidases.5–8 These studies indicated that auxin gradients would not only be formed by transport machinery, but also modulated by complicated processes involving local biosynthesis and inactivation of auxin. Visualization of cellular auxin gradients using auxin-responsive reporter lines has been widely utilized to dissect the role of auxin gradient formation in physiological response of plant.9–11 As an alternative approach, using an artificial auxin gradient system would benefit studies of the positional impact of auxin levels on diverse type of cells and tissue in plants.
A caged compound is an inactivated bioactive molecule and can rapidly release the original active molecule upon photolysis of a photo-cleavable protecting group (caging group) with optical devices.12,13 With caged compounds, the uncaging reaction can be spatiotemporally controlled at cellular level by precious control of light-irradiation. Additionally, the active molecule is intracellularly released, which can be repeated at any point during an experiment without physical perturbing. The intracellular levels of the released active molecule can be precisely controlled by the rate of uncaging.12,14 We previously demonstrated that 2′,5′-dimethoxyphenyl-2-nitrobenzyl (DMPNB)-caged auxins were useful tools to establish an artificial auxin gradient.15 DMPNB-caged auxin displayed useful features and advantages in comparison with classical methods of auxin application, such as a solution, a waxy paste, or an agar block containing auxin. Plant cells generally show higher metabolic capacity than mammal cells. Therefore, conventional caging groups used for mammalian biology are readily hydrolyzed by plant enzymes to release auxins without photolysis.15 DMPNB-caged auxin was designed to be resistant against esterases and to have considerable stability both in vitro and in vivo. DMPNB-caged auxins enable the manipulation of cellular auxin levels within a single cell by fine control of optical devices, suggesting that an artificial auxin gradient can be achieved at a desired position by means of a caged auxin system.
In this study, we synthesized MNI (4-methoxy-7-nitroindolinyl)-caged IAA, NAA and 2,4-D in which the carboxylic acid was protected by a secondary amine of MNI caging group.
MNI-caged auxins showed higher in vivo stability than DMPNB-caged auxins. Additionally, MNI-caged auxins can be used for two-photon uncaging system, as the MNI-caging group was originally developed to be a photolabile group for the two-photon excitation system,16 and the MNI-caged L-glutamate and MNI-caged-D-aspartate are widely used as commercial reagent for two-photon uncaging in neuroscience.13 Two-photon uncaging takes advantage of the high spatial resolution in comparison with one-photon uncaging, because photolysis was occurred at the cross-section of the two-photon excitation (Fig. 1A).17 Additionally, near infrared light (760 nm) used in two-photon excitation can reach into deeper tissue compared with UV light in one-photon excitation (360 nm). However, higher concentrations of caged molecules are required due to the low uncaging efficiency by two-photon excitation. We herein demonstrate the high stability of MNI-caged auxins in Arabidopsis plants and the photo-triggered spatiotemporal controls of auxin response in roots by one-photon excitation system.
Figure 1.
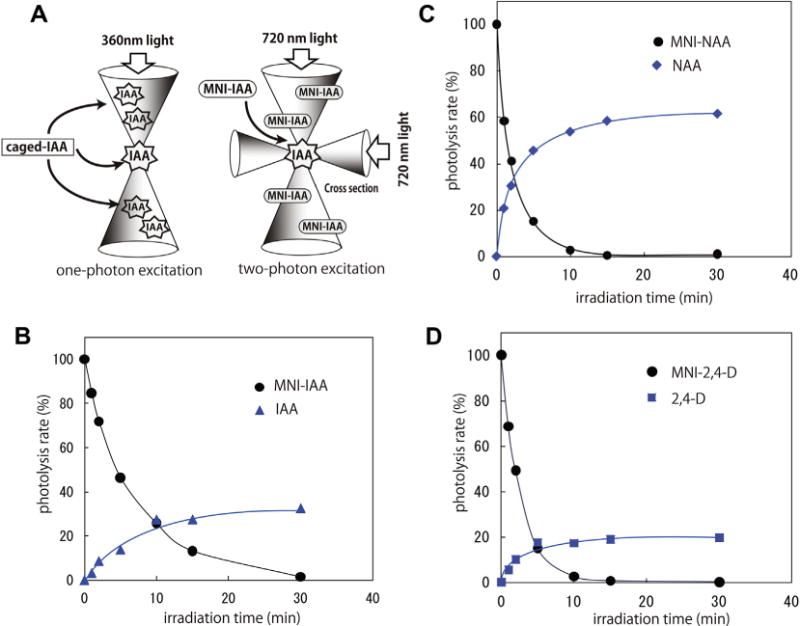
In vitro uncaging of MNI-auxins. (A) A schematic diagram for one-photon and two-photon excitation. (B–D) In vitro uncaging rates of MNI-caged auxins. (B) MNI-IAA, (C) MNI-NAA (D) MNI-NAA, 100 μM MNI-caged auxin solution (85% aqueous EtOH) was irradiated by a fluorophotometer (365 nm), and photolyzed solution were analyzed by HPLC at regular intervals. The amount of released auxins was plotted as the relative uncaged yield (%) from the initial amount of caged auxins.
Synthesis of MNI-caged auxins
Napthalene-1-acetic acid (NAA) and 2,4-dichlorophenoxy acetic acid (2,4-D), two well characterized synthetic auxins, and the natural auxin IAA are (Scheme 1) widely used for physiological experiments. NAA and 2,4-D exhibited a different transport profile from IAA. The transport profiles of the two typical synthetic auxins have been extensively characterized.3,4 PIN carrier proteins export NAA to outside of the cell, but NAA is not actively imported into the cell. In contrast, 2,4-D is imported by AUX1, but is not actively exported.4 Therefore, these two synthetic auxins have been used as diagnostic tools for influx or efflux auxin transport machinery.
Scheme 1.
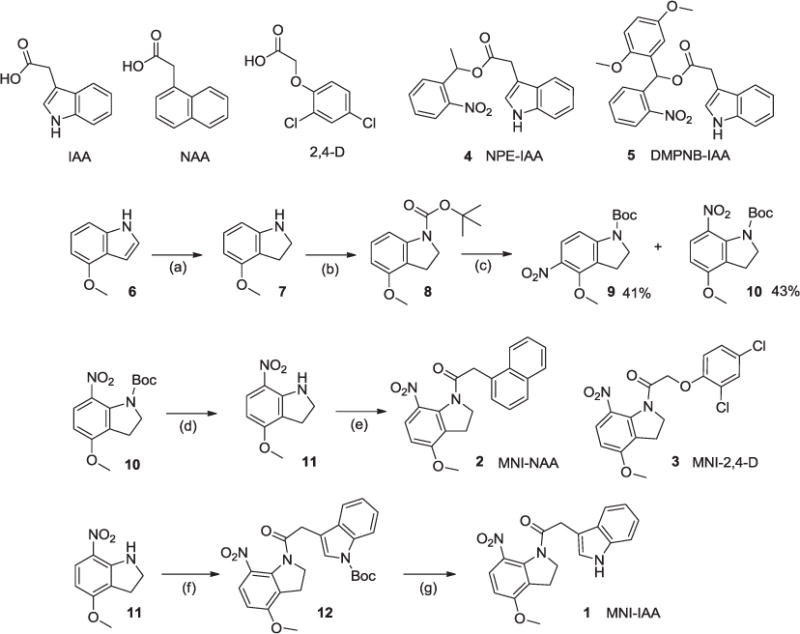
Reagents and conditions: (a) NaBH3CN in AcOH, rt, 3 h (98%); (b) di-tert-butyl dicarbonate, triethylamine, rt, 3 h (93%); (c) AgNO3, AcCl in CH3CN, rt, 2 h (41% for 9 and 43% for 10); (d) TFA in CH2Cl2, 0 °C, 0.5 h then rt, 3 h (93%); (e) 1-NAA or 2,4-D, SOCl2, in toluene, 75 °C, 8 h (56% for 2 and 63% for 3); (f) N-tert-butoxycarbonyl-indole 3-acetic acid, SOCl2, in toluene, 70 °C, 6 h (81%); (g) TFA in CH2Cl2, 0 °C, 0.5 h then rt, 3 h (94%).
As the initial step of auxin response, auxin is perceived by TIR1-Aux/IAA receptor complex in Arabidopsis. The crystal structures of the auxin-bound TIR1-Aux/IAA receptor complex revealed that aromatic ring and carboxylic acid in auxin molecule are essential for the affinity to the binding site of the receptor complex.18 Based on the structural information, caging of the carboxylic acid in auxin would diminish the binding activity of auxin molecules to yield physiologically inert caged auxin molecules.
Plant cells seem to show higher esterase activities and the sterically accessible ester linkage in caged compound, such as NPE-IAA (4) are readily hydrolyzed in planta. In our previous studies, DMPNB-caged auxin was designed to be esterase-resistant caged auxin.15 The introduction of the bulky 1,4-dimethoxyphenyl (DMP) ring into the 2-nitrobenzyl caging group, effectively blocked the hydrolysis of the ester of caged auxin by the plant esterases and consequently enhanced the stability of the caged auxin in vivo. However, the DMPNB-caged auxins were still slightly hydrolyzed in planta to release auxins during long-term cultivation. Therefore, we applied MNI caging group to the caged auxin system. In MNI-caged auxin, the carboxylic acid in auxin is caged by ternary amide with MNI group. Thus, MNI-caged auxins are expected to be completely inert to esterases and are resistant to general peptidases. Furthermore, in principle, MNI-caged auxin can be applicable to two-photon uncaging system as MNI caging group has been originally developed for two-photon excitation.17
The synthetic scheme of MNI-auxins is shown in Scheme 1. 4-Methoxyindole (6) was reduced by cyanoborohydride in acetic acid to give 4-methoxyindoline (7). This indoline was protected with tert-butoxycarbonyl (Boc) group by the treatment with di-tert-butyl dicarbonate and triethylamine in THF.19 The Boc-indoline (8) was nitrated with AgNO3 and acetyl chloride in CH3CN to yield a mixture of the 5- and 7-mononitro regioisomers (9 and 10), (in 1:1 ratio). 1-N-Boc-7-nitoro-4-methoxyindoline (10) was then deprotected with trifluoroacetic acid in CH2Cl2 to afford 7-nitoro-4-methoxyindoline (11). IAA was reacted with thionyl chloride and 7-nitoro-4-methoxyindoline (11) in toluene at 70 °C for 6 h to yield amido (12). This 1-N-Boc-MNI-IAA (12) was deprotected with trifluoroacetic acid to give a MNI-caged IAA (1) as pale yellow oil. Two typical synthetic auxins, NAA and 2,4-D were also caged by photolabile MNI group. MNI-caged NAA (2) and MNI-caged 2,4-D (3) were synthesized by the condensation of 7-nitoro-4-methoxyindoline (11) and synthetic auxins in the presence of thionyl chloride as shown in Scheme 1.
MNI-caged auxins are uncaged in vitro by light-irradiation
To estimate the uncaging efficiency of MNI-caged auxins, 100 μM MNI-caged auxins in the aqueous ethanol solution (85% EtOH) was light-irradiated by fluorometer (365 nm) and the photolyzed solution at regular intervals was analyzed by HPLC (Fig. 1B). MNI-IAA was photolyzed to release original IAA molecule in proportion to irradiation time until 5 min and MNI-IAA was completely consumed after 30 min irradiation. The release yield of IAA from MNI-IAA was ca. 30% at 30 min exposure. MNI-NAA and MNI-2,4-D was also rapidly photolyzed and released original auxins (Fig. 1C and D). Both MNI-NAA and MNI-2,4-D were completely disappeared by 15-min irradiation. The release yield of NAA and 2,4-D was reached to ca. 60% and 20% at 30-min exposure in our experimental condition, respectively.
MNI-caged auxins are highly stable in plant cells
Next, we validated the uncaging efficiency of MNI-caged auxin in culture medium and evaluated in vivo stability. The transgenic Arabidopsis auxin-responsive DR5::GUS reporter line was used for the evaluation of auxin level that was released from the MNI-caged auxins. The DR5::GUS reporter line contains the β-glucuronidase (GUS) reporter gene under the control of the synthetic auxin-responsive promoter DR5, and the expression of GUS reporter enzyme was specifically controlled by auxin in a dose-dependent manner.20 The auxin-induced GUS enzyme activity was estimated by the histochemical staining using chromogenic substrate X-Gluc and was fluorometrically quantified using 4-methyl umbelliferyl-β-D-glucuronide as a fluorogenic substrate. For the measurement of the uncaging efficiency in the culture medium, MNI-caged auxins (20 μM) in GM liquid medium was irradiated by using a UV lamp (360 nm), and the 5-days old DR5::GUS seedling was cultured in the photolyzed medium for 5 h in the dark (Fig. 2A–C, light irradiated). For the evaluation for intracellular stability of caged auxin, the DR5::GUS seedlings were incubated for 5 h (Fig. 2A–C, without light) and 24 h (Fig. 2D) in GM medium containing caged auxins in the dark. In this stability assay, caged auxin molecule would be constantly diffused from medium into the plant cells. The intracellular caged auxin would be converted to original auxin molecule, if caged auxin is acceptable substrate for the metabolic conversion catalyzed in planta.
Figure 2.
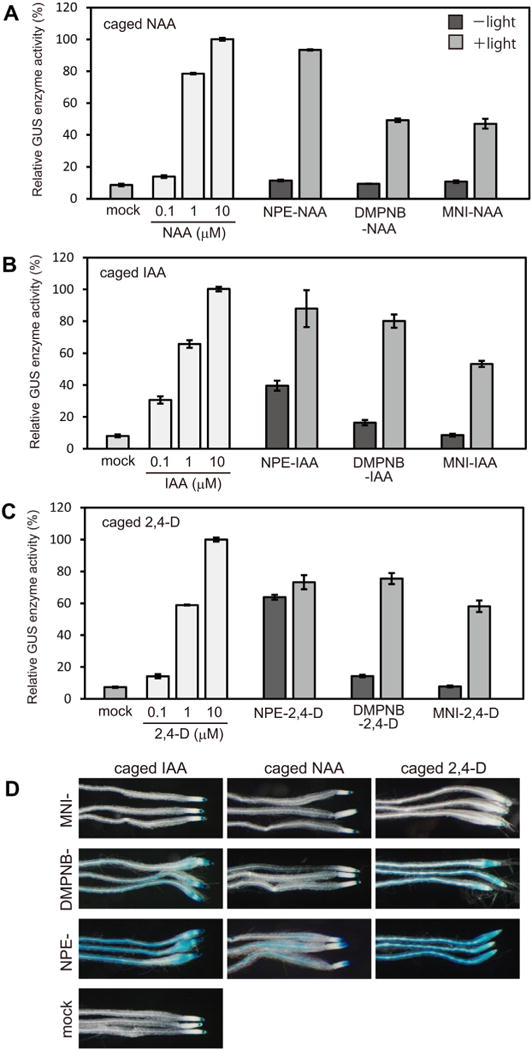
MNI-caged auxins are highly stable in planta and are uncaged by light-irradiation. (A–C) The caged auxins (20 μM) in the GM culture media were photolyzed by light irradiation (350–400 nm). 5-days-old Arabidopsis auxin-responsive DR5::GUS seedlings were incubated in the photolyzed medium (+light) or non-irradiated medium (−light) for 5 h in the dark. The GUS reporter enzyme was induced to the released auxins, and quantified by a fluorometer with a fluorescent substrate. The GUS activity was shown as normalized values to 100% values of 10 μM auxin treatment. Bar represents SD (n = 15). (D) DR5::GUS seedling was incubated with 20 μM caged auxin for 24 h in the dark without photolysis and then auxin-inducible DR5::GUS activity visualized by histochemical β-D-glucuronidase (GUS) staining.
In previous Letter, esterase resistant DMPNB-caged auxins were demonstrated to be more stable in plant cell than 2-nitrophenylethyl (NPE)-caged auxin. NPE group is widely used as conventional caging group in mammal biology, but NPE-IAA and NPE-2,4-D were unstable in plants. Consistent with previous Letters, DMPNB-caged auxins released original auxins upon photolysis, thereby induced DR5::GUS expression (Fig. 2A–C). Similarly, MNI-auxins in culture medium were uncaged by light to release original auxins and then lead to activation of DR5::GUS expression. However, uncaging efficiency of MNI-caged auxins was lower than those of DMPNB-caged auxins. Without photolysis, NPE-caged auxins activated DR5::GUS reporter expression after 5 h incubation (Fig. 2A–C). Contrary, DMPNB-caged auxin and MNI-caged auxins (20 μM) did not activate DR5::GUS expression by 5-h incubation without photolysis, suggesting that both DMPNB- and MNI-caged auxins were stable in the cell. However, after 24 h incubation without photolysis, DMPNB-caged IAA and DMPNB-caged 2,4-D were hydrolyzed to induce DR5::GUS expression visualized by blue staining with X-gluc substrate (Fig. 2D). In the control root, DR5:: GUS reporter gene has been expressed at root tip where endogenous IAA is accumulated (Figs. 2D and 3, mock). Similarly, MNI-caged auxin did not induce any DR5::GUS expression on root after 24 h incubation (Fig. 2D). Our results suggest that MNI-auxins were highly stable in planta and were resistant to the enzymatic hydrolysis by plant amidases.
Figure 3.
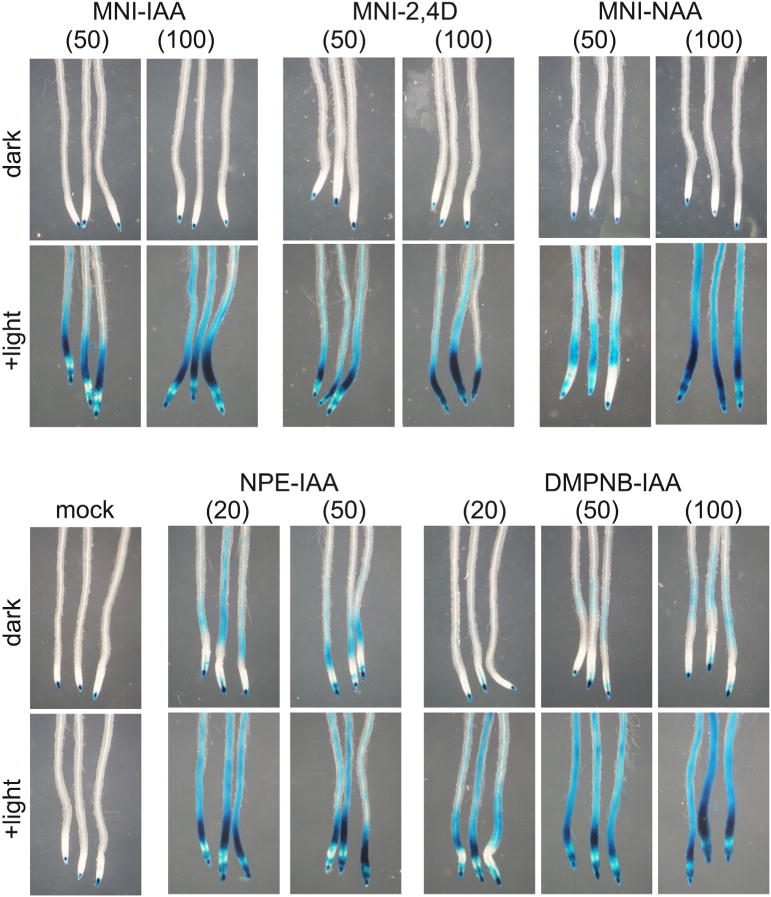
Uncaging of intracellular caged auxin in planta. 5-days-old DR5::GUS seedling was incubated in caged auxin solution for 30 min and then the seedlings were washed with fresh medium. After loading of caged auxins, the seedlings were placed on agar medium plate. The intracellular caged auxins were uncaged by light irradiation and then the seedling was incubated for 5 h in the dark. The auxin-induced GUS enzyme activity was visualized by histochemical blue staining. The values in parenthesis represent the concentration (μM) of caged auxin solution used for loading.
We next examined the manipulation of intracellular auxin levels by using MNI-auxins (Fig. 3). After the incorporation of MNI-caged auxins into the cells, intracellular MNI-caged auxins would be uncaged to directly release IAA inside the cells. To load the MNI-caged auxins to the cell, the 5-days-old DR5::GUS seedlings were incubated in 50 and 100 μM MNI-caged auxin solution (liquid medium) for 30 min, and then MNI-caged auxins outside the cells were washed out with fresh medium. The seedlings were placed on the glass slide covered with agar plate medium, and the roots were irradiated with a fluorescence microscope equipped with high pressure mercury lamp (>UV350 nm pass filter) for a few seconds. The DR5::GUS seedlings on the glass slide were then cultured for additional 5 h in the dark.
MNI-caged auxins were uncaged intracellularly to release auxins and activated the DR5::GUS expression, but no GUS reporter expression was observed without photolysis at 100 μM loading condition (Fig. 3). Contrary, NPE-caged auxins and DMPNB-caged auxins released auxin by light irradiation. However, over 50 μM loading condition, esterase-resistant DMPNB-caged auxins were slightly hydrolyzed without photolysis. This result clearly demonstrated that the MNI-caged auxin can be highly stable in the plant cell and could modulate the endogenous auxin level by controlling light irradiation.
MNI-caged auxins alter the auxin-regulated physiological responses in Arabidopsis plants
To examine consequences of spatiotemporal manipulation of intercellular auxin level in planta, we used transgenic Arabidopsis DII-VENUS line expressing a translational fusion protein of the Aux/IAA auxin-interaction domain (DII) and yellow fluorescent protein variant (VENUS) expressed under a constitutive 35S promoter.10 Auxin binds to TIR1/AFB auxin receptors and promotes the interaction between DII-VENUS and SCFTIR1 E3 ligase complex. DII-VENUS fusion protein is ubiquitinated in an auxin-dependent manner and rapidly decomposed via the ubiquitin–proteasome pathway. The amount of DII-VENUS protein in cell indirectly reflects the intracellular auxin concentration.10 The DII-VENUS seedlings were treated with auxin biosynthesis inhibitors, L-kynurenine (5 μM)21 and yucasin (50 μM)22 for 5 h. Because native DII-VENUS fluorescent image in root is already a reflect of endogenous IAA distribution. The DII-VENUS protein was uniformly accumulated and distributed after the inhibition of endogenous IAA production by the inhibitors. The DII-VENUS seedlings were then incubated with MNI-IAA for 20 min and then the root was immediately irradiated by light (Fig. 4).
Figure 4.
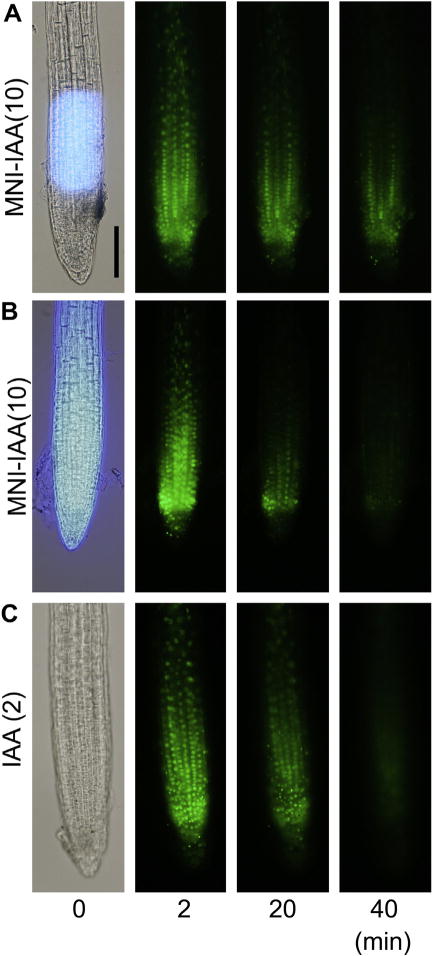
Spatiotemporal manipulation of cellular auxin level in planta. 5-days-old DII-VENUS root was incubated in 10 μM MNI-IAA solution for 20 min. The roots were immediately irradiated by light (360 nm) as a spot (A) or whole root (B). The roots were immersed in 2 μM IAA (C). The degradation of nuclear-localized DII-VENUS protein was monitored at 2, 20, and 40 min after exposure and IAA treatment. Bar represents 100 μm.
The time series displayed that the VENUS fluorescence in the nucleus disappeared by whole root irradiation of MNI-IAA treated root (Fig. 4B) and IAA treatment (Fig. 4C), whereas the degradation of DII-VENUS fluorescent signals in MNI-IAA treated root was limited within at light-irradiated area (Fig. 4A). Thus, MNI-caged auxin system can spatiotemporally manipulate intracellular auxin levels in planta by controlling light illumination.
We next investigated the light-control of auxin-regulated physiological responses (Fig. 5). Auxin represses primary root growth that is a typical rapid response to exogenous auxin. The Arabidopsis seedlings were vertically placed on agar medium after loading MNI-IAA into the roots. The seedling was vertically incubated in the dark for 5 h after light irradiation (5 min, UV350–360 nm). UV light alone did not affect the primary root growth (Fig. 5A). MNI-IAA inhibited the root growth after light irradiation (Fig. 5A), but did not without the irradiation.
Figure 5.
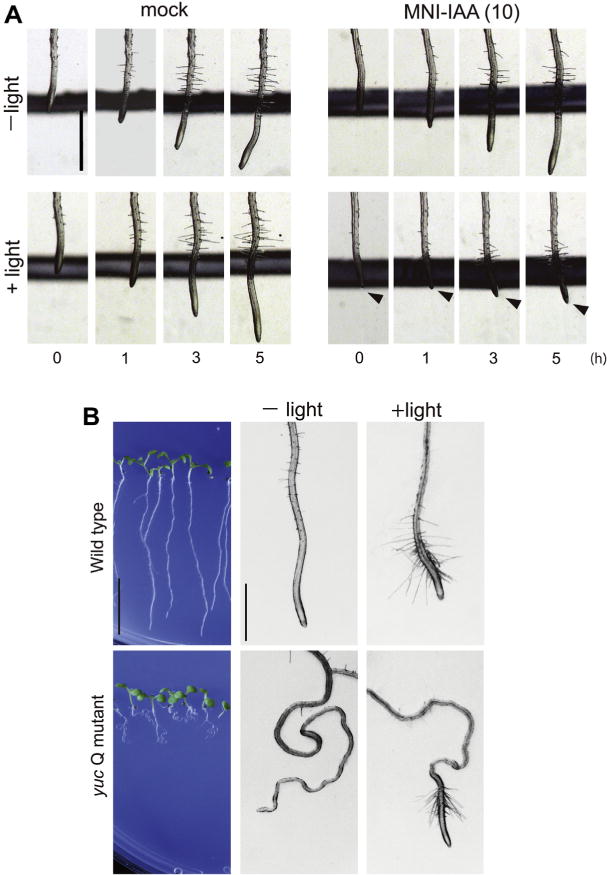
Manipulation of physiological auxin response by light irradiation using MNI-IAA. (A) 5-days-old seedlings were incubated in 10 μM MNI-IAA solution for 20 min. After washing with fresh medium, seedlings were irradiated on the agar medium plate for 5 min with light (350–400 nm). After photolysis, the seedlings were cultured vertically in the dark. Photograph was recorded at regular intervals. (B) 6-days-old yuc Q (yuc 3 5 7 8 9) mutant and wild type seedling were incubated in 50 μM MNI-IAA solution for 20 min and then uncaged with light. The seedlings were vertically cultured in the dark after photolysis for 15 h. Bar represent 10 mm and 1 mm each.
IAA is biosynthesized from tryptophan by two enzymes, the tryptophan aminotransferase TAA1 and YUCCA (YUC) enzymes in the Indole-3-pyruvate (IPA) pathway. YUCCA, flavin-monooxygenases catalyze the conversion of IPA to IAA and function as a rate-limiting enzyme in IPA pathway. Arabidopsis has 11 YUC family genes and the disruption of root expressing 5 YUC genes (yucca 3 5 7 8 9) showed severe auxin deficient root phenotypes.6 Especially, yucca quintuple (yuc Q) mutants show clear defects in root gravitropism and root hair formation.6 MNI-IAA was loaded to wild-type and yuc Q mutant plants and then intracellular MNI-IAA was uncaged on the GM agar plate by light irradiation. MNI-IAA promoted the root hair formation in wild-type root by photolysis. Furthermore, the defects in root hair formation and root gravitropic response (winding root) in yuc Q mutants were recovered by uncaging of MNI-IAA, suggesting that the MNI-IAA can manipulate endogenous auxin levels without toxic effects of uncaged MNI group on root development. This result demonstrated that the physiological auxin responses can be controlled by a light using the MNI-caged auxin system.
Caged auxin system is a promising approach to establish an artificial auxin gradient in a defined spatiotemporal manner. To investigate a physiological function of a spatiotemporal regulation of auxin gradient modulated by polar auxin transport, it is crucial to manipulate the cellular distribution of auxin. Two-photon uncaging system takes significant advantages of the high spatial resolution.13,17 However, cellular level of caged molecule in two-photon system requires considerably higher concentration than one-photon system. Therefore, caged molecule in two-photon system must be inert to metabolic pathways in cell and be non-toxic to the cell. MNI-caged auxins would satisfy these requirements to perform the two-photon uncaging by two-photon fluorescent microscopy. Very recently, DMPNB-caged auxin derivative in combination with auxin-inducible degradation system demonstrated light-controlled spatiotemporal regulation of conditional proteolysis in mammal cell. MNI-caged auxins might be directly applicable to these mammal system.23 We anticipate that MNI-caged auxins would be useful tools for spatiotemporal regulation for auxin gradient leading to the control of plant development.
Supplementary Material
Acknowledgments
This work was supported by Grants-in-Aid for Scientific Research (Japan) 15K01828 and 25114518 to K.H. and NIH (United States) GM114660 to Y.Z.
Footnotes
Supplementary data
Supplementary data (the experimental procedures and the spectroscopic data of new compounds) associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.bmcl.2015.09.001.
References and notes
- 1.Hayashi K. Plant Cell Physiol. 2012;53:965. doi: 10.1093/pcp/pcs035. [DOI] [PubMed] [Google Scholar]
- 2.Enders TA, Strader LC. Am J Bot. 2015;102:180. doi: 10.3732/ajb.1400285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adamowski M, Friml J. Plant Cell. 2015;27:20. doi: 10.1105/tpc.114.134874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petrasek J, Friml J. Development. 2009;136:2675. doi: 10.1242/dev.030353. [DOI] [PubMed] [Google Scholar]
- 5.Korasick DA, Enders TA, Strader LC. J Exp Bot. 2013;64:2541. doi: 10.1093/jxb/ert080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Q, Dai X, De-Paoli H, Cheng Y, Takebayashi Y, Kasahara H, Kamiya Y, Zhao Y. Plant Cell Physiol. 2014;1072:55. doi: 10.1093/pcp/pcu039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanaka K, Hayashi K, Natsume M, Kamiya Y, Sakakibara H, Kawaide H, Kasahara H. Plant Cell Physiol. 2014;55:218. doi: 10.1093/pcp/pct173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pencik A, Simonovik B, Petersson SV, Henykova E, Simon S, Greenham K, Zhang Y, Kowalczyk M, Estelle M, Zazimalova E, Novak O, Sandberg G, Ljung K. Plant Cell. 2013;25:3858. doi: 10.1105/tpc.113.114421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayashi K, Nakamura S, Fukunaga S, Nishimura T, Jenness MK, Murphy AS, Motose H, Nozaki H, Furutani M, Aoyama T. Proc Natl Acad Sci USA. 2014;111:11557. doi: 10.1073/pnas.1408960111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brunoud G, Wells DM, Oliva M, Larrieu A, Mirabet V, Burrow AH, Beeckman T, Kepinski S, Traas J, Bennett MJ, Vernoux T. Nature. 2012;482:103. doi: 10.1038/nature10791. [DOI] [PubMed] [Google Scholar]
- 11.Peer WA, Jenness MK, Murphy AS. Physiol Plant. 2014;151:97. doi: 10.1111/ppl.12182. [DOI] [PubMed] [Google Scholar]
- 12.Ellis-Davies GC. Nat Methods. 2007;4:619. doi: 10.1038/nmeth1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellis-Davies GCR. Beilstein J Org Chem. 2013;9:64. doi: 10.3762/bjoc.9.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayashi K, Hashimoto K, Kusaka N, Yamazoe A, Fukaki H, Tasaka M, Nozaki H. Bioorg Med Chem Lett. 2006;16:2470. doi: 10.1016/j.bmcl.2006.01.103. [DOI] [PubMed] [Google Scholar]
- 15.Kusaka N, Maisch J, Nick P, Hayashi K, Nozaki H. ChemBioChem. 2009;10:2195. doi: 10.1002/cbic.200900289. [DOI] [PubMed] [Google Scholar]
- 16.Matsuzaki M, Ellis-Davies GCR, Nemoto T, Miyashita Y, Iino M, Kasai H. Nat Neurosci. 2001;4:1086. doi: 10.1038/nn736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ellis-Davies GC. ACS Chem Neurosci. 2011;2:185. doi: 10.1021/cn100111a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. Nature. 2007;446:640. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 19.Fedoryak OD, Sul JY, Haydon PG, Ellis-Davies GC. Chem Commun. 2005;3664 doi: 10.1039/b504922a. [DOI] [PubMed] [Google Scholar]
- 20.Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Plant Cell. 1997;9:1963. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He W, Brumos J, Li H, Ji Y, Ke M, Gong X, Zeng Q, Li W, Zhang X, An F, Wen X, Li P, Chu J, Sun X, Yan C, Yan N, Xie DY, Raikhel N, Yang Z, Stepanova AN, Alonso JM, Guo H. Plant Cell. 2011;23:3944. doi: 10.1105/tpc.111.089029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishimura T, Hayashi K, Suzuki H, Gyohda A, Takaoka C, Sakaguchi Y, Matsumoto S, Kasahara H, Sakai T, Kato J, Kamiya Y, Koshiba T. Plant J. 2014;77:352. doi: 10.1111/tpj.12399. [DOI] [PubMed] [Google Scholar]
- 23.Delacour Q, Li C, Plamont MA, Billon-Denis E, Aujard I, Le Saux T, Jullien L, Gautier A. ACS Chem Biol. 2015 doi: 10.1021/acschembio.5b00069. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


