Abstract
To elucidate the molecular mechanisms of stomatal opening and closure, we performed a genetic screen using infrared thermography to isolate stomatal aperture mutants. We identified a mutant designated low temperature with open-stomata 1 (lost1), which exhibited reduced leaf temperature, wider stomatal aperture, and a pale green phenotype. Map-based analysis of the LOST1 locus revealed that the lost1 mutant resulted from a missense mutation in the Mg-chelatase I subunit 1 (CHLI1) gene, which encodes a subunit of the Mg-chelatase complex involved in chlorophyll synthesis. Transformation of the wild-type CHLI1 gene into lost1 complemented all lost1 phenotypes. Stomata in lost1 exhibited a partial ABA-insensitive phenotype similar to that of rtl1, a Mg-chelatase H subunit missense mutant. The Mg-protoporphyrin IX methyltransferase (CHLM) gene encodes a subsequent enzyme in the chlorophyll synthesis pathway. We examined stomatal movement in a CHLM knockdown mutant, chlm, and found that it also exhibited an ABA-insensitive phenotype. However, lost1 and chlm seedlings all showed normal expression of ABA-induced genes, such as RAB18 and RD29B, in response to ABA. These results suggest that the chlorophyll synthesis enzymes, Mg-chelatase complex and CHLM, specifically affect ABA signaling in the control of stomatal aperture and have no effect on ABA-induced gene expression.
Electronic supplementary material
The online version of this article (doi:10.1007/s10265-014-0636-0) contains supplementary material, which is available to authorized users.
Keywords: ABA, Arabidopsis thaliana, Infrared thermography, Mg-chelatase I subunit 1, Mg-protoporphyrin IX methyltransferase, Stomatal guard cells
Introduction
Stomatal pores in the plant epidermis, each surrounded by a pair of guard cells, regulate gas exchange between plants and the atmosphere to control processes such as CO2 uptake for photosynthesis and transpiration for water loss regulation (Shimazaki et al. 2007). Stomatal opening is induced by light, whereas stomatal closure is induced by the phytohormone abscisic acid (ABA), which is synthesized in response to drought stress (Assmann and Shimazaki 1999; Schroeder et al. 2001). ABA-induced stomatal closure is driven by an efflux of K+ from guard cells through outward-rectifying K+ channels in the plasma membrane. Activation of K+ channels requires depolarization of the plasma membrane; this depolarization is achieved mainly through anion channels in the plasma membrane (Joshi-Saha et al. 2011; Kim et al. 2010; Negi et al. 2008; Schroeder et al. 1987; Vahisalu et al. 2008).
Recently, the PYR/PYL/RCAR (Pyrabactin resistance/Pyrabactin resistance 1-like/Regulatory component of ABA receptor) family of proteins was identified as an ABA receptor in Arabidopsis thaliana and ABA recognition by PYR/PYL/RCAR family proteins was shown to activate SnRK2 family protein kinases through inactivation of the central negative regulator type 2C protein phosphatases (PP2Cs) (Cutler et al. 2010; Ma et al. 2009; Park et al. 2009; Santiago et al. 2009). A pyr1/pyl1/pyl2/pyl4 quadruple mutant exhibited strong ABA-insensitive phenotypes for seed germination, root growth, gene expression (Park et al. 2009), and stomatal opening and closing (Nishimura et al. 2010) indicating functional redundancy among the PYR/PYL/RCAR family proteins. ABA has been suggested to activate SLOW ANION CHANNEL-ASSOCIATED 1 (SLAC1), which is thought to be a slow-type anion channel (Negi et al. 2008; Vahisalu et al. 2008), via PYR/PYL/RCAR-PP2Cs-SnRK2 modules followed by depolarization of the plasma membrane (Geiger et al. 2009; Lee et al. 2009). In addition to PYR/PYL/RCAR, several candidate ABA receptors have been reported including the Mg-chelatase H subunit (CHLH) (Du et al. 2012; Shen et al. 2006; Wu et al. 2009), G-protein coupled receptor 2 (GCR2) (Liu et al. 2007), and G-protein coupled receptor-type G proteins (GTG1 and GTG2) (Pandey et al. 2009). However, it was also reported that CHLH is not an ABA receptor (Müller and Hansson 2009; Tsuzuki et al. 2011). Therefore, the question of whether CHLH indeed functions as an ABA receptor remains controversial. Likewise, GCR2 is also controversial (Klingler et al. 2010).
CHLH is a multifunctional protein involved in chlorophyll synthesis (Bollivar 2006; Masuda 2008), plastid-to-nucleus retrograde signaling (Mochizuki et al. 2001), and ABA signaling. In chlorophyll synthesis, CHLH functions as a subunit of Mg-chelatase consisting of three subunits I, D, and H that are encoded by the CHLI, CHLD, and CHLH genes, respectively. Mg-chelatase catalyzes the insertion of Mg2+ into protoporphyrin IX, which is the first step of the chlorophyll synthesis pathway after diverging from the tetrapyrrole biosynthesis pathway. In the next step, Mg-protoporphyrin IX is methylated by Mg-protoporphyrin IX methyltransferase (CHLM) and four subsequent catalytic reactions produce chlorophyll a (Bollivar 2006; Masuda 2008).
Recently, the rapid transpiration in detached leaves 1 (rtl1) mutant bearing a novel missense mutation in CHLH was isolated from ethyl methanesulfonate (EMS)-treated Arabidopsis thaliana using a screen for mutants with altered stomatal aperture (Tsuzuki et al. 2011). Stomatal movement in the rtl1 mutant was insensitive to ABA, but the effects of ABA on seed germination and root growth were normal. CHLI1, a major Mg-chelatase I subunit isoform in Arabidopsis (Huang and Li 2009), has also been reported to affect ABA signaling. Stomata of chli1, a knockout mutant of CHLI1, showed an ABA-insensitive phenotype (Tsuzuki et al. 2011). Du et al. (2012) also reported that the stomata of cs, a T-DNA insertion knockdown mutant of CHLI1 and CHLI-RNAi lines showed an ABA-insensitive phenotype. These results indicated that the Mg-chelatase complex, including CHLH and CHLI1, is likely to affect the ABA-signaling pathway in guard cells. However, the molecular mechanism by which CHLH and CHLI1 mediate ABA-signaling in guard cells remains largely unknown.
The molecular mechanisms of signaling pathways that control stomatal opening and closure are largely unknown. Several screens for stomatal aperture mutants using infrared thermography under drought, low or high CO2, or blue light-illumination conditions identified mutants that showed a reduced temperature phenotype due to wide stomatal aperture or an elevated temperature phenotype due to constricted stomatal aperture. These studies identified important components involved in the regulation of stomatal opening or closure such as OPEN STOMATA (OST) 1 and 2 (Merlot et al. 2002, 2007; Mustilli et al. 2002), HIGH LEAF TEMPERATURE 1 (HT1) (Hashimoto et al. 2006), SLAC1 (Negi et al. 2008), STOMATAL CARPENTER 1 (SCAP1) (Negi et al. 2013), BLUE LIGHT SIGNALING 1 (BLUS1) (Takemiya et al. 2013), and PROTON ATPASE TRANSLOCATION CONTROL 1 (PATROL1) (Hashimoto-Sugimoto et al. 2013) indicating that infrared thermography is a potent tool for identification of stomatal aperture mutants.
In this study, we performed a screen using infrared thermography under normal growth conditions to elucidate molecular mechanisms of stomatal opening and closure and isolated a mutant designated low temperature with open-stomata (lost) 1. The lost1 mutant had a novel missense mutation in the CHLI1 gene and exhibited a partial ABA-insensitive phenotype similar to that of a CHLH mutant. Moreover, we showed that a subsequent enzyme in chlorophyll synthesis, CHLM, also affects ABA signaling in the control of stomatal aperture. Our data suggest that chlorophyll synthesis enzymes may play an important role in ABA signaling in stomatal guard cells.
Materials and methods
Plant materials and growth conditions
All Arabidopsis (Arabidopsis thaliana) mutants, glabra1 (gl1), phototropin2-1 (phot2 mutant), chli1, chlm, npq2, gun2-1 and gun3-1 were in the Col background and were grown in soil under a 16-h fluorescent light (6:00 A.M. to 10:00 P.M.; 50 µmol m−2 s−1)/8-h dark cycle at 24 °C at a relative humidity of 55–70 % in a growth room (Kinoshita et al. 2001; Mochizuki et al. 2008). The T-DNA insertional mutants chli1 (SAIL_230_D11) and chlm (SALK_110265) were obtained from the Arabidopsis Biological Resource Center (ABRC, Columbus, OH, USA). Homozygous chli1 mutant plants were identified by PCR using the T-DNA left-border primer, LB1, and a CHLI1 gene-specific primer. Homozygous chlm mutant plants were identified by PCR using the T-DNA left border primer, LBb1.3, and a CHLM gene-specific primer. Primers are shown in Table S1.
Mutant screening using infrared thermography
Ten thousand phot2 (M1) seeds were treated with ethyl methanesulfonate (EMS; Sigma-Aldrich) and EMS-mutagenized M2 seeds were prepared as described previously (Lightner and Caspar 1998). Approximately 80 M2 plants were grown for 2–3 weeks in soil in each pot. Leaf temperatures of the mutants were measured in a growth room using a TVS-500EX infrared thermography instrument (NEC Avio Infrared Technologies Co., Ltd.) and analyzed with the Avio Thermography Studio software. Leaf temperature measurements were taken in the centers of the mature leaves (11–25 leaves/mutant line). Individual mutant candidates with a leaf temperature more than 1 °C below the temperatures of the leaves of surrounding plants were selected. The candidates were further selected by re-measurement of leaf temperature in the M3 generation. The selected mutants were grown for 4 weeks and subjected to stomatal aperture measurement. We isolated three cold mutants designated as low temperature with open-stomata (lost) 1, lost2, and lost3.
Identification of the lost1, lost2, and lost3 loci
To identify the mutant loci, we performed genetic mapping in the lost mutants. The mutants were crossed with plants of the Landsberg erecta (Ler) ecotype and F2 plants were obtained. F2 plants with visible mutant phenotypes including pale green leaves for lost1 and reduced leaf temperature, wilting, and dark-green rosette leaves for lost2 and lost3 were selected and genomic DNA was isolated from the individual mutant plants (13–20 plants/mutant). Genetic mapping was performed using simple sequence length polymorphism (SSLP) markers and cleaved amplified polymorphism (CAPS) markers.
Measurement of stomatal aperture
Stomatal aperture was measured as described previously (Inoue et al. 2008) with minor modifications. Fully expanded rosette leaves were harvested in the dark from 5 to 7-week-old plants. The leaves were blended in a Waring blender equipped with an MC1 mini container (Waring Commercial) in 35 mL of MilliQ water. The epidermal fragments were collected on nylon mesh and rinsed with MilliQ water. The epidermal tissues were incubated in basal buffer (5 mM MES-Bistrispropane, 50 mM KCl, and 0.1 mM CaCl2, pH 6.5) and were irradiated with red/blue mixed light [blue light (Stick-B-32; EYELA, Tokyo, Japan) at 10 µmol m−2 s−1 combined with background red light (LED-R; EYELA) at 50 µmol m−2 s−1] for 2.5 h at 24 °C in the presence of 1, 10, or 20 µM ABA or equal volumes of dimethyl sulfoxide (DMSO). Stomatal apertures were measured microscopically in the abaxial epidermis by focusing on the inner lips of stomata. All measurements of stomatal response to light were performed between 11:00 A.M. and 3:00 P.M. To measure the apertures under growth conditions, the epidermal fragments were isolated as described above at zeitgeber time (ZT) 5–9 and were immediately subjected to microscopic measurement.
Measurement of chlorophyll contents
The chlorophyll contents of rosette leaves from 5- to 6-week-old plants were determined as described previously (Tsuzuki et al. 2011).
Construction of plant transformation vector
To complement the lost1 mutant with the wild-type CHLI1 gene, we constructed a gene transfer vector bearing the genomic CHLI1 gene under the control of the native CHLI1 promoter. The genomic CHLI1 gene, extending from −1,063 to 2,576 bp of the genomic CHLI1 locus including 5′ and 3′ noncoding sequences, was amplified by PCR from wild-type (Col-0) genomic DNA using primers shown in Table S1. The amplified 3,639-bp DNA fragment was treated with EcoRI and cloned into the gene transfer vector pCAMBIA1300 using the EcoRI site. The resulting vector was verified by DNA sequencing.
Transformation of Arabidopsis
The gene transfer vector was introduced into the Agrobacterium tumefaciens strain GV3101 and used to transform the lost1 mutant (Clough and Bent 1998). The transformed plants were selected on MS plates containing 1 % (w/v) sucrose and 50 µg ml−1 hygromycin. Complementation testing was performed using independent homozygous transgenic lines from the T3 generation.
RT–PCR and quantitative RT–PCR
Expression of CHLI1 (At4g18480) and CHLM (At4g25080) in the lost1 and chlm mutants, respectively, was determined by RT–PCR. Total RNA was extracted from rosette leaves of four- to six-week-old plants using an RNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA), according to the manufacturer’s instructions. First-strand cDNAs were synthesized from 0.4 to 1 µg of total RNA using a Takara PrimeScript II First Strand cDNA Synthesis Kit (Takara, Tokyo, Japan) with oligo(dT)12–18 primer. The cDNA fragments were amplified by PCR using specific primers. TUB2 (At5g62690) was amplified by PCR to serve as an internal standard.
We determine the expression of the CHLI1, CHLH (At5g13630), and CHLM genes in guard-cell protoplasts (GCPs) of wild type (Col-0) using RT–PCR. GCPs were isolated from 5-week-old Col-0 plants as described previously (Ueno et al. 2005). Total RNA was extracted and first strand cDNAs were prepared from the GCPs as described above. CHLI1, CHLM, CHLH, and TUB2 cDNAs were amplified by PCR using specific primers shown in Table S1.
We used quantitative RT–PCR to determine the expression of the ABA-responsive genes RAB18 (At5g66400) and RD29B (At5g52300) in response to ABA in phot2, lost1, wild type (Col-0), chli1, and chlm plants. Two-week-old seedlings were incubated in liquid MS medium (pH 5.8) containing 50 µM ABA or an equal volume of DMSO for 3 h at 24 °C under light (50 µmol m−2 s−1). Total RNA was extracted and first strand cDNAs were prepared from the ABA-treated plants as described above. Quantitative RT–PCR was performed using Power SYBR Green PCR Master Mix and a StepOne™ Real-Time PCR system (Applied Biosystems, Carlsbad, CA, USA), as reported previously (Takahashi et al. 2012). RAB18, RD29B, and TUB2 cDNAs were amplified by PCR using specific primers (Table S1). Relative quantification was performed using the comparative cycle threshold method and the relative amount of the amplified RAB18 or RD29B product was normalized to that of TUB2, which served as an internal control.
Immunoblotting
Immunoblotting was performed as described previously (Tsuzuki et al. 2011) with minor modifications. Rosette leaves were harvested from 6-week-old plants and ground in extraction buffer (50 mM MOPS-KOH, pH 7.5, 100 mM NaCl, 2.5 mM EDTA, 2 mM DTT, 1 mM phenylmethylsulfonyl fluoride, and 200 nM leupeptin) using a mortar and pestle. Fifty micrograms of protein were loaded onto a gel and separated by SDS–polyacrylamide gel electrophoresis. Polyclonal antibodies against CHLH (Tsuzuki et al. 2011) and monoclonal antibody against actin (Sigma-Aldrich) were used at a 3,000-fold dilution.
Results
Isolation of stomatal aperture mutants by infrared thermography
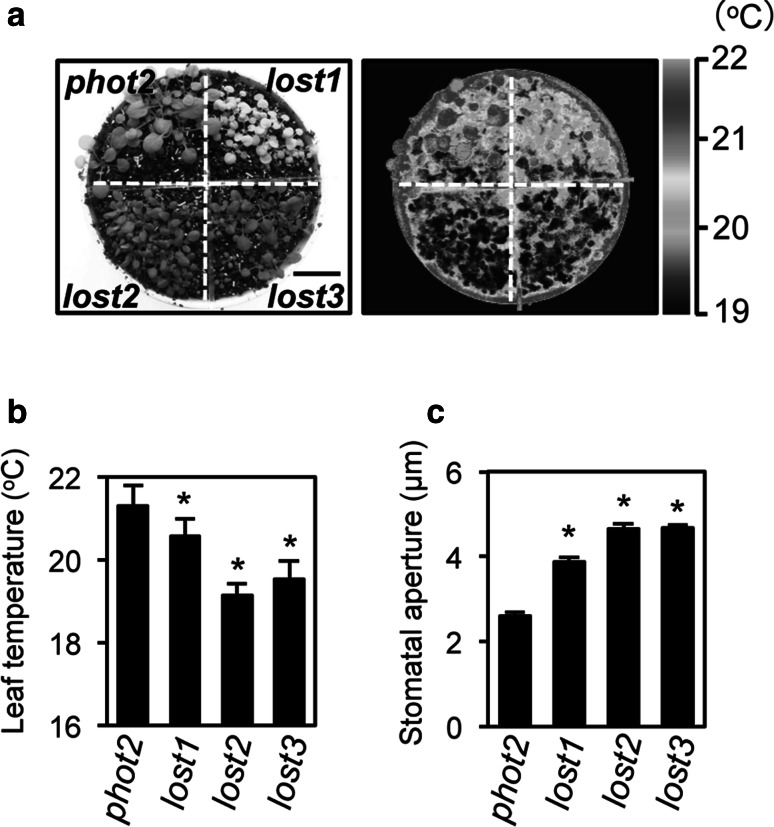
To elucidate the mechanisms of stomatal opening and closure, we performed a screen to identify stomatal aperture mutants using leaf temperature as measured by infrared thermography under illumination as an indicator of stomatal aperture. We selected mutants from a population of M2 plants generated by EMS-treatment of Arabidopsis thaliana. The phot2 mutant was used as a reference mutant background to avoid redundant effects of the blue light receptors, phot1 and phot2, in the blue light-induced stomatal opening (Kinoshita et al. 2001). We isolated three mutants that exhibited reduced leaf temperature that were designated as low temperature with open-stomata (lost) 1, lost2, and lost3. Average leaf temperatures of the lost1, lost2, and lost3 mutants were reduced by 0.7, 2.2, and 1.8 °C, respectively, relative to the control phot2 mutant (Fig. 1a, b), with significantly wider stomatal aperture compared to phot2 under growth conditions (Fig. 1c).
Fig. 1.
Leaf temperature and stomatal aperture in lost mutants. a Thermal image (right) and the corresponding bright-field image (left) of lost mutants and the control phot2 mutant. The images of the 3-week-old plants were taken under illuminated growth conditions. The scale bar indicates 2 cm. b Leaf temperatures of the lost and phot2 mutants were calculated from quantification of the thermal image in a. Values are means of 11–25 leaves with standard deviations (SDs). Asterisks indicate a significant statistical difference relative to the control phot2 by the t test (P < 0.01). The experiments were repeated on three occasions with similar results. c Stomatal aperture of the lost and phot2 mutants under illuminated growth conditions. Epidermal fragments were isolated from the plants at ZT5–9 and immediately subjected to measurement of stomatal aperture. Values are means of three independent experiments with standard errors (SEs) (n = 25). Asterisks indicate a significant statistical difference relative to the control phot2 by the t test (P < 0.01)
lost2 and lost3 are ABA-deficient mutants
As shown in Fig. 1, lost2 and lost3 showed strong leaf temperature and stomatal aperture phenotypes. We performed map-based cloning of the LOST2 and LOST3 loci and found that the lost2 mutation was caused by a missense mutation (Gly169 to Asp) in the ABA DEFICIENT 2 (ABA2, At1g52340) gene, which encodes short-chain dehydrogenase/reductase. The lost3 mutation was due to a nonsense mutation (Gln575 to stop codon) in the ABA DEFICIENT 3 (ABA3, At1g16540) gene, which encodes molybdenum cofactor sulfurase (Fig. S1) (Nambara and Marion-Poll 2005). To determine if the lost2 and lost3 mutants were ABA-deficient mutants, we examined NaCl-tolerance during seed germination (Léon-Kloosterziel et al. 1996). Seeds of the control phot2 mutant did not germinate in the presence of 200 mM NaCl, but lost2 and lost3 seeds germinated as well as did seeds of the ABA-deficient mutant npq2 (aba1) (Niyogi et al. 1998) (Fig. S1e) indicating that lost2 and lost3 are likely to be ABA-deficient mutants. Note that aba2 and aba3 were isolated previously as mutants showing low temperature and open-stomata phenotypes using infrared thermography (Merlot et al. 2002) indicating that the screening approach used in this study is effective for the isolation of stomatal aperture mutants.
Low leaf temperature and open stomata phenotypes in lost1 leaves are caused by a missense mutation in CHLI1
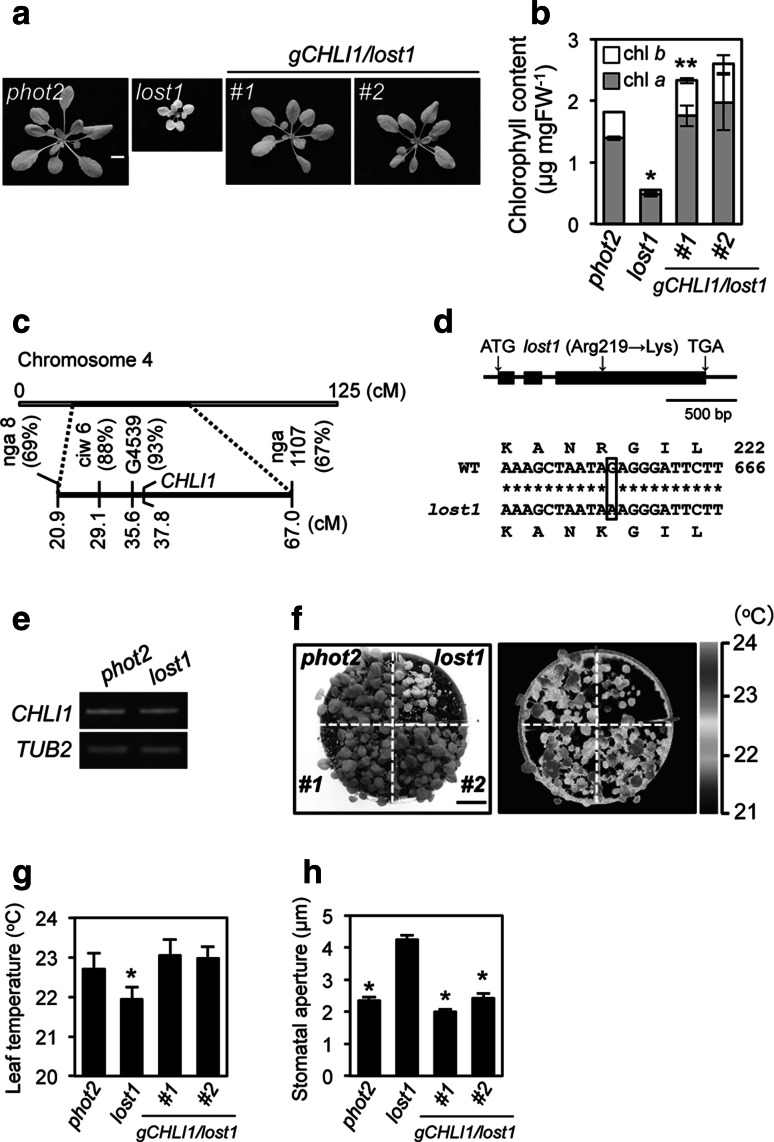
The lost1 mutant was recessive and showed semi-dwarf and pale green phenotypes with decreased chlorophyll content (Fig. 2a, b). Based on map-based cloning, the LOST1 locus showed strong linkage to the CAPS marker G4539 on chromosome 4 (Fig. 2c). We searched The Arabidopsis Information Resource (TAIR) database and found that Mg-chelatase I subunit 1 (CHLI1, At4g18480) is located close to G4539 and that chli1 mutants are known to show a pale green phenotype. Sequence analysis of genomic DNA from lost1 plants revealed that the lost1 mutation results from a single nucleotide substitution from G to A at nucleotide 656 in the CHLI1 gene, which causes a missense mutation from Arg to Lys at amino acid 219 (Fig. 2d). Transformation of the wild-type CHLI1 gene under its own promoter into lost1 complemented the semi-dwarf, pale green, low leaf temperature, and open stomata lost1 phenotypes (Fig. 2) indicating that the lost1 phenotypes were due the mutation in the CHLI1 gene.
Fig. 2.
A missense mutation in CHLI1 gene is responsible for the lost1 phenotype. a Typical phenotypes in control phot2, lost1, and two independent complementation lines gCHLI1/lost1 (#1 and #2). Plants were grown in soil under illuminated growth conditions for 5 weeks. The scale bar represents 1 cm. b Chlorophyll contents of phot2, lost1, and gCHLI1/lost1 (#1 and #2) rosette leaves. Chlorophyll contents were calculated based on fresh weight (FW) of leaves. Chlorophyll a and chlorophyll b contents are indicated by gray and white colors, respectively. Values are means of three independent experiments with SDs. Asterisks indicate a significant difference in total chlorophyll content relative to the control phot2 by the t test (*P < 0.01 and **P < 0.05). c Mapping analysis of the LOST1 locus. Numbers in parentheses indicate percentages of no recombination in 40 chromosomes. The LOST1 locus was close to CAPS marker G4539 and Mg-chelatase I subunit 1 (CHLI1). d Determination of the mutation in the lost1 mutant. The genomic structure of the CHLI1 gene on chromosome 4 is shown (upper). Black boxes and bars indicate exons and introns, respectively. Partial CHLI1 cDNA sequences and the deduced amino acid sequences from wild-type and lost1 are shown (lower). The lost1 mutant results from a G to A nucleotide substitution in the third exon. The nucleotide change causes the substitution of Lys for Arg at amino acid 219. Nucleotide and amino acid numbers are indicated in the right. Asterisks indicate the same nucleotide of the CHLI1 gene in wild type and lost1. e CHLI1 expression analyzed by RT–PCR in lost1 and the control phot2. Total RNA was extracted from 4-week-old seedlings. TUB2 was used as an internal standard for cDNA amounts. All PCRs were performed with 25 cycles. The experiments were repeated on three occasions with similar results. f Thermal image (right) and the corresponding bright-field image (left) of control phot2, lost1, and gCHLI1/lost1 (#1 and #2). The images were obtained and are presented as in Fig. 1a. g Leaf temperatures of control phot2, lost1, and gCHLI1/lost1 (#1 and #2). Leaf temperature was calculated from quantification of the infrared image f as described in Fig. 1b. Values are means of 12–16 leaves with SDs. Asterisk indicates a significant difference in leaf temperature relative to the control phot2 by the t test (P < 0.01). The experiments were repeated on three occasions with similar results. h Stomatal apertures under illuminated growth conditions in control phot2, lost1, and gCHLI1/lost1 (#1 and #2). Apertures were measured and presented as described in Fig. 1c. Values are means of 25 stomata measurements with SEs. Asterisks indicate a significant difference in stomatal aperture relative to lost1 by the t test (P < 0.01). The experiments were repeated on three occasions with similar results
Stomata of lost1 exhibit a partially ABA-insensitive phenotype
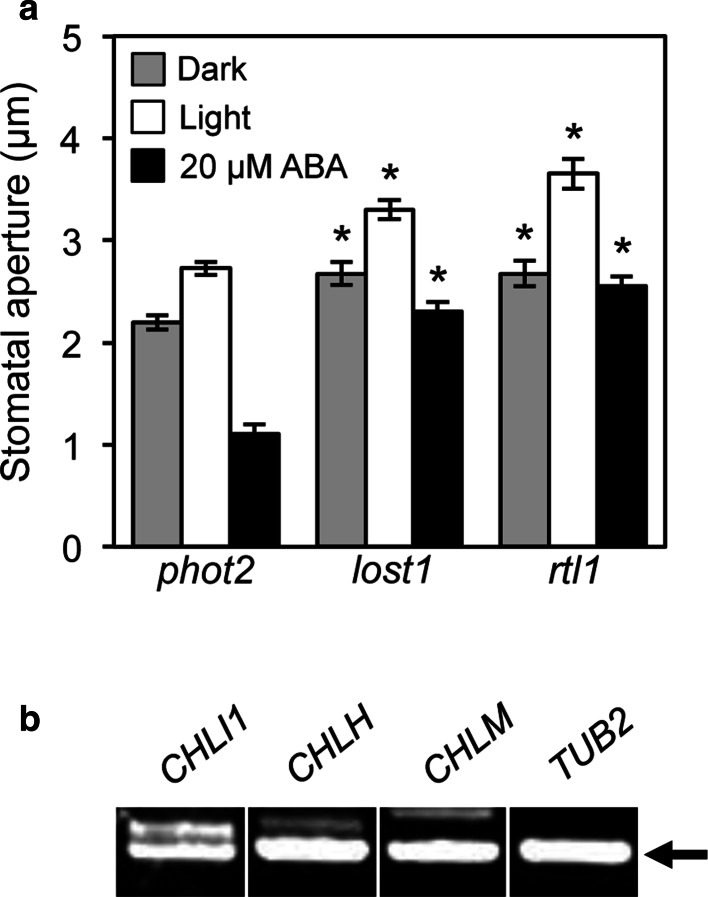
Mg-chelatase consists of three subunits I, D, and H (Gibson et al. 1995; Willows et al. 1996). The Mg-chelatase H subunit (CHLH) was shown to affect ABA signaling in stomatal guard cells (Du et al. 2012; Legnaioli et al. 2009; Shen et al. 2006; Tsuzuki et al. 2011; Wu et al. 2009). Moreover, chli1 mutants also showed an ABA-insensitive phenotype in stomatal guard cells (Du et al. 2012; Tsuzuki et al. 2011) (Fig. S2a). We examined ABA-sensitivity in lost1 relative to rapid transpiration in detached leaves 1 (rtl1), a missense mutant of CHLH (Fig. 3a) (Tsuzuki et al. 2011). The control phot2 plants showed light-induced stomatal opening, but in the presence of ABA (20 µM), phot2 stomata exhibited reduced aperture than under dark. The lost1 plants showed wider stomatal aperture than phot2 under both dark and light conditions. ABA suppressed light-induced stomatal opening in lost1 and rtl1, but the stomatal apertures were not reduced as in phot2 (Fig. 3a), indicating that lost1 is a partial ABA-insensitive mutant with respect to stomatal opening. It should be noted that a CHLI1-knockout and CHLI1-knockdown mutants showed a complete ABA-insensitive phenotype for stomatal movement (Fig. S2a) (Tsuzuki et al. 2011; Du et al. 2012), indicating that the Arg219 to Lys missense mutation in lost1 is a weak mutation for ABA-sensitivity. In addition, we tested ABA contents in leaves of lost1 and rtl1 (Fig. S4). However, there was no significant difference between lost1 and the control plant as well as rtl1 in our condition, in which an ABA-deficient mutant npq2 showed significant difference in ABA content, suggesting that the open-stomata phenotype of lost1 is not due to decrease of ABA content.
Fig. 3.
Stomatal phenotype in lost1 and rtl1 and expression of CHLI1, CHLH, and CHLM in guard cells. a Effect of ABA on light-induced stomatal opening in lost1, rtl1, and control phot2 mutants. Epidermal peels from dark-adapted plants in buffer containing 20 µM ABA or equal volumes of DMSO were irradiated with 10 µmol m−2 s−1 blue light and 50 µmol m−2 s−1 red light. Values are means of 25 stomata measurements with SEs. Asterisks indicate a significant difference in each treatment relative to phot2 by the t test (P < 0.01). The experiments were repeated on three occasions with similar results. b Expression of chlorophyll synthetic enzyme, CHLI1, CHLH, and CHLM, in wild-type (Col) guard cells. CHLI1, CHLH, and CHLM mRNA amounts were analyzed by RT–PCR. Total RNA was extracted from GCP isolated from 5-week-old wild type plants. TUB2 was used as an internal standard for cDNA amounts. All PCRs were performed with 35 cycles. Bands amplified by RT–PCR are indicated by an arrow. The experiments were repeated on three occasions with similar results
We also examined the expression of CHLI1 and CHLH in wild-type guard cell protoplasts (GCPs) by RT–PCR. As shown in Fig. 3b, CHLI1 and CHLH expression was detected in GCPs. Consistent with this, the available expression database, Arabidopsis eFP Browser, also indicates that CHLI1 and CHLH are expressed in guard cells. These results suggest that Mg-chelatase might affect ABA signaling within guard cells. Further investigations will be needed to clarify this by the complementation experiments using guard cell-specific promoter, such as GC1 (Kinoshita et al. 2011; Wang et al. 2014; Yang et al. 2008).
ABA induces expression of ABA-responsive genes in lost1
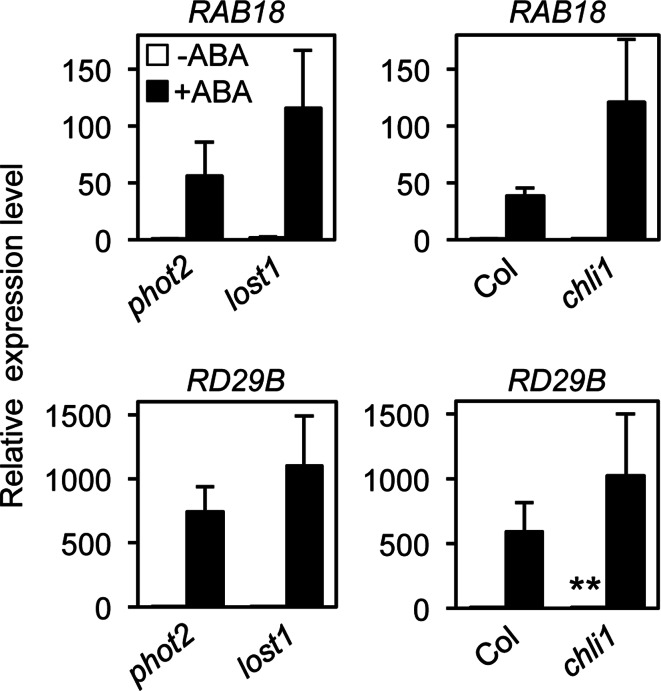
We performed quantitative RT–PCR to determine the expression levels of representative ABA-responsive genes, RAB18 and RD29B (Fujii et al. 2007; Leonhardt et al. 2004) in seedlings (Fig. 4). Incubation of phot2 seedlings in 50 µM ABA for 3 h induced RAB18 and RD29B expression. The expression levels of these genes also increased in lost1 seedlings in response to ABA. Consistent with this, even a strong chli1 allele also showed ABA-induced gene expression similar to that in background Col, suggesting that CHLI1 has no effect on ABA-induced gene expression. In addition, seed germination and root growth in lost1 showed normal ABA sensitivity similar to that in the control phot2 and rtl1 (Fig. S3) (Tsuzuki et al. 2011). These results indicated that the ABA-insensitive phenotype in lost1 is specific for stomatal movement.
Fig. 4.
ABA-induced gene expression in Mg-chelatase subunit mutants. Seedlings of lost1 and chli1 mutants were treated with 50 µM ABA or equal volumes of DMSO for 3 h. Relative mRNA levels of the ABA-induced RAB18 and RD29B genes were quantified by quantitative RT–PCR. Values are means of three independent experiments with SDs. Asterisks indicate a significant difference in each treatment relative to the control plants by the t test (P < 0.05). TUB2 was used as an internal standard for cDNA amounts
CHLM also affects ABA signaling
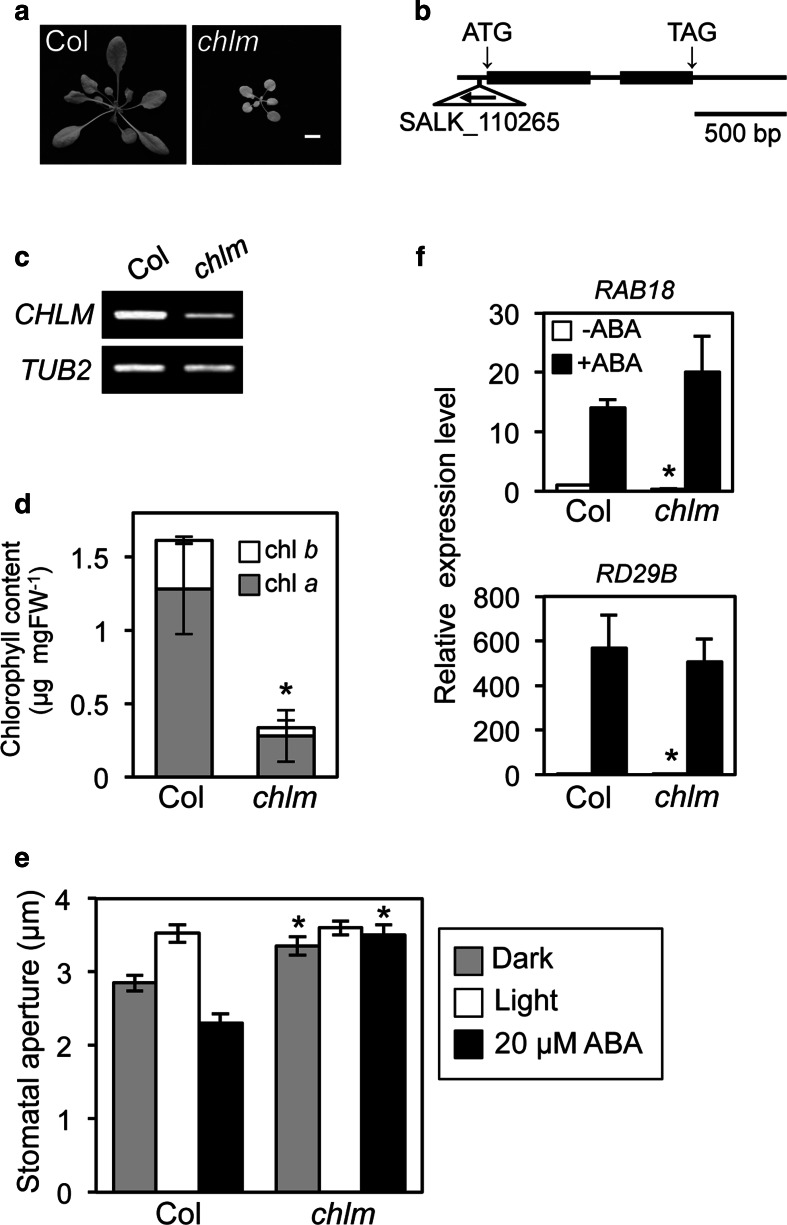
We examined ABA-sensitivity in the chlm mutant, a knockdown mutant of the Mg-protoporphyrin IX methyltransferase (CHLM) gene, to investigate whether other chlorophyll synthesis enzymes affect ABA signaling in guard cells. CHLM is an enzyme that acts after Mg-chelatase in the chlorophyll synthesis pathway (Bollivar 2006; Masuda 2008). Semi-dwarf and pale green phenotypes similar to lost1 and rtl1 phenotypes were seen in chlm plants and CHLM expression was greatly reduced (Fig. 5a–d). Stomata in chlm showed wider stomatal aperture in the dark than in background Col, and an almost equivalent aperture under illumination. Interestingly, chlm showed a completely ABA-insensitive phenotype with respect to stomatal movement (Fig. 5e). CHLM expression was detected in GCPs. Consistent with this, the available expression database, Arabidopsis eFP Browser, also indicates that CHLM is expressed in guard cells. These results suggest that CHLM probably affects ABA signaling within guard cells (Fig. 3b). In contrast, chlm plants showed normal ABA-induced expression of the RAB18 and RD29B genes (Fig. 5f) and ABA-dependent suppression of seed germination and root growth as well as rtl1 (Fig. S3) (Tsuzuki et al. 2011), indicating that CHLM specifically affects ABA signaling in stomatal movement.
Fig. 5.
The chlm mutant exhibited an ABA-insensitive phenotype for stomatal response. a Typical phenotypes of wild type (Col) and chlm plants. Plants were grown in soil under illuminated growth conditions for 5 weeks. The scale bar represents 1 cm. b Schematic structure of the CHLM gene and the locus of T-DNA insertion in the chlm mutant. Exons and introns are indicated by black boxes and bars, respectively. The T-DNA insertion was located at −41 bp in the 5′UTR of the CHLM gene. c Analysis of CHLM and TUB2 expression by RT–PCR in chlm and wild-type plants. Total RNA was extracted from 6-week-old seedlings. TUB2 was used as an internal standard for cDNA amounts. All PCRs were performed with 30 cycles. The experiments were repeated on three occasions with similar results. d Chlorophyll contents of chlm and wild-type rosette leaves. Chlorophyll contents were calculated based on fresh weight (FW) of leaves. Chlorophyll a and chlorophyll b contents are indicated by gray and white colors, respectively. Values are means of three independent experiments with SDs. Asterisk indicates a significant difference in total chlorophyll content relative to the wild type by the t test (P < 0.01). e Inhibition of light-induced stomatal opening by ABA in the chlm mutant. Stomatal apertures were measured and presented as described in Fig. 3a. Values are means of 25 stomatal measurements with SEs. Asterisks indicate a significant difference in each treatment relative to the wild type by the t test (P < 0.01). The experiments were repeated on three occasions with similar results. f ABA-induced gene expression in the chlm mutant. Relative mRNA levels of ABA-induced genes were quantified and presented as described in Fig. 4. Values are means of three independent experiments with SDs. Asterisks indicate a significant difference in each treatment relative to the wild type by the t test (P < 0.01). TUB2 was used as an internal standard for cDNA amounts
Mg-chelatase subunit and CHLM mutants accumulate CHLH protein in leaves
CHLH protein was reported to accumulate to high levels in CHLI1, CHLH, and CHLM mutants (Huang and Li 2009; Pontier et al. 2007; Tsuzuki et al. 2011). We examined the amount of CHLH protein in the leaves of the lost1, rtl1, chli1, and chlm mutants. Consistent with previous reports, all mutants showed high accumulation of CHLH protein (Fig. S5). The amount of CHLH protein in these mutants was four- to ten-fold higher than in control plants.
Discussion
Infrared thermal imaging is a useful tool for isolation of stomatal aperture mutants (Merlot et al. 2002). In this study, we isolated three mutants that exhibited low leaf temperature and open stomata phenotypes. These mutants were designated low temperature with open-stomata (lost) 1, lost2, and lost3. The lost2 and lost3 mutants were deemed to be ABA-deficient mutants with mutations in the ABA2 and ABA3 genes, respectively (Figs.1, S1). The aba2 and aba3 mutants were isolated in a previous study using infrared thermography (Merlot et al. 2002) indicating that the screening approach used in this study is reliable.
The lost1 mutant contained an Arg219 to Lys missense mutation in Mg-chelatase I subunit 1 (CHLI1) and showed a partial ABA-insensitive phenotype for stomatal movement and reduced chlorophyll content. Furthermore, transformation of the wild-type CHLI1 gene into lost1 complemented these phenotypes (Fig. 2). These results indicate that CHLI1 is responsible for the lost1 phenotypes. Mg-chelatase consists of three subunits I, D, and H (Gibson et al. 1995; Willows et al. 1996). The Arabidopsis genome has two CHLI genes, CHLI1 and CHLI2, and CHLI1 is thought to be the major isoform of Mg-chelatase I subunits in Arabidopsis (Huang and Li 2009). CHLI1 belongs to the AAA+ (ATPases associated with various cellular activities)-family of ATPases and the ATPase activity drives the Mg insertion reaction (Masuda 2008). Arg219 is located close to an ATP-binding Walker B motif in CHLI1 (Fodje et al. 2001), thus the lost1 mutation affects Mg-chelatase activity suggesting that Mg-chelatase activity is important not only for chlorophyll synthesis but also for ABA signaling in guard cells.
ABA-induced expression of ABA-responsive genes, such as RD29A and MYB2, was inhibited in the leaves of CHLH knockdown mutants (Shen et al. 2006) suggesting that CHLH is a positive regulator for ABA-responsive genes. In contrast, a recent investigation indicated that CHLH mutants such as cch and rtl1 showed normal ABA-induced expression of the ABA-responsive RAB18 and RD29B genes in guard cells (Tsuzuki et al. 2013). Therefore, whether CHLH mediates ABA-induced expression of ABA-responsive genes remains controversial. In this study, we examined the effect of CHLI1 on ABA-induced expression of ABA-responsive genes in seedlings by quantitative RT-PCR and found that the CHLI1 mutants, lost1 and chli1, had no effect on ABA-induced expression of the ABA-responsive RAB18 and RD29B genes (Fig. 4). In addition, seed germination and root growth in the lost1 missense mutant showed normal ABA sensitivity as in the control phot2 (Fig. S3). Taken together, these results indicate that Mg-chelatase complex including CHLH and CHLI1 probably has no effect on ABA-induced expression of ABA-responsive genes or ABA-dependent inhibition of seed germination and root growth.
Mg-chelatase catalyzes the insertion of Mg2+ into protoporphyrin IX, which is the first step of the chlorophyll synthesis pathway after the branch point from the tetrapyrrole biosynthesis pathway. Therefore, we next examined whether Mg-protoporphyrin IX methyltransferase (CHLM), which is a subsequent enzyme in chlorophyll synthesis, is involved in the regulation of stomatal movement. Interestingly, chlm, a knockdown mutant of CHLM, also showed an ABA-insensitive phenotype for stomatal response (Fig. 5e). In addition, the chlm mutant showed normal ABA-induced expression of the ABA-responsive RAB18 and RD29B genes and ABA-dependent inhibition of seed germination and root growth (Figs. 5f, S3). Taken together, these results indicate that CHLM also specifically affects ABA signaling in stomatal movement but not in gene expression or seed germination and root growth. To our knowledge, this is the first evidence that CHLM affects ABA signaling in stomatal movement.
Interestingly, lost1, chli1, and chlm accumulated the endogenous CHLH protein (Fig. S5), but these mutants showed ABA-insensitive phenotype in stomatal movement (Figs. 3, S2a). On the other hand, CHLH overexpression in wild-type plants showed ABA-hypersensitive phenotype in stomatal movement (Tsuzuki et al. 2013). These results suggest that CHLH protein accumulation is not sufficient for complementation of the ABA-insensitive phenotype of these mutants, and that CHLI1 and CHLM may be required in addition to CHLH for a proper ABA response in stomatal movement. Further investigation will be needed to clarify whether CHLH over-accumulates in guard cells of these mutants, since the present data obtained from leaves (Fig. S5). CHLM antisense transgenic tobacco plants showed reduced Mg-chelatase activity (Alawady and Grimm 2005). Therefore, it is possible that decreased CHLM expression in the chlm-knockdown mutant affects ABA signaling by impairing Mg-chelatase activity. Further investigation is needed to clarify how CHLM affects ABA signaling via Mg-chelatase activity in guard cells. It is noteworthy that cytosolic Ca2+ is suggested to be important for the regulation of CHLH-mediated ABA signaling in guard cells (Tsuzuki et al. 2011). Therefore, further investigations of intercellular Ca2+ changes in guard cells of CHLH, CHLI1, and CHLM mutants will provide important information on the physiological role and molecular mechanism of CHLH, CHLI1, and CHLM in the ABA-signaling in stomatal guard cells.
Note that ch1-2, which is a loss-of-function mutant of chlorophyll a oxygenase converting from chlorophyll a to chlorophyll b showed normal ABA-response in stomatal movement (Shen et al. 2006). However, whether four enzymes (Mg-protoporphyrin IX monomethyl ester cyclase, 3,8 divinyl-protochlorophyllide 8 vinyl reductase, protochlorophyllide reductase, and chlorophyll synthase) subsequent to CHLM in the chlorophyll synthesis pathway from Mg-protoporphyrin IX monomethyl ester to chlorophyll a (Bollivar 2006; Masuda 2008) are involved in the regulation of stomatal movement is still unknown. Therefore, the effects of these four enzymes on stomatal movement should be examined.
Protoporphyrin IX is catalyzed not only by Mg-chelatase in the chlorophyll synthesis pathway, but also by the enzymes in phytochromobilin synthesis pathway (Mochizuki et al. 2010; Tanaka et al. 2011). It is possible that the phytochromobilin synthesis pathway also affects ABA-signaling in guard cells. Then, we examined stomatal phenotype in gun2-1 and gun3-1, mutants of heme oxygenase and phytochromobilin synthase in the phytochromobilin synthesis, respectively (Mochizuki et al. 2008). Interestingly, these mutants showed open-stomata and partial ABA-insensitive phenotypes (Fig. S2b). These results suggest that the phytochromobilin synthesis pathway also affects ABA-signaling in guard cells, although Shen et al. (2006) showed normal ABA-induced stomatal closure in gun2 and gun3 mutants. It should be noted that gun2-1 and gun3-1 mutants are phytochromobilin-deficient mutants and have no functional phytochromes (Parks and Quail 1991). However, it has been reported that phyB acts as a positive regulator for light-induced stomatal opening (Wang et al. 2010). Therefore, open-stomata phenotype in gun2-1 and gun3-1 is unlikely due to deficient of phytochromes. Further investigation will be needed to clarify how the phytochromobilin synthesis pathway affects ABA-signaling in guard cells.
Electronic supplementary material
Fig. S1 lost2 and lost3 mutation loci. a Mapping analysis of the LOST2 locus. Numbers in parentheses indicate percentages of no recombination in 40 chromosomes. The LOST2 locus is located close to SSLP marker ciw1 and ABA DEFICIENT 2 (ABA2). b Determination of the mutation in the lost2 mutant gene. The ABA2 gene genomic structure on chromosome 1 is shown (upper). Black boxes and bars indicate exons and introns, respectively. Partial ABA2 cDNA sequences and the deduced amino acid sequences from the wild type and lost2 are shown (lower). The lost2 mutant results from a G to A nucleotide substitution in the second exon. The nucleotide change causes the substitution of Asp for Gly at amino acid 169. Nucleotide and amino acid numbers are indicated on the right. Asterisks indicate the same nucleotide of the ABA2 gene in the wild type and lost2. c Mapping analysis of the LOST3 locus. Numbers in parenthesis indicate the percentage of no recombination in 26 chromosomes. The LOST3 locus is located close to SSLP marker F20D23 and ABA DEFICIENT 3 (ABA3). d Determination of the mutation in the lost3 mutant gene. The genomic structure of the ABA3 gene on chromosome 1 is shown (upper) as described in Figure S1b. Partial ABA3 cDNA sequences and the deduced amino acid sequences in wild type and lost3 are shown (lower) as described in b. The lost3 mutant contains a C to T nucleotide substitution in exon 16. The nucleotide change results in the substitution of a stop codon for a glutamine codon at residue 575. e Effect of NaCl on lost2, lost3, npq2, and control phot2 seed germination. The seed germination assay was performed as described previously (Leon-Kloosterziel et al. 1996) with some modifications. Approximately 25 sterilized seeds were placed on a plate containing MS medium (pH 5.8), 2 % sucrose, and 0.8 % agar with or without 200 mM NaCl. The plate was kept at 4 °C for three days, then incubated at 24 °C under illuminated growth conditions for 1 week. The germination rate was determined from the percentage of germinated seeds with green cotyledons. The experiments were repeated on three occasions with similar results. N.D., not detected (TIFF 2519 kb)
Fig. S2 Effect of ABA on light-induced stomatal opening in chli1 (a), gun2 (b), gun3 (b), and the wild type. Epidermal peels in buffer containing 20 µM ABA or equal volumes of DMSO were irradiated with 10 µmol m−2 s−1 blue light and 50 µmol m−2 s−1 red light. Values are means of 25 stomata measurements with SEs. Asterisks indicate a significant difference in each treatment relative to the wild type by the t test (P < 0.01). The experiments were repeated on three occasions with similar results (TIFF 1843 kb)
Fig. S3 ABA sensitivity of seed germination and root growth in Mg-chelatase subunit and CHLM mutants. The seed germination and root growth assay was performed as described previously (Tsuzuki et al. 2011) with some modifications. For seed germination assay, seeds were placed on a plate containing MS medium (pH 5.8), 2 % sucrose, 0.8 % agar, and 1 µM ABA or equal volumes of DMSO. The plate was kept at 4 °C for three days and then incubated at 24 °C under illuminated growth conditions. a Effect of ABA on germination of lost1 and control phot2, chlm, and wild-type seeds. Ten-day-old lost1, rtl1, control phot2, chlm, and wild-type plants grown on MS medium containing 1 µM ABA or equal volumes of DMSO. The scale bar represents 1 cm. b The germination rate was determined by the percentage of germinated seeds (n = 50) showing radicle emergence at the indicated days after the start of stratification. Values are means of three independent experiments with SDs. For root growth assay, four-day-old seedlings grown on a MS plate under light were transferred to MS medium containing 20 µM ABA or equal volumes of DMSO. c Effect of ABA on root growth of lost1 and control phot2, chlm, and wild-type seeds. Ten-day-old lost1, rtl1, control phot2, chlm, and wild-type plants grown on MS medium containing 20 µM ABA or equal volumes of DMSO. The scale bar represents 1 cm. d The root length was measured at 7 days after transferring to the medium containing ABA. Values are means of 25 seedlings measurements with SEs. The experiments were repeated on three occasions with similar results (TIFF 13147 kb)
Fig. S4 ABA contents of phot2, lost1, rtl1, wild type (Col) and npq2 in rosette leaves. The ABA concentrations in rosette leaves of four- to seven-week-old plants were measured with Phytodetek ABA Test Kit (Agdia), as described previously (Kim et al. 2009) with some modifications. For ABA extraction, 200 mgFW tissue was ground in liquid nitrogen and suspended with 250 µl of MilliQ water, and agitated over night at 4 °C. The suspension was centrifuged at 4,000 rpm for 20 min, and the supernatant was collected into a clean tube and dried under vacuum. The dry residue was dissolved with 24 µl of MilliQ water, and diluted by 1:25 in TBS. The ABA concentration was determined with a Phytodetek ABA Test Kit (Agdia). The absorbance at 405 nm was detected using microplate reader (ARVO X3, ParkinElmer). Values are means of three independent experiments with SDs. Asterisk indicates a significant difference in ABA content relative to the control plant by the t-test (P < 0.01).Kim MJ, Shin R, Schachtman DP (2009) A nuclear factor regulates abscisic acid responses in Arabidopsis. Plant Physiol 151:1433–1445 (TIFF 646 kb)
Fig. S5 Immunoblot analysis of CHLH protein in Mg-chelatase subunit and CHLM mutants. Each lane was loaded with 50 µg of protein extracted from rosette leaves of lost1, rtl1, phot2, chli1, chlm, and wild-type plants. Actin protein served as an internal control. The experiments were repeated on three occasions with similar results (TIFF 640 kb)
Acknowledgments
We thank Dr. Takuya Matsuo (Nagoya University) for technical advice on measurement of ABA contents. This work was supported in part by Grants in Aid for Scientific Research (22119005 and 21227001) from the Ministry of Education, Culture, Sports, Science, and Technology (T.K.) and by the Advanced Low Carbon Technology Research and Development Programme from the Japan Science and Technology Agency (T.K.).
References
- Alawady AE, Grimm B. Tobacco Mg protoporphyrin IX methyltransferase is involved in inverse activation of Mg porphyrin and protoheme synthesis. Plant J. 2005;41:282–290. doi: 10.1111/j.1365-313X.2004.02291.x. [DOI] [PubMed] [Google Scholar]
- Assmann SM, Shimazaki K. The multisensory guard cell: stomatal responses to blue light and abscisic acid. Plant Physiol. 1999;119:809–815. doi: 10.1104/pp.119.3.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollivar DW. Recent advances in chlorophyll biosynthesis. Photosynth Res. 2006;90:173–194. doi: 10.1007/s11120-006-9076-6. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- Du SY, Zhang XF, Lu Z, Xin Q, Wu Z, Jiang T, Lu Y, Wang XF, Zhang DP. Roles of the different components of magnesium chelatase in abscisic acid signal transduction. Plant Mol Biol. 2012;80:519–537. doi: 10.1007/s11103-012-9965-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodje MN, Hansson A, Hansson M, Olsen JG, Gough S, Willows RD, Al-Karadaghi S. Interplay between an AAA module and an integrin I domain may regulate the function of magnesium chelatase. J Mol Biol. 2001;311:111–122. doi: 10.1006/jmbi.2001.4834. [DOI] [PubMed] [Google Scholar]
- Fujii H, Verslues PE, Zhu JK. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell. 2007;19:485–494. doi: 10.1105/tpc.106.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, Stange A, Marten I, Bauer H, Ache P, Matschi S, Liese A, Al-Rasheid KAS, Romeis T, Hedrich R. Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc Natl Acad Sci USA. 2009;106:21425–21430. doi: 10.1073/pnas.0912021106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson LCD, Willows RD, Kannangara CG, von Wettstein D, Hunter CN. Magnesium-protoporphyrin chelatase of Rhodobacter sphaeroides: reconstitution of activity by combining the products of the bchH, -I, and -D genes expressed in Escherichia coli. Proc Natl Acad Sci USA. 1995;92:1941–1944. doi: 10.1073/pnas.92.6.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Negi J, Young J, Israelsson M, Schroeder JI, Iba K. Arabidopsis HT1 kinase controls stomatal movements in response to CO2. Nature Cell Biol. 2006;8:391–397. doi: 10.1038/ncb1387. [DOI] [PubMed] [Google Scholar]
- Hashimoto-Sugimoto M, Higaki T, Yaeno T, Nagami A, Irie M, Fujimi M, Miyamoto M, Akita K, Negi J, Shirasu K, Hasezawa S, Iba K. A Munc13-like protein in Arabidopsis mediates H+-ATPase translocation that is essential for stomatal responses. Nat Commun. 2013;4:2215. doi: 10.1038/ncomms3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Li H. Arabidopsis CHLI2 can substitute for CHLI1. Plant Physiol. 2009;150:636–645. doi: 10.1104/pp.109.135368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S, Kinoshita T, Matsumoto M, Nakayama K, Doi M, Shimazaki K. Blue light-induced autophosphorylation of phototropin is a primary step for signaling. Proc Natl Acad Sci USA. 2008;105:5626–5631. doi: 10.1073/pnas.0709189105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi-Saha A, Valon C, Leung J. Abscisic acid signal off the STARting block. Mol Plant. 2011;4:562–580. doi: 10.1093/mp/ssr055. [DOI] [PubMed] [Google Scholar]
- Kim TH, Böhmer M, Hu H, Nishimura N, Schroeder JI. Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu Rev Plant Biol. 2010;61:561–591. doi: 10.1146/annurev-arplant-042809-112226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Doi M, Suetsugu N, Kagawa T, Wada M, Shimazaki K. phot1 and phot2 mediate blue light regulation of stomatal opening. Nature. 2001;414:656–660. doi: 10.1038/414656a. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Ono N, Hayashi Y, Morimoto S, Nakamura S, Soda M, Kato Y, Ohnishi M, Nakano T, Inoue S, Shimazaki K. FLOWERING LOCUS T regulates stomatal opening. Curr Biol. 2011;21:1232–1238. doi: 10.1016/j.cub.2011.06.025. [DOI] [PubMed] [Google Scholar]
- Klingler JP, Batelli G, Zhu JK. ABA receptors: the START of a new paradigm in phytohormone signalling. J Exp Bot. 2010;61:3199–3210. doi: 10.1093/jxb/erq151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Lan W, Buchanan BB, Luan S. A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc Natl Acad Sci USA. 2009;106:21419–21424. doi: 10.1073/pnas.0910601106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnaioli T, Cuevas J, Mas P. TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. EMBO J. 2009;28:3745–3757. doi: 10.1038/emboj.2009.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI. Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell. 2004;16:596–615. doi: 10.1105/tpc.019000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léon-Kloosterziel KM, Gil MA, Ruijs GJ, Jacobsen SE, Olszewski NE, Schwartz SH, Zeevaart JA, Koornneef M. Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J. 1996;10:655–661. doi: 10.1046/j.1365-313X.1996.10040655.x. [DOI] [PubMed] [Google Scholar]
- Lightner J, Caspar T (1998) Seed Mutagenesis of Arabidopsis. In: Martinez-Zapater JM, Salinas J (eds) Arabidopsis protocols, Humana Press, New York, pp 91–103
- Liu X, Yue Y, Li B, Nie Y, Li W, Wu WH, Ma L. A G protein-coupled receptor is a plasma membrane receptor for the plant hormone abscisic acid. Science. 2007;315:1712–1716. doi: 10.1126/science.1135882. [DOI] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- Masuda T. Recent overview of the Mg branch of the tetrapyrrole biosynthesis leading to chlorophylls. Photosynth Res. 2008;96:121–143. doi: 10.1007/s11120-008-9291-4. [DOI] [PubMed] [Google Scholar]
- Merlot S, Mustilli AC, Genty B, North H, Lefebvre V, Sotta B, Vavasseur A, Giraudat J. Use of infrared thermal imaging to isolate Arabidopsis mutants defective in stomatal regulation. Plant J. 2002;30:601–609. doi: 10.1046/j.1365-313X.2002.01322.x. [DOI] [PubMed] [Google Scholar]
- Merlot S, Leonhardt N, Fenzi F, Valon C, Costa M, Piette L, Vavasseur A, Genty B, Boivin K, Müller A, Giraudat J, Leung J. Constitutive activation of a plasma membrane H+-ATPase prevents abscisic acid-mediated stomatal closure. EMBO J. 2007;26:3216–3226. doi: 10.1038/sj.emboj.7601750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J. Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc Natl Acad Sci USA. 2001;98:2053–2058. doi: 10.1073/pnas.98.4.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N, Tanaka R, Tanaka A, Masuda T, Nagatani A. The steady-state level of Mg-protoporphyrin IX is not a determinant of plastid-to-nucleus signaling in Arabidopsis. Proc Natl Acad Sci USA. 2008;105:15184–15189. doi: 10.1073/pnas.0803245105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N, Tanaka R, Grimm B, Masuda T, Moulin M, Smith AG, Tanaka A, Terry MJ. The cell biology of tetrapyrroles: a life and death struggle. Trends Plant Sci. 2010;15:488–498. doi: 10.1016/j.tplants.2010.05.012. [DOI] [PubMed] [Google Scholar]
- Müller AH, Hansson M. The barley magnesium chelatase 150-kd subunit is not an abscisic acid receptor. Plant Physiol. 2009;150:157–166. doi: 10.1104/pp.109.135277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell. 2002;14:3089–3099. doi: 10.1105/tpc.007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A. Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol. 2005;56:165–185. doi: 10.1146/annurev.arplant.56.032604.144046. [DOI] [PubMed] [Google Scholar]
- Negi J, Matsuda O, Nagasawa T, Oba Y, Takahashi H, Kawai-Yamada M, Uchimiya H, Hashimoto M, Iba K. CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature. 2008;452:483–486. doi: 10.1038/nature06720. [DOI] [PubMed] [Google Scholar]
- Negi J, Moriwaki K, Konishi M, Yokoyama R, Nakano T, Kusumi K, Hashimoto-Sugimoto M, Schroeder JI, Nishitani K, Yanagisawa S, Iba K. A Dof transcription factor, SCAP1, is essential for the development of functional stomata in Arabidopsis. Curr Biol. 2013;23:479–484. doi: 10.1016/j.cub.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Sarkeshik A, Nito K, Park SY, Wang A, Carvalho PC, Lee S, Caddell DF, Cutler SR, Chory J, Yates JR, Schroeder JI. PYR/PYL/RCAR family members are major in vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J. 2010;61:290–299. doi: 10.1111/j.1365-313X.2009.04054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi KK, Grossman AR, Björkman O. Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. Plant Cell. 1998;10:1121–1134. doi: 10.1105/tpc.10.7.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Nelson DC, Assmann SM. Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell. 2009;136:136–148. doi: 10.1016/j.cell.2008.12.026. [DOI] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, Alfred SE, Bonetta D, Finkelstein R, Provart NJ, Desveaux D, Rodriguez PL, McCourt P, Zhu JK, Schroeder JI, Volkman BF, Cutler SR. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks BM, Quail PH. Phytochrome-deficient hy1 and hy2 long hypocotyl mutants of Arabidopsis are defective in phytochrome chromophore biosynthesis. Plant Cell. 1991;3:1177–1186. doi: 10.1105/tpc.3.11.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontier D, Albrieux C, Joyard J, Lagrange T, Block MA. Knock-out of the magnesium protoporphyrin IX methyltransferase gene in Arabidopsis. Effects on chloroplast development and on chloroplast-to-nucleus signaling. J Biol Chem. 2007;282:2297–2304. doi: 10.1074/jbc.M610286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J, Rodrigues A, Saez A, Rubio S, Antoni R, Dupeux F, Park SY, Márquez JA, Cutler SR, Rodriguez PL. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 2009;60:575–588. doi: 10.1111/j.1365-313X.2009.03981.x. [DOI] [PubMed] [Google Scholar]
- Schroeder JI, Raschke K, Neher E. Voltage dependence of K channels in guard-cell protoplasts. Proc Natl Acad Sci USA. 1987;84:4108–4112. doi: 10.1073/pnas.84.12.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder JI, Allen GJ, Hugouvieux V, Kwak JM, Waner D. Guard cell signal transduction. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:627–658. doi: 10.1146/annurev.arplant.52.1.627. [DOI] [PubMed] [Google Scholar]
- Shen Y, Wang X, Wu F, Du S, Cao Z, Shang Y, Wang X, Peng C, Yu X, Zhu S, Fan R, Xu Y, Zhang D. The Mg-chelatase H subunit is an abscisic acid receptor. Nature. 2006;443:823–826. doi: 10.1038/nature05176. [DOI] [PubMed] [Google Scholar]
- Shimazaki K. Doi M, Assmann SM, Kinoshita T Light regulation of stomatal movement. Annu Rev Plant Biol. 2007;58:219–247. doi: 10.1146/annurev.arplant.57.032905.105434. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Hayashi K, Kinoshita T. Auxin activates the plasma membrane H+-ATPase by phosphorylation during hypocotyl elongation in Arabidopsis. Plant Physiol. 2012;159:632–641. doi: 10.1104/pp.112.196428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemiya A, Sugiyama N, Fujimoto H, Tsutsumi T, Yamauchi S, Hiyama A, Tada Y, Christie JM, Shimazaki K. Phosphorylation of BLUS1 kinase by phototropins is a primary step in stomatal opening. Nat Commun. 2013;4:2094. doi: 10.1038/ncomms3094. [DOI] [PubMed] [Google Scholar]
- Tanaka R, Kobayashi K, Masuda T. Tetrapyrrole metabolism in Arabidopsis thaliana. Arabidopsis Book. 2011;9:e0145. doi: 10.1199/tab.0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuzuki T, Takahashi K, Inoue S, Okigaki Y, Tomiyama M, Hossain MA, Shimazaki K, Murata Y, Kinoshita T. Mg-chelatase H subunit affects ABA signaling in stomatal guard cells, but is not an ABA receptor in Arabidopsis thaliana. J Plant Res. 2011;124:527–538. doi: 10.1007/s10265-011-0426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuzuki T, Takahashi K, Tomiyama M, Inoue S, Kinoshita T (2013) Overexpression of the Mg-chelatase H subunit in guard cells confers drought tolerance via promotion of stomatal closure in Arabidopsis thaliana. Front Plant Sci 4:440 [DOI] [PMC free article] [PubMed]
- Ueno K, Kinoshita T, Inoue S, Emi T, Shimazaki K. Biochemical characterization of plasma membrane H+-ATPase activation in guard cell protoplasts of Arabidopsis thaliana in response to blue light. Plant Cell Physiol. 2005;46:955–963. doi: 10.1093/pcp/pci104. [DOI] [PubMed] [Google Scholar]
- Vahisalu T, Kollist H, Wang YF, Nishimura N, Chan WY, Valerio G, Lamminmäki A, Brosché M, Moldau H, Desikan R, Schroeder JI, Kangasjärvi J. SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature. 2008;452:487–491. doi: 10.1038/nature06608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang FF, Lian HL, Kang CY, Yang HQ. Phytochrome B is involved in mediating red light-induced stomatal opening in Arabidopsis thaliana. Mol Plant. 2010;3:246–259. doi: 10.1093/mp/ssp097. [DOI] [PubMed] [Google Scholar]
- Wang Y, Noguchi K, Ono N, Inoue S, Terashima I, Kinoshita T. Overexpression of plasma membrane H+-ATPase in guard cells promotes light-induced stomatal opening and enhances plant growth. Proc Natl Acad Sci USA. 2014;111:533–538. doi: 10.1073/pnas.1305438111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willows RD, Gibson LC, Kanangara CG, Hunter CN, von Wettstein D. Three separate proteins constitute the magnesium chelatase of Rhodobacter sphaeroides. Eur J Biochem. 1996;235:438–443. doi: 10.1111/j.1432-1033.1996.00438.x. [DOI] [PubMed] [Google Scholar]
- Wu FQ, Xin Q, Cao Z, Liu ZQ, Du SY, Mei C, Zhao CX, Wang XF, Shang Y, Jiang T, Zhang XF, Yan L, Zhao R, Cui ZN, Liu R, Sun HL, Yang XL, Su Z, Zhang DP. The magnesium-chelatase H subunit binds abscisic acid and functions in abscisic acid signaling: new evidence in Arabidopsis. Plant Physiol. 2009;150:1940–1954. doi: 10.1104/pp.109.140731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Costa A, Leonhardt N, Siegel RS, Schroeder JI. Isolation of a strong Arabidopsis guard cell promoter and its potential as a research tool. Plant Method. 2008;4:6. doi: 10.1186/1746-4811-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 lost2 and lost3 mutation loci. a Mapping analysis of the LOST2 locus. Numbers in parentheses indicate percentages of no recombination in 40 chromosomes. The LOST2 locus is located close to SSLP marker ciw1 and ABA DEFICIENT 2 (ABA2). b Determination of the mutation in the lost2 mutant gene. The ABA2 gene genomic structure on chromosome 1 is shown (upper). Black boxes and bars indicate exons and introns, respectively. Partial ABA2 cDNA sequences and the deduced amino acid sequences from the wild type and lost2 are shown (lower). The lost2 mutant results from a G to A nucleotide substitution in the second exon. The nucleotide change causes the substitution of Asp for Gly at amino acid 169. Nucleotide and amino acid numbers are indicated on the right. Asterisks indicate the same nucleotide of the ABA2 gene in the wild type and lost2. c Mapping analysis of the LOST3 locus. Numbers in parenthesis indicate the percentage of no recombination in 26 chromosomes. The LOST3 locus is located close to SSLP marker F20D23 and ABA DEFICIENT 3 (ABA3). d Determination of the mutation in the lost3 mutant gene. The genomic structure of the ABA3 gene on chromosome 1 is shown (upper) as described in Figure S1b. Partial ABA3 cDNA sequences and the deduced amino acid sequences in wild type and lost3 are shown (lower) as described in b. The lost3 mutant contains a C to T nucleotide substitution in exon 16. The nucleotide change results in the substitution of a stop codon for a glutamine codon at residue 575. e Effect of NaCl on lost2, lost3, npq2, and control phot2 seed germination. The seed germination assay was performed as described previously (Leon-Kloosterziel et al. 1996) with some modifications. Approximately 25 sterilized seeds were placed on a plate containing MS medium (pH 5.8), 2 % sucrose, and 0.8 % agar with or without 200 mM NaCl. The plate was kept at 4 °C for three days, then incubated at 24 °C under illuminated growth conditions for 1 week. The germination rate was determined from the percentage of germinated seeds with green cotyledons. The experiments were repeated on three occasions with similar results. N.D., not detected (TIFF 2519 kb)
Fig. S2 Effect of ABA on light-induced stomatal opening in chli1 (a), gun2 (b), gun3 (b), and the wild type. Epidermal peels in buffer containing 20 µM ABA or equal volumes of DMSO were irradiated with 10 µmol m−2 s−1 blue light and 50 µmol m−2 s−1 red light. Values are means of 25 stomata measurements with SEs. Asterisks indicate a significant difference in each treatment relative to the wild type by the t test (P < 0.01). The experiments were repeated on three occasions with similar results (TIFF 1843 kb)
Fig. S3 ABA sensitivity of seed germination and root growth in Mg-chelatase subunit and CHLM mutants. The seed germination and root growth assay was performed as described previously (Tsuzuki et al. 2011) with some modifications. For seed germination assay, seeds were placed on a plate containing MS medium (pH 5.8), 2 % sucrose, 0.8 % agar, and 1 µM ABA or equal volumes of DMSO. The plate was kept at 4 °C for three days and then incubated at 24 °C under illuminated growth conditions. a Effect of ABA on germination of lost1 and control phot2, chlm, and wild-type seeds. Ten-day-old lost1, rtl1, control phot2, chlm, and wild-type plants grown on MS medium containing 1 µM ABA or equal volumes of DMSO. The scale bar represents 1 cm. b The germination rate was determined by the percentage of germinated seeds (n = 50) showing radicle emergence at the indicated days after the start of stratification. Values are means of three independent experiments with SDs. For root growth assay, four-day-old seedlings grown on a MS plate under light were transferred to MS medium containing 20 µM ABA or equal volumes of DMSO. c Effect of ABA on root growth of lost1 and control phot2, chlm, and wild-type seeds. Ten-day-old lost1, rtl1, control phot2, chlm, and wild-type plants grown on MS medium containing 20 µM ABA or equal volumes of DMSO. The scale bar represents 1 cm. d The root length was measured at 7 days after transferring to the medium containing ABA. Values are means of 25 seedlings measurements with SEs. The experiments were repeated on three occasions with similar results (TIFF 13147 kb)
Fig. S4 ABA contents of phot2, lost1, rtl1, wild type (Col) and npq2 in rosette leaves. The ABA concentrations in rosette leaves of four- to seven-week-old plants were measured with Phytodetek ABA Test Kit (Agdia), as described previously (Kim et al. 2009) with some modifications. For ABA extraction, 200 mgFW tissue was ground in liquid nitrogen and suspended with 250 µl of MilliQ water, and agitated over night at 4 °C. The suspension was centrifuged at 4,000 rpm for 20 min, and the supernatant was collected into a clean tube and dried under vacuum. The dry residue was dissolved with 24 µl of MilliQ water, and diluted by 1:25 in TBS. The ABA concentration was determined with a Phytodetek ABA Test Kit (Agdia). The absorbance at 405 nm was detected using microplate reader (ARVO X3, ParkinElmer). Values are means of three independent experiments with SDs. Asterisk indicates a significant difference in ABA content relative to the control plant by the t-test (P < 0.01).Kim MJ, Shin R, Schachtman DP (2009) A nuclear factor regulates abscisic acid responses in Arabidopsis. Plant Physiol 151:1433–1445 (TIFF 646 kb)
Fig. S5 Immunoblot analysis of CHLH protein in Mg-chelatase subunit and CHLM mutants. Each lane was loaded with 50 µg of protein extracted from rosette leaves of lost1, rtl1, phot2, chli1, chlm, and wild-type plants. Actin protein served as an internal control. The experiments were repeated on three occasions with similar results (TIFF 640 kb)