Abstract
Vitamin B2 (riboflavin) is essential for metabolic functions and is synthesized by many bacteria, yeast, and plants, but not by mammals and other animals, which must acquire it from the diet. In mammals, modified pyrimidine intermediates from the microbial biosynthesis of riboflavin are recognized as signature biomarkers of microbial infection. This recognition occurs by specialized lymphocytes known as mucosal associated invariant T (MAIT) cells. The major histocompatibility class I-like antigen-presenting molecule, MR1, captures these pyrimidine intermediates, but only after their condensation with small molecules derived from glycolysis and other metabolic pathways to form short-lived antigens. The resulting MR1-Ag complexes are recognized by MAIT cell antigen receptors (αβ T cell receptors (TCRs)), and the subsequent MAIT cell immune responses are thought to protect the host from pathogens at mucosal surfaces. Here, we review our understanding of how these novel antigens are generated and discuss their interactions with MR1 and MAIT TCRs.
Keywords: bacterial metabolism, folate, innate immunity, riboflavin, vitamin, Ag presentation, MAIT cells, MR1, T cell recognition
MAIT Cells
T cells are lymphocytes that mediate a range of immune functions such as killing infected host cells and producing cytokines and other factors that regulate immunity and inflammation. T cells generally recognize peptide antigens (Ags)6 or lipid-based Ags complexed to specialized Ag-presenting molecules, MHC and CD1, respectively, that interact with an Ag-specific αβ T cell receptor (TCR) (1, 2). An expansive repertoire of TCRs is generated by the random rearrangement of V, D, and J gene segments and by pairing of TCR α- and β-chains, allowing specific recognition of a diverse range of Ags. Mucosal associated invariant T (MAIT) cells are a specialized subset of αβ T cells originally identified in CD4− CD8− (double negative) human blood lymphocytes expressing a dominant invariant TCR α-chain gene rearrangement, TRAV1-2-TRAJ33 (based on IMGT, or Vα7.2-Jα33 based on Arden nomenclature (3)) (4, 5) with less frequent usage of TRAJ12 and TRAJ20 (6–9). The MAIT TCR α-chain contains a complementarity-determining region (CDR) 3 loop of constant length with two variable amino acids in the V-J junction (4) located at the base of this loop (10, 11). MAIT cells are evolutionarily conserved in mammals with the canonical TCR α-chain composed of homologous TRAV and TRAJ elements also found in mice, cattle (5) and sheep (12). TRAV1-2 is not exclusive to MAIT cells, with some MHC-I-restricted T cells (13, 14) and CD1b-restricted germline-encoded, mycolyl lipid-reactive (GEM) T cells also using this TRAV gene segment (15).
The β-chain of MAIT cells has no apparent restrictions in Jβ usage, but there is a bias toward the use of human TRBV20 and TRBV6 segments (or Vβ2 and Vβ13 based on Arden nomenclature) and, similarly in mice, to TRBV19 and TRBV13 (or Vβ6 and Vβ8 based on Arden nomenclature), both being the murine orthologous segments of human TRBV6 (4, 5, 9). Furthermore, recent TCR sequencing revealed a marked oligoclonality of MAIT TCR CDR3β regions and a bias in the length of mostly between 11 and 14 amino acids (9).
MR1 Presentation of Antigens Derived from Vitamin B2 or B9 Synthesis
MAIT cell development and Ag-specific activation depend on expression of the Ag-presenting molecule MR1 (MHC-related protein-1 (MR1)) (16), an MHC class I-like molecule (17) that assembles with β2-microglobulin to form a heterodimer (18, 19). MR1 is monomorphic (20) and is the most highly conserved MHC-like gene across diverse mammalian species, with 90% sequence identity between mouse and humans (21, 22), allowing for considerable species cross-reactivity of MAIT cells (23).
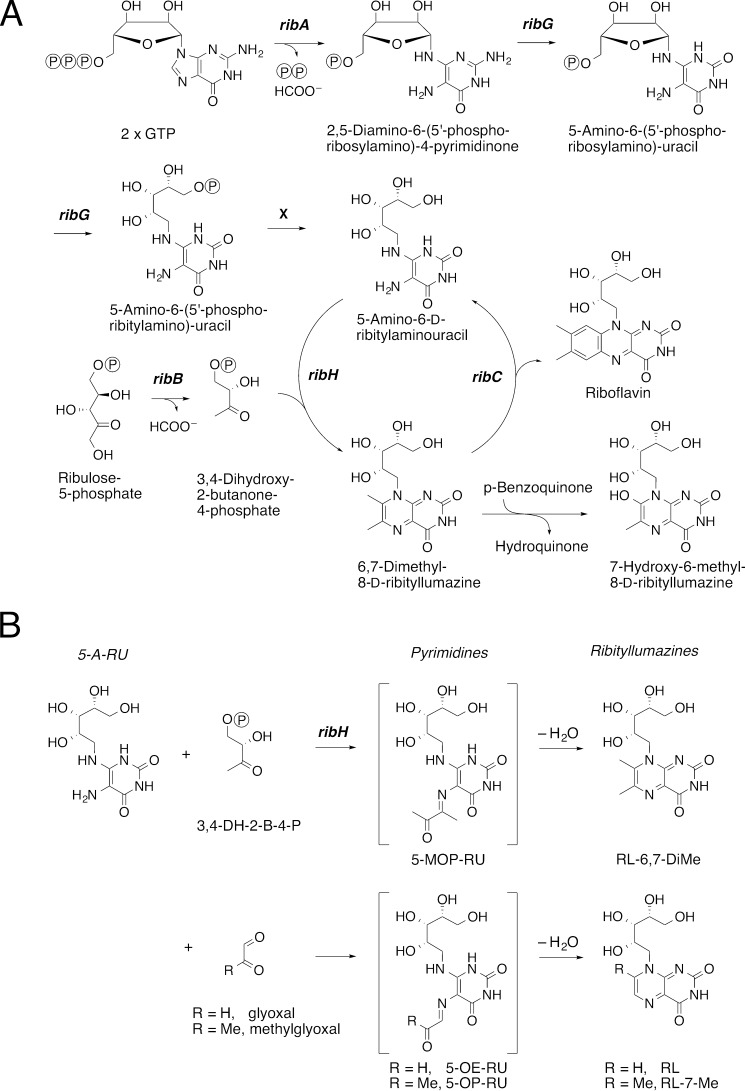
Until recently, the exact nature of the Ag presented by MR1 to T cells was unknown. It was initially postulated that the monomorphic nature of MR1 and the constrained MAIT TCR usage reflected a limited diversity in MR1 ligands recognized by MAIT cells in a pattern recognition-like manner (24). Notably, although human MAIT cells respond to a wide variety of bacteria such as Escherichia coli, Pseudomonas aeruginosa, Klebsiella pneumoniae, Staphylococcus aureus, Staphylococcus epidermidis, Salmonella enterica serovar Typhimurium, Mycobacterium smegmatis, Mycobacterium tuberculosis, Mycobacterium abscessus, Candida albicans, Candida glabrata, and Saccharomyces cerevisiae, they are not activated by viruses and certain strains of Listeria, Enterobacter, and Streptococcus (6, 25–27). The canonical pathway of riboflavin biosynthesis involves a number of enzymes essential for ligand production (Fig. 1A), and those bacteria that do not activate MAIT cells have defects in the riboflavin synthesis pathway (26), consistent with derivation of MR1 ligands from riboflavin synthesis. In some bacteria, the riboflavin synthesis pathway is regulated by an operon (Rib) such that the presence of flavin mononucleotides, or riboflavin itself, inhibits riboflavin synthesis and impairs generation of ligands that activate MAIT cells (28, 29). Moreover, in two bacterial strains, S. Typhimurium and Lactococcus lactis, MAIT cell activation was dependent on the enzymatic production of 5-amino-6-d-ribitylaminouracil (5-A-RU), an intermediate in riboflavin biosynthesis that is present in diverse bacteria and yeast as well as plants (29). Mutations in enzymes essential to the production of 5-A-RU, but not enzymes required later in riboflavin synthesis (Fig. 1A), prevented the production of activating MAIT cell ligands by these bacteria (29). This finding was recently verified in mutants of E. coli (30). Although 5-A-RU plays an important role in MAIT cell activation, MR1 could not be refolded efficiently with 5-A-RU alone (29, 31).
FIGURE 1.
Derivation of MR1 ligands from riboflavin synthesis. A, schematic display of the riboflavin biosynthesis pathway. ribH, lumazine synthase; X, hypothetical phosphatase. B, chemical formation of pyrimidines and ribityllumazines from condensation of small metabolites with 5-A-RU. 3,4-DH-2-B-4-P, 3,4-dihydroxy-2-butanone-4-phosphate.
Despite the link between MAIT cell activation and the biosynthesis of vitamin B2, the first MR1-binding ligand to be identified was 6-formylpterin (6-FP), a photodegradation product of vitamin B9 (folic acid) (26). It was identified by LC-MS analysis of the eluted material from recombinant MR1 molecules assembled in folic acid-containing medium (26). However, MR1-6-FP complexes did not activate MAIT cells, leading us to a wider search for the natural activating ligand (26). When recombinant MR1 was loaded with filtered bacterial cultures of S. Typhimurium grown in folate-deficient medium, LC-MS indicated that the main chemical component from the eluted material had a molecular formula of C12H18N4O7 (26). A search for compounds structurally related to riboflavin biosynthetic intermediates matching this formula included (i) a dihydrogen-reduced product of 7-hydroxy-6-methyl-8-d-ribityllumazine (RL-6-Me-7-OH, C12H16N4O7), a metabolite of the key intermediate 6,7-dimethyl-8-d-ribityllumazine (RL-6,7-DiMe, C13H16N4O6, Fig. 1, A and B) and (ii) the dihydrogen-reduced 6-hydroxymethyl-8-d-ribityllumazine (rRL-6-CH2OH, C12H18N4O7), the latter of which had no known link to riboflavin biosynthetic intermediates apart from structural similarity. Indeed, all three compounds (RL-6,7-DiMe, RL-6-Me-7-OH, and rRL-6-CH2OH) activated MAIT cells in an MR1-dependent manner in cellular assays, albeit with varied levels of potency (26).
MR1 assembled with RL-6-Me-7-OH sufficiently well in vitro for the MAIT TCR-MR1-RL-6-Me-7-OH structure to be solved by x-ray crystallography (10), although the recovered complexes were very unstable and made tetramer production challenging (8). At this point, rRL-6-CH2OH seemed to be the most potent activator of MAIT cells in vitro and ex vivo (26). However, wild-type MR1 could not be refolded with rRL-6-CH2OH, so that a crystal structure could not be obtained for MR1 complexed with rRL-6-CH2OH. A low resolution (3.3 Å) crystal structure of a human MAIT TCR and humanized bovine MR1 complexed with a heterogeneous extract from E. coli was later reported, but the structure of the bound Ags remains unclear (32). Therefore we took a genetic and biochemical approach to identifying the nature of the ligands captured by MR1 from the riboflavin synthesis pathway in bacteria.
Formation of MAIT Cell Ligands through the Interaction of Different Metabolic Pathways
A key step in riboflavin biosynthesis is the condensation of 5-A-RU with 3,4-dihydroxy-2-butanone-4-phosphate (3,4-DH-2-B-4-P) to generate RL-6,7-DiMe (Fig. 1A). This reaction proceeds via a putative intermediate, 5-(1-methyl-2-oxopropylideneamino)-6-d-ribitylaminouracil (5-MOP-RU), which spontaneously undergoes ring closure via dehydration to form RL-6,7-DiMe (33, 34) (Fig. 1B). This reaction can be catalyzed by lumazine synthase (RibH), but RL-6,7-DiMe can also be generated in the absence of an enzyme (33, 34). Because 5-MOP-RU and the related compound 5-(2-oxopropylideneamino)-6-d-ribitylaminouracil (5-OP-RU) differ only by a single methyl group, we conceived that 5-OP-RU might be formed as a short-lived Ag via the analogous condensation of 5-A-RU with methylglyoxal, en route to cyclization to the corresponding lumazine 7-methyl-8-d-ribityllumazine (RL-7-Me) (Fig. 1B). In attempts to generate potential MAIT cell activators, we therefore chemically combined the key riboflavin biosynthetic intermediate 5-A-RU with glyoxal and methylglyoxal, small molecules abundantly formed in a number of metabolic pathways including glycolysis (35). When these reaction mixtures were used to refold MR1, the ligands identified in the cleft were surprisingly the chemically unstable pyrimidine intermediates 5-(2-oxoethylideneamino)-6-d-ribitylaminouracil (5-OE-RU) and 5-OP-RU, instead of the corresponding and relatively more stable ribityl lumazines, 8-d-ribityllumazine (RL) and RL-7-Me, respectively. These Ags, 5-OE-RU and 5-OP-RU, are normally unstable in water, but were encapsulated and stabilized by MR1 through Schiff base formation (see below) and an extensive hydrogen-bonding network as evidenced in high resolution (2.1–2.6 Å) crystal structures (29). Both Ags activated MAIT cells and formed stable MR1 tetramers that stained MAIT cells with great specificity in the case of both human (29) and mouse MAIT cells (36). Furthermore, they were identical to the ligands recovered from recombinant MR1 assembled with supernatant from S. Typhimurium, L. lactis, and E. coli, as determined by high resolution LC-MS (29). Other researchers have since also confirmed the identity of these MAIT cell-activating ligands, 5-OP-RU and 5-OE-RU (30). The structural features of their binding revealed the basis for the capture of these intermediates by MR1 as discussed below (37). By contrast, when 5-A-RU was combined with 2,3-butanedione, the resultant mixture did not activate MAIT cells or refold with MR1, although we are uncertain whether 5-MOP-RU reached a significant concentration before its rapid cyclization to give RL-6,7-DiMe. This inability of MR1 to bind to an essential intermediate of riboflavin biosynthesis may be critical in directing MAIT cell activation.
Structural Basis of MR1-Antigen Binding and MAIT Cell TCR Recognition
Consistent with its sequence similarity to an MHC-I molecule, MR1 has a structure that overall is analogous to MHC-I, but in which the Ag is contained within an aromatic cradle formed between the α1- and α2-helices sitting atop an antiparallel β-sheet (Fig. 2A) (26). The cradle is lined with both charged and hydrophobic residues, of which a large number are aromatic, thus providing an ideal environment for the capture of small molecules (26). A number of high resolution MAIT TCR-MR1 ternary structures determined by x-ray crystallography have allowed the examination of the intermolecular interactions involved in the capture and presentation of this novel class of Ags by MR1 and revealed a number of interesting features (2, 9, 24, 28, 30).
FIGURE 2.
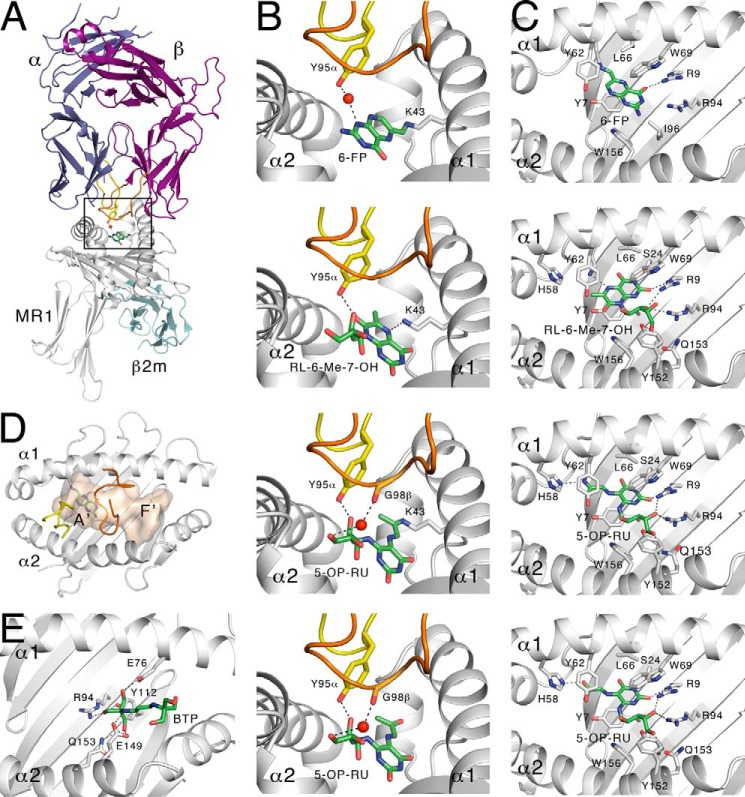
Structural characterization of MR1-binding ligands derived from vitamin B synthetic intermediates. A, ternary structure of MR1 presenting 6-FP to MAIT TCR. Boxed area represents sections shown in more detail in B. B, contact between the MR1 bound antigens 6-FP, RL-6-Me-7-OH, 5-OP-RU, or non-covalently bound 5-OP-RU, with the MAIT TCR. C, contacts sequestering 6-FP, RL-6-Me-7-OH, 5-OP-RU, or non-covalently bound 5-OP-RU within the MR1-binding cleft. D, solvent-accessible area within the MR1-binding cleft with the A′- and F′-pockets labeled. The CDR3α (yellow) and CDR3β (orange) of the MAIT TCR are shown. E, Bistris propane bound within the F′-pocket of MR1 (from PDB ID: 4PJX). All dashed black lines represent hydrogen bonds, and red spheres represent water molecules.
The most unusual feature of MR1 was identified from the initial MR1-6-FP structure and subsequent analysis, where the Ag was covalently bound to MR1 through a Schiff base formed between the formyl group of 6-FP and Lys-43 of MR1 (Fig. 2B) (26). This covalent bond demonstrates a strong association between MR1 and the Ag, despite its inability to activate MAIT cells. The Lys-43 residue is located at the base of the cleft, and thus the small 6-FP molecule is also buried deeply within the comparatively large solvent-accessible MR1 cavity (Fig. 2B) (26). Consequently, the crystal structure of MR1-6-FP bound to a MAIT TCR demonstrated the relative inaccessibility of the compound in this location for TCR recognition (10). The TCR bound orthogonally to the main axis of the Ag-binding cleft and centrally over the cradle, placing the variable regions of the TCR on the surface of MR1 immediately above the compound (Fig. 2A) (10). The Tyr-95 residue from the CDR3α loop extended down into the cleft but only formed a single indirect link between 6-FP and the TCR, through a water-mediated hydrogen bond (Fig. 2B) (10).
The crystal structure of a MAIT TCR bound to complexes of MR1-RL-6-Me-7-OH provided initial insight into the mechanism of MAIT cell activation, while noting that RL-6-Me-7-OH only weakly activated MAIT cells (10). In this structure, the TCR was bound to MR1 in an identical fashion to the non-activating MR1-6-FP complexes, but made a direct hydrogen bond with the ribityl moiety of the Ag through the highly conserved Tyr-95 (Fig. 2B), explaining the differential MAIT cell responses. Notably, RL-6-Me-7-OH cannot form a Schiff base with MR1, as it does not have a carbonyl group. The larger RL-6-Me-7-OH Ag also occupied a more expansive region of the cavity, making numerous contacts within the cleft to correctly orient the compound for T cell recognition in the absence of Schiff base bond formation (Fig. 2C). The structures of MAIT TCRs complexed with MR1-5-OE-RU and MR1-5-OP-RU showed the same orientation of the ribityl moiety so that the Ag could also form a direct contact with Tyr-95 of the MAIT TCR (Fig. 2B) (10). However, in contrast to RL-6-Me-7-OH binding to MR1, both 5-OE-RU and 5-OP-RU were able to form a Schiff base bond with Lys-43 through a reactive carbonyl group (29). Furthermore, the CDR3β loop of the TCR β-chain now made an additional contact with both pyrimidine adducts through its own water-mediated hydrogen bond (Fig. 2B) (29). Affinities between MR1-5-OP-RU or MR1-5-OE-RU complexes and MAIT TCRs measured by surface plasmon resonance were comparable with conventional TCRs recognizing peptide-MHC complexes (Kdeq ∼ 0.5–10 μm) (31).
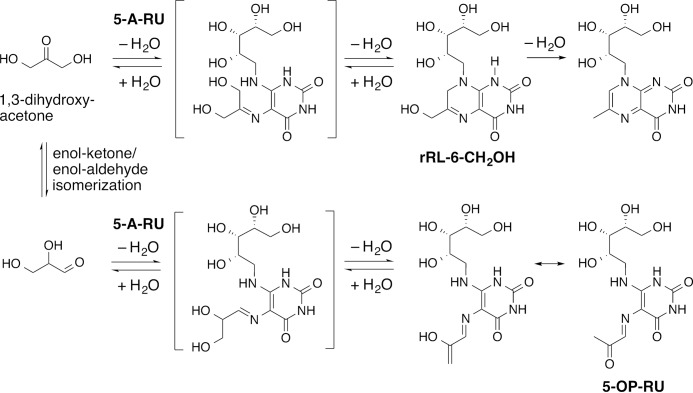
Notably, although rRL-6-CH2OH could not be refolded with wild-type MR1, it could be loaded into pre-folded MR1 molecules containing a K43A mutation that prohibits formation of a Schiff base with Ag. This crystal structure also revealed that the trapped ligand was the monocyclic 5-OP-RU (Fig. 2B) (29). This bound ligand had an LC-MS/MS fragmentation pattern that was indistinguishable from rRL-6-CH2OH (28, 29) and is primed for cyclization via intramolecular condensation of the free amine with the ketone. Consistent with these findings, a trace amount of 5-OP-RU was often present in synthetic samples of rRL-6-CH2OH (Fig. 3).7 The structure of a MAIT TCR-MR1(K43A)-5-OP-RU complex clearly demonstrated that the interactions within the MR1 cleft were sufficient to stabilize the 5-OP-RU pyrimidine adduct and prevent ligand cyclization without the contribution of Lys-43 to capture the reactive carbonyl group via Schiff base formation (Fig. 2C) (29). Although the structure confirmed that the covalent Schiff base interaction is not required by MR1 for Ag capture or activation of MAIT cells, MR1-Ag stability was reduced markedly in the absence of the Schiff base (17 °C lower half-maximum melting temperature by thermostability) (31).
FIGURE 3.
Synthesis of rRL-6-CH2OH and the possible formation of the byproduct 5-OP-RU.
MAIT Cell Antigen Diversity
Although 5-OP-RU and 5-OE-RU are potent MAIT cell Ags, the full range of naturally produced 5-A-RU adducts has not yet been defined. It is possible that other small molecules in addition to glyoxal and methylglyoxal may also arise as physiological byproducts potentially generating distinct variation in ligands in different microbes, or metabolic stages of the microbe and host. For example, although 5-OE-RU was the dominant species in the supernatant of E. coli, this was a subdominant species in the supernatant of S. Typhimurium, indicating that the bacterial source can impact on the relative abundance of the two distinct ligands (29).
The variability of MAIT TCR β-chain usage and the original discovery of several MAIT cell ligands recognized by MAIT cells suggest that there may be additional natural Ags that stimulate MAIT cells (26). As discussed earlier, in the dominant MAIT TCR α-chain, residue Tyr-95 forms a hydrogen bond with the ribityl moiety of activating ligands that is pivotal in MAIT cell activation (8, 10, 11, 31, 38). However, other less frequent TRAJ regions have been sequenced and tentatively assigned to MAIT cells, two of which do not encode Tyr-95 (TRAJ9, TRAJ39) (25). They may not represent functional MAIT TCRs, perhaps reflecting incomplete allelic exclusion at the TRA locus, but alternatively, it is possible either that they are reactive with known activating Ags in a manner distinct to MAIT TCRs encoding TRAJ Tyr-95 or that they recognize novel MAIT cell Ags. Structural studies have shown that MAIT TCR heterogeneity, especially in the CDR3β loop, can fine-tune MR1 recognition in an Ag-dependent manner (31), highlighting the possibility that MAIT cells might have the capacity to discriminate between diverse ligands in a MAIT cell subset-specific manner. Gold et al. (25) have recently observed distinct MAIT TCR repertoire mobilization in response to diverse pathogens that could reflect the existence of discrete pathogen-associated Ags presented by MR1. This possibility is yet to be confirmed with biochemical evidence. On the other hand, the limited variation in the MAIT TCR α-chain and oligoclonality in the CDR3β loop partly reflect structural requirements for MR1 recognition because both regions are critical as illuminated in crystal structures and mutagenesis analysis (10, 11, 38). Another contributing factor to the limited MAIT TCR repertoire might be preferential TCR rearrangements and α- and β-chain pairing. To this end, it has been confirmed that the canonical TRAV1-2-TRAJ33 amino acid sequence is produced efficiently from multiple TCR nucleotide sequences that can arise from multiple recombination events, a process referred to as convergent recombination (39). Interestingly, in Vα19i Cα−/−MR1−/− mice, a significant population of “MAIT-like” T cells develops, apparently selected by classical MHC-I molecules but reactive to MAIT Ags presented by MR1+ Ag-presenting cells (40). This raises the possibility that the variability of MAIT TCR β-chain and Jα usage might also be driven by avoidance of self-reactivity to polymorphic MHC-I and MHC-II molecules by the relatively fixed MAIT TCR.
The x-ray crystal structures of MR1-Ag complexes also raise the possibility of additional, novel MR1 Ags. All structures to date have shown the Ag to be bound within the MR1 A′-pocket (Fig. 2D, equivalent to the MHC-I pocket that binds the N-terminal peptide residue), which contains both Lys-43 for Schiff base formation and the residues that interact directly with the ribityl moiety. However, the F′-pocket (equivalent to the MHC-I pocket that binds the C-terminal residue of peptides), which is shallower than the A′-pocket, has already shown a predisposition to presenting small molecules (Fig. 2D). A number of the published MAIT TCR-MR1 structures deposited to the Protein Data Bank (PDB) contain the buffer compounds, Bistris propane or glycerol (e.g. 4PJX and 4PJE), used in the crystallization conditions, located within this cleft (31). Bistris propane makes a number of hydrogen bonds with charged residues in the F′-pocket (Fig. 2E) and, although these molecules are unlikely to be MAIT cell Ags themselves and are relatively distal from the canonical MAIT TCR docking region, this does provide a tantalizing glimpse of what naturally occurring MR1 ligands within this F′-pocket may resemble.
It is possible, but not obvious, that distinct MR1 ligands might be derived from additional microbial pathways other than riboflavin biosynthesis. However, the genetic correlation between the riboflavin biosynthetic pathway, ligand production, and MAIT cell activation suggests this is a dominant source of Ag. This, in turn, implies that the basic scaffold of any additional microbial Ags will derive from 5-A-RU. Indeed, tetramers of MR1-5-OP-RU or MR1-5-OE-RU bind to most if not all MAIT cells (8), suggesting that 5-OP-RU and 5-OE-RU represent universal and highly potent MAIT cell Ags.
MR1 Expression and Ligand Capture
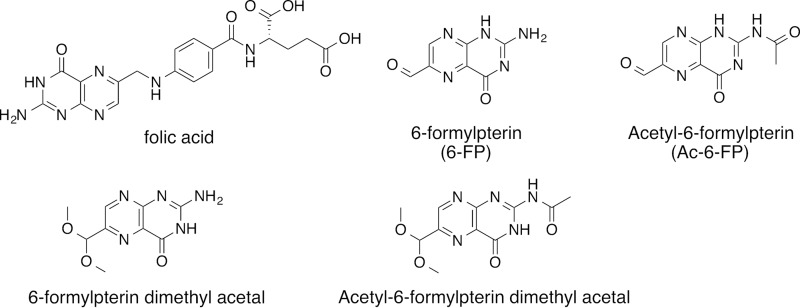
MR1 transcripts and intracellular protein are ubiquitously expressed (17, 21), whereas MR1 is expressed at low levels at the cell surface and is more readily detectable in the presence of ligand (26, 31, 41). Studies prior to the discovery of MR1 ligands showed that intracellularly MR1 is primarily found in the endoplasmic reticulum, where it associates with all known members of the peptide-loading complex, including calnexin, calreticulin, ERp57, TAP (transporter associated with antigen processing), and tapasin, although not all residues known to interact with the loading complex are present in MR1, thus pointing toward a novel manner of loading complex interaction (19). However, MAIT cell activation by endogenous ligand was shown to be independent of chaperoning by the MHC loading complex (16, 42). Instead, the endocytic compartment, including MHC class II chaperones Ii and DM, which promote endosomal trafficking, was shown to be important for MAIT cell activation in this system (42). Notably, MR1 can present both extracellular (supernatant) and infection-related Ag. The precise mechanisms by which bacterial Ags are loaded onto MR1 and the location of this event in each case remain to be understood. It is also still unclear whether MR1 Ag presentation is mediated by a specialized Ag-presenting cell in the periphery, as both myeloid cells as well as non-myeloid cells, such as epithelial cell lines, including A549 (6), BEAS-2B (43), and HeLa (44), can act as Ag-presenting cells in vitro. Both 6-FP (26) and its synthetic analogue acetyl-6-formylpterin (Ac-6-FP) (Fig. 4) (31) potently up-regulate cell surface expression of MR1 and act as competitive inhibitors of MAIT cell activation by 5-OP-RU, the latter being the superior competitive inhibitor. When comparing the kinetics of MR1 up-regulation, Ac-6-FP induced a more rapid and more prolonged increase in MR1 levels at the cell surface and higher maximum surface expression levels of MR1 were reached (31). The difference could reflect the lower stability of MR1-6-FP as compared with MR1-Ac-6-FP (8 °C lower half-maximum melting temperature by thermostability) (31). This difference in stability was associated with additional hydrogen bonding between the acetyl group of Ac-6-FP and Arg-94 of MR1, although we cannot rule out effects related to the intrinsic stability of the ligands themselves or their relative cellular uptake. Additional analogues of 6-FP that up-regulate MR1 and competitively inhibit MAIT cell activation have recently been reported, namely the dimethyl acetals of 6-FP and Ac-6-FP (30) (Fig. 4). Both possess these activities despite being unable to form a Schiff base with MR1, although partial hydrolysis of the acetal could generate an aldehyde capable of reacting with Lys-43 (30). MAIT cells in human PBMCs and MAIT cell reporter cell lines are activated at nm concentrations of Ag (31), suggesting that a small number of cell surface MR1-5-OP-RU complexes might be required for MAIT cell activation. Consistent with this notion, competitive inhibition of 5-OP-RU by the most potent inhibitor, Ac-6-FP, requires formation of stable MR1-antagonist complexes and 106-fold molar inhibitor excess for complete competitive inhibition of activation by 5-OP-RU (31).
FIGURE 4.
Folic acid and its structurally related MR1 ligands.
In summary, MR1 has sufficient plasticity to accommodate a range of chemical entities, including those identified to date, such as the bicyclic pterins (6-FP, Ac-6-FP), monocyclic pyrimidine derivatives (5-OP-RU, 5-OE-RU), and bicyclic lumazines (RL-6,7-DiMe, RL-6-Me-7-OH) (10, 26, 29, 31), all of which are derived from vitamin B2 (riboflavin) or B9 (folic acid) biosynthesis. It is not yet known whether distinct classes of ligands can be accommodated in the A′ versus the more solvent-exposed F′-pockets of MR1. Further studies are in progress to map the full complement of potential MR1-binding ligands that can activate or inhibit MAIT cells, and to characterize the immunological and pharmacological consequences of MR1 recognition of small heterocyclic molecules and their subsequent presentation as Ags for MAIT cell activation. These ongoing studies promise to reveal new insights into the molecular basis of MAIT cell-mediated immune responses in physiology and disease.
This work was supported by Australian Research Council Grants E140100011 and LE110100106 and National Health and Medical Research Council (NHMRC) of Australia Grants 1016629, 1013667, 1062889, and 1063587. The authors declare that they have no conflicts of interest with the contents of this article.
A. J. Corbett, S. B. Eckle, R. W. Birkinshaw, L. Liu, O. Patel, J. Mahony, Z. Chen, R. Reantragoon, B. Meehan, H. Cao, N. A. Williamson, R. A. Strugnell, D. Van Sinderen, J. Y. Mak, D. P. Fairlie, L. Kjer-Nielsen, J. Rossjohn, and J. McCluskey, unpublished results.
- Ag
- antigen
- MAIT
- mucosal associated invariant T
- TCR
- T cell receptor
- CDR
- complementarity-determining region
- MR1
- MHC-related protein-1
- 5-A-RU
- 5-amino-6-d-ribitylaminouracil
- 6-FP
- 6-formylpterin
- Ac-6-FP
- acetyl-6-formylpterin
- 5-MOP-RU
- 5-(1-methyl-2-oxopropylideneamino)-6-d-ribitylaminouracil
- 5-OE-RU
- 5-(2-oxoethylideneamino)-6-d-ribitylaminouracil
- 5-OP-RU
- 5-(2-oxopropylideneamino)-6-d-ribitylaminouracil
- rRL-6-CH2OH
- 6-hydroxymethyl-8-d-ribityllumazine
- RL-6,7-DiMe
- 6,7-dimethyl-8-d-ribityllumazine
- RL-6-Me-7-OH
- 7-hydroxy-6-methyl-8-d-ribityllumazine
- RL-7-Me
- 7-methyl-8-d-ribityllumazine
- RL
- 8-d-ribityllumazine
- rRL
- dihydrogen-reduced RL
- DiMe
- dimethyl
- Bistris propane
- 1,3-bis(tris(hydroxymethyl)methylamino)propane
- TRAV
- T cell receptor alpha variable
- TRBV
- T cell receptor β variable
- TRAJ
- T cell receptor alpha junctional
- TRBJ
- T cell receptor β junctional.
References
- 1.Bhati M., Cole D. K., McCluskey J., Sewell A. K., and Rossjohn J. (2014) The versatility of the αβ T-cell antigen receptor. Protein Sci. 23, 260–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rossjohn J., Gras S., Miles J. J., Turner S. J., Godfrey D. I., and McCluskey J. (2015) T cell antigen receptor recognition of antigen-presenting molecules. Annu. Rev. Immunol. 33, 169–200 [DOI] [PubMed] [Google Scholar]
- 3.Scaviner D., and Lefranc M. P. (2000) The human T cell receptor α variable (TRAV) genes. Exp. Clin. Immunogenet. 17, 83–96 [DOI] [PubMed] [Google Scholar]
- 4.Porcelli S., Yockey C. E., Brenner M. B., and Balk S. P. (1993) Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4−8− α/β T cells demonstrates preferential use of several V β genes and an invariant TCR α chain. J. Exp. Med. 178, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tilloy F., Treiner E., Park S. H., Garcia C., Lemonnier F., de la Salle H., Bendelac A., Bonneville M., and Lantz O. (1999) An invariant T cell receptor α chain defines a novel TAP-independent major histocompatibility complex class Ib-restricted α/β T cell subpopulation in mammals. J. Exp. Med. 189, 1907–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gold M. C., Cerri S., Smyk-Pearson S., Cansler M. E., Vogt T. M., Delepine J., Winata E., Swarbrick G. M., Chua W. J., Yu Y. Y., Lantz O., Cook M. S., Null M. D., Jacoby D. B., Harriff M. J., Lewinsohn D. A., Hansen T. H., and Lewinsohn D. M. (2010) Human mucosal associated invariant T cells detect bacterially infected cells. PLoS Biol. 8, e1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gold M. C., and Lewinsohn D. M. (2013) Co-dependents: MR1-restricted MAIT cells and their antimicrobial function. Nat. Rev. Microbiol. 11, 14–19 [DOI] [PubMed] [Google Scholar]
- 8.Reantragoon R., Corbett A. J., Sakala I. G., Gherardin N. A., Furness J. B., Chen Z., Eckle S. B., Uldrich A. P., Birkinshaw R. W., Patel O., Kostenko L., Meehan B., Kedzierska K., Liu L., Fairlie D. P., Hansen T. H., Godfrey D. I., Rossjohn J., McCluskey J., and Kjer-Nielsen L. (2013) Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells. J. Exp. Med. 210, 2305–2320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lepore M., Kalinichenko A., Colone A., Paleja B., Singhal A., Tschumi A., Lee B., Poidinger M., Zolezzi F., Quagliata L., Sander P., Newell E., Bertoletti A., Terracciano L., De Libero G., and Mori L. (2014) Parallel T-cell cloning and deep sequencing of human MAIT cells reveal stable oligoclonal TCRβ repertoire. Nat. Commun. 5, 3866 , [DOI] [PubMed] [Google Scholar]
- 10.Patel O., Kjer-Nielsen L., Le Nours J., Eckle S. B., Birkinshaw R., Beddoe T., Corbett A. J., Liu L., Miles J. J., Meehan B., Reantragoon R., Sandoval-Romero M. L., Sullivan L. C., Brooks A. G., Chen Z., Fairlie D. P., McCluskey J., and Rossjohn J. (2013) Recognition of vitamin B metabolites by mucosal-associated invariant T cells. Nat. Commun. 4, 2142. [DOI] [PubMed] [Google Scholar]
- 11.Reantragoon R., Kjer-Nielsen L., Patel O., Chen Z., Illing P. T., Bhati M., Kostenko L., Bharadwaj M., Meehan B., Hansen T. H., Godfrey D. I., Rossjohn J., and McCluskey J. (2012) Structural insight into MR1-mediated recognition of the mucosal associated invariant T cell receptor. J. Exp. Med. 209, 761–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldfinch N., Reinink P., Connelley T., Koets A., Morrison I., and Van Rhijn I. (2010) Conservation of mucosal associated invariant T (MAIT) cells and the MR1 restriction element in ruminants, and abundance of MAIT cells in spleen. Vet. Res. 41, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tynan F. E., Borg N. A., Miles J. J., Beddoe T., El-Hassen D., Silins S. L., van Zuylen W. J., Purcell A. W., Kjer-Nielsen L., McCluskey J., Burrows S. R., and Rossjohn J. (2005) High resolution structures of highly bulged viral epitopes bound to major histocompatibility complex class I. Implications for T-cell receptor engagement and T-cell immunodominance. J. Biol. Chem. 280, 23900–23909 [DOI] [PubMed] [Google Scholar]
- 14.Tynan F. E., Reid H. H., Kjer-Nielsen L., Miles J. J., Wilce M. C., Kostenko L., Borg N. A., Williamson N. A., Beddoe T., Purcell A. W., Burrows S. R., McCluskey J., and Rossjohn J. (2007) A T cell receptor flattens a bulged antigenic peptide presented by a major histocompatibility complex class I molecule. Nat. Immunol. 8, 268–276 [DOI] [PubMed] [Google Scholar]
- 15.Van Rhijn I., Kasmar A., de Jong A., Gras S., Bhati M., Doorenspleet M. E., de Vries N., Godfrey D. I., Altman J. D., de Jager W., Rossjohn J., and Moody D. B. (2013) A conserved human T cell population targets mycobacterial antigens presented by CD1b. Nat. Immunol. 14, 706–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Treiner E., Duban L., Bahram S., Radosavljevic M., Wanner V., Tilloy F., Affaticati P., Gilfillan S., and Lantz O. (2003) Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature 422, 164–169 [DOI] [PubMed] [Google Scholar]
- 17.Hashimoto K., Hirai M., and Kurosawa Y. (1995) A gene outside the human MHC related to classical HLA class I genes. Science 269, 693–695 [DOI] [PubMed] [Google Scholar]
- 18.Yamaguchi H., and Hashimoto K. (2002) Association of MR1 protein, an MHC class I-related molecule, with β2-microglobulin. Biochem. Biophys. Res. Commun. 290, 722–729 [DOI] [PubMed] [Google Scholar]
- 19.Miley M. J., Truscott S. M., Yu Y. Y., Gilfillan S., Fremont D. H., Hansen T. H., and Lybarger L. (2003) Biochemical features of the MHC-related protein 1 consistent with an immunological function. J. Immunol. 170, 6090–6098 [DOI] [PubMed] [Google Scholar]
- 20.Parra-Cuadrado J. F., Navarro P., Mirones I., Setién F., Oteo M., and Martínez-Naves E. (2000) A study on the polymorphism of human MHC class I-related MR1 gene and identification of an MR1-like pseudogene. Tissue Antigens 56, 170–172 [DOI] [PubMed] [Google Scholar]
- 21.Riegert P., Wanner V., and Bahram S. (1998) Genomics, isoforms, expression, and phylogeny of the MHC class I-related MR1 gene. J. Immunol. 161, 4066–4077 [PubMed] [Google Scholar]
- 22.Yamaguchi H., Hirai M., Kurosawa Y., and Hashimoto K. (1997) A highly conserved major histocompatibility complex class I-related gene in mammals. Biochem. Biophys. Res. Commun. 238, 697–702 [DOI] [PubMed] [Google Scholar]
- 23.Huang S., Martin E., Kim S., Yu L., Soudais C., Fremont D. H., Lantz O., and Hansen T. H. (2009) MR1 antigen presentation to mucosal-associated invariant T cells was highly conserved in evolution. Proc. Natl. Acad. Sci. U.S.A. 106, 8290–8295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hansen T. H., Huang S., Arnold P. L., and Fremont D. H. (2007) Patterns of nonclassical MHC antigen presentation. Nat. Immunol. 8, 563–568 [DOI] [PubMed] [Google Scholar]
- 25.Gold M. C., McLaren J. E., Reistetter J. A., Smyk-Pearson S., Ladell K., Swarbrick G. M., Yu Y. Y., Hansen T. H., Lund O., Nielsen M., Gerritsen B., Kesmir C., Miles J. J., Lewinsohn D. A., Price D. A., and Lewinsohn D. M. (2014) MR1-restricted MAIT cells display ligand discrimination and pathogen selectivity through distinct T cell receptor usage. J. Exp. Med. 211, 1601–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kjer-Nielsen L., Patel O., Corbett A. J., Le Nours J., Meehan B., Liu L., Bhati M., Chen Z., Kostenko L., Reantragoon R., Williamson N. A., Purcell A. W., Dudek N. L., McConville M. J., O'Hair R. A., Khairallah G. N., Godfrey D. I., Fairlie D. P., Rossjohn J., and McCluskey J. (2012) MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 491, 717–723 [DOI] [PubMed] [Google Scholar]
- 27.Le Bourhis L., Martin E., Péguillet I., Guihot A., Froux N., Coré M., Lévy E., Dusseaux M., Meyssonnier V., Premel V., Ngo C., Riteau B., Duban L., Robert D., Huang S., Rottman M., Soudais C., and Lantz O. (2010) Antimicrobial activity of mucosal-associated invariant T cells. Nat. Immunol. 11, 701–708 [DOI] [PubMed] [Google Scholar]
- 28.Burgess C., O'Connell-Motherway M., Sybesma W., Hugenholtz J., and van Sinderen D. (2004) Riboflavin production in Lactococcus lactis: potential for in situ production of vitamin-enriched foods. Appl Environ. Microbiol. 70, 5769–5777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corbett A. J., Eckle S. B., Birkinshaw R. W., Liu L., Patel O., Mahony J., Chen Z., Reantragoon R., Meehan B., Cao H., Williamson N. A., Strugnell R. A., Van Sinderen D., Mak J. Y., Fairlie D. P., Kjer-Nielsen L., Rossjohn J., and McCluskey J. (2014) T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature 509, 361–365 [DOI] [PubMed] [Google Scholar]
- 30.Soudais C., Samassa F., Sarkis M., Le Bourhis L., Bessoles S., Blanot D., Hervé M., Schmidt F., Mengin-Lecreulx D., and Lantz O. (2015) In vitro and in vivo analysis of the Gram-negative bacteria-derived riboflavin precursor derivatives activating mouse MAIT cells. J. Immunol. 194, 4641–4649 [DOI] [PubMed] [Google Scholar]
- 31.Eckle S. B., Birkinshaw R. W., Kostenko L., Corbett A. J., McWilliam H. E., Reantragoon R., Chen Z., Gherardin N. A., Beddoe T., Liu L., Patel O., Meehan B., Fairlie D. P., Villadangos J. A., Godfrey D. I., Kjer-Nielsen L., McCluskey J., and Rossjohn J. (2014) A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells. J. Exp. Med. 211, 1585–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.López-Sagaseta J., Dulberger C. L., Crooks J. E., Parks C. D., Luoma A. M., McFedries A., Van Rhijn I., Saghatelian A., and Adams E. J. (2013) The molecular basis for Mucosal-Associated Invariant T cell recognition of MR1 proteins. Proc. Natl. Acad. Sci. U.S.A. 110, E1771–E1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bacher A., Eberhardt S., Fischer M., Kis K., and Richter G. (2000) Biosynthesis of vitamin B2 (riboflavin). Annu. Rev. Nutr. 20, 153–167 [DOI] [PubMed] [Google Scholar]
- 34.Cushman M., Yang D., Gerhardt S., Huber R., Fischer M., Kis K., and Bacher A. (2002) Design, synthesis, and evaluation of 6-carboxyalkyl and 6-phosphonoxyalkyl derivatives of 7-oxo-8-ribitylaminolumazines as inhibitors of riboflavin synthase and lumazine synthase. J. Org. Chem. 67, 5807–5816 [DOI] [PubMed] [Google Scholar]
- 35.Wang Y., and Ho C. T. (2012) Flavour chemistry of methylglyoxal and glyoxal. Chem. Soc. Rev. 41, 4140–4149 [DOI] [PubMed] [Google Scholar]
- 36.Rahimpour A., Koay H. F., Enders A., Clanchy R., Eckle S. B., Meehan B., Chen Z., Whittle B., Liu L., Fairlie D. P., Goodnow C. C., McCluskey J., Rossjohn J., Uldrich A. P., Pellicci D. G., and Godfrey D. I. (2015) Identification of phenotypically and functionally heterogeneous mouse mucosal-associated invariant T cells using MR1 tetramers. J. Exp. Med. 212, 1095–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Birkinshaw R. W., Kjer-Nielsen L., Eckle S. B., McCluskey J., and Rossjohn J. (2014) MAITs, MR1 and vitamin B metabolites. Curr. Opin. Immunol. 26, 7–13 [DOI] [PubMed] [Google Scholar]
- 38.Young M. H., U'Ren L., Huang S., Mallevaey T., Scott-Browne J., Crawford F., Lantz O., Hansen T. H., Kappler J., Marrack P., and Gapin L. (2013) MAIT cell recognition of MR1 on bacterially infected and uninfected cells. PLoS One 8, e53789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greenaway H. Y., Ng B., Price D. A., Douek D. C., Davenport M. P., and Venturi V. (2013) NKT and MAIT invariant TCRα sequences can be produced efficiently by VJ gene recombination. Immunobiology 218, 213–224 [DOI] [PubMed] [Google Scholar]
- 40.Sakala I. G., Kjer-Nielsen L., Eickhoff C. S., Wang X., Blazevic A., Liu L., Fairlie D. P., Rossjohn J., McCluskey J., Fremont D. H., Hansen T. H., and Hoft D. F. (2015) Functional heterogeneity and antimycobacterial effects of mouse mucosal-associated invariant T cells specific for riboflavin metabolites. J. Immunol. 195, 587–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chua W. J., Kim S., Myers N., Huang S., Yu L., Fremont D. H., Diamond M. S., and Hansen T. H. (2011) Endogenous MHC-related protein 1 is transiently expressed on the plasma membrane in a conformation that activates mucosal-associated invariant T cells. J. Immunol. 186, 4744–4750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang S., Gilfillan S., Kim S., Thompson B., Wang X., Sant A. J., Fremont D. H., Lantz O., and Hansen T. H. (2008) MR1 uses an endocytic pathway to activate mucosal-associated invariant T cells. J. Exp. Med. 205, 1201–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harriff M. J., Cansler M. E., Toren K. G., Canfield E. T., Kwak S., Gold M. C., and Lewinsohn D. M. (2014) Human lung epithelial cells contain Mycobacterium tuberculosis in a late endosomal vacuole and are efficiently recognized by CD8+ T cells. PLoS One 9, e97515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Le Bourhis L., Dusseaux M., Bohineust A., Bessoles S., Martin E., Premel V., Coré M., Sleurs D., Serriari N. E., Treiner E., Hivroz C., Sansonetti P., Gougeon M. L., Soudais C., and Lantz O. (2013) MAIT cells detect and efficiently lyse bacterially-infected epithelial cells. PLoS Pathog. 9, e1003681. [DOI] [PMC free article] [PubMed] [Google Scholar]