Background: The CORVET and HOPS complexes regulate endosomal cargo trafficking but have not been well characterized in mammals.
Results: A detailed analysis of subunit interactions within the mammalian CORVET, HOPS, and VIPAS39/VPS33B complexes.
Conclusion: Tethering complexes have adapted to the higher complexity of trafficking in mammalian cells.
Significance: This work provides a detailed architectural insight into the mammalian endosomal tethering complexes.
Keywords: endosome, intracellular trafficking, membrane fusion, Rab, transport
Abstract
Trafficking of cargo through the endosomal system depends on endosomal fusion events mediated by SNARE proteins, Rab-GTPases, and multisubunit tethering complexes. The CORVET and HOPS tethering complexes, respectively, regulate early and late endosomal tethering and have been characterized in detail in yeast where their sequential membrane targeting and assembly is well understood. Mammalian CORVET and HOPS subunits significantly differ from their yeast homologues, and novel proteins with high homology to CORVET/HOPS subunits have evolved. However, an analysis of the molecular interactions between these subunits in mammals is lacking. Here, we provide a detailed analysis of interactions within the mammalian CORVET and HOPS as well as an additional endosomal-targeting complex (VIPAS39-VPS33B) that does not exist in yeast. We show that core interactions within CORVET and HOPS are largely conserved but that the membrane-targeting module in HOPS has significantly changed to accommodate binding to mammalian-specific RAB7 interacting lysosomal protein (RILP). Arthrogryposis-renal dysfunction-cholestasis (ARC) syndrome-associated mutations in VPS33B selectively disrupt recruitment to late endosomes by RILP or binding to its partner VIPAS39. Within the shared core of CORVET/HOPS, we find that VPS11 acts as a molecular switch that binds either CORVET-specific TGFBRAP1 or HOPS-specific VPS39/RILP thereby allowing selective targeting of these tethering complexes to early or late endosomes to time fusion events in the endo/lysosomal pathway.
Introduction
The endocytic pathway is a dynamic system in which vesicles are continuously fusing, moving, and budding in order to deliver their cargo to the correct compartment. After internalization, endocytic cargo is delivered to the early endosome (EE)3 from which it can be recycled or degraded (1). Cargo destined for degradation such as internalized nutrients, activated growth receptors, and endocytosed pathogens are targeted to the late endosomes (LEs) and lysosomes where the acidic environment and resident proteases allow for degradation. Rab5 (EE) and Rab7 (LE) are master regulators of endosomal transport and fusion and regulate cargo flux through the endocytic system in conjunction with multisubunit motor and tethering complexes. In yeast, EE and LE tethering is regulated by the CORVET (class C core vacuole/endosome tethering) and HOPS (homotypic fusion and vacuole protein sorting) complexes, respectively, which have been characterized in high detail (2–8). Both complexes consist of a shared core (vps16, vps18, vps11, vps33) that associates with CORVET-specific (vps3 and vps8) or HOPS-specific (vps39 and vps41) subunits. These different subunits target the complexes to membranes by interaction with respectively vps21 (yeast Rab5) and Ypt7 (yeast Rab7) (6, 7). In addition, vps41 can also bind lipids (9). Apparent homologues of CORVET and HOPS subunits in mammals have been implicated in endosomal maturation (10), EE fusion (11), and fusion of lysosomes with late endosomes, phagosomes or autophagosomes (12–15). Based on sequence alignment, homologs of all eight yeast HOPS and CORVET subunits are present in mammalian cells with the addition of two novel homologues; VIPAS39 (also known as SPE-39 or VIPAR) and VPS33B. Recently it was shown that mammalian VPS8 and TGFBRAP1 are the vps8 and vps3 homologues (11), and the mammalian CORVET, therefore, consists of VPS8, TGFBRAP1, VPS18, VPS16, VPS33A, and VPS11, whereas HOPS consists of VPS41, VPS39, VPS18, VPS16, VPS33A, and VPS11. Although VPS33B was initially shown to interact with HOPS subunits (16), more recent data suggest that VPS33B and VIPAS39 form a separate complex (12).
The organization of these mammalian complexes has only partly been addressed with seemingly conflicting results and hampered the interpretation of functional experiments in which the roles of specific or multiple units are addressed (17–27). Furthermore, it is not known what drives membrane specificity of these complexes in mammals. Here we provide an extensive map of the mammalian CORVET, HOPS, and VPS33B-VIPAS39 complexes describing their intersubunit interactions and membrane recruitment. In agreement with Wartosch et al. (12) we find that VIPAS39 and VPS33B are not part of the CORVET or HOPS complex but assemble in a distinct complex. We find that the CORVET complex is prevented from recruitment to LE (in line with its function at the EE). Within the HOPS complex there are multiple RILP binding modules, and pathogenic (arthrogryposis-renal dysfunction-cholestasis (ARC) syndrome) mutations in VPS33B disrupt VPS33B-VIPAS39 complex assembly or RILP-dependent LE recruitment. Within the shared CORVET/HOPS core, VPS11 can bind TGFBRAP1 (CORVET) as well as VPS39 (HOPS), and these subunits likely compete for binding to VPS11 in the core of the CORVET-HOPS complex, thereby driving the membrane-specific assembly and tethering capability of these complexes.
Experimental Procedures
Reagents
Rabbit anti-GFP and rabbit anti-mRFP antibodies were generated in-house using purified His-mRFP or His-GFP recombinant proteins, respectively. Cross-reactivity has been excluded by Western blot analyses with various mRFP- or GFP-labeled fusion proteins. Other antibodies used were: mouse anti-CD63 (28), mouse anti-EEA1 (ab2900; Abcam), mouse anti-V5 and anti-V5-HRP (R96025, R96125; Invitrogen), anti-Myc (2278P; Cell Signaling), anti-Myc-HRP (NB600-302H; Novus), anti-HA (12013819001, Roche Applied Science), anti-HA-HRP (ab1190; Abcam), anti-FLAG (M2) and anti-FLAG-HRP (F3165, A8592; Sigma), anti-VPS33A (C1C3) (GTX119416; GeneTex), anti-VPS33b (12195-1-AP, Proteintech), anti-VPS11 (19140-1-AP; Proteintech), anti-TGFBRAP1 (SC-13134 Santa Cruz), anti-VPS41 (13869-1-AP, Proteintech), anti-VPS8 (HPA036871, Sigma), anti-VPS16 (17776-1-AP; Proteintech), and anti-VIPAS39 was a gift of V. Faundez (Center for Translational Social Neuroscience, Emory University, Atlanta). Fluorescent and HRP-conjugated secondary antibodies were obtained from Invitrogen.
Constructs
RAB7 and RILP (29) and full-length VPS constructs and GFP-VPS33b L30P (30)have been described previously. GFP-VPS16 and GFP-VPS18 were gifts from Chengyu Liang (Department of Molecular Microbiology and Immunology, University of Southern California, Los Angeles). VPS33a was a gift of V. Faundez (Center translational social neuroscience, Emory University, Atlanta), Vps39 was a gift of J. Bonifacino (National Institutes of Health, Bethesda, MD). HA-VPS8 was purchased from Origene and inserted into pcDNA3.2-HA/DEST vector (Invitrogen) using Gateway recombination cloning. Truncation constructs were generated by PCR using these respective cut sites, donor vectors, forward primer, and reverse primers: VPS33b 1–437, Xho1-BamH1 GFP-C1, cccaCTCGAGCCATGGCTTTTCCCCATCG and cccaGGATCCTCACAGATTGGAGAAGGTTAGCA; VPS33b 438–617, EcoR1-BamH1 GFP-C1, CCCAGAATTCCCTGCGAAGAGCTGGGCTCCT and CCCAGGATCCTCAGGCTTTCACCTCACTCA; mVPS16 1–516, EcoR1-Asp-718 GFP-C1, cccaGAATTCCATGGACTGTTACACTGCGAA and cccaGGTACCTCAACCAGGCGTGTCACCCAGCT; mVPS16 1–420, EcoR1-BamH1 GFP-C1, cccaGAATTCCATGGACTGCTACACGGCGAA and cccaGGATCCTCAGTGCACGAAGCTGTCGGGTG; mVPS16 517–835, EcoR1-BamH1 GFP-C1, cccaGAATTCCTCTTACTCCGACATTGCTGC and cccaGGATCCTCATTGTGCCCTGGCCCGTTGAA; mVPS16 517–839, EcoR1-EcoR1 GFP-C1, cccaGAATTCCTCTTACTCCGACATTGCTGC and agaattcTCACTTCTTCTGGGCTTGTG; VPS41 1–790, Xho1-BamH1 GFP-C1, cccaCTCGAGCCATGGCGGAAGCAGAGGAGCA and GGATCCTCAGATGTTCTCCTCATCAACAA; VPS41 1–571, Xho1-BamH1 GFP-C1, cccaCTCGAGCCATGGCGGAAGCAGAGGAGCA and cccaGGATCCTCAAAGCATGTCAACAGCTTTCT; VPS41 712–854, BglII-EcoR1 GFP-C1, cccaAGATCTGGCTTGTTAAACAACATTGG and cccaGAATTCCTATTTTTTCATCTCCAAAA; VPS11 1–773, Xho1-EcoR1 GFP-C1, cccaCTCGAGCCATGGCGGCCTACCTGCAGTG and cccaGaattcTCAGTCCCTGATGACGGAGA; VPS11 774–940, EcoR1-BamH1 GFP-C1, cccagaattccTACCTGGTCCAAAAACTACA and cccaGGATCCTCAAGTGCCCCTCCTGGAGTGCA; VPS11 774–812, EcoR1-BamH1 GFP-C1, cccagaattccTACCTGGTCCAAAAACTACA and cccaGGATCCTCAGGCCTTGAGCTCTTGGATCT; VPS11 774–859, EcoR1-BamH1 GFP-C1, cccagaattccTACCTGGTCCAAAAACTACA and cccaGGATCCTCAGGTGGGGCAGTCAGCATCAC; VPS11 859–940, EcoR1-BamH1 GFP-C1, cccaGAATTCCACCTGCCTCCCTGAAAACCG and cccaGGATCCTCAAGTGCCCCTCCTGGAGTGCA; VPS18 1–743, EcoR1-BamH1 GFP-C1, CCCAGAATTCCATGGCGTCCATCCTGGATGA and CCCAGGATCCTTAATCAGGAAAGAAGGGCA; VPS18 1–612, EcoR1-BamH1 GFP-C1, CCCAGAATTCCATGGCGTCCATCCTGGATGA and cccaGGATCCTCAATCTACAAGCTGGCGGGGGAT; VPS18 500–612, EcoR1-BamH1 GFP-C1, cccaGAATTCCAGCCGGCTTGGGGCTCTGCA and cccaGGATCCTCAATCTACAAGCTGGCGGGGGAT; VPS18 743–973, EcoR1-BamH1 GFP-C1, CCCAGAATTCCGATTTCGTCACCATCGACCA and cccaGGATCCTCACAGCCAACTGAGCTGCTCCT; VPS18 854–973, EcoR1-BamH1 GFP-C1, cccaGAATTCCGGCACTGTGGAGCCCCAGGA and cccaGGATCCTCACAGCCAACTGAGCTGCTCCT; VPS39 551–761, EcoR1-BamH1 GFP-C1, cccaGAATTCCCTGCATTTGATTTTCTCCTA and cccaGGATCCTCATAGTTCCAGCTTGATTGG; VPS39 761–869, EcoR1-BamH1 GFP-C1, cccaGAATTCCCTACTGGAGCCAAAAGCCAA and cccaGGATCCTCAATTGGGGTATCTTGCAAATG.
Cell Culture and Microscopy
MelJuSo cells were cultured in Iscove's modified Dulbecco's medium (Invitrogen) supplemented with 8% FCS in a 5% CO2-humidified culture hood at 37 °C. HEK293 cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 8% FCS in a 5% CO2 humidified culture hood at 37 °C. All specimens were analyzed by confocal laser-scanning microscopes (TCS-SP1, TCS-SP2, or AOBS; Leica) equipped with HCX Plan-Apochromat 63× NA 1.32 and HCX Plan-Apochromat lbd.bl 63× NA 1.4 oil-corrected objective lenses (Leica) using LCS (Leica) acquisition software or Deltavision wide field microscope (Applied Precision) with a 100/1.4A immersion objective. Widefield images were deconvolved using SoftWorx software (Applied Precision).
Transfection
Expression constructs were transfected using Effectene reagents (Qiagen) according to the manufacturer's instructions. For silencing, cells were transfected with Dharmafect1 (Thermo Fisher Scientific) with siRNAs (ON-TARGETplus SMARTpool) against VPS16, VPS33B, VPS33A, SPE-39, or control siRNA (Thermo Fisher Scientific).
Microscopy Sample Preparation
Transfected cells were fixed 24 h post-transfection with 4% formaldehyde in PBS for 15 min and permeabilized for 10 min with 0.05% Triton X-100 in PBS at room temperature. Nonspecific binding of antibodies was blocked by 0.5% BSA in PBS for 40 min, after which cells were incubated with primary antibodies in 0.5%BSA in PBS for 1 h at room temperature. After washing 3 times with PBS, primary antibodies were visualized with Alexa-Fluor secondary antibody conjugates antibodies in 0.5% BSA in PBS for 30 min at room temperature (Invitrogen). After three washes with PBS samples were mounted in Vectashield mounting medium (Vector Laboratories).
Protein Immunoprecipitation
MelJuSo cells were washed with ice-cold PBS and scraped into cell lysis buffer (50 mm Tris-HCl, 150 mm NaCl, 5 mm MgCl2, 1% Nonidet P-40, 10% glycerol, pH 7.4) supplemented with complete EDTA-free protease inhibitor mixture. Cell lysates were obtained by incubation on ice for 10 min followed by centrifugation for clearing. The supernatants were incubated for 2 h with respective antibodies followed by capture with protein G-Sepharose 4 FF resin and washed extensively with wash-buffer (50 mm Tris, 150 mm NaCl, 5 mm MgCl2, 10% glycerol, pH 7.4). For GFP-tagged pulldown, GFP-TRAP beads were used (Chromotek). All experiments were repeated multiple times. To compose the figure panels, inserts from a single experiment containing all the experimental conditions (and all the transfected constructs) were composed as described in the figure legends.
Statistical Analysis
For calculation of correlation coefficients the signal intensity over a vector was plotted using the plot profile tool in ImageJ (http://imagej.nih.gov) in the respective fluorescence channels. Correlation coefficient for colocalization assays were calculated from plot profiles measuring intensity for the indicated fluorescence channels over a vector through the cells (as in van der Kant et al. (30)). Correlation coefficients for the two plots were calculated in Excel using the function,
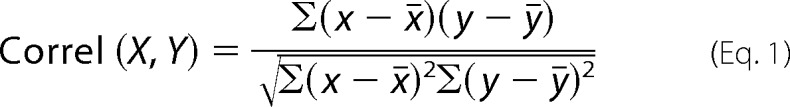 |
Statistical testing was performed in Graphpad Prism 6 using a one-way analysis of variance with Tukey's multiple comparison test for significance.
Results
Characterization and Definition of the Mammalian CORVET, HOPS, and VPS33B-VIPAS39 Complex
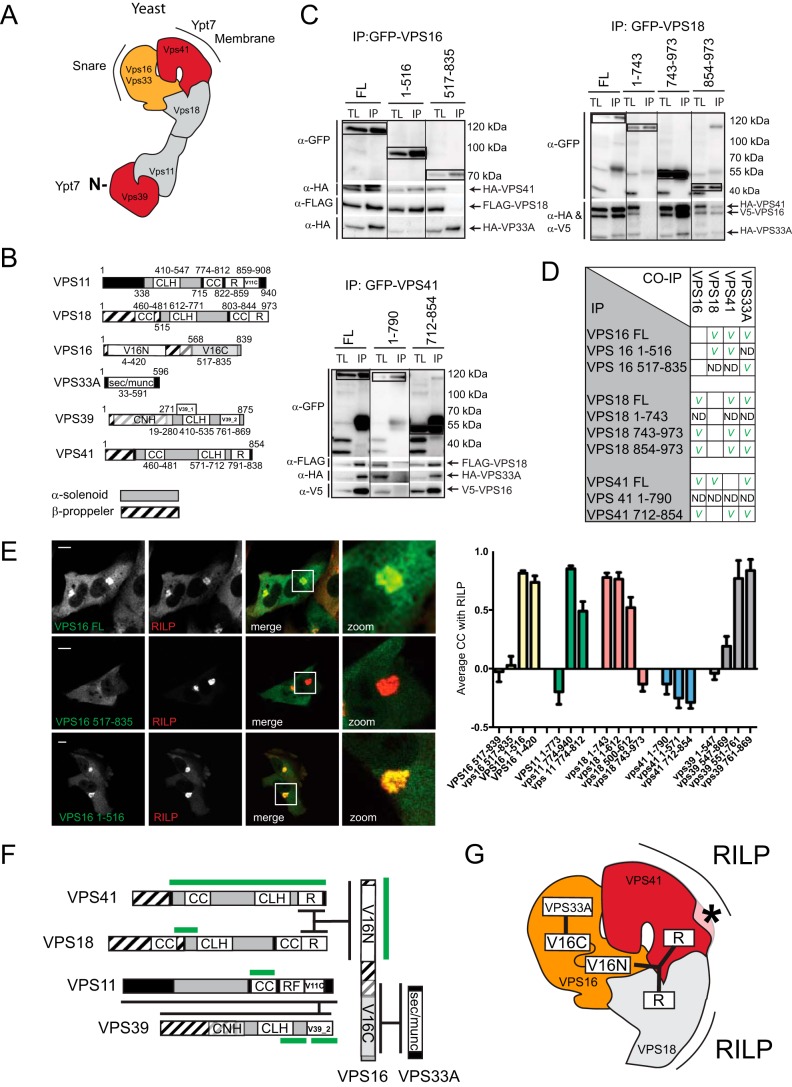
The yeast CORVET and HOPS complexes are assembled from eight different proteins (a Vps16/Vps33/Vps18/Vps11 core with Vps3/Vps8 or Vps39/Vps41). Mammals do not possess 8, but 10 homologues to the yeast CORVET/HOPS subunits. Two mammalian homologues for yeast Vps33 exist (VPS33A and VPS33B) as well as an additional protein with weak homology to Vps16 (VIPAS39). To probe the interactions between the 10 putative CORVET/HOPS subunits and determine the topology of the different subunits in the CORVET-HOPS complexes we performed an extensive array of co-immunoprecipitations (IP) in human MelJuSo cells with antibodies against endogenous VIPAS39, VPS33B, VPS11, VPS16, VPS8, and VPS33A (Fig. 1A) (antibodies specific for other components were either not available or did not work for IP). We observed extensive interactions between VPS11, VPS16, VPS8, TGFBRAP1, and VPS33A. In contrast, VIPAS39 and VPS33B interacted with each other but not with any of the other canonical CORVET/HOPS subunits in line with recent data (12).
FIGURE 1.
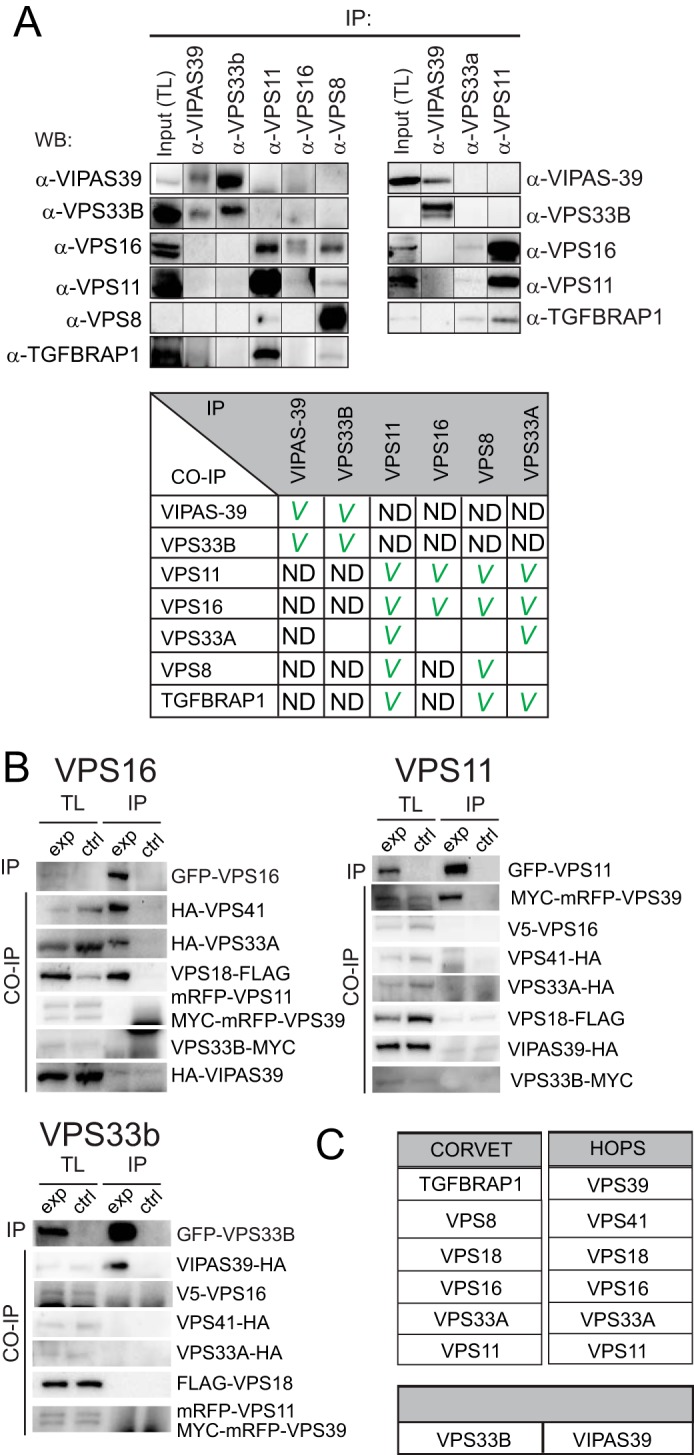
Biochemical definition of mammalian CORVET, HOPS, and VPS33B-VIPAS39. A, MelJuSo lysates were immunoprecipitated (IP) by the indicated antibodies and analyzed by WB using indicated antibodies. Each panel represents an independent experiment. Upper left panel, MelJuSo cell lysates were generated (at which point a total lysate (TL) fraction was taken) and divided over five fractions, and an immunoprecipitation was performed on each lysate using one of the indicated antibodies (generating five experimental conditions, horizontal axis). The five experimental conditions were run on the same blot with the same exposures for each detection antibody. A separate gel was run for each detection antibody (on the vertical axis). From these blots cutouts were taken and grouped to compose the figure panel. Top right panel, same as the left panel, with three experimental conditions. The table (lower panel) summarizes the results from the IP experiments. A checkmark indicates interaction detected. ND = not detected; empty boxes, not tested. B, lysates of MelJuSo cells expressing eight tagged (experimental condition (exp)) or seven tagged subunits (control condition (ctrl)) not-expressing the GFP-tagged subunit) were immunoprecipitated (IP) with anti-GFP (to pull down VPS16, VPS11, or VPS33B) and analyzed by WB. Shown are total lysates for experimental and control lanes and the immunoprecipitated fraction for experimental and control lanes. (C) Summary of experimental data A-E, VIPAS39 and VPS33B do not interact with HOPS complex subunits.
To further map the HOPS complex and define possible interactions between VIPAS39 and VPS33B with HOPS components that we could not map in endogenous pulldown experiments, we ectopically co-expressed all six putative HOPS complex subunits as well as VPS33B and VIPAS39 as tagged proteins. We previously showed that expression levels of these tagged proteins in our systems are low (comparable to endogenous) and that the tagged subunits incorporate into functional complexes (30).
We pulled down GFP-VPS16, GFP-VPS11, or GFP-VPS33B and detected the associated proteins by Western blotting with antibodies against the different epitope tags (Fig. 1B). GFP-VPS16 specifically interacted with VPS41, VPS18, and VPS33A. In addition, GFP-VPS11 interacted with VPS39. In contrast to the endogenous co-IP (Fig. 1A), no interactions between the two groups (VPS16/41/18/33a and VPS11/39) were observed. A pulldown experiment with GFP-VPS11 also failed to detect interactions between GFP-VPS11 and endogenous VPS16/VPS33a (data not shown). This may reflect a limitation of the overexpression system or an interference with normal interactions by the associated tags. In line with the endogenous IP (Fig. 1A), we again observed strong interactions between VPS33B and VIPAS39 but not with any other subunit of the HOPS complex. This suggests that mammalian HOPS consist of VPS11/VPS16/VPS18/VPS33A/VPS39/VPS41, whereas VPS33B-VIPAS39 assembles in a distinct complex (Fig. 1C). Furthermore, the observation that tagged subunits recapitulate in large part endogenous interactions indicates we can use these constructs for further detailed mapping of the respective complexes.
A High Detail Interaction Map of the Mammalian HOPS Complex
The yeast HOPS complex resembles a seahorse-shaped complex with a head (Vps16/Vps33/Vps41/Vps18) and a tail (Vps11/Vps39), and interactions within the complex have been extensively mapped (6, 7) (Fig. 2A). No structure for the mammalian HOPS complex is available, yet our experiments (Fig. 1) suggest that interactions within the head (VPS16/VPS33/VPS41/VPS18) and tail (VPS39/VPS11) are conserved from yeast to mammal. To study the interactions within the proposed head of the mammalian HOPS complex, we expressed tagged truncation mutants of a particular subunit (domain organizations based on STRING 9.1 depicted in Fig. 2B and assessed the interactions with the three other head proteins by co-IP experiments (Fig. 2C and summarized in Fig. 2D). We observed that VPS16 is a central protein in the head as its C terminus binds VPS33A, whereas the N-terminal segment binds to VPS41 and VPS18. The Ring finger (R) domains of VPS18 and VPS41 interact with the N terminus of VPS16 possibly acting in a tripartite interaction.
FIGURE 2.
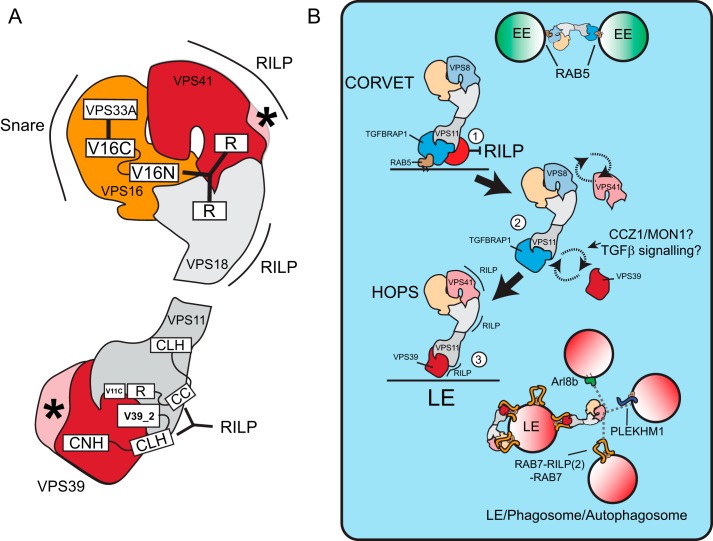
Interactions within the mammalian HOPS complex. A, structure of the yeast HOPS complex. Yeast HOPS interacts with Ypt-7 via Vps41 and the N terminus of Vps39. B, domain organization of the mammalian HOPS complex subunit orthologs. CLH, clathrin heavy chain repeat; CC, coiled coil; R, ring finger; V11C, PFAM VPS11 C terminus; V16N, PFAM VPS11 N terminus; V16C, PFAM VPS11 C terminus; V39_1, VPS39 domain 1; V39_2, VPS39 domain 2; CNH, citron homology. C, lysates of MelJuSo cells co-expressing tagged VPS constructs as indicated were immunoprecipitated (IP) with anti-GFP antibodies and analyzed by WB using anti-HA, anti-FLAG, anti-V5, and anti-GFP antibodies as indicated. Within each panel, experimental conditions were run on the same blot with the same exposures for each detection antibody, and cutouts were taken and grouped for presentation purposes. D, summarized results of Fig. 2C. E, MelJuSo cells expressing GFP-VPS16 constructs (green) and mRFP-RILP (red). Scale bars: 10 μm. Graphs show average correlation coefficient (CC) ± S.E. n > 25, between different VPS truncation constructs and RILP. F, detailed map of domain interactions and membrane targeting modules (domains required for RILP binding are in green) within the mammalian HOPS complex. G, interactions in the head domain of the mammalian HOPS complex superimposed on the known structure of the yeast HOPS complex. Asterisk indicates functional divergence (absence of lipid binding motif in mammalian VPS41) between yeast and mammalian HOPS.
Different from yeast, the mammalian HOPS complex is not recruited to membranes by RAB7 (Ypt7 in yeast) but by binding to the RAB7 effector RILP that has no apparent ortholog in yeast (30, 32). To evaluate membrane targeting of the mammalian HOPS complex, we mapped the minimal domains for membrane recruitment of distinct subunits by expressing tagged truncation mutants and assessed their recruitment to RILP-containing LE (Fig. 2E). In addition to VPS41 (32), we observed that small domains of various HOPS subunits could be recruited to RILP (summarized in Fig. 2, F and G). This suggests that RILP could act as a scaffold in the assembly of the HOPS complex or that interactions within the mammalian HOPS complex are further stabilized via secondary interactions of its subunits with RILP. In assembly, our data suggest that interactions within the HOPS complex are reasonably conserved from yeast to mammals, yet the complex has significantly evolved to accommodate a shift from RAB7 to binding to RAB7 effector RILP instead, possibly reflecting higher order regulation such as the concomitant binding of the dynein-motor complex (30).
VPS11 as the Core Receptor for HOPS- and CORVET-specific Subunits
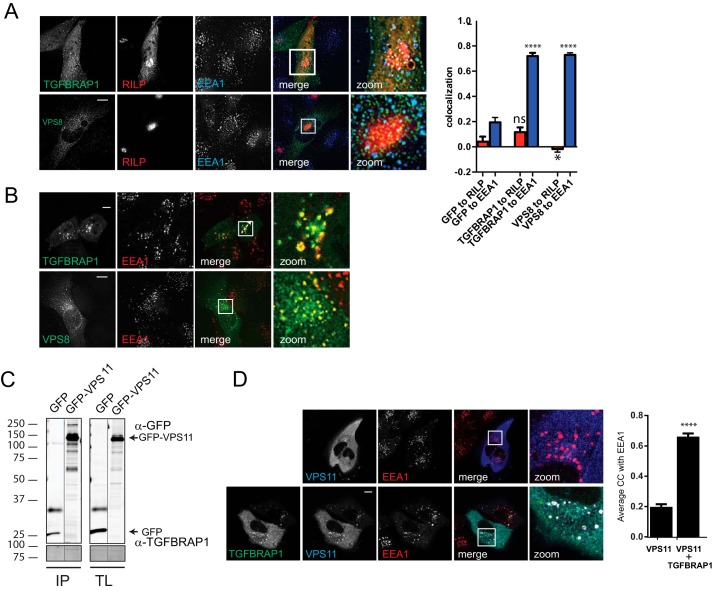
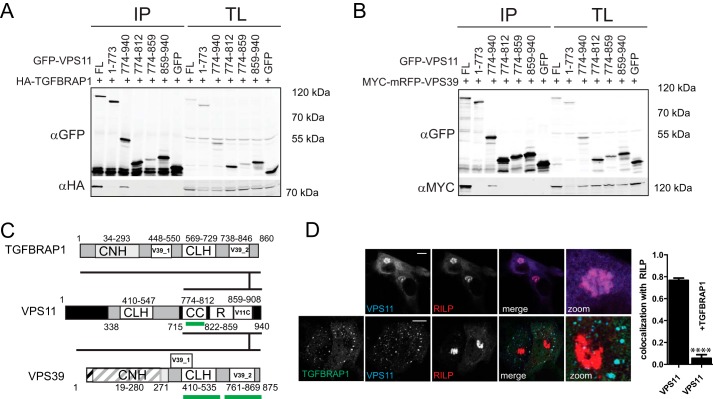
We have shown that TGFBRAP1 and VPS8 endogenously bind core CORVET/HOPS subunits including VPS11 and VPS16 (Fig. 1A) and that VPS11 and VPS16 are recruited to LE by RILP (Ref. 30; Fig. 2E). This argued that overexpression of RILP via their secondary interactions with VPS11 or VPS16 might attract VPS8 and TGFBRAP1 to LE. However, when RILP and VPS8 or TGFBRAP1 were co-expressed, the latter were not recruited to RILP but remained localized to EE (Fig. 3A) similar to VPS8 and TGFBRAP1 expression alone (Fig. 3B). This indicates an active mechanism that prevents recruitment of TGFBRAP1 and VPS8 to LE even in the presence of RILP. We, therefore, asked how CORVET- versus HOPS-specific protein binding to the core of the complex is regulated to assure correct targeting and sequential activity in endosomal maturation. Because TGFBRAP1 and VPS39 have high sequence homology, their competitive binding to the same subunit in the CORVET/HOPS core suggests a mechanism for sequential binding. A likely candidate for a common binding partner in the core of the CORVET-HOPS complex would be VPS11, as we already identified interactions between VPS11 and VPS39 (Fig. 1). In line with our endogenous IP with VPS11 antibodies, an interaction between TGFBRAP1 and GFP-VPS11 was observed (Fig. 3C), and co-expression of VPS11 and TGFBRAP1 resulted in the recruitment of cytosolic GFP-VPS11 to EE (Fig. 3D). To map the interactions between TGFBRAP1 and VPS39 with VPS11, we expressed tagged truncation mutants of VPS11 and assessed putative interactions (Fig. 4, A and B). TGFBRAP1 and VPS39 bound to the same domain in VPS11 (Fig. 4, A and B), implying that these two proteins might compete for VPS11 binding (Fig. 4C). As TGFBRAP1 binds VPS11 and VPS11 binds RILP, we argued that a RILP-VPS11-TGFBRAP1 interaction could exist that could induce (erroneous) recruitment of TGFBRAP1 to LE. However, when VPS11 was co-expressed with TGFBRAP1 and RILP, VPS11 was exclusively recruited to EE and no longer recruited to LE (even though high levels of RILP were present) (Fig. 4D). This suggests a regulatory mechanism in which binding of TGFBRAP1 to VPS11 in the CORVET complex prevents VPS11 binding to RILP, thereby blocking the targeting of VPS11 to LE and premature assembly of the HOPS complex on LE. Such a mechanism would safeguard a sequential assembly of the CORVET and HOPS and thereby time the fusion events of EE and LE.
FIGURE 3.
VPS11 interacts with CORVET specific subunits. A, MelJuSo cells co-expressing mRFP-RILP (red) and GFP-TGFBRAP1 or HA-VPS8 (green) were fixed, stained with antibodies against EEA1 (and anti-HA for VPS8), and imaged by CLSM. Scale bars: 10 μm. Graphs show average correlation coefficient (CC) ± S.E n > 25, for RILP or EEA1 and GFP, TGFBRAP1, or VPS8. Asterisks indicate significance to GFP control (****, p ≤ 0.0001; * p ≤ 0.05; ns = not significant). B, MelJuSo cells co-expressing GFP-TGFBRAP1 or HA-VPS8 were fixed, stained with antibodies against EEA1 (and anti-HA for VPS8), and imaged by CLSM. Scale bars: 10 μm. C, lysates of MelJuSo cells expressing GFP or GFP-VPS11 were immunoprecipitated (IP) with anti-GFP and analyzed by WB using anti-GFP and anti-TGFBRAP1 antibodies. Experimental conditions were run on the same blot with the same exposures for each detection antibody, and cutouts were taken and grouped for presentation purposes. TL, total lysate. D, MelJuSo cells expressing mRFP-VPS11 (blue) or co-expressing mRFP-VPS11 (blue) and GFP-TGFBRAP1 (green) were fixed, stained with antibodies against EEA1 (red), and imaged by CLSM. Scale bars: 10 μm. Graphs show average correlation coefficient (CC) ± S.E n > 25, between VPS11 and EEA1 ± ectopically expressed TGFBRAP1 (****, p ≤ 0.0001).
FIGURE 4.
CORVET- and HOPS-specific assembly via VPS11. A and B, lysates of HEK293 cells co-expressing GFP-VPS11 constructs (as indicated) and HA-TGFBRAP1 (A) or MYC-mRFP-VPS39 (B) constructs were immunoprecipitated (IP) with anti-GFP before SDS-PAGE and WB and probed with anti-GFP and anti-HA or anti-MYC antibodies. TL, total lysate. C, detailed map of interactions between TGFBRAP1, VPS39, and VPS11. TGFBRAP1 and VSP11 both bind to the same region in VPS11, indicating competitive binding. Domains that contribute to RILP binding are indicated in green. D, MelJuSo cells expressing GFP-VPS11 (blue) and HA-RILP (red) ± GFP-TGFBRAP1 (green) were fixed, stained with antibodies against HA and FLAG and imaged by CLSM. Scale bars: 10 μm. Graphs show average correlation coefficient (CC) ± S.E (n > 25) between VPS11 and RILP in the ± ectopically expressed TGFBRAP1 (****, p ≤ 0.0001).
ARC Mutations Specifically Disrupt VPS33B-VIPAS39 Interactions or Complex Recruitment to Late Endosomes by RILP
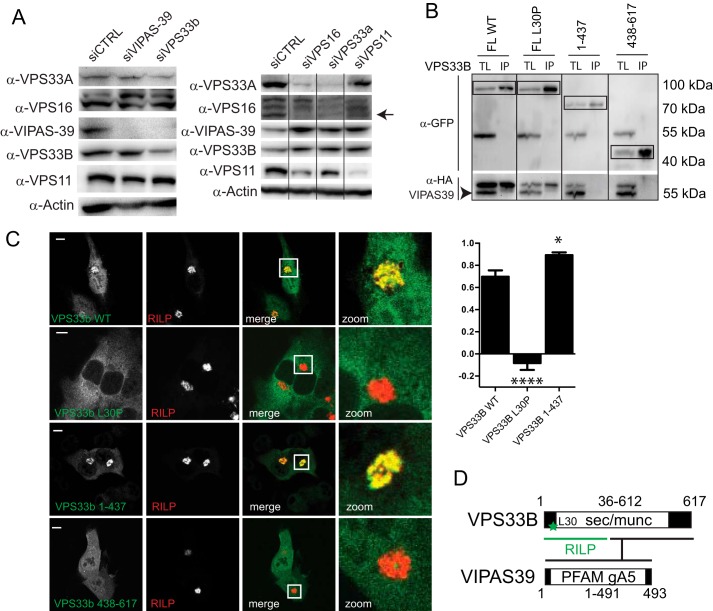
Published data regarding the role of VPS33B and VIPAS39 as members of the HOPS complex are seemingly conflicting (12, 16). Our endogenous IP data (Fig. 1A) as well as our IPs with tagged subunits (Fig. 1B) indicated that VPS33B and VIPAS39 did not interact with other HOPS subunits. To further substantiate the existence of a separate VPS33B-VIPAS39 complex, we depleted CORVET/HOPS subunits by siRNA and assessed the effects on the stability of their endogenous interaction partners by Western blot (WB) (Fig. 5A). Silencing of VPS16 significantly decreased VPS33A and VPS11 protein levels, whereas VPS33B and VIPAS39 levels did not decrease (Fig. 5A). Similarly, silencing of VPS33B compromised VIPAS39 levels without affecting VPS16, VPS33A, or VPS11 (Fig. 5A). These findings reinforce the notion that VPS33B and VIPAS39 assemble into a separate complex.
FIGURE 5.
VPS33B and VIPAS39 are not HOPS complex subunits and mutations in VPS33B differentially affect VPS33B interactions with VIPAS39 and RILP. A, lysates of MelJuSo cells silenced for indicated HOPS subunits were analyzed by WB using anti-VPS16, anti-VPS33B, anti-VIPAS39, anti-VPS11, anti-VPS33, and anti-actin (as the loading control) antibodies as indicated. Within each panel, experimental conditions were run on the same blot with the same exposures for each detection antibody, and cutouts were taken and grouped for presentation purposes. B, Immunoprecipitates (IP) with anti-GFP from lysates of MelJuSo cells co-expressing GFP-VPS33B mutants and HA-VIPAS39 (as indicated) were analyzed by WB using anti-GFP and anti-HA antibodies. Experimental conditions were run on the same blot with the same exposures for each detection antibody, and cutouts were taken and grouped for presentation purposes. TL, total lysate. C, MelJuSo cells expressing GFP-VPS33B constructs (green) and mRFP-RILP (red) were fixed and imaged by CLSM. Scale bars: 10 μm. Correlation coefficient was calculated from plot profiles measuring RILP intensity and GFP intensity over a vector through the cells. Graphs show the average correlation coefficient ± S.E (n > 25) between VPS33B constructs and RILP (*, p ≤ 0.05; ****, p ≤ 0.0001, ns = not significant, when compared with VPS33B wt). D, summary of VPS33B domains involved in interactions with RILP (green) and VIPAS39. Asterisks indicate Leu-30 (L30) residue (mutated in ARC syndrome) required for VPS33B recruitment to RILP.
Interestingly, autosomal recessive mutations in VPS33B or VIPAS39 cause ARC syndrome (23, 33), a fatal multisystem disorder characterized by defects in apical transport in polarized cells. In line with this, VPS33B and VIPAS39 have previously been shown to function at RAB11 positive recycling endosomes (21, 22). Under steady-state conditions VPS33B and VIPAR do not significantly localize to LE (22), but we have shown that these proteins can be recruited to LE by RILP (30). In line with a function of VPS33B and VIPAR at the LE, it was recently shown that VPS33B is important for the maturation of the α-granule, a specialized late endosomal compartment (34). Therefore, we investigated whether pathogenic mutations in VPS33B could still be recruited to LE by RILP. We studied two pathogenic mutations of VPS33B, a single amino acid substitution mutant (VPS33B L30P) and a truncation mutant (VPS33B 1–437), and mapped the effect on VPS33B-VIPAS39 interactions as well as RILP-dependent membrane recruitment. The truncated mutant of VPS33B (1–437) did not interact with VIPAS39 (Fig. 5B; Ref. 22) but was still recruited to LE by RILP, indicating that VPS33B can bind RILP independent of VIPAS39 (Fig. 5C). The L30P mutation in VPS33B had a different effect; although VPS33B L30P still interacted with VIPAS39 (Fig. 5B), it failed to be recruited to LE by RILP (Fig. 5C; Ref. 30). Thus, our results indicate that two specific mutations in VPS33B, as found in ARC patients, affect assembly with VIPAS39 (VPS33B 1–437) or LE recruitment of the VPS33B-VIPAS39 complex (VPS33B L30P) by RILP (Fig. 5D).
Discussion
We have investigated the interactions between the proposed mammalian subunits of the multisubunit CORVET and HOPS tethering complexes and found, with modifications, that the organization of these complexes is largely conserved from yeast to mammals (Fig. 6A). As in yeast, the mammalian HOPS complex consists of VPS16, VPS11, VPS33A, VPS18, VPS41, and VPS39. We confirm that TGFBRAP1 and VPS8 are the mammalian CORVET-specific subunits (with TGFBRAP1 as the yeast vps3 ortholog) (11, 35). In contrast to previous data (16), however, our data indicate that VIPAS39 and VPS33B, two CORVET/HOPS subunit homologues not present in yeast, are not part of CORVET/HOPS as also suggested by others (12, 21, 36). We cannot formally exclude the occurrence of transient interactions of VPS33B and/or VIPAS39 with CORVET/HOPS or the existence of VPS33B-VIPAS39-HOPS interactions in other cellular systems. However, the existence of a VPS33/VIPAS39 complex independent of other core subunits might explain why only mutations in VPS33B and VIPAS39 but not any of other HOPS subunits are associated to ARC syndrome (21–23, 33) and why all HOPS complex subunits, but not VPS33B and VIPAS39, are required for Ebola-virus infection (19). Conservation of these separate complexes is not apparent in all organisms. For example, in Caenorhabditis elegans the two VPS33 homologues (VPS33.1 and VPS33.2) are present in HOPS (VPS33.1) and CORVET (VPS33.2) (37). It is unclear, however, whether VPS33.1 and VPS33.2 are direct orthologous for mammalian VPS33A and VPS33B or reflect other evolutionary divergence. Similar to mammals, Drosophila VPS33A (Carnation) and VPS33B have distinct functions (38) and Drosophila VPS16 interacts with VPS33A, whereas Drosophila VIPAS39 interacts with VPS33B (39). However, Drosophila VIPAS39 also interacted with VPS18, suggesting that VIPAS39-VPS33B in Drosophila can interact with CORVET/HOPS core subunits (39). In mice, mutations in VPS33A yielded hypopigmentation and mild platelet deficiency but did not cause ARC, indicating that VPS33A and VPS33B functions do not completely overlap in mice (40). This suggests that VPS33B-VIPAS39 interactions might have gradually diverted from that of CORVET/HOPS during mammalian evolution. Yet, functions of VIPAS39 seem conserved between different organisms. For example, the observation that Drosophila lacking VIPAS39 (dVPS16) has a profound defect in phagosomal acidification is in line with our observation that VIPAS39 functions at LE compartments (30). Here we report that ARC-syndrome mutations in VPS33B differentially affect the binding of VPS33b to VIPAS39 or membrane recruitment of VPS33B to RILP. This consolidates the finding that VPS33B-VIPAS39 function at LEs (34) as well as recycling endosomes (16, 21, 22) and that both membrane binding and VIPAS39 binding are needed for correct VPS33B function.
FIGURE 6.
Model of membrane binding specificity of the CORVET and HOPS complex and their conversion during maturation. A, model of the mammalian HOPS complex superimposed on the yeast structure, depicting RILP-binding domains. Asterisks indicate poorly conserved regions in the N terminus of VPS39 and loss of the lipid binding motif of VPS41, which have altered membrane targeting in the mammalian complex. CLH, clathrin heavy chain repeat; CC, coiled coil; R, ring finger; CNH, citron homology. B, the CORVET complex binds to RAB5 on EE and within this complex, TGFBRAP1 binding to VPS11 prevent association of VPS11 to RILP (1). 2, to allow RAB7-RILP binding, TGFBRAP1 is replaced by VPS39, possibly controlled by CCZ1/MON1 and TGFβ signaling. Replacement of VPS8 with VPS41 adds an additional RILP binding motif, completing the conversion of CORVET to HOPS complex from RAB5- to RAB7- during endosomal maturation (3). Both the head and tail of mammalian HOPS can bind the homodimer RAB7-RILP. There may be two conditions; an inactive conformation where HOPS binds back to RILP on the same vesicle failing to contact a fusion partner and an active conformation in which RILP-HOPS contacts other vesicles (LE, phagosomes or autophagosomes containing HOPS interactors such as ARL8b, PLEKHM1, or RILP) for tethering and subsequent fusion.
We show that HOPS subunits have evolved to accommodate binding to the RAB7 effector RILP (for a mammalian model superimposed on the yeast structure see Fig. 3G). The N terminus of yeast vps39 that binds Ypt7 is poorly conserved in the mammalian VPS39 ortholog, which might explain why the mammalian HOPS complex has shifted to bind RAB7 effectors such as RILP (30, 32) and PLEKHM1 (14, 15). The HOPS complex binds RILP on either side of the complex (VPS39/VPS11 in the tail and VPS41/VPS18 in the head) and may bridge RILP molecules on opposing vesicles. Because VPS33A in the head of HOPS also binds to SNAREs (41), a connection between two vesicles will then support tethering and subsequent fusion. It is interesting to note that the HOPS complex can also bind the GTPase Arl8b on lysosomes (20, 42) and PLEKHM1 on LE, phagosomes, and autophagosomes (14, 15), possibly tethering these vesicles with RILP-containing organelles (Fig. 6B). RAB7-RILP concomitantly binds the HOPS complex and the dynein-motor complex for retrograde transport (30), whereas the Arl8b effector SKIP also concomitantly regulates HOPS binding and anterograde transport by kinesin (42), indicating that timing of vesicle tethering is likely coupled to their transport.
We found that binding of TGFBRAP1 to VPS11 prevents the interaction of VPS11 with RILP and erroneous recruitment of CORVET complexes to RILP-bearing LE. This might provide an important regulatory step in the conversion of CORVET to HOPS complexes (Fig. 5) where TGFBRAP1 in CORVET would have to release VPS11, thereby allowing VPS11 to bind VPS39 and recruit the entire HOPS complex to RAB7-RILP during RAB5 (EEs) to RAB7 (LE) conversion. Specific targeting of tethering complexes by modulation of targeting subunits was recently also shown for GARP and EARP tethering on Golgi and EEs, respectively, indicating this might be a common mechanism in the regulation of vesicular fusion (43). TGFBRAP1 to VPS39 exchange might further be regulated by proteins such as MON1-CCZ1 (Fig. 6B), which are recruited by CORVET, interact with VPS39, and act as activators of RAB7 (44–46) and signaling pathways such as the TGFβ pathway, which has been shown to intersect with TGFBRAP1 and VPS39 function (47–49).
In conclusion, we describe the molecular architecture of the mammalian CORVET, HOPS, and VIPAS39-VPS33B complexes. We find that different ARC syndrome mutations affect VIPAS39-VPS33B or VPS33B-RILP interactions, indicating that this complex has a function at the LE. Through mapping of the CORVET and HOPS complexes we found that within the shared CORVET/HOPS core, VPS11 is a molecular switch that, depending on its interacting proteins (TGFBRAP1 or VPS39), determines targeting of CORVET and HOPS to EE and LE.
Author Contributions
R. v d K. designed and performed the experiments for Figs. 1–4, analyzed the data, and wrote the paper. C. T. H. J. designed and performed the experiments for Fig. 3, analyzed the data, and wrote the paper. R. H. W. (Fig. 4) and J. B. (Fig. 2E) performed the experiments. L. J. cloned the constructs used in the manuscript. J. K. and J. N. designed the experiments and wrote the paper.
Acknowledgments
We thank C. Liang, J. Bonifacino, V. Faundez, and S. Zlatic for kindly providing reagents. We thank Romain Galmes and Peter van der Sluijs for valuable discussions.
This work was supported by a European Research Council Advanced Grant and a TOP grant from The Netherlands Organization for Scientific Research, Chemical Sciences. The authors declare that they have no conflicts of interest with the contents of this article.
- EE
- early endosome
- LE
- late endosome
- RILP
- RAB7 interacting lysosomal protein
- ARC
- arthrogryposis-renal dysfunction-cholestasis
- IP
- immunoprecipitation
- WB
- Western blot
- CLSM
- confocal laser-scanning microscope.
References
- 1.Stenmark H. (2009) Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10, 513–525 [DOI] [PubMed] [Google Scholar]
- 2.Wickner W. (2010) Membrane fusion: five lipids, four SNAREs, three chaperones, two nucleotides, and a Rab, all dancing in a ring on yeast vacuoles. Annu. Rev. Cell Dev. Biol. 26, 115–136 [DOI] [PubMed] [Google Scholar]
- 3.Sato T. K., Rehling P., Peterson M. R., and Emr S. D. (2000) Class C Vps protein complex regulates vacuolar SNARE pairing and is required for vesicle docking/fusion. Mol. Cell 6, 661–671 [DOI] [PubMed] [Google Scholar]
- 4.Wurmser A. E., Sato T. K., and Emr S. D. (2000) New component of the vacuolar class C-Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion. J. Cell Biol. 151, 551–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakamura N., Hirata A., Ohsumi Y., and Wada Y. (1997) Vam2/Vps41p and Vam6/Vps39p are components of a protein complex on the vacuolar membranes and involved in the vacuolar assembly in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 272, 11344–11349 [DOI] [PubMed] [Google Scholar]
- 6.Plemel R. L., Lobingier B. T., Brett C. L., Angers C. G., Nickerson D. P., Paulsel A., Sprague D., and Merz A. J. (2011) Subunit organization and Rab interactions of Vps-C protein complexes that control endolysosomal membrane traffic. Mol. Biol. Cell 22, 1353–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ostrowicz C. W., Bröcker C., Ahnert F., Nordmann M., Lachmann J., Peplowska K., Perz A., Auffarth K., Engelbrecht-Vandré S., and Ungermann C. (2010) Defined subunit arrangement and rab interactions are required for functionality of the HOPS tethering complex. Traffic 11, 1334–1346 [DOI] [PubMed] [Google Scholar]
- 8.Bröcker C., Kuhlee A., Gatsogiannis C., Balderhaar H. J., Hönscher C., Engelbrecht-Vandré S., Ungermann C., and Raunser S. (2012) Molecular architecture of the multisubunit homotypic fusion and vacuole protein sorting (HOPS) tethering complex. Proc. Natl. Acad. Sci. U.S.A. 109, 1991–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cabrera M., Langemeyer L., Mari M., Rethmeier R., Orban I., Perz A., Bröcker C., Griffith J., Klose D., Steinhoff H.-J., Reggiori F., Engelbrecht-Vandré S., and Ungermann C. (2010) Phosphorylation of a membrane curvature-sensing motif switches function of the HOPS subunit Vps41 in membrane tethering. J. Cell Biol. 191, 845–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rink J., Ghigo E., Kalaidzidis Y., and Zerial M. (2005) Rab conversion as a mechanism of progression from early to late endosomes. Cell 122, 735–749 [DOI] [PubMed] [Google Scholar]
- 11.Perini E. D., Schaefer R., Stöter M., Kalaidzidis Y., and Zerial M. (2014) Mammalian CORVET is required for fusion and conversion of distinct early endosome subpopulations. Traffic 15, 1366–1389 [DOI] [PubMed] [Google Scholar]
- 12.Wartosch L., Günesdogan U., Graham S. C., and Luzio J. P. (2015) Recruitment of VPS33A to HOPS by VPS16 is required for lysosome fusion with endosomes and autophagosomes. Traffic 16, 727–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang P., Nishimura T., Sakamaki Y., Itakura E., Hatta T., Natsume T., and Mizushima N. (2014) The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Mol. Biol. Cell 25, 1327–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McEwan D. G., Popovic D., Gubas A., Terawaki S., Suzuki H., Stadel D., Coxon F. P., Miranda de Stegmann D., Bhogaraju S., Maddi K., Kirchof A., Gatti E., Helfrich M. H., Wakatsuki S., Behrends C., Pierre P., and Dikic I. (2015) PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol. Cell 57, 39–54 [DOI] [PubMed] [Google Scholar]
- 15.McEwan D. G., Richter B., Claudi B., Wigge C., Wild P., Farhan H., McGourty K., Coxon F. P., Franz-Wachtel M., Perdu B., Akutsu M., Habermann A., Kirchof A., Helfrich M. H., Odgren P. R., Van Hul W., Frangakis A. S., Rajalingam K., Macek B., Holden D. W., Bumann D., and Dikic I. (2015) PLEKHM1 regulates Salmonella-containing vacuole biogenesis and infection. Cell Host Microbe 17, 58–71 [DOI] [PubMed] [Google Scholar]
- 16.Zhu G.-D., Salazar G., Zlatic S. A., Fiza B., Doucette M. M., Heilman C. J., Levey A. I., Faundez V., and L'hernault S. W. (2009) SPE-39 family proteins interact with the HOPS complex and function in lysosomal delivery. Mol. Biol. Cell 20, 1223–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chirivino D., Del Maestro L., Formstecher E., Hupé P., Raposo G., Louvard D., and Arpin M. (2011) The ERM proteins interact with the HOPS complex to regulate the maturation of endosomes. Mol. Biol. Cell 22, 375–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang C., Lee J.-S., Inn K.-S., Gack M. U., Li Q., Roberts E. A., Vergne I., Deretic V., Feng P., Akazawa C., and Jung J. U. (2008) Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat. Cell Biol. 10, 776–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carette J. E., Raaben M., Wong A. C., Herbert A. S., Obernosterer G., Mulherkar N., Kuehne A. I., Kranzusch P. J., Griffin A. M., Ruthel G., Dal Cin P., Dye J. M., Whelan S. P., Chandran K., and Brummelkamp T. R. (2011) Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 477, 340–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garg S., Sharma M., Ung C., Tuli A., Barral D. C., Hava D. L., Veerapen N., Besra G. S., Hacohen N., and Brenner M. B. (2011) Lysosomal trafficking, antigen presentation, and microbial killing are controlled by the Arf-like GTPase Arl8b. Immunity 35, 182–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith H., Galmes R., Gogolina E., Straatman-Iwanowska A., Reay K., Banushi B., Bruce C. K., Cullinane A. R., Romero R., Chang R., Ackermann O., Baumann C., Cangul H., Cakmak Celik F., Aygun C., Coward R., Dionisi-Vici C., Sibbles B., Inward C., Kim C. A., Klumperman J., Knisely A. S., Watson S. P., and Gissen P. (2012) Associations among genotype, clinical phenotype, and intracellular localization of trafficking proteins in ARC syndrome. Hum. Mutat. 33, 1656–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cullinane A. R., Straatman-Iwanowska A., Zaucker A., Wakabayashi Y., Bruce C. K., Luo G., Rahman F., Gürakan F., Utine E., Ozkan T. B., Denecke J., Vukovic J., Di Rocco M., Mandel H., Cangul H., Matthews R. P., Thomas S. G., Rappoport J. Z., Arias I. M., Wolburg H., Knisely A. S., Kelly D. A., Müller F., Maher E. R., and Gissen P. (2010) Mutations in VIPAR cause an arthrogryposis, renal dysfunction, and cholestasis syndrome phenotype with defects in epithelial polarization. Nat. Genet. 42, 303–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gissen P., Johnson C. A., Morgan N. V., Stapelbroek J. M., Forshew T., Cooper W. N., McKiernan P. J., Klomp L. W., Morris A. A., Wraith J. E., McClean P., Lynch S. A., Thompson R. J., Lo B., Quarrell O. W., Di Rocco M., Trembath R. C., Mandel H., Wali S., Karet F. E., Knisely A. S., Houwen R. H., Kelly D. A., and Maher E. R. (2004) Mutations in VPS33B, encoding a regulator of SNARE-dependent membrane fusion, cause arthrogryposis-renal dysfunction-cholestasis (ARC) syndrome. Nat. Genet. 36, 400–404 [DOI] [PubMed] [Google Scholar]
- 24.Lo B., Li L., Gissen P., Christensen H., McKiernan P. J., Ye C., Abdelhaleem M., Hayes J. A., Williams M. D., Chitayat D., and Kahr W. H. (2005) Requirement of VPS33B, a member of the Sec1/Munc18 protein family, in megakaryocyte and platelet α-granule biogenesis. Blood 106, 4159–4166 [DOI] [PubMed] [Google Scholar]
- 25.van Luijn M. M., Kreft K. L., Jongsma M. L., Mes S. W., Wierenga-Wolf A. F., van Meurs M., Melief M.-J., der Kant R. v., Janssen L., Janssen H., Tan R., Priatel J. J., Neefjes J., Laman J. D., and Hintzen R. Q. (2015) Multiple sclerosis-associated CLEC16A controls HLA class II expression via late endosome biogenesis. Brain 138, 1531–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tornieri K., Zlatic S. A., Mullin A. P., Werner E., Harrison R., L'hernault S. W., and Faundez V. (2013) Vps33b pathogenic mutations preferentially affect VIPAS39/SPE-39-positive endosomes. Hum. Mol. Genet. 22, 5215–5228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pols M. S., van Meel E., Oorschot V., ten Brink C., Fukuda M., Swetha M. G., Mayor S., and Klumperman J. (2013) hVps41 and VAMP7 function in direct TGN to late endosome transport of lysosomal membrane proteins. Nat. Commun. 4, 1361. [DOI] [PubMed] [Google Scholar]
- 28.Vennegoor C., Calafat J., Hageman P., van Buitenen F., Janssen H., Kolk A., and Rümke P. (1985) Biochemical characterization and cellular localization of a formalin-resistant melanoma-associated antigen reacting with monoclonal antibody NKI/C-3. Int. J. Cancer 35, 287–295 [DOI] [PubMed] [Google Scholar]
- 29.Jordens I., Fernandez-Borja M., Marsman M., Dusseljee S., Janssen L., Calafat J., Janssen H., Wubbolts R., and Neefjes J. (2001) The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr. Biol. 11, 1680–1685 [DOI] [PubMed] [Google Scholar]
- 30.van der Kant R., Fish A., Janssen L., Janssen H., Krom S., Ho N., Brummelkamp T., Carette J., Rocha N., and Neefjes J. (2013) Late endosomal transport and tethering are coupled processes controlled by RILP and the cholesterol sensor ORP1L. J. Cell Sci. 126, 3462–3474 [DOI] [PubMed] [Google Scholar]
- 31.Deleted in proof
- 32.Lin X., Yang T., Wang S., Wang Z., Yun Y., Sun L., Zhou Y., Xu X., Akazawa C., Hong W., and Wang T. (2014) RILP interacts with HOPS complex via VPS41 subunit to regulate endocytic trafficking. Sci. Rep. 4, 7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gissen P., Johnson C. A., Gentle D., Hurst L. D., Doherty A. J., O'Kane C. J., Kelly D. A., and Maher E. R. (2005) Comparative evolutionary analysis of VPS33 homologues: genetic and functional insights. Hum. Mol. Genet. 14, 1261–1270 [DOI] [PubMed] [Google Scholar]
- 34.Bem D., Smith H., Banushi B., Burden J. J., White I. J., Hanley J., Jeremiah N., Rieux-Laucat F., Bettels R., Ariceta G., Mumford A. D., Thomas S. G., Watson S. P., and Gissen P. (2015) VPS33B regulates protein sorting into and maturation of α-granule progenitor organelles in mouse megakaryocytes. Blood 126, 133–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lachmann J., Glaubke E., Moore P. S., and Ungermann C. (2014) The Vps39-like TRAP1 is an effector of Rab5 and likely the missing Vps3 subunit of human CORVET. Cell. Logist. 4, e970840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Graham S. C., Wartosch L., Gray S. R., Scourfield E. J., Deane J. E., Luzio J. P., and Owen D. J. (2013) Structural basis of Vps33A recruitment to the human HOPS complex by Vps16. Proc. Natl. Acad. Sci. U.S.A. 110, 13345–13350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Solinger J. A., and Spang A. (2014) Loss of the Sec1/Munc18-family proteins VPS-33.2 and VPS-33.1 bypasses a block in endosome maturation in Caenorhabditis elegans. Mol. Biol. Cell 25, 3909–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akbar M. A., Ray S., and Krämer H. (2009) The SM protein Car/Vps33A regulates SNARE-mediated trafficking to lysosomes and lysosome-related organelles. Mol. Biol. Cell 20, 1705–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pulipparacharuvil S., Akbar M. A., Ray S., Sevrioukov E. A., Haberman A. S., Rohrer J., and Krämer H. (2005) Drosophila Vps16A is required for trafficking to lysosomes and biogenesis of pigment granules. J. Cell Sci. 118, 3663–3673 [DOI] [PubMed] [Google Scholar]
- 40.Suzuki T., Oiso N., Gautam R., Novak E. K., Panthier J.-J., Suprabha P. G., Vida T., Swank R. T., and Spritz R. A. (2003) The mouse organellar biogenesis mutant buff results from a mutation in Vps33a, a homologue of yeast vps33 and Drosophila carnation. Proc. Natl. Acad. Sci. U.S.A. 100, 1146–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lobingier B. T., and Merz A. J. (2012) Sec1/Munc18 protein Vps33 binds to SNARE domains and the quaternary SNARE complex. Mol. Biol. Cell 23, 4611–4622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khatter D., Raina V. B., Dwivedi D., Sindhwani A., Bahl S., and Sharma M. (2015) The small GTPase Arl8b regulates assembly of the mammalian HOPS complex to lysosomes. J. Cell Sci. 128, 1746–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schindler C., Chen Y., Pu J., Guo X., and Bonifacino J. S. (2015) EARP is a multisubunit tethering complex involved in endocytic recycling. Nat. Cell Biol. 17, 639–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nordmann M., Cabrera M., Perz A., Bröcker C., Ostrowicz C., Engelbrecht-Vandré S., and Ungermann C. (2010) The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr. Biol. 20, 1654–1659 [DOI] [PubMed] [Google Scholar]
- 45.Poteryaev D., Datta S., Ackema K., Zerial M., and Spang A. (2010) Identification of the switch in early-to-late endosome transition. Cell 141, 497–508 [DOI] [PubMed] [Google Scholar]
- 46.Kinchen J. M., and Ravichandran K. S. (2010) Identification of two evolutionarily conserved genes regulating processing of engulfed apoptotic cells. Nature 464, 778–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Felici A., Wurthner J. U., Parks W. T., Giam L. R.-Y., Reiss M., Karpova T. S., McNally J. G., and Roberts A. B. (2003) TLP, a novel modulator of TGF-β signaling, has opposite effects on Smad2- and Smad3-dependent signaling. EMBO J. 22, 4465–4477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Messler S., Kropp S., Episkopou V., Felici A., Würthner J., Lemke R., Jerabek-Willemsen M., Willecke R., Scheu S., Pfeffer K., and Wurthner J. U. (2011) The TGF-β signaling modulators TRAP1/TGFBRAP1 and VPS39/Vam6/TLP are essential for early embryonic development. Immunobiology 216, 343–350 [DOI] [PubMed] [Google Scholar]
- 49.Wurthner J. U., Frank D. B., Felici A., Green H. M., Cao Z., Schneider M. D., McNally J. G., Lechleider R. J., and Roberts A. B. (2001) Transforming growth factor-β receptor-associated protein 1 is a Smad4 chaperone. J. Biol. Chem. 276, 19495–19502 [DOI] [PubMed] [Google Scholar]