Abstract
Mitochondrial oxidative phosphorylation produces most of the energy in aerobic cells by coupling respiration to the production of ATP. Mitochondrial uncouplers, which reduce the proton gradient across the mitochondrial inner membrane, create a futile cycle of nutrient oxidation without generating ATP. Regulation of mitochondrial dysfunction and associated cellular bioenergetics has been recently identified as a promising target for anticancer therapy. Here, we show that SR4 is a novel mitochondrial uncoupler that causes dose-dependent increase in mitochondrial respiration and dissipation of mitochondrial membrane potential in HepG2 hepatocarcinoma cells. These effects were reversed by the recoupling agent 6-ketocholestanol but not cyclosporin A and were nonexistent in mitochondrial DNA-depleted HepG2 cells. In isolated mouse liver mitochondria, SR4 similarly increased oxygen consumption independent of adenine nucleotide translocase and uncoupling proteins, decreased mitochondrial membrane potential, and promoted swelling of valinomycin-treated mitochondria in potassium acetate medium. Mitochondrial uncoupling in HepG2 cells by SR4 results in the reduction of cellular ATP production, increased ROS production, activation of the energy-sensing enzyme AMPK, and inhibition of acetyl-CoA carboxylase and mammalian target of rapamycin signaling pathways, leading to cell cycle arrest and apoptosis. Global analysis of SR4-associated differential gene expression confirms these observations, including significant induction of apoptotic genes and down-regulation of cell cycle, mitochondrial, and oxidative phosphorylation pathway transcripts at 24 h post-treatment. Collectively, our studies demonstrate that the previously reported indirect activation of AMPK and in vitro anticancer properties of SR4 as well as its beneficial effects in both animal xenograft and obese mice models could be a direct consequence of its mitochondrial uncoupling activity.
Introduction
Hepatocellular carcinoma (HCC)2 is the most common and severe form of liver cancer, accounting for about 80–90% of primary liver cancers and 5% of all human cancers. More than 600,000 deaths are attributed to HCC every year with 2:1 ratio for men versus women (1, 2). HCC is a primary cancer of hepatocytes that most typically occurs in the setting of known risk factors, including cirrhosis and chronic hepatitis B virus or hepatitis C virus infections (2, 3), although recently, several lines of evidence suggest that type 2 diabetes is also an independent risk factor for HCC development (4). HCC is an aggressive tumor and represents a major health problem as its incidence is increasing. Systemic chemotherapies have proven ineffective against advanced HCC, so it typically leads to death within 6–20 months (5). Hepatocarcinogenesis is a multistep process involving inflammation, hyperplasia, and dysplasia that finally leads to malignant transformation. In recent years, mitochondria have been found to provide a novel targeting site for new anticancer drugs (known as “mitocans”) that can selectively kill cancer cells without affecting normal cells (6, 7). However, to date, little is known regarding the role of mitochondrial functions, such as redox regulation and oxidative phosphorylation (OxPhos), in HCC progression and survival. The other important question that has not yet been systematically addressed is whether hepatocarcinoma cells rely more on OxPhos or glycolysis. In this context, new candidate drugs capable of targeting multiple critical nodes of HCC signaling assume significance.
The primary role of mitochondria is the generation of ATP through a complex process of controlled substrate degradation and oxygen consumption known as OxPhos (8, 9). Oxidation of nutrient molecules, such as carbohydrates, lipids, and amino acids, yields electrons in the form of reduced hydrogen carriers NADH+ and FADH2. These reduced cofactors donate electrons to a series of enzyme complexes embedded in the inner mitochondrial membrane known as the electron transport chain (ETC) (10). The transfer of electrons along the respiratory chain is accompanied by pumping of protons (H+) across the inner mitochondrial membrane, which results in transmembrane differences in proton concentration (gradient). The proton-motive force is subsequently used to drive the synthesis of ATP, as H+ flows passively back into the matrix through proton pores formed by ATP synthase (1, 8, 9). Thus, ATP is synthesized by coupling electron transport and H+ pumping to phosphorylation of ADP. However, not all of the energy available in the electrochemical gradient is coupled to ATP synthesis. Some of the energy is consumed by proton leak reactions, by which protons pumped out of the matrix are able to reflow back along the proton gradient through proton conductance pathways in the inner membrane that bypass the ATP synthase (11, 12). As a result, the energy derived from the metabolic oxidation reaction is dissipated as heat. This nonproductive proton leak, termed “mitochondrial uncoupling,” is physiologically important and accounts for 20–30% of the basal metabolic rate (13, 14). Mitochondrial uncoupling also plays an important role in the reprogramming of cancer cell metabolism (15).
It is now clear that mitochondria are not only the site of OxPhos and energy metabolism but also play key roles in cell signaling, redox regulation, and apoptosis (16). Given the central roles of mitochondria in the regulation of fundamental cellular functions and bioenergetics, it is not surprising that these organelles have been implicated in multiple aspects of cancer development and tumor progression. Several lines of evidence support the hypothesis that cancer is primarily a disease of energy metabolism, and mitochondrial dysfunction has been found to be associated with the development of several human cancers (17, 18). Cancer cells often exhibit some type of mitochondrial dysfunction, including mitochondrial DNA mutations, alterations in energy metabolism, elevated reactive oxygen species (ROS) generation, and increased mitochondrial membrane potential (MMP). These changes suggest a biochemical basis for preferentially targeting cancer cells to provide therapeutic selectivity (19–22). In particular, pharmacological use of synthetic and naturally occurring uncouplers, with their potential to compromise mitochondrial functions and affect cellular bioenergetics and metabolism, have lately shown promising anticancer effects (23–27). Mitochondrial uncouplers generally belong to one of two general classes: (a) protonophore uncouplers (protein-independent uncoupling) that transport protons across the inner mitochondrial membrane, dissipating the transmembrane proton gradient required for the coupling between electron transport and OxPhos, and (b) non-protonophores (protein-mediated uncoupling) that activate proton leak through protein complexes such as the adenine nucleotide translocase (ANT) or uncoupling proteins (UCPs) (28, 29).
Previously, our laboratory has identified several novel dichlorophenyl urea compounds with potent anticancer activities. The lead compound SR4 showed promising antitumor activities in in vitro cell cultures and in vivo animal xenograft models, including leukemia, lung cancers, and melanoma (30–32). SR4 was shown to indirectly activate the energy-sensing enzyme AMPK (33), but the mechanism remained unknown. In this study, we identify a novel mitochondrial uncoupling property of SR4, which is directly associated with its anticancer properties. Studies were performed in both hepatic carcinoma (HepG2) cells and mitochondria isolated from mouse liver, and the uncoupling potency of SR4 was compared with that of the classic prototype protonophore uncoupler, carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP). We proposethat SR4 uncouples mitochondrial respiration, resulting in a decrease in cellular ATP and a change in AMP/ATP ratio, which promotes AMPK activation and inhibition of mTOR signaling. In addition, the loss of membrane potential also induces mitochondrial swelling and ROS production. These effects of SR4 in cellular bioenergetics and metabolism lead to modulation of a number of genes involved in mitochondrial dysfunction, OxPhos, and most anabolic processes, resulting in inhibition of cell proliferation, induction of cell cycle arrest, and apoptosis.
Experimental Procedures
Chemicals and Reagents
SR4 was synthesized according to a previously validated protocol at the Chemical GMP Synthesis Facility, Translational Medicinal Chemistry Laboratory, Beckman Research Institute of the City of Hope (30). FCCP, antimycin A, rotenone, 6-ketocholestanol (6-KCH), cyclosporin A (CSA), bongkrekic acid, N,N,N′,N′-tetramethyl-p-phenylenediamine (TMPD), valinomycin, ascorbic acid, malic acid, succinate, MitoTempo, guanosine diphosphate (GDP), N-acetyl-l-cysteine (NAC), fatty acid/endotoxin-free bovine serum albumin (BSA), tetramethylrhodamine ester (TMRE), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT), and crystal violet (CV) were purchased from Sigma. Sodium pyruvate and glutamine were purchased from Thermo Fisher Scientific. Both SR4 and FCCP were dissolved in DMSO at 50 mm stock solution. Antibodies against AMPKα, phospho-AMPKα (Thr172), acetyl-CoA carboxylase, phospho-acetyl-CoA carboxylase (Ser79), raptor, phospho-raptor (Ser792), 4E-BP, phospho-4E-BP (Thr37/46), TSC2, phospho-TSC2 (Ser1387), and β-actin were obtained from Cell Signaling Technology (Danvers, MA). UCP-2 siRNA, AMPKα1/α2 siRNA, and their corresponding non-sense control siRNAs were from Thermo Fisher Scientific and Santa Cruz Biotechnology, Inc. (Dallas, TX), respectively. The universal Mycoplasma detection kit was procured from ATCC (Manassas, VA).
Cell Culture
HepG2, H358 human lung cancer, and A2058 human melanoma cell lines were obtained from ATCC and were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum and 1% penicillin/streptomycin antibiotics (ATCC, Manassas, VA) at 37 °C in a humidified atmosphere of 5% CO2. Cells were subcultured every 2–3 days at ∼70–80% confluence. Primary mouse hepatocytes (a gift from Dr. Bangan L. Stiles, University of Southern California, Los Angeles, CA) were cultured in DMEM supplemented with 10% FBS, 5 μg/ml insulin (Sigma), and 10 ng/ml epidermal growth factor (Invitrogen). HepG2 cells lacking mitochondrial DNA (ρ0 cells) were generated by chronic exposure to low doses of ethidium bromide, as described by Hashiguchi and Zhang-Akiyama (34). ρ0 mitochondria can maintain electrochemical potential across the inner membrane by a mechanism coupled to ATP hydrolysis, making them net consumers of ATP in cells and an excellent tool for studying mitochondrial function (34). The cells were immediately expanded and frozen after being obtained and restarted every 3–4 months from a frozen vial of the same batch of cells, and no additional authentication was done on these cells. All cell lines were free of Mycoplasma infection tested by the universal Mycoplasma detection kit.
Cell Proliferation and Viability Assay
Cells were seeded into 96-well plates at density of 10,000 cells/well for HepG2, H358, and A2058 cells and 5,000 cells/well for primary mouse hepatocytes. Cells were incubated overnight at 37 °C prior to drug treatment. Drugs were diluted in cell culture medium and added to each well at the indicated concentrations. Cell viability was measured 48 h later using MTT or CV staining (0.5% (w/v) in 50% methanol). Absorbance was measured using a Tecan microplate reader (Tecan Infinite M200 Pro, Tecan Group Ltd., Männedorf, Switzerland). Cell viability of drug-treated cells was expressed as a percentage of control cells (i.e. cells treated with equivalent concentrations of the DMSO vehicle). The final concentration of DMSO exposed to the cells was <0.1% (v/v) for the duration of the experiment.
Cellular ATP, AMP, and ADP Measurement
ATP generation from HepG2 cells treated with the test compounds at various time points (0–8 h) was measured using the luminescent ATP detection assay kit (Abcam, Cambridge, MA) and expressed as the percentage of DMSO-treated cells at the same time points. In addition, cellular levels of AMP, ATP, and ADP were measured 1 h after treatment as described previously (33).
Mitochondria Isolation
Mitochondria were isolated from the livers of 10-week-old male C57BL/6 mice. Mice were euthanized by CO2 inhalation followed by cervical dislocation. Livers were removed, minced with scissors, and immediately placed in ∼10 volumes of ice-cold mitochondria isolation medium (250 mm sucrose, 10 mm Tris-HCl, 1 mm EGTA, 1% fatty acid/endotoxin-free BSA, pH 7.4). The presence of BSA in the medium allowed the endogenous free fatty acids to be chelated from the homogenate suspension. The tissues were homogenized using 25–30 strokes in a Potter-Elvehjem tissue grinder (Sigma). Following centrifugation at 800 × g for 10 min at 4 °C, the fats/lipids were carefully aspirated, and the remaining supernatant was filtered through sterile 0.40-μm nylon mesh membrane and centrifuged at 8,000 × g for 10 min at 4 °C. The supernatant and any white debris were removed, and the mitochondrial pellet was resuspended in ice-cold mitochondrial assay solution (MAS buffer: 70 mm sucrose, 220 mm mannitol, 10 mm KH2PO4, 5 mm MgCl2, 2 mm HEPES, 1 mm EGTA, 0.2% fatty acid-free BSA, pH 7.2), and the centrifugation was repeated. The final pellet was resuspended in a minimal volume of MAS buffer. The mitochondrial protein concentration was determined by the DC protein assay kit (Bio-Rad) with BSA as a standard. The isolated mitochondria were placed on ice and used within 3 h.
Measurements of Mitochondrial Respiration in Whole Cells
The oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were measured using a Seahorse XF24 or XF96 flux analyzers (Seahorse Biosciences, North Billerica, MA). Cells were seeded in DMEM in either Seahorse 24 or 96-well tissue culture plates at a density of 5 × 103 cells/well (96-well) or 1–1.2 × 104 cells/well (24-well) and allowed to adhere for 24 h. Prior to the assay, the medium was changed to DMEM containing 25 mm glucose, 1 mm pyruvate, and 1 mm glutamine with 0.2% fatty acid/endotoxin-free BSA (w/v) (pH 7.4), and the cells were equilibrated for 30 min at 37 °C. Test compounds were injected during the assay, and OCR and ECAR were measured using 2–3-min measurement periods. Similar sets of experiments were performed in the presence of the mitochondria recoupler agent 6-KCH or the mitochondrial permeability transition (MPT) pore blocker cyclosporin A, where each compound was injected into the assay medium prior to SR4 or FCCP injection. In separate experiments with HepG2 and HepG2 ρ0 cells, a modified Mito stress test was performed accordingly per Seahorse Bioscience protocols (35). Briefly, test compounds were injected after oligomycin (ATP synthase inhibitor) treatment followed by injection of rotenone/antimycin A (complex I and III inhibitor, respectively). OCR was measured throughout the different injections of the stressors and test compounds (indicated by arrows in the figures).
Measurements of Mitochondrial Respiration in Isolated Mitochondria
Isolated mouse liver mitochondria (5 or 2 μg/well) were seeded on Seahorse 24 or 96-well plate, respectively, and respiration (OCR) was measured on mitochondria respiring either on pyruvate (10 mm) and malate (2 mm) or succinate (10 mm) and rotenone (2 μm) using a Seahorse XF24 or XF96 flux analyzer according to Rogers et al. (36).
Mitochondrial Membrane Depolarization Measurements
HepG2 cells (4 × 105/well in 6-well plates) cultured overnight were preincubated with the fluorescent indicator of MMP TMRE (200 nm) or DMSO control for 30 min at 37 °C in CO2 incubator. Cells were washed with prewarmed normal DMEM, and test compounds were added with fresh medium for the indicated times. Cells were then trypsinized, collected, centrifuged for 5 min at 800 × g, and washed once in prewarmed PBS plus 0.2% BSA. HepG2 cells were then resuspended in the same buffer prior to flow cytometric analysis. A total of 30,000 cells were counted per sample, and data were analyzed by a CyAn ADP flow cytometer with excitation/emission of 488 nm/572 nm (Beckman Coulter, Inc., Brea, CA). For measurements of membrane potential in isolated mouse liver mitochondria, the organelles were preincubated with 200 nm TMRE in MASSRO buffer (MAS buffer supplemented with 10 mm succinate, 1 μm rotenone, and 1 μm oligomycin) for 20 min at 25 °C. The mitochondria were centrifuged for 5 min at 3,000 × g and resuspended in MASSRO buffer. Mitochondria were then added to MASSRO buffer containing the indicated concentrations of SR4, FCCP, or DMSO vehicle control and incubated at room temperature for 15 min. The mitochondria were then centrifuged for 5 min at 3,000 × g. The supernatant was then removed and placed into a black clear bottom 96-well plate (100 μl/well). TMRE fluorescence was measured with a Tecan microplate reader using an excitation/emission of 545 nm/580 nm.
Mitochondrial Swelling Assay
Mitochondrial swelling was measured in non-respiring mouse liver mitochondria incubated in buffered isotonic potassium acetate in the presence of valinomycin as described (37, 38). Briefly, isolated liver mitochondria were added to 1 ml of isotonic acetate buffer (145 mm potassium acetate, 5 mm Tris-HCl, 0.5 mm EDTA, 5 μm valinomycin, and 1 μm rotenone, pH 7.4) in a cuvette at a final concentration of 0.5 mg/ml mitochondrial protein. Absorbance of the mitochondrial suspension before and after the addition of test compounds was measured every 10 s using a Varian Cary 300 spectrophotometer (Agilent Technologies, Santa Clara, CA). Swelling was recorded from the decrease in absorbance at 600 nm and expressed as percentage from initial reading after the addition of mitochondria.
Effects on Electron Flow
Electron flow assays were performed using the methods described by Rogers et al. (36). Briefly, 2 μg of mitochondrial protein in MAS buffer was loaded into a Seahorse XF96 tissue culture plate and centrifuged at 2,000 × g for 15 min at 4 °C. Prior to the assay, mitochondria were incubated at 37 °C for 10 min in MAS buffer containing 10 mm pyruvate, 2 mm malate, and 5 μm SR4 or FCCP. Rotenone (2 μm), succinate (10 mm), antimycin A (4 μm), TMPD (100 μm), and ascorbate (10 mm) were added sequentially as indicated in Fig. 4G.
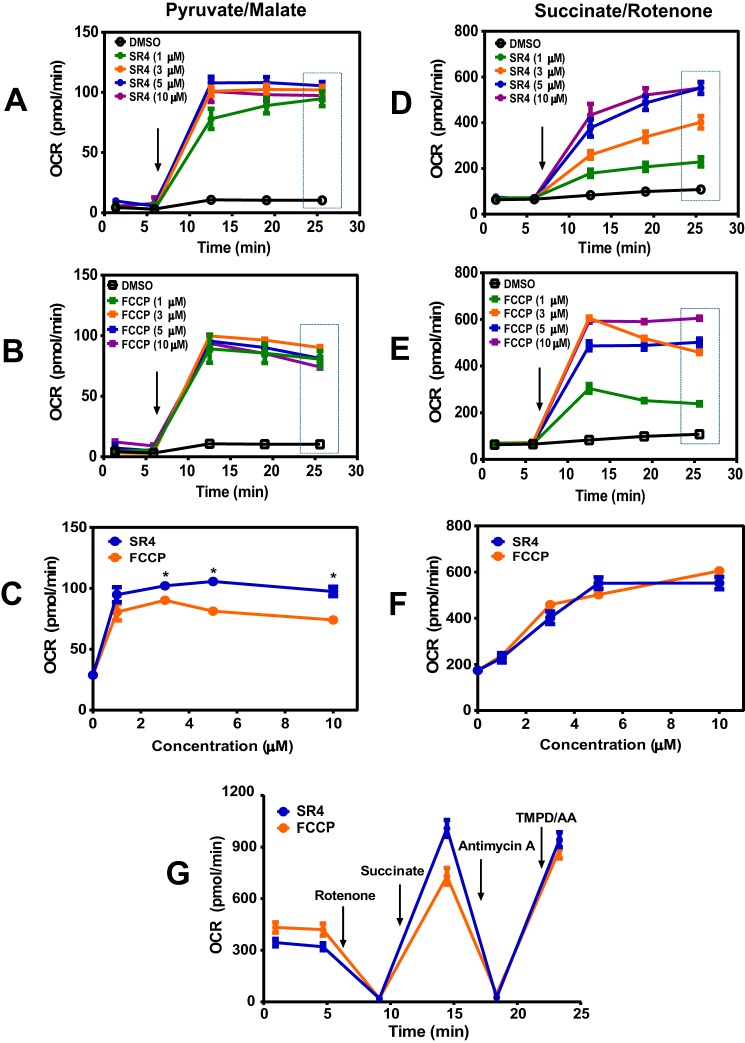
FIGURE 4.
SR4 causes mitochondrial uncoupling in isolated mouse liver mitochondria. OCR of isolated mouse liver mitochondria respiring on pyruvate/malate (A–C) or succinate in the presence of rotenone (D--F) was measured following treatment with increasing concentrations of SR4 or FCCP. Arrow, time of the addition of test compounds. The dotted box (a, b, d, and e) indicates the OCR values depicted in c and f. G, effects of SR4 and FCCP on electron flow. Isolated mouse liver mitochondria respiring on pyruvate and malate in the presence of SR4 (5 μm) or FCCP (5 μm) were treated sequentially with rotenone (4 μm), succinate (10 mm), antimycin A (4 μm), and the electron donors TMPD (100 μm) and ascorbate (10 mm). OCR was measured after injection of each compound as indicated by arrows. OCR values are representative rates of n = 6–8 wells/treatment/experiment from three separate experiments. *, p < 0.05 versus FCCP by two-way ANOVA with Sidak's post hoc test. Error bars, S.E.
ANT and UCP Inhibition Assays
HepG2 cells were plated in XF96 tissue culture plates (5,000 cells/well) and incubated overnight. Just before initiation of the XF assay, cells were washed twice with 200 μl/well MAS buffer, permeabilized with 1.0 nm XF PMP (Seahorse Bioscience), and simultaneously provided 4 mm ADP, 10 mm succinate, and 1 μm rotenone to obtain state 3 respiration, as described previously (38). The plate was immediately placed into the XF96 instrument, and the assay was initiated. OCR was recorded, followed by sequential injections of the ANT inhibitor carboxyatractyloside (CAT) (3 μg/ml) or the UCP inhibitor GDP (20 mm) and then with SR4 or FCCP, with measurements taken after each injection. Similar assays were performed in isolated mouse liver mitochondria (2 μg/well in XF96 plates), without the PMP addition step.
UCP-2 and AMPK siRNA Knockdown
HepG2 cells were reverse transfected with 100 nm human siRNA targeting either UCP-2 or AMPK α1/α2 or with the non-silencing control using LipofectamineTM RNAiMAX (Invitrogen) in serum-free culture medium without antibiotics according to the manufacturer's recommendations for 12 h. Growth medium was then added back to the cells, which were cultured for an additional 48 h. The expression level of target proteins in transfected cells was always tested by Western blots. Only cells with target protein significant knockdown were used for the experiments.
Cell Cycle Analyses
HepG2 cells were plated in 6-well tissue culture plates (4 × 105/well) and incubated overnight. Medium was replaced with fresh DMEM with or without test compounds for 12 h. After incubation, the cells were trypsinized, harvested, and fixed with 70% ethanol at 4 °C overnight. After removal of ethanol by washing with ice-cold PBS, cells were resuspended in 1 ml of propidium iodide (PI) staining solution (4% PI, 2 μg/ml RNase in PBS) and incubated for 15 min at 37 °C. Fluorescence-activated cell sorting (FACS) analysis was performed using the CyAn ADP cytometer, and the cell cycle distribution was analyzed using the Mod Fit LT software version 3.2 (Verity Software House, Topsham, ME). The experiments were repeated at least three times.
Annexin V-PI Assay of Apoptotic Cells
Apoptosis was determined by FACS analysis using the Annexin V-FITC apoptosis detection kit (Thermo Fisher Scientific) as described by the manufacturer. HepG2 cells were plated in 6-well tissue culture plates (4 × 105 cells/well) and incubated overnight. Medium was replaced with fresh DMEM containing the testing agents at the indicated final concentrations. After 24 h of treatment, cells were trypsinized and centrifuged at 1,000 rpm for 5 min. The cell pellets were then washed twice with 1 ml of ice-cold PBS. The cells were resuspended in 100 μl of binding buffer and were stained with 5 μl of Annexin V-FITC solution and 5 μl of PI solution for 20 min at room temperature in the dark. Then the samples were diluted with 400 μl of 1× binding buffer, processed for data acquisition, and analyzed by CyAn ADP flow cytometer. Approximately 50,000 counts were made for each sample. The percentage distribution of necrotic (R1, top left quadrant), late apoptotic (R2, top right quadrant), normal/viable (R3, bottom left quadrant), and early apoptotic cells (R4, bottom right quadrant) was calculated using Summit software (version 4.2, Cytomation Inc., Fort Collins, CO) (see Fig. 8). The experiments were repeated at least three times.
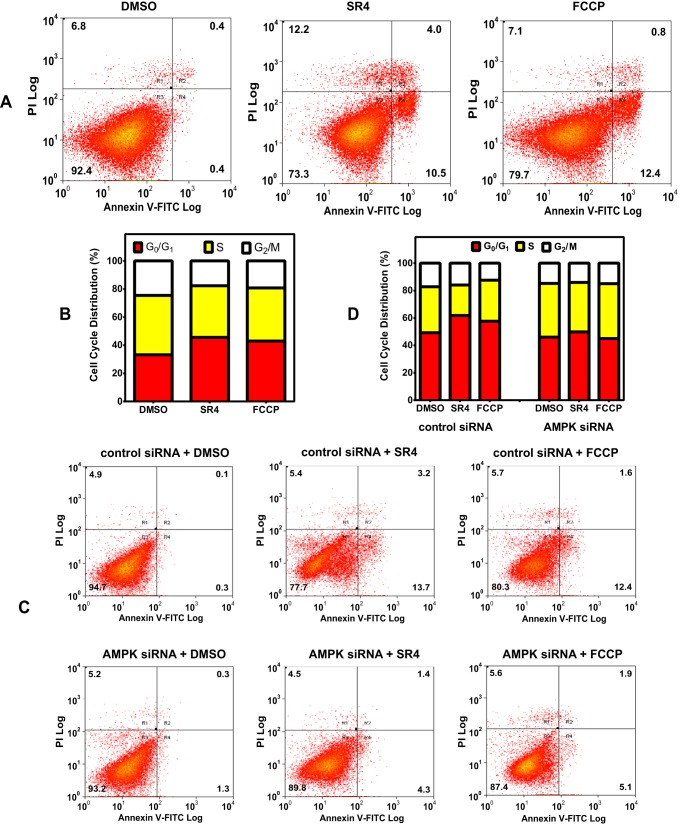
FIGURE 8.
SR4 and FCCP induces cell cycle arrest and apoptosis. A, representative cytograms of Annexin V-PI double staining. HepG2 cells were incubated with or without test compounds for 24 h, washed, and harvested. The cells were then fixed and double-stained with Annexin V-FITC and PI and analyzed by flow cytometry. The percentage distribution of normal/viable (R3, bottom left quadrant), early apoptotic (R4, bottom right quadrant), late apoptotic (R2, top right quadrant), and necrotic cells (R1, top left quadrant) was calculated using Summit software. B, representative cytogram data of cell cycle arrest of HepG2 cells exposed to either 5 μm SR4 or FCCP for 12 h. Cell cycle distribution was measured using flow cytometry, and the cell number in each cell cycle phase was calculated and expressed as an overall percentage. C and D, effects of AMPK siRNA knockdown on SR4-mediated cell cycle arrest and apoptosis. HepG2 cells were transfected with control or AMPKα1/α2 siRNA and then treated with either DMSO (control) or 5 μm SR4 or FCCP. Cell cycle distribution and apoptosis were measured using flow cytometry as noted above.
Mitochondrial ROS Production
Mitochondrial ROS generation in HepG2 was assessed by MitoSoxTM Red (Thermo Fisher Scientific), a fluorogenic dye developed and validated for highly selective detection of superoxide in the mitochondria of living cells. Briefly, HepG2 cells (4 × 105 cells/well) were seeded in a 6-well plate and incubated overnight. Twenty-four hours after plating, cells were treated with test compounds as indicated in each experiment. After washing with prewarmed PBS, cells were incubated with fresh medium containing 5 μm MitoSox Red for 15 min at 37 °C. The fluorescence intensity was measured at excitation/emission of 530 nm/590 nm using flow cytometry (CyAn ADP). The cells-only sample (no MitoSox dye added) was used as the background. Approximately 30,000 counts were made for each sample. Experiments were repeated three times, and at least duplicate samples were included in each experiment.
Protein Extraction and Western Blotting
HepG2 cells treated with test compounds for the indicated times were harvested and washed twice with ice-cold PBS. Total cytosolic proteins were extracted with cell lysis buffer (Cell Signaling Technology), and the protein concentration was determined using the DC protein assay kit. Equal amounts of proteins (25 μg) were loaded onto 4–15% Criterion TGX gels (Bio-Rad), resolved by SDS-PAGE, and then transferred to nitrocellulose membranes for immunoblotting. Membranes were blocked with 5% nonfat dry milk in Tris-buffered saline containing 0.05% Tween 20 before incubation overnight at 4 °C with primary antibodies as listed above. Immunoreactive proteins were visualized by peroxidase-labeled secondary antibodies and the ECL system (Western Lightning chemiluminescence reagent, PerkinElmer Life Sciences). Equal loading of proteins was confirmed by stripping and reprobing the membranes with β-actin antibodies.
RNA Isolation and Sequencing
HepG2 cells (4 × 105/well in 6-well plates) were treated with 5 μm SR4 or vehicle (DMSO), and RNA was isolated 4 and 24 h post-treatment. Three biological replicates were prepared for each time point as well as control untreated cells. After treatment, total RNA was prepared using the RNeasy Mini kit (Qiagen) according to the manufacturer's instructions and eluted in 50 μl of RNase/DNase-free water, and the initial concentration and purity were assessed by a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). Prior to sequencing, RNA quality was also assessed by microfluidic capillary electrophoresis using an Agilent 2100 Bioanalyzer and the RNA 6000 Nano Chip kit (Agilent Technologies, Santa Clara, CA). Sequencing libraries were prepared with the TruSeq RNA Sample Prep kit V2 (Illumina, San Diego, CA) according to the manufacturer's protocol with minor modifications. Briefly, ribosomal RNA was removed from 500 ng of total RNA using the RiboZero kit (Illumina), and the resulting RNA was ethanol-precipitated. Pellets were resuspended in 17 μl of Elute/Prime/Fragment Mix (Illumina), and first-strand cDNA synthesis was performed using DNA polymerase I and RNase H. cDNA was end-repaired, 3′-end-adenylated, and universal adapter-ligated, followed by 10 cycles of PCR using the Illumina PCR primer mixture and Phusion DNA polymerase (Illumina). Libraries were purified with Agencourt AMPure XP beads, validated with Agilent Bioanalyzer 2100, and quantified with Qubit (Life Technologies, Inc.). Libraries were sequenced on Illumina Hiseq 2500 with single-end 40-bp reads. Reads were aligned using TopHat (version 2.0.8b) to human genome hg19. Read alignment statistics are provided in Table 1.
TABLE 1.
Read alignment statistics for RNA-seq
Single end reads were aligned using TopHat (version 2.0.8b) to human genome hg19.
| Sample | Total read count | Total aligned reads | Percent aligned | Transcript coverage |
|---|---|---|---|---|
| % | % | |||
| Control-1 | 29,578,056 | 25,710,329 | 86.90 | 69.50 |
| Control-2 | 22,247,533 | 19,306,982 | 86.80 | 70.80 |
| Control-3 | 45,920,173 | 43,849,025 | 95.50 | 69.70 |
| SR4-4hrs-1 | 25,029,799 | 21,666,068 | 86.60 | 70.80 |
| SR4-4hrs-2 | 24,189,061 | 20,958,949 | 86.60 | 71.00 |
| SR4-4hrs-3 | 23,686,524 | 20,606,769 | 87.00 | 72.30 |
| SR4-24hrs-1 | 49,270,437 | 46,702,323 | 94.80 | 70.60 |
| SR4-24hrs-2 | 42,364,128 | 36,646,166 | 86.50 | 71.40 |
| SR4-24hrs-3 | 35,890,942 | 31,257,089 | 87.10 | 70.70 |
Quantitative RT-PCR (qRT-PCR) Validation
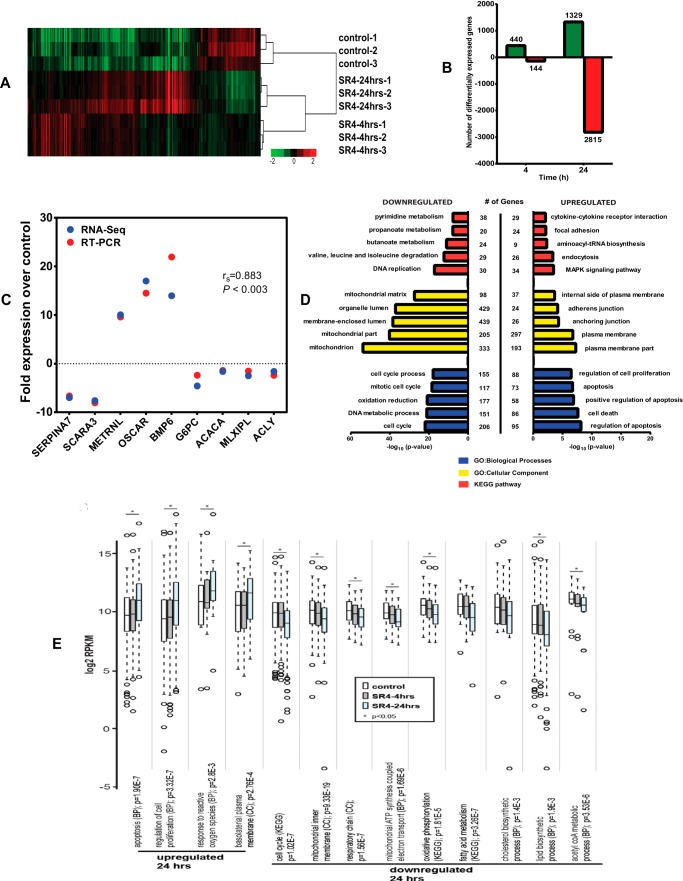
To confirm the results obtained from RNA-seq analysis, several up- and down-regulated genes at 24 h after SR4 treatment were selected and analyzed by qRT-PCR. First strand cDNA was prepared using the High Capacity cDNA reverse transcription kit (Life Technologies). Prevalidated primer pairs for OSCAR, ACLY, ACACA, SCARA3, BMP6, G6PC, METRNL, SERPINA7, and MLXIPL were purchased from Bio-Rad (PrimePCRTM PCR primers). qRT-PCR was performed on three independent samples per treatment using the ABI-7500 fast real-time PCR system (Life Technologies) and Power SYBR Green master mix. After initial incubation for 2 min at 50 °C, the cDNA was denatured at 95 °C for 10 min, followed by 40 cycles of PCR (95 °C for 15 s, 60 °C for 60 s). For statistical tests related to RNA-seq differential gene expression, gene ontology, and qRT-PCR correlations with RNA-seq, see below.
Statistical Methods
RNA-seq, qRT-PCR, and Gene Ontology
Differential gene expression was identified from standard Partek workflow (Partek Genomics Suite version 6.6, Partek, Inc.) using ANOVA, with step-up false discovery rate multiple testing correction p value <0.05 and requiring a >1.5-fold change between each time point and control samples. Gene ontology for the 24-h SR4 treatment for the up- or down-regulated genes was analyzed for functional enrichment using the Database for Annotation, Visualization, and Integrated Discovery (DAVID; version 6.7) (39). Databases included GOTERM_BP_FAT (biological process), GOTERM_CC_FAT (cellular component), and the Kyoto Encyclopedia of Genes and Genomes (KEGG_PATHWAY). For inclusion, terms required an EASE score of p < 0.005. Fold changes in gene expression between control and 24-h SR4 treatment were derived from the comparative CT method (40) with β-actin as an internal control. Correlation between the expression values detected by RNA-seq (normalized log2 RPKM fold change) and qRT-PCR (mean -fold change) for the nine genes tested were estimated by calculating Spearman's ρ correlation in the Prism version 6.0 software (GraphPad Software, Inc., La Jolla, CA). For expression distribution via box plots, average RPKM values for genes falling within each ontology term were log2-converted, and box plots were generated with BoxPlotR software (41). Mann-Whitney U tests were used to assess statistical significance between time points (two-tailed; *, p < 0.05).
Mitochondrial Measurements, ROS, Intracellular ATP Levels, AMP/ATP Ratios, and MTT/CV Assays
TMRE measurements were taken from at least two independent experiments with triplicate flow cytometry counts (n ≥ 6) to determine mean fluorescence intensity. Statistical significance was determined by one-way ANOVA and Dunnett's post hoc multiple comparison test (*, p < 0.05; **, p < 0.01), as compared with control samples. For remaining experiments, at least two independent biological replicates with triplicate measurements (n ≥ 6 measurements) were taken. Additional details can be found in corresponding figure legends. Data are presented as mean ± S.E. Statistical significance from two-factor experiments (e.g. drug × dose) was assessed with a two-way ANOVA and Sidak's post hoc multiple comparison test (*, p < 0.05). Unpaired Student's t tests were also used for TMRE, MTT, and intracellular AMP/ATP ratios (*, p < 0.05). Statistical tests were conducted using Prism version 6.0 software.
Results
SR4 Increases Mitochondrial Respiration in Whole Cells and Isolated Mitochondria
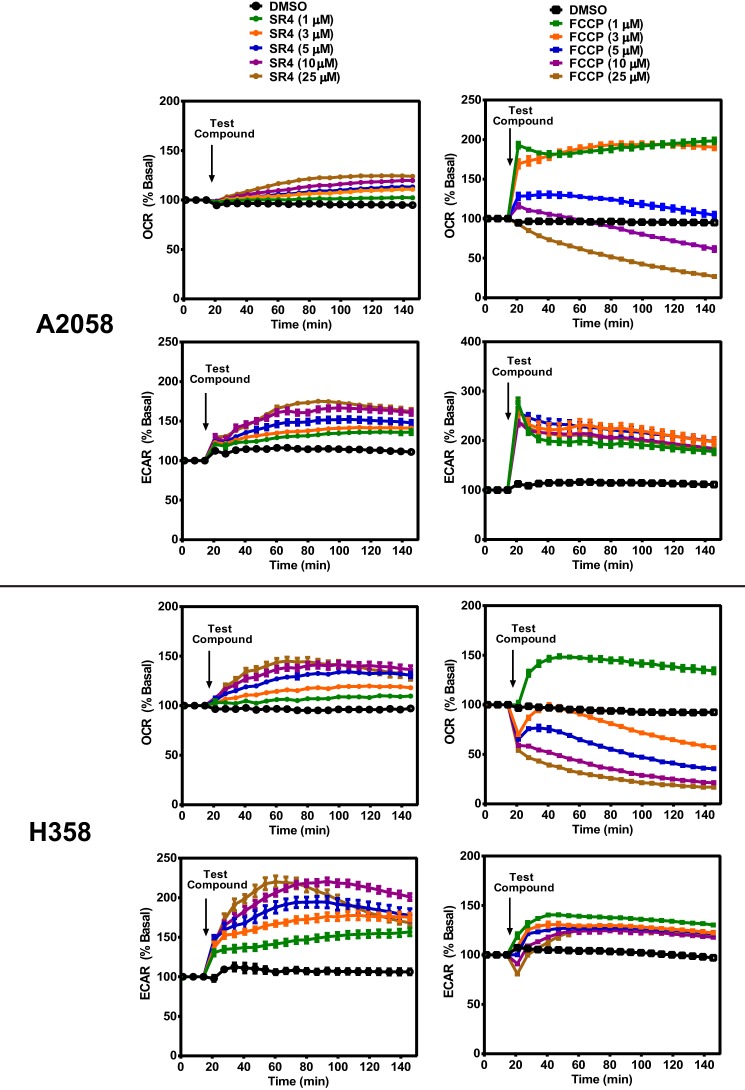
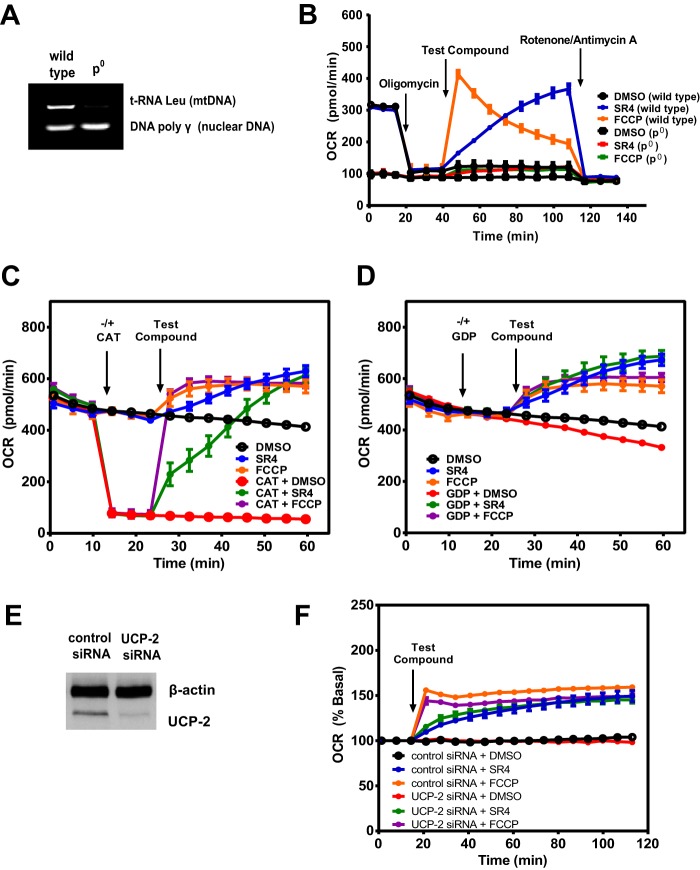
To initially assess the mitochondrial uncoupling properties of SR4, we used Seahorse XF 24/96 flux analyzers to measure its effects on mitochondrial respiration. Increased OCR is one of the consequences of uncoupling, and as shown in Fig. 1A, SR4 increased OCR in a time- and dose-dependent manner in HepG2 cells. Compared with the prototype uncoupler FCCP, which caused an abrupt increase in OCR at lower concentrations and decreases OCR at higher (>5 μm) concentrations, SR4 at higher concentrations (up to 25 μm) was able to maintain uncoupled respiration at a high rate in HepG2 cells (Fig. 1, A and B) and other cancer cell lines, including A2058 human melanoma and H358 human lung cancer (Fig. 2). Additionally, SR4 also dose-dependently increased the cellular ECAR, a measure of both glycolysis (proton co-transport with lactate out of the cell) and nutrient oxidation (the hydration of CO2 to form HCO3− and H+) at higher concentration ranges than FCCP (Figs. 1 (C and D) and 2). These data provide initial evidence that SR4 is an uncoupler of mitochondrial respiration.
FIGURE 1.
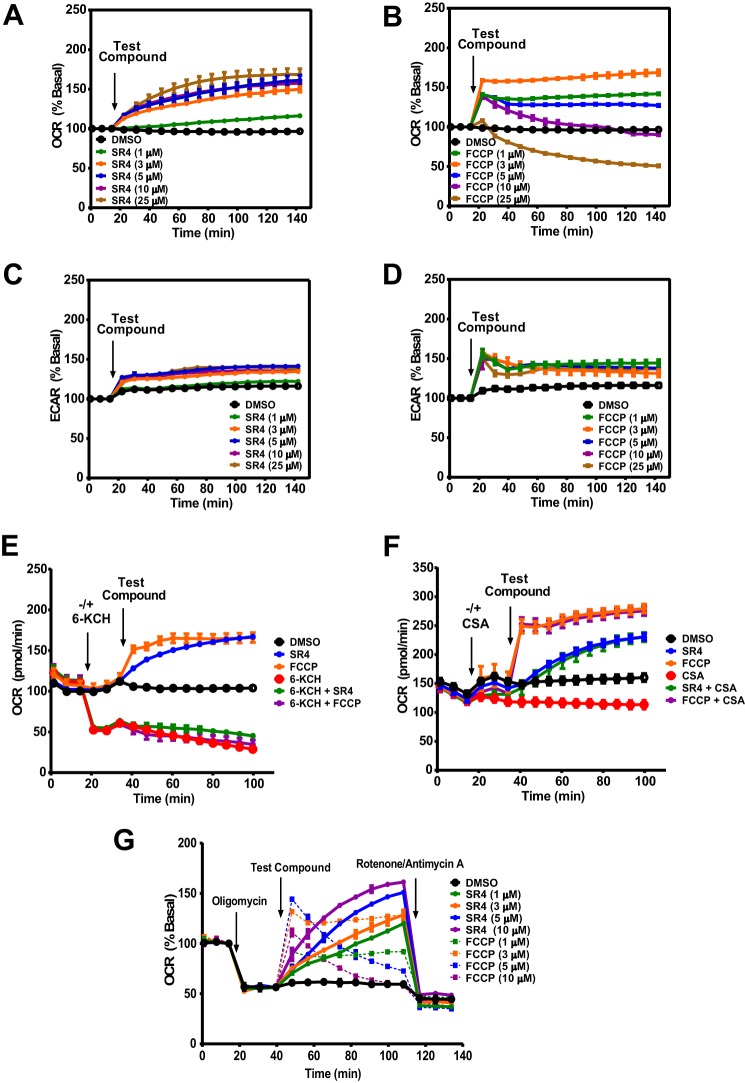
Effects of SR4 or FCCP on mitochondrial respiration in HepG2 cells. A–D, OCR and ECAR in HepG2 cells treated with various concentrations (0–25 μm) of test compounds as measured using Seahorse XF flux analyzers. E and F, OCR rates of HepG2 cells treated with SR4 or FCCP in the presence/absence of 6-KCH and CSA. 6-KCH (200 mm) and CSA (1 μm) were added to the assay system prior to injection of SR4 (5 μm) or FCCP (3 μm). G, OCR of HepG2 cells treated with SR4 or FCCP in the presence of oligomycin. Cells were sequentially treated with oligomycin (1 μm), the indicated concentrations of SR4 or FCCP (uncoupler), and rotenone (1 μm) plus antimycin A (1 μm), as shown by arrows. OCR and ECAR values are representative rates of n = 6–8 wells/treatment/experiment from three separate experiments. Error bars, S.E.
FIGURE 2.
OCR and ECAR in A2058 human melanoma and H358 human lung cancer cells treated with various concentrations (0–25 μm) of test compounds as measured using Seahorse XF flux analyzers. OCR values are representative rates of n = 6–8 wells/treatment/experiment from two separate experiments. Error bars, S.E.
To further characterize putative SR4-induced uncoupling, HepG2 cells were incubated with the documented mitochondria recoupler 6-KCH, (42) and the MPT blocker CSA prior to exposure to SR4 or FCCP. 6-KCH completely prevented both SR4- and FCCP-stimulated OCR increase (Fig. 1E). In contrast, pretreatment with CSA did not inhibit the increase in OCR induced by either SR4 or FCCP (Fig. 1F), indicating that the uncoupling effects of both compounds were not mediated by the MPT.
To establish that SR4 is a bona fide uncoupler, we performed the Seahorse Mito stress test assay using sequential additions of oligomycin (ATP synthase inhibitor), SR4 or FCCP, and rotenone/antimycin A (complex I and complex III inhibitor, respectively). We investigated whether SR4 could stimulate oxygen consumption in the presence of the ATP synthase inhibitor oligomycin. Again, using FCCP as a positive control, we found that SR4 dose- and time-dependently increased mitochondrial respiration in the presence of oligomycin in HepG2 (Fig. 1G), suggesting that SR4 indeed uncouples mitochondrial respiration.
To verify that SR4 acts directly on mitochondrial respiration, we measured oxygen consumption in mitochondrial DNA-depleted (ρ0) HepG2 cells (Fig. 3A) as well as freshly isolated mouse liver mitochondria and treated them with either SR4 or FCCP. As expected, both compounds failed to increase OCR in HepG2 ρ0 cells compared with wild type HepG2 (Fig. 3B). In isolated liver mitochondria, SR4 dose- and time-dependently increased OCR of mitochondria respiring on pyruvate and malate (Fig. 4, A and B) and exhibited significantly greater uncoupling effects at higher (>3 μm) concentrations than FCCP (Fig. 4C). Similar dose- and time-dependent effects were obtained with succinate-driven respiration (Fig. 4, D and E), although this time, both SR4 and FCCP exhibited comparable OCR increases at all concentrations tested (Fig. 4F). Additionally, both SR4 and FCCP did not alter mitochondrial electron flow in mitochondria respiring on pyruvate and malate and sequentially treated with rotenone, succinate, antimycin A, and the complex IV electron donor system TMPD and ascorbate. As demonstrated in Fig. 4G, the addition of rotenone inhibited respiration in uncoupled mitochondria respiring on the complex I-linked substrates pyruvate and malate. Succinate was added next to enable complex II-dependent respiration and then antimycin A to block respiration. Because both SR4 and FCCP-mediated respiration were inhibited by antimycin A, these data demonstrate that neither compound can donate electrons from succinate to cytochrome c or complex IV. The addition of the electron donor system of TMPD and ascorbate was able to fully rescue respiration at complex IV in the presence of both SR4 and FCCP. Taken together, these results provide convincing evidence of the uncoupling effects of SR4 on the fully functional mitochondrial electron transport chain.
FIGURE 3.
Effects of SR4 and FCCP on ρ0 cells and permeability transition pore (PTP) proteins in HepG2. OCR in wild type and ρ0 cells was measured after sequential injections (as indicated by arrows) of metabolic stressors (1 μm oligomycin, 5 μm SR4 or 3 μm FCCP, and 1 μm rotenone plus 1 μm antimycin A). The tRNA Leu mitochondrial gene was used as indicator of mitochondrial DNA depletion (A and B). SR4 increases OCR independent of ANT or UCPs. Freshly isolated mouse liver mitochondria respiring on succinate (10 mm), rotenone (2 μm), and ADP (4 mm) were treated with the ANT inhibitor CAT (3 μg/ml) (C) or UCP inhibitor guanosine diphosphate (GDP, 20 mm) (D), followed by either 5 μm SR4 or FCCP. OCR was measured after injection of each compound as indicated by arrows. The increase in OCR induced by either SR4 or FCCP is not affected by knockdown of UCP-2. UCP-2 knockdown by siRNA was assessed by Western blotting (E), and OCR was measured in control siRNA and UCP-2 knockdown HepG2 cells treated with SR4 (5 μm) or FCCP (3 μm) (F). OCR values are representative rates of n = 6–8 wells/treatment/experiment from 2–3 independent experiments. Error bars, S.E.
SR4 Uncoupling Activity Is Independent of ANT and UCPs
Mitochondrial uncoupling can also be mediated by proteins in the inner membrane of the mitochondria, including ANT or UCPs (29). ANT is specifically susceptible to thiol modification leading to opening of the MPT pore (43), whereas superoxide can activate UCPs (44). To determine whether ANT was necessary for SR4-mediated uncoupling, we pretreated freshly isolated mouse liver mitochondria with the ANT inhibitor CAT prior to treatment with either SR4 or FCCP and measured OCR using the Seahorse XF assay methods. We observed that either compound stimulated OCR in both isolated liver mitochondria (Fig. 3C) and permeabilized HepG2 cells (data not shown), even in the presence of CAT, suggesting that their uncoupling mechanism is independent of ANT. To assess whether SR4 modulates UCP to uncouple respiration, we pretreated mitochondria with the general UCP inhibitor GDP before SR4 or FCCP injection and measured the OCR. Both compounds were able to increase OCR despite the presence of GDP (Fig. 3D). We also knocked down UCP-2 via siRNA transfection (Fig. 3E). Both SR4 and FCCP increased OCR at similar rates in control as well as in UCP-2 knockdown HepG2 (Fig. 3F). These results indicate that the uncoupling of mitochondrial respiration by SR4 is also not mediated through UCPs.
SR4 Induces MMP and Mitochondrial Swelling
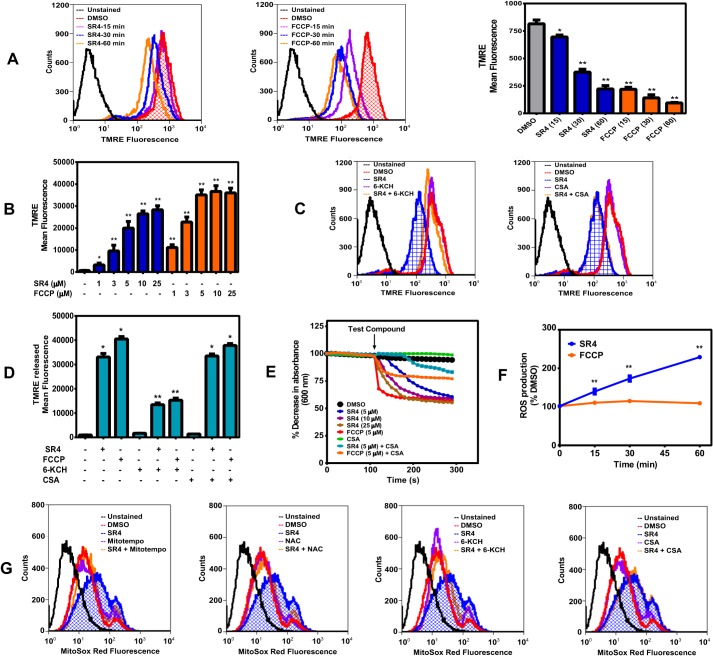
Increasing proton leak into mitochondria is expected to induce MMP and mitochondrial swelling, the latter a reflection of MPT. We observed a time-dependent decrease in the membrane potential of HepG2 cells treated with 5 μm SR4, similarly with equimolar concentration of FCCP, as measured by TMRE fluorescence (Fig. 5A). In isolated mouse liver mitochondria, both SR4 and FCCP showed a concentration-dependent decrease in membrane potential, as demonstrated by increasing amounts of preloaded TMRE released into the medium after treatment with the test compounds for 30 min (Fig. 5B). Pretreatment with 6-KCH, but not with the MPT blocker CSA, alleviated the SR4-mediated decrease in membrane potential in HepG2 cells and isolated mouse liver mitochondria (Fig. 5, C and D). These data suggest that SR4 disrupts MMP via its uncoupling effects.
FIGURE 5.
SR4 decreases membrane potential, induces mitochondrial swelling, and stimulates ROS production. Shown is time-dependent depolarization of the mitochondrial membrane as a consequence of SR4 or FCCP exposure. HepG2 cells were treated with 5 μm SR4 or FCCP from 0 to 60 min. After incubation, cells were then rinsed and stained with the fluorescent dye TMRE, and the overall fluorescence was analyzed by flow cytometry (excitation/emission 488 nm/572 nm). The bar graph is from three independent flow cytometry experiments. A, SR4 depolarizes isolated mouse liver mitochondria. TMRE released from isolated mouse liver mitochondria respiring on succinate (10 mm) in the presence of rotenone (1 μm) was measured as described under “Experimental Procedures” following treatment with increasing concentrations of SR4 and FCCP. The bar graph is from two independent experiments with n = 6. *, p < 0.05; **, p < 0.01 versus DMSO control by one-way ANOVA with Dunnett's post hoc test (B). C and D, effects of 6-KCH and CSA on SR4-mediated loss of membrane potential. HepG2 cells or freshly isolated mouse liver mitochondria were preloaded with TMRE (200 nm) for 30 min before treatment with 5 μm SR4 or FCCP alone or together with 200 μm 6-KCH or 1 μm CSA. Overall fluorescence was analyzed by flow cytometry (excitation/emission, 488 nm/572 nm) in HepG2 cells (C) or measurements of the released TMRE in isolated mitochondria (D), respectively. The bar graph is from two independent experiments with n = 8. *, p < 0.05 versus DMSO control; **, p < 0.01 versus SR4 or FCCP alone (unpaired t test). SR4 induces proton-dependent mitochondrial swelling (E). Mitochondrial membrane swelling was detected as a decrease in absorbance at 600 nm following the addition of SR4 or FCCP alone or in the presence of CSA (1 μm) on isolated mouse liver mitochondria respiring on succinate (10 mm) in the presence of rotenone (1 μm) and valinomycin (5 μm) in potassium acetate buffer. The arrow indicates time of the addition of test compounds into the mitochondria solution. Data were from three separate experiments with different mitochondria. F, SR4 promotes ROS production in HepG2. Cells were treated with SR4 or FCCP from 0 to 60 min, and superoxide production was measured from MitoSox Red fluorescence by flow cytometry. ROS production was expressed as a percentage of DMSO-treated cells based on mean fluorescence intensity of each treatment. *, p < 0.05; **, p < 0.01 versus FCCP by two-way ANOVA with Sidak's post hoc test. G, effects of 6-KCH, CSA, and antioxidants on SR4-mediated ROS production in HepG2. Cells were pretreated with 6-KCH (200 mm), CSA (1 μm), and MitoTempo (5 μm) or NAC (2 mm) for 15 min before the addition of SR4 (5 μm). Cells were incubated for another 30 min, and superoxide production was detected by flow cytometry of MitoSox Red fluorescence. Error bars, S.E.
In order to further characterize SR4-mediated uncoupling, we performed mitochondrial swelling assays in potassium acetate buffer treated with valinomycin. The K+ ionophore valinomycin was incubated with non-energized mitochondria to drive the movement of H+-acetate across the mitochondrial membrane in the protonated form generating a proton gradient. Under these conditions, mitochondria do not swell, because acetate can cross the membrane only as undissociated acetic acid, and the transmembrane passage of potassium in the form of the K+-valinomycin complex generates a charge imbalance, preventing further permeation of K+ (37). Intramitochondrial accumulation of potassium acetate becomes possible only if H+ can be exported from the inner compartment, thus enabling the influx of K+, resulting in mitochondria swelling. As shown in Fig. 5E, mitochondrial swelling was observed in the presence of either SR4 or the classical protonophore FCCP, which was prevented by the addition of CSA. These results suggest that SR4 could be functioning as a protonophore uncoupler.
SR4 Increases Mitochondrial ROS Production
Previous studies have shown that some protonophore uncouplers can increase ROS production, whereas others do not (23, 38, 45). We assessed the ROS production in HepG2 cells using MitoSoxTM Red fluorescence. Treatment of SR4 caused a temporal increase in superoxide production in HepG2 cells, with a >200% increase after 1-h incubation with the compound. In contrast, treatment of cultured cells with FCCP showed minimal ROS production (Fig. 5F). The SR4-mediated increase in superoxide generation was prevented by the antioxidants MitoTempo and NAC as well as 6-KCH but not with the MPT blocker CSA (Fig. 5G). Collectively, these observations indicate that ROS production induced by SR4 was associated directly with uncoupling and not through MPT opening.
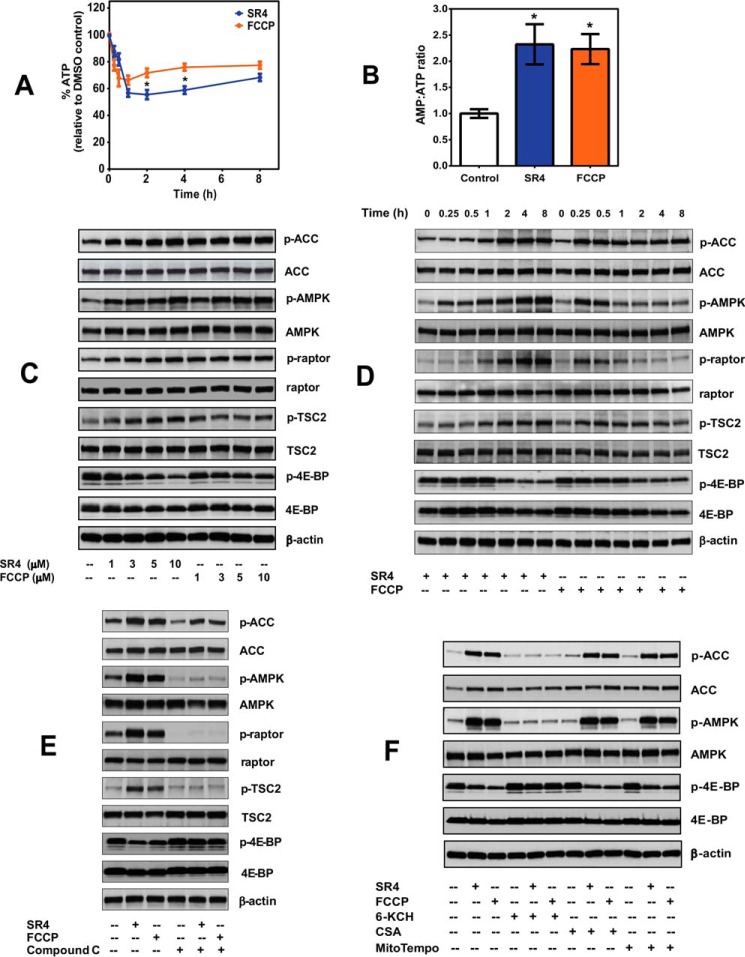
SR4 Decreases ATP Production and Activates the AMPK-mTOR Signaling Pathways
Mitochondrial uncoupling creates a futile cycle of nutrient oxidation without generating ATP due to proton leak and loss of proton motive force. Both SR4 and FCCP significantly decreased intracellular ATP production within 1 h (45 and 35%, respectively) in HepG2 cells (Fig. 6A), leading to increased AMP/ATP ratio (Fig. 6B). Such energy deficiency triggers activation of the energy-sensing enzyme AMPK, where we observed a dose- and time-dependent activation by both SR4 and FCCP (Fig. 6, C and D). This AMPK activation coincided well with the kinetics of OCR and MMP in intact HepG2 for both compounds (Figs. 1 (A and B) and 5A). Using equimolar concentrations of 5 μm, FCCP activated AMPK readily within 15 min and then slowly declined through time, whereas SR4 treatment activated AMPK progressively with prolonged exposure to the compound. Phosphorylation of acetyl-CoA carboxylase, one of the principal downstream targets of AMPK, was also elevated by SR4 or FCCP treatment and mimicked the temporal and dose-dependent activation of AMPK by the two compounds. Moreover, both SR4 and FCCP inhibited the mTORC1 signaling pathway, as demonstrated by a dose- and time-dependent increase in the phosphorylation of raptor and TSC2, and decreased phosphorylation of the mTOR downstream effector 4E-BP1 (Fig. 6, C and D). Treatment with the AMPK inhibitor Compound C prevented AMPK activation and mTOR inhibition for both compounds (Fig. 6E). Co-incubation with recoupler 6-KCH also prevented AMPK activation by either SR4 or FCCP, whereas pretreatment with MPT blocker CSA and antioxidant MitoTempo had no effects (Fig. 6F). Collectively, these results strongly indicate that AMPK activation is downstream of the uncoupling activity of SR4 and is independent of its ROS production and MPT opening.
FIGURE 6.
SR4 decreases intracellular ATP production and modulates AMPK-mTOR signaling in HepG2. A, total intracellular ATP production in cells treated with either 5 μm SR4 or FCCP was measured by bioluminescence assay and expressed as a percentage of time-matched vehicle (DMSO) control. *, p < 0.05 versus FCCP by two-way ANOVA with Sidak's post hoc test. B, intracellular AMP/ATP ratios in HepG2 cells following 1 h treatment with either 5 μm SR4 or FCCP. *, p < 0.05 versus DMSO control (unpaired t test). C and D, representative Western blots analyses showing dose- and time-dependent modulation of the AMPK-mTOR signaling pathways in HepG2 cells. E, effects of the AMPK inhibitor Compound C on SR4 and FCCP-mediated AMPK-mTOR signaling. Cells were pretreated for 30 min with Compound C, followed by the addition of either 5 μm SR4 or FCCP for 4 h. F, effects of 6-KCH, CSA, and MitoTempo on SR4/FCCP-mediated AMPK activation. HepG2 cells were pretreated with 6-KCH (200 mm), CSA (1 μm), and MitoTempo (5 μm) for 15 min before the addition of SR4 (5 μm) or FCCP (5 μm). For all blots, total cell lysates treated without or with test compounds for the indicated times were resolved under electrophoresis and immunoblotted with antibodies against phosphorylated and total AMPK, acetyl-CoA carboxylase, raptor, TSC2, 4E-BP, and β-actin, which served as an internal control. Error bars, S.E.
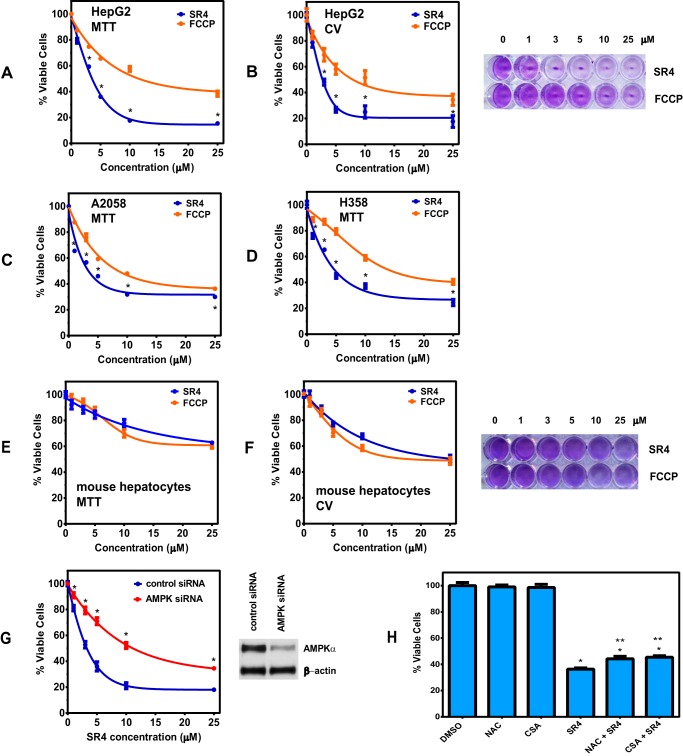
SR4 Induces Cytotoxicity, Cell Cycle Arrest, and Apoptosis
Mitochondrial uncoupling by chemical uncouplers has been shown to have beneficial anticancer properties by inhibiting proliferation and promoting cell cycle arrest and apoptosis (23–27). Using both MTT and CV staining assays, we observed that SR4 and FCCP are both cytotoxic and inhibit proliferation of HepG2 in a dose- and time-dependent manner, with IC50 values of ∼3.5 and 11.0 μm and 3.0 and 8.0 μm for MTT and CV staining, respectively, after 48-h treatment (Fig. 7, A and B). Both compounds were also cytotoxic to other human cancer cells (Fig. 7, C and D), but SR4 exhibited less cytotoxicity (IC50 of >20 μm) to normal cells, such as primary mouse hepatocytes (Fig. 7, E and F).
FIGURE 7.
SR4 decreases cell viability in HepG2 and other cancer cells but exhibited less cytotoxicity to normal cells. Cells were treated with various concentrations of either SR4 or FCCP compounds. After a 48-h exposure, cell viability was assessed by an MTT assay (A) and CV staining (B). *, p < 0.05 versus FCCP by two-way ANOVA with Sidak's post hoc test. C–F, viability of A2058 human melanoma and H358 human lung cancer cells by MTT assay (C and D) and primary mouse hepatocytes after 48-h exposure to 5 μm SR4 or FCCP by MTT (E) and CV staining (F). G, effects of AMPK knockdown on SR4-mediated cytotoxicity in HepG2 cells. HepG2 cells were transfected with control or AMPKα1/α2 siRNA and then treated with either DMSO (control) or SR4 for 48 h before cell viability was measured by an MTT assay. *, p < 0.05 versus control siRNA viability by two-way ANOVA with Sidak's post test. Inset, expressions of AMPKα and β-actin in those cells were also assessed by Western blot analyses. H, effects of NAC and CSA on SR4-mediated cytotoxicity in HepG2 cells. Cells were pretreated with NAC (2 mm) or CSA (1 μm) for 30 min before the addition of SR4 (5 μm). After 48 h, cell viability was measured by MTT. *, p < 0.05 versus DMSO control; **, p < 0.05 versus SR4 alone control (t test). Error bars, S.E.
To determine the effects of AMPK inhibition on the cytotoxicity of SR4 to HepG2, we knocked down AMPKα1/α2 by siRNA transfection prior to SR4 treatment. As shown in Fig. 7G, AMPK knockdown significantly suppressed the cytotoxic effects of SR4, with IC50 values of ∼3.8 and 11.5 μm for control siRNA- and AMPK siRNA-treated HepG2 cells, respectively. In addition, pretreatment of the antioxidant NAC and MPT blocker CSA only slightly increased the cell viability of SR4-treated HepG2 cells (Fig. 7H), suggesting that most of the cytotoxic effects of SR4 were mediated via AMPK activation.
The anti-proliferative effects of both compounds were associated with G0/G1 cell cycle arrest and induction of apoptosis in HepG2 cells (Fig. 8, A and B). As expected, knockdown of AMPK significantly prevented the cell cycle arrest induced by both SR4 and FCCP in HepG2 cells, as evident by a decrease in cells at the G0/G1 phase concurrent with an increase in cells in the S phase (Fig. 8C). Similarly, AMPK knockdown also prevented uncoupler-mediated apoptosis (Fig. 8D), further confirming that activation of AMPK is involved in the cytotoxicity of both uncouplers in HepG2 cells.
Uncoupling by SR4 Affects a Number of Genes Involved in Apoptosis, Cell Cycle Regulation, and Mitochondria and Energy Metabolism
Because we observed that SR4 uncoupling of OxPhos affects overall cellular bioenergetics and influences cell growth and proliferation of HepG2 cells, we reasoned that such responses would be reflected additionally in global transcriptional responses to SR4. To this end, SR4 induced differential gene expression was quantified at 4 and 24 h post-treatment, and the latter time point was interrogated for functional enrichment by gene ontology.
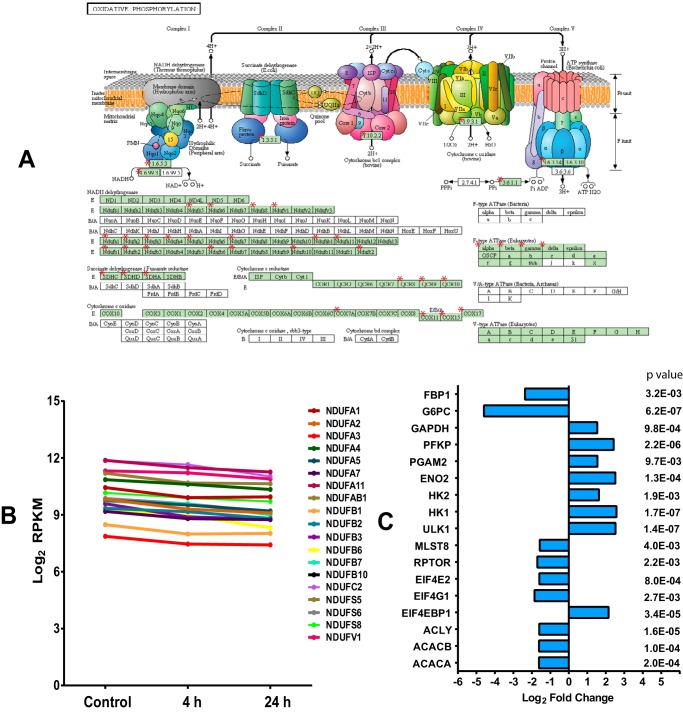
SR4 treatment of HepG2 cells showed tight group clustering when subjected to hierarchical clustering analysis (Fig. 9A). A total of 584 genes (440 up-regulated and 144 down-regulated) were differentially expressed after a 4-h exposure to SR4, whereas 4,144 genes (1,329 up-regulated and 2,815 down-regulated) were differentially expressed after 24 h of SR4 treatment (Fig. 9B). qRT-PCR on a random selection of nine genes showing differential accumulation levels confirmed both the direction and the magnitude of changes as shown by the Spearman ρ correlation value of 0.883 (p < 0.003) (Fig. 9C). Consistent with SR4-mediated induction of apoptosis and inhibition of cell proliferation, the highest ranked biological process ontology terms were regulation of apoptosis (96 genes, p = 7.04E−9) and cell cycle (208 genes, p = 1.07E−22) for up- and down-regulated genes, respectively, and this process probably involves the up-regulation of ROS responses (13 genes, p = 2.81E−03) and MAPK signaling (34 genes, p = 3.41E−04), as well as down-regulation of DNA replication (30 genes, p = 5.85E−18) (Fig. 9, D and E, and supplemental Table 1). Additionally, as a reflection of SR4 uncoupling activity, we observed significant down-regulation of mitochondria associated genes (mitochondrion (CC); 333 genes, p = 1.57E−54), including those products localized to the inner mitochondrial membrane and participants of the respiratory chain. These results complement clear down-regulation of OxPhos as well as downstream elements of mTOR signaling inhibition, including protein, fatty acid, and cholesterol biosynthesis (Fig. 9E and supplemental Table 1). The oxidative phosphorylation pathway (40 genes, p = 1.81E−5) in particular is expanded in Fig. 10A, which illustrates the down-regulation of components of all five ETC complexes, specifically complex I genes, where 19 of the total 44 subunit genes were impacted (Fig. 10B). Indeed, inhibition of mitochondrial electron transport coupled to ATP synthesis (23 genes, p = 1.69E−6) appears to be a major consequence of SR4 activity.
FIGURE 9.
SR4-associated differential gene expression and corresponding gene ontology. A, heat map of differentially expressed genes subsequent to SR4 treatment. Log2-transformed RPKM values were used for clustering using cluster 3.0 and Java Tree View version 1.1.6r4. Genes were mean-centered, and the average linkage method was selected for clustering. Samples clearly subcluster according to biological replicate. B, RNA-seq differential genes were quantified by time point post-SR4 treatment. Differential gene expression was identified from standard Partek workflow (Partek Genomics Suite version 6.6, Partek) using ANOVA, with step-up false discovery rate multiple testing correction p value <0.05 and requiring a >1.5-fold change between each time point and control samples. C, positive correlation between RNA-seq and qRT-PCR results. For nine randomly selected genes, the mean -fold change by qRT-PCR was plotted alongside log2 RPKM -fold changes from RNA-seq. Spearman's ρ correlation was 0.883 (p < 0.003). D, top five KEGG pathways and gene ontology biological process (GO-BP) and cellular component (CC) categories enriched by 24 h SR4 treatment in HepG2 cells. For redundant terms with identical gene lists, only the first term was listed within the figure (see supplemental Tables 1 and 2 for complete ontology results). E, box plots of selected functional gene groups enriched by ontology analysis. Expression distributions for genes falling within selected terms for control cells and those sampled 4 and 24 h after SR4 treatment are given. Center lines, medians; box limits, 25th and 75th percentiles with whiskers extended 1.5 times interquartile range from those percentiles (R software); outliers are shown as open circles (Tukey). Mann-Whitney U tests were used to assess statistical significance (*, p < 0.05).
FIGURE 10.
SR4 treatment down-regulates mitochondrial and OxPhos gene expression and modulates genes involved in AMPK-mTOR signaling. Oxidative phosphorylation pathway (KEGG) members down-regulated 24 h after SR4 treatment (A) are highlighted with red asterisks (51). Complex I gene expression values are expanded with their log2 RPKM expression values at 4 and 24 h post-treatment with SR4 (B). Shown are the effects of SR4 on selected genes associated with AMPK-mTOR signaling, including lipid metabolism, glycolysis, and gluconeogenesis. C, log2 -fold change values of each gene after 24-h exposure to 5 μm SR4.
Discussion
HCC is a major global public health problem due to the rising incidence and high mortality in both developing and developed countries. To date, sorafenib (a small molecule kinase inhibitor) is the only standard drug therapy for patients with advanced HCC with modest effectiveness at prolonging patients' overall survival for around 2–3 months (46). In addition, the tolerance and resistance to sorafenib in some patients further limit the clinical efficacy of sorafenib, with most phase III trials failing to reach their primary end points. The efficacy of systemic chemotherapy therapies is limited in patients because of their cirrhotic liver, potentially poor hepatic reserve, and the chemoresistance of the tumor (47, 48). Given the limited efficacy of sorafenib, there is still a need for a treatment of advanced HCC.
An emerging concept in the field of cancer metabolism is the importance of mitochondrial metabolism. Compared with normal cells, cancer cells show alterations in many cellular processes, including energy metabolism. Mitochondria are highly sensitive to stress and respond dynamically to the changes in their cellular microenvironment, and they continuously go through fusion and fission events. This process, termed mitochondrial dynamics, is essential for the maintenance of mitochondrial bioenergetic functions (49, 50). A comprehensive understanding of mitochondrial biology in cancer cells and the interaction between cellular metabolism and drug action is essential in developing mitochondria-targeted agents for cancer treatment.
Recently, we have shown that SR4 is a novel small molecule with promising therapeutic potential against leukemia, melanoma, and lung cancer and is very effective in both syngeneic and nude mice xenograft models along with regulating multiple nodes of cancer signaling like AMPK-mTOR, cell cycle arrest, apoptosis, cell proliferation, and angiogenesis (30–32). SR4 is a multitargeting agent with no overt toxicity for normal tissues when administered orally, as observed in our previous experiments. In the present study, we investigated the molecular mechanisms of SR4-induced cell death in HepG2 cells in relation to mitochondrial dysfunction and bioenergetics, which we hypothesize as upstream signals for its anticancer properties based on earlier observations of indirect AMPK activation (33). We found that SR4 dose- and time-dependently increased the mitochondrial respiration (increased OCR) and promoted collapse of mitochondrial potential in both intact HepG2 cells and isolated mouse liver mitochondria. These effects were reversed by the addition of the recoupler 6-KCH. SR4 also induced mitochondrial membrane swelling and decreased cellular ATP production. These results suggest that SR4 acts as an uncoupler because these events have been associated with the effects of several well known uncouplers, such as FCCP (which we used as a positive control in our experiments), CCCP, and 2,4-dinitrophenol.
The mechanism of action of many uncouplers is not yet fully understood. Uncouplers translocate protons from the intermembrane space to the matrices when they cross the membrane in their protonated form. This is followed by the translocation of their deprotonated form to the outer side of the membrane to bind another proton, thus “short circuiting” the coupling between the electron transport and phosphorylation reactions and preventing ATP synthesis. For induction of protonophore (protein-independent) uncoupling, an acid-dissociable group, bulky hydrophobic moiety, and strong electron-withdrawing groups are required (28). Based on our mitochondria swelling experiments on potassium acetate medium treated with the K+ ionophore valinomycin, SR4 exhibited “protonophoric” properties similarly with the structurally different FCCP, a prototype protonophore. Earlier data indicate that even the classical protonophores, such as FCCP and 2,4-dinitrophenol, may involve interaction with inner membrane proteins for their uncoupling mechanisms (13). We did not observe restoration of mitochondrial coupling in the presence of CAT and GDP, inhibitors of the inner mitochondrial membrane proteins ANT and UCP, respectively. On the other hand, both SR4- and FCCP-mediated uncoupling were significantly inhibited by 6-KCH, a documented mitochondria recoupler (42). The mechanism of action of 6-KCH is not well understood but was suggested to incorporate into the mitochondrial membrane and induce a strong asymmetrical increase in the membrane dipole potential (52). 6-KCH was also shown to preferentially inhibit protonophore uncouplers, which cross the membrane in the form of an anion, but had no effects on other uncouplers that cross the membrane in the form of a dimer of anionic and protonated species (53). Future structure-activity studies will be required to identify the precise uncoupling mechanisms of SR4, which could focus on SR4-mediated effects on proton conductance in artificial bilipid layer membranes and/or SR4 targeting preferences for unique lipid/protein compositions that exist within the mitochondrial inner membrane.
The present study also investigated the effects of mitochondrial uncoupling on downstream molecular signaling events in cancer proliferation. As observed with other uncouplers, the decrease in cellular ATP production by SR4 or FCCP triggers the activation of AMPK. Here, we also show that uncoupling of OxPhos affects mTOR signaling. An association of AMPK and mTOR signaling pathways in cancer growth and proliferation is well documented (54, 55). Indeed, inhibition of AMPK signaling with Compound C during the mitochondrial uncoupling stimulus abolished the inhibitory effect of SR4 and FCCP on mTOR signaling. Moreover, siRNA-mediated AMPK knockdown significantly reversed the cytotoxic effects of SR4 on HepG2. We observed similar inhibitory effects of SR4 on mTOR in previous studies with 3T3-L1 adipocyte (33) and lung cancer cells (31) using both Compound C and AMPK knockdown. In the lung cancer studies, similar AMPK knockdown decreased the cytotoxic effects of SR4, concomitant with decreased inhibition of mTOR signaling. Although the effects of SR4 on the AMPK-mTOR pathway were not directly identified in the RNA-seq analyses by gene ontology or KEGG pathway analysis, a number of key genes associated with the known downstream effects of AMPK activation and mTOR inhibition were modulated by the compound, including those for decreased lipid and cholesterol synthesis (ACACA, ACACB, and ACLY), inhibition of protein synthesis (EIF4EBP1, EIF4G1, and EIF4E2) and gluconeogenesis (G6PC and FBP1), and induction of autophagy/mitophagy (ULK1) (Fig. 10C and supplemental Table 2). In addition, there was reduced expression of mTORC1 subunit genes (RPTOR and MLST8). Taken together, these new data confirm that the AMPK-mTOR pathway is one of the important downstream signaling targets of SR4-mediated uncoupling associated with its anticancer properties.
Both metabolic flux measurements from Seahorse assays and RNA sequence analysis revealed that mitochondrial uncoupling of SR4 in HepG2 leads to diversion from OxPhos to glycolytic phenotype, as evident from the immediate increase in ECAR upon SR4 exposure; down-regulation of numerous OxPhos genes (Fig. 10A) and transcriptional up-regulation of several key glycolytic genes (HK1, HK2, ENO2, PGAM2, PFKP, and GAPDH) with simultaneous down-regulation of gluconeogenic genes (G6PC and FBP1) (Fig 10C and supplemental Table 2); and down-regulation of isoleucine, leucine and valine degradation (KEGG pathway, p < 4.9E−13; supplemental Tables 1 and 2), the latter of which are metabolized in the mitochondria into products (acetyl-CoA or succinyl-CoA) that feed into the tricarboxylic acid cycle. Recent works support these observations by demonstrating that the decreased ATP synthesis in response to mitochondrial uncoupling results in a preferential oxidation of non-glucose carbon sources to maintain mitochondrial function and an increase in lactate generation under aerobic conditions (15, 56).
SR4 treatment results in most anabolic pathways being shut down, leading to widespread effects on many cellular components, biological processes, and signaling systems (Fig. 9, D and E, and supplemental Tables 1 and 2). Previous comprehensive gene studies using microarrays in rhabdomyosarcoma cells exposed to FCCP showed that the uncoupler mimics the hypoxic environment, where a sudden drop in ATP production as a result of loss of OxPhos inhibits protein synthesis and induces G1-S cell cycle arrest and DNA damage and ultimately apoptosis (57). Here, such global effects were also evident in HepG2 as SR4 severely impacted a number of metabolic processes vital to cell growth and proliferation. Of the seven “uncoupler molecular marker” genes identified in the aforementioned FCCP study, six of these high -fold change genes were also impacted by SR4 in the same direction (up or down), including DDIT3, PCNA, SARS, MTHFD1, HSPE1 and ASNS (supplemental Table 2), suggesting that uncoupling may have some universal effects independent of cell type. Further studies using other uncouplers and cancer cell types will be needed to confirm these observations.
Considering that both SR4 and FCCP more or less affected the same downstream pathways, such as induction of mitochondrial membrane depolarization and MPT (as shown by mitochondria swelling), AMPK-mTOR signaling, cell cycle arrest, and apoptosis, as well differential gene expression of several “uncoupler marker genes,” as noted above, it is tempting to speculate that more potent cytotoxic effects of SR4 compared with FCCP across a wide range of human cancer cell types could be attributed to its ability to induce ROS. Interestingly, we observe that ROS production following SR4 exposure occurs rapidly in HepG2 and is independent of AMPK activation and MPT pore opening. Although the superoxide anion is not harmful when ROS production is limited within the constraints of the cellular redox system, because the cells must maintain moderate ROS to sustain cellular signaling, an acute and substantial ROS increase may have a much more immediate effect and commit the cells to undergo apoptosis. In cancer cells, the increase in ROS generation leads to up-regulation of antioxidant defense to maintain the ROS levels below the toxic threshold. As such, cancer cells are more dependent on the antioxidant system and more vulnerable to further oxidative stress induced by exogenous ROS-generating agents that will probably cause elevation of ROS above the threshold level, leading to cell death (58). Indeed, many chemotherapeutic drugs, including some mitocans, are aimed at increasing mitochondrial ROS levels to promote apoptosis (6, 7). How are ROS generated by SR4? Although we have not specifically addressed this question in the current study, ROS can be produced as a result of opening the MPT because this can allow antioxidant molecules, such as glutathione, to exit the mitochondria, reducing the organelles' ability to neutralize ROS. However, such scenario is unlikely because treatment of MPT blockers like CSA and bongkrekic acid prior to SR4 failed to inhibit ROS production. Also, the hypothesis that mitochondrial NADPH mediated superoxide production can also be ruled out because we did not observe SR4 to induce oxidation of NADPH in isolated liver mitochondria. Interestingly, pretreatment with 6-KCH significantly prevented ROS generation, suggesting that the ROS increase is directly associated with the uncoupling mechanism. A recent study on the anesthetic agent bupivacaine demonstrated that drug-induced mitochondrial uncoupling and ROS production can occur in HepG2 cells via inhibition of complex I and III and reverse electron transfer (59). Surprisingly, we also observed that SR4 inhibited complex I activity in crude mitochondria extracts of HepG2. Because macromolecules within the mitochondria are more prone to ROS-induced damage due to their close proximity to the source of ROS, the damage exerted by ROS on mitochondrial components may lead to a higher degree of mitochondrial dysfunction and, in turn, to higher ROS production, leading to a vicious cycle of ROS amplification (60). As evident by our RNA-seq analysis, SR4 significantly down-regulated the expression of various components of the mitochondrial ETC, primarily complex I genes (Fig. 10, A and B), as well as an important ROS scavenger SCARA3 (scavenger receptor class A, member 3) (supplemental Table 1). SCARA3 (also referred to as the cellular stress response (CSR) gene) has been shown to deplete ROS and thus play an important role in protecting cells from oxidative stress (61). FCCP did not produce any increase in ROS, and based on recent microarray gene analysis, no associations between FCCP and any components the ETC as well as ROS scavenger genes were identified (57). Although speculative at this time, the activity of SR4 on the ETC components could be one of its unique characteristics that differentiate it from other uncouplers, such as FCCP, allowing it to have increased cytotoxicity in cancer cells.
Mitochondria are the checkpoint of the intrinsic pathway of apoptosis, and, therapeutically, mitochondrial uncoupling is an attractive drug target for chemotherapy due to its ability to induce MPT, leading to the release of pro-apoptotic factors. These factors result in the assembly of the apoptosome and commitment of the cell to apoptosis. To our knowledge, this is the first study that confirms the potential of uncouplers as anticancer therapy, although we have previously shown SR4 to inhibit lung cancer and melanoma progression in xenograft animal models (31, 32). Our results also suggest that SR4 is a useful probe for studying mitocan-based processes for the treatment as well as prevention of HCC through targeting critical cellular processes in its pathogenesis and progression. However, future pharmacokinetic analysis will be required to determine whether SR4 is suitable for long term treatment of pathologies that require constant exposure. Using pharmacological agents to uncouple all mitochondria throughout the body may be a risky treatment, because it might compromise energy homeostasis in other tissues, such as heart and brain, causing irreversible damage. For uncouplers to work safely, these compounds should cause uncoupling that increases very little as their concentration rises, potentially widening the difference between therapeutic and toxic doses and giving a wide therapeutic window (62). Recently, however, targeted uncoupling showing precise spatial and temporal control of mitochondrial function have been demonstrated, leading to the selective uncoupling of either individual or small groups of them within a cell (63, 64). Such technology, if adapted to SR4 in the future, will increase its safety and potentially lower the effective maximum dose required for its cytotoxic effects on tumors.
In summary, we report the identification of SR4 as a new chemotype mitochondrial uncoupler. SR4 is highly potent and demonstrates a greater effective dynamic range than the accepted standard uncoupler FCCP. One of the most important advantages of SR4 for an enhanced anticancer effect is that it induces loss of membrane potential and causes increased ROS production. These properties enable a sustained maximal rate of mitochondrial respiration with reduced ATP production and subsequent generation of free radicals that can further damage mitochondria and other cellular components of ROS-sensitive cancer cells.
Conclusions
Because cancer is a multistage process, it requires multi-targeted approaches for the development of an effective treatment regimen. HCC is a very aggressive tumor with notoriously poor prognosis once the disease becomes metastatic. Despite recent advances in the treatment of HCC, only a subset of patients respond, and such responses are usually of limited durability, less than 1 year. This poor response rate has necessitated the identification and incorporation of novel pathways and new approaches to enhance the activity of established targeted therapies. In the current studies, SR4 treatment effectively uncouples OxPhos, activates AMPK, and induces MPT and ROS production, leading to cell cycle arrest and apoptosis in HepG2 cells. This is a novel finding and helps to delineate how SR4 exerts potent anticancer effects in HCC. In a previously published study (31, 32), we found that the oral administration of SR4 was well tolerated without any overt toxicity along with inducing effective tumor inhibition in both syngeneic and nude mouse xenograft models of melanoma and lung cancer. Thus, we believe that SR4 is a novel mitochondrial uncoupling agent that elicits desirable responses across multiple cancer types and warrants additional study to standardize dose, routes of administration, organ targeting, and bioavailability in humans.
Author Contributions
J. L. F. contributed to research data and wrote the manuscript. J. S. and J. D. T. contributed to research data. G. W. R. and D. H. contributed to reviewing/editing the manuscript. C. W. contributed to data analysis and interpretation of data. A. D. R. and S. A. contributed to discussion and to reviewing/editing the manuscript. S. S. S. contributed to research data and wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We are grateful to Dr. Brian Armstrong (Microscope Core Laboratory, Beckman Research Institute of the City of Hope) and Lucy Brown (Analytical Cytometry Core, Beckman Research Institute of the City of Hope) for technical assistance with the fluorescence microscope and flow cytometry analyses, respectively. We also thank Lu Yang and Xiwei Wu (Functional Genomics Core) for help in RNA sequencing and analyses and Timothy Synold (Analytical Pharmacology Core) for providing expertise in AMP/ATP quantification.
This work was supported by Isaac and Jacquelin Moradi. This work was also supported by the Department Chair (Prof. Arthur Riggs) and Beckman Research Institute of the City of Hope. G. W. R. is an employee of Seahorse Bioscience, which manufactures the XF instruments used in this study. The other authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Tables 1 and 2.
- HCC
- hepatocellular carcinoma
- ANT
- adenine nucleotide translocase
- CAT
- carboxyatractyloside
- CSA
- cyclosporin A
- ECAR
- extracellular acifidication rates
- ETC
- electron transport chain
- FCCP
- carbonyl cyanide p trifluoromethoxyphenylhydrazone
- MMP
- mitochondrial membrane permeabilization
- MPT
- mitochondrial permeability transition
- OCR
- oxygen consumption rate
- OxPhos
- oxidative phosphorylation
- ROS
- reactive oxygen species
- RPKM
- reads per kilobase of gene per million mapped fragments
- TMPD
- N,N,N′,N′-tetramethyl-p-phenylenediamine
- UCP
- uncoupling protein
- 6-KCH
- 6-ketocholestanol
- NAC
- N-acetyl-l-cysteine
- TMRE
- tetramethylrhodamine ester
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide
- CV
- crystal violet
- AMPK
- AMP-dependent kinase
- MAS
- mitochondrial assay solution
- PI
- propidium iodide
- qRT-PCR
- quantitative RT-PCR
- ANOVA
- analysis of variance
- RNA-seq
- RNA sequencing
- ρ0 cells
- HepG2 cells lacking mitochondrial DNA.
References
- 1.Venook A. P., Papandreou C., Furuse J., and de Guevara L. L. (2010) The incidence and epidemiology of hepatocellular carcinoma: a global and regional perspective. Oncologist 15, 5–13 [DOI] [PubMed] [Google Scholar]
- 2.Dhanasekaran R., Limaye A., and Cabrera R. (2012) Hepatocellular carcinoma: current trends in worldwide epidemiology, risk factors, diagnosis, and therapeutics. Hepat. Med. 4, 19–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.El-Serag H. B. (2012) Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology 142, 1264–1273.e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang P., Kang D., Cao W., Wang Y., and Liu Z. (2012) Diabetes mellitus and risk of hepatocellular carcinoma: a systematic review and meta-analysis. Diabetes Metab. Res. Rev. 28, 109–122 [DOI] [PubMed] [Google Scholar]
- 5.Cancer of the Liver Italian Program (CLIP) investigators (1998) A new prognostic system for hepatocellular carcinoma: a retrospective study of 435 patients. Hepatology 28, 751–755 [DOI] [PubMed] [Google Scholar]
- 6.Neuzil J., Dong L.F., Rohlena J., Truksa J., and Ralph S.J. (2013) Classification of mitocans, anticancer drugs acting on mitochondria. Mitochondrion 13, 199–208 [DOI] [PubMed] [Google Scholar]
- 7.Rohlena J., Dong L.F., Ralph S.J., and Neuzil J. (2011) Anticancer drugs targeting the mitochondrial electron transport chain. Antioxid. Redox Signal. 15, 2951–2974 [DOI] [PubMed] [Google Scholar]
- 8.Papa S., Martino P.L., Capitanio G., Gaballo A., De Rasmo D., Signorile A., and Petruzzella V. (2012) The oxidative phosphorylation system in mammalian mitochondria. Adv. Exp. Med. Biol. 942, 3–37 [DOI] [PubMed] [Google Scholar]
- 9.Mitchell P. (1966) Chemiosmotic coupling in oxidative and photosynthetic phosphorylation. Biol. Rev. 41, 445–502 [DOI] [PubMed] [Google Scholar]
- 10.Schultz B.E., and Chan S.I. (2001) Structures and proton-pumping strategies of mitochondrial respiratory enzymes. Annu. Rev. Biophys. Biomol. Struct. 30, 23–65 [DOI] [PubMed] [Google Scholar]
- 11.Brand M.D., Chien L. F., Ainscow E. K., Rolfe D. F. S., and Porter R. K. (1994) The causes and functions of mitochondrial proton leak. Biochim. Biophys. Acta 1187, 132–139 [DOI] [PubMed] [Google Scholar]
- 12.Divakaruni A. S., and Brand M. D. (2011) The regulation and physiology. The regulation and physiology of mitochondrial proton leak. Physiology 26, 192–205 [DOI] [PubMed] [Google Scholar]
- 13.Skulachev V.P. (1998) Uncoupling: new approaches to an old problem of bioenergetics. Biochim. Biophys. Acta 1363, 100–124 [DOI] [PubMed] [Google Scholar]
- 14.Busiello R. A., Savarese S., and Lombardi A. (2015) Mitochondrial uncoupling proteins and energy metabolism. Front. Physiol. 10.3389/fphys.2015.00036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Samudio I., Fiegl M., and Andreeff M. (2009) Mitochondrial uncoupling and the Warburg effect: molecular basis for the reprogramming of cancer. Cancer Res. 69, 2163–2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan D. C. (2006) Mitochondria: dynamic organelles in disease, aging, and development. Cell 125, 1241–1252 [DOI] [PubMed] [Google Scholar]
- 17.Seyfried T. N., and Shelton L. M. (2010) Cancer as a metabolic disease. Nutr. Metab. (Lond.) 7, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Modica-Napolitano J. S., and Singh K. K. (2004) Mitochondrial dysfunction in cancer. Mitochondrion 4, 755–762 [DOI] [PubMed] [Google Scholar]
- 19.Boland M. L., Chourasia A. H., and Macleod K. F. (2013) Mitochondrial dysfunction in cancer. Front. Oncol. 3, 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang F., Ogasawara M.A., and Huang P. (2010) Small mitochondria-targeting molecules as anticancer agents. Mol. Aspects Med. 31, 75–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wen S., Zhu D., and Huang P. (2013) Targeting cancer cell mitochondria as a therapeutic approach. Future Med. Chem. 5, 53–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fulda S., Galluzzi L., and Kroemer G. (2010) Targeting mitochondria for cancer therapy. Nat. Rev. Drug Discov. 9, 447–464 [DOI] [PubMed] [Google Scholar]
- 23.Han Y. H., Kim S. W., Kim S. H., Kim S. Z., and Park W. H. (2008) 2,4-Dinitrophenol induces G1 phase arrest and apoptosis in human pulmonary adenocarcinoma Calu-6 cells. Toxicol. In Vitro 22, 659–670 [DOI] [PubMed] [Google Scholar]
- 24.Pardo-Andreu G. L., Nuñez-Figueredo Y., Tudella V. G., Cuesta-Rubio O., Rodrigues F. P., Pestana C. R., Uyemura S. A., Leopoldino A. M., Alberici L. C., and Curti C. (2011) The anti-cancer agent nemorosone is a new potent protonophoric mitochondrial uncoupler. Mitochondrion 11, 255–263 [DOI] [PubMed] [Google Scholar]
- 25.Reis F. H., Pardo-Andreu G. L., Nuñez-Figueredo Y., Cuesta-Rubio O., Marín-Prida J., Uyemura S. A., Curti C., and Alberici L. C. (2014) Clusianone, a naturally occurring nemorosone regioisomer, uncouples rat liver mitochondria and induces HepG2 cell death. Chem. Biol. Interact. 212, 20–29 [DOI] [PubMed] [Google Scholar]
- 26.Han Y. H., Moon H. J., You B. R., Kim S. Z., Kim S. H., and Park W. H. (2009) Effects of carbonyl cyanide p-(trifluoromethoxy) phenylhydrazone on the growth inhibition in human pulmonary adenocarcinoma Calu-6 cells. Toxicology 265, 101–117 [DOI] [PubMed] [Google Scholar]
- 27.Jara J. A., Castro-Castillo V., Saavedra-Olavarría J., Peredo L., Pavanni M., Jaña F., Letelier M.E., Parra E., Becker M. I., Morello A., Kemmerling U., Maya J. D., and Ferreira J. (2014) Antiproliferative and uncoupling effects of delocalized, lipophilic, cationic gallic acid derivatives on cancer cell lines: validation in vivo in singenic mice. J. Med. Chem. 57, 2440–2454 [DOI] [PubMed] [Google Scholar]
- 28.Terada H. (1990) Uncouplers of oxidative phosphorylation. Environ. Health Perspect. 87, 213–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Starkov A. A. (2006) Protein-mediated energy-dissipating pathways in mitochondria. Chem. Biol. Interact. 163, 133–144 [DOI] [PubMed] [Google Scholar]
- 30.Figarola J. L., Weng Y., Lincoln C., Horne D., and Rahbar S. (2012) Novel dichlorophenyl urea compounds inhibit proliferation of human leukemia HL-60 cells by inducing cell cycle arrest, differentiation and apoptosis. Invest. New Drugs 30, 1413–1425 [DOI] [PubMed] [Google Scholar]
- 31.Singhal S. S., Figarola J., Singhal J., Nagaprashantha L., Berz D., Rahbar S., and Awasthi S. (2013) Novel compound 1,3-bis(3,5-dichlorophenyl) urea inhibits lung cancer progression. Biochem. Pharmacol. 86, 1664–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singhal S. S., Figarola J., Singhal J., Leake K., Nagaprashantha L., Lincoln C., Gabriel Gugiu B., Horne D., Jove R., Awasthi S., and Rahbar S. (2012) 1,3-Bis(3,5-dichlorophenyl) urea compound “COH-SR4” inhibits proliferation and activates apoptosis in melanoma. Biochem. Pharmacol. 84, 1419–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Figarola J. L., and Rahba S. (2013) Small-molecule COH-SR4 inhibits adipocyte differentiation via AMPK activation. Int. J. Mol. Med. 31, 1166–1176 [DOI] [PubMed] [Google Scholar]
- 34.Hashiguchi K., and Zhang-Akiyama Q.M. (2009) Establishment of human cell lines lacking mitochondrial DNA. Methods Mol. Biol. 554, 383–391 [DOI] [PubMed] [Google Scholar]
- 35.Nicholls D. G., Darley-Usmar V. M., Wu M., Jensen P. B., Rogers G. W., and Ferrick D. A. (2010) Bioenergetic profile experiment using C2C12 myoblast cells. J. Vis. Exp. 46, e2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rogers G. W., Brand M. D., Petrosyan S., Ashok D., Elorza A. A., Ferrick D. A., and Murphy A. N. (2011) High throughput microplate respiratory measurements using minimal quantities of isolated mitochondria. PLoS One 6, e21746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shchepinova M. M., Denisov S. S., Kotova E. A., Khailova L. S., Knorre D. A., Korshunova G. A., Tashlitsky V. N., Severin F. F., and Antonenko Y. N. (2014) Dodecyl and octyl esters of fluorescein as protonophores and uncouplers of oxidative phosphorylation in mitochondria at submicromolar concentrations. Biochim. Biophys. Acta 1837, 149–158 [DOI] [PubMed] [Google Scholar]
- 38.Kenwood B. M., Weaver J. L., Bajwa A., Poon I. K., Byrne F. L., Murrow B. A., Calderone J. A., Huang L., Divakaruni A. S., Tomsig J. L., Okabe K., Lo R. H., Cameron Coleman G., Columbus L., Yan Z., Saucerman J. J., Smith J. S., Holmes J. W., Lynch K. R., Ravichandran K. S., Uchiyama S., Santos W. L., Rogers G. W., Okusa M. D., Bayliss D. A., Hoehn K. L. (2014) Identification of a novel mitochondrial uncoupler that does not depolarize the plasma membrane. Mol. Metab. 3, 114–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang da W., Sherman B.T., and Lempicki R.A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmittgen T.D., and Livak K.J. (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1118 [DOI] [PubMed] [Google Scholar]
- 41.Spitzer M., Wildenhain J., Rappsilber J., and Tyers M. (2014) BoxPlotR: a web tool for generation of box plots. Nat. Methods 11, 121–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Starkov A. A., Bloch D. A., Chernyak B. V., Dedukhova V. I., Mansurova S. E., Severina I. I., Simonyan R. A., Vygodina T. V., and Skulachev V. P. (1997) 6-Ketocholestanol is a recoupler for mitochondria, chromatophores and cytochrome oxidase proteoliposomes. Biochim. Biophys. Acta 1318, 159–172 [DOI] [PubMed] [Google Scholar]
- 43.Costantini P., Belzacq A. S., Vieira H. L., Larochette N., de Pablo M. A., Zamzami N., Susin S. A., Brenner C., and Kroemer G. (2000) Oxidation of a critical thiol residue of the adenine nucleotide translocator enforces Bcl-2-independent permeability transition pore opening and apoptosis. Oncogene 19, 307–314 [DOI] [PubMed] [Google Scholar]
- 44.McStay G. P., Clarke S. J., and Halestrap A. P. (2002) Role of critical thiol groups on the matrix surface of the adenine nucleotide translocase in the mechanism of the mitochondrial permeability transition pore. Biochem. J. 367, 541–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brennan J. P., Southworth R., Medina R. A., Davidson S. M., Duchen M. R., and Shattock M. J. (2006) Mitochondrial uncoupling, with low concentration FCCP, induces ROS-dependent cardioprotection independent of KATP channel activation. Cardiovasc. Res. 72, 313–321 [DOI] [PubMed] [Google Scholar]
- 46.Llovet J. M., Ricci S., and Mazzaferro V., Hilgard P., Gane E., Blanc J. F., de Oliveira A. C., Santoro A., Raoul J. L., Forner A., Schwartz M., Porta C., Zeuzem S., Bolondi L., Greten T. F., Galle P. R., Seitz J. F., Borbath I., Häussinger D., Giannaris T., Shan M., Moscovici M., Voliotis D., Bruix J., and SHARP Investigators Study Group (2008) Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 359, 378–390 [DOI] [PubMed] [Google Scholar]
- 47.Marin J. J. G., Castaño B., Martinez-Becerra P., Rosales R., and Monte M. J. (2008) Chemotherapy in the treatment of primary liver tumours. Cancer Ther. 6, 711–728 [Google Scholar]
- 48.Llovet J. M., and Hernandez-Gea V. (2014) Hepatocellular carcinoma: reasons for phase III failure and novel perspectives on trial design. Clin. Cancer Res. 20, 2072–2079 [DOI] [PubMed] [Google Scholar]
- 49.Archer S.L. (2013) Mitochondrial dynamics: mitochondrial fission and fusion in human diseases. N. Engl. J. Med. 369, 2236–2251 [DOI] [PubMed] [Google Scholar]
- 50.Benard G., Bellance N., Jose C., and Rossignol R. (2011) in Mitochondrial Dynamics and Neurodegeneration (Lu B., ed) pp. 47–68, Springer, Netherlands [Google Scholar]
- 51.Kanehisa M., Goto S., Sato Y., Kawashima M., Furumichi M., and Tanabe M. (2014) Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 42, D199–D205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weng Z., Luo Y., Yang X., Greenhaw J. J., Li H., Xie L., Mattes W. B., and Shi Q. (2015) Regorafenib impairs mitochondrial functions, activates AMP-activated protein kinase, induces autophagy, and causes rat hepatocyte necrosis. Toxicology 327, 10–21 [DOI] [PubMed] [Google Scholar]
- 53.Starkov A. A., Dedukhova V. I., and Skulachev V. P. (1994) 6-Ketocholestanol abolishes the effect of the most potent uncouplers of oxidative phosphorylation in mitochondria. FEBS Lett. 355, 305–308 [DOI] [PubMed] [Google Scholar]
- 54.Inoki K., Kim J., and Guan K.L. (2012) AMPK and mTOR in cellular energy homeostasis and drug targets. Annu. Rev. Pharmacol. Toxicol. 52, 381–400 [DOI] [PubMed] [Google Scholar]
- 55.Chen S., Zhu X., Lai X., Xiao T., Wen A., and Zhang J. (2014) Combined cancer therapy with non-conventional drugs: all roads lead to AMPK. Mini Rev. Med. Chem. 14, 642–654 [DOI] [PubMed] [Google Scholar]
- 56.Vélez J., Hail N. Jr., Konopleva M., Zeng Z., Kojima K., Samudio I., and Andreeff M. (2013) Mitochondrial uncoupling and the reprograming of intermediary metabolism in leukemia cells. Front. Oncol. 3, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuruvilla S., Qualls C. W. Jr., Tyler R. D., Witherspoon S. M., Benavides G. R., Yoon L. W., Dold K., Brown R. H., Sangiah S., and Morgan K. T. (2003) Effects of minimally toxic levels of carbonyl cyanide p-(trifluoromethoxy) phenylhydrazone (FCCP), elucidated through differential gene expression with biochemical and morphological correlations. Toxicol. Sci. 73, 348–361 [DOI] [PubMed] [Google Scholar]
- 58.Trachootham D., Alexandre J., and Huang P. (2009) Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat. Rev. Drug Discov. 8, 579–591 [DOI] [PubMed] [Google Scholar]
- 59.Cela O., Piccoli C., Scrima R., Quarato G., Marolla A., Cinnella G., Dambrosio M., and Capitanio N. (2010) Bupivacaine uncouples the mitochondrial oxidative phosphorylation, inhibits respiratory chain complexes I and III and enhances ROS production: results of a study on cell cultures. Mitochondrion 10, 487–496 [DOI] [PubMed] [Google Scholar]
- 60.Zorov D. B., Juhaszova M., and Sollott S. J. (2006) Mitochondrial ROS-induced ROS release: an update and review. Biochim. Biophys. Acta 1757, 509–517 [DOI] [PubMed] [Google Scholar]
- 61.Han H. J., Tokino T., and Nakamura Y. (1998) CSR, a scavenger receptor-like protein with a protective role against cellular damage caused by UV irradiation and oxidative stress. Hum. Mol. Genet. 7, 1039–1046 [DOI] [PubMed] [Google Scholar]
- 62.Lou P. H., Hansen B. S., Olsen P. H., Tullin S., Murphy M. P., and Brand M. D. (2007) Mitochondrial uncouplers with an extraordinary dynamic range. Biochem. J. 407, 129–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chalmers S., Caldwell S. T., Quin C., Prime T. A., James A. M., Cairns A. G., Murphy M. P., McCarron J. G., and Hartley R. C. (2012) Selective uncoupling of individual mitochondria within a cell using a mitochondria-targeted photoactivated protonophore. J. Am. Chem. Soc. 134, 758–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McQuaker S. J., Quinlan C. L., Caldwell S. T., Brand M. D., and Hartley R. C. (2013) A prototypical small-molecule modulator uncouples mitochondria in response to endogenous hydrogen peroxide production. Chembiochem 14, 993–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.