Background: The role of a conserved serine residue in glutamate transporters is not fully understood.
Results: Mutation of this conserved serine residue changed substrate and cation selectivity and affinity.
Conclusion: The hydroxyl side chain of this serine residue plays a crucial role in coupling the sodium and substrate fluxes.
Significance: The direct coupling of substrate and cation fluxes described herein may be relevant for other ion-coupled transporters.
Keywords: excitotoxicity, molecular dynamics, mutagenesis, neurotransmitter transport, structure-function, GLT-1, SLC1A2, glutamate transporter, excitatory neurotransmission, neurotoxicity, structure/function studies
Abstract
Glutamate transporters maintain synaptic concentration of the excitatory neurotransmitter below neurotoxic levels. Their transport cycle consists of cotransport of glutamate with three sodium ions and one proton, followed by countertransport of potassium. Structural studies proposed that a highly conserved serine located in the binding pocket of the homologous GltPh coordinates l-aspartate as well as the sodium ion Na1. To experimentally validate these findings, we generated and characterized several mutants of the corresponding serine residue, Ser-364, of human glutamate transporter SLC1A2 (solute carrier family 1 member 2), also known as glutamate transporter GLT-1 and excitatory amino acid transporter EAAT2. S364T, S364A, S364C, S364N, and S364D were expressed in HEK cells and Xenopus laevis oocytes to measure radioactive substrate transport and transport currents, respectively. All mutants exhibited similar plasma membrane expression when compared with WT SLC1A2, but substitutions of serine by aspartate or asparagine completely abolished substrate transport. On the other hand, the threonine mutant, which is a more conservative mutation, exhibited similar substrate selectivity, substrate and sodium affinities as WT but a lower selectivity for Na+ over Li+. S364A and S364C exhibited drastically reduced affinities for each substrate and enhanced selectivity for l-aspartate over d-aspartate and l-glutamate, and lost their selectivity for Na+ over Li+. Furthermore, we extended the analysis of our experimental observations using molecular dynamics simulations. Altogether, our findings confirm a pivotal role of the serine 364, and more precisely its hydroxyl group, in coupling sodium and substrate fluxes.
Introduction
Glutamate is the major excitatory neurotransmitter in the nervous system. To terminate the synaptic action of this neurotransmitter and to avoid excitotoxic, prolonged, extracellular accumulation of glutamate, it is crucial that glutamate is removed rapidly from the synaptic cleft after release. The removal is accomplished by high-affinity glutamate transporters (EAATs)5 from the SLC1 solute carrier family (1–3). SLC1A2 (GLT-1, EAAT2), which is located on the perisynaptic side of astrocytes, is responsible for 90% of glutamate clearance in the mammalian central nervous system (4). Because high extracellular glutamate concentration causes excitotoxicity, SLC1A2 dysfunction has been associated with numerous neurological diseases such as amyotrophic lateral sclerosis, epilepsy, or Alzheimer disease (1, 5–7). Each transport cycle of glutamate transporters consists of the cotransport of glutamate, three sodium ions, and one proton followed by the countertransport of one potassium ion (8–10). Glutamate transporters mediate two distinct types of substrate-induced steady-state currents. The first is the inward-directed “coupled” current reflecting the net electrogenic translocation of two positive charges across the membrane during glutamate translocation, based on the coupling stoichiometry described above. The second is the thermodynamically “uncoupled” current carried by Cl− ions. This anion conductance is a pore-mediated permeation process with two conducting states, a sodium-activated conductance or leak conductance, and a sodium-dependent substrate-gated conductance (11–16).
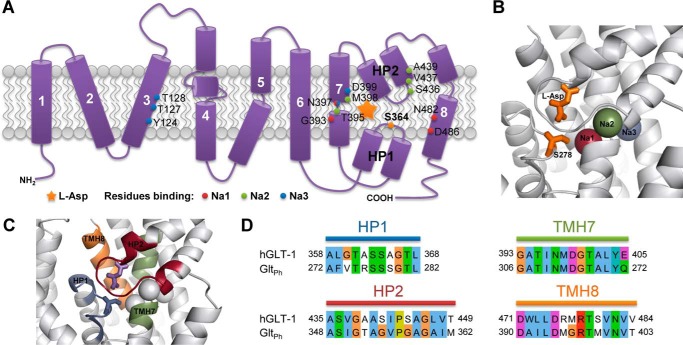
Ten years ago, a high-resolution crystal structure of GltPh, an archaeal glutamate transporter homologue from Pyrococcus horikoshii, revealed a trimeric conformation with each monomer being a functional unit (17). This assembly was observed for the eukaryotic glutamate transporters as well (18–21). Each protomer possesses eight transmembrane segments (TM) and two re-entrant helical hairpins (HP1 and HP2), one between TMs 6 and 7 and one between TMs 7 and 8 (Fig. 1A). In addition, TMs 7 and 8 and both hairpin loops are part of the transporter binding pocket (17, 22) (Fig. 1, B and C). Although GltPh shares only ∼36% sequence identity with eukaryotic EAAT transporters (22), the binding site and its neighborhood are highly conserved (∼64% sequence identity, see Fig. 1D) (17), making GltPh suitable as a model system for studying human SLC1A2.
FIGURE 1.
Amino acid residues involved in substrate and sodium ion binding of the glutamate transporter SLC1A2 and its archaeal homologue GltPh. A, schematic model of human SLC1A2 topology, representing the eight transmembrane helices and hairpin loops HP1 and HP2. The location of the bound substrate is marked by a star. Residues involved in sodium ion binding according to GltPh x-ray structures are indicated and color-coded. Note that Asn-397 is binding both Na1 and Na3. The herein studied Ser-364 residue is marked in orange within HP1. B, structure of the substrate binding site in GltPh (PDB ID: 2NWX). Sodium ions, bound l-aspartate substrate, and Ser-278 (corresponding to Ser-364 in human SLC1A2) are shown and color-coded according to panel A. The location of Na3 is taken from Bastug et al. (29). C, protein regions considered to be in the neighborhood of the substrate binding site are highlighted in color. D, conservation of the substrate binding site and its neighborhood as shown on panel C between human SLC1A2 (hGLT-1) and GltPh. Sequence identity of the selected regions is 64.2%, as opposed to 36% for the full-length proteins.
Due to their large anomalous scattering signal, thallium (Tl+) ions have been used to visualize the Na+ binding sites (22). Two Tl+ binding sites were identified and proposed to be sodium sites, although GltPh has been shown to translocate three Na+ ions per transport cycle (23). Functional evidence obtained on mutants of the amino acid Asp-455 of SLC1A1 (Asp-405 in GltPh and Asp-487 in SLC1A2), which was proposed to be part of Na1 site, confirmed the results obtained with thallium (24). Localization of the third bound sodium ion was attempted using theoretical and simulation studies. Although some studies placed Na3 near the bound substrate (25), other studies indicated that Asp-312 in GltPh, as well as the corresponding residue in the human glutamate transporters (Asp-399 in SLC1A2), serves as coordinating residue (26–29).
Serine 278 of GltPh, which is located in the first re-entrant loop, has been proposed to coordinate the sodium ion 1 (Na1) via its hydroxyl group and the substrates by its main chain nitrogen (22). This serine residue is conserved in eukaryotic glutamate transporters and corresponds to the Ser-364 of the glial human isoform SLC1A2. Although a cysteine substitution of this serine residue was found to block d-aspartate transport by SLC1A2 (30), and several simulation studies mention that the serine-rich loop, including serine 364, plays a role in substrate recognition and binding (31–33), no further experimental investigations were accomplished to elucidate the exact role of this serine in the SLC1A2 transport mechanism.
Therefore, we aimed to investigate the role of Ser-364 and especially of its hydroxyl group in substrate and/or sodium binding and transport in human SLC1A2. To answer these questions, we generated several Ser-364 mutants of SLC1A2 (S364A, S364T, S364C, S364D, and S364N) and studied their substrate and cation transport properties. Interestingly, the S364T mutant was very similar to the WT counterpart. However, other mutants, such as S364A and S364C, which had reduced but significant transport activities, showed changes in substrate and ion selectivity. In addition, our findings were placed in a dynamic structural context using molecular dynamics simulations performed on the homologous GltPh transporter. Overall we present evidence that Ser-364 and in particular its side chain hydroxyl group is required for the substrate and cation selectivity. Therefore, this serine residue would play an important role in coupling the sodium and substrate fluxes.
Experimental Procedures
Chemicals and Reagents
Unless specified otherwise, all chemicals and reagents were obtained from Sigma-Aldrich. Densitometric quantifications were performed using the National Institutes of Health software, ImageJ.
Generation and Subcloning of SLC1A2 Mutants
Human SLC1A2 was cloned in pTracer-CMV2 vector (Life Technologies) and used as a parental construct for site-directed mutagenesis using the QuikChange site-directed mutagenesis kit (Agilent Technologies). SLC1A2 wild-type and mutants were then subcloned into the Pol1 vector for Xenopus laevis oocyte expression (34).
Cell Culture and Protein Expression in HEK293 Cells and in X. laevis Oocytes
HEK293 cells were obtained from ATCC and maintained at 37 °C in a humidified 5% CO2 incubator in DMEM (Invitrogen) supplemented with 10% fetal bovine serum and 100 units/ml penicillin/streptomycin mixtures (Invitrogen). HEK293 cells were grown on poly-d-lysine-coated 6-well plates or 96-well plates. On the next day, cells were transiently transfected with 3 μg of indicated cDNA for 6-well plates or 0.2 μg for 96-well plates using Lipofectamine 2000 (Invitrogen) and incubated for 24 h.
Defolliculated stage V-VI oocytes were micro-injected with ∼20 ng of each cDNA-derived cRNA using the mMESSAGE mMACHINE T7 kit (Ambion) and then maintained for 72 h at 18 °C in modified Barth's medium (88 mm NaCl, 1 mm KCl, 2.4 mm NaHCO3, 0.82 mm MgSO4, 0.66 mm NaNO3, 0.75 mm CaCl2, 10 mm Na-HEPES) supplemented with antibiotics. Functional studies were performed 3 days after micro-injection.
Surface Biotinylation and Immunoblotting
Cell surface biotinylation experiments were essentially conducted as described earlier (35). Briefly, cells were rinsed with PBS, and surface proteins were biotinylated by incubation with 1.5 mg/ml sulfo-NHS-SS-biotin for 60 min with horizontal motion at 4 °C. After labeling, cells were washed with quenching buffer (PBS containing 1 mm MgCl2, 0.1 mm CaCl2, and 100 mm glycine) and then rinsed once with PBS. Next, cells were lysed in radioimmunoprecipitation assay buffer (150 mm NaCl, 5 mm EDTA, 1% Triton X-100, 0.5% deoxycholate, 0.1% SDS, 50 mm Tris·HCl, pH 7.4) containing fresh protease inhibitors (Roche Applied Science), and lysates were cleared by centrifugation. Cell lysates of equivalent amounts of protein were equilibrated overnight with streptavidin-agarose beads at 4 °C. Beads were washed sequentially with solution A (100 mm NaCl, 5 mm EDTA, 50 mm Tris·HCl, pH 7.4) three times, solution B (500 mm NaCl, 50 mm Tris·HCl, pH 7.4) two times, and solution C (50 mm Tris·HCl, pH 7.4) once. Biotinylated proteins were then released by heating to 95 °C with 2× Laemmli buffer, resolved on SDS-polyacrylamide gels, and transferred onto Immobilon-P membrane blots (Millipore). After sequential incubations of the blots with primary and secondary antibodies, proteins were revealed by chemiluminescence using ECL solution (GE Healthcare). Primary antibodies were used at a 1/1,000 dilution and obtained from the following sources: rabbit polyclonal α-SLC1A2 (PA5-17099; Thermo Scientific), mouse monoclonal α-Na+/K+ ATPase α-1 (05-369; Millipore), and rabbit polyclonal anti-actin (I-19; Santa Cruz Biotechnology). The secondary antibodies were used at a dilution of 1/4,000 for the HRP-conjugated goat anti-mouse IgG (172-1011; Bio-Rad) and 1/20,000 for the HRP-conjugated goat anti-rabbit IgG (W4011; Promega).
l-[3H]Glutamate, l-[3H]Aspartate, and d-[3H]Aspartate Uptake in HEK Cells
HEK293 cells transfected in 96-well plates for 24 h as described above were washed once with uptake buffer (140 mm NaCl, 2.5 mm KCl, 1 mm CaCl2, 1 mm MgCl2, 1.2 mm K2HPO4, 100 mm glucose, and 10 mm HEPES, pH 7.4). Then, 100 μl of uptake buffer containing the indicated concentrations of cold substrate and 0.05 μCi of tritiated substrate (PerkinElmer) were added into each well. Uptake was carried out at room temperature during 10 min. Uptake was stopped by three washes of ice-cold uptake buffer. After the addition of 100 μl of scintillation fluid (PerkinElmer) to each well and 1 h of agitation to allow complete cell lysis, the uptake of 3H-substrate was determined using the TopCount microplate scintillation and luminescence counter (PerkinElmer).
In sodium concentration-response and cation selectivity experiments, NaCl was replaced by equimolar concentrations of choline chloride and lithium chloride, respectively. For each condition, cpm values of at least three wells were averaged, and the numbers were transformed into influx rates and expressed as the percentage of WT maximal uptake, using the following equation.
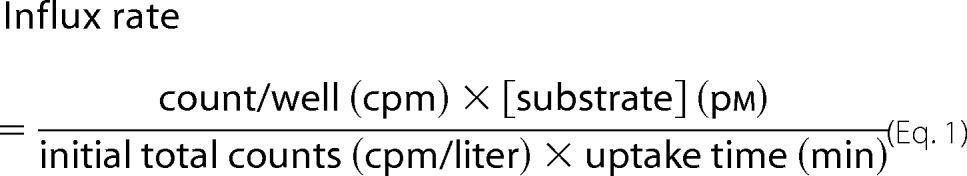 |
Experimental results of influx rates were fitted to the Michaelis-Menten equation using GraphPad Prism 5 to obtain Km values.
Electrophysiology
Two-electrode voltage clamp experiments were conducted at room temperature 3 days after glutamate transporter cRNA injection. Oocytes were placed in the recording chamber and impaled with two glass microelectrodes, containing 3 m KCl (resistance between 0.5 and 2 milliohms). Currents were measured at a holding potential of −50 mV with an OC-725 amplifier (Warner Instruments), filtered at 1 kHz, and digitized at 2000 Hz with a Digidata 1440 data acquisition system (Axon Instruments). Data were captured using pClamp 10 (Axon Instruments). Oocytes were bathed in oocyte buffer (100 mm NaCl, 2 mm KCl, 1 mm CaCl2, 1 mm MgCl2, 10 mm HEPES, pH 7.5). Substrate-evoked steady-state currents were measured by applying an I-V protocol consisting of 300-ms steps from the holding potential (−50 mV) to voltages between −100 mV and +40 mV in increments of 10 mV.
Data Analysis
All current-voltage relations correspond to the steady-state substrate-induced net currents ((Isubstrate+buffer) − (Ibuffer)) analyzed by Clampfit 10.2 (Axon Instruments). Due to variations in expression levels between oocytes, data were normalized to the current induced by 5 mm l-glutamate at −100 mV in oocyte buffer (Inorm).
Statistics
Statistical analysis was done using Student's t test or one-way analysis of variance to correct for multiple comparisons, as appropriate. All statistical tests were two-sided, and a p value < 0.05 was considered statistically significant.
Computational Analysis
Molecular dynamics simulations were based on the structure of the GltPh archaeal homologue. Starting from the membrane-embedded orientation of the outward-open conformation (Protein Data Bank (PDB) ID: 2NWW) taken from the Orientations of Proteins in Membranes (OPM) database (36), the cocrystallized l-threo-β-benzoylaspartate inhibitor was removed, and the l-Asp substrate and the Na1 ion was placed in the binding site based on structural superposition with the outward-occluded state (PDB ID: 2NWX). The Na3 was introduced according to Bastug et al. (29). The CHARMM-GUI (37, 38) server was used to solvate the system in a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) bilayer, to mimic the archaeal membrane of the crystallized protein, and the equilibration protocol supplied by CHARMM-GUI was carried out, with six cycles of gradually decreasing coordinate restraints. To help further equilibration, 1 million steps of simulation (corresponding to 500 ps) were additionally performed with a 0.5 kcal/mol/Å2 harmonic restraint around the initial coordinates of all heavy atoms. Production simulations were performed using NAMD 2.9 (39) on the UBELIX computing cluster at the University of Bern. Hydrogen bonds for the subsequent analysis were defined as having at most 2.5 Å distance between acceptor and hydrogen atoms, with α ≥ 120° and β ≥ 90°, where α is the donor-hydrogen-acceptor angle, β is the hydrogen-acceptor-COG angle, and COG is defined as the center of geometry of the first covalent neighbors of the acceptor atom. The analysis of the hydrogen bond network was performed using custom scripts on 300 uniformly selected frames along the 60-ns trajectories of each system.
Results
Substrate Transport and Surface Expression of the Serine 364 Mutants
To investigate the role of the conserved serine located at position 364 of human SLC1A2, this residue was mutated to amino acids with shorter or larger side chains such as alanine, aspartate, or asparagine. Moreover, substitutions by cysteine (which possesses a side chain of similar size) and threonine (which belongs to the same class of polar uncharged amino acids) were also made.
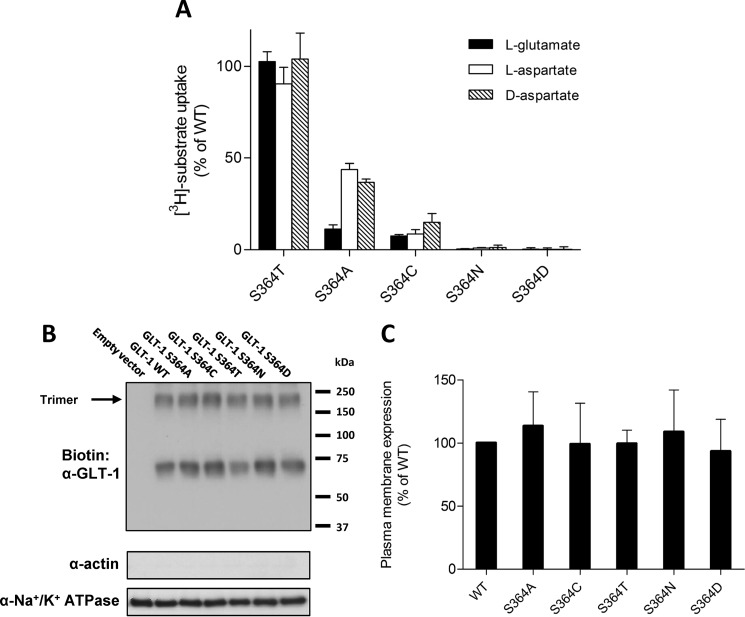
First, we expressed each mutant in HEK cells and measured the transport of l-[3H]glutamate, l-[3H]aspartate, and d-[3H]aspartate in the presence of 1 mm cold substrate. No measurable transport was observed for any substrate when analyzing the S364D and S364N substitutions. The S364A mutants exhibited moderate uptake of l- and d-aspartate and no uptake of l-glutamate (Figs. 2A and 3). In contrast, transport of each substrate was drastically reduced for the cysteine mutant, and no clear substrate selectivity was observed. S364T displayed similar substrate uptake characteristics as SLC1A2-WT (Figs. 2A and 3). To show that the loss or reduction in activity was not due to lower expression of the different mutants in the plasma membrane, we compared their plasma membrane expression with that of WT using a surface biotinylation approach (Fig. 2, B and C). Surface expression of all mutant transporters was similar to WT as shown by SDS-PAGE. Note that we detect glycosylated monomers and presumably trimers of SLC1A2 at ∼70 and ∼210 kDa, respectively.
FIGURE 2.
Transport and plasma membrane expression of WT and Ser-364 mutant SLC1A2. A, uptake of 1 mm l-[3H]glutamate (black bars), d-[3H]aspartate (empty bars), and l-[3H]aspartate (hatched bars) by HEK cells expressing the indicated mutants. The uptake of each substrate was corrected for the background uptake by empty vector-transfected HEK cells. Uptake is represented as the percentage of the SLC1A2-WT. The data are means ± S.D. of three independent experiments done in triplicates. Plasma membrane expression of SLC1A2-WT and the indicated mutants was assessed by immunoblotting of the labeled plasma membrane protein by sulfo-NHS-LC-biotin isolated on streptavidin-agarose beads. B, a representative experiment is shown. The absence of actin signal confirms the purity of the membrane fraction, Na+/K+ ATPase was used as equal loading control. C, quantification of plasma membrane expression from three independent experiments. For each substrate, data were normalized to the uptake of SLC1A2-WT transfected HEK cells in each individual experiment and are represented as the percentage of SLC1A2-WT.
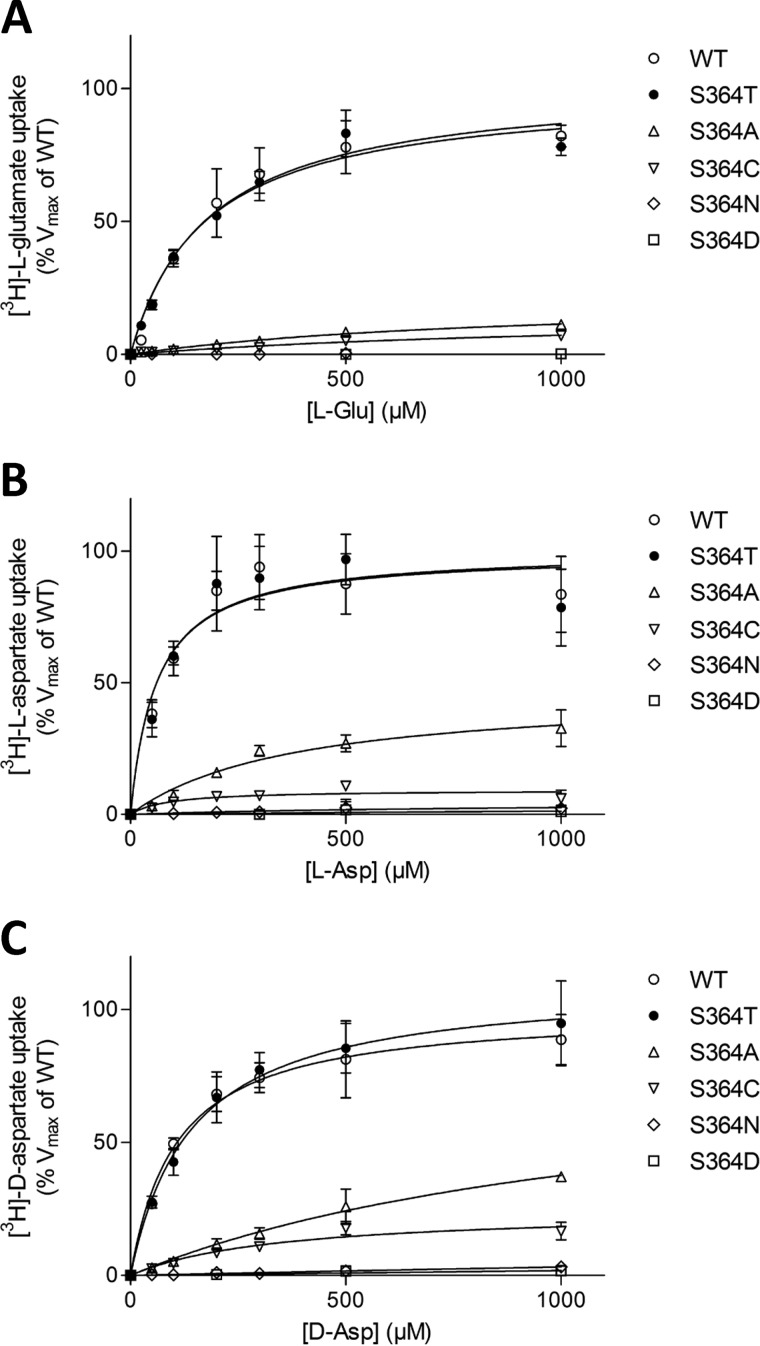
FIGURE 3.
Substrate concentration-dependent uptake of 3H-substrates by SLC1A2-WT and Ser-364 mutants. A–C, l-[3H]glutamate (A), l-[3H]aspartate (B), and d-[3H]aspartate (C) uptake kinetics by HEK cells expressing WT and Ser-364 mutants SLC1A2. For each concentration of each individual substrate, the background uptake measured for the empty vector-transfected HEK control cells was subtracted. For each substrate, data were normalized to the maximal uptake of SLC1A2-WT expressing HEK cells in each experiment and are represented as the percentage of Vmax SLC1A2-WT. The data are means ± S.D. of at least three distinct experiments performed in triplicates.
Substrate Affinities and Selectivity of the Serine 364 Mutants
To determine the affinity of each substrate for SLC1A2-WT and serine 364 mutants, substrate uptake was measured in the presence of different concentrations of the indicated substrates (Fig. 3). For those mutants that exhibited significant substrate uptake, Km values were determined (Table 1). In comparison with WT, the serine to threonine substitution exhibits similar affinities for all substrates. In contrast, the alanine and cysteine substitutions clearly increased the Km values for all substrates by 6–18- and 4–7-fold, respectively (Table 1). Moreover, as already stated, when compared with SLC1A2-WT, the alanine mutant displayed larger uptakes of l- and d-aspartate in the presence of 1 mm substrate, when compared with l-glutamate (Figs. 2A and 3).
TABLE 1.
Apparent affinities obtained from substrate-induced 3H-substrate uptake by SLC1A2-WT and Ser-364 mutants
Km values from uptake experiments in HEK cells expressing SLC1A2-WT and the indicated Ser-364 mutants are shown below. Km values were calculated from data obtained from three independent experiments and are represented as means ± S.D. (*, p < 0.05, mutant versus WT).
| SLC1A2 | l-Glutamate | l-Aspartate | d-Aspartate |
|---|---|---|---|
| μm | μm | μm | |
| WT | 158.1 ± 28.7 | 48.1 ± 26.5 | 91.1 ± 40.2 |
| S364T | 161.8 ± 12.1 | 53.6 ± 15.3 | 115.1 ± 51.28 |
| S364A | 900.7 ± 84.5* | 882.6 ± 642.2* | 1151.2 ± 398.0* |
| S364C | 1080.0 ± 52.0* | 188.8 ± 66.1* | 350.1 ± 91.6* |
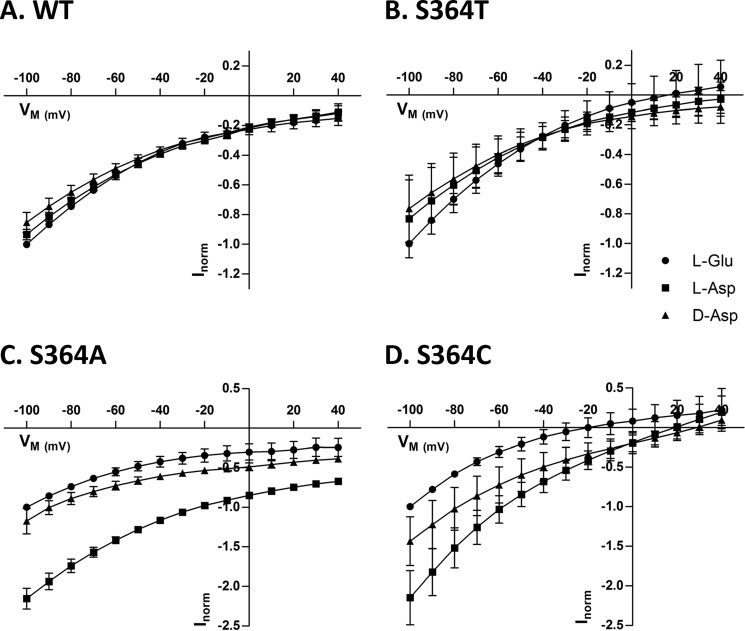
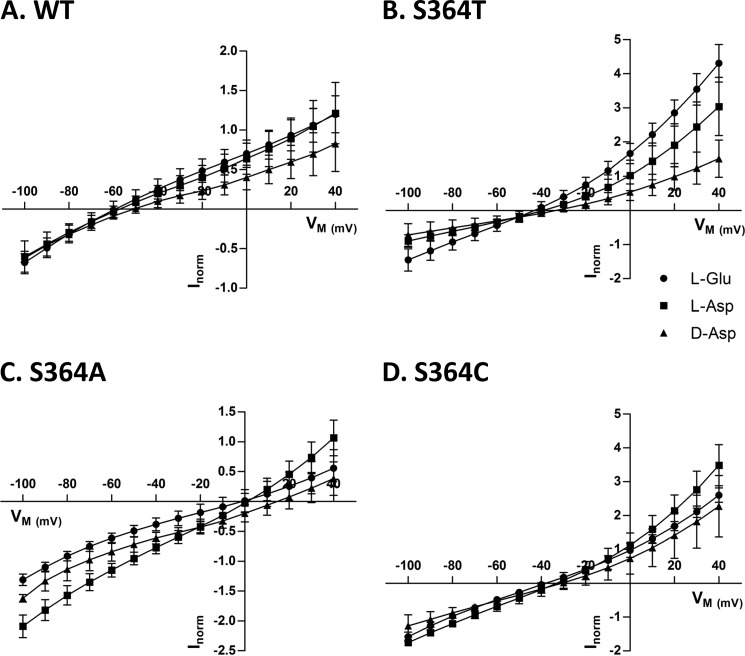
In oocytes expressing SLC1A2-WT, saturating concentrations of l-glutamate, l-aspartate, and d-aspartate induced currents of similar amplitude and voltage dependence (Fig. 4A). Substrate-induced currents were measured for S364T, S364A, and S364C for all substrates (Fig. 4, B–D), but not for S364N and S364D. The S364T mutant was similar to WT with regard to substrate selectivity and voltage dependence (Fig. 4B). However, with respect to the S364A and S364C mutants, the maximal current elicited by l-aspartate was the highest and the current for d-aspartate was higher than the one evoked by l-glutamate (Fig. 4, C and D). Taken together these findings suggest that the hydroxyl group of the serine 364 is required for substrate binding and selectivity.
FIGURE 4.
Voltage dependence of substrate-induced steady-state currents by SLC1A2-WT and Ser-364 mutants. A–D, steady-state currents induced by 5 mm l-[3H]glutamate (circles), l-[3H]aspartate (squares), and d-[3H]aspartate (triangles) in X. laevis oocytes expressing SLC1A2-WT and the indicated Ser-364 mutants were measured using the voltage protocol described under “Experimental Procedures.” For each substrate, currents were corrected for the background current observed without substrate and normalized to currents obtained with 5 mm l-glutamate (Inorm) and plotted against the holding potential (Vm). The data are means ± S.D. of 3–5 oocytes. Currents at −100 mV induced by 5 mm l-glutamate ranged from −163 to −265 nA for WT, from −98 to −549 nA for S364T, from −38 to −147 nA for S364A, and from −49 to −111 nA for S364C.
Anion Conductance of SLC1A2-WT, S364A, S364T, and S364C
Another important transport characteristic of glutamate transporters is their anion conductance, which is also gated by substrates (11, 13, 40). This specific conductance is measurable upon replacement of 20 mm of NaCl by an equimolar concentration of NaSCN (42), because the permeability for SCN− is ∼70-fold higher than for Cl− (14, 43). At positive potentials, the outward currents reflect the entry of SCN− into the oocytes. In SLC1A2-WT and mutants, replacement of 20 mm NaCl by NaSCN abolished the inward rectification of the currents induced by all three substrates tested (Fig. 5). In the presence of SCN−, oocytes expressing the WT transporter exhibited similar l-glutamate-, l-aspartate-, and d-aspartate-induced currents, except at positive potentials, where the d-aspartate-induced currents tended to be slightly smaller (Fig. 5A). However, in contrast to WT, at positive potentials, the S364T mutant displayed the largest outward currents with l-glutamate, and currents induced by l-aspartate were higher than with d-aspartate (Fig. 5B). The S364A and S364C mutants exhibited similar l-glutamate- and d-aspartate-induced currents at positive potentials, which had a tendency to be smaller than in the presence of l-aspartate (Fig. 5, C and D). These observations support a role of serine 364 for substrate selectivity. In addition, when compared with WT, in the presence of SCN−, reversal potentials of the S364T and S364C mutants were slightly shifted to −40 mV, and even a larger change to 0 mV was observed for S364A, suggesting a smaller contribution of the anion conductance to the total current in the Ser-364 mutants (Fig. 5).
FIGURE 5.
Substrate-evoked anion conductance by WT and S364A, S364T, and S364C mutants. A–D, steady-state currents evoked by 5 mm l-[3H]glutamate (circles), l-[3H]aspartate (squares), and d-[3H]aspartate (triangles) in X. laevis oocytes expressing SLC1A2-WT or the indicated Ser-364 mutants were measured using the voltage clamp protocol described under “Experimental Procedures.” Currents were measured in oocyte buffer, where 20 mm NaCl was replaced by an equimolar concentration of NaSCN. For each substrate, currents were corrected with respect to the background current observed without substrate (Inorm), normalized to the currents obtained with 5 mm l-glutamate in SCN−-free solution (Inorm), and plotted against the holding potential (Vm). The data are means ± S.D. of 3–5 oocytes. Currents at −100 mV induced by 5 mm l-glutamate in SCN−-containing buffer ranged from −100 to −190 nA for WT, from −169 to −625 nA for S364T, from −43 to −188 nA for S364A, and from −114 to −159 nA for S364C.
Sodium Dependence and Cation Selectivity of the SLC1A2-WT and Serine 364 Mutants
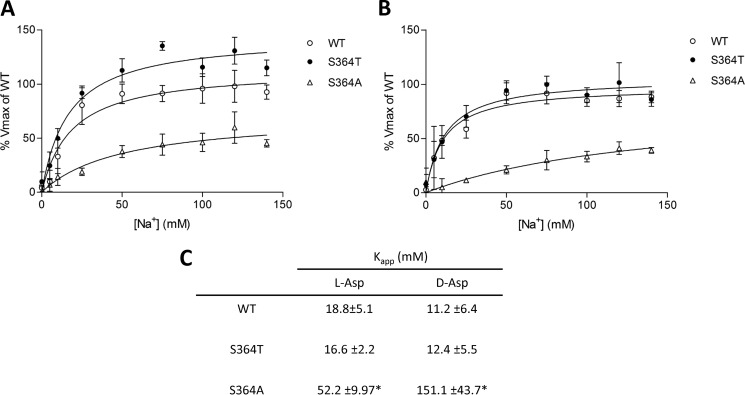
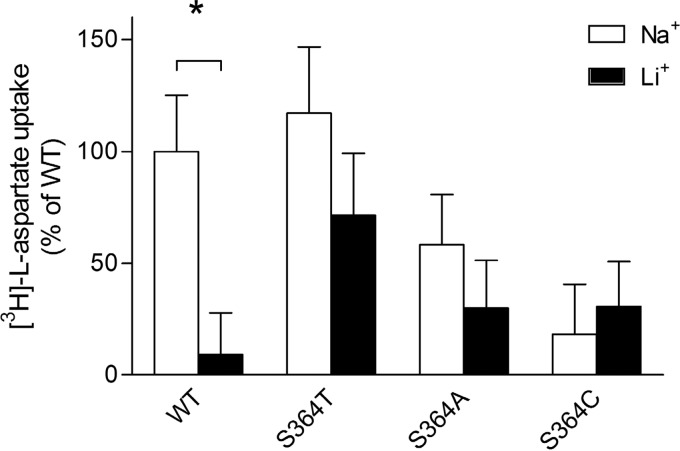
To investigate whether serine 364 is important for sodium binding and transport, we examined the sodium dependence of WT, S364T, and S364A and the cation selectivity of the WT, S364T, S364A, and S364C transporters. Because the transport capabilities of the S364C mutant were already greatly reduced in the presence of 140 mm Na+ (Figs. 2A and 3), we were not able to measure the sodium dependence of this particular mutant. In addition, very low l-glutamate transport activity could be measured for S364A. Therefore, we looked specifically at the sodium dependence of l- and d-aspartate transport. In comparison with WT and S364T (which have similar apparent affinities for sodium), the S364A mutant had a 3-fold lower apparent affinity for sodium in the presence of l-aspartate and a 14-fold lower one in the presence of d-aspartate (Fig. 6). To compare cation selectivity of WT and mutants, we performed uptake of 2 mm l-aspartate, which is transported by all mutants tested in normal uptake buffer and in an uptake buffer where Na+ was replaced by equimolar concentrations of Li+. SLC1A2-WT exhibited very low transport of l-aspartate when Na+ was replaced by Li+ (Fig. 7). However, the threonine, alanine, and cysteine mutants displayed no significant difference of l-aspartate uptake in Na+- and in Li+-containing buffer. These results indicate that serine 364 is crucial for determining apparent Na+ affinity and cation selectivity.
FIGURE 6.
Sodium concentration-dependence of l-[3H]- and d-[3H]aspartate uptake by SLC1A2-WT and S364A and S364T mutants. A and B, sodium dependence of 2 mm of l-[3H]aspartate (A) and d-[3H]aspartate (B) uptake by HEK cells expressing SLC1A2-WT and S364A and S364T mutants. For each concentration of each individual substrate, the background uptake obtained for empty vector-transfected HEK control cells was subtracted. For each substrate, data were normalized to the maximal uptake of SLC1A2-WT expressing HEK cells in each individual experiment and are represented as the percentage of the Vmax of SLC1A2-WT. C, apparent affinities for sodium for WT, S364T, and S364A in the presence of 2 mm of the indicated substrates. The data are means ± S.D. of three individual experiments performed in triplicates. (*, p < 0.01, S364T versus WT).
FIGURE 7.
Cation selectivity of l-[3H]aspartate uptake by SLC1A2-WT and mutants. The Li+ and Na+ dependence of 2 mm l-[3H]aspartate uptake by SLC1A2-WT and the indicated Ser-364 mutants are shown. For each cation and substrate, the uptake was corrected against the background uptake obtained for empty vector-transfected HEK control cells. For each construct, the data were normalized to the uptake measured in the presence of Na+ and represented as the percentage of WT. The data are means ± S.D. of two distinct experiments with 10 wells per condition. (*, p < 0.01, uptake in Li+ versus Na+).
Computational Studies Based on the Structure of the Archaeal Homologue GltPh
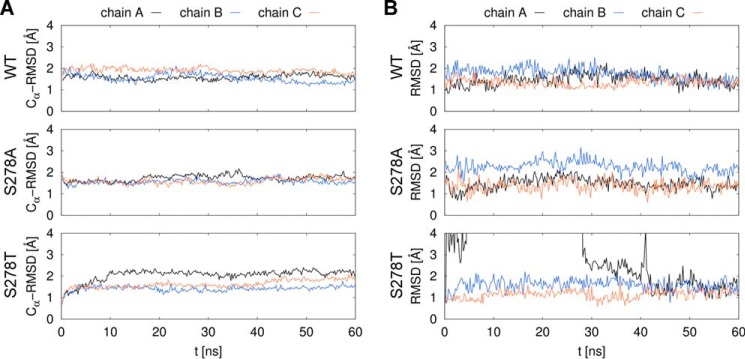
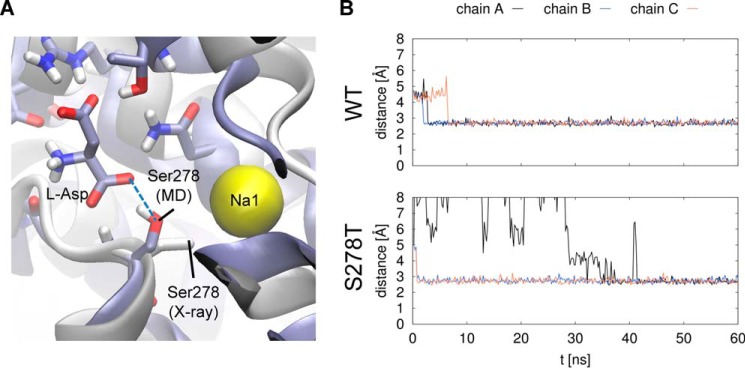
To support and further explain our experimental findings, we performed molecular dynamics simulations to investigate the atomic contacts of the Ser-364 side chain (Ser-278 in GltPh) in the homologous GltPh transporter. Simulation systems for the WT and S278A and S278T mutants were generated. Each simulated system contained the protein in a trimeric arrangement, as crystallized, referred to as protein chains A, B, and C. As each protomer of the trimer was shown to function independently (19, 20, 22), they can be considered as three independent copies of the same protein. The differences between the initial coordinates of each protomer were negligible, but they underwent a different time evolution due to the random starting velocities. After equilibration, all three protein chains in the trimer structure remained within 2–2.5 Å root mean square deviation values from the initial coordinates, indicating overall stability of the system (Fig. 8A). Each protein chain contains a single substrate molecule that finds equilibrium after a transient period, close to the initial coordinates observed in the x-ray structure (Fig. 8B). In the case of the S278A mutant, we observed persistently higher root mean square deviation values for the l-aspartate substrate bound to protein chain B, which were due to a trans/gauche conformational change in the substrate. With regard to S278T, we witnessed a transient unbinding event in chain A. However, the system relaxed into a near-native state by the end of the simulation.
FIGURE 8.
Stability of the protein chains and the substrate molecules in GltPh simulations. Molecular dynamics simulations of 60 ns were performed for the WT and two mutant (S278A, S278T) GltPh proteins. Ser-278 corresponds to Ser-364 in human SLC1A2. Individual transporters in the homotrimer are labeled as chain A, B, and C. Root mean square deviation (RMSD) values of coordinates from the initial state are shown for the protein Cα atoms (A) and the l-aspartate substrate atoms (B). A transient unbinding event of the substrate can be seen in chain A of the S278T system on panel B, after which the substrate returns into a near-native state at around 42 ns.
Given that our simulation system is overall stable, we wanted to investigate the role of the hydroxyl group of the Ser-278 side chain, corresponding to serine 364 in human SLC1A2, shown to be important for substrate binding and transport by our experimental data. Based on observations of the x-ray structure alone, one would conclude that Ser-278 is coordinating the Na1 ion at a relatively unfavorable distance (3.6 Å), without any apparent role in substrate binding. In contrast, the Ser-278 side chain in our simulations shows an active engagement in coordinating the substrate. Further analysis of the distance between the protein hydroxyl group of the serine (WT) or threonine (S278T) residues and the substrate shows that apart from the transient unbinding event described earlier, the contacts between the substrate and the protein are maintained in both WT and S278T systems throughout the simulation (Fig. 9). Because the contacts remain within the hydrogen-bonding distance (<3.5 Å), we find that the Ser-278 and Thr-278 side chains bind the carboxyl group of the substrate via stable hydrogen-bond interactions.
FIGURE 9.
Coordination of the l-Asp substrate by the hydroxyl group of the protein Ser-278 side chain. A, the predominant conformation of the Ser-278 side chain observed in our molecular dynamics (MD) simulations of GltPh (blue) overlaid with the x-ray conformation (gray, taken from PDB structure 2NWX). Substrate (l-aspartate) and bound sodium (Na1) are indicated. The stable hydrogen bond observed in our simulations between the Ser-278 side chain and the substrate is marked by a blue dashed line. B, the distance of the protein Ser-278 Oγ (WT)/Thr-278 Oγ1 (S278T) atom to the closest carboxyl oxygen of the l-Asp substrate was calculated over the simulation trajectory for each protein chain of the trimer. Note that the transient unbinding event in chain A of the S278T system is also visible as seen in Fig. 8B.
Because our experiments show that the WT and S278T transporters have a similar substrate affinity (Table 1) but different selectivity toward cations (Fig. 7), we wanted to investigate how the side chain at position 278 affects the way cations modulate the transport cycle. Because the binding site seems to contain several structural water molecules according to our simulations, and given that different cations are known to affect the structure of water differently (44), we performed a systematic analysis of water configurations linking the Na1 ion and the substrate. In each of the WT, S278A, and S278T systems, such water-mediated interaction networks were evaluated, and the distinct network configurations were identified and ranked according to their frequency of occurrence. The two most frequent configurations for each system are presented in Fig. 10. We found that in WT, both of the two most frequent scenarios involve a single water molecule mediating interactions between the Na1 ion and the hydroxyl group of Ser-278 (Fig. 10, A and B). This configuration is highly stable and present in over 78% of total simulation time (sum of panels A and B in Fig. 10). In the absence of the hydroxyl group, its role is taken by water molecules as seen in the S278A system (Fig. 10, C and D). In contrast, in the S278T system, the involvement of the protein hydroxyl group in this chain of interactions is not fully recovered (Fig. 10, E and F). In particular, the WT-like network is only second in frequency, appearing in 17% of total simulation time (Fig. 10F). Also counting other, less frequent configurations, the occurrence of a single-water-mediated contact between the Na1 and the protein is still below 31% in the S278T system, which is significantly lower than in the case of WT.
FIGURE 10.
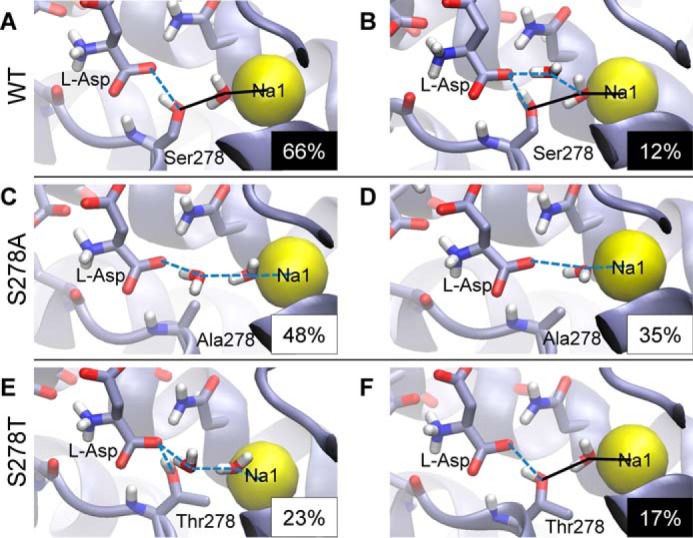
Distinct interaction networks between the l-aspartate substrate and the Na1 ion occurring in molecular dynamics simulations, identified using hydrogen-bond analysis. Individual configurations were ranked according to the frequency of their occurrence, shown in the lower right-hand corner of each panel as a percentage of total simulation time. The two most often encountered configurations are shown for simulation trajectories of the WT (A and B), S278A (C and D), and S278T (E and F) proteins. Lines indicate hydrogen bond and ion coordination interactions. The frequency percentage is shown in inverse for configurations where a WT-like single-water-mediated interaction pattern between Na1 and the protein was found (A, B, and F). The contacts forming the pattern are drawn as solid black lines. Such contacts are stable and abundant in the WT system (over 78% total occurrence as seen on panels A and B), whereas their overall frequency is less than 31% in the S278T system, including the configuration on panel F and others not shown in the figure.
Discussion
Substitution of Ser-364, which is conserved throughout prokaryotes and eukaryotes, to cysteine in SLC1A2 has been described to drastically reduce d-aspartate uptake (30), but the role of this particular amino acid in transport function has not been elucidated thus far. Our findings demonstrated that Ser-364 is functionally replaceable by threonine and partially by alanine or cysteine, in particular when measuring l-aspartate or d-aspartate transport (Figs. 2A and 3). The detailed study of substrate and cation selectivity of the mutants we generated provided evidence that Ser-364 is involved in substrate binding (Figs. 2–5 and Table 1) as well as in determining apparent cation binding and selectivity (Figs. 6 and 7). Such changes in substrate selectivity were reported when replacing other conserved amino acids in the glutamate transporter isoform SLC1A1 (EAAC1/EAAT3) (42, 45). For SLC1A2-WT, using radiolabeled substrate uptake assays, we obtained similar Km values for l-aspartate and d-aspartate and a 2–3-fold higher value with l-glutamate, which is in agreement with a previous study performed with COS-7 cells (46). Interestingly, the more conservative substitution S364T presented features that were very similar to those of WT, such as substrate affinities and selectivity and the affinity for sodium (Figs. 2A, 3, 4, and 6 and Table 1). However, the cation selectivity was different, because we observed a much larger transport rate for l-aspartate with S364T in the presence of Li+ (Fig. 7), as well as the substrate selectivity for the gating of the anion conductance at positive potentials (Fig. 5). Similar changes in cation selectivity that were observed for l-glutamate, l-aspartate, and d-aspartate as substrates have already been described for S440G (47). We found that S364D and S364N were not functional (Figs. 2A and 3). The absence of activity of S364D is probably due to the negative charge introduced and/or the increased length of the side chain. A longer side chain could also explain the absence of function observed in S364N. Because Ser-364 seems to be involved in substrate coordination and shaping apparent sodium affinity, the lack of activity of these mutants can be due either to the impaired substrate or Na+ binding to SLC1A2. Surprisingly, substitution by a shorter amino acid such as alanine or by one of similar size but without a hydroxyl group did not completely block SLC1A2 transport activity (Figs. 2A, 3, 4, and 5). Nevertheless, S364A exhibits a higher substrate transport capacity than S364C but lower affinities for l-aspartate and d-aspartate (Fig. 3 and Table 1). In addition, both mutants showed different substrate selectivities when compared with WT, independently of the presence of SCN− (Figs. 4 and 5).
Substrate selectivity for S364A and S364C decreases as follows: l-aspartate > d-aspartate > l-glutamate. These observations can be explained by the longer side chain of l-glutamate, which might need to be better stabilized by the hydroxyl group of Ser-364 to be transported. Strikingly, when compared with WT, we observed a shift in reversal potential of all the mutants, in the presence of all the substrates, when using SCN−-containing buffer. These values shifted to the one observed for SLC1A2-WT in SCN−-free buffer, and the shift was particularly pronounced for S364A (Figs. 4 and 5). A similar change in reversal potential has been described, after treatment by a reducing agent, for the SLC1A1 cross-linked mutant W441C/K269C, which is only capable of facilitating the substrate-gated anion conductance (48). Therefore, the reversal potential shift can be explained by a smaller contribution of the anion conductance to the total current in S364A when compared with SLC1A2-WT, suggesting a potential role of serine 364 in the anion selectivity of the uncoupled sodium-dependent current of the transporter. Indeed recently, it has been demonstrated that residues on the tip of HP1 are lining the anion permeation pore, suggesting that mutations in this region such as S364A might affect anion conductance as well (41).
Taken together our findings suggest a pivotal role of the serine 364 and more precisely its hydroxyl group in coupling Na+ and substrate fluxes. We further explored these findings using molecular dynamics simulations performed on the homologous archaeal protein, GltPh.
Molecular dynamics simulations showed that Ser-278 (corresponding to Ser-364 in SLC1A2) is indeed an important structural element contributing to substrate binding. First, we observed that the orientation of the Ser-278 side chain in the x-ray structure is transient and that Ser-278 forms a stable hydrogen bond with the carboxyl group of the substrate. A loss of these interactions, as in S278A, predicts a lower binding affinity of the substrate, which is in line with the experimental results presented in Table 1. The trans/gauche conformational change of the substrate observed in chain B of the S278A system (Fig. 8B) can be an indication that a larger conformational space is available to the bound substrate when the hydrogen bond with Ser-278 is missing. This entropic penalty may also impact the turnover rate of the transport by hindering the substrate in reaching the conformation necessary for the commencement of the transport cycle. This can result in a reduced rate of transport even under saturating substrate concentrations, as seen in Figs. 3 and 6. The affinity and also the transport activity can be recovered by the S278T mutation (Table 1), which is explained by the recovery of the hydrogen bond between the protein and the substrate, as shown in Fig. 9. In addition, analysis of the hydrogen bonds in the binding site shows the presence of an intricate network of bonds linking the Na1 ion and the substrate. In the WT transporter, a highly stable link mediated by a single water molecule between Na1 and the hydroxyl group of Ser-278 is observed. We hypothesize that this network is responsible for determining the effect of the cotransported ion on the transport cycle. In the case of the S278A and S278T mutants, this network is either completely lost or only partially recovered, which correlates with the loss of ion selectivity observed in these systems (Fig. 7). It should be emphasized that the recovery of WT-like contacts between the protein and the substrate is independent of the recovery of the water-mediated interaction network between the protein and the Na1 ion. This is in line with our observation that the S278T mutant recovers transport activity but loses selectivity for the cotransported cation. Note that our simulations do not readily hint at the mechanism by which the bound ion species at the Na1 location affects the transport cycle. We speculate that the ion exerts its effect by reorganizing the water structure in the binding site, thus affecting protein conformation and either promoting or interfering with the closure of hairpin loop HP2, which is required for the commencement of the transport cycle.
In summary, our results demonstrate that the residue Ser-364 of SLC1A2 is involved in substrate as well as cation binding and selectivity. Moreover, this residue might play a key role in the anion selectivity of the uncoupled anion conductance. Molecular dynamics analysis revealed an intricate network of bonds linking the Na1 ion to the protein and the substrate. This network is likely responsible for determining the effect of the cotransported ion on the transport cycle. To conclude, our experimental results and molecular dynamics analysis suggest a pivotal role of the serine 364 and more precisely its hydroxyl group in coupling sodium and substrate fluxes.
Author Contributions
A. S., N. M., and M. A. H. conceived the idea for the project, coordinated the study and wrote most of the paper. A. S. and N. M. conducted most of the experiments and analyzed the results. G. G. conducted all molecular dynamics analyses and wrote part of the paper. J. P.-G. performed electrophysiological measurements in X. laevis oocytes and analyzed the results. All authors reviewed the results and approved the final version of the manuscript.
This work was in part supported by the Swiss National Science Foundation (SNSF) through the National Center of Competence in Research (NCCR) TransCure and by the Marie Curie Actions International Fellowship Program (IFP) TransCure (to G. G. and J. P.-G.). The authors declare that they have no conflicts of interest with the contents of this article.
- EAAT
- excitatory amino acid transporter
- TM
- transmembrane segment
- HP
- hairpin.
References
- 1.Kanai Y., and Hediger M. A. (2003) The glutamate and neutral amino acid transporter family: physiological and pharmacological implications. Eur. J. Pharmacol. 479, 237–247 [DOI] [PubMed] [Google Scholar]
- 2.Rothstein J. D., Dykes-Hoberg M., Pardo C. A., Bristol L. A., Jin L., Kuncl R. W., Kanai Y., Hediger M. A., Wang Y., Schielke J. P., and Welty D. F. (1996) Knockout of glutamate transporters reveals a major role for astroglial transport in excitotoxicity and clearance of glutamate. Neuron 16, 675–686 [DOI] [PubMed] [Google Scholar]
- 3.Kanai Y., Clémençon B., Simonin A., Leuenberger M., Lochner M., Weisstanner M., and Hediger M. A. (2013) The SLC1 high-affinity glutamate and neutral amino acid transporter family. Mol. Aspects Med. 34, 108–120 [DOI] [PubMed] [Google Scholar]
- 4.Maragakis N. J., and Rothstein J. D. (2004) Glutamate transporters: animal models to neurologic disease. Neurobiol. Dis. 15, 461–473 [DOI] [PubMed] [Google Scholar]
- 5.Sheldon A. L., and Robinson M. B. (2007) The role of glutamate transporters in neurodegenerative diseases and potential opportunities for intervention. Neurochem. Int. 51, 333–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trotti D., Aoki M., Pasinelli P., Berger U. V., Danbolt N. C., Brown R. H. Jr., and Hediger M. A. (2001) Amyotrophic lateral sclerosis-linked glutamate transporter mutant has impaired glutamate clearance capacity. J. Biol. Chem. 276, 576–582 [DOI] [PubMed] [Google Scholar]
- 7.Trotti D., Rolfs A., Danbolt N. C., Brown R. H. Jr., and Hediger M. A. (1999) SOD1 mutants linked to amyotrophic lateral sclerosis selectively inactivate a glial glutamate transporter. Nat. Neurosci. 2, 427–433; Correction (1999) Nat. Neurosci. 2, 848 [DOI] [PubMed] [Google Scholar]
- 8.Zerangue N., and Kavanaugh M. P. (1996) Flux coupling in a neuronal glutamate transporter. Nature 383, 634–637 [DOI] [PubMed] [Google Scholar]
- 9.Kavanaugh M. P., Bendahan A., Zerangue N., Zhang Y., and Kanner B. I. (1997) Mutation of an amino acid residue influencing potassium coupling in the glutamate transporter GLT-1 induces obligate exchange. J. Biol. Chem. 272, 1703–1708 [DOI] [PubMed] [Google Scholar]
- 10.Levy L. M., Warr O., and Attwell D. (1998) Stoichiometry of the glial glutamate transporter glt-1 expressed inducibly in a Chinese hamster ovary cell line selected for low endogenous Na+-dependent glutamate uptake. J. Neurosci. 18, 9620–9628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fairman W. A., Vandenberg R. J., Arriza J. L., Kavanaugh M. P., and Amara S. G. (1995) An excitatory amino-acid transporter with properties of a ligand-gated chloride channel. Nature 375, 599–603 [DOI] [PubMed] [Google Scholar]
- 12.Arriza J. L., Eliasof S., Kavanaugh M. P., and Amara S. G. (1997) Excitatory amino acid transporter 5, a retinal glutamate transporter coupled to a chloride conductance. Proc. Natl. Acad. Sci. U.S.A. 94, 4155–4160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wadiche J. I., Amara S. G., and Kavanaugh M. P. (1995) Ion fluxes associated with excitatory amino-acid-transport. Neuron 15, 721–728 [DOI] [PubMed] [Google Scholar]
- 14.Melzer N., Biela A., and Fahlke C. (2003) Glutamate modifies ion conduction and voltage-dependent gating of excitatory amino acid transporter-associated anion channels. J. Biol. Chem. 278, 50112–50119 [DOI] [PubMed] [Google Scholar]
- 15.Watzke N., Bamberg E., and Grewer C. (2001) Early intermediates in the transport cycle of the neuronal excitatory amino acid carrier EAAC1. J. Gen. Physiol. 117, 547–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borre L., and Kanner B. I. (2001) Coupled, but not uncoupled, fluxes in a neuronal glutamate transporter can be activated by lithium ions. J. Biol. Chem. 276, 40396–40401 [DOI] [PubMed] [Google Scholar]
- 17.Yernool D., Boudker O., Jin Y., and Gouaux E. (2004) Structure of a glutamate transporter homologue from Pyrococcus horikoshii. Nature 431, 811–818 [DOI] [PubMed] [Google Scholar]
- 18.Koch H. P., and Larsson H. P. (2005) Small-scale molecular motions accomplish glutamate uptake in human glutamate transporters. J. Neurosci. 25, 1730–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grewer C., Balani P., Weidenfeller C., Bartusel T., Tao Z., and Rauen T. (2005) Individual subunits of the glutamate transporter EAAC1 homotrimer function independently of each other. Biochemistry 44, 11913–11923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koch H. P., Brown R. L., and Larsson H. P. (2007) The glutamate-activated anion conductance in excitatory amino acid transporters is gated independently by the individual subunits. J. Neurosci. 27, 2943–2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leary G. P., Stone E. F., Holley D. C., and Kavanaugh M. P. (2007) The glutamate and chloride permeation pathways are colocalized in individual neuronal glutamate transporter subunits. J. Neurosci. 27, 2938–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boudker O., Ryan R. M., Yernool D., Shimamoto K., and Gouaux E. (2007) Coupling substrate and ion binding to extracellular gate of a sodium-dependent aspartate transporter. Nature 445, 387–393 [DOI] [PubMed] [Google Scholar]
- 23.Groeneveld M., and Slotboom D. J. (2010) Na+:aspartate coupling stoichiometry in the glutamate transporter homologue GltPh. Biochemistry 49, 3511–3513 [DOI] [PubMed] [Google Scholar]
- 24.Teichman S., Qu S., and Kanner B. I. (2009) The equivalent of a thallium binding residue from an archeal homolog controls cation interactions in brain glutamate transporters. Proc. Natl. Acad. Sci. U.S.A. 106, 14297–14302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larsson H. P., Wang X., Lev B., Baconguis I., Caplan D. A., Vyleta N. P., Koch H. P., Diez-Sampedro A., and Noskov S. Y. (2010) Evidence for a third sodium-binding site in glutamate transporters suggests an ion/substrate coupling model. Proc. Natl. Acad. Sci. U.S.A. 107, 13912–13917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tao Z., Zhang Z., and Grewer C. (2006) Neutralization of the aspartic acid residue Asp-367, but not Asp-454, inhibits binding of Na+ to the glutamate-free form and cycling of the glutamate transporter EAAC1. J. Biol. Chem. 281, 10263–10272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tao Z., Rosental N., Kanner B. I., Gameiro A., Mwaura J., and Grewer C. (2010) Mechanism of cation binding to the glutamate transporter EAAC1 probed with mutation of the conserved amino acid residue Thr101. J. Biol. Chem. 285, 17725–17733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang Z. J., and Tajkhorshid E. (2010) Identification of the third Na+ site and the sequence of extracellular binding events in the glutamate transporter. Biophys. J. 99, 1416–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bastug T., Heinzelmann G., Kuyucak S., Salim M., Vandenberg R. J., and Ryan R. M. (2012) Position of the third Na+ site in the aspartate transporter GltPh and the human glutamate transporter, EAAT1. PLoS One 7, e33058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grunewald M., and Kanner B. I. (2000) The accessibility of a novel reentrant loop of the glutamate transporter GLT-1 is restricted by its substrate. J. Biol. Chem. 275, 9684–9689 [DOI] [PubMed] [Google Scholar]
- 31.Shrivastava I. H., Jiang J., Amara S. G., and Bahar I. (2008) Time-resolved mechanism of extracellular gate opening and substrate binding in a glutamate transporter. J. Biol. Chem. 283, 28680–28690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gu Y., Shrivastava I. H., Amara S. G., and Bahar I. (2009) Molecular simulations elucidate the substrate translocation pathway in a glutamate transporter. Proc. Natl. Acad. Sci. U.S.A. 106, 2589–2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grazioso G., Limongelli V., Branduardi D., Novellino E., De Micheli C., Cavalli A., and Parrinello M. (2012) Investigating the mechanism of substrate uptake and release in the glutamate transporter homologue GltPh through metadynamics simulations. J. Am. Chem. Soc. 134, 453–463 [DOI] [PubMed] [Google Scholar]
- 34.Bergeron M. J., Bürzle M., Kovacs G., Simonin A., and Hediger M. A. (2011) Synthesis, maturation, and trafficking of human Na+-dicarboxylate cotransporter NaDC1 requires the chaperone activity of cyclophilin B. J. Biol. Chem. 286, 11242–11253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simonin A., and Fuster D. (2010) Nedd4–1 and β-arrestin-1 are key regulators of Na+/H+ exchanger 1 ubiquitylation, endocytosis, and function. J. Biol. Chem. 285, 38293–38303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lomize M. A., Lomize A. L., Pogozheva I. D., and Mosberg H. I. (2006) OPM: orientations of proteins in membranes database. Bioinformatics 22, 623–625 [DOI] [PubMed] [Google Scholar]
- 37.Jo S., Kim T., Iyer V. G., and Im W. (2008) CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 [DOI] [PubMed] [Google Scholar]
- 38.Wu E. L., Cheng X., Jo S., Rui H., Song K. C., Dávila-Contreras E. M., Qi Y., Lee J., Monje-Galvan V., Venable R. M., Klauda J. B., and Im W. (2014) CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem. 35, 1997–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., and Schulten K. (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wadiche J. I., Arriza J. L., Amara S. G., and Kavanaugh M. P. (1995) Kinetics of a human glutamate transporter. Neuron 14, 1019–1027 [DOI] [PubMed] [Google Scholar]
- 41.Machtens J. P., Kortzak D., Lansche C., Leinenweber A., Kilian P., Begemann B., Zachariae U., Ewers D., de Groot B. L., Briones R., and Fahlke C. (2015) Mechanisms of anion conduction by coupled glutamate transporters. Cell 160, 542–553 [DOI] [PubMed] [Google Scholar]
- 42.Rosental N., and Kanner B. I. (2010) A conserved methionine residue controls the substrate selectivity of a neuronal glutamate transporter. J. Biol. Chem. 285, 21241–21248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wadiche J. I., and Kavanaugh M. P. (1998) Macroscopic and microscopic properties of a cloned glutamate transporter chloride channel. J Neurosci 18, 7650–7661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marcus Y. (2009) Effect of ions on the structure of water: structure making and breaking. Chem. Rev. 109, 1346–1370 [DOI] [PubMed] [Google Scholar]
- 45.Teichman S., Qu S., and Kanner B. I. (2012) Conserved asparagine residue located in binding pocket controls cation selectivity and substrate interactions in neuronal glutamate transporter. J. Biol. Chem. 287, 17198–17205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arriza J. L., Fairman W. A., Wadiche J. I., Murdoch G. H., Kavanaugh M. P., and Amara S. G. (1994) Functional comparisons of three glutamate transporter subtypes cloned from human motor cortex. J. Neurosci. 14, 5559–5569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y., and Kanner B. I. (1999) Two serine residues of the glutamate transporter GLT-1 are crucial for coupling the fluxes of sodium and the neurotransmitter. Proc. Natl. Acad. Sci. U.S.A. 96, 1710–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shabaneh M., Rosental N., and Kanner B. I. (2014) Disulfide cross-linking of transport and trimerization domains of a neuronal glutamate transporter restricts the role of the substrate to the gating of the anion conductance. J. Biol. Chem. 289, 11175–11182 [DOI] [PMC free article] [PubMed] [Google Scholar]