Background: Enzymes involved in the biosynthesis of bacterial cell walls are validated antibiotic targets, including penicillin binding proteins (PBPs).
Results: We unveiled the permissible structural composition of unnatural d-amino acids in labeling bacterial surfaces via PBPs.
Conclusion: The chemical nature of fluorescent d-amino acids governs their effectiveness as probes of peptidoglycan biosynthesis.
Significance: Chemical probes of PBPs can be powerful tools to decipher cell wall assembly.
Keywords: amino acid, Bacillus, bacteria, bacterial conjugation, cell surface, cell wall, peptide biosynthesis, peptide chemical synthesis, peptides, peptidoglycan
Abstract
Peptidoglycan is an essential and highly conserved mesh structure that surrounds bacterial cells. It plays a critical role in retaining a defined cell shape, and, in the case of pathogenic Gram-positive bacteria, it lies at the interface between bacterial cells and the host organism. Intriguingly, bacteria can metabolically incorporate unnatural d-amino acids into the peptidoglycan stem peptide directly from the surrounding medium, a process mediated by penicillin binding proteins (PBPs). Metabolic peptidoglycan remodeling via unnatural d-amino acids has provided unique insights into peptidoglycan biosynthesis of live bacteria and has also served as the basis of a synthetic immunology strategy with potential therapeutic implications. A striking feature of this process is the vast promiscuity displayed by PBPs in tolerating entirely unnatural side chains. However, the chemical space and physical features of this side chain promiscuity have not been determined systematically. In this report, we designed and synthesized a library of variants displaying diverse side chains to comprehensively establish the tolerability of unnatural d-amino acids by PBPs in both Gram-positive and Gram-negative organisms. In addition, nine Bacillus subtilis PBP-null mutants were evaluated with the goal of identifying a potential primary PBP responsible for unnatural d-amino acid incorporation and gaining insights into the temporal control of PBP activity. We empirically established the scope of physical parameters that govern the metabolic incorporation of unnatural d-amino acids into bacterial peptidoglycan.
Introduction
Peptidoglycan is a mesh-like biopolymer that maintains bacterial cell shape and serves to counter intracellular turgor pressure (1). Monomeric peptidoglycan units are composed of the disaccharides N-acetylglucosamine and N-acetylmuramyl linked to a short stem pentapeptide (2–7). Quite uniquely, the stem peptide is composed of various d-amino acids that can differ slightly in composition on an organism-dependent basis (Fig. 1A). The assembly of peptidoglycan relies on several enzymes working in concert. Of great importance are the penicillin binding proteins (PBPs),2 which are tasked with the assembly and maturation of the peptidoglycan, including glycosyl transfer, transpeptidation (TP), and carboxypeptidation (CP) (8, 9). PBP TP domains are tasked with cross-linking neighboring stem peptides to increase structural strength and rigidity of the peptidoglycan, a vital process for the viability of bacterial cells. Because of their essential role in the life cycle of bacteria, small molecules that inactivate PBP TPs (e.g. β-lactams) display powerful antibiotic activities (10). The first step in PBP TP cross-linking entails removal of the terminal d-alanine to yield a covalent acyl intermediate between the stem peptide and the PBP active site serine. Acyl transfer of the covalent intermediate by an amino group on meso-diaminopimelic acid (or l-lysine), results in an amide bond cross-link. Given the importance of PBPs as drug targets, it is surprising that the specific role of various PBPs in cell wall assembly remains the subject of active research.
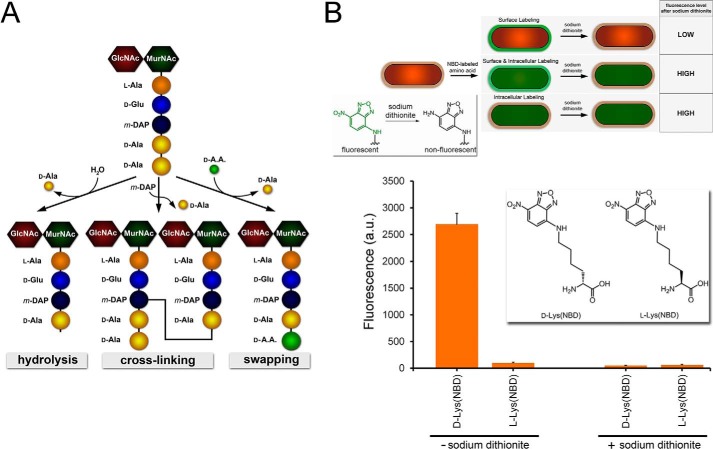
FIGURE 1.
A, schematic showing the monomeric unit of peptidoglycan. The pentapeptide is processed by PBPs CP and TP to generate three possible products, all resulting from the acyl intermediate following the release of the terminal d-Ala. B, schematic demonstrating the three possible labeling locations within the bacterial cell. Only extracellular labeling would lead to complete reduction of the signal in the presence of the reducing agent sodium dithionite. Inset, reduction of the nitro group on NBD by sodium dithionite quenches the green fluorescence. B. subtilis cells were labeled with either d-Lys(NBD) or l-Lys(NBD) overnight. Cells were analyzed by flow cytometry. Data are represented as mean ± S.D. (n = 3).
The peptidoglycan stem peptide is an integral component of the cell wall. As such, its composition and structure are linked to cell wall stiffness and susceptibility to binding partners, features that have important implications in drug resistance to cell wall antibiotics and host immunity. Recently, it has been demonstrated that changes to levels of cross-linking of the stem peptide chains by PBP TP correlate with methicillin resistance in Staphylococcus aureus (11). In another example, the replacement of the fifth-position d-alanine with d-lactic acid in the stem peptide of Enterococcus faecium and Enterococcus faecalis directly renders these pathogens resistant to the antibiotic vancomycin (12). Alteration of the amide backbone to an ester backbone on the stem peptide is sufficient to reduce the affinity of vancomycin for the stem peptide by 3 orders of magnitude (13). Additionally, peptidoglycan stem peptides play a central role in the recognition of pathogenic bacteria by the human immune system. Nucleotide-binding oligomerization-like receptors sense motifs within the peptidoglycan, activating response mechanisms (14). Clearly, structural changes to the stem peptide can potentially have profound physiological consequences. The recent demonstration that peptidoglycan stem peptides are prone to chemical remodeling by unnatural d-amino acids from the growth medium reveals an additional mode of stem peptide modification.
PBP TP activity is fundamental to normal bacterial physiology. However, to date, it has not been possible to conclusively determine which PBPs may be most critical for TP activity in live cells. Redundancy within PBPs can further compound the difficulties in establishing the role of PBP in peptidoglycan biosynthesis. Recent work has demonstrated that S. aureus can survive despite genomic deletion of seven of the nine total PBPs (15). Likewise, strains of Bacillus subtilis lacking PBP1, PPB2c, PBP2d, and PBP4 (either alone or in various combinations) display little to no change in peptidoglycan cross-linking ratios (16). Genetic deletion of PBP2b in B. subtilis is lethal, suggesting that it may play a primary role in TP activity (17). Together, these examples highlight the limitations of using genetic manipulations alone to track PBP enzymatic activity. Chemical probes that report on the activity of PBPs in live bacterial cells can complement genetics-based models by providing critical information on enzymatic activity with spatial and temporal resolution (18–21). A more comprehensive examination of PBP TP activity may have a profound effect on our understanding of cell wall biosynthesis and drug lead development.
Metabolic stem peptide remodeling via synthetic peptidoglycan mimetics has paved the way for a novel class of PBP chemical probes. Unnatural d-amino acids facilitate the tracking of new peptidoglycan production, and they have revealed the presence of peptidoglycan in Chlamydia trachomatis for the first time, a subject of intense debate for over 50 years (18, 20, 22–24). Supplementation of unnatural d-amino acids to the culture medium leads to their incorporation onto bacterial cell surfaces by mischarging at the TP step, generating a swapped terminal amino acid (Fig. 1A) (25). Swapping of the terminal d-alanine with unnatural d-amino acids from the medium is a robust method to decorate bacterial surfaces with diverse chemical handles (22, 23, 26–32). For example, our research group recently introduced a class of d-amino acid hapten conjugates designed to recruit endogenous antibodies to the bacterial surface as a novel immunomodulatory strategy (33, 34). Prior studies involving unnatural d-amino acids to remodel bacterial surfaces suggest that the PBP TP domains possess extraordinary promiscuity in tolerating diverse amino acid side chains. However, to date, no systematic correlation between incorporation efficiency and side chain structural features (e.g. charge, size, and polarity) of unnatural d-amino acids had been established. Here we quantitatively determined the side chain tolerability of unnatural d-amino acids by bacterial PBP TP in live cells. We anticipate that a comprehensive map of the structural elements tolerated by PBP TPs may guide future designs of unnatural side chains of d-amino acids for mechanistic investigations and therapeutic interventions. Additionally, nine B. subtilis mutant strains lacking PBPs reported to possess TP activity were probed with fluorescent d-amino acids.
Experimental Procedures
Materials
All amino acids were purchased from Chem-Impex. All other chemicals were purchased from Sigma-Aldrich or Fisher Scientific and used without further purification. The bacterial strains that were used for these experiments were as follows: B. subtilis NCIB 3610, B. subtilis Δdac, B. subtilis ΔpbpA, B. subtilis ΔpbpC, B. subtilis ΔpbpD, B. subtilis ΔpbpF, B. subtilis ΔpbpG, B. subtilis ΔpbpH, B. subtilis ΔpbpI, B. subtilis ΔPonA, Staphylococcus epidermidis NRS 101, Escherichia coli MG1665, S. aureus ATCC 25923, S. aureus SCO1, and E. faecalis ATCC 29212.
d-Amino Acid Fluorescent Labeling
Bacterial cells were grown overnight at 37 °C in LB. The following day, the medium was replaced with LB supplemented with fluorescently labeled d-amino acids. The bacteria were grown in the presence of the fluorescent d-amino acid for either 4 h or overnight, depending on the experiment. Following incubation, the bacteria were harvested, washed three times with PBS, and fixated with a 4% formaldehyde solution. All experiments were run in triplicate from independent bacterial cell cultures and analyzed using flow cytometry on a BDFacs Canto II (BD Biosciences) equipped with a 488-nm argon laser (L1) and a 530/30 band-pass filter (FL1). The fluorescence data are expressed as mean arbitrary fluorescence units of the 10,000 events that were counted for each data set and gated to include all healthy bacteria. The data were analyzed using FACSDiva version 6.1.1.
Sodium Dithionite Quenching of Fluorescently Labeled Bacteria
B. subtilis cells were grown overnight at 37 °C with shaking in LB medium supplemented with either 100 μm d-Lys(NBD) or 100 μm l-Lys(NBD). The following morning, the cells were washed three times with PBS. The bacteria were then subjected to incubation with 5 mm sodium dithionite for 5 min. The bacteria were then washed again three more times with PBS, fixated with a 4% formaldehyde solution, and analyzed via flow cytometry.
Sulfhydryl Fluorescent Labeling of Bacterial Surface
Compounds SH-1, SH-4, and SH-6 were bought from Chem-Impex and used without any further purification. B. subtilis ΔdacA cells were incubated for 4 h at 37 °C with shaking in LB medium supplemented with 500 μm of each sulfhydryl compound. The cells were harvested and washed three times in a 5 mm dithiothreitol solution to reverse any oxidation that may have occurred overnight. The cells were then washed an additional five times with PBS. The cells were then incubated in PBS containing 50 μm of the NBD-l-Lys-(maleimide) conjugate at 37 °C for 30 min. The cells were then washed three more times with PBS and fixated with a 4% formaldehyde solution. Following fixation, the cells were analyzed via flow cytometry. To confirm that labeling occurred because of the presence of the incorporated sulfhydryl group, cells were incubated with 50 μm N-ethylmaleimide for 30 min following the overnight incubation with the sulfhydryl compounds as described previously. The cells were then washed three times and incubated with NBD-l-Lys-(maleimide), washed, and analyzed as described previously. To confirm that cell labeling occurred on the cell surface, the labeled cells were subjected to treatment with sodium dithionite to quench fluorescence from the NBD fluorophore as described previously.
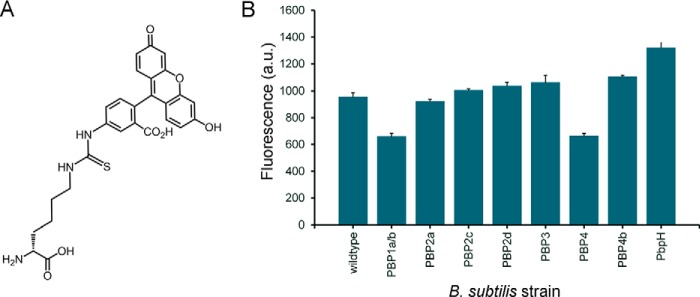
d-Lys(FITC)-OH Pulse Labeling of B. subtilis Mutant Strains
B. subtilis mutants were grown in LB medium at 37 °C overnight with shaking. The following morning, the A600 of all the mutants was diluted to A600 = 0.1. The bacteria were then grown in LB medium until reaching either early log (A600 = 0.3), mid-log (A600 = 0.6), or stationary phase (A600 > 1.0). At the respective phase, d-Lys(FITC) was spiked into the solution to achieve a final concentration of 1 mm in LB medium. The cells were then incubated for 15 min in the presence of the d-amino acid fluorophore. Following incubation, the cells were washed three times with PBS, fixated with a 4% formaldehyde solution, and then analyzed via flow cytometry or imaged with a Nikon Eclipse TE2000-U microscope at ×100 magnification.
Results
The replacement of the terminal d-alanine in the peptidoglycan stem peptide with unnatural d-amino acids is mediated by a PBP TP reaction. Although PBP TP promiscuity is unusual in its ability to accommodate a number of naturally and unnaturally occurring d-amino acids as substrates, the expansive substrate scope was highlighted prominently by the successful incorporation of d-amino acids displaying entirely non-native side chains (e.g. large hydrophobic fluorophores and biorthogonal handles) (22, 27, 29). However, the relative incorporation levels of d-amino acids across a range of side chains have not been reported. We set out to systematically assess the permissible chemical space of the d-amino acid side chain in live bacterial cells. We hypothesized that inherent PBP active site constraints could be discerned by evaluating labeling levels with a library of unnatural d-amino acids.
First, we evaluated a set of two amino acid derivatives (d-Lys(NBD) and l-Lys(NBD)) for the ability to label the peptidoglycan of B. subtilis (Fig. 1B). B. subtilis was chosen as our initial model organism because of its wide use in unnatural d-amino acid studies and its compatibility with genetic manipulation (35). The small fluorophore NBD was conjugated to the side chain to monitor incorporation of the d-amino acid into the bacterial cell wall (see supplemental Experimental Procedures). We have shown previously that d-Lys(NBD) is incorporated within the peptidoglycan stem peptide by a combination of peptidoglycan isolation and mass spectrometry (34). To show that the fluorescence signal measured by flow cytometry is reflective of the d-amino acid incorporated within the cell wall, we performed a fluorescence quenching assay. Following the labeling of B. subtilis cells with either d-Lys(NBD) or l-Lys(NBD), cells were exposed to a short treatment of the reducing agent sodium dithionite. Sodium dithionite is known to reduce the NBD nitro group to produce a non-fluorescent aryl amine derivative (Fig. 1B) (36). The poor lipid bilayer permeation of the highly polar dithionite species limits the quenching of the NBD moiety to the extracellular space of the cell (37). We reasoned that complete quenching of the fluorescence signal following treatment with sodium dithionite would be consistent with extracellular incorporation of the NBD moiety (Fig. 1B). Intracellular accumulation of the unnatural d-amino acid or a combination of intracellular/extracellular accumulation would be expected to yield a fluorescence signal above basal levels. B. subtilis cells were labeled with d-Lys(NBD) and analyzed by flow cytometry with and without sodium dithionite treatment (Fig. 1B). As we observed previously, the fluorescent d-Lys(NBD) efficiently labeled B. subtilis cells. The exposure of labeled bacterial cells to sodium dithionite reduced fluorescence signals down to basal levels (similar to unlabeled cells), a result that is consistent with the site-selective incorporation of the NBD label on the exterior of the cell. Next, the same procedure was performed with the enantiomer l-Lys(NBD). As expected, minimal B. subtilis labeling was observed in the absence of sodium dithionite. The stereospecificity of the labeling is indicative of incorporation of d-Lys(NBD) by PBP TP (25). Together, these results establish that NBD-conjugated d-amino acids can report on the incorporation of unnatural d-amino acids into bacterial peptidoglycan.
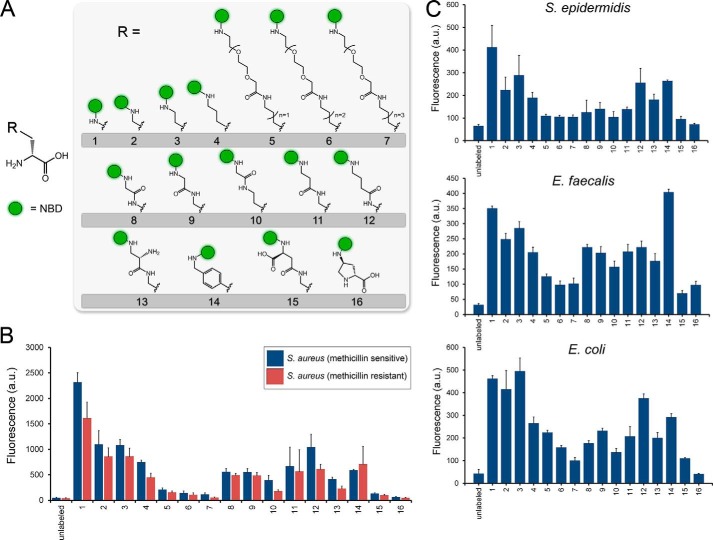
Next we set out to comprehensively assess the PBP TP tolerability of the side chain modifications. We synthesized a library of 16 unnatural d-amino acid derivatives with diverse side chains (Fig. 2A). For all variants, NBD was conjugated covalently onto a side chain amino group to track incorporation and has been demonstrated previously to be a suitable fluorescent probe for tracking peptidoglycan biosynthesis (28, 29). The side chains were chosen to encompass a wide-scoping set of chemical space by including structural elements that vary in length, bulkiness, flexibility, polarity, and charge. With the NBD-modified d-amino acids assembled, we performed a series of labeling experiments that quantitatively established the relative incorporation of these variants. Initially, we determined incorporation levels of the unnatural d-amino acids in the human pathogen S. aureus by incubating the bacteria with each d-amino acid variant and assessing incorporation by flow cytometry (Fig. 2B). Clear preferences for side chain composition emerge from this first set of data. Not unexpectedly, the overall size of the side chain has a strong influence on incorporation efficiency. The variant with the smallest sidechain, 1, displayed the highest incorporation levels. The endogenous substrate (most commonly d-alanine) for PBP TP has a small side chain, a feature that is likely matched by the cavity in the TP active site. The elongation of the side chain by methylene units from 1-4 highlights the correlation between side chain length and labeling levels. There is approximately a 3.5-fold decrease in surface labeling from d-diaminopropionoic acid (1) to d-lysine (4). Further elongation of the side chain by conjugation to flexible/polar PEG spacers (5-7) reduced incorporation to near basal levels. From these results, it can be concluded that there is considerable tolerance in linear tethers connecting the backbone to the NBD moiety even in instances when the side chain is vastly different from d-alanine.
FIGURE 2.
A, metabolic labeling of bacterial surfaces with d-lysine conjugated to the small fluorophore NBD. B, labeling of S. aureus cells (methicillin-sensitive and -resistant) was performed with all the members of the d-amino acid NBD conjugates. Cells were incubated overnight at 100 μm and analyzed by flow cytometry. Data are represented as mean ± S.D. (n = 3). C, S. epidermidis and E. faecalis cells were labeled with the panel of NBD-conjugated d-amino acids for 4 h at stationary phase at 100 μm. E. coli cells were labeled with the panel of NBD-conjugated d-amino acids for 4 h at stationary phase at 500 μm. Cells were analyzed by flow cytometry, and data are represented as mean ± S.D. (n = 3).
The combination of linker length and location of an internal amide bond was assessed among derivatives 8-12. Amidation of d-diaminopropionoic acid with glycine to yield 8 was a well tolerated modification. This particular variant is important because of the combination of small side chain size and an easy point of acylation to various conjugates of interest (e.g. fluorophores). Minimal discrimination was observed on the basis of the location of the tether amide bond for similarly sized tethers (variants 10, 11, and 12). Three additional side chain attributes were assessed among derivatives 13-16: side chain charge, tolerance of aromatic group, and secondary amino group. The positively charged 13 and the negatively charged 15 have the same tether as the uncharged 11. The introduction of a negative charge drastically reduced labeling levels, whereas a positive charge was permissible. Perhaps surprisingly, the aromatic side chain in 14 proved to be compatible with surface labeling. Finally, it was observed that the pyrrolidine ring of d-proline abolished incorporation altogether. Steric constraints imposed by a secondary amine (the nucleophilic species in the TP reaction) may kinetically disfavor acylation onto the peptidoglycan-anchored tetrapeptide.
The incorporation profile of methicillin-resistant S. aureus cells was also obtained. Despite the established difference in PBPs between methicillin-resistant and methicillin-sensitive S. aureus, a minimal difference in labeling profile was observed (Fig. 2B) (38). There appears to be a slight decrease in labeling across some of the d-amino acid derivatives, an intriguing finding that will be the subject of future investigations. Having established the incorporation profile of one human pathogen, we sought to gain a similar insight into the side chain preference for additional organisms. The incorporation profile of S. epidermidis displayed similar labeling preferences as S. aureus, a finding that is consistent with a shared genus (Fig. 2C). Next we set out to evaluate the incorporation profile of a distinct Gram-positive pathogen. E. faecalis has emerged as a problematic pathogen because of increasing instances of drug resistance. Although many of the trends are similar, there are some prominent deviations (Fig. 2C). The aromatic variant 14 labeled E. faecalis cell surfaces most efficiently. The incorporation profile of the Gram-negative E. coli was also obtained. Despite differences in the architecture of the peptidoglycan, the trend remained in line with the Gram-positive organisms evaluated. Interestingly, d-proline has been shown previously in vitro to be incorporated onto peptidoglycan monomers by E. coli PBP1A (25). The lack of labeling with the similar d-proline-based 16 may reflect altered substrate specificity in live E. coli cells.
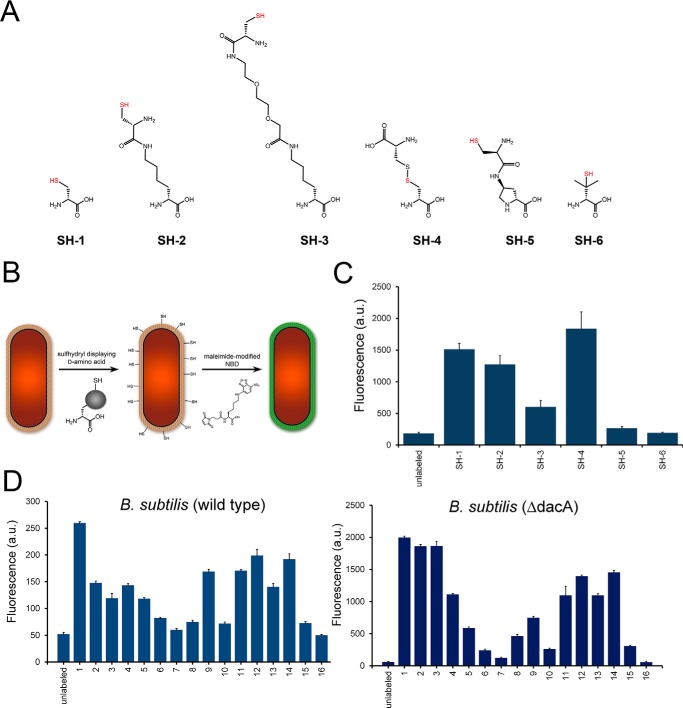
The conjugation of the NBD fluorophore to d-amino acids variants 1-16 was critical in facilitating quantitative measurement of surface labeling. Although its small size is expected to only minimally contribute to the trends in incorporation profiles, it still prevented our ability to assess the incorporation of small side chains. It has been demonstrated previously that d-cysteine can be incorporated into the stem peptide of bacterial peptidoglycan through a variety of methods (39, 40). We devised an alternative strategy to measure labeling efficiency to corroborate our findings with NBD-conjugated d-amino acids on the basis of sulfhydryl-maleimide coupling (Fig. 3A). B. subtilis cells were treated with the sulfhydryl-displaying variants (SH-1 to SH-6), exposed to a short treatment with dithiothreitol to reverse oxidation, and detected with an NBD-maleimide conjugate. The incubation of d-cysteine (SH-1) served to establish the conditions for cell surface labeling. Incubation of cells with SH-1 led to a marked increase in cellular fluorescence (39). Remarkably, the d-lysine derivative SH-2 labeled nearly as well as the much smaller SH-1. These results highlight the unusual tolerance of accommodating large side chains equally well as small side chains. The addition of a PEG chain (SH-3) resulted in significantly lower levels of cell surface labeling. Clearly, the tolerance of side chain bulk has some limitations, consistent with our NBD-conjugated d-amino acids. Interestingly, d-cysteine (SH-4) was incorporated to high levels despite its larger size. Perhaps it represents a homologated analog of the endogenous TP substrate m-DAP. Display of the sulfhydryl onto a d-proline resulted in no surface labeling, consistent with 16. Lastly, the Food and Drug Administration-approved agent d-penicillamine (SH-6) was chosen to evaluate the tolerance of steric bulk near the N terminus. A complete lack of incorporation was observed for cells treated with SH-6, a strong indication that side chain bulk near the backbone can interfere with incorporation.
FIGURE 3.
A, chemical structures of B. subtilis cells were labeled with the panel of sulfhydryl containing d-amino acids. Cells were incubated for 4 h at stationary phase at 500 μm, incubated with NBD-l-Lys(maleimide), and analyzed by flow cytometry. Data are represented as mean ± S.D. (n = 3). D, wild-type and ΔdacA mutant B. subtilis cells were labeled with the panel of NBD-conjugated d-amino acids. Cells were incubated for 4 h at stationary phase at 100 μm and analyzed by flow cytometry. Data are represented as mean ± S.D. (n = 3).
We further confirmed that all of the observed fluorescent signals originate from the incorporation of exogenous d-amino acid sulfhydryl compounds by incubating sulfhydryl-labeled cells with N-ethylmaleimide (a nonfluorescent compound) prior to exposing the cells to NBD-maleimide conjugate. The exposure of the cells to N-ethylmaleimide reduced fluorescent signals to basal levels, confirming that the observed fluorescent signals are a result of cellular incorporation of the d-amino acid sulfhydryl compounds (data not shown). Additionally, to confirm that these compounds were located in the peptidoglycan, we again took advantage of the cell-impermeable compound sodium dithionite. Sulfhydryl-labeled cells were first incubated with NBD-maleimide conjugate, followed by a second incubation with sodium dithionite, which resulted in the quenching of fluorescent signals to background levels (data not shown.) These two results in conjunction indicate that fluorescence is due to the incorporation of the sulfhydryl displaying d-amino acids into the bacterial peptidoglycan.
Next the incorporation profile was obtained for wild-type B. subtilis (Fig. 3D). As with other organisms, there is a size dependence in incorporation efficiency, and additional structural preferences appear to be mostly conserved. However, not all the trends were identical. For example, there is a marked difference in labeling levels between 8 and 9 in wild-type B. subtilis, a feature that is not shared by any of the other Gram-positive organisms we evaluated. The incorporation profile of a mutant B. subtilis strain lacking the gene dacA was also obtained. PBP5 (the protein product of dacA) is a PBP that possesses d,d-CP activity. PBP5 hydrolyzes the fifth position d-amino acid, which, in most organisms, is d-alanine. Overall, incorporation levels were elevated in B. subtilis (ΔdacA) ∼10-fold for most of the d-amino acid variants compared with wild-type B. subtilis. There are two possible explanations for the increase in d-amino acid incorporation. One possibility is that an increased abundance of terminal d-alanine results in higher levels of PBP TP activity and, consequently allows greater mischarging with unnatural d-amino acids. Hydrolysis of the stem peptide by PBP5 serves to trim it to a tetrapeptide, a product that is no longer a substrate for d,d-TP. Second, PBP5 may act to remove unnatural d-amino acids already incorporated onto the fifth position of the stem peptide. There is some evidence showing that the CP activity of PBP5 removes unnatural d-amino acids from the fifth position on the stem peptide (24). The use of a d-amino acid-based dipeptide displaying either an alkyne or an azido group on the side chain led to the installment of unnatural d-amino acid at the fifth position initiated at the intracellular MurF ligase. The incorporation by MurF decouples the process from competing PBP TP activity and strongly suggests that PBP5 CP activity also displays side chain promiscuity. Along these lines, we hypothesize that differences in relative incorporation between wild-type and ΔdacA B. subtilis strains may correlate with inherent PBP5 CP preferences for side chains on the terminal position of the stem peptide. For example, surface labeling of 1 is greater than 2-fold compared with 3 in wild-type B. subtilis, whereas no significant difference is observed in B. subtilis (ΔdacA). The decreased labeling of 3 in wild-type B. subtilis may reflect a decreased retention within the stem peptide because of PBP5 CP activity.
From our NBD library scan, it is evident that PBPs have distinct structural side chain preferences of the unnatural d-amino acid. Aside from the side chain, variations within the d-amino acid scaffold can be accommodated at the C terminus. Interestingly, conversion of the unnatural d-amino acid C terminus from a carboxylic acid to a carboxamide has been shown previously to block PBP TP on the modified stem peptide (41). Likewise, we have demonstrated previously that entirely unnatural changes to the C terminus can modulate labeling efficiency (42). It is conceivable that carboxamide modification also renders the stem peptide resistant to CP, a feature that would result in elevated labeling levels in wild-type strains that express PBP5. We set out to establish the effect of d-amino acid amidation on additional PBP-null B. subtilis strains (Fig. 4). Clearly, the simple amidation of the C terminus led to larger variations in incorporation on a PBP-dependent manner, a finding that supports our previous findings. The relative incorporation (to wild-type cells) also revealed key differences in how individual PBPs may be processing amidated d-amino acids. Amidation of the d-amino acid caused a decrease in labeling in cells deficient of PbpH, contrary to the carboxylic acid variant.
FIGURE 4.
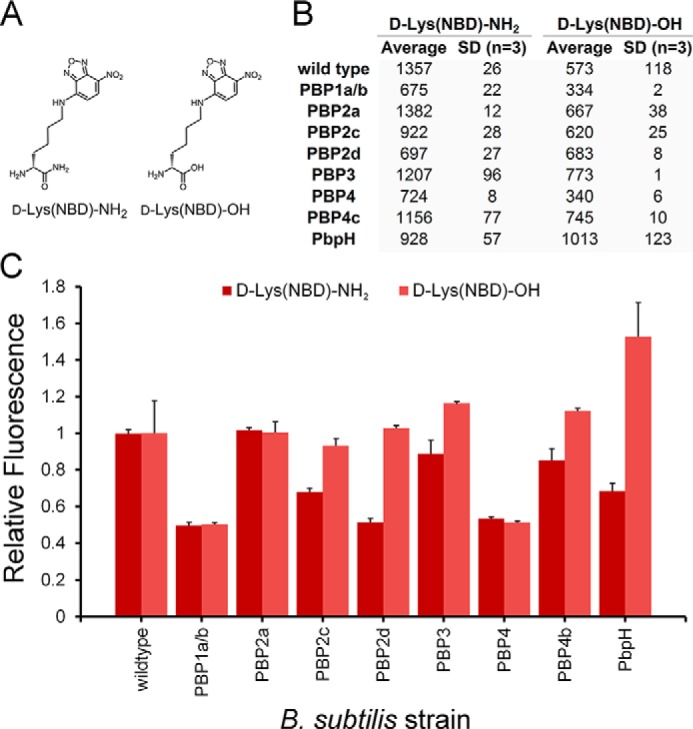
A, chemical structures of d-Lys(NBD)-OH and d-Lys(NBD)-NH2. B, B. subtilis cells were labeled with the panel of NBD-conjugated d-amino acids. Cells were incubated overnight at 100 μm and analyzed by flow cytometry. Data are represented as mean ± S.D. (n = 3). C, data are represented as mean relative to the wild type + S.D. (n = 3).
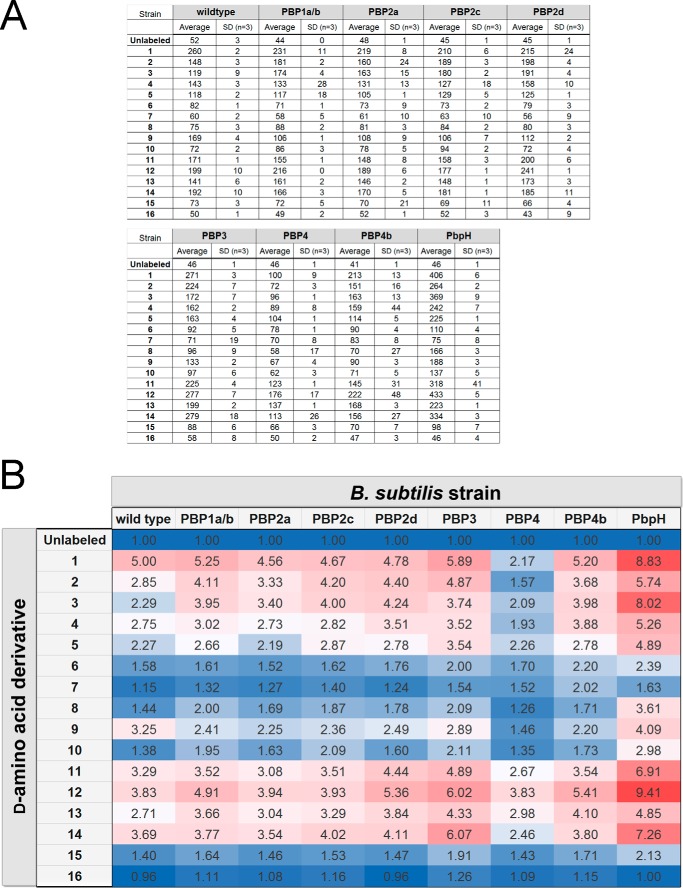
Having established comprehensive incorporation profiles for a number of organisms, we next sought to apply our d-amino acid library toward the activity profiling of individual PBP with TP activity in B. subtilis. We anticipated that the fluorescently labeled d-amino acids could form the basis of a class of chemical probes that reveal insights into PBP activity. Surprisingly, to date, the specific PBP responsible for TP activity has not been established conclusively. In B. subtilis, it has been shown recently in vitro that PBP1a/b can catalyze the installment of unnatural d-amino acids onto stem peptides (41). Although instrumental in deciphering the mechanism of unnatural d-amino acid incorporation, it is unclear whether the same PBP would be the primary protein responsible for this process in live bacteria. In all, eight single PBP TP deletion mutant B. subtilis strains were subjected to evaluation. Overall, it appears that most of the PBP-null strains result in a minimal change in labeling across all 16 d-amino acid variants (Fig. 5). A closer inspection reveals subtle but significant differences in several strains that may reflect distinctions in TP active sites. In the case of the PBP2c-null incorporation profile, there is increased incorporation of 2 and 3 but lower incorporation of 9 relative to the wild-type strain. PBP3-null cells labeled considerably better with 12 and 14 but displayed lower incorporation of 9. Interestingly, significant increases in incorporation were observed upon deletion of PBP4b or PbpH. Perhaps the deletion of a PBP leads to a compensatory activity for a complementary PBP whose TP activity is able to more efficiently mischarge with unnatural d-amino acids. Most importantly, a prominent decrease in incorporation across the board was observed for PBP4-null cells. From these results, we propose that PBP4 is the primary protein responsible for the incorporation of unnatural d-amino acids on B. subtilis peptidoglycan stem peptides.
FIGURE 5.
A, mutant strains of B. subtilis lacking specified PBPs were labeled with NBD-conjugated d-amino acids. Cells were incubated for 4 h at stationary phase at 100 μm and analyzed by flow cytometry. Data are represented as mean ± S.D. (n = 3). B, heat map of labeling a series of B. subtilis mutants with the NBD-conjugated d-amino acids. (Data were derived from A). The heat map of incorporation shows relative fluorescence levels to unlabeled cells. Red cells indicate elevated incorporation, and blue cells indicate lower incorporation.
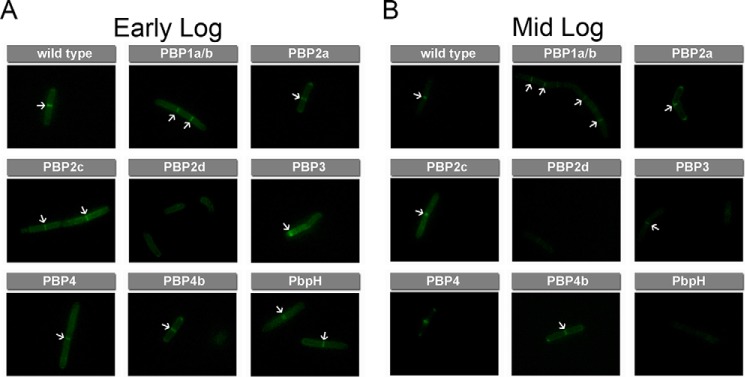
Cell wall biosynthesis and cell division require the orchestration of PBPs within the cell surface. Localization of PBPs and defined regulation of their activity is crucial for the proper assembly of newly synthesized peptidoglycan and septation (41). We hypothesized that we could gain insights into the temporal control of PBP TP activity by leveraging the combination of fluorescently labeled d-amino acids with the panel of PBP-null B. subtilis strains. During various stages of cell growth and division, there may be distinct cellular requirements in terms of cell wall-modifying PBPs. Regulation of PBPs may not be found at the DNA/RNA level and, therefore, may not be trackable by traditional techniques (e.g. RT-PCR). Instead, we propose that d-amino acid activity-based probes could be used to monitor potentially subtle changes in PBP TP activities. The quantification of probe incorporation at different growth stages of B. subtilis should allow for sensitive measurements PBP TP activity. We sampled B. subtilis at three distinct stages (early log, mid-log, and stationary phase) by a pulse incubation with d-Lys(FITC)-OH, another suitable fluorescent probe for monitoring peptidoglycan biosynthesis (28, 29). The change in the fluorophore from NBD to FITC did not appreciably alter relative incorporation within the panel of PBP mutants, whereas, at the same time, it allowed for better quantification of signals in comparison with NBD because of the increased brightness provided by the FITC fluorophore (Fig. 6). For wild-type B. subtilis, there is substantially greater incorporation at the early log phase. By the mid-log stage, all labeling levels are approximately halved. Finally, there is another 4-fold reduction at stationary phase (Fig. 7). The relative labeling (to wild-type cells) within each stage reveals PBP-dependent patterns (Fig. 8). For most mutants, there is a relative decrease in labeling at stationary phase compared with the two other phases. For example, PBP1a/b and PBP2c mutant cells label half as efficiently in the stationary phase, whereas there is almost no distinguishable difference between early and mid-log phases. Other mutants (e.g. PBP2d and PBP4b) damped the difference across growth phases, an indication that they may not play a significant role in PBP TP. Primarily septal labeling was observed for all B. subtilis strains, including the wild type (Fig. 9A). A slight deviation from this pattern was observed in PBP2d-null cells at early log phase, in which labeling was observed throughout the entire cell surface at approximately equal intensity, with no punctate fluorescence at the septal interface. Analysis of the labeling at mid-log phase revealed an intriguing switch in labeling for PBP4-null cells (Fig. 9B). PBP TP activity in PBP4-null cells is observed through the entire cell at early log phase. However, by mid-log phase, the labeling is heavily concentrated at the septal regions. These results suggest temporal control in PBP TP activity that is linked with the growth phase of the cells.
FIGURE 6.
A, chemical structures of d-Lys(FITC)-OH. B, B. subtilis cells were labeled with the panel of NBD-conjugated d-amino acids. Cells were incubated for 4 h at stationary phase at 100 μm and analyzed by flow cytometry. Data are represented as mean ± S.D. (n = 3).
FIGURE 7.
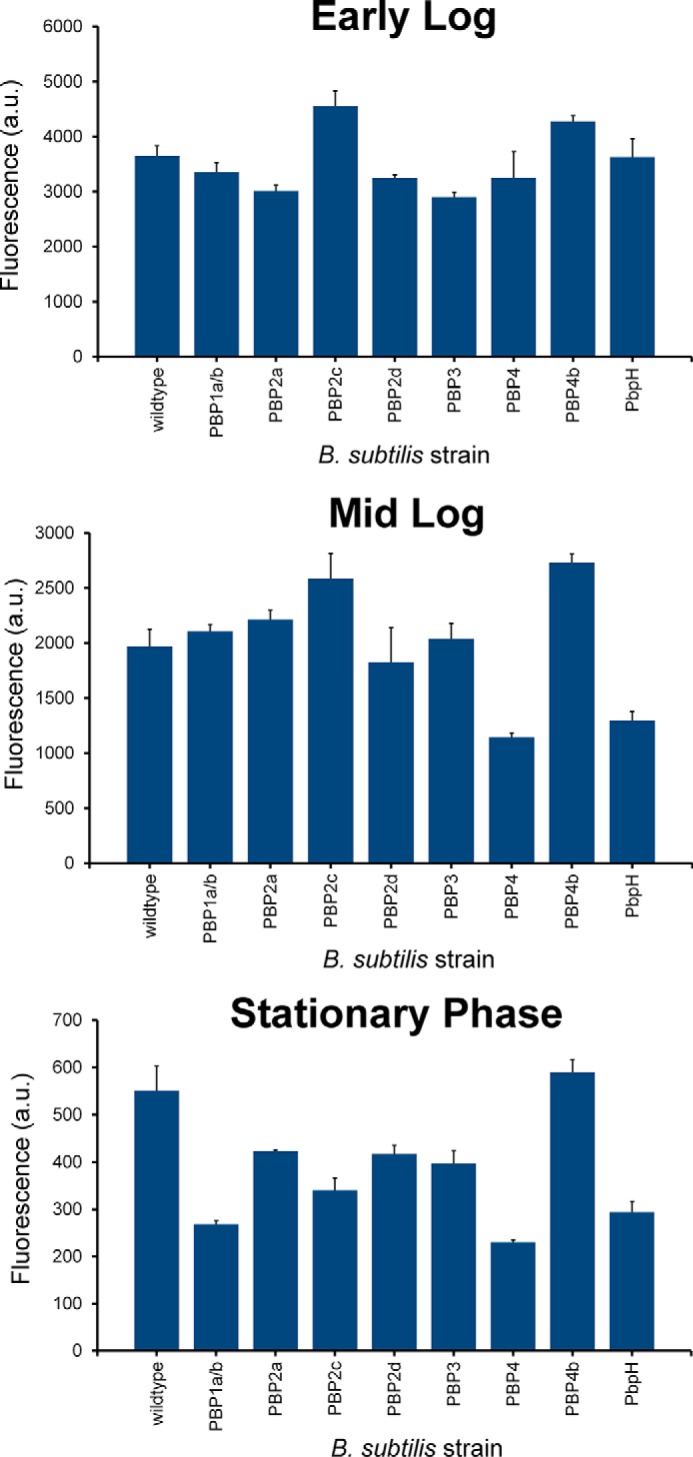
Pulse labeling of mutant strains of B. subtilis lacking specified PBPs with d-Lys(FITC)-OH. Cells were incubated for 15 min at specified phases at 1 mm and analyzed by flow cytometry. Data are represented as mean ± S.D. (n = 3).
FIGURE 8.
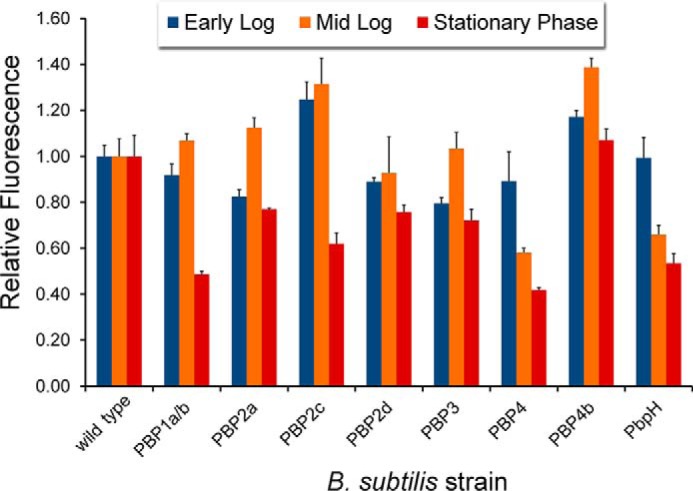
Pulse labeling of mutant strains of B. subtilis lacking specified PBPs with d-Lys(FITC)-OH. Cells were incubated for 15 min at specified phases at 1 mm and analyzed by flow cytometry. Data are represented as mean relative to wild type + S.D. (n = 3).
FIGURE 9.
A and B, pulse labeling of mutant strains of B. subtilis lacking specified PBPs with d-Lys(FITC)-OH. Cells were incubated for 15 min at early log (A) and mid-log phase (B) at 1 mm and analyzed by fluorescence microscopy. Arrows are indicative of increased septal labeling.
Discussion
In the short time since the initial discovery that unnatural d-amino acids can be incorporated onto bacterial peptidoglycan, this mode of surface remodeling has spurred peptidoglycan probes and potentially unique antimicrobial strategies. Our scan represents the most comprehensive analysis of d-amino acid incorporation to date, and it may give unique insights into the physiological function of this remodeling process. We anticipate that this thorough evaluation of the incorporation of unlabeled d-amino acids can shed some light onto aspects that control peptidoglycan remodeling. Although there are a few examples of d-amino acids being generated and utilized in eukaryotic organisms, it was believed that its existence in bacteria was limited to being a building block for non-ribosomal peptide synthesis and the core component of the peptidoglycan. The discovery that many bacterial species are capable of generating diverse d-amino acids that are expelled into the surrounding medium suggests that this process may have specific physiological consequences (43, 44). However, there has not been any conclusive physiological role assigned to peptidoglycan remodeling via unnatural d-amino acids.
A striking feature of the replacement of bacterial surface-bound d-alanine with unnatural d-amino acids is the vast promiscuity displayed by PBPs in tolerating entirely unnatural side chains. However, the chemical space and physical features of the side chain have not been mapped out systematically. In this report, we assembled a series of 16 d-amino acid variants with diverse side chains. For all the variants, the small fluorophore NBD was covalently conjugated onto an amino group. The side chains were chosen to encompass a diverse set of chemical space by including structural elements that vary in length, bulkiness, flexibility, polarity, and charge. With the NBD-modified d-amino acids in hand, we measured incorporation levels using flow cytometry in a number of bacteria, including B. subtilis, S. epidermidis, E. faecalis, S. aureus, and E. coli. In addition to serving as guides for the design of unnatural side chains of d-amino acids, our scan represents the most comprehensive analysis of d-amino acid incorporation to date, and it may give unique insights into the physiological function of this remodeling process. In addition, we evaluated labeling with unnatural d-amino acids for a series of PBP-null B. subtilis mutants and found specific mutations that modulate the incorporation.
Author Contributions
J. M. F. designed, performed, and analyzed all experiments. M. M. P. conceived and coordinated the study and wrote the paper. D. K. provided the mutant strains and wrote the paper. All authors analyzed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Anthony Iacoviello for assistance with conducting some of the labeling experiments.
This work was supported in part by National Institutes of Health Grant R01-GM113172 (to D. K.) and by Lehigh University. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Experimental Procedures.
- PBP
- penicillin binding protein
- TP
- transpeptidase/transpeptidation
- CP
- carboxypeptidase/carboxypeptidation
- NBD
- 4-chloro-7-nitro-2,1,3-benzoxadiazole.
References
- 1.Vollmer W., Blanot D., and de Pedro M. A. (2008) Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32, 149–167 [DOI] [PubMed] [Google Scholar]
- 2.Frère J. M., and Joris B. (1985) Penicillin-sensitive enzymes in peptidoglycan biosynthesis. Crit. Rev. Microbiol. 11, 299–396 [DOI] [PubMed] [Google Scholar]
- 3.van Heijenoort J. (2001) Recent advances in the formation of the bacterial peptidoglycan monomer unit. Nat. Prod. Rep. 18, 503–519 [DOI] [PubMed] [Google Scholar]
- 4.Bugg T. D., and Walsh C. T. (1992) Intracellular steps of bacterial cell wall peptidoglycan biosynthesis: enzymology, antibiotics, and antibiotic resistance. Nat. Prod. Rep. 9, 199–215 [DOI] [PubMed] [Google Scholar]
- 5.Scheffers D. J., and Pinho M. G. (2005) Bacterial cell wall synthesis: new insights from localization studies. Microbiol. Mol. Biol. Rev. 69, 585–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barreteau H., Kovac A., Boniface A., Sova M., Gobec S., and Blanot D. (2008) Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol. Rev. 32, 168–207 [DOI] [PubMed] [Google Scholar]
- 7.Typas A., Banzhaf M., Gross C. A., and Vollmer W. (2012) From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 10, 123–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sauvage E., Kerff F., Terrak M., Ayala J. A., and Charlier P. (2008) The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 32, 234–258 [DOI] [PubMed] [Google Scholar]
- 9.Spratt B. G. (1975) Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc. Natl. Acad. Sci. U.S.A. 72, 2999–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waxman D. J., and Strominger J. L. (1983) Penicillin-binding proteins and the mechanism of action of β-lactam antibiotics. Annu. Rev. Biochem. 52, 825–869 [DOI] [PubMed] [Google Scholar]
- 11.Loskill P., Pereira P. M., Jung P., Bischoff M., Herrmann M., Pinho M. G., and Jacobs K. (2014) Reduction of the peptidoglycan crosslinking causes a decrease in stiffness of the Staphylococcus aureus cell envelope. Biophys. J. 107, 1082–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arthur M., and Courvalin P. (1993) Genetics and mechanisms of glycopeptide resistance in enterococci. Antimicrob. Agents Chemother. 37, 1563–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allen N. E., LeTourneau D. L., and Hobbs J. N. Jr. (1997) Molecular interactions of a semisynthetic glycopeptide antibiotic with d-alanyl-d-alanine and d-alanyl-d-lactate residues. Antimicrob. Agents Chemother. 41, 66–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fritz J. H., Ferrero R. L., Philpott D. J., and Girardin S. E. (2006) Nod-like proteins in immunity, inflammation and disease. Nat. Immunol. 7, 1250–1257 [DOI] [PubMed] [Google Scholar]
- 15.Reed P., Atilano M. L., Alves R., Hoiczyk E., Sher X., Reichmann N. T., Pereira P. M., Roemer T., Filipe S. R., Pereira-Leal J. B., Ligoxygakis P., and Pinho M. G. (2015) Staphylococcus aureus survives with a minimal peptidoglycan synthesis machine but sacrifices virulence and antibiotic resistance. PLoS Pathog. 11, e1004891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McPherson D. C., and Popham D. L. (2003) Peptidoglycan synthesis in the absence of class A penicillin-binding proteins in Bacillus subtilis. J. Bacteriol. 185, 1423–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daniel R. A., Williams A. M., and Errington J. (1996) A complex four-gene operon containing essential cell division gene pbpB in Bacillus subtilis. J. Bacteriol. 178, 2343–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siegrist M. S., Aditham A. K., Espaillat A., Cameron T. A., Whiteside S. A., Cava F., Portnoy D. A., and Bertozzi C. R. (2015) Host actin polymerization tunes the cell division cycle of an intracellular pathogen. Cell Rep. 11, 499–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kocaoglu O., Calvo R. A., Sham L. T., Cozy L. M., Lanning B. R., Francis S., Winkler M. E., Kearns D. B., and Carlson E. E. (2012) Selective penicillin-binding protein imaging probes reveal substructure in bacterial cell division. ACS Chem. Biol. 7, 1746–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gautam S., Kim T., and Spiegel D. A. (2015) Chemical probes reveal an extraseptal mode of cross-linking in Staphylococcus aureus. J. Am. Chem. Soc. 137, 7441–7447 [DOI] [PubMed] [Google Scholar]
- 21.Tiyanont K., Doan T., Lazarus M. B., Fang X., Rudner D. Z., and Walker S. (2006) Imaging peptidoglycan biosynthesis in Bacillus subtilis with fluorescent antibiotics. Proc. Natl. Acad. Sci. U.S.A. 103, 11033–11038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siegrist M. S., Whiteside S., Jewett J. C., Aditham A., Cava F., and Bertozzi C. R. (2013) (D)-Amino acid chemical reporters reveal peptidoglycan dynamics of an intracellular pathogen. ACS Chem. Biol. 8, 500–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacquier N., Frandi A., Pillonel T., Viollier P. H., Viollier P., and Greub G. (2014) Cell wall precursors are required to organize the chlamydial division septum. Nat. Commun. 5, 3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liechti G. W., Kuru E., Hall E., Kalinda A., Brun Y. V., VanNieuwenhze M., and Maurelli A. T. (2014) A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature 506, 507–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lupoli T. J., Tsukamoto H., Doud E. H., Wang T. S., Walker S., and Kahne D. (2011) Transpeptidase-mediated incorporation of d-amino acids into bacterial peptidoglycan. J. Am. Chem. Soc. 133, 10748–10751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cava F., de Pedro M. A., Lam H., Davis B. M., and Waldor M. K. (2011) Distinct pathways for modification of the bacterial cell wall by non-canonical d-amino acids. EMBO J. 30, 3442–3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shieh P., Siegrist M. S., Cullen A. J., and Bertozzi C. R. (2014) Imaging bacterial peptidoglycan with near-infrared fluorogenic azide probes. Proc. Natl. Acad. Sci. U.S.A. 111, 5456–5461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuru E., Tekkam S., Hall E., Brun Y. V., and Van Nieuwenhze M. S. (2015) Synthesis of fluorescent d-amino acids and their use for probing peptidoglycan synthesis and bacterial growth in situ. Nat. Protoc. 10, 33–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuru E., Hughes H. V., Brown P. J., Hall E., Tekkam S., Cava F., de Pedro M. A., Brun Y. V., and VanNieuwenhze M. S. (2012) In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent d-amino acids. Angew. Chem. Int. Ed. Engl. 51, 12519–12523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fleurie A., Lesterlin C., Manuse S., Zhao C., Cluzel C., Lavergne J. P., Franz-Wachtel M., Macek B., Combet C., Kuru E., VanNieuwenhze M. S., Brun Y. V., Sherratt D., and Grangeasse C. (2014) MapZ marks the division sites and positions FtsZ rings in Streptococcus pneumoniae. Nature 516, 259–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monteiro J. M., Fernandes P. B., Vaz F., Pereira A. R., Tavares A. C., Ferreira M. T., Pereira P. M., Veiga H., Kuru E., VanNieuwenhze M. S., Brun Y. V., Filipe S. R., and Pinho M. G. (2015) Cell shape dynamics during the staphylococcal cell cycle. Nat. Commun. 6, 8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacKenzie D. A., Sherratt A. R., Chigrinova M., Kell A. J., and Pezacki J. P. (2015) Bioorthogonal labelling of living bacteria using unnatural amino acids containing nitrones and a nitrone derivative of vancomycin. Chem. Commun. 51, 12501–12504 [DOI] [PubMed] [Google Scholar]
- 33.Fura J. M., and Pires M. M. (2015) d-amino carboxamide-based recruitment of dinitrophenol antibodies to bacterial surfaces via peptidoglycan remodeling. Biopolymers 104, 351–359 [DOI] [PubMed] [Google Scholar]
- 34.Fura J. M., Sabulski M. J., and Pires M. M. (2014) d-amino acid mediated recruitment of endogenous antibodies to bacterial surfaces. ACS Chem. Biol. 9, 1480–1489 [DOI] [PubMed] [Google Scholar]
- 35.Sonenshein A. L., Hoch J. A., and Losick R. (1993) Bacillus subtilis and other Gram-positive bacteria: biochemistry, physiology, and molecular genetics, American Society for Microbiology, Washington, D.C. [Google Scholar]
- 36.McIntyre J. C., and Sleight R. G. (1991) Fluorescence assay for phospholipid membrane asymmetry. Biochemistry 30, 11819–11827 [DOI] [PubMed] [Google Scholar]
- 37.Mukhopadhyay K., Whitmire W., Xiong Y. Q., Molden J., Jones T., Peschel A., Staubitz P., Adler-Moore J., McNamara P. J., Proctor R. A., Yeaman M. R., and Bayer A. S. (2007) In vitro susceptibility of Staphylococcus aureus to thrombin-induced platelet microbicidal protein-1 (tPMP-1) is influenced by cell membrane phospholipid composition and asymmetry. Microbiology 153, 1187–1197 [DOI] [PubMed] [Google Scholar]
- 38.Utsui Y., and Yokota T. (1985) Role of an altered penicillin-binding protein in methicillin- and cephem-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 28, 397–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Pedro M. A., Quintela J. C., Höltje J. V., and Schwarz H. (1997) Murein segregation in Escherichia coli. J. Bacteriol. 179, 2823–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schouten J. A., Bagga S., Lloyd A. J., de Pascale G., Dowson C. G., Roper D. I., and Bugg T. D. (2006) Fluorescent reagents for in vitro studies of lipid-linked steps of bacterial peptidoglycan biosynthesis: derivatives of UDPMurNAc-pentapeptide containing d-cysteine at position 4 or 5. Mol. Biosyst. 2, 484–491 [DOI] [PubMed] [Google Scholar]
- 41.Lebar M. D., May J. M., Meeske A. J., Leiman S. A., Lupoli T. J., Tsukamoto H., Losick R., Rudner D. Z., Walker S., and Kahne D. (2014) Reconstitution of peptidoglycan cross-linking leads to improved fluorescent probes of cell wall synthesis. J. Am. Chem. Soc. 136, 10874–10877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pidgeon S. E., Fura J. M., Leon W., Birabaharan M., Vezenov D., and Pires M. M. (2015) Metabolic profiling of bacteria by unnatural C-terminated d-amino acids. Angew. Chem. Int. Ed. Engl. 54, 6158–6162 [DOI] [PubMed] [Google Scholar]
- 43.Lam H., Oh D. C., Cava F., Takacs C. N., Clardy J., de Pedro M. A., and Waldor M. K. (2009) d-amino acids govern stationary phase cell wall remodeling in bacteria. Science 325, 1552–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alvarez L., Espaillat A., Hermoso J. A., de Pedro M. A., and Cava F. (2014) Peptidoglycan remodeling by the coordinated action of multispecific enzymes. Microb. Drug Resist. 20, 190–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.