Abstract
The HAT-B enzyme complex is responsible for acetylating newly synthesized histone H4 on lysines K5 and K12. HAT-B is a multisubunit complex composed of the histone acetyltransferase 1 (Hat1) catalytic subunit and the Hat2 (rbap46) histone chaperone. Hat1 is predominantly localized in the nucleus as a member of a trimeric NuB4 complex containing Hat1, Hat2, and a histone H3-H4 specific histone chaperone called Hif1 (NASP). In addition to Hif1 and Hat2, Hat1 interacts with Asf1 (anti-silencing function 1), a histone chaperone that has been reported to be involved in both replication-dependent and -independent chromatin assembly. To elucidate the molecular roles of the Hif1 and Asf1 histone chaperones in HAT-B histone binding and acetyltransferase activity, we have characterized the stoichiometry and binding mode of Hif1 and Asf1 to HAT-B and the effect of this binding on the enzymatic activity of HAT-B. We find that Hif1 and Asf1 bind through different modes and independently to HAT-B, whereby Hif1 binds directly to Hat2, and Asf1 is only capable of interactions with HAT-B through contacts with histones H3-H4. We also demonstrate that HAT-B is significantly more active against an intact H3-H4 heterodimer over a histone H4 peptide, independent of either Hif1 or Asf1 binding. Mutational studies further demonstrate that HAT-B binding to the histone tail regions is not sufficient for this enhanced activity. Based on these data, we propose a model for HAT-B/histone chaperone assembly and acetylation of H3-H4 complexes.
Keywords: acetyltransferase, gene regulation, histone acetylation, histone chaperone, protein complex
Introduction
Histone acetylation has been linked to diverse functions including transcriptional activation, gene silencing, DNA repair, cell cycle progression, chromatin maturation and dynamics, and nucleosome assembly (1, 2). There are two main classes of enzymes responsible for acetylation of histones: type A and type B histone acetyltransferases. Type A histone acetyltransferases are nuclear enzymes that acetylate histones in the context of chromatin, whereas type B acetyltransferases are responsible for the acetylation of newly synthesized histones H3 and H4. Although histone H3 is only moderately acetylated upon its synthesis (∼30%) (3, 4), newly synthesized histone H4 is highly acetylated on lysine residues K5 (H4K5) and K12 (H4K12) (3, 5, 6). These modifications are transient, and once transported across the nucleus, the histones are deacetylated during the course of chromatin maturation (7). Although acetylation of H4K5/12 is highly conserved throughout evolution, it has been shown to be nonessential through mutagenic analysis, in which replacing these residues with arginine (mimicking the constitutively unacetylated state) has no effect on cell growth in yeast (8). Thus the evolutionarily conserved function of H4K5/K12 acetylation remains unknown.
The HAT-B2 enzyme complex is responsible for acetylating newly synthesized histone H4 on lysines K5 and K12 (9–13). HAT-B is a multisubunit complex composed of histone acetyltransferase 1 (Hat1), the catalytic subunit, and Hat2 (rbap46), a histone chaperone that increases the catalytic activity of Hat1 by ∼10-fold (14). HAT-B is the only known member of the type B histone acetyltransferases that exhibits specificity toward free H4 but is unable to acetylate nucleosomal H4 substrates (15). Previous studies have demonstrated that Hat1 is essential in mammals, because a Hat1 knock-out mouse shows neonatal lethality (16). Although HAT-B was originally identified in the cytoplasm, more recent studies demonstrate that Hat1 is predominantly localized in the nucleus as a member of a trimeric NuB4 complex containing Hat1, Hat2, and Hif1 (NASP) (9, 17, 18). The protein Hif1, similar to Hat2, is a histone H3-H4 specific chaperone and was initially identified as a component of the HAT-B complex where it has been hypothesized to be responsible for recruiting histones H3-H4 to HAT-B. Deletion experiments have shown that the loss of Hif1 in combination with K9R and K14R mutations in H3 is comparable to deletion of Hat1, which results in defects of telomeric silencing and DNA double-stranded break repair (19). The association of the histone chaperone Hif1 with HAT-B was the first direct evidence implicating a role for Hat1 in the chromatin assembly process (17). In addition to Hif1 and Hat2, Hat1 has also been shown to interact with another histone chaperone, Asf1 (anti-silencing function 1) (20, 21). Asf1 has been reported to be involved in both replication-dependent and -independent chromatin assembly where it can either assemble or disassemble chromatin in DNA replication, repair, and transcription (22–26). Significantly, unlike most enzymes that are involved in transient interactions with their substrate, Hat1 remains associated with H4 through much of histone H3-H4 assembly and deposition (3).
Although HAT-B is known to associate with both Hif1 and Asf1, the modes of histone chaperone binding to HAT-B and their effects on HAT-B acetyltransferase activity have not been studied. To elucidate the molecular roles of the Hif1 and Asf1 histone chaperones in HAT-B acetyltransferase activity, we have extensively characterized the enzymatic activity of HAT-B on histone protein substrates in the presence and absence of both the Hif1 and Asf1 chaperones. Previous studies have used a histone H4 peptide to measure the enzyme efficiency of Hat1 alone (27). We show here that the full-length histone H3-H4 heterodimer is a much better protein substrate for both HAT-B and Hat1 alone, independent of Hif1 and Asf1 binding. Mutational analysis indicates that HAT-B contacts to the histone tail regions are not sufficient for this enhanced activity. In addition, we determine that both Hif1 and Asf1 bind independently and with different stoichiometries to HAT-B/histone H3-H4 complexes, whereby Hif1 binds directly to Hat2 and Asf1 binds HAT-B through contacts with H3-H4. These data have allowed us to propose a binding mode for the interactions of Hif1 and Asf1 with the HAT-B complex and further enhance our understanding of the roles of Hat1, Hat2, Hif1, and Asf1 in H3-H4 assembly and acetylation.
Experimental Procedures
Protein Preparation
Asf1 and the full-length histone H3-H4 heterodimers were generated as described previously (25, 28). The “tail-less” histone constructs (H3 residues 45–135 and H4 residues 20–102) were generated from human H3.3 and H4 cDNA by PCR amplification followed by ligation into the BamHI/XhoI sites of Escherichia coli protein expression vector pET Duet (Novagen). Protein expression was performed as previously described (28). The gene encoding full-length Hat1, Hat2, and Hif1 proteins were subcloned from Saccharomyces cerevisiae genomic DNA. Hat1 DNA was ligated into a pFastBac vector encoding an N-terminal GST fusion protein followed by a TEV protease cleavage site. Hat2 DNA was ligated into a pFastBac vector encoding an N-terminal hexahistidine tag followed by a TEV protease cleavage site. Hif1 DNA was ligated into both a pFastBac vector with no tag and a N-terminal hexahistidine tag followed by a TEV protease cleavage site. These plasmids were then used to generate recombinant baculovirus, which were then either used to transfect or cotransfect Sf9 insect cells for protein or protein complex overexpression. Cells were harvested 48-h post-transfection/cotransfection and then subjected to lysis by sonication in PBS buffer (150 mm NaCl, 8.3 mm Na2HPO4, 1.85 mm NaH2PO4, pH 7.4) supplemented with protease inhibitor mixture. After centrifugation, the supernatant was incubated with either GST or nickel-nitrilotriacetic acid affinity resin for 1 h and poured onto a column followed by washing with 20 column volumes of PBS buffer supplemented with 20 mm imidazole (nickel-nitrilotriacetic acid purification). GST affinity columns were eluted with PBS-glutathione (20 mm glutathione) buffer and nickel-nitrilotriacetic acid columns were eluted with PBS-imidazole (250 mm imidazole) buffer. Proteins and protein complexes were then dialyzed at 4 °C overnight in the presence of TEV protease to cleave N-terminal GST and/or hexahistidine tags. Additional purification was performed the following day using a 5-ml HiTrapQ anion exchange column (in 20 mm Tris pH 7.9 buffer with a NaCl gradient from 0 to 1 m over 20 column volumes) and Superdex 200 gel filtration chromatography (in 20 mm Tris, pH 7.9, 500 mm NaCl). The protein was then concentrated to ∼200 μm using a Millipore protein concentrator, flash frozen in liquid nitrogen, and stored at −80 °C until further use.
Hat2 mutants were constructed by site-directed mutagenesis (Q5 Hot start polymerase; New England Biolabs). The mutant constructs were all verified by DNA sequencing. Protein expression was performed as described above for the wild-type proteins.
Generation of Protein Complexes
To generate the Hat2/Hif1 protein complex, N-terminal His-tagged constructs of both full-length Hat2 and Hif1 were purified separately on nickel-nitrilotriacetic acid resin as described above. These proteins were than mixed at equimolar concentrations and incubated on ice for 10 min. The complex was then subjected to Superdex 200 gel filtration chromatography (in 20 mm Tris, pH 7.9, 500 mm NaCl). The same protocol was used to determine that Asf1 is not capable of binding HAT-B in the absence of the histones.
To prepare NuB4/H3-H4, a molar excess of H3-H4 was incubated on ice with NuB4 for 10 min. This complex was then subjected to Superdex 200 gel filtration chromatography (in 20 mm Tris, pH 7.9, 500 mm NaCl). To prepare HAT-B/Asf1/H3-H4, a molar excess of Asf1/H3-H4 was incubated on ice with HAT-B for 10 min. This complex was then subjected to Superdex 200 gel filtration chromatography (in 20 mm Tris, pH 7.9, 500 mm NaCl). To prepare NuB4/Asf1/H3-H4, a molar excess of Asf1/H3-H4 was incubated on ice with NuB4 for 10 min. This complex was then subjected to Superdex 200 gel filtration chromatography (in 20 mm Tris, pH 7.9, 500 mm NaCl). These proteins were immediately used for the sedimentation equilibrium experiments discussed below.
Analytical Ultracentrifugation
Sedimentation equilibrium analytical ultracentrifugation experiments were performed at 4 °C with absorbance optics at 280 nm using a Beckman Optima XL-I analytical ultracentrifuge. We used a four-hole rotor containing six-channel centerpieces with quartz windows, spinning at 6,000, 9,000, 12,000, and 18,000 rpm. Protein samples were analyzed at A280 = 0.8, 0.4, and 0.2 in gel filtration buffer (20 mm Tris, pH 7.9, 500 mm NaCl). The data for each speed were collected in quadruplicate. The viscosity of the samples were estimated using Sedenterp (29, 30), and the most representative runs were included to calculate a theoretical molecular masses using the program HeteroAnalysis.
HAT Enzyme Assay
We employed a previously described enzyme assay with 14C-labeled ACoA as a radioactive tracer (31). All assays were performed with 100 nm enzyme at 20 °C for 20 min in 20 mm Tris (pH 7.9) and 500 mm NaCl at saturating concentrations of 14C-labeled ACoA (500 μm) enzyme in a 50-μl reaction volume. A time course was used to determine the linear range of the enzyme in the presence of saturating substrate (500 μm H3-H4 histones or an H4 peptide containing residues 1–19). The reaction was quenched by spotting 20 μl of the solution onto P81 negatively charged ion exchange paper squares (Millipore) and immediately washed with 10 mm HEPES pH 7.5 buffer. Washes were repeated three times for 1 min to remove excess 14C-ACoA. The papers were then dipped in acetone and allowed to dry before being placed into vials containing 5 ml of scintillation fluid. The samples were analyzed using a Beckman liquid scintillation counter. The Km and Vmax of the histone and peptide substrates were then calculated by varying the concentration of substrate. All reactions were performed in duplicate, and radioactive counts were converted to molar units using a standard curve generated by spotting known concentrations of 14C-ACoA onto P81 paper and allowing the papers to dry before being placed into scintillation fluid for counting.
Results
Hif1 and Asf1 Bind Independently and with Different Stoichiometries to HAT-B/Histone H3-H4 Complexes
A previous report employing Δhat2 yeast strains demonstrated that Hat2 is essential for Hif1 association with Hat1, although this study did not address whether this association was mediated by direct Hif1 binding to Hat1, Hat2, or both proteins (19). To elucidate the interactions that Hif1 makes with Hat1 and Hat2, we purified recombinant Hat1, Hat2, and Hif1 and performed size exclusion chromatography (SEC) to determine whether Hif1 can bind directly to either Hat1 or Hat2 alone or requires both Hat1 and Hat2 for binding. In agreement with previous experiments, we demonstrated that Hif1 is unable to bind Hat1 in the absence of Hat2 (data not shown). However, when Hat2 was incubated with excess Hif1 and SEC was performed, there was a shift in elution volume to higher molecular mass. SDS-PAGE analysis revealed that Hif1 and Hat2 comigrate to form a stable protein complex (Fig. 1A). These experiments were performed at 0.5 m NaCl, and therefore nonspecific electrostatic interactions are unlikely. These data indicated that Hif1 can bind to Hat2 in the absence of Hat1, suggesting that in the NuB4 complex, Hif1 binds directly to Hat2 with little or no interaction with Hat1.
FIGURE 1.
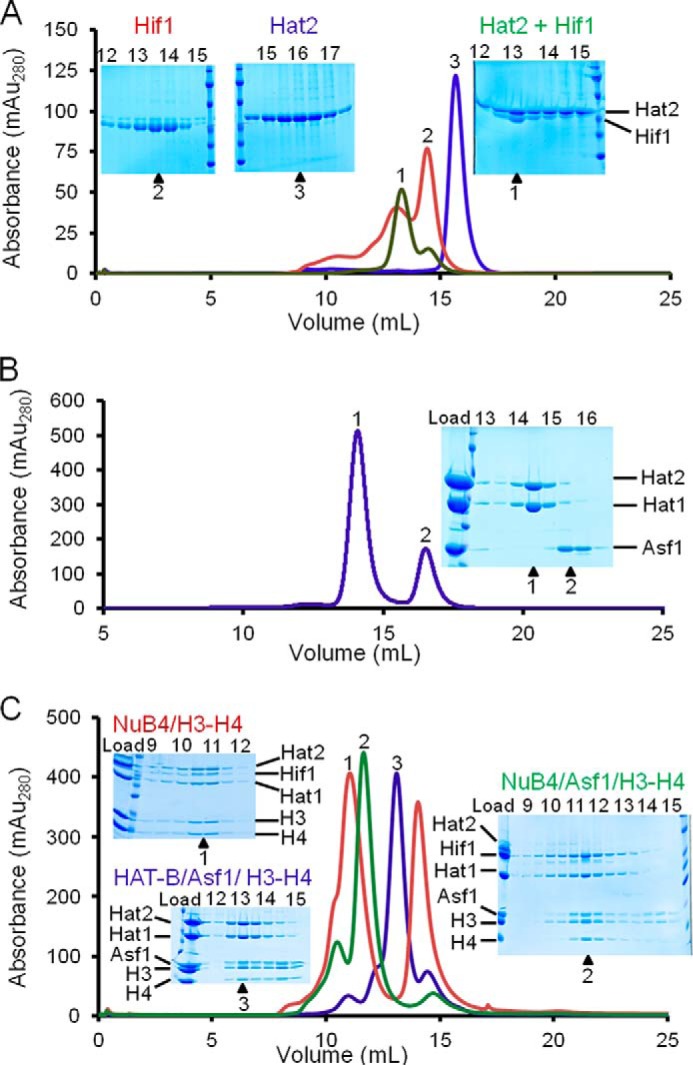
Preparation of Hat1, histone chaperones, and their complexes. Proteins and complexes are analyzed by SEC at protein/complex concentrations of between 1 and 10 μm, and corresponding SDS-PAGE analysis and elution volumes (insets) are indicted. A, Hif1 binding to Hat2 with SEC chromatograms overlaid and color-coded for Hif1 (red), Hat2 (blue), and Hat2 + Hat1 (green) with the corresponding SDS-PAGE analysis. B, SEC of a preformed complex of Asf1, Hat1, and Hat2 with the corresponding SDS-PAGE analysis. C, Hif1 and Asf1 binding modes to HAT-B (Hat1/Hat2) analyzed by SEC and corresponding SDS-PAGE analysis. The following complexes were preformed and analyzed by SEC and SDS-PAGE, and overlaid chromatographs are color-coded as indicated. Trace 1, HAT-B with Hif1 and H3-H4 (NuB4/H3-H4, red); trace 2, HAT-B with Hif1, Asf1, and H3-H4 (NuB4/Asf1/H3-H4, green); trace 3, HAT-B with Asf1 and H3-H4 (HAT-B/Asf1/H3-H4, blue). We note that Hif1 staining on SDS-PAGE appears substoichiometric with other associated Hat1 complexes, suggesting that Hif1 either stains anomalously weakly or partially dissociates from the complex during SEC.
To determine the mode of binding of the histone chaperone Asf1 to the HAT-B complex, we performed similar SEC experiments to determine whether Asf1 was capable of binding either Hat1 alone, Hat2 alone or requires both Hat1 and Hat2 for binding. These experiments were all performed at 0.5 m NaCl to avoid nonspecific electrostatic interactions. When Asf1 was incubated with either Hat1 or Hat2 alone or the HAT-B complex (containing both Hat1 and Hat2) followed by SEC, a stable complex with Asf1 could not be detected (Fig. 1B). In contrast, when HAT-B and Asf1 were incubated in the presence of the H3-H4 histone complex, followed by SEC, the results show that a stable interaction did occur (Fig. 1C). These data indicate that under these conditions, Asf1 does not directly interact with either Hat1 or Hat2 and indeed is only involved in the HAT-B complex through contacts made to the histones.
Although Hif1 and Asf1 can bind to the HAT-B/H3-H4 histone complex, it was not known whether Hif1 and Asf1 can bind independently or simultaneously to HAT-B. To investigate this issue, we analyzed HAT-B/histone chaperone complexes using SEC. We formed the following complexes: 1) HAT-B with Asf1 and H3-H4 (HAT-B/Asf1/H3-H4), 2) HAT-B with Hif1 and H3-H4 (NuB4/H3-H4), and 3) the six-protein complex HAT-B with Hif1, Asf1, and H3-H4 (NuB4/Asf1/H3-H4). Each of these complexes were formed by generating an equal molar amount of purified recombinant Hat1, Hat2, and/or Hif1 and mixing either the HAT-B or NuB4 complexes with an excess of either Asf1/H3-H4 or an excess of H3-H4. After a 10-min incubation, the samples were run on SEC and SDS-PAGE analysis was performed. These data indicate that all of the combinations of protein complexes can be stably formed in vitro (Fig. 1C). Interestingly, there was an obvious shift in elution volume to higher molecular mass in the five-protein complex without Asf1 (NuB4/H3-H4) compared with the six-protein complex containing Asf1 (NuB4/Asf1/H3-H4). This apparent higher molecular mass was possibly due to the addition of a second copy of Hif1/H3-H4 being present in the complex, and this apparent higher molecular mass was also observed in the analytical ultracentrifugation (AUC) experiments discussed in detail below.
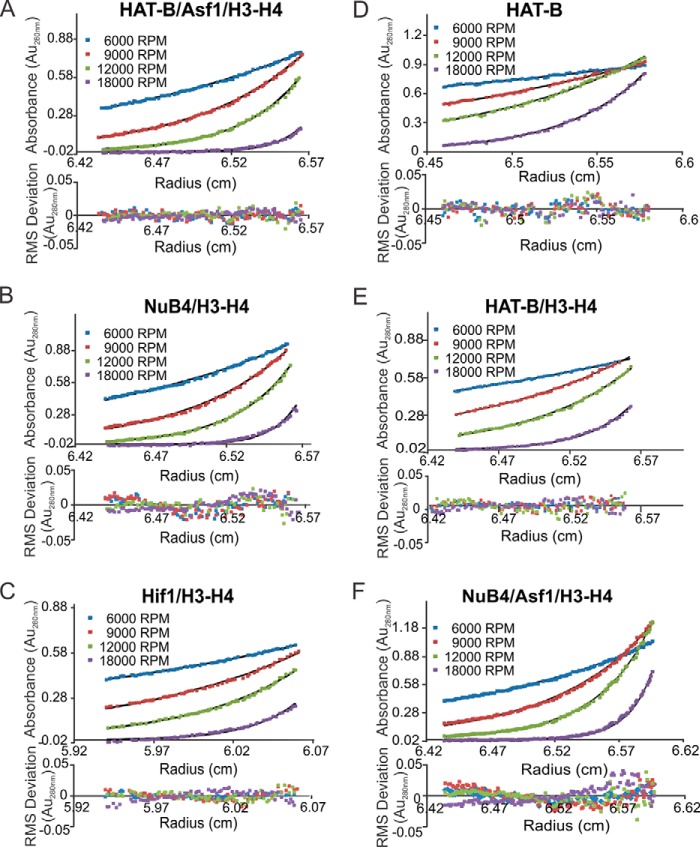
The stoichiometry of HAT-B complexes with histone chaperones has not been investigated. To address this issue, we performed AUC using sedimentation equilibrium analysis (Fig. 2). The five protein HAT-B/Asf1/H3-H4 complex has a calculated molecular mass of 130 kDa (Fig. 2A and Table 1). This number is very close to the theoretical molecular mass of 135 kDa assuming a 1:1:1:1:1 stoichiometric interaction. The five-protein NuB4/H3-H4 complex has a calculated molecular mass of 240 kDa (Fig. 2B and Table 1). Significantly, the calculated molecular mass is higher than the theoretical molecular mass of 159 kDa for a stoichiometry of 1:1:1:1:1. However, it is possible that there is an alternate stoichiometric arrangement that would also be in agreement with an early elution volume during the SEC experiments. For example, two copies of Hif1 bound to two copies of the H3-H4 dimer would allow for a predicted molecular mass of 235 kDa, which is much closer to the experimentally calculated molecular mass. This proposed stoichiometry is indeed in agreement with previous experiments performed on the human ortholog of Hif1 (sNASP) that used AUC to show sNASP homodimerizes and binds two copies of H3-H4 heterodimers (21). To verify this, we performed sedimentary equilibrium experiments on Hif1 bound to histones H3-H4. These data show a calculated molecular mass of 135 kDa (Fig. 2C and Table 1), which is very close to the theoretical molecular mass of two copies of Hif1 binding to two copies of the histones H3-H4 (147 kDa). In addition, when AUC performed on HAT-B alone (Fig. 2D and Table 1) or HAT-B with the H3-H4 histone complex (Fig. 2E and Table 1), the calculated molecular masses all agree with a 1:1 stoichiometry of Hat1 and Hat2. These data confirm that only in the presence of Hif1 do we observe a molecular mass that corresponds to higher stoichiometric configurations. Taken together, these experiments are consistent with the model that the five-protein NuB4/H3-H4 complex contains Hat1, Hat2, and two copies of Hif1 that bind to two H3-H4 histone heterodimers. The six-protein NuB4/Asf1/H3-H4 complex (Fig. 2F and Table 1) has an experimental molecular mass that ranges between 179 kDa (0.1–0.2 A280) and 205 kDa at higher concentration (0.3–0.9 A280). The calculated molecular mass at low A280 is almost identical to the theoretical molecular mass of 178 kDa assuming a 1:1:1:1:1:1 stoichiometric interaction. The calculated molecular mass at higher A280 could be indicative of self-association at higher protein concentrations, or it is possible there was an equilibrium between multiple protein complexes when all six proteins are present. For example, if an equilibrium existed between NuB4/H3-H4 (235 kDa) and NuB4/Asf1/H3-H4 (179 kDa), the apparent molecular mass might represent an average between the two species (207 kDa), which is very close to the experimental molecular mass at higher protein concentrations (205 kDa). Furthermore, we observe a higher molecular mass species on the size exclusion chromatogram of NuB4/Asf1/H3-H4, indicating the presence of multiple species. In fact, this higher molecular mass species elutes at the same volume as NuB4/H3-H4 (Fig. 1C).
FIGURE 2.
Analytical ultracentrifugation sedimentation equilibrium analysis of Hat1 protein complexes. Sedimentation equilibrium analytical ultracentrifugation experiments were performed at complex absorbances of 0.2, 0.5, and 0.8 Au280 nm for A–E and 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, and 0.9 Au280 nm for F, with centrifugations speeds of 6,000 rpm (blue), 9,000 rpm (red), 12,000 rpm (green), and 18,000 rpm (purple). Global fits to the data were carried out using the program heteroanalysis to generate sedimentation profiles (top panels) and the corresponding residuals (bottom panels). Molecular masses of single species complexes were determined using an ideal fitting model on the data. The model fits are shown for each centrifugation speed at one representative concentration (0.5 Au280 nm) as indicated for the following complexes: HAT-B/Asf1/H3-H4 (A), NuB4/H3-H4 (B), Hif1/H3-H4 (C), HAT-B (D), HAT-B/H3-H4 (E), and NuB4/Asf1/H3-H4 (F).
TABLE 1.
Summary of analytical ultracentrifugation sedimentation equilibrium data of protein complexes
| Complex | Theoretical mass | Experimental mass | Average root mean square deviation | Proposed stoichiometry | Molecular mass of proposed |
|---|---|---|---|---|---|
| Da | Da | Au280 nm | Da | ||
| HAT-B | 88,900 | 84,700 | 0.00843 | Hat1 (1), Hat2 (1) | 88,900 |
| HAT-B/H3-H4 | 116,000 | 126,000 | 0.00712 | Hat1 (1), Hat2 (1), H3-H4 (1/2) | 116,000 |
| Hif1/H3-H4 | 73,500 | 135,000 | 0.00609 | Hif1 (2), H3-H4 (2) | 147,000 |
| NuB4 | 136,000 | 131,000 | 0.00573 | Hat1 (1), Hat2 (1), Hif1 (1) | 136,000 |
| HAT-B/Asf1/H3-H4 | 135,000 | 130,000 | 0.00592 | Hat1 (1), Hat2 (1), Asf1 (1), H3-H4 (1) | 135,000 |
| NuB4/H3-H4 | 159,000 | 240,000 | 0.00890 | Hat1 (1), Hat2 (1), Hif1 (2), H3-H4 (2) | 236,000 |
| NuB4/Asf1/H3-H4 | 179,000 | 179,000/206,000 | 0.00371/0.00411 | Hat1 (1), Hat2 (1), Hif1 (1), Asf1 (1), H3-H4 (1) | 179,000 |
Taken together, these data demonstrate that the histone chaperones Asf1 and Hif1 can bind to HAT-B either independently or simultaneously. In addition, it appears that when Asf1 is present, it exists as a monomer bound to one H3-H4 histone heterodimer. In contrast, the data presented here and by others (21) demonstrate that two copies of Hif1 bind to two copies of H3-H4 histones (either two heterodimers or one heterotetramer) within the NuB4/H3-H4 complex.
Binding of the Hif1 and Asf1 Histone Chaperones Does Not Significantly Influence HAT-B-mediated Histone H4 Acetylation Activity
Previous studies have demonstrated that Hat2 association significantly increasing the catalytic efficiency of Hat1. These experiments were performed by measuring the activity of wild-type and Δhat2 yeast cells on a histone H4 substrate. These data demonstrate that there is an approximately 10-fold enhancement in the overall activity when wild-type cells are compared with Δhat2 yeast strains (19). Additionally, there were conflicting data for how Hif1 affects activity of Hat1 where one study showed Δhif1 yeast strains exhibit no change in overall activity toward a histone H4 substrate in vivo (19), whereas a second study demonstrated that the addition of the human orthologs NASP to the HAT-B complex enhanced the activity of Hat1 toward H4 (21). Furthermore, no information has been reported on how the histone chaperone Asf1 affects the catalysis of Hat1. To more quantitatively determine the roles of both Hif1 and Asf1 on the enzymatic activity of Hat1, we performed extensive Michaelis-Menten enzyme kinetic experiments using a histone H3-H4 heterodimer substrate.
With recombinant Hat1, HAT-B (Hat1 and Hat2), NuB4 (HAT-B and Hif1), and histone H3-H4 heterodimers on hand, an activity assay using 14C-labeled ACoA as a radioactive tracer was employed to measure the enzyme activity of 100 nm Hat1, HAT-B, or NuB4 against varying concentrations of H3-H4 in the presence of saturating (500 μm) 14C isotopically labeled ACoA (Fig. 3A). As expected, the addition of Hat2 to Hat1 in the HAT-B complex enhanced the catalytic efficiency (kcat/Km) by ∼4-fold. This increase in efficiency was caused by a modest increase in Km for histones (from 11 μm for Hat1 alone to 18 μm for HAT-B) and a more significant increase in kcat (from 16 min−1 for Hat1 alone to 85 min−1 for HAT-B). The addition of Hif1 to the HAT-B complex resulted in essentially no change in the Km for histones (from 18 μm for HAT-B alone to 19 μm for NuB4) and a modest increase in the kcat (from 85 min−1 for HAT-B alone to 94 min−1 for NuB4), resulting in no overall change in catalytic efficiency (4.9 μm−1 min−1 for both HAT-B and NuB4). Together, these studies demonstrate that Hif1 binding does not significantly influence the catalytic efficiency of the HAT-B complex toward H4 acetylation.
FIGURE 3.
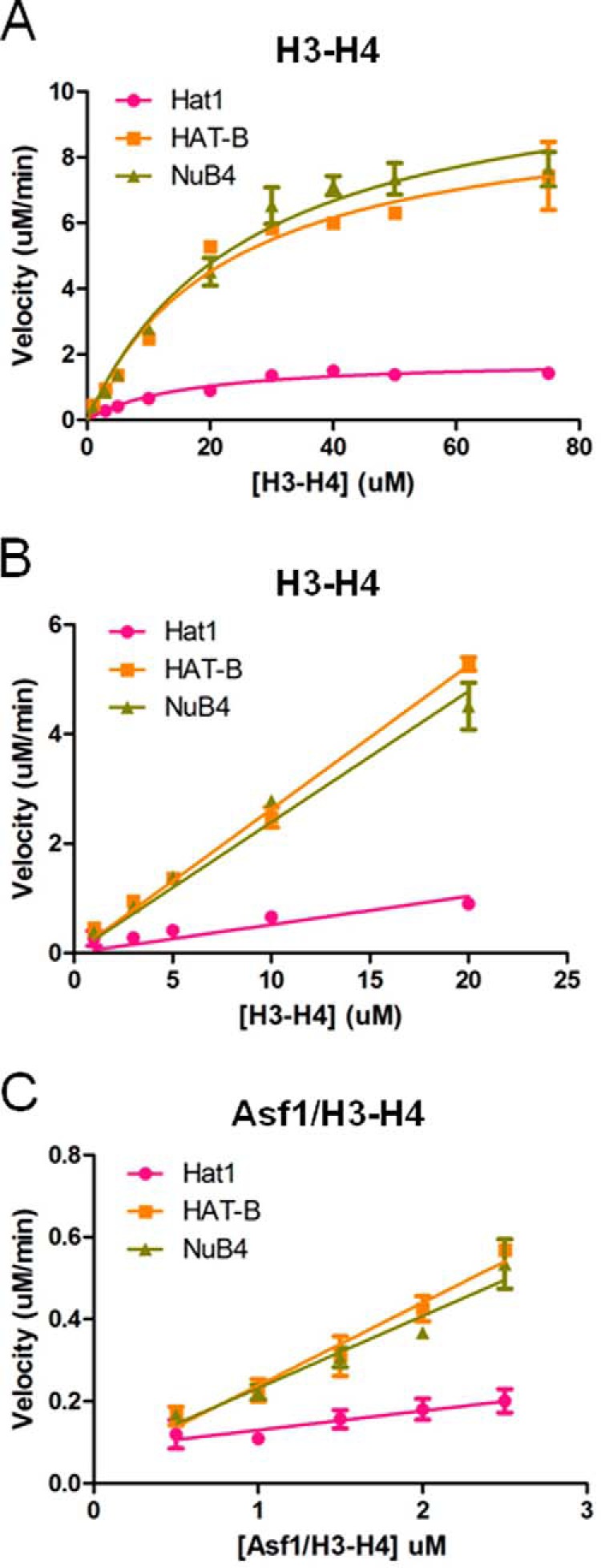
Activity of Hat1 and Hat1-containing complexes as a function of Hif1 and Asf1 histone chaperones. Enzyme kinetic analysis comparing the activity of Hat1 (red), HAT-B (yellow), and NuB4 (green) on H3-H4 substrate (top and middle panels) and Asf1/H3-H4 substrate (bottom panels). A, the data for Hat1 or Hat1-containing complexes with varying concentrations of H3-H4 (1–75 μm) are fit to Michaelis-Menten kinetics (GraphPad Prism 5) to calculate steady state parameters. B, the data for Hat1 or Hat1-containing complexes varying amounts of H3-H4 (1–20 μm) are fit to the linear portion of the rate data (GraphPad Prism 5) to calculate kcat/Km. C, the data for Hat1 or Hat1-containing complexes with varying concentrations of Asf1/H3-H4 (0.5–2.5 μm)) are fit to the linear portion of the rate data (GraphPad Prism 5) to calculate kcat/Km. All reactions were performed using 100 nm enzyme at saturating levels of 14C-labeled acetyl CoA (500 μm), and all measurements were carried out in duplicate.
To determine the role of Asf1 on enzyme activity, we measured the kcat/Km for Hat1, HAT-B, or NuB4 using varying concentrations of the substrate Asf1 bound to histones H3-H4 in the presence of saturating 14C-ACoA. Because we were unable to produce large enough quantities of Asf1 bound to histones H3-H4 to do a complete Km and kcat analysis, we measured the catalytic efficiency (kcat/Km) using the slope of the linear region in the Michaelis-Menten plot. To directly compare Asf1/H3-H4 to H3-H4 alone as substrates for Hat1, HAT-B, and NuB4 we analyzed only at the linear region of both plots (Fig. 3, B and C). These data indicate that the addition of Asf1 has a modest effect on enzyme efficiency (2.0 μm−1 min−1 for HAT-B against full-length H3-H4 compared with 2.6 μm−1 min−1 for HAT-B against Asf1/H3-H4) (Table 2). Based on this observation, we conclude that Asf1 binding does not contribute to HAT-B-mediated histone H4 acetylation. This is consistent with a more important role for Asf1 in mediating histone deposition by the HAT-B complex.
TABLE 2.
Summary of measured steady state kinetic parameters for Hat1 and complexes
ND, not determined.
| Complex | Vmax | Km | kcat | kcat/Km |
|---|---|---|---|---|
| μm/min−1 | μm | min−1 | μm−1 min−1 | |
| Substrate H3-H4 | ||||
| Hat1 | 1.59 ± 0.12 | 11.1 ± 3.2 | 15.9 ± 1.2 | 1.43 ± 0.39 |
| HAT-B | 8.56 ± 0.52 | 17.6 ± 3.3 | 85.3 ± 5.2 | 4.86 ± 1.56 |
| NuB4 | 9.44 ± 0.65 | 19.3 ± 4.0 | 94.4 ± 6.5 | 4.90 ± 1.63 |
| Substrate H4 (1–19) peptide | ||||
| Hat1 | 1.72 ± 0.16 | 275.4 ± 59.9 | 17.2 ± 1.6 | 0.063 ± 0.025 |
| HAT-B | 6.14 ± 0.19 | 87.2 ± 9.4 | 61.4 ± 1.9 | 0.71 ± 0.21 |
| NuB4 | 7.59 ± 0.24 | 82.8 ± 8.4 | 75.9 ± 2.4 | 0.92 ± 0.28 |
| Substrate Asf1/H3-H4 determined using slope | ||||
| Hat1 | ND | ND | ND | 0.47 ± 0.14 |
| HAT-B | ND | ND | ND | 2.01 ± 0.19 |
| NuB4 | ND | ND | ND | 1.75 ± 0. 21 |
| Substrate H3-H4 determined using slope | ||||
| Hat1 | ND | ND | ND | 0.52 ± 0.05 |
| HAT-B | ND | ND | ND | 2.63 ± 0.05 |
| NuB4 | ND | ND | ND | 2.39 ± 0.09 |
Full-length Histone Substrates Are Required for Robust Enzymatic Activity
To our knowledge, the enzymatic analysis that we present here is the first performed on Hat1, HAT-B, and NuB4 using full-length histone H3-H4 heteromeric substrates. Previous experiments have measured the activity of Hat1 on a histone H4 peptide and reported a kcat of ∼4.2 s−1 and a Km of ∼21 μm (27). To our knowledge, there are no previous kinetic experiments on the HAT-B complex. Using the knowledge that Hat2 makes extensive interactions with both H3 and H4 (32), we hypothesized that full-length histones would be much better substrates than an N-terminal histone H4 tail peptide. To test this hypothesis, we performed extensive Michaelis-Menten kinetics on Hat1, HAT-B, and NuB4 using a histone H4 peptide (residues 1–19) and compared that to the complete enzymatic analysis of the full-length histone H3-H4 substrate (Fig. 4, A and B). These experiments demonstrate that there is a remarkable difference in both Km and kcat when comparing the peptide to the full-length histone protein substrates. The overall increase in catalytic efficiency for the full-length histone substrate over the histone peptide is ∼23-fold for Hat1 alone, ∼7-fold for HAT-B, and ∼5-fold for NuB4. These large changes in catalytic efficiency are mainly due to large decreases in Km with more modest increases in kcat for the full-length histone proteins relative to the histone peptide. To ensure that the increase in enzyme efficiency that we observed was specific for the H4 tail, we measured the activity of HAT-B against a tail-less construct of H3-H4 (Fig. 4C) and observed no nonspecific acetylation of the histone core. Together, these data suggest that histone regions outside of the histone tails contribute to histone H4 tail acetylation by the Hat1-containing HAT-B and NuB4 complexes.
FIGURE 4.
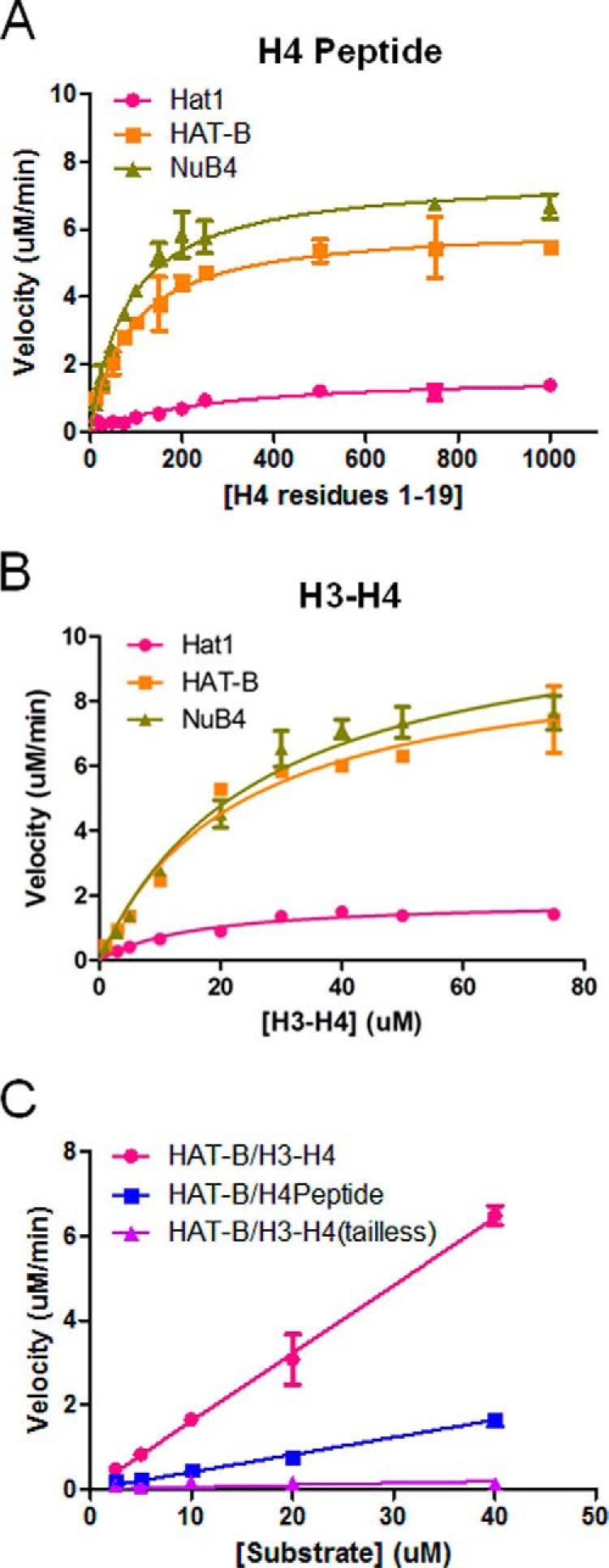
Activity of Hat1 and Hat1-containing complexes against histone H4 substrates. A and B, the data for Hat1 (red) or Hat1-containing complexes (yellow and green) at varying concentrations of H4 peptide (residues 1–19, 2.5–1000 μm) (A) or H3-H4 histones (1–75 μm) (B) are fit to Michaelis-Menten kinetics (GraphPad Prism 5) to calculate steady state parameters. C, the data for HAT-B at varying concentrations of H4 peptide (blue), H3-H4 (pink), or tail-less H3-H4 (H3(45–135)-H4(20–120)) (purple) residues H3-H4 histones (1–75 μm) are fit to the linear portion of the rate data (GraphPad Prism 5) to calculate kcat/Km. All reactions were performed using 100 nm enzyme at saturating concentrations of 14C-labeled acetyl CoA (500 μm), and all kinetic measurements were carried out in duplicate.
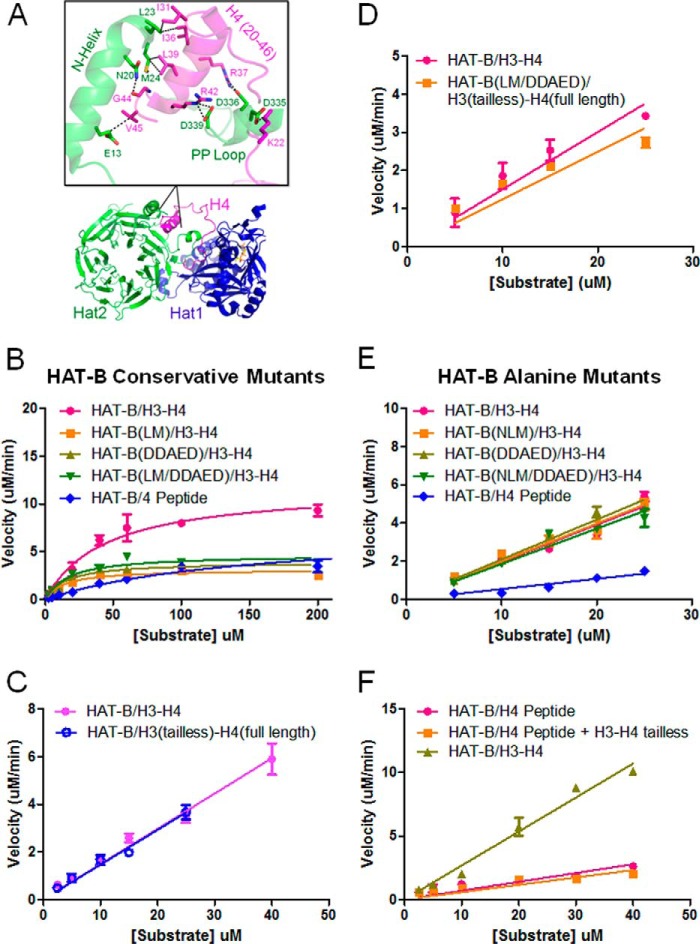
To better understand the regions of the histones that are critical for robust enzymatic activity, we used the existing crystal structures of Hat2 bound to H4(1–48) (33) and HAT-B bound to H4(2–49) and a H3 N-terminal peptide (32) as a guide for potential interactions (Fig. 5A). The Hat2/H4(1–48) structure reveals extensive contacts of an N-terminal helix and a second α-helix termed the PP-loop (33) of Hat2 with a helix of H4 composed of residues 25–41. These interactions were shown to be essential for H4 binding to Hat2 by mutagenic analysis. To determine whether these contacts are important for the enhanced enzyme efficiency of HAT-B, we generated the same mutations to the yeast Hat2 protein (Hat2 DDAED: containing D335N + D336N + E338Q + D339N and Hat2 LM: containing L23D and M24K) and measured the catalytic efficiency toward the full-length histone substrates. Surprisingly, these mutants had little effect on the overall catalytic efficiency when compared with the wild-type protein (Fig. 5B), indicating that these interactions are not important for HAT-B acetylation of histone H4. These data also show no change in catalytic efficiency using full-length H3 or a tail-less construct of H3 and therefore indicate that the N-terminal tail on H3 does not contribute to HAT-B acetylation of histone H4 (Fig. 5C). In addition, we measured the activity of the Hat2 DDAED + LM mutant protein against the H3-H4 heterodimer that lacks residues 1–44 on the H3 tail. These data show that a combination of the loss of the tail on H3 in combination with the mutants to Hat2 again does not affect the overall catalytic efficiency (Fig. 5D). Additionally, we generated mutations that were structurally disruptive to further abolish the interaction between Hat2 and histone H4 (Hat2 DDAED to 5× alanine) and Hat2 NLM (containing N20A, L23Y, and M24A). These mutants had no effect on the overall catalytic efficiency when compared with the wild-type protein (Fig. 5E), further indicating that these interactions are not important for HAT-B acetylation of histone H4. To address the possibility that the H3-H4 core domain somehow allosterically stimulates the catalytic activity of the HAT-B complex for histone H4 tail acetylation, we measured the catalytically activity of HAT-B toward the histone H4 tail in the presence of the tail-less H3-H4 heteromeric core added in trans (Fig. 5F). We found that there was no change in the activity when comparing the peptide alone versus the peptide histone H4 tail in the presence of the tail-less H3-H4 heterodimeric core. Taken together with our earlier observations that full-length H3-H4 histone heterodimer is a better substrate for HAT-B than an H4 peptide, these data suggest that HAT-B binding to the histone tail regions is not sufficient for the preferential acetylation activity of HAT-B for the H3-H4 histone heterodimer over the histone H4 tail.
FIGURE 5.
Mutational analysis of HAT-B acetylation of the H3-H4 heterodimer. A, crystal structure of HAT-B bound to H4 (32) with the binding region of Hat2 and H4 enlarged to highlight key interacting residues. B, enzyme kinetics of Hat2 structurally conservative mutations. HAT-B and HAT-B containing Hat2 mutants with saturating levels of 14C-labeled acetyl CoA and varying concentrations of either H3-H4 (1–75 μm) or H4 peptide (2.5–1000 μm) are fit to Michaelis-Menten kinetics (GraphPad Prism 5) to calculate steady state parameters. C, enzyme kinetics of HAT-B with saturating concentrations of 14C-labeled acetyl CoA with either H3-H4 (pink) or tail-less H3(45–135)-H4 (H3tl-H4) (blue) substrates (0.5–25 μm) are fit to the linear portion of the rate data (GraphPad Prism 5) to calculate kcat/Km. D, enzyme kinetics of HAT-B with either H3-H4 compared with Hat2 mutants on tail-less H3tl-H4 substrates as described for C. E, enzyme kinetics of Hat2 structurally disruptive mutations as described for C. F, enzyme kinetics of wild-type HAT-B varying both H4 peptide added in trans with tail-less H3(45–135)-H4(20–102) (H3tl-H4tl; 0.5–40 μm) is fit to the linear portion of the rate data (GraphPad Prism 5) to calculate kcat/Km. All reactions were performed using 100 nm enzyme at saturating concentrations of 14C-labeled acetyl CoA (500 μm), and all kinetic measurements were carried out in duplicate.
Discussion
The HAT-B histone acetyltransferase is the enzyme complex responsible for acetylation of newly synthesized histone H4 molecules on K5 and K12 (9–13), an activity that is evolutionarily conserved from yeast to human. HAT-B has also been demonstrated by coimmunoprecipitation and native complex purification to associate with the histone chaperones Hif1 (sNASP) and Asf1, possibly to coordinate histone deposition by these proteins (17). The biochemical nature of the HAT-B/histone chaperone complexes and the roles played by Hif1 and Asf1, as well as the H3-H4 histone complex in HAT-B-mediated histone H4 acetylation, has not previously been studied.
To investigate the molecular properties of the interactions between the histone chaperones with HAT-B, we performed a series of experiments to determine the mode of binding between Asf1 and Hif1 with HAT-B. We performed size exclusion chromatography and concluded that Hif1 can bind directly to Hat2 in the absence of Hat1. This is the first direct demonstration that Hat1 does not significantly contribute to the HAT-B/Hif1 interface. Additionally, our sedimentation equilibrium data indicate that Hif1 in the presence of the histones is most likely binding HAT-B as a dimer that contains two copies of the histone molecules H3-H4. We also performed similar experiments on the histone chaperone Asf1 in the presence of HAT-B to determine whether there are any direct interactions with either Hat1 and/or Hat2. These data indicate that Asf1 requires the histone molecules H3 and H4 to associate with HAT-B, suggesting that H3-H4 functions as a bridge between Asf1 and HAT-B. Such a mode of binding is consistent with previous reported experiments on the fungal-specific Rtt109 histone acetyltransferase that also associates with Asf1 to acetylate H3K56, through bridging interactions of H3-H4 (34). Furthermore, we have used SEC and AUC to determine that Hif1 and Asf1 can bind to HAT-B either simultaneously or separately to form stable protein complexes. Simultaneous binding of Asf1 and Hif1 to the HAT-B/H3-H4 histone complex is consistent with previous pulldown studies from cells (21).
Based on our equilibrium sedimentation experiments, we conclude that in the presence of histones, the NuB4/H3-H4 complex contains HAT-B as a heterodimer bound to two copies of Hif1/H3-H4. This is in contrast to the HAT-B/Asf1/H3-H4 complex where Asf1 binds HAT-B as a monomer bound to one copy of the H3-H4 heterodimer. Additionally, when both Asf1 and Hif1 are present, an experimental molecular mass represents a 1:1:1:1:1:1 binding of all six proteins. Based on these data, we have proposed a model for HAT-B/histone chaperone assembly and acetylation on H3-H4 complexes HAT-B in the histone deposition pathway (Fig. 6). Taken together, the structurally distinct Hif1 and Asf1 histone chaperones bind through distinct modes and independently to HAT-B/H3-H4 complexes.
FIGURE 6.
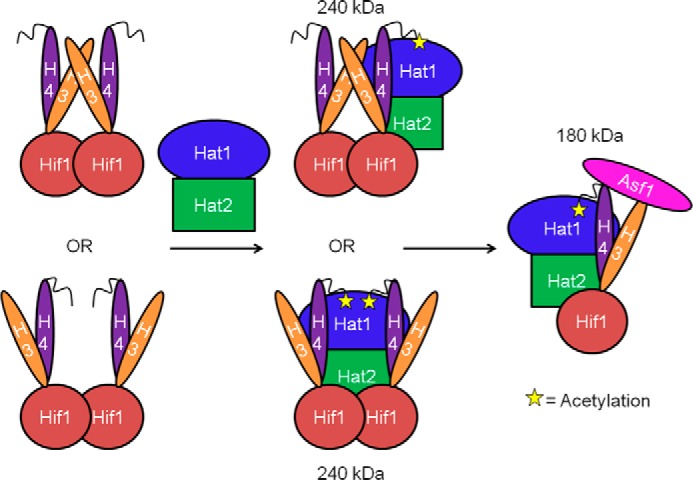
A model for participation of Hat1, Hat2, Hif1, and Asf1 in histone H3-H4 deposition. Hif1 is responsible for recruiting newly synthesized histones H3-H4 heteromers into HAT-B for acetylation. It is likely that Hif1 forms a dimer and binds to two copies of an H3-H4 dimer (as two heterodimers or one heterotetramer). Hif1 then brings the histones to the HAT-B complex for acetylation where the tails and cores of H3 and H4 make direct contact with both Hat1 and Hat2. This results in a complex with one Hat1, one Hat2, and two Hif1/H3-H4 molecules. After acetylation, Asf1 binds to the tetrameric interface of H3-H4 prior to H3-H4 transport into the nucleus for histone deposition into chromatin.
Additionally, Michaelis-Menten enzyme kinetic experiments of HAT-B in the presence or absence of the Hif1 or Asf1 histone chaperones reveal that Hif1 and Asf1 do not significantly contribute to the overall catalytic efficiency of HAT-B for histone H4 acetylation. Based on this observation, we propose that the role of Hif1 and Asf1 is strictly related to transport of the histones either into or out of the HAT-B complex to be later used for histone deposition. Having narrowed down the enhancement of Hat1 catalytic activity to Hat2, we wanted to further dissect its molecular contribution to histone H4 acetylation. We first performed experiments to compare the kinetics of HAT-B using either a peptide substrate (H4 residues 1–19) or full-length H3-H4 histone heterodimer. These data reveal a remarkable difference in a lower Km when comparing the peptide to the full-length proteins, indicating that the full-length histones are much better substrates than the N-terminal H4 peptide. We then used crystal structure information of Hat2 or HAT-B complexes bound to histone H4 N-terminal fragments to help dissect the histone contributions to H4 tail acetylation. Remarkably, we found that mutation of residues that mediate Hat2 interaction with H4 did not have a significant effect on histone H4 tail acetylation. Significantly, a ∼23-fold increase in catalytic efficiency is observed when comparing the activity of Hat1 alone on either full-length histone substrates versus the H4 N-terminal peptide. This suggests that the full-length histones make important contacts with Hat1 for enhanced activity. These data, together with our observations that full-length H3-H4 is a better substrate than the histone H4 peptide for HAT-B, suggest that the core region of H3-H4 likely contributes to histone H4 tail acetylation.
Based on our experiments we propose a model to place Hif1 and Asf1 within the HAT-B complex along the histone assembly and acetylation pathway (Fig. 6). Previous studies have demonstrated the majority of H4 bound to Asf1 is acetylated on K5 and K12 (3, 21, 24), and therefore the NuB4 complex (Hat1, Hat2, and Hif1) likely acts upstream of Asf1. We hypothesize that Hif1 is responsible for recruiting newly synthesized histones H3-H4 heterodimers into HAT-B for acetylation, with the recruitment occurring through Hif1-Hat2 interaction. Based on our data and previous studies (21), two subunits of Hif1 form a complex with either two H3-H4 heterodimers or a single H3-H4 heterotetramer. This is consistent with previous studies demonstrating that the human ortholog of Hif1, sNASP, dimerizes to form a complex with two H3-H4 dimers (21). After Hif1 binds to H3-H4, it then brings the histones to the HAT-B complex for acetylation where the tails and cores of H3 and H4 make direct contact with both Hat1 and Hat2. This results in a complex with one Hat1, one Hat2, and two Hif1/H3-H4 molecules. Because Asf1 has been shown to bind H3-H4 heterodimers (25, 26) and act downstream of NuB4 (3, 21, 24), it is likely that NuB4 also binds H3-H4 heterodimers. Therefore, after HAT-B-mediated histone H4 acetylation, Asf1 likely comes in and binds to the tetrameric interface of H3-H4 as previously described (25, 26). This complex must then be transported into the nucleus where the histones likely interact with additional chaperones to be primed for deposition onto replicating chromatin. The process of histone deposition is poorly understood, but the CAF-1 histone chaperone complex has been shown to assist in generation of the H3-H4 tetramer for histone deposition into chromatin (35). Taken together, the data presented here progress our understanding for how Hat1, Hat2, Hif1, and Asf1 contribute to acetylation-dependent histone H3-H4 assembly.
Author Contributions
A. H. and M. D. R. performed the experiments described in the manuscript. A. H. prepared manuscript figures and text. R. M. designed and supervised experiments by A. H. and M. D. R. and revised manuscript text. All authors read and approved the submitted manuscript.
Acknowledgments
We thank Cheryl McCullough, Yadilette Rivera-Colón, Robert Magin, John Domsic, and Adam Olia for helpful discussions.
This work was supported by National Institutes of Health Grants R01 GM060293 and P01 AG031862 (to R. M.), Training Grant F32CA180504 (to A. H.), Grant P30 CA010815 (to the proteomics core facility at the Wistar Institute), and Grant P30 CA016520 (to the University of Pennsylvania DNA Sequencing Facility at the Perelman School of Medicine). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- HAT-B
- histone acetyltransferase type B
- Hat
- histone acetyltransferase
- Asf1
- anti-silencing function 1
- SEC
- size exclusion chromatography
- AUC
- analytical ultracentrifugation
- ACoA
- acetyl coenzyme A.
References
- 1.Brownell J. E., and Allis C. D. (1996) Special HATs for special occasions: linking histone acetylation to chromatin assembly and gene activation. Curr. Opin. Genet. Dev. 6, 176–184 [DOI] [PubMed] [Google Scholar]
- 2.Lee K. K., and Workman J. L. (2007) Histone acetyltransferase complexes: one size doesn't fit all. Nat. Rev. Mol. Cell Biol. 8, 284–295 [DOI] [PubMed] [Google Scholar]
- 3.Parthun M. R. (2012) Histone acetyltransferase 1: More than just an enzyme? Biochim. Biophys. Acta. 1819, 256–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cousens L. S., and Alberts B. M. (1982) Accessibility of newly synthesized chromatin to histone acetylase. J. Biol. Chem. 257, 3945–3949 [PubMed] [Google Scholar]
- 5.Benson L. J., Gu Y., Yakovleva T., Tong K., Barrows C., Strack C. L., Cook R. G., Mizzen C. A., and Annunziato A. T. (2006) Modifications of H3 and H4 during chromatin replication, nucleosome assembly, and histone exchange. J. Biol. Chem. 281, 9287–9296 [DOI] [PubMed] [Google Scholar]
- 6.Chicoine L. G., Schulman I. G., Richman R., Cook R. G., and Allis C. D. (1986) Nonrandom utilization of acetylation sites in histones isolated from Tetrahymena: evidence for functionally distinct H4 acetylation sites. J. Biol. Chem. 261, 1071–1076 [PubMed] [Google Scholar]
- 7.Annunziato A. T., and Seale R. L. (1983) Histone deacetylation is required for the maturation of newly replicated chromatin. J. Biol. Chem. 258, 12675–12684 [PubMed] [Google Scholar]
- 8.Ma X. J., Wu J., Altheim B. A., Schultz M. C., and Grunstein M. (1998) Deposition-related sites K5/K12 in histone H4 are not required for nucleosome deposition in yeast. Proc. Natl. Acad. Sci. U.S.A. 95, 6693–6698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruiz-García A. B., Sendra R., Galiana M., Pamblanco M., Pérez-Ortín J. E., and Tordera V. (1998) HAT1 and HAT2 proteins are components of a yeast nuclear histone acetyltransferase enzyme specific for free histone H4. J. Biol. Chem. 273, 12599–12605 [DOI] [PubMed] [Google Scholar]
- 10.Sobel R. E., Cook R. G., and Allis C. D. (1994) Non-random acetylation of histone H4 by a cytoplasmic histone acetyltransferase as determined by novel methodology. J. Biol. Chem. 269, 18576–18582 [PubMed] [Google Scholar]
- 11.Mingarro I., Sendra R., Salvador M. L., and Franco L. (1993) Site specificity of pea histone acetyltransferase B in vitro. J. Biol. Chem. 268, 13248–13252 [PubMed] [Google Scholar]
- 12.Richman R., Chicoine L. G., Collini M. P., Cook R. G., and Allis C. D. (1988) Micronuclei and the cytoplasm of growing Tetrahymena contain a histone acetylase activity which is highly specific for free histone H4. J. Cell Biol. 106, 1017–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiegand R. C., and Brutlag D. L. (1981) Histone acetylase from Drosophila melanogaster specific for H4. J. Biol. Chem. 256, 4578–4583 [PubMed] [Google Scholar]
- 14.Parthun M. R., Widom J., and Gottschling D. E. (1996) The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell 87, 85–94 [DOI] [PubMed] [Google Scholar]
- 15.Kleff S., Andrulis E. D., Anderson C. W., and Sternglanz R. (1995) Identification of a gene encoding a yeast histone H4 acetyltransferase. J. Biol. Chem. 270, 24674–24677 [DOI] [PubMed] [Google Scholar]
- 16.Nagarajan P., Ge Z., Sirbu B., Doughty C., Agudelo Garcia P. A., Schlederer M., Annunziato A. T., Cortez D., Kenner L., and Parthun M. R. (2013) Histone acetyl transferase 1 is essential for mammalian development, genome stability, and the processing of newly synthesized histones H3 and H4. PLoS Genet. 9, e1003518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ai X., and Parthun M. R. (2004) The nuclear Hat1p/Hat2p complex: a molecular link between type B histone acetyltransferases and chromatin assembly. Mol. Cell 14, 195–205 [DOI] [PubMed] [Google Scholar]
- 18.Ge Z., Wang H., and Parthun M. R. (2011) Nuclear Hat1p complex (NuB4) components participate in DNA repair-linked chromatin reassembly. J. Biol. Chem. 286, 16790–16799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poveda A., Pamblanco M., Tafrov S., Tordera V., Sternglanz R., and Sendra R. (2004) Hif1 is a component of yeast histone acetyltransferase B, a complex mainly localized in the nucleus. J. Biol. Chem. 279, 16033–16043 [DOI] [PubMed] [Google Scholar]
- 20.Qin S., and Parthun M. R. (2002) Histone H3 and the histone acetyltransferase Hat1p contribute to DNA double-strand break repair. Mol. Cell Biol. 22, 8353–8365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campos E. I., Fillingham J., Li G., Zheng H., Voigt P., Kuo W. H., Seepany H., Gao Z., Day L. A., Greenblatt J. F., and Reinberg D. (2010) The program for processing newly synthesized histones H3.1 and H4. Nat. Struct. Mol. Biol. 17, 1343–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Groth A., Corpet A., Cook A. J., Roche D., Bartek J., Lukas J., and Almouzni G. (2007) Regulation of replication fork progression through histone supply and demand. Science 318, 1928–1931 [DOI] [PubMed] [Google Scholar]
- 23.Natsume R., Eitoku M., Akai Y., Sano N., Horikoshi M., and Senda T. (2007) Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature 446, 338–341 [DOI] [PubMed] [Google Scholar]
- 24.Jasencakova Z., Scharf A. N., Ask K., Corpet A., Imhof A., Almouzni G., and Groth A. (2010) Replication stress interferes with histone recycling and predeposition marking of new histones. Mol. Cell 37, 736–743 [DOI] [PubMed] [Google Scholar]
- 25.English C. M., Adkins M. W., Carson J. J., Churchill M. E., and Tyler J. K. (2006) Structural basis for the histone chaperone activity of Asf1. Cell 127, 495–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.English C. M., Maluf N. K., Tripet B., Churchill M. E., and Tyler J. K. (2005) ASF1 binds to a heterodimer of histones H3 and H4: a two-step mechanism for the assembly of the H3-H4 heterotetramer on DNA. Biochemistry 44, 13673–13682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu H., Moshkina N., Min J., Zeng H., Joshua J., Zhou M. M., and Plotnikov A. N. (2012) Structural basis for substrate specificity and catalysis of human histone acetyltransferase 1. Proc. Natl. Acad. Sci. U.S.A. 109, 8925–8930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daniel Ricketts M., Frederick B., Hoff H., Tang Y., Schultz D. C., Singh Rai T., Grazia Vizioli M., Adams P. D., and Marmorstein R. (2015) Ubinuclein-1 confers histone H3.3-specific-binding by the HIRA histone chaperone complex. Nat. Commun. 6, 7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lebowitz J., Lewis M. S., and Schuck P. (2002) Modern analytical ultracentrifugation in protein science: a tutorial review. Protein Sci. 11, 2067–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harding S. E., Rowe A. J., and Horton J. C. (1992) Analytical ultracentrifugation in biochemistry and polymer science, Royal Society of Cambridge, Cambridge, UK [Google Scholar]
- 31.Yan Y., Harper S., Speicher D. W., and Marmorstein R. (2002) The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate. Nat. Struct. Biol. 9, 862–869 [DOI] [PubMed] [Google Scholar]
- 32.Li Y., Zhang L., Liu T., Chai C., Fang Q., Wu H., Agudelo Garcia P. A., Han Z., Zong S., Yu Y., Zhang X., Parthun M. R., Chai J., Xu R. M., and Yang M. (2014) Hat2p recognizes the histone H3 tail to specify the acetylation of the newly synthesized H3/H4 heterodimer by the Hat1p/Hat2p complex. Genes Dev. 28, 1217–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murzina N. V., Pei X. Y., Zhang W., Sparkes M., Vicente-Garcia J., Pratap J. V., McLaughlin S. H., Ben-Shahar T. R., Verreault A., Luisi B. F., and Laue E. D. (2008) Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure. 16, 1077–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han J., Zhou H., Li Z., Xu R. M., and Zhang Z. (2007) Acetylation of lysine 56 of histone H3 catalyzed by RTT109 and regulated by ASF1 is required for replisome integrity. J. Biol. Chem. 282, 28587–28596 [DOI] [PubMed] [Google Scholar]
- 35.Winkler D. D., Zhou H., Dar M. A., Zhang Z., and Luger K. (2012) Yeast CAF-1 assembles histone (H3-H4)2 tetramers prior to DNA deposition. Nucleic Acids Res. 40, 10139–10149 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]