Abstract
There are two major stem cell populations in the intestinal crypt region that express either Bmi1 or Lgr5; however, it has been shown that other populations in the crypt can regain stemness. In this study, we demonstrate that the transcription factor NK2 homeobox 2 (Nkx2.2) is expressed in enteroendocrine cells located in the villus and crypt of the intestinal epithelium and is coexpressed with the stem cell markers Bmi1 and Lgr5 in a subset of crypt cells. To determine whether Nkx2.2-expressing enteroendocrine cells display cellular plasticity and stem cell potential, we performed genetic lineage tracing of the Nkx2.2-expressing population using Nkx2.2Cre/+;R26RTomato mice. These studies demonstrated that Nkx2.2+ cells are able to give rise to all intestinal epithelial cell types in basal conditions. The proliferative capacity of Nkx2.2-expressing cells was also demonstrated in vitro using crypt organoid cultures. Injuring the intestine with irradiation, systemic inflammation, and colitis did not enhance the lineage potential of Nkx2.2-expressing cells. These findings demonstrate that a rare mature enteroendocrine cell subpopulation that is demarcated by Nkx2.2 expression display stem cell properties during normal intestinal epithelial homeostasis, but is not easily activated upon injury.
Keywords: Bmi1, enteroendocrine cells, Lgr5, Nkx2.2, stem cells
the mammalian intestine consists of a small and large intestine that are each lined by a single-cell epithelium. The epithelium of the small intestine is organized into crypt and villus compartments, whereas the large intestine only contains crypts. Crypts invaginate into the underlying mesenchyme and contain stem cells and proliferating transit-amplifying cells. Villi protrude into the gut lumen and consist of differentiated cell types. The intestinal epithelium contains five terminally differentiated cell populations: absorptive enterocytes and secretory Paneth, goblet, tuft, and enteroendocrine cells. The hormone-producing enteroendocrine cells represent only 1% of the cells in the intestinal epithelium, but express at least 15 different types of hormones (23, 29) and represent the largest endocrine system in the body. Most enteroendocrine cells can be identified by chromogranin A (Chga) expression.
The intestinal epithelium contains a high self-renewal capacity. Turnover time is ∼4–5 days and is the fastest turnover rate in mammals (9, 39). Stem cells in the crypt give rise to transit-amplifying cells that differentiate into the absorptive and secretory epithelial cell types around the crypt-villus border. During differentiation, Paneth cells migrate down toward the crypt base, whereas the other differentiated cell types migrate up toward the villus tip, where they undergo apoptosis and are shed into the gut lumen (39).
A significant number of studies have identified and characterized two major stem cell populations in the crypt of the small intestine. The highly proliferating crypt base columnar cells (CBCs) are located at the base of the crypt between the Paneth cells and are marked by the leucine-rich repeat containing G protein-coupled receptor 5 (Lgr5) (4, 8). The other stem cell population is found at or near the +4 position, is slow cycling and label-retaining, and is demarcated by the expression of the polycomb ring finger oncogene Bmi1 (3, 28, 31). Recently, however, Bmi1 transcripts were also detected in Lgr5+ CBCs (25). Interestingly, interconversion between the two Lgr5+ and Bmi1+ stem cell populations has been demonstrated (36), allowing the intestine to regenerate in the absence of Lgr5+ cells (38). Furthermore, several studies have shown that there is additional plasticity in the intestine. Label-retaining secretory progenitors committed to the Paneth or enteroendocrine cell lineages, as well as delta-like 1-expressing secretory progenitor cells, can gain stemness upon intestinal damage (7, 40). In addition, label-retaining Paneth cells can proliferate and begin to express the stem cell marker Bmi1 (30) in injury conditions. Similarly, enteroendocrine cells in the crypt express stem cell markers (33) and can be activated after irradiation in vitro (41).
The homeodomain transcription factor NK2 homeobox 2 (Nkx2.2) is expressed in the pancreas, ventral neural tube, and intestine. Nkx2.2 is required for cell fate decisions in the pancreatic islet and cell patterning in the ventral neural tube (6, 35). In the intestine, expression of Nkx2.2 is first detected at embryonic day (E) 15.5 and maintained throughout adulthood, where Nkx2.2 is expressed in enteroendocrine cells in the crypt as well as in the villus (11, 42). In the crypt, some Nkx2.2+ cells coexpress the enteroendocrine progenitor cell marker neurogenin 3 (Ngn3) (42), a gene that is essential for the development of intestinal and pancreatic endocrine cells (14, 18). Nkx2.2 is also critical for determining cell fate decisions of enteroendocrine cell lineages in the intestine downstream of Ngn3; Nkx2.2 null mice display a reduction in most enteroendocrine cell populations, with the exception of ghrelin-positive cells, which are upregulated (11, 42). Other intestinal epithelial cell types are unaffected by the loss of Nkx2.2 (11). Lineage-tracing analysis showed that all enteroendocrine cell populations derive from Ngn3+ cells. Furthermore, Ngn3 lineage-tracing revealed that Ngn3 is expressed in a rare multipotent cell that can give rise to whole crypt-villus units, in addition to rare single Paneth and goblet cells (32). In this study, we demonstrate that a subpopulation of Nkx2.2+ enteroendocrine cells expresses the stem cell markers Bmi1 and Lgr5. Lineage tracing with Nkx2.2 confirms that a rare subset of Nkx2.2-expressing mature enteroendocrine cells has expanded lineage potential, but cannot be activated upon intestinal injury.
MATERIALS AND METHODS
Animals.
Mice were housed and treated in accordance with the animal care protocol (AAAG3206) approved by Columbia University's Institutional Animal Care and Use Committee. Mice were maintained on a C57BL/6J background (The Jackson Laboratory). Lgr5EGFP/+ [B6.129P2-Lgr5tm1(cre/ERT2)Cle/J] (4), Bmi1EGFP/+ (kindly provided by Prof. Dr. Weissman) (17) and R26RTomato [B6.Cg-Gt(ROSA)26-Sortm14(CAG−tdTomato)Hze/J] (21) mice were obtained from The Jackson Laboratory. Nkx2.2Cre/+ and Nkx2.2LacZ/+ mice were described previously (1, 2).
The Nkx2.2cCre knock-in line was derived utilizing recombination-mediated cassette exchange, using Nkx2.2LCA acceptor cells (1). Specifically, a DNA construct with COOH-terminal Cre (cCre)-T2A (43) inserted at the 5′ ATG start codon of the Nkx2.2 coding sequence was generated to allow coordinate expression of Nkx2.2 and cCre (Supplemental Fig. S1A; Supplemental material for this article is available online at the Journal website). The modified Nkx2.2 locus and a hygromycin selection cassette were flanked by Lox66 and Lox2272 elements. Subsequent recombination-mediated cassette exchange targeting was performed in the Nkx2.2LCA embryonic stem (ES) cells, as previously described (1). The Ngn3nCre knock-in allele was similarly derived (Supplemental Fig. S1B). First, an acceptor ES cell line carrying the Ngn3LCA allele was derived using traditional homologous recombination-based targeting. The Ngn3LCA allele has Lox66 inserted 3.5 kb upstream of the transcription initiation site of Ngn3 and Lox2272 1 kb downstream of the Ngn3 polyA signal. Mice carrying the Ngn3LCA allele alone have no detectable phenotype, suggesting that the insertion of Lox sites near the Ngn3 locus does not interfere with Ngn3 expression. The NH2-terminal Cre (nCre)-T2A (43) coding sequence was inserted at the ATG of the Ngn3 gene. The presence of T2A allows the coordinated expression of nCre and Ngn3 from the targeted allele. A DNA construct with Lox66, 3.5-kb Ngn3 5′ region, nCre-T2A-Ngn3 coding region and polyA signal, and Lox2272 was then produced. Cre-mediated cassette exchange was then performed to derive ES cells carrying the Ngn3nCre knock-in allele. Blastocyst injections were performed for the production of mice. The Nkx2.2cCre/+;Ngn3nCre/+;R26RTomato (Ai9) mice were generated through interbreeding. For genotyping the Nkx2.2cCre/+ allele, the following primers were used: 5′-CTGGAAGGGCGTGCTCCAGGCT-3′ and 5′-GCTCGCTCCAACCTGGGCCATT-3′ (wild type = 499 bp, Nkx2.2cCre = 610 bp). To genotype the Ngn3nCre/+ allele, the following primers were used: 5′-GACTTGAGCAGGGACCGTCTCT-3′ and 5′-CTCAGAGAGGGAAACGGCTTGT-3′ (wild type = 217 bp, Ngn3nCre = 442 bp) (Supplemental Fig. S1C).
For timed pregnancies, noon on the day a vaginal plug was observed was considered to be E0.5. Unless otherwise indicated, adult mice were analyzed at 6 wk of age.
Animal treatments.
Whole body irradiation (6 and 12 Gy) of mice was performed with a low-dose irradiator for small animals (Atomic Energy of Canada Gammacell 40 Cesium Unit with a dose rate of 1 Gy/min). Mice were irradiated at 6–7 wk of age and killed 7 days after irradiation. For systemic inflammation studies, 8-wk-old mice were intraperitoneally injected with 2.5 mg/kg lipopolysaccharide (LPS) (from E. coli O111:B4; Millipore) on day 0 and day 7, and dissected 7 days after the last injection on day 14. Acute colitis in 8-wk-old mice was induced by 3% dextran sodium sulfate (DSS) (colitis grade, molecular mass 36–50 kDa; MP Biomedicals, no. 0216011025) in the drinking water for 4 days. Mice were left to recover for 14 days with normal drinking water and were analyzed on day 18.
Histology and immunofluorescence.
Intestinal samples were cut longitudinally, flushed with cold phosphate-buffered saline (PBS), rolled to “swiss rolls” (24), and fixed overnight in 4% paraformaldehyde at 4°C. Samples were then cryo-preserved with 30% sucrose, cryo-embedded in Tissue-Tek O.C.T (Fisher Scientific, no. 14-373-65), and cryo-sectioned into 5-μm sections. For immunofluorescence staining, sections were incubated for 15 min in 0.3% H2O2, washed in PBS, and blocked for 30 min at room temperature with 10% donkey serum (Fisher Scientific, no. NC9624464) in PBT (PBS with 0.3% Triton). Sections were incubated overnight at 4°C with the following primary antibodies diluted in 5% donkey serum in PBT: chicken anti-β-galactosidase (1:200, Abcam, ab9361); rabbit anti-Chga (1:500-1,000, ImmunoStar, no. 20085); goat anti-Chga (1:100, Santa Cruz, sc-1488); fluorescein Dolichos biflorus agglutinin (1:100, Vector Laboratories, FL-1031); rabbit anti-doublecortin-like kinase 1 (Dclk1) (1:10, Abgent, no. AP7219b); goat anti-fatty acid binding protein (FABP) 2/intestinal type FABP (10 μg/ml, R&D, no. AF1486); rabbit anti-green fluorescent protein (GFP) (1:100, Novus, no. NB600-308); rabbit anti-lysozyme (1:200, Dako, no. A0099). After being washed with PBT, sections were incubated with appropriate secondary antibodies diluted in 5% donkey serum in PBT for 2 h at room temperature. Secondary antibodies were conjugated with Alexa488, Alexa594, Alexa647, Cy5, and DyLight649 (1:200, Jackson ImmunoResearch). The Tomato signal was detected by direct fluorescence of the protein. Images were acquired with either a confocal microscope (Zeiss LSM710; software “Zen 2012”) or a fluorescence stereomicroscope (Leica MZ16F; software “QCapturePro v5.1”). Hematoxylin and eosin (H&E) staining was performed according to the standard staining procedure (12).
Intestinal organoid cultures.
Mouse crypt cultures were prepared as described previously (16, 22), with minor modifications. Small intestine of 6-wk-old Nkx2.2Cre/+;R26RTomato mice was isolated (10 cm as measured from the pyloric sphincter), cut longitudinally, and washed in cold Dulbecco's phosphate-buffered saline (D-PBS) (Fisher Scientific, no. MT-21-031-CV). Villi were scraped off using a razor blade, and the tissue was cut into ∼5-mm pieces. The tissue was washed thoroughly several times with cold D-PBS and incubated in 5 mM EDTA in D-PBS for 60 min on ice. Tissue fragments were resuspended with a 10-ml pipette in 10% fetal bovine serum (Gemini Bio Products, no. 100–106). The supernatant enriched in crypts was centrifuged at 175 g for 5 min at 4°C, resuspended in 10-ml basal medium (Advanced DMEM/F12, Invitrogen, no. 12634010) supplemented with 10 mM HEPES (Invitrogen, no. 15630080), 2 mM GlutaMAX (Invitrogen, no. 35050061), and 100 U/ml penicillin + 100 μg/ml streptomycin (Invitrogen, no. 15140122). The suspension was centrifuged at 112 g for 5 min at 4°C, resuspended in 5 ml of the basal medium, and passed through a 100-μm cell strainer (Fisher Scientific, no. 352360). Afterwards, the crypt fractions were centrifuged at 175 g for 5 min at 4°C. The crypts were then embedded in Matrigel (Fisher Scientific, no. 356231) and seeded in drops in a prewarmed 48-well plate. Crypt-Matrigel drops were solidified at 37°C and afterwards overlaid with 300 μl of basal medium that has the following supplements: 1 × N2 supplement (Invitrogen, no. 17502048), 1 × B27 supplement (Invitrogen, no. 17504044), 1 mM N-acetylcysteine (Sigma, no. A9165-5G), 10 mM nicotinamide (Sigma, no. N0636-100G), 250 ng/ml amphotericin B (Invitrogen, no. 15240-062), 100 ng/ml human Noggin (Fisher Scientific, no. 6057-NG-025/CF), 1 μg/ml mouse R-Spondin 1 (Fisher Scientific, no. 3474-RS-050), and 50 ng/ml mouse epidermal growth factor (Invitrogen, no. PMG8041). Medium was renewed every other day. D-PBS without calcium and magnesium was used for all steps of the crypt isolation.
For single-cell organoid cultures, whole crypts were isolated as described above, but dissociated with TrypleE Express (Invitrogen, no. 12604013), including 1 mg/ml DNaseI (Roche, no. 10104159001) and 10 μM Y-27632 (Sigma, no. Y0503), at 37°C. Dissociated cells were centrifuged, resuspended in 2% fetal bovine serum, including 10 μM Y-27632 and DAPI, passed through a 40-μm cell strainer (Fisher Scientific, no. 352340), and sorted by fluorescence-activated cell sorting (FACS) using a BD FacsAria Cell Sorter. Viable Tomato+ single cells were collected, pelleted, and seeded in Matrigel drops in a prewarmed 48-well plate (500-1,000 cells/well). The Matrigel-cell drops were solidified at 37°C and afterwards overlaid with 300 μl basal medium, including the supplements stated above, but with additional 500 ng/ml Jagged 1 FC (R&D, no. 599-JG-100), 100 ng/ml Wnt3a (R&D, no. 1324-WN-010), and 10 μM Y-27632.
Data analysis.
The location of Nkx2.2+ cells in the intestinal crypt was defined by analyzing a total of 100 well-sectioned crypts from the small intestine of three mice. To determine the percentage of Lgr5-enhanced GFP (EGFP+) crypts that contain Nkx2.2+/Lgr5+ double-positive cells, 100 well-sectioned Lgr5-EGFP+ crypts of the small intestine were analyzed per sample (n = 3). To determine the percentage of Nkx2.2+/Bmi1+ or Nkx2.2+/Chga+/Bmi1+ coexpressing cells per total Bmi1-EGFP+ or total Nkx2.2+ cells, a total of 300 Bmi1-EGFP+ or Nkx2.2+ cells were analyzed from three mice.
The ratio of organoids that contain active Nkx2.2-expressing stem cells in the crypt culture was calculated by counting the total number of organoids grown on day 7 in culture and counting organoids that have at least one Tomato-stained cell cluster. All organoids were included in the analysis, even organoids that do not show the typical organoid structure with several buds growing out at day 7 in culture, or organoids that are small, round, or are completely Tomato-labeled.
Quantification of Nkx2.2 lineage-traced villi in the small intestine or crypts in the large intestine in treated vs. nontreated mice was performed using three random “swiss roll” sections of the small or large intestine per mouse. Each villus or crypt that had at least one Tomato-stained cell cluster (at least two adjacent Tomato+ cells) was counted. Tomato-stained villi or crypts that were not sectioned in its full length, but rather into two parts, were counted as two separate villi or crypts.
Values are expressed as means ± SE. Statistical analysis was performed using a two-tailed unpaired Student's t-test. Results were considered significant when P < 0.05.
RESULTS
Nkx2.2 is expressed in intestinal stem cells of adult mice.
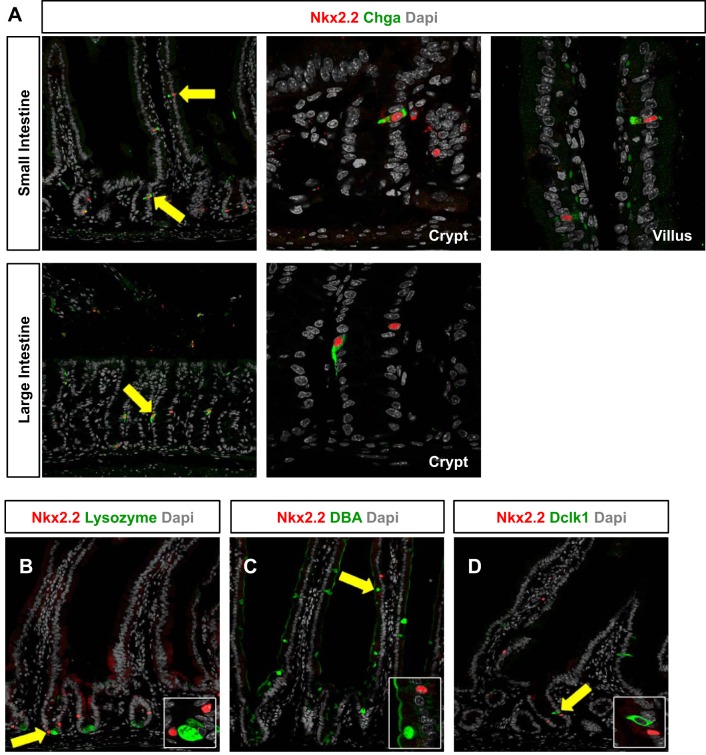
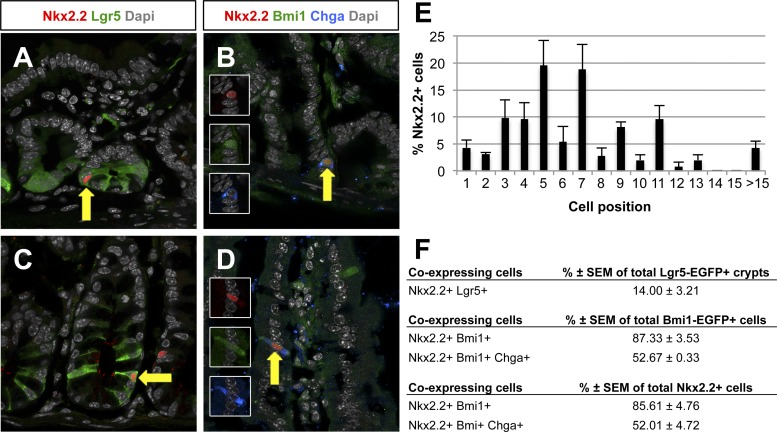
Nkx2.2 is expressed in the intestinal crypt and villus epithelium of embryonic and adult mice. Nkx2.2 is exclusively expressed in enteroendocrine cells, which is confirmed by coexpression analysis with several enteroendocrine hormones and the enteroendocrine cell marker Chga in the small and large intestine (11, 42) (Fig. 1A). Nkx2.2 expression could not be detected in the other intestinal epithelial cell types, including the Paneth, goblet, and tuft cells (Fig. 1, B–D). However, since Nkx2.2 is expressed in the crypt region where the stem cell populations are located, we assessed whether Nkx2.2 is coexpressed with the stem cell markers Lgr5 and Bmi1. Nkx2.2+ cells were found in almost every position of the crypt, but were predominantly located at the +5 and +7 position of the crypt (Fig. 2, A–E). Due to the absence of suitable antibodies for immunohistochemical staining, Nkx2.2LacZ/+ knock-in mice (1) were crossed to Lgr5EGFP/+ or Bmi1EGFP/+ mice (4, 17) to detect their respective coexpression. Consistent with its position in the crypt, Nkx2.2 can be found in a rare subset of Lgr5+ cells in the crypt of the small and large intestine of 6- to 7-wk-old mice (Fig. 2, A and C). 14 ± 3.21% of Lgr5-EGFP+ crypts in the small intestine contained Nkx2.2+/Lgr5+ coexpressing cells (Fig. 2F). More frequently, Nkx2.2 was expressed in Bmi1+ cells in the small intestine of 6-wk-old mice. Interestingly, Nkx2.2+/Bmi1+ coexpressing cells were detected not only in the crypt (Fig. 2B), but also in the villus (Fig. 2D). In the crypt of the small intestine, 87.33 ± 3.53% of Bmi1-EGFP+ cells coexpressed Nkx2.2+/Bmi1+ (Fig. 2F). More than one-half of these Nkx2.2+/Bmi1+ double-positive cells also expressed the enteroendocrine cell marker Chga (Fig. 2, B, D, and F; 52.67 ± 0.33%), suggesting that a subset of Nkx2.2+ enteroendocrine cells may retain stem cell potential.
Fig. 1.
NK2 homeobox 2 (Nkx2.2) is expressed in enteroendocrine cells in crypts and villi of the intestinal epithelium of adult mice. A: immunofluorescence of the small and large intestine of 5- to 6-wk-old Nkx2.2LacZ/+ mice shows coexpression of Nkx2.2 with the enteroendocrine cell marker chromogranin A (Chga) in crypts and villi. Costaining of Nkx2.2 with lysozyme-expressing Paneth cells (B), Dolichos biflorus agglutinin (DBA) lectin-marked goblet cells (C), and doublecortin-like kinase 1 (Dclk1)-expressing tuft cells (D) could not be detected in the small intestine. Nkx2.2 was detected by β-galactosidase antibody staining (Nkx2.2LacZ/+). Confocal images are shown. Magnification: ×20 (A; crypt and villus) and ×63 (B–D; insets from arrow). Dapi, 4,6-diamidino-2-phenylindole.
Fig. 2.
Nkx2.2 is coexpressed with leucine-rich repeat containing G protein-coupled receptor 5 (Lgr5) and Bmi1 polycomb ring finger oncogene (Bmi1). Immunofluorescence analysis of the small intestine of 6- to 7-wk-old Nkx2.2LacZ/+;Lgr5EGFP/+ and Nkx2.2LacZ/+;Bmi1EGFP/+ mice is shown. Nkx2.2 is detected in rare Lgr5-enhanced green fluorescent protein (EGFP)-expressing cells in the small (A) and large (C) intestine (arrows), but in many Bmi1-EGFP-positive cells in crypts (B) as well as villi (D) of the small intestine. Some of the Nkx2.2+/Bmi1-EGFP+ cells also express the enteroendocrine cell marker Chga (B and D, arrows). Nkx2.2 was detected by β-galactosidase antibody staining (Nkx2.2LacZ/+). Confocal images are shown. Magnification: ×63. E: analysis of the location Nkx2.2+ cells in the crypt of the small intestine. F: analysis of Nkx2.2+/Lgr5+, Nkx2.2+/Bmi1+, and Nkx2.2+/Bmi1+/Chga+ cell numbers in the small intestine.
Nkx2.2-expressing cells give rise to all intestinal epithelial cell types.
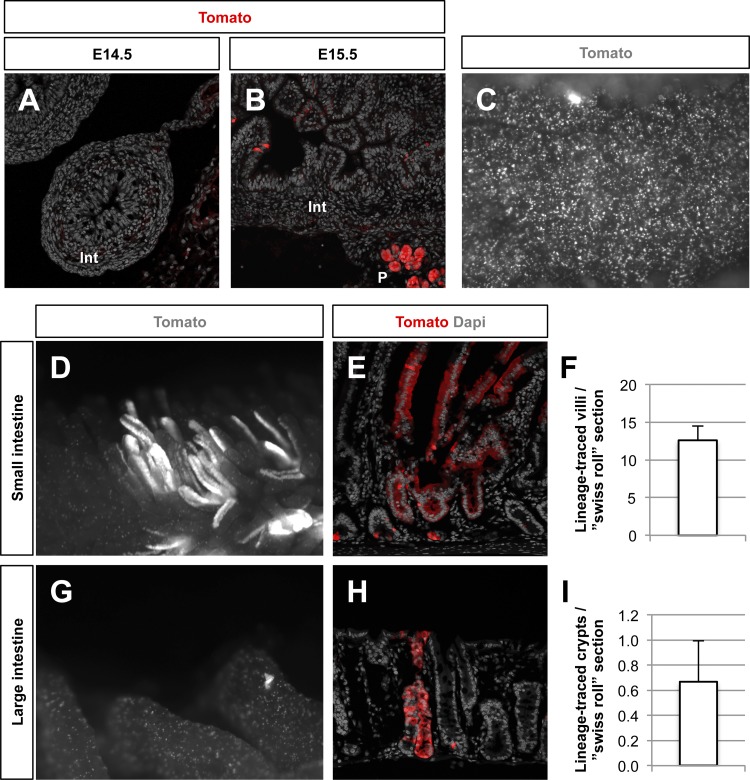
Given the coexpression of Nkx2.2 with the stem cell markers Bmi1 and Lgr5 in a subset of intestinal crypt cells, we sought to determine the stem cell capacity of Nkx2.2-expressing cells using a constitutive Cre-loxP genetic lineage-tracing approach. The constitutive Nkx2.2Cre/+ knock-in mouse line (2) was crossed to R26RTomato reporter mice (21) (Nkx2.2Cre/+;R26RTomato) and analyzed at the embryonic stages E14.5 and E15.5 to define the initiation of Cre enzyme activity. Once the Cre becomes activated, all Nkx2.2-expressing cells and their descendants are labeled with the Tomato reporter. At E14.5, there were no Tomato-labeled cells detected in the intestinal epithelium (Fig. 3A). Single Tomato-stained cells were detected as early as E15.5, consistent with the beginning of Nkx2.2 expression in the intestinal epithelium at this early stage of development (11) (Fig. 3B). To confirm the efficiency of the Cre enzyme, we analyzed the crypt region in 6-wk-old mice and found that the majority of crypts contain Tomato-labeled cells (Fig. 3C).
Fig. 3.
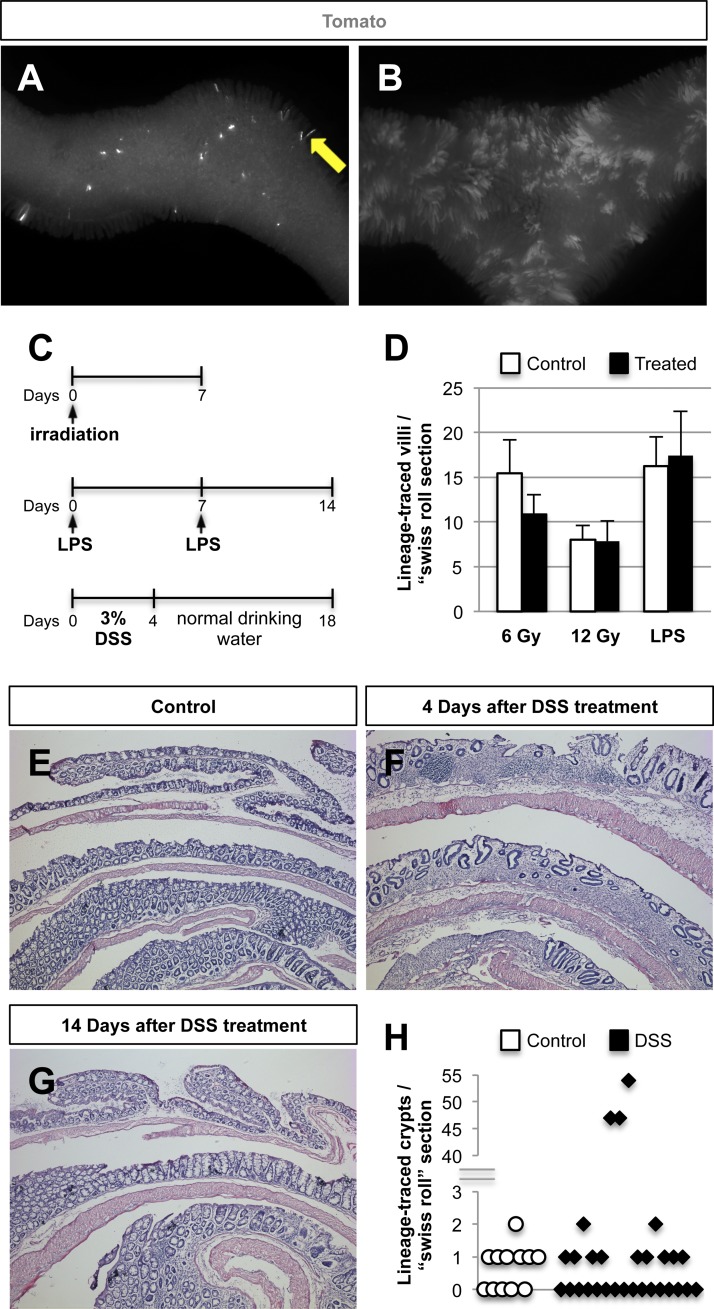
Lineage-tracing of Nkx2.2-expressing cells in the murine intestinal epithelium. Activity of Nkx2.2Cre is verified by Tomato staining in sections of embryonic day (E) 14.5 (A) and E15.5 (B) Nkx2.2Cre/+;R26RTomato embryos. Tomato-stained cells can be found as early as E15.5 in the intestine (Int). P, pancreas. C: in 6-wk-old mice, Tomato-stained cells are detected in almost every crypt. Whole-mount images (D and G) and sections (E and H) from the small (D and E) and large (G and H) intestine of 6-wk-old Nkx2.2Cre/+;R26RTomato mice show single Tomato+ cells as well as whole crypts and villi being Tomato labeled. Magnification: ×4 (C, D, and G) and ×20 (A, B, E, and H). Quantification of Tomato-stained villi per “swiss roll” section in the small intestine (F; n = 12) and Tomato-labeled crypts per “swiss roll” section in the large intestine (I; n = 4) are shown.
To assess the stem cell potential of Nkx2.2-expressing enteroendocrine cells, whole-mount analysis of the small and large intestine of adult Nkx2.2Cre/+;R26RTomato mice was performed. We detected scattered, single Tomato+ cells throughout the intestine. In addition, entire crypt-villus units within the small intestine and crypts within the large intestine were sporadically labeled with the Tomato reporter (Fig. 3, D–I), indicating that a rare subset of Nkx2.2-expressing enteroendocrine cells had stem cell potential. On average, we detected 12.61 ± 1.91 Nkx2.2 lineage-traced villi in each “swiss roll” (24) section of the whole small intestine of 7- to 10-wk-old mice (Fig. 3F), whereas in the large intestine we identified 0.67 ± 0.33 Nkx2.2 lineage-traced crypts per “swiss roll” section of 10-wk-old mice (Fig. 3I).
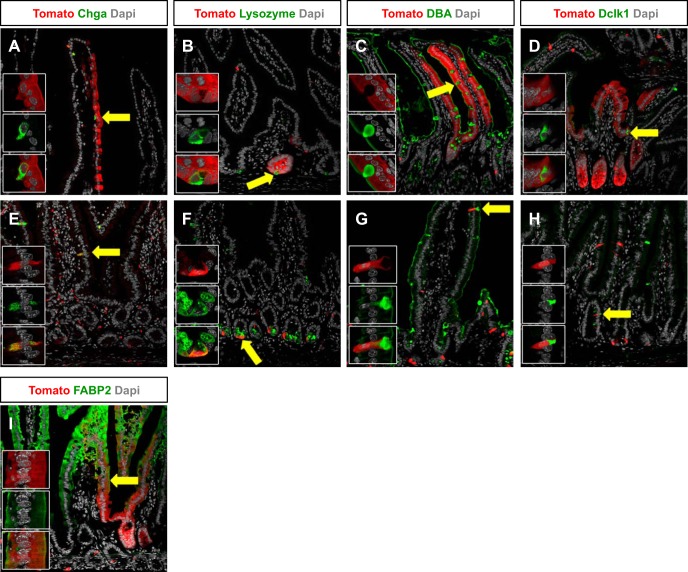
To determine whether all intestinal epithelial cell lineages could be derived from Nkx2.2-expressing cells, we performed immunofluorescence staining with the Tomato reporter and specific markers of the different intestinal epithelial cell types, including enteroendocrine, Paneth, goblet, and tuft cells, as well as enterocytes. Within crypt-villus units that were entirely labeled with the Tomato tracer, we were able to detect costaining of each of the epithelial cell types, including Chga-expressing enteroendocrine cells, lysozyme-expressing Paneth cells, Dolichos biflorus agglutinin lectin-marked goblet cells, Dclk1-expressing tuft cells, and Fabp2-expressing enterocytes (Fig. 4, A–D and I). As expected from the Nkx2.2 expression analysis, the majority of single Tomato+ cells represent Nkx2.2-expressing enteroendocrine cells, identified by Chga expression (Figs. 1A and 4E) (11, 42). Furthermore, small populations of single Paneth, goblet, and tuft cells were also labeled with the Tomato reporter (Fig. 4, F–H). These data show that all cell types of the intestinal epithelium can be derived from a rare subset of Nkx2.2-expressing enteroendocrine cells.
Fig. 4.
Nkx2.2 lineage tracing labels all intestinal epithelial cell types. Immunofluorescence of the small intestine of 6-wk-old Nkx2.2Cre/+;R26RTomato demonstrates that enteroendocrine cells marked by Chga are Tomato positive (A and E; arrow). Lysozyme-expressing Paneth cells (B and F; arrow), goblet cells that are marked by DBA (C and G; arrow), rare Dclk1-expressing tuft cells (D and H; arrow), and fatty acid binding protein 2 (Fabp2)-expressing enterocytes (I; arrow) are stained with Tomato as well. Confocal images are shown. Magnification: ×20 (A–I) and ×63 (insets from arrow).
Enteroendocrine progenitor cells expressing Nkx2.2 and Ngn3 have low stem cell potential.
Interestingly, lineage tracing with the enteroendocrine progenitor cell marker Ngn3 has been reported to show a similar labeling pattern as Nkx2.2 lineage tracing; individual cells as well as rare crypt-villus units are stained with the reporter gene in the small intestine (32) (Fig. 3, D and E). Since Nkx2.2 and Ngn3 are coexpressed in a subset of crypt cells in adult mice (42), we hypothesized that these coexpressing cells could represent the enteroendocrine cell population that possesses stem cell potential. We used a dimerizable Cre system that allows for cell-specific expression of two inactive Cre peptides (nCre and cCre) that can efficiently dimerize and reconstitute Cre activity when coexpressed in the same cell (43). Nkx2.2cCre/+;Ngn3nCre/+ knock-in mice were crossed to R26RTomato reporter mice to assess whether Nkx2.2+/Ngn3+ double-positive cells demarcated a specialized enteroendocrine progenitor population that can give rise to all intestinal epithelial cell types. Similar to the lineage tracing of Nkx2.2-expressing cells, we could detect scattered, individual Tomato-labeled cells from the Nkx2.2+/Ngn3+ coexpressing lineages throughout the small intestine. However, there were few entirely lineage-labeled crypt-villus units compared with Nkx2.2-derived lineages (Figs. 3, D–F, and 5). The lower number of completely lineage-labeled crypt-villus units in Nkx2.2cCre/+;Ngn3nCre/+;R26RTomato animals is not likely due to the lower efficiency of the dimerizable Cre system, since the amount of single Tomato-labeled cells appear to be similar in Nkx2.2Cre/+;R26RTomato and Nkx2.2cCre/+;Ngn3nCre/+;R26RTomato mice (Figs. 3D and 5A). Furthermore, the efficiency of Cre-mediated recombination in the pancreas of Nkx2.2cCre/+;Ngn3nCre/+ mice is >95% (data not shown). Therefore, the data suggest that the Nkx2.2+/Ngn3+ coexpressing cells do not represent a specialized enteroendocrine cell population with elevated stem cell potential.
Fig. 5.
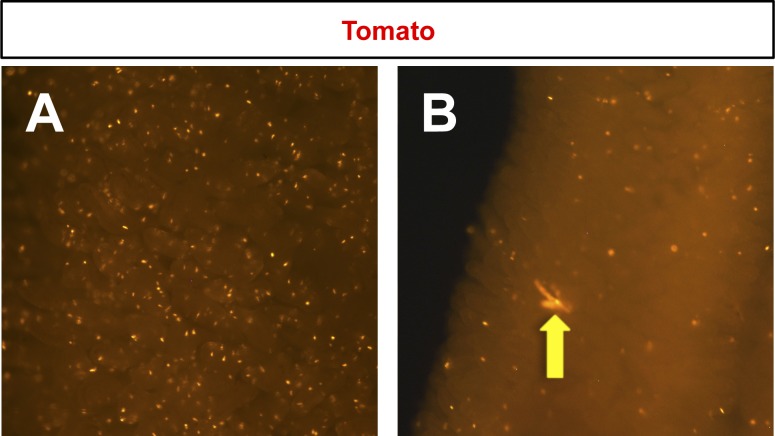
Lineage-labeling of Nkx2.2+/neurogenin 3+ (Ngn3+) coexpressing cells. Whole-mount images of the small intestine of 6-wk-old Nkx2.2cCre/+;Ngn3nCre/+;R26RTomato mice show single Tomato-stained cells (A and B) and rare Nkx2.2+/Ngn3+ lineage-labeled crypt-villus units (B, arrow). N = 2. Magnification: ×2.5.
Postmitotic Nkx2.2+ cells can form organoids in vitro.
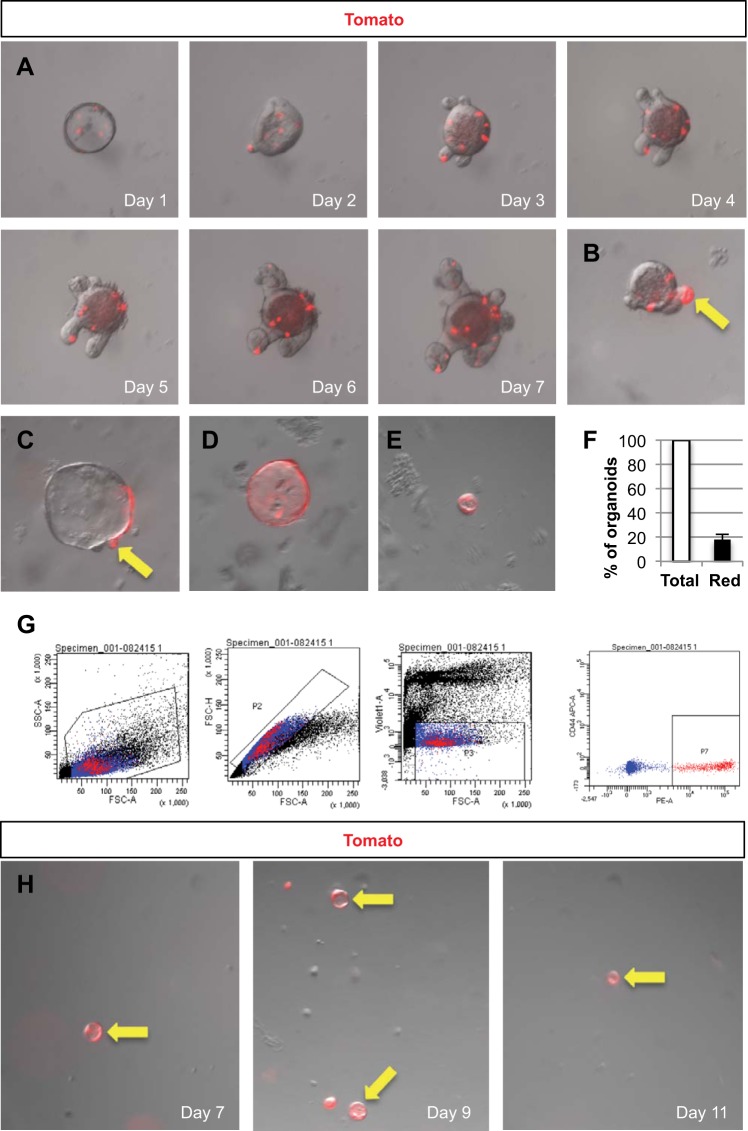
To confirm that a subpopulation of Nkx2.2-expressing enteroendocrine cells are multipotent and to exclude the possibility that the few Nkx2.2 lineage-labeled crypt-villus units are due to spurious expression, whole crypt organoids were cultured from 6-wk-old Nkx2.2Cre/+;R26RTomato mice. Tomato-labeling of the organoids was observed over a 7-day time course. At day 7, 18 ± 4% of the organoids contained clusters of Tomato-stained cells (Fig. 6), indicating that the Tomato clusters can arise from Nkx2.2+ cells. Some budding crypts were also Tomato-labeled (Fig. 6, B and C, arrows), showing that Nkx2.2+ cells also give rise to crypts. In addition, a few organoids were completely Tomato-labeled (Fig. 6, D and E), indicating that either the isolated crypt was already entirely Tomato+ at the time of crypt isolation, or a stem cell was labeled and gave rise to other cells of the organoid during the course of the experiment. To assess the lineage potential of single Nkx2.2+ enteroendocrine cells, we isolated single Tomato+ cells via FACS (Fig. 6G) from the intestine of 6-wk-old to 7-mo-old Nkx2.2Cre/+;R26RTomato mice and cultured these single Tomato+ cells for up to 11 days. Remarkably, single cells from both young and old animals were able to form small organoids that were entirely Tomato-labeled (Fig. 6H), indicating that either an actively expressing Nkx2.2+ cell or an Nkx2.2 descendant cell can give rise to organoids. It is interesting to note that the organoids did not develop the typical budding crypts, but formed spheres that were similar in size and shape to spheroids present in the whole crypt organoid culture (Fig. 6, E and H). These combined in vitro data suggest that a subset of Nkx2.2-expressing cells have the potential to replicate and form organoids in culture.
Fig. 6.
Nkx2.2 lineage-tracing in small intestinal crypt organoid cultures. Crypts were isolated from 6- to 7-wk-old Nkx2.2Cre/+;R26RTomato mice and cultured for 7 days. A: an image of a single representative organoid was taken every day over the time course of the crypt culture. Organoids show scattered Tomato-stained cells. After 7 days in culture, some organoids developed crypts that are entirely Tomato-labeled (B and C, arrows), whereas other organoids are completely Tomato red (D and E). Magnification: ×10. F: quantification of organoids with Tomato-stained cell clusters in proportion to the total amount of organoids in the culture (n = 5). G: viable single Tomato+ cells were isolated via fluorescence-activated cell sorting from the small intestine of 6-wk-old to 7-mo-old Nkx2.2Cre/+;R26RTomato mice and grown for up to 11 days into organoids. H: small, round organoids grew that were entirely Tomato-labeled. FSC, forward-scattered light; PE, phycoerythrin.
Nkx2.2-expressing cells with stem cell potential do not respond to injury of the intestinal epithelium.
Similar to the limited multipotent properties of Nkx2.2+ cells in vitro, the majority of Nkx2.2Cre/+;R26RTomato mice analyzed only showed sporadic labeling of entire crypt-villus units. However, we occasionally detected extensive expansion of Nkx2.2 lineage-labeled villi (Fig. 7, A and B), indicating that a subset of Nkx2.2-expressing enteroendocrine cells can be mobilized to repopulate the intestine under certain physiological conditions. To analyze whether injury of the intestine can activate these Nkx2.2-expressing cells with stem cell properties, we performed 6- and 12-Gy whole body irradiation on 6- to 7-wk-old Nkx2.2Cre/+;R26RTomato mice (6 Gy: n = 3, 12 Gy: n = 5). Following one dose of radiation, mice were killed after 7 days (Fig. 7C) and Tomato-labeled, Nkx2.2 lineage-traced villi quantified. We observed no difference in the amount of Tomato+ villi in the small intestine of 6- or 12-Gy irradiated mice compared with nonirradiated controls (Fig. 7D).
Fig. 7.
No change in the amount of whole Nkx2.2 lineage-traced villi after irradiation, LPS, and dextran sodium sulfate (DSS) treatment. A and B: whole-mount images of the small intestine of 7-mo-old Nkx2.2Cre/+;R26RTomato mice. A: in most mice, rare entire crypt-villus units in the small intestine are Tomato labeled (arrow). B: occasionally, the amount of entirely Tomato-stained villi increased (n = 1). C: schematic overview of the used intestinal injury models. D: quantification of Tomato-stained villi in the small intestine of 6-Gy, 12-Gy, and LPS-treated mice compared with control mice shows no significant differences (6 Gy: n = 3, 12 Gy: n = 5, LPS: n = 4, 3 “swiss” roll sections analyzed per sample). Hematoxylin and eosin stained sections of the large intestine of control (E) and DSS-treated mice (F and G) confirm that colitis is apparent 4 days after the last day of DSS treatment (F), and the large intestine is regenerated 14 days after the last day of DSS (G). Magnification: ×1 (A and B) and ×5 (E–G). H: quantification of Tomato-stained crypts in the large intestine of DSS-treated mice compared with control mice does not show a significant change in most samples. However, the large intestine of one mouse shows a dramatic increase in Nkx2.2 lineage-labeled crypts (control: n = 4, DSS: n = 10, 3 “swiss” roll sections analyzed per sample).
To examine whether inflammation induces the activity of the subset of Nkx2.2-expressing enteroendocrine cells that have expanded lineage potential, mice were treated with either LPS or DSS. Intraperitoneal injection of the endotoxin LPS triggers a systemic inflammatory response (10), whereas administration of 3% DSS in the drinking water induces colitis (26). Eight-week-old Nkx2.2Cre/+;R26RTomato mice (n = 4) were injected with 2.5 mg/kg LPS on days 0 and 7 and dissected 1 wk after the last injection on day 14, and then we examined the number of Tomato-labeled villi in the small intestine (Fig. 7C). Systemic inflammation induced by LPS did not alter the number of Nkx2.2 lineage-traced villi in the small intestine compared with PBS-injected controls (Fig. 7D). Colitis in 8-wk-old Nkx2.2Cre/+;R26RTomato mice (control: n = 4, DSS: n = 10) was induced by 3% DSS in the drinking water for 4 days after which the mice were switched to normal drinking water until day 18 when they were killed (Fig. 7C). Successful induction of colitis was confirmed by analyzing H&E-stained sections of the large intestine of a mouse treated for 4 days with 3% DSS and left untreated for another 4 days before analysis. The intestinal epithelium shows common features of colitis, including the disappearance of crypts (27) (Fig. 7F). To demonstrate that the epithelium of the large intestine completely regenerated at the end of the experiment (day 18; 14 days after the last day of DSS treatment), we analyzed the tissue histology of the large intestine by H&E staining. The intestinal epithelium of the large intestine from DSS-treated mice after the recovery period was indistinguishable from those of control mice that received normal drinking water over the entire course of the experiment (Fig. 7, E and G). Most DSS-treated mice did not show an increase in Tomato-labeled crypts in the large intestine. However, large-intestine sections of one mouse exhibited elevated numbers of Tomato+ crypts, suggesting that colitis might enhance activation of Nkx2.2+ cells (Fig. 7H). However, since only one mouse responded with increased number of Tomato+ crypts after inflammation, we hypothesize that additional insults need to be combined with inflammation to activate the subset of Nkx2.2-expressing enteroendocrine cells with increased lineage potential.
Taken together, these data show that Nkx2.2+ enteroendocrine cells that have multipotent properties do not play a dominant role in intestinal epithelial homeostasis after injury, but might be activated by inflammation. Since we observe sporadic expansion of the Nkx2.2 lineages (Fig. 7, A and B), it is possible that a combination of physiological assaults is necessary to activate the multipotent Nkx2.2-expressing enteroendocrine population.
Nkx2.2 is dispensible for the pluripotency of intestinal stem cells.
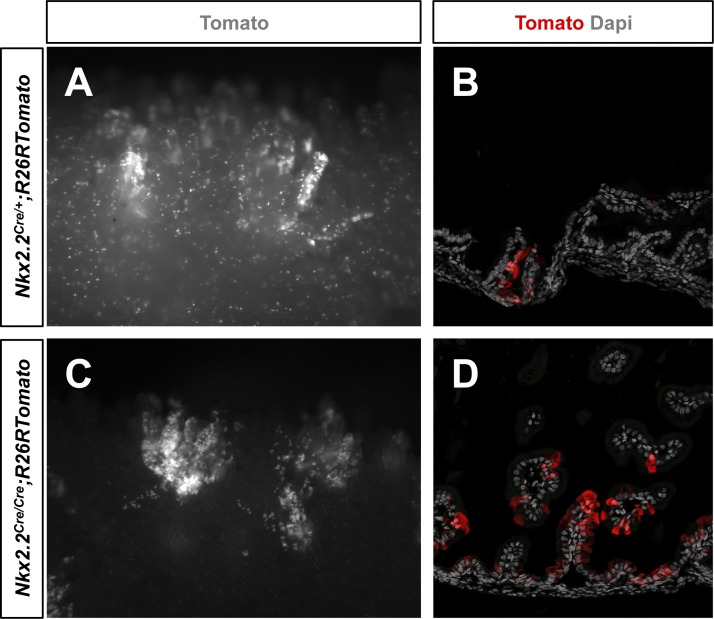
To investigate whether Nkx2.2 is important for the function of stem cells, we deleted Nkx2.2 using homozygous Nkx2.2Cre/Cre knock-in mice. Since these mice die shortly after birth, the intestines of newborn animals were analyzed for Nkx2.2 lineage tracing. We detected fewer single Tomato-labeled cells in Nkx2.2Cre/Cre mice (Fig. 8, A and C), consistent with the reduction of enteroendocrine cell populations in Nkx2.2 knockout mice (11). However, similar to heterozygous Nkx2.2Cre/+ mice, entire Tomato+ crypt-villus units were observed (Fig. 8), which seemed to appear by a similar frequency (data not shown), suggesting that Nkx2.2 is dispensable for pluripotency of this stem cell-like population.
Fig. 8.
Nkx2.2 appears to be dispensable for pluripotency. Tomato-stained villi were found by whole-mount imaging (A and C) and in sections (B and D) of the small intestine of newborn Nkx2.2Cre/+;R26RTomato (A and B) and mutant Nkx2.2Cre/Cre;R26RTomato (C and D) mice. Magnification: ×4 (A and C) and ×20 (B and D).
DISCUSSION
Two stem cell populations have been identified in the small intestine of mice. The highly proliferating CBCs express Lgr5 and reside in the crypt base of both the small and large intestine (4). The Bmi1+ stem cell population, on the other hand, is mainly found at the +4 position of the crypt in the proximal small intestine (31). Recent data indicate that other cell populations in the crypt can contribute to the stem cell pool, as secretory progenitor cells, and subpopulations of Paneth and enteroendocrine cells become activated and gain stem cell characteristics upon intestinal damage (7, 30, 40, 41). These findings suggest that there are still unanswered questions regarding the stem cell populations within the intestine. In particular, it remains elusive whether differentiated cell populations in the intestinal crypt can contribute to the stem cell pool under normal intestinal homeostasis, and which particular physiological conditions activate them.
In this study, we utilized a constitutive Nkx2.2 lineage-tracing approach to demonstrate that a subset of Nkx2.2-expressing mature enteroendocrine cells has stem cell properties. The Nkx2.2Cre mice have the Cre gene inserted into the endogenous Nkx2.2 genomic locus so that Cre faithfully recapitulates Nkx2.2 expression (2). In this study, we confirmed that Cre is accurately expressed in the Nkx2.2 expression domain of the intestine and marks all Nkx2.2 derivative cells with the reporter gene. Although there are limitations to this constitutive lineage-tracing technique, since you cannot control the timing of Cre activation, the high efficiency of constitutive lineage-tracing allowed us to identify a rare Nkx2.2-expressing enteroendocrine population that has expanded lineage potential. An inducible Nkx2.2-Cre system would have provided a more refined analysis of the prospective stem cell; however, these mice were not available for our studies. Furthermore, an inducible Nkx2.2 lineage-tracing strategy may have failed to identify the rare subset of enteroendocrine cells that displayed multipotent properties due to the lower efficiency associated with inducible Cre alleles.
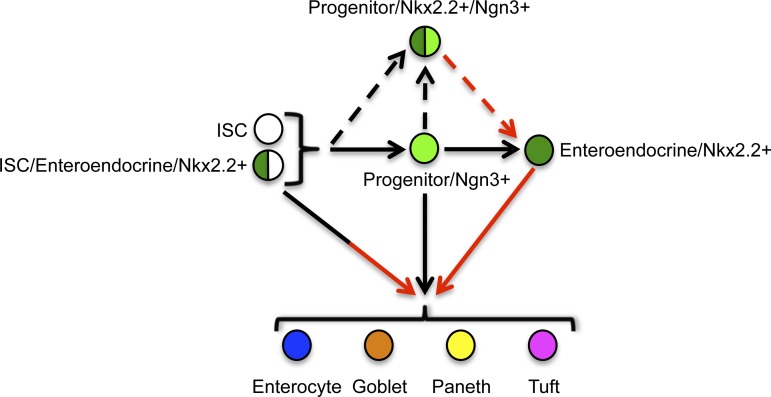
While single Ngn3-expressing enteroendocrine progenitor cells also have the capacity to give rise to differentiated epithelial cell types other than enteroendocrine populations (32), we demonstrate that the Nkx2.2+/Ngn3+ coexpressing enteroendocrine progenitors do not show enriched stem cell capacity. This would suggest that single Nkx2.2+ cells that have stem cell potential do not represent the Nkx2.2-expressing enteroendocrine progenitors, but are a more mature enteroendocrine population, as schematized in the model (Fig. 9).
Fig. 9.
Nkx2.2-expressing mature enteroendocrine cells can give rise to all cell types of the intestinal epithelium. The Lgr5+ and Bmi1+ intestinal stem cells (ISC; white) are known to give rise to all intestinal epithelial cell types, including enterocytes (blue), goblet cells (orange), Paneth cells (yellow), tuft cells (pink), and enteroendocrine cells (green). In addition, it has been shown that enteroendocrine cells express Nkx2.2 and are derived from Ngn3-expressing progenitor cells (light green). Lineage tracing with Ngn3 has further demonstrated that Ngn3+ enteroendocrine progenitor cells can give rise to all intestinal epithelial cell types. In this study, we showed that some Nkx2.2-expressing enteroendocrine cells coexpress Lgr5 or Bmi1 (green/white). Furthermore, we demonstrated that Nkx2.2-expressing mature enteroendocrine cells can give rise to all intestinal epithelial cell types. In contrast, Nkx2.2+/Ngn3+ coexpressing progenitor cells (green/light green) are not able to give rise to all intestinal cell types. Black arrows, published by others; dashed arrows, hypothetical; red arrows, shown in this study.
We demonstrate that Nkx2.2 is expressed in a rare subset of Lgr5+ CBCs in the small and large intestine. This is consistent with a recent report that identified a rare Lgr5+ enteroendocrine cell population (15). However, expression of Nkx2.2 was mainly detected in the Bmi1+ stem cells of the proximal small intestine. This is supported by the predominant localization of Nkx2.2 to the +5 to +7 position of the crypt, where +4 (Bmi1+) stem cells can be found as well (5). Interestingly, costaining with Chga revealed that the majority of Nkx2.2+/Bmi1+ coexpressing cells are enteroendocrine cells. It is possible that retention of Bmi1 expression in this differentiated cell population is indicative of sustained stem cell capacity. The lineage-tracing experiments in vivo and in vitro confirmed that few Nkx2.2-expressing enteroendocrine cells possess stem cell potential, as only a subpopulation of Nkx2.2+ cells gives rise to all intestinal cell types within an entire crypt-villus unit during normal intestinal homeostasis. Another possible explanation for the occasional Tomato+ crypt-villus units could be the high plasticity of intestinal tissue, such that a rare subset of Nkx2.2-expressing enteroendocrine cells dedifferentiates to a more multipotent progenitor state. It is unlikely that the labeled Nkx2.2-derived lineages are initiated from an early endodermal cell that gave rise to a stem cell during fetal development, since Nkx2.2 expression has never been observed in undifferentiated endoderm-derived intestinal lineages. Furthermore, several of the Nkx2.2 lineage-labeled budding crypts that formed in the in vitro whole crypt organoid cultures were labeled after the initiation of the adult-derived organoid cultures. Interestingly, in addition to entire crypt-villus units being lineage labeled, we identified several isolated Nkx2.2 lineage-traced goblet, Paneth, and tuft cells. This observation is similar to that previously reported for lineage-tracing with Ngn3 in the intestine (32). By using β-galactosidase as a reporter for Nkx2.2 expression in the intestine of Nkx2.2LacZ/+ mice, we were unable to detect any goblet, Paneth, and tuft cells expressing Nkx2.2(LacZ). Therefore, we conclude that the isolated Nkx2.2 lineage-traced cells found in Nkx2.2Cre/+;R26RTomato mice are progeny from a cell that formerly expressed Nkx2.2 rather than actively expressing Nkx2.2. It is unlikely that the expression pattern of the Nkx2.2LacZ/+ allele differs from the Nkx2.2Cre/+ allele, since both are knock-in alleles under the same promoter and enhancer elements. The function of the transcription factor Nkx2.2, however, does not appear important for maintenance of pluripotency of Nkx2.2-expressing stemlike cells, since entirely Tomato-labeled crypt-villus units were still visible after ablation of Nkx2.2.
Consistent with our findings, several studies in other tissues as well as in the intestine have revealed that differentiated cells have the potential to serve as stem cells. In the proximal tubule of the kidney, differentiated epithelial cells have been shown to dedifferentiate following injury. These cells proliferate and express stem cell markers to repair the tissue (20). In the stomach, mature Troy+ chief cells act as reserve stem cells to generate all cell lineages of the gastric epithelium (34). Furthermore, secretory cells in the lung airway epithelium are able to convert into functional stem cells in vivo. Following ablation of the airway stem cells, the secretory cells start proliferating, and lineage-tracing analysis confirms that the secretory cells can dedifferentiate into basal stem cells (37). In the intestine, it was previously demonstrated that mature enteroendocrine cells can express stem cell markers (33, 41). Analyses of transgenic mice that express GFP under the promoter for the hormone cholecystokinin (CCK) showed that CCK-GFP+ cells in the crypt express not only the enteroendocrine cell marker Chga, but also several stem cell markers, including Lgr5. These cells were negative for the proliferation markers Ki-67 and phospho-Histone H3 (33), indicating that the cells were either mature or quiescent, but not actively proliferating. Furthermore, it has been shown that intestinal epithelial cells expressing high levels of SRY box 9 (Sox9) express markers for terminally differentiated enteroendocrine cells (13). Flow cytometry and microarray analysis demonstrated that these Sox9High enteroendocrine cells also express the +4 stem cell marker Bmi1. Only after radiation injury are these Sox9High enteroendocrine cells able to form organoids in vitro (41), suggesting that they are able to contribute to the regeneration process, but are not important during normal intestinal epithelial homeostasis. In contrast to these observations, our data show that Nkx2.2-expressing mature enteroendocrine cells are sporadically activated to become stem cells during normal intestinal epithelial homeostasis, as well as during regeneration of the intestinal epithelium. However, their activity could not be enhanced by the used intestinal injury models. Although Nkx2.2 is coexpressed with the Lgr5 and Bmi1 stem cell markers, it is surprising that intestinal injury by irradiation does not enhance the activity of the Nkx2.2-expressing cells. Recent publications have shown that ablation of the Lgr5+ stem cells with either 12-Gy irradiation or diphtheria toxin results in an expansion of the Bmi1+ lineage (38, 44). Therefore, one would expect that there would be more entire Nkx2.2 lineage-traced crypt villus units after 12-Gy irradiation, since Nkx2.2 is coexpressed with Bmi1 in 87% of the total Bmi1+ cells. Instead, it seems the Nkx2.2-expressing cells are radiation resistant.
The intestine is a tissue with high self-renewal capacity that constantly responds to changing environmental stimuli, including harsh conditions and continual changes in nutrients and microorganisms. To maintain robust intestinal homeostasis, it is likely that the intestine has developed multiple backup mechanisms beyond the two well-characterized stem cell populations to ensure organ integrity. We hypothesize that a subset of differentiated cell populations, such as Nkx2.2+/Bmi1+ enteroendocrine cells, retain stem cell-like characteristics so that they can easily be reactivated in response to changing physiological conditions to supplement, or replace when necessary, the activities of the canonical stem cell populations.
Our data demonstrate that the Nkx2.2-expressing enteroendocrine cells that have expanded lineage potential are present in the small and large intestine, but that their activity is not greatly enhanced by the intestinal injury models analyzed in this study. Given the robustness of the Lgr5+ and Bmi+ canonical stem cells, it is likely that activation of a reserve stem cell population may require additional or more extreme environmental insults, suggesting that the stem cell-like Nkx2.2+ enteroendocrine cells follow the “two-hit-hypothesis” (19). Further studies are needed to better define which conditions alone or in combination influence the cellular plasticity and lineage potential of the subset of Nkx2.2-expressing enteroendocrine cells that have increased intestinal cell lineage potential.
GRANTS
This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases Grant R01 DK-082590 (L. Sussel) and American Diabetes Association Grant 7-11-MN-61 (L. Sussel and S. Gross). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: S.G. and L.S. conception and design of research; S.G., D.B., J.L., and S.A. performed experiments; S.G. analyzed data; S.G. and L.S. interpreted results of experiments; S.G. prepared figures; S.G. drafted manuscript; S.G., D.B., J.L., S.A., G.G., T.C.W., and L.S. edited and revised manuscript; S.G. and L.S. approved final version of manuscript.
Supplementary Material
ACKNOWLEDGMENT
We thank Yoku Hayakawa and Ashlesha Muley from the Wang laboratory for technical advice and helpful discussions. We thank Lisa Mazinski at New York University for help with slide scanning. We thank the Flow Cytometry Core at Columbia University for assistance with cell sorting by FACS. We thank the Sussel laboratory for critical reading of the manuscript.
Present address of S. Asfaha: Victoria Research Laboratory, Rm A4-140, 800 Commissioners Road East, University of Western Ontario, London, Ontario, Canada N6A 5W9.
REFERENCES
- 1.Arnes L, Leclerc K, Friel JM, Hipkens SB, Magnuson MA, Sussel L. Generation of Nkx2.2:lacZ mice using recombination-mediated cassette exchange technology. Genesis 50: 612–624, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balderes DA, Magnuson MA, Sussel L. Nkx2.2:Cre knock-in mouse line: a novel tool for pancreas- and CNS-specific gene deletion. Genesis 51: 844–851, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker N, van de Wetering M, Clevers H. The intestinal stem cell. Genes Dev 22: 1856–1864, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449: 1003–1007, 2007. [DOI] [PubMed] [Google Scholar]
- 5.Barker N, van Oudenaarden A, Clevers H. Identifying the stem cell of the intestinal crypt: strategies and pitfalls. Cell Stem Cell 11: 452–460, 2012. [DOI] [PubMed] [Google Scholar]
- 6.Briscoe J, Sussel L, Serup P, Hartigan-O'Connor D, Jessell TM, Rubenstein JL, Ericson J. Homeobox gene Nkx2.2 and specification of neuronal identity by graded Sonic hedgehog signalling. Nature 398: 622–627, 1999. [DOI] [PubMed] [Google Scholar]
- 7.Buczacki SJ, Zecchini HI, Nicholson AM, Russell R, Vermeulen L, Kemp R, Winton DJ. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature 495: 65–69, 2013. [DOI] [PubMed] [Google Scholar]
- 8.Cheng H, Leblond CP. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. I. Columnar cell. Am J Anat 141: 461–479, 1974. [DOI] [PubMed] [Google Scholar]
- 9.Cheng H, Leblond CP. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. V. Unitarian theory of the origin of the four epithelial cell types. Am J Anat 141: 537–561, 1974. [DOI] [PubMed] [Google Scholar]
- 10.Copeland S, Warren HS, Lowry SF, Calvano SE, Remick D,. Inflammation, and the host response to injury. I. Acute inflammatory response to endotoxin in mice and humans. Clin Diagn Lab Immunol 12: 60–67, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desai S, Loomis Z, Pugh-Bernard A, Schrunk J, Doyle MJ, Minic A, McCoy E, Sussel L. Nkx2.2 regulates cell fate choice in the enteroendocrine cell lineages of the intestine. Dev Biol 313: 58–66, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer AH, Jacobson KA, Rose J, Zeller R. Hematoxylin and eosin staining of tissue and cell sections. CSH Protoc 2008: pdb.prot4986, 2008. [DOI] [PubMed] [Google Scholar]
- 13.Formeister EJ, Sionas AL, Lorance DK, Barkley CL, Lee GH, Magness ST. Distinct SOX9 levels differentially mark stem/progenitor populations and enteroendocrine cells of the small intestine epithelium. Am J Physiol Gastrointest Liver Physiol 296: G1108–G1118, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gradwohl G, Dierich A, LeMeur M, Guillemot F. Neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci U S A 97: 1607–1611, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grun D, Lyubimova A, Kester L, Wiebrands K, Basak O, Sasaki N, Clevers H, van Oudenaarden A. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature 525: 251–255, 2015. [DOI] [PubMed] [Google Scholar]
- 16.Hayakawa Y, Jin G, Wang H, Chen X, Westphalen CB, Asfaha S, Renz BW, Ariyama H, Dubeykovskaya ZA, Takemoto Y, Lee Y, Muley A, Tailor Y, Chen D, Muthupalani S, Fox JG, Shulkes A, Worthley DL, Takaishi S, Wang TC. CCK2R identifies and regulates gastric antral stem cell states and carcinogenesis. Gut 64: 544–553, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hosen N, Yamane T, Muijtjens M, Pham K, Clarke MF, Weissman IL. Bmi-1-green fluorescent protein-knock-in mice reveal the dynamic regulation of bmi-1 expression in normal and leukemic hematopoietic cells. Stem Cells 25: 1635–1644, 2007. [DOI] [PubMed] [Google Scholar]
- 18.Jenny M, Uhl C, Roche C, Duluc I, Guillermin V, Guillemot F, Jensen J, Kedinger M, Gradwohl G. Neurogenin3 is differentially required for endocrine cell fate specification in the intestinal and gastric epithelium. EMBO J 21: 6338–6347, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A 68: 820–823, 1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kusaba T, Lalli M, Kramann R, Kobayashi A, Humphreys BD. Differentiated kidney epithelial cells repair injured proximal tubule. Proc Natl Acad Sci U S A 111: 1527–1532, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13: 133–140, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahe MM, Aihara E, Schumacher MA, Zavros Y, Montrose MH, Helmrath MA, Sato T, Shroyer NF. Establishment of gastrointestinal epithelial organoids. Curr Protoc Mouse Biol 3: 217–240, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.May CL, Kaestner KH. Gut endocrine cell development. Mol Cell Endocrinol 323: 70–75, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moolenbeek C, Ruitenberg EJ. The “Swiss roll”: a simple technique for histological studies of the rodent intestine. Lab Anim 15: 57–59, 1981. [DOI] [PubMed] [Google Scholar]
- 25.Munoz J, Stange DE, Schepers AG, van de Wetering M, Koo BK, Itzkovitz S, Volckmann R, Kung KS, Koster J, Radulescu S, Myant K, Versteeg R, Sansom OJ, van Es JH, Barker N, van Oudenaarden A, Mohammed S, Heck AJ, Clevers H. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent “+4” cell markers. EMBO J 31: 3079–3091, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okayasu I, Hatakeyama S, Yamada M, Ohkusa T, Inagaki Y, Nakaya R. A novel method in the induction of reliable experimental acute and chronic ulcerative colitis in mice. Gastroenterology 98: 694–702, 1990. [DOI] [PubMed] [Google Scholar]
- 27.Perse M, Cerar A. Dextran sodium sulphate colitis mouse model: traps and tricks. J Biomed Biotechnol 2012: 718617, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Potten CS, Kovacs L, Hamilton E. Continuous labelling studies on mouse skin and intestine. Cell Tissue Kinet 7: 271–283, 1974. [DOI] [PubMed] [Google Scholar]
- 29.Rindi G, Leiter AB, Kopin AS, Bordi C, Solcia E. The “normal” endocrine cell of the gut: changing concepts and new evidences. Ann N Y Acad Sci 1014: 1–12, 2004. [DOI] [PubMed] [Google Scholar]
- 30.Roth S, Franken P, Sacchetti A, Kremer A, Anderson K, Sansom O, Fodde R. Paneth cells in intestinal homeostasis and tissue injury. PLoS One 7: e38965, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet 40: 915–920, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schonhoff SE, Giel-Moloney M, Leiter AB. Neurogenin 3-expressing progenitor cells in the gastrointestinal tract differentiate into both endocrine and non-endocrine cell types. Dev Biol 270: 443–454, 2004. [DOI] [PubMed] [Google Scholar]
- 33.Sei Y, Lu X, Liou A, Zhao X, Wank SA. A stem cell marker-expressing subset of enteroendocrine cells resides at the crypt base in the small intestine. Am J Physiol Gastrointest Liver Physiol 300: G345–G356, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stange DE, Koo BK, Huch M, Sibbel G, Basak O, Lyubimova A, Kujala P, Bartfeld S, Koster J, Geahlen JH, Peters PJ, van Es JH, van de Wetering M, Mills JC, Clevers H. Differentiated Troy+ chief cells act as reserve stem cells to generate all lineages of the stomach epithelium. Cell 155: 357–368, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sussel L, Kalamaras J, Hartigan-O'Connor DJ, Meneses JJ, Pedersen RA, Rubenstein JL, German MS. Mice lacking the homeodomain transcription factor Nkx2.2 have diabetes due to arrested differentiation of pancreatic beta cells. Development 125: 2213–2221, 1998. [DOI] [PubMed] [Google Scholar]
- 36.Takeda N, Jain R, LeBoeuf MR, Wang Q, Lu MM, Epstein JA. Interconversion between intestinal stem cell populations in distinct niches. Science 334: 1420–1424, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tata PR, Mou H, Pardo-Saganta A, Zhao R, Prabhu M, Law BM, Vinarsky V, Cho JL, Breton S, Sahay A, Medoff BD, Rajagopal J. Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature 503: 218–223, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tian H, Biehs B, Warming S, Leong KG, Rangell L, Klein OD, de Sauvage FJ. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature 478: 255–259, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Flier LG, Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu Rev Physiol 71: 241–260, 2009. [DOI] [PubMed] [Google Scholar]
- 40.van Es JH, Sato T, van de Wetering M, Lyubimova A, Nee AN, Gregorieff A, Sasaki N, Zeinstra L, van den Born M, Korving J, Martens AC, Barker N, van Oudenaarden A, Clevers H. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat Cell Biol 14: 1099–1104, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Landeghem L, Santoro MA, Krebs AE, Mah AT, Dehmer JJ, Gracz AD, Scull BP, McNaughton K, Magness ST, Lund PK. Activation of two distinct Sox9-EGFP-expressing intestinal stem cell populations during crypt regeneration after irradiation. Am J Physiol Gastrointest Liver Physiol 302: G1111–G1132, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang YC, Gallego-Arteche E, Iezza G, Yuan X, Matli MR, Choo SP, Zuraek MB, Gogia R, Lynn FC, German MS, Bergsland EK, Donner DB, Warren RS, Nakakura EK. Homeodomain transcription factor NKX2.2 functions in immature cells to control enteroendocrine differentiation and is expressed in gastrointestinal neuroendocrine tumors. Endocr Relat Cancer 16: 267–279, 2009. [DOI] [PubMed] [Google Scholar]
- 43.Xu Y, Xu G, Liu B, Gu G. Cre reconstitution allows for DNA recombination selectively in dual-marker-expressing cells in transgenic mice. Nucleic Acids Res 35: e126, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan KS, Chia LA, Li X, Ootani A, Su J, Lee JY, Su N, Luo Y, Heilshorn SC, Amieva MR, Sangiorgi E, Capecchi MR, Kuo CJ. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci U S A 109: 466–471, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.