Abstract
Patients with systemic lupus erythematosus (SLE) and primary Sjögren’s syndrome (pSS) are typically characterized by the presence of autoantibodies and an IFN-signature. The strength of the IFN-signature positively correlates with disease severity, suggesting that type I IFNs are active players in these diseases. BAFF is a cytokine critical for development and proper selection of B cells, and the targeting of BAFF has emerged as a successful treatment strategy of SLE. Previous reports have suggested that BAFF expression is directly induced by type I IFNs, but the precise mechanism for this remains unknown. In this article, we demonstrate that BAFF is a bona fide ISG and that IFN regulatory factors (IRFs) control the expression of BAFF. We identify IRF1 and IRF2 as positive regulators of BAFF transcription and IRF4 and IRF8 as potent repressors; in addition, we have mapped the precise binding site for these factors in the BAFF promoter. IFN-β injections induced BAFF expression mainly in neutrophils and monocytes, and BAFF expression in neutrophils from pSS patients strongly correlated with the strength of the IFN-signature. In summary, we show that BAFF expression is directly induced by type I IFNs via IRF1 and IRF2, whereas IRF4 and IRF8 are negative regulators of BAFF expression. These data suggest that type I IFN blockade in SLE and pSS patients will lead to downregulation of BAFF and a consequential reduction of autoreactive B cell clones and autoantibodies.
Introduction
Systemic lupus erythematosus (SLE) and primary Sjögren’s syndrome (pSS) are systemic autoimmune diseases that strongly associate with a hyperactivated type I IFN system. This manifests as the overexpression of hundreds of IFN-stimulated genes (ISGs) in peripheral blood cells, the so-called IFN-signature, which positively correlates with disease activity (1–5). SLE and pSS patients are also characterized by the presence of autoantibodies against intracellular Ags, indicating that the B cell selection in these patients is not functioning properly. BAFF is a cytokine that is important for the development and selection of B cells. Mice that lack BAFF have a dramatic reduction in the numbers of follicular and marginal zone B cells, whereas mice that overexpress BAFF develop symptoms of systemic autoimmunity because of escape of autoreactive clones and increased T cell help in germinal centers (6). Interestingly, the levels of autoantibodies in pSS patients correlate with the levels of BAFF, suggesting that BAFF also plays an active role in human systemic autoimmune diseases (7). Increased BAFF levels have been suggested to play a role in both SLE and pSS (8, 9). Indeed, because of the increased BAFF levels in serum of SLE and pSS patients, biologics have been developed to block BAFF (10, 11). BAFF blockade using a human anti-BAFF mAb (belimumab) is the first U.S. Food and Drug Administration–approved treatment of SLE using a biologic (12); in addition, promising phase II clinical trials are completed for belimumab in pSS (11). Interestingly, BAFF expression is induced in vivo by type I IFNs, although the exact molecular mechanisms for this remain unknown (13, 14). Understanding how the expression of BAFF is related to the activity of the type I IFN system in SLE and pSS might give important clues to the underlying mechanisms of these diseases. Two previous reports have analyzed the promoter of the BAFF gene and identified putative transcription factor binding sites (TFBS) in the proximal promoter region (15, 16). Woo et al. (16) identified a putative IFN-stimulated response element (ISRE) site but failed to validate it by mutational analysis; furthermore, the reported ISRE site is not conserved between species, indicating that it is a nonfunctional site. Surprisingly, Moon et al. (15) did not identify any ISRE site in the promoter of BAFF. To clarify the mechanism for how type I IFNs may drive the expression of BAFF, we set out to verify that BAFF is directly induced by type I IFNs as a bona fide ISG and to identify genuine TFBS in the BAFF promoter. We first treated multiple sclerosis (MS) patients de novo with IFN-β (Avonex) and quantified the BAFF expression in sorted immune cell subsets before and after treatment. We also sorted immune cell subsets from pSS patients and healthy control subjects followed by quantification of BAFF expression in neutrophils, monocytes, and lymphocytes. BAFF was strongly induced in neutrophils after IFN-β treatment; in addition, BAFF expression correlated with the activity of the type I IFN system in neutrophils from pSS patients. To identify well-conserved TFBS in the promoter of BAFF, we used phylogenetic footprinting and identified a highly conserved ISRE site adjacent to the transcriptional start site (TSS) in the BAFF gene. By using luciferase reporter assays we could identify that IFN regulatory factor (IRF) 1 and IRF2 as potent activators of BAFF expression, and that specific mutations of the identified ISRE site completely abolished the induction of the BAFF by IRF1 and IRF2. Furthermore, we identified IRF4 and IRF8 as strong repressors of BAFF expression and as antagonists to IRF1. We have thus demonstrated that type I IFNs drive the expression of BAFF via the binding of IRFs to a novel ISRE site in the BAFF promoter. In summary, these results might contribute to understanding how BAFF is involved in type I IFN–associated systemic autoimmune diseases.
Materials and Methods
Patients
Blood samples were taken from eight treatment-naive MS patients (one male and seven female patients) before and 18 h after their first Avonex (IFN-β-1a) injection (30 μg/0.5 ml). Blood samples were taken from 21 autoantibody-positive pSS patients who were diagnosed according to the revised European-American consensus criteria. This study was approved by the Regional Ethical Ethics Committee, Stockholm, and all subjects gave informed, written consent.
Cell sorting
Blood samples were subjected to Ficoll–Hypaque (GE Healthcare) separation. First, the PBMC fraction was isolated according to the manufacturer’s protocol; then the remaining gradient was removed without disturbing the granulocyte/erythrocyte pellet. Twenty-five milliliters PBS (Sigma-Aldrich) and 25 ml 3% dextran (VWR) solution was added and the tube was inverted 10 times before it was incubated for 18–20 min at room temperature. The upper, straw-colored fraction was then removed and centrifuged for 10 min at 1000 rpm. Remaining erythrocytes were lysed by adding distilled water for 1 min, followed by addition of PBS to restore isotonicity. PBMCs were stained with CD14-allophycocyanin-Cy7, CD3-FITC, and CD19-allophycocyanin, and neutrophils were stained with CD15-PE (all Abs were from BD Biosciences). During flow cytometry a live gate was applied and populations were detected based on their respective fluorophore. The cells were sorted using a MoFlo flow cytometry cell sorter (Becton Coulter). To isolate CD14+ monocytes for in vitro stimulation using a blocking anti-IFNAR2 Ab, we first isolated PBMCs using Ficoll–Hypaque as described earlier. We then used magnetically labeled anti-CD14 beads (Miltenyi) to isolate human monocytes.
Stimulation of human monocytes with plasma
We cultured human THP-1 monocytic leukemia cells and primary human CD14+ monocytes in 24-well plates (0.5 ml, 106 cells/well). Cells were stimulated in triplicates for 18 h with plasma taken from MS patients before and after Avonex treatment (50 μl plasma/well). To block type I IFN signaling, we pretreated cells with 5 μg/ml of a blocking anti-IFNAR2 Ab (clone MMHAR-2; PBL Assay Science) for 45 min before addition of plasma. Eighteen hours poststimulation the cells were collected and lysed in TRIzol (Life Technologies) for RNA isolation and gene expression analysis using TaqMan quantitative RT-PCR.
Gene expression analysis
TRIzol (Life Technologies) and chloroform (Sigma-Aldrich) were used to extract total RNA from cells sorted from peripheral blood of patients. A total of 50 μg glycogen (Life Technologies) was added during the extraction to enhance RNA yield. RNA quality and quantity were determined using Nano Drop ND-1000 spectrophotometer (Thermo Scientific). cDNA was synthesized using iScript cDNA Synthesis Kit (Bio-Rad) according to the manufacturer’s protocol, and concentrations were within the range 3–10 ng/μl. Amplification was performed using TaqMan probes; MX1: Hs00895608, BAFF: Hs00198106, OAS2: Hs00942643_m1, ACTB: Hs01060665, HPRT: Hs01003267 (Applied Biosystems). Relative expression was calculated using the ΔCt method using HPRT or ACTB for normalization. Gene expression data for BAFF (TNFSF13B) in PBMCs of pSS patients and healthy control subjects were obtained from Gene Expression Omnibus (GSE48378) and analyzed using Multi Experiment Viewer (MeV) and GraphPad Prism software.
BAFF promoter analysis
Using phylogenetic footprinting (rVista 2.0 and ConSite), we searched for conserved TFBS in the −3 kg bp (kb) to +0.5 kb region of the human BAFF (TNFSF13B) gene. We identified a strongly conserved ISRE site adjacent to the TSS (+1) of BAFF. We cloned a 1-kb fragment of the BAFF gene (−867 to +150) and inserted it into the pGL4.14-luc2 plasmid to generate a BAFF reporter construct (BAFF-luc). To verify that the identified ISRE site was functional, we mutated two nucleotides in the ISRE site (mutated BAFF promoter [BAFF-mut]). We used X-tremeGENE 9 (Roche) to transfect HEK293T cells in 24-well plates with these reporter constructs together with IRF constructs and pRL-TK-Renilla for normalization. Luminescence was measured 24 h posttransfection using the Modulus Single Tube Reader (Promega). Relative luminescence unit is reported as firefly (Photinus) luciferase signal divided by Renilla luciferase signal.
Plasmids
The IRF-encoding plasmids were generated as previously described (17) by inserting the coding cDNA of IRFs into the pFLAG-CMV-6c plasmid (Sigma) using TOPO-based PCR cloning. BAFF-luc was generated by PCR cloning of a 1-kb promoter region from the RP23-9206 bacterial artificial chromosome (BACPAC Resources Center, CHORI) that was inserted into pGL4.14-luc2 (Promega). The BAFF-mut was generated by gene synthesis (MWG Eurofins) and inserted into the pEX-K4 vector. The BAFF-mut sequence was amplified by PCR using AccuPrime Pfx DNA polymerase (Invitrogen) and inserted into the pGL4.14 plasmid (Promega) using BglII and XhoI (Thermo Scientific). All plasmids were verified by Sanger sequencing. Plasmid sequences are available on request.
Statistics
The GraphPad Prism software was used for statistical tests and for generating graphs. The analysis of expression data from before and after treatment with Avonex was performed using the Wilcoxon matched-pairs test. BAFF expression levels in pSS patients versus healthy control subjects were analyzed using Student t test. Pearson correlation was used to calculate the correlation between expression levels of BAFF and MX1. Expression data from cells treated with human plasma were analyzed using Student t test.
Results
BAFF production in neutrophils and monocytes is driven by type I IFN in vivo
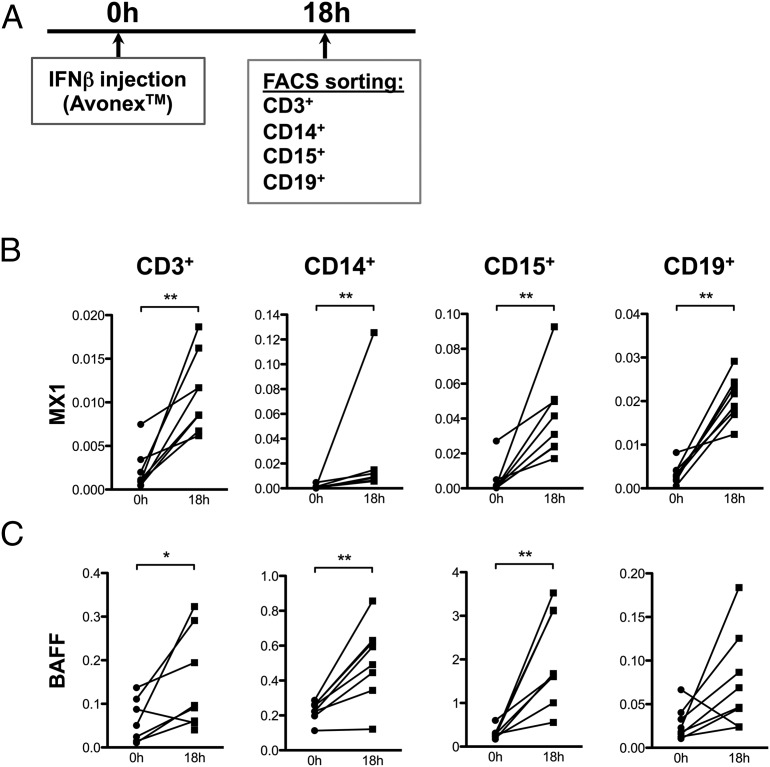
To determine how type I IFNs influence BAFF expression in different immune cell subsets in humans, we sorted peripheral blood cells from treatment-naive MS patients before and after de novo treatment with IFN-β. Peripheral blood was collected before treatment and 18 h posttreatment. After Ficoll separation, T cells (CD3+), B cells (CD19+), and monocytes (CD14+) were sorted from the interphase, whereas neutrophils (CD15+) were sorted from the granulocyte fraction (Fig. 1A). We first quantified the expression of the canonical ISG MX1 before and after Avonex treatment to verify that IFN-β triggered a response in patients and control subjects (Fig. 1B). IFN-β treatment also triggered a strong induction of BAFF expression, primarily in neutrophils, demonstrating that BAFF is a typical ISG (Fig. 1C). BAFF was also induced in monocytes and in T cells. These data establish that IFN-β induces the expression of BAFF primarily in CD15+ neutrophils in vivo in humans.
FIGURE 1.
IFN-β induces BAFF in vivo primarily in monocytes and neutrophils. (A) Peripheral blood was collected from eight MS patients before and 18 h after de novo IFN-β treatment (Avonex). Cells were sorted by flow cytometry to isolate pure T cells (CD3+), B cells (CD19+), monocytes (CD14+), and neutrophils (CD15+), and then prepared for gene expression analysis. (B and C) Expression levels of MX1 and BAFF were quantified in the different cell populations before and after IFN-β treatment using quantitative RT-PCR. The expression levels were normalized to ACTB. Statistics were calculated using the Wilcoxon matched-pairs test (*p ≤ 0.05, **p ≤ 0.01).
BAFF expression is directly induced by type I IFN and positively correlates with the IFN-signature in pSS patients
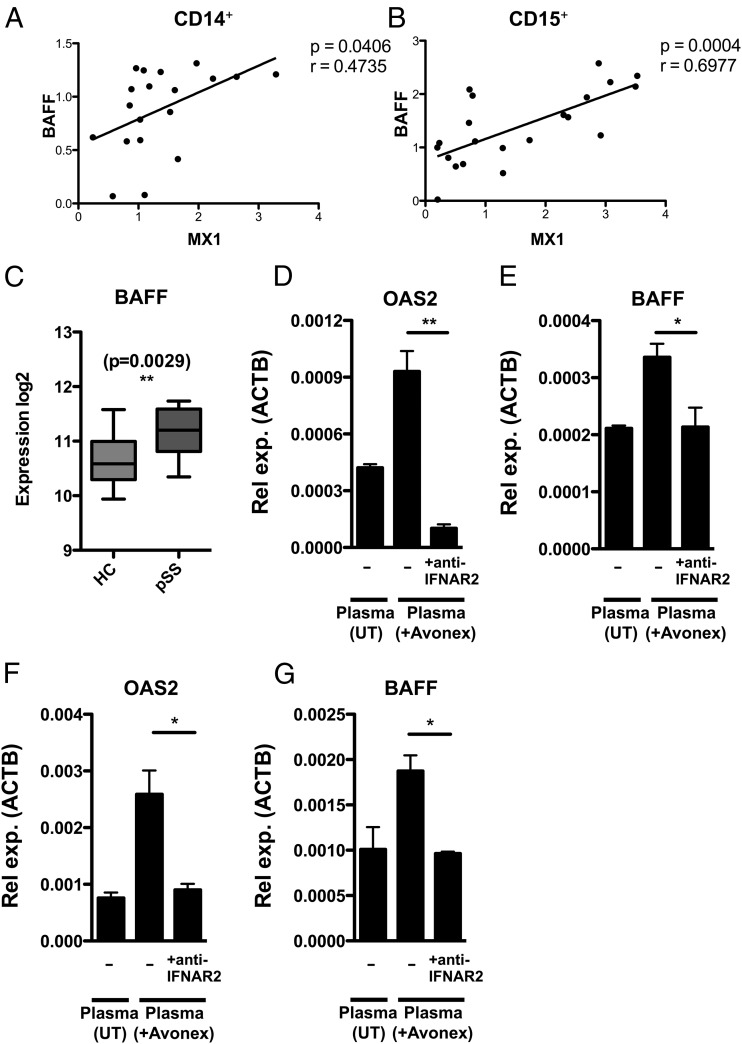
It has previously been reported that BAFF levels are increased in serum from patients with SLE and pSS, and that BAFF levels correlate with the IFN-signature in monocytes from pSS patients (3, 7, 18, 19). To address this in additional cell types, we used flow cytometry to sort CD3+, CD19+, CD14+, and CD15+ cells from peripheral blood of patients with pSS and healthy control subjects, followed by gene expression analysis. The expression levels of BAFF in both monocytes and neutrophils positively correlated with expression levels of the type I IFN surrogate marker MX1 (Fig. 2A, 2B). The correlation between BAFF expression and activity of the type I IFN system was, however, strongest in neutrophils. We could also show that BAFF expression is increased in PBMCs from pSS patients compared with PBMCs from healthy control subjects, this by mining public expression data (GSE48378) (Fig. 2C). Because IFN-β (Avonex) triggered BAFF expression in vivo, and BAFF levels positively correlated with MX1, we hypothesized that the production of BAFF is directly controlled by type I IFNs. To verify that IFN-β directly induces BAFF expression, we stimulated monocytes in vitro with plasma from Avonex-treated MS patients while blocking the IFNAR receptor using a blocking anti-IFNAR2 Ab (Fig. 2D–G). Plasma from Avonex-treated MS patients induced expression of the ISG OAS2 and of BAFF in THP-1 monocytic leukemia cells (Fig. 2D–E) and in primary human CD14+ monocytes (Fig. 2F, 2G). In contrast, plasma from the same patients before Avonex treatment did not induce the expression of OAS2 and BAFF. Importantly, blocking the IFNAR receptor with an anti-IFNAR2 Ab completely abolished the induction of OAS2 and BAFF, demonstrating that type I IFN directly causes the induction of BAFF.
FIGURE 2.
BAFF expression is directly induced by type I IFNs. (A and B) Flow cytometry was used to isolate monocytes (CD14+) and neutrophils (CD15+) from pSS patients followed by gene expression analysis of BAFF and MX1. BAFF expression levels correlated with MX1 expression levels in monocytes (n = 19) and neutrophils (n = 21). Expression levels were normalized to HPRT. Pearson correlation test was used to calculate p and r values. (C) To determine BAFF expression in immune cells of pSS patients, we mined gene expression data from 11 pSS patients and 17 healthy control subjects (HC), and found that BAFF expression was higher in PBMCs from pSS patients. Student t test was used to calculate the p value. To verify that type I IFN is a direct inducer of BAFF in monocytes, we treated THP-1 cells (D and E) and primary human CD14+ monocytes (F and G) with plasma from Avonex-treated MS patients while blocking type I IFN signaling with an anti-IFNAR2 Ab (clone MMHAR-2). Plasma from patients before Avonex treatment (UT) did not induce expression of OAS2 or BAFF, whereas plasma from the same patients after Avonex treatment (18 h posttreatment) induced the expression of OAS2 and BAFF. Importantly, pretreating cells with an anti-IFNAR2 Ab completely abolished the induction of OAS2 and BAFF, demonstrating that type I IFN directly induces BAFF. One representative data from three experiments are shown (n = 3). Expression levels were normalized to ACTB, and statistics were calculated with Student t test (*p ≤ 0.05, **p ≤ 0.01).
Identification of a novel ISRE site in the BAFF promoter
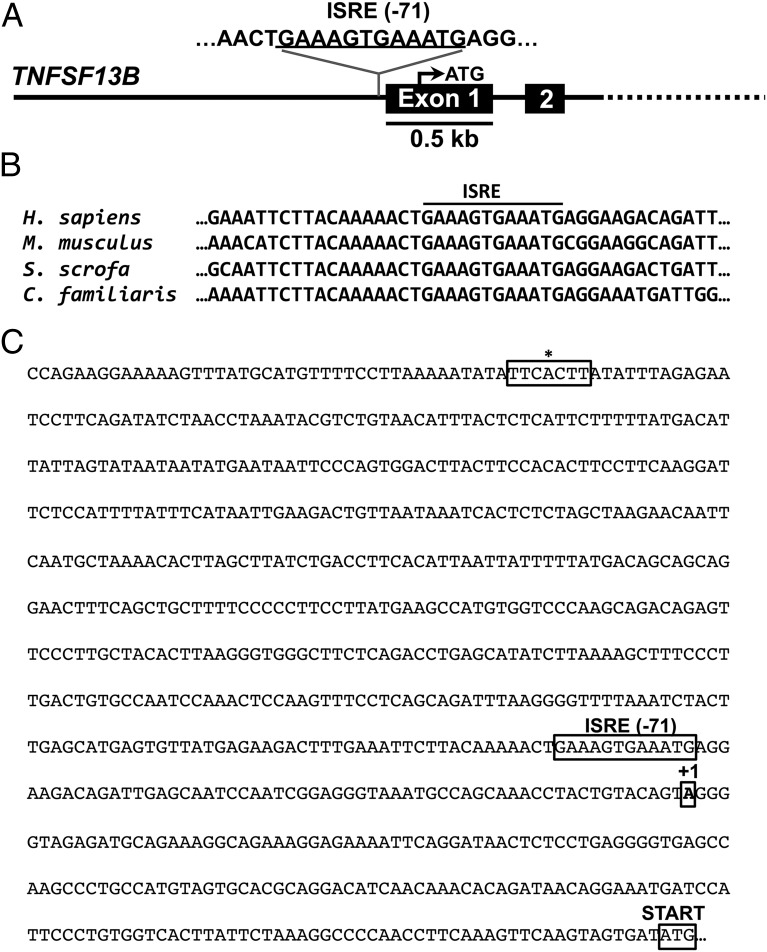
Because type I IFN directly induced BAFF expression in monocytes, and BAFF expression is positively correlated with the IFN-signature in pSS patients, we reasoned that type I IFN is likely to drive the expression of BAFF via JAK/STAT signaling. Earlier studies of the BAFF promoter have indeed reported γ activated sites (GAS) and ISRE sites, but importantly these sites are not conserved and were not functionally validated by mutational analysis (15, 16). This could be because sequence conservation of the identified TFBS was not considered or because updated position weight matrix–based prediction tools were not used for TFBS prediction. To clarify this, we first used position weight matrix–based phylogenetic footprinting tools (rVista 2.0 and ConSite) to identify conserved GAS or ISRE sites in the promoter of BAFF (Fig. 3A, 3C). By comparing the genomic sequence of the proximal BAFF promoter between different species, we identified a highly conserved ISRE site adjacent to the TSS (Fig. 3B). Importantly, this ISRE site was not identified by two previous studies on the BAFF promoter (15, 16). In contrast with Woo et al. (16), we did not identify any conserved GAS site in the BAFF promoter (20). This could be because Woo et al. (16) did not consider sequence conservation while predicting TFBS.
FIGURE 3.
Identification of a conserved ISRE site in the human BAFF promoter. (A) Using phylogenetic footprinting (rVista 2.0 and ConSite), we identified an ISRE site consisting of two GAAA core elements in the human BAFF gene (TNFSF13B). (B) The ISRE site is conserved between species, strongly indicating that it is a functional transcription factor-binding site. (C) The identified ISRE is located −71 bp upstream of the TSS (+1) in contrast with a previously suggested nonconserved ISRE site (*) located more upstream in the gene.
BAFF expression is controlled by IRFs
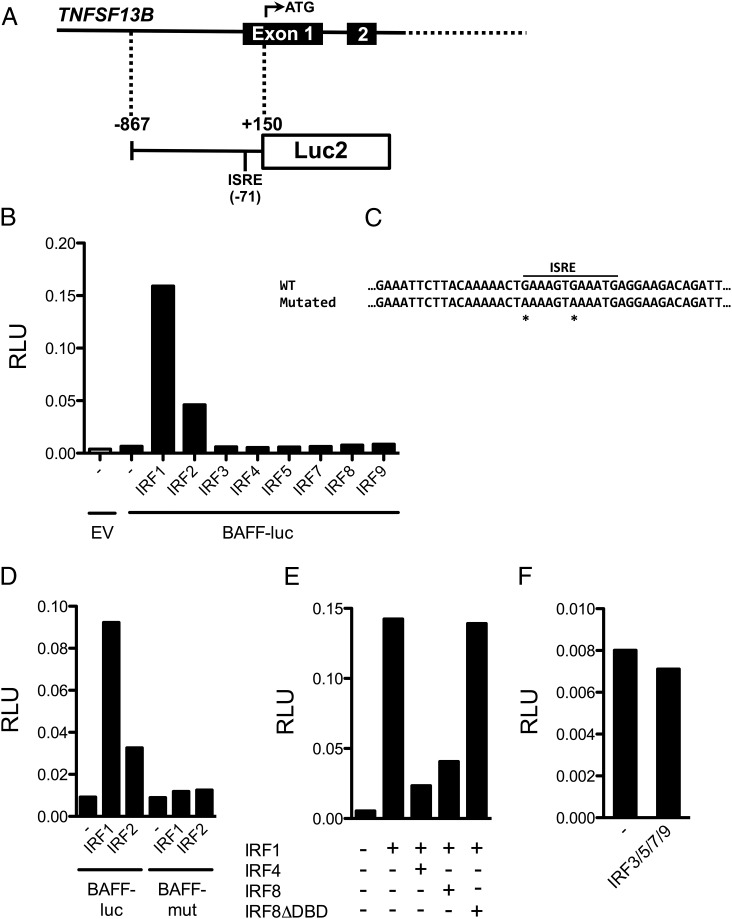
To verify the functionality of the identified ISRE site, we first generated a luciferase reporter construct (BAFF-luc) containing a 1000-bp fragment of the BAFF gene including the core promoter, the TSS, and the ISRE site (Fig. 4A). Type I IFNs activate the transcription factors IRF1, IRF2, and IRF9 followed by their binding to ISRE sites. To identify IRFs that regulate the expression of BAFF, we performed BAFF-luc reporter assays after cotransfection with a panel of IRFs. Only IRF1 and IRF2 could potently activate the BAFF-luc reporter (Fig. 4B). We also generated a BAFF-mut construct where the two core GAAA elements of the identified ISRE site were altered to AAAA (Fig. 4C). Importantly, IRF1 and IRF2 failed to activate luciferase expression from the BAFF-mut construct, demonstrating the functionality of the identified ISRE site (Fig. 4D). Previous reports have demonstrated that IRF4 and IRF8 are potent repressors of the expression of ISGs (17), and to test whether this also applies to BAFF, we cotransfected the BAFF-luc construct with IRF1 together with IRF4 or IRF8. Indeed, both IRF4 and IRF8 were potent repressors of IRF1-dependent activation of the BAFF promoter (Fig. 4E). Cotransfecting the BAFF-luc reporter together with IRF1 and IRF8 without DNA-binding domain, we demonstrated that the repressive effect of IRF8 is dependent on DNA binding (Fig. 4E). Transfection with a pool of IRF3, IRF5, IRF7, and IRF9 did not affect the activity of the BAFF-luc reporter (Fig. 4F). In summary, we have identified a novel functionally validated ISRE site in the promoter of BAFF that is regulated by several members of the IRF transcription factor family.
FIGURE 4.
BAFF expression is directly controlled by IRFs. (A) A reporter construct was generated where a BAFF gene promoter fragment (−867 to +150) drives the expression of the luciferase gene (BAFF-luc). (B) HEK293T cells were cotransfected with the BAFF-luc reporter and different IRF transcription factors, and this identified IRF1 and IRF2 as potent inducers of BAFF expression. (C) To confirm that the identified ISRE site was responsible for the IRF-induced BAFF expression, we generated a mutant of the BAFF reporter where the two GAAA core elements were altered into AAAA (BAFF-mut). Mutated nucleotides are marked with asterisks. (D) HEK293T cells were cotransfected with either the BAFF-luc reporter or the BAFF-mut reporter together with IRF1 or IRF2. IRF1 and IRF2 could not induce expression from the BAFF-mut reporter, demonstrating that the identified ISRE site is a functional site. (E) HEK293T cells were cotransfected with the BAFF-luc reporter and IRF1 alone or together with IRF4, IRF8, or IRF8 lacking the DNA-binding domain (IRF8ΔDBD). IRF4 and IRF8 were potent repressors of IRF1-dependent BAFF expression. This repression was dependent on DNA binding. (F) Cotransfecting HEK293T cells with IRF3, IRF5, IRF7, and IRF9 did not affect the activity of the BAFF-luc reporter. All bar charts show the mean of three technical replicates. All experiments were performed at least twice with the same results. EV, empty vector.
Discussion
BAFF levels are increased in serum of patients with pSS and SLE; however, the exact mechanism for this increase in BAFF is not known (7, 18, 19, 21). Interestingly, several reports have linked BAFF expression to type I IFNs, and it has been reported that BAFF expression is increased in human immune cells and serum after IFN-β treatment (13, 14, 22, 23). These reports, however, did not use flow cytometry to isolate highly pure cell populations and did not explicitly compare and report BAFF levels in isolated cells before and after IFN-β treatment. Also, these reports failed to identify and validate any regulatory elements in the BAFF promoter. To demonstrate that BAFF is a direct target gene of type I IFNs, and to identify genuine TFBS in the BAFF promoter, we first isolated highly pure cell subsets and directly compared BAFF expression before and after IFN-β treatment of humans. We also isolated pure immune cell subsets from pSS patients and healthy control subjects to compare BAFF expression. In this article, we demonstrate that BAFF is directly induced by type I IFN in vivo in humans, and that the level of BAFF expression in neutrophils from pSS patients is positively correlated to the activity of the type I IFN system. Furthermore, we demonstrate that the induction of BAFF expression is directly due to type I IFN signaling via the transcription factors IRF1 and IRF2. We identified an ISRE site adjacent to the TSS of the BAFF gene, and functionally validated this ISRE site by mutating the GAAAGTGAAA sequence into AAAAGTAAAA. Indeed, this mutation of the ISRE site completely abolished the induction of the BAFF reporter by IRF1 and IRF2. In addition, the expression of BAFF was blocked by the myeloid- and lymphoid-specific transcription factors IRF4 and IRF8. A mutant of IRF8, lacking the DNA-binding domain, was unable to repress IRF1, indicating that the repression of BAFF by IRF8 was mediated by direct competition for the ISRE site. The strong repression of BAFF mediated by IRF4 and IRF8 suggests that BAFF is tightly regulated by these two transcription factors; indeed, a previous report has identified IRF8 as a repressor of BAFF expression in B cells (24). In summary, our data demonstrate that BAFF expression is directly downstream of type I IFN signaling, and that members of the IRF family tightly regulate the expression of BAFF. Because increased BAFF expression is caused by type I IFNs, it is likely that novel IFN blockers will lead to a reduction in type I IFN signaling and a concomitant reduction of BAFF levels as previously has been suggested (25). Understanding the precise mechanisms whereby type I IFNs control BAFF expression might contribute to revealing the role of BAFF in systemic autoimmune diseases.
Acknowledgments
We thank Dr. Susanna Brauner for assistance with recruiting Avonex-treated MS patients and Dr. Marika Kvarnström for assistance with recruiting patients with Sjögren’s syndrome.
This work was supported by the Karolinska Institutet, the Swedish Research Council, the Center of Excellence for Research on Inflammation and Cardiovascular Disease, and by the Temacentrum för Inflammationssjukdomar, Karolinska Institutet.
- BAFF-luc
- BAFF reporter construct
- BAFF-mut
- mutated BAFF promoter
- GAS
- γ activated site
- IRF
- IFN regulatory factor
- ISG
- IFN-stimulated gene
- ISRE
- IFN-stimulated response element
- MS
- multiple sclerosis
- pSS
- primary Sjögren’s syndrome
- SLE
- systemic lupus erythematosus
- TFBS
- transcription factor binding site
- TSS
- transcriptional start site.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Bennett L., Palucka A. K., Arce E., Cantrell V., Borvak J., Banchereau J., Pascual V. 2003. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J. Exp. Med. 197: 711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baechler E. C., Batliwalla F. M., Karypis G., Gaffney P. M., Ortmann W. A., Espe K. J., Shark K. B., Grande W. J., Hughes K. M., Kapur V., et al. 2003. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc. Natl. Acad. Sci. USA 100: 2610–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brkic Z., Maria N. I., van Helden-Meeuwsen C. G., van de Merwe J. P., van Daele P. L., Dalm V. A., Wildenberg M. E., Beumer W., Drexhage H. A., Versnel M. A. 2013. Prevalence of interferon type I signature in CD14 monocytes of patients with Sjogren’s syndrome and association with disease activity and BAFF gene expression. Ann. Rheum. Dis. 72: 728–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gottenberg J. E., Cagnard N., Lucchesi C., Letourneur F., Mistou S., Lazure T., Jacques S., Ba N., Ittah M., Lepajolec C., et al. 2006. Activation of IFN pathways and plasmacytoid dendritic cell recruitment in target organs of primary Sjögren’s syndrome. [Published erratum appears in 2006 Proc. Natl. Acad. Sci. USA. 103: 5242.] Proc. Natl. Acad. Sci. USA 103: 2770–2775. [DOI] [PMC free article] [PubMed]
- 5.Mavragani C. P., Crow M. K. 2010. Activation of the type I interferon pathway in primary Sjogren’s syndrome. J. Autoimmun. 35: 225–231. [DOI] [PubMed] [Google Scholar]
- 6.Mackay F., Figgett W. A., Saulep D., Lepage M., Hibbs M. L. 2010. B-cell stage and context-dependent requirements for survival signals from BAFF and the B-cell receptor. Immunol. Rev. 237: 205–225. [DOI] [PubMed] [Google Scholar]
- 7.Mariette X., Roux S., Zhang J., Bengoufa D., Lavie F., Zhou T., Kimberly R. 2003. The level of BLyS (BAFF) correlates with the titre of autoantibodies in human Sjögren’s syndrome. Ann. Rheum. Dis. 62: 168–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varin M. M., Le Pottier L., Youinou P., Saulep D., Mackay F., Pers J. O. 2010. B-cell tolerance breakdown in Sjögren’s syndrome: focus on BAFF. Autoimmun. Rev. 9: 604–608. [DOI] [PubMed] [Google Scholar]
- 9.Vincent F. B., Morand E. F., Schneider P., Mackay F. 2014. The BAFF/APRIL system in SLE pathogenesis. Nat. Rev. Rheumatol. 10: 365–373. [DOI] [PubMed] [Google Scholar]
- 10.Dooley M. A., Houssiau F., Aranow C., D’Cruz D. P., Askanase A., Roth D. A., Zhong Z. J., Cooper S., Freimuth W. W., Ginzler E. M., BLISS-52 and -76 Study Groups 2013. Effect of belimumab treatment on renal outcomes: results from the phase 3 belimumab clinical trials in patients with SLE. Lupus 22: 63–72. [DOI] [PubMed] [Google Scholar]
- 11.Mariette X., Seror R., Quartuccio L., Baron G., Salvin S., Fabris M., Desmoulins F., Nocturne G., Ravaud P., De Vita S. 2015. Efficacy and safety of belimumab in primary Sjögren’s syndrome: results of the BELISS open-label phase II study. Ann. Rheum. Dis. 74: 526–531. [DOI] [PubMed] [Google Scholar]
- 12.Sanz I., Yasothan U., Kirkpatrick P. 2011. Belimumab. Nat. Rev. Drug Discov. 10: 335–336. [DOI] [PubMed] [Google Scholar]
- 13.Hedegaard C. J., Sellebjerg F., Krakauer M., Hesse D., Bendtzen K., Nielsen C. H. 2011. Interferon-beta increases systemic BAFF levels in multiple sclerosis without increasing autoantibody production. Mult. Scler. 17: 567–577. [DOI] [PubMed] [Google Scholar]
- 14.Krumbholz M., Faber H., Steinmeyer F., Hoffmann L. A., Kümpfel T., Pellkofer H., Derfuss T., Ionescu C., Starck M., Hafner C., et al. 2008. Interferon-beta increases BAFF levels in multiple sclerosis: implications for B cell autoimmunity. Brain 131: 1455–1463. [DOI] [PubMed] [Google Scholar]
- 15.Moon E. Y., Park H. 2007. B cell activating factor (BAFF) gene promoter activity depends upon co-activator, p300. Immunobiology 212: 637–645. [DOI] [PubMed] [Google Scholar]
- 16.Woo S. J., Im J., Jeon J. H., Kang S. S., Lee M. H., Yun C. H., Moon E. Y., Song M. K., Kim H. H., Han S. H. 2013. Induction of BAFF expression by IFN-γ via JAK/STAT signaling pathways in human intestinal epithelial cells. J. Leukoc. Biol. 93: 363–368. [DOI] [PubMed] [Google Scholar]
- 17.Sjöstrand M., Ambrosi A., Brauner S., Sullivan J., Malin S., Kuchroo V. K., Espinosa A., Wahren-Herlenius M. 2013. Expression of the immune regulator tripartite-motif 21 is controlled by IFN regulatory factors. J. Immunol. 191: 3753–3763. [DOI] [PubMed] [Google Scholar]
- 18.Groom J., Kalled S. L., Cutler A. H., Olson C., Woodcock S. A., Schneider P., Tschopp J., Cachero T. G., Batten M., Wheway J., et al. 2002. Association of BAFF/BLyS overexpression and altered B cell differentiation with Sjögren’s syndrome. J. Clin. Invest. 109: 59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheema G. S., Roschke V., Hilbert D. M., Stohl W. 2001. Elevated serum B lymphocyte stimulator levels in patients with systemic immune-based rheumatic diseases. Arthritis Rheum. 44: 1313–1319. [DOI] [PubMed] [Google Scholar]
- 20.Ehret G. B., Reichenbach P., Schindler U., Horvath C. M., Fritz S., Nabholz M., Bucher P. 2001. DNA binding specificity of different STAT proteins. Comparison of in vitro specificity with natural target sites. J. Biol. Chem. 276: 6675–6688. [DOI] [PubMed] [Google Scholar]
- 21.Ittah M., Miceli-Richard C., Eric Gottenberg J., Lavie F., Lazure T., Ba N., Sellam J., Lepajolec C., Mariette X. 2006. B cell-activating factor of the tumor necrosis factor family (BAFF) is expressed under stimulation by interferon in salivary gland epithelial cells in primary Sjögren’s syndrome. Arthritis Res. Ther. 8: R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gandhi K. S., McKay F. C., Schibeci S. D., Arthur J. W., Heard R. N., Stewart G. J., Booth D. R. 2008. BAFF is a biological response marker to IFN-beta treatment in multiple sclerosis. J. Interferon Cytokine Res. 28: 529–539. [DOI] [PubMed] [Google Scholar]
- 23.Singh M. K., Scott T. F., LaFramboise W. A., Hu F. Z., Post J. C., Ehrlich G. D. 2007. Gene expression changes in peripheral blood mononuclear cells from multiple sclerosis patients undergoing beta-interferon therapy. J. Neurol. Sci. 258: 52–59. [DOI] [PubMed] [Google Scholar]
- 24.Shin D. M., Lee C. H., Morse H. C., III 2011. IRF8 governs expression of genes involved in innate and adaptive immunity in human and mouse germinal center B cells. PLoS One 6: e27384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao Y., Richman L., Higgs B. W., Morehouse C. A., de los Reyes M., Brohawn P., Zhang J., White B., Coyle A. J., Kiener P. A., Jallal B. 2009. Neutralization of interferon-alpha/beta-inducible genes and downstream effect in a phase I trial of an anti-interferon-alpha monoclonal antibody in systemic lupus erythematosus. Arthritis Rheum. 60: 1785–1796. [DOI] [PubMed] [Google Scholar]