Abstract
Advances in cancer research and personalized medicine will require significant new bridging infrastructures, including more robust biorepositories that link human tissue to clinical phenotypes and outcomes. In order to meet that challenge, four cancer centers formed the TIES Cancer Research Network, a federated network that facilitates data and biospecimen sharing among member institutions. Member sites can access pathology data that is de-identified and processed with the TIES natural language processing system, which creates a repository of rich phenotype data linked to clinical biospecimens. TIES incorporates multiple security and privacy best practices that, combined with legal agreements, network policies and procedures, enable regulatory compliance. The TIES Cancer Research Network now provides integrated access to investigators at all member institutions, where multiple investigator-driven pilot projects are underway. Examples of federated search across the network illustrate the potential impact on translational research, particularly for studies involving rare cancers, rare phenotypes, and specific biologic behaviors. The network satisfies several key desiderata including local control of data and credentialing, inclusion of rich phenotype information, and applicability to diverse research objectives. The TIES Cancer Research Network presents a model for a national data and biospecimen network.
Keywords: Federated Data Sharing, Biorepositories, Translational Research, Biomedical Informatics, Natural Language Processing
Introduction
Obtaining access to sufficient numbers of annotated human tissues remains a significant impediment to translational cancer research (1) and is needed to advance cancer care towards precision medicine (2). Cancer researchers have been at the forefront of developing biomedical data and resource sharing consortia (3–7), but these have typically employed centralized models in which a single institution acts as central broker between requesting researchers and contributing institutions. This centralized approach has several advantages, including ease of distribution using a single point of contact and relatively uncomplicated information technology requirements. However, centralization becomes precarious as the number of organizations increases. Transfer of data to one location creates a potential central source of failure. Centralization also limits the adoption of institution-specific policy choices. A national-scale data and biospecimen sharing network may not be achievable using a centralized model.
Federated networks offer an alternative that may adapt more readily to increasing scale. In this model, each institution controls its own data and resources, and the network facilitates exchange among parties. A recent commentary by Kohane and Mandl (8) outlined the importance of self-organizing federated networks for biomedical research, as well as some of the challenges. Successful clinical data sharing networks outside of cancer research are already being developed using federated models (9–12). Previous large-scale biomedical informatics efforts such as the Cancer Biomedical Informatics Grid (caBIG) (13) and the Biomedical Informatics Research Network (BIRN) (14) have contributed to informatics advances supporting federated models of data and biospecimen exchange. Two of our institutions have previously collaborated on small-scale demonstration projects that validated the potential of this approach (15, 16). However, most previous efforts to develop federated data and biospecimen networks have focused more on information technology needs, and less on the complex regulatory, legal, security, privacy, and workflow requirements to support such infrastructures. This is perhaps the reason that few previous efforts have progressed beyond the pilot stage.
Diversity across diseases, resource types, and collection methodology is an important aspect of such a national network. A broader, general system would be more useful than a plethora of disease-specific networks. A national network should also accommodate access to multiple resource types (e.g. formalin fixed paraffin embedded samples, fresh frozen specimens, and images) and support retrospective as well as prospective research.
We previously developed the Text Information Extraction System (TIES) (17), an open-source computer-based system. TIES uses natural language processing (NLP) methods to automate annotation of tissue samples using text-based electronic medical records. Until recently, this system was used only locally by cancer centers to provide research access to investigators at their individual institutions.
Building on this work, we established the TIES Cancer Research Network (TCRN) (18), a multi-institutional, collaborative federated research network that provides de-identified clinical data and associated biospecimens to investigators from member institutions. In this manuscript, we describe the regulatory and organizational principles, operating standards, and technical foundations of TCRN as a model for future large-scale cancer data and biospecimen sharing networks.
Organization of the TIES Cancer Research Network
The TIES Cancer Research Network is composed of Georgia Regents University (GRU) Cancer Center, Roswell Park Cancer Institute (RPCI), University of Pennsylvania Abramson Cancer Center (ACC), and UPMC CancerCenter/University of Pittsburgh Cancer Institute (UPCI). Each institution brings unique strengths to the network and expands TCRN’s geographic and demographic diversity. SPOREs in ovarian cancer, melanoma, head and neck cancer, and lung cancer add further diversity by providing centers of expertise that can utilize the network for collaborative efforts across institutional boundaries.
The TCRN Executive Committee governs the TCRN network, its member institutions, and its users. The Executive Committee approves policies and processes for operating the network, advances the use of the network, and considers requests for new member organizations. The Executive Committee is composed of representatives of each of the member sites, including faculty with primary responsibilities for biobanking and cancer informatics. The Executive Committee maintains a Policies and Processes Subcommittee, which is responsible for drafting policies, procedures, and recommendations for consideration by the Executive Committee. Additionally, this subcommittee provides a forum for trans-network communication on regulatory matters.
Participation in TCRN requires member institutions to sign the publicly available (18) TCRN Network Agreement with the University of Pittsburgh. The Network Agreement includes Data Use Terms and an agreement to use the Universal Biological Materials Transfer Agreement (UBMTA) for transfer of biomaterials. Prior to transfer of materials, point-to-point UBMTAs are executed and cost-recovery mechanisms are determined. The Network Agreement includes an instrument of adherence to enable the inclusion of new member organizations.
Ensuring Regulatory Compliance
TCRN supports regulatory compliance through a number of interlocking mechanisms. Our approach is based on earlier qualitative research on the complex regulatory requirements of biomedical data grids (19). Each institution obtains an exempt approval from each institution’s IRB to create and maintain its TIES node. All member institutions have determined that subsequent use of TCRN data by investigators is Non-Human Subjects Research and is therefore IRB exempt. Consequently, IRB review is not required for TCRN studies that use only data. Investigators who wish to access tissue must provide proof of local IRB review and approval.
TCRN is governed by a set of recommendations, policies, and standard operating procedures. These documents (Table 1), which are all publicly available (18), cover aspects of membership, deployment, testing, and use of the system. Policies and procedures are developed and refined by the TCRN committees. TCRN member institutions agree to abide by these policies and implement defined procedures under the terms of the Network Agreement.
Table 1.
Standard Operating Policies (SOPs), Procedures and Recommendations.
| SOP or Recommendation |
Purpose | Elements |
|---|---|---|
| Governance | To ensure that all TCRN members participate in governing the network |
|
| Validating Quality of De-Identification | To ensure that TCRN members achieve an acceptable level of text de-identification |
|
| Verifying Eligibility of Users | To ensure that only eligible investigators from member institutions are granted TCRN access |
|
| Study Registration and Authorization | To outline the requirements and procedure for TCRN study registration and authorization |
|
| Procedure for Increasing Level of Access to TCRN (Step Up Requests) | To outline the process for users to increase their level of access to the network |
|
| Auditing of Users and Searches | To ensure that all users accessing TCRN are using the network appropriately and assess value of TCRN |
|
| Incident Reporting | To describe incidents that may arise in TCRN and how members must report and respond to them |
|
| Recommendation for Member Institutions on Establishing Approval Bodies for External Users | To suggest a streamlined and consistent process by which external collaborators gain access to institutional data |
|
All current SOPs and Recommendations are available at http://ties.pitt.edu/tcrn
Despite this incorporated governance structure, member institutions have freedom to make local decisions. This greater autonomy, which supports the variability between institutions, is a crucial advantage of the federated model. One example of this is the approval of registered studies. TCRN provides guidance about the composition of approval bodies and what institutions might consider in their processes. However, member institutions can design and implement unique processes.
Another regulatory mechanism utilized by the TCRN is the creation of distinct user roles that limit users’ access to defined categories of information. TIES controls access to the data based on these user roles. Four separate software portals provide access to specific functions. Administrators and Honest Brokers (20) operate exclusively within the context of their own institutional TIES nodes. The Administration Portal enables TIES Administrators to manage local user accounts. The Honest Broker Portal enables institutionally-recognized neutral third parties to search for identified data and process orders at their own institutions. Honest brokers and administrators also grant approved researchers access to their institutions’ data and/or tissues. Researchers and Preliminary Users may operate across multiple institutions, given appropriate permissions. The Preliminary User Portal allows users to obtain aggregate information in the form of charts and tables, but does not show de-identified records. The Researcher Portal allows approved researchers to search for de-identified data, create case sets, and potentially submit tissue orders.
TCRN users are only approved within the context of a specific study to reduce the possibility of unauthorized use of the network. Researchers may have multiple studies in TIES, and studies may list multiple researchers, including collaborators at other institutions. Users may be approved for different access levels on different studies. Before accessing TIES, TCRN users must indicate the specific study under which they are searching.
The TCRN also addresses privacy regulations by handling identity provisioning, study registration, and study approval at the local level. Using a federated model, TCRN authorizes institutions to vet their local users and provide credentials to access TCRN. The local TIES Administrator is responsible for verifying that potential local users are eligible to use TCRN. Additionally, each member institution controls access to its own data. Incoming requests from external investigators are automatically routed to requested organizations, which can approve or reject study requests. Organizations may set the researcher’s access level based on local requirements and TCRN policies.
Natural Language Processing Annotation of Specimens
At the heart of the TCRN is an automated process for annotating tissues using natural language processing (NLP). All TCRN members automatically process their pathology records using the TIES NLP annotation software to create a retrospective index to all tissues collected through clinical care. These may include FF and FFPE tissues, and will typically include a bulk load of all historical pathology reports as well as ongoing processing of reports as they accrue in the clinical system. Optionally, they may also tag specific cases that are associated with prospectively collected materials available in biobanking systems.
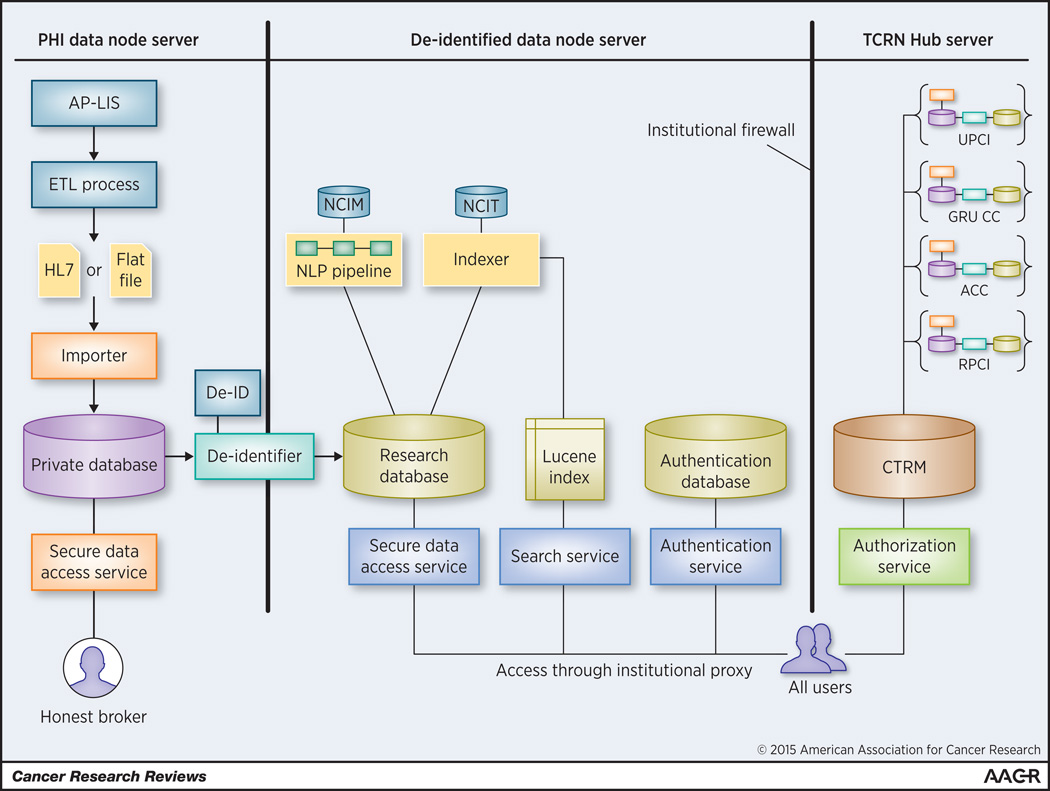
TIES is an open-source suite of datastores, software services, and client applications, written in the Java programming language (17). TIES provides all necessary infrastructure for creating large repositories of processed clinical documents linked to tissue samples, and making them available to cancer researchers. TIES uses three primary datastores (Figure 1): [1] the private datastore contains protected health information (PHI); [2] the research datastore contains de-identified, annotated text; and [3] the collaborative datastore contains metadata about studies. Each institution hosts one private and one research datastore. The network hosts a single collaborative datastore across member institutions.
Figure 1.
Architecture of the TIES Cancer Research Network.
TIES services include data preparation services, that operate locally, and information retrieval services, that operate both locally and across the network. Data preparation services include: [1] an acquisition service that transfers data from the Anatomic Pathology Laboratory Information system and Tissue Banking Systems to the private datastore; [2] a de-identification service that de-identifies text to HIPAA “safe-harbor” standard and loads the research datastore; [3] an NLP pipeline service that leverages NCI terminology to create semantic and syntactic annotations on text; and [4] an indexing service that uses Apache Lucene (21) to index documents for fast retrieval over a hierarchy of concepts, based on the NCI Thesaurus ontology. Information retrieval services include: [1] a query matching service, [2] a search service, and [3] data services, all of which act sequentially to match user-entered text to concepts, correct spelling errors, convert the user query to the Lucene query language, and to deliver documents through a secure, encrypted conduit.
The TIES NLP pipeline service produces NLP annotations by performing sequential tokenization, parsing, noun phrasing, concept recognition, semantic filtering, negation and uncertainty detection, and shallow discourse reasoning. Previous evaluation of TIES has shown that it is associated with high precision across multiple query types (17).
Unique Identifiers
As part of the data loading process, institutions use their Enterprise Master Patient Index (EMPI) and other resources to ensure that all documents related to a single individual are appropriately aggregated, despite potential differences in names, medical record numbers and social security numbers. Each patient within an individual institution’s TIES data store is assigned a Universally Unique Identifier (UUID), generated using the Java UUID Generator library (JUG). UUIDs are 128 bit alphanumeric tokens that can be created without central coordination to enable unique identifiers across distributed networks. While UUIDs are not guaranteed to be unique, the likelihood of the same UUID being generated is exceedingly low. Importantly, we made no attempt to match individual patients across institutions. Thus, it is possible that one individual could appear as two or more separate individuals if their health care was delivered at multiple TCRN participating organizations.
Security and Privacy
Privacy and security policies of TCRN are implemented through TIES and operate both locally and across the network. Two foundational principles apply: [1] identified data are maintained separately and are available only to individuals with appropriate permissions, and [2] identified data never leaves the institution. Identified data are accessible only from behind an institution’s firewall, where TIES datastores are deployed. In order to ensure the security of TIES and maintain the privacy of PHI, TIES uses the Globus Toolkit 4.2.2 GSI Grid Security Infrastructure (22) (Java implementation) to encrypt communications, authorize and authenticate users, and manage user credentials.
Automated text de-identification is a key component of the privacy controls of the TIES system. All text documents in the research datastore have been de-identified to the HIPAA “Safe Harbor” standard (23). Although TIES can be used with any de-identification software, all current TCRN members have chosen to use the commercial De-ID system (24). De-ID has been shown to remove 99% of the HIPAA Safe Harbor identifiers while maintaining 95% of non-PHI information (25). For further security, all communications are encrypted using the Globus Security Infrastructure asymmetric public private key cryptography (RSA 1024). De-ID also maintains privacy by creating tokens for individuals and shifting dates using a random offset. It preserves temporal relationships by substituting the same token for each instance of the same name, and by using a standard offset for all dates in a given document. TIES extends these mechanisms to preserve longitudinal relationships across documents.
Authorization and authentication are also essential to security in the TCRN. TIES includes the infrastructure to protect resource access based on account type. An external user can access data at another site only if the institution explicitly grants access to specific services on a specific study. Each user has a fully qualified distinguished name, and use of a password with sufficient security attributes is required. Message content passed between sites contains embedded user information so each server knows who requests a resource. All usernames, passwords, and private and public keys are stored within the database of the user’s organization.
As an additional layer of protection, TCRN users must agree to quarantine reports that contain identifiers missed during automated de-identification. Documents are quarantined easily with a single mouse click. Honest brokers can view these as part of TCRN quality assurance procedures. Quarantined reports are manually or automatically scrubbed before being returned to the active pool of records.
Auditing is also performed on a regular basis to ensure regulatory compliance. TIES maintains comprehensive audit logs that are used during the TCRN user auditing procedure. Audit logs include authentications, searches performed, and documents accessed. For every log entry, TIES captures the user, user role, activity type, date and time, and the protocol under which the activity was performed.
Current Status and Pilot Projects
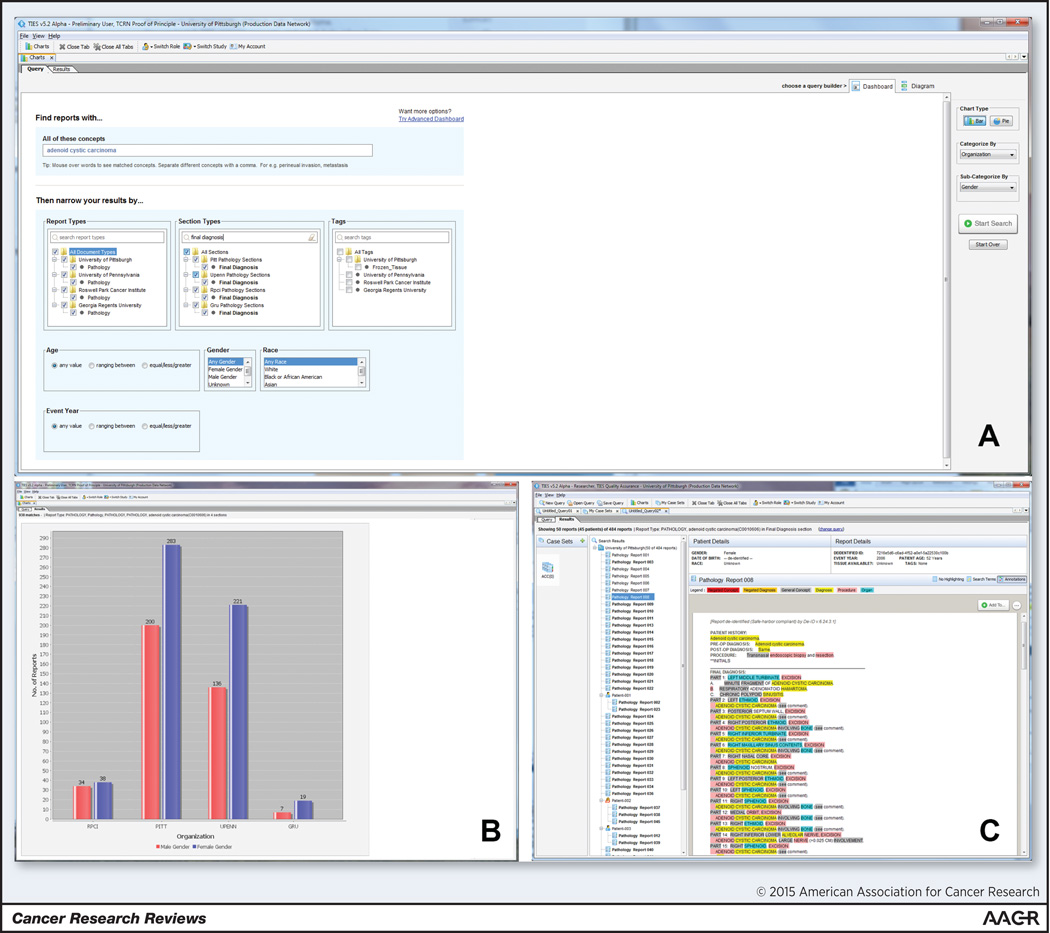
TCRN now has active nodes at four current member institutions, making it possible to search across 5.5 million cases and 2.3 million patients. Figure 2 shows an example query across the network (A) with aggregate results at each institution (B), and a de-identified text document with NLP annotations fitting these criteria (C).
Figure 2.
Researcher portal showing query for cases of Adenoid Cystic Carcinoma (2A) with aggregate case counts by TCRN site and gender (2B) and a specific de-identified, annotated document (2C).
Table 2 describes the data encompassed by the network. Counts and statistics are shown on the total number of patients and cases for fifteen of the most common cancers (26) and fifteen uncommon neoplasms randomly selected from two rare cancer lists (27,28). Advantages of the federated approach include the ability to increase sample size for retrospective studies and to generalize across more diverse populations.
Table 2.
TCRN Case Statistics for Numbers of Patients and Cases (A) and the Number of Cases of Rare Tumors (B) and Common Cancer Categories (C) based on final diagnosis.
| A. Case Statistics | GRU | RPCI | ACC | UPCI | Total |
|---|---|---|---|---|---|
| Patients | 76,404 | 72,376 | 465,717 | 1,840,156 | 2,454,653 |
| Pathology Cases | 157,316 | 156,555 | 857,681 | 4,588,017 | 5,759,569 |
| B. Rare Tumors | GRU | RPCI | ACC | UPCI | Total |
|---|---|---|---|---|---|
| Adenoid Cystic Carcinoma | 41 | 88 | 404 | 509 | 1,042 |
| Adrenocortical Carcinoma | 5 | 20 | 59 | 63 | 147 |
| Alveolar Soft Part Sarcoma | 3 | 15 | 10 | 25 | 53 |
| Angioimmunoblastic Lymphadenopathy |
12 | 35 | 58 | 84 | 189 |
| Chordoma | 5 | 14 | 124 | 245 | 388 |
| Follicular Dendritic Cell Sarcoma | 2 | 2 | 8 | 13 | 25 |
| Merkel Cell Carcinoma | 9 | 72 | 165 | 196 | 442 |
| Ovarian Granulosa Cell Tumor | 4 | 10 | 23 | 34 | 71 |
| Phaeochromocytoma | 15 | 38 | 272 | 164 | 489 |
| Pleomorphic Xanthoastrocytoma | 2 | 5 | 12 | 53 | 72 |
| Pseudomyxoma Peritonei | 6 | 36 | 46 | 129 | 217 |
| Rhabdomyosarcoma | 34 | 70 | 86 | 270 | 460 |
| Sebaceous Adenocarcinoma | 13 | 33 | 26 | 94 | 166 |
| Sinonasal Undifferentiated Carcinoma |
2 | 6 | 31 | 27 | 66 |
| Thymoma | 13 | 45 | 433 | 210 | 701 |
| C. Common Cancer Categories | GRU | RPCI | ACC | UPCI | Total |
|---|---|---|---|---|---|
| Bladder Carcinoma | 345 | 1,618 | 3,873 | 6,711 | 12,547 |
| Breast Carcinoma | 1,143 | 9,605 | 28,262 | 37,691 | 76,701 |
| Colorectal Carcinoma | 465 | 2,530 | 6,898 | 11,608 | 21,501 |
| Endometrial Carcinoma | 394 | 1,815 | 3,707 | 7,706 | 13,622 |
| Esophageal Carcinoma | 63 | 1,477 | 2,452 | 3,514 | 7,506 |
| Hepatic Carcinoma | 153 | 633 | 2,912 | 5,720 | 9,418 |
| Lung Carcinoma | 820 | 4,264 | 10,208 | 17,955 | 33,247 |
| Lymphoma | 1,387 | 6,795 | 10,605 | 15,689 | 34,476 |
| Malignant Glial Neoplasm | 242 | 292 | 2,198 | 4,943 | 7,675 |
| Malignant Melanoma | 335 | 2,675 | 5,180 | 7,068 | 15,258 |
| Ovarian Carcinoma | 503 | 2,872 | 4,659 | 6,446 | 14,480 |
| Pancreatic Carcinoma | 162 | 740 | 1,866 | 3,622 | 6,390 |
| Prostate Carcinoma | 903 | 3,612 | 18,867 | 19,445 | 42,827 |
| Renal Cell Carcinoma | 364 | 1,319 | 3,183 | 10,950 | 15,816 |
| Thyroid Carcinoma | 474 | 1,236 | 7,681 | 12,387 | 21,778 |
As an important first step after deployment of the system, TCRN measured the adequacy of de-identification across all institutions. Following established TCRN policy for evaluating the quality of de-identification, all institutions ran a set of programs that were designed to identify potential errors in the de-identification process. Sites quarantined the results, manually classified the quarantined documents, and identified and remediated the causes of true errors (29). This iterative process included specific searches for patient names and medical record numbers.
Deployment efforts at all four institutions accompanied TCRN development. All sites maintain local web portals that investigators use to [1] request TIES access, [2] download the Java client, [3] access regulatory requirements, and [4] access user manuals and documentation. Training materials include video introductions to all aspects of TIES. At UPCI, where the system was developed, TIES is widely used by researchers. As other institutions complete the QA process, their investigators are adopting TIES. Deployment to research communities is an active process, and we anticipate a growing cadre of TCRN researchers.
As part of an NCI-funded initiative, the TCRN consortium currently supports multiple pilot projects. Two current projects exemplify the diversity of translational research TCRN supports:
Identifying cohorts with rare phenotypes
Investigators at one TCRN founder institution (RPCI) sought to identify a cohort of patients with small ulcerating breast cancers, in order to determine whether they may represent a unique subset within the T4 stage grouping. Rare features such as skin ulceration can be difficult to find using free text search because negative mentions (e.g. “no evidence of ulceration”) are far more common than positive mentions. NLP increases the accuracy of searches for these cases by removing negative mentions, making detailed correlations and comparisons more feasible. Small numbers of cases can be mitigated by inclusion of cases from multiple cancer centers. For this study, TCRN data from UPCI were combined with outcomes data from the Cancer Registry, allowing investigators at RPCI to more than double the cohort they were able to obtain from a combination of other institutions.
Biospecimen requests by progression pattern
At another TCRN founder institution (UPCI), investigators are using retrospective TCRN cases to evaluate spatial domain low-coherence quantitative phase microscopy (SL-QPM) (30) as an ‘optical biomarker’ for risk stratification and prediction of cancer progression in patients with Barrett’s esophagus (BE). To test their methodology, the investigators sought FFPE tissue from biopsies of BE patients who were not found to have any dysplasia during index endoscopy surveillance, yet had high-grade dysplasia or esophageal adenocarcinoma on follow-up endoscopy more than one year later. Searching across both the University of Pittsburgh and University of Pennsylvania, the TIES indexing system provided these investigators with the ability to issue complex language-based temporal queries. Such queries would be nearly impossible to perform within typical electronic medical record systems. The latest version of TIES used for TCRN directly supports the use of de-identified virtual microscopy slides in addition to de-identified data.
Discussion
We describe a fully implemented federated data and biospecimen sharing network for supporting cross-institutional collaboration in cancer research. TCRN incorporates de-identified clinical documents processed using NLP technologies. Around this core technology, we have developed a federated query system, approval process, and policies and procedures to operate the network. TCRN provides a unique infrastructure for accelerating translational cancer research in the era of personalized medicine.
TCRN differs from other recent data and biospecimen resource sharing initiatives (3–7) in the use of de-identified clinical text, which requires NLP methods to characterize phenotype. Investigators using retrospective tissue specimens typically require detailed information, which is usually available only through such clinical documents. The system we have developed makes it possible to identify cases despite the many complexities of medical language. Additionally, our system enables temporal queries that support identification of cohorts not easily found through other means. For example, TIES has been used to identify patients with dysplastic nevi followed by melanoma years later, and to identify patients with multiple neoplasms associated with familial cancer syndromes.
Federation is a second key factor differentiating TCRN from other data and biospecimen-sharing networks with similar goals. Benefits of federation include the ability of each cancer center to control its own data and access to those data. Another advantage is that TCRN leverages the local institution as the authority in vetting individual identity and credentials of investigators. Importantly, federation provides a means to easily expand the network without sustaining increased burden to a centralized human or technical infrastructure. The TCRN Network Agreement has been crafted to specifically support this kind of expansion as additional institutions seek to become members. TCRN policies and processes that govern admission of new member institutions provide a clear path to a larger and more inclusive network.
As a result of our collaboration, we have learned a great deal about the practical aspects of federated data and tissue sharing. Thoughtful sociotechnical implementation is needed to yield control to member institutions where they require autonomy, yet promote standardization wherever possible. Strong and consistent governance coupled with open lines of communication among partners are important factors for success (8,31).
Recent NIH and NCI initiatives (32) have underscored the need for innovative data infrastructures to drive discovery science. Future advances in cancer research and precision medicine will require significant new national infrastructure, including more robust biorepositories that link human tissue to phenotypes and outcomes (1), especially as distinctions among molecular subtypes become increasingly refined. We envision that the TIES Cancer Research Network could provide the foundation for a national federated network for cancer data and biospecimen sharing, a goal that now seems within reach.
Acknowledgments
This work was supported by the National Cancer Institute at the National Institutes of Health (R01 CA132672 and U24 CA180921). This project used the UPCI Tissue and Research Pathology Services that is supported in part by award P30CA047904. We are deeply indebted to the many individuals at our Institutional Review Boards, Offices of Research, and legal counsel who worked together to create the foundation for this network. We also gratefully acknowledge Lucy Cafeo in the Department of Biomedical Informatics at University of Pittsburgh and Lisa Middleton at Georgia Regents University for their expert preparation and review of the manuscript.
Footnotes
The authors disclose no potential conflicts of interest.
REFERENCES
- 1.Scott CT, Caulfield T, Borgelt E, Illes J. Personal medicine--the new banking crisis. Nat Biotechnol. 2012;30(2):141–147. doi: 10.1038/nbt.2116. [DOI] [PubMed] [Google Scholar]
- 2.Hamburg MA, Collins FS. The path to personalized medicine. N Engl J Med. 2010;363(4):301–304. doi: 10.1056/NEJMp1006304. [DOI] [PubMed] [Google Scholar]
- 3.Amin W, Parwani AV, Schmandt L, Mohanty SK, Farhat G, Pople AK, et al. National Mesothelioma Virtual Bank: a standard based biospecimen and clinical data resource to enhance translational research. BMC Cancer. 2008;8:236. doi: 10.1186/1471-2407-8-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amin W, Singh H, Dzubinski LA, Schoen RE, Parwani AV. Design and utilization of the colorectal and pancreatic neoplasm virtual biorepository: An early detection research network initiative. J Pathol Inform. 2010;1:22. doi: 10.4103/2153-3539.70831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack CR, Jagust W, et al. Ways toward an early diagnosis in Alzheimer's disease: the Alzheimer's Disease Neuroimaging Initiative (ADNI) Alzheimers Dement. 2005;1(1):55–66. doi: 10.1016/j.jalz.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel AA, Gilbertson JR, Showe LC, London JW, Ross E, Ochs MF, et al. A novel cross-disciplinary multi-institute approach to translational cancer research: lessons learned from Pennsylvania Cancer Alliance Bioinformatics Consortium (PCABC) Cancer Inform. 2007;3:255–274. [PMC free article] [PubMed] [Google Scholar]
- 7.Qualman SJ, France M, Grizzle WE, LiVolsi VA, Moskaluk CA, Ramirez NC, et al. Establishing a tumour bank: banking, informatics and ethics. Br J Cancer. 2004;90(6):1115–1119. doi: 10.1038/sj.bjc.6601678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mandl KD, Kohane IS. Federalist principles for healthcare data networks. Nature Biotechnology. 2015;33(4):360–363. doi: 10.1038/nbt.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amin W, Tsui FR, Borromeo C, Chuang CH, Espino JU, Ford D, et al. PaTH: towards a learning health system in the Mid-Atlantic region. Journal of the American Medical Informatics Association: JAMIA. 2014;21(4):633–636. doi: 10.1136/amiajnl-2014-002759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McMurry AJ, Gilbert CA, Reis BY, Chueh HC, Kohane IS, Mandl KD. A self-scaling, distributed information architecture for public health, research, and clinical care. Journal of the American Medical Informatics Association: JAMIA. 2007;14(4):527–533. doi: 10.1197/jamia.M2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohno-Machado L, Agha Z, Bell DS, Dahm L, Day ME, Doctor JN, et al. pSCANNER: patient-centered Scalable National Network for Effectiveness Research. Journal of the American Medical Informatics Association: JAMIA. 2014;21(4):621–626. doi: 10.1136/amiajnl-2014-002751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weber GM, Murphy SN, McMurry AJ, Macfadden D, Nigrin DJ, Churchill S, et al. The Shared Health Research Information Network (SHRINE): a prototype federated query tool for clinical data repositories. Journal of the American Medical Informatics Association: JAMIA. 2009;16(5):624–630. doi: 10.1197/jamia.M3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buetow KH. An infrastructure for interconnecting research institutions. Drug Discov Today. 2009;14(11–12):605–610. doi: 10.1016/j.drudis.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helmer KG, Ambite JL, Ames J, Ananthakrishnan R, Burns G, Chervenak AL, et al. Enabling collaborative research using the Biomedical Informatics Research Network (BIRN) Journal of the American Medical Informatics Association: JAMIA. 2011;18(4):416–422. doi: 10.1136/amiajnl-2010-000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drake TA, Braun J, Marchevsky A, Kohane IS, Fletcher C, Chueh H, et al. A system for sharing routine surgical pathology specimens across institutions: the Shared Pathology Informatics Network. Hum Pathol. 2007;38(8):1212–1225. doi: 10.1016/j.humpath.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 16.Reis SE, Patterson PD, Fitzgerald GA, Ford D, Sherwin RS, Solway J, et al. The sharing partnership for innovative research in translation (SPIRiT) consortium: a model for collaboration across CTSA sites. Clin Transl Sci. 2013;6(2):85–87. doi: 10.1111/cts.12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crowley RS, Castine M, Mitchell K, Chavan G, McSherry T, Feldman M. caTIES: a grid based system for coding and retrieval of surgical pathology reports and tissue specimens in support of translational research. Journal of the American Medical Informatics Association: JAMIA. 2010;17(3):253–264. doi: 10.1136/jamia.2009.002295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The TIES Cancer Research Network (TCRN) Homepage. < http://ties.pitt/edu/tcrn>.
- 19.Manion FJ, Robbins RJ, Weems WA, Crowley RS. Security and privacy requirements for a multi-institutional cancer research data grid: an interview-based study. BMC Med Inform Decis Mak. 2009;9:31. doi: 10.1186/1472-6947-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dhir R, Patel AA, Winters S, Bisceglia M, Swanson D, Aamodt R, et al. A multidisciplinary approach to honest broker services for tissue banks and clinical data: a pragmatic and practical model. Cancer. 2008;113(7):1705–1715. doi: 10.1002/cncr.23768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lucene homepage. < https://lucene.apache.org/>.
- 22.Globus Security Infrastructure. < http://toolkit.globus.org/toolkit/security/>.
- 23.Guidance Regarding Methods for De-identification of Protected Health Information in Accordance with the Health Insurance Portability and Accountability Act (HIPAA) Privacy Rule. < http://www.hhs.gov/ocr/privacy/hipaa/understanding/coveredentities/De-identification/guidance.html>.
- 24.De-ID System homepage. < http://www.de-idata.com/>.
- 25.Gupta D, Saul M, Gilbertson J. Evaluation of a deidentification (De-Id) software engine to share pathology reports and clinical documents for research. Am J Clin Pathol. 2004;121(2):176–186. doi: 10.1309/E6K3-3GBP-E5C2-7FYU. [DOI] [PubMed] [Google Scholar]
- 26.American Cancer Society Facts and Figures. < http://www.cancer.org/research/cancerfactsstatistics/>.
- 27.Cancer Research UK Rare Cancers List. < http://www.cancerresearchuk.org/about-cancer/type/rare-cancers/rare-cancers-name/%20?openFull=1#list>.
- 28.Rare Cancer Alliance. < http://www.rare-cancer.org/info/raw-adult-list.php>.
- 29.TIES Quality Assurance Plan. http://ties.dbmi.pitt.edu/wp-content/uploads/2014/02/TIESQualityAssurancePlan.pdf.
- 30.Wang P, Bista RK, Khalbuss WE, Qiu W, Uttam S, Staton K, et al. Nanoscale nuclear architecture for cancer diagnosis beyond pathology via spatial-domain low-coherence quantitative phase microscopy. J Biomed Opt. 2010;15(6):066028. doi: 10.1117/1.3523618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaught J, Kelly A, Hewitt R. A review of international biobanks and networks: success factors and key benchmarks. Biopreserv Biobank. 2009;7(3):143–150. doi: 10.1089/bio.2010.0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372(9):793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]