Abstract
Many complex traits, as drug response, are associated with changes in biological pathways rather than being caused by single gene alterations. Here, a predictive framework is presented in which gene expression data are recoded into activity statuses of signal transduction circuits (sub-pathways within signaling pathways that connect receptor proteins to final effector proteins that trigger cell actions). Such activity values are used as features by a prediction algorithm which can efficiently predict a continuous variable such as the IC50 value. The main advantage of this prediction method is that the features selected by the predictor, the signaling circuits, are themselves rich-informative, mechanism-based biomarkers which provide insight into or drug molecular mechanisms of action (MoA).
Predicting the best treatment strategy from genomic information is a core goal of precision medicine. In particular, the ability to predict drug response is crucial to establish proper dosages and identify individuals at increased risk for adverse effects1. Different attempts to create predictive models of drug response produced gene expression signatures for different aspects of the effect of the drug over the cell culture2,3,4,5. Similar attempts were made for responses to chemical compounds6,7,8. However, the prediction accuracy of biomarkers such as gene expression signatures has been questioned in numerous occasions because of their low reproducibility across different studies9 and their lack of robustness when challenged with different samples10,11. Apart from technical or methodological problems, the main difficulty in predicting complex traits, such as drug response, come from the fact that they cannot be understood using methods that ignore the complex gene interactions that may drive cellular response12,13. Therefore, the knowledge of the underlying molecular mechanism of action (MoA) by which the drug affects cell functionality constitutes a critical step in the prediction of drug responses14. With this purpose, several authors have tried to combine biomarkers over biological modules related to cell functionality, such as protein networks15,16 or biological pathways17. Particularly relevant in drug response are signaling pathways, which provide a formal representation of the processes by which the cell triggers specific functional activities in the cell in response to particular stimulus through different circuits of intermediate gene products. Interestingly, the activity of these signaling circuits can directly be related to cell functionalities. Different repositories, such as KEGG18, Reactome19 and others, contain abundant and detailed information about signaling pathways.
Conventional methods (pathway topology PT-based algorithms20) use the topological relationships between the proteins within a pathway to compute a score related to its global activation status from gene expression values21,22,23,24 or from mutations25. Seeking for a more detailed description of pathway activity, more sophisticated approaches aimed to discover any type of activated sub-network within pathways26,27,28. In particular, more recent methods specifically focus on the estimation of the activity of those sub-networks that can directly be related to cell activity: the signaling circuits that receive a stimulus and trigger a response29,30,31. Preferably, the activity of such stimulus-response signaling circuit could be inferred from (phospho)proteomic and chemoproteomic experiments32. However, the production of these types of data still results relatively complex and consequently very few datasets are available. Nevertheless, abundant gene expression data on comparative studies of drugs33,34,35,36 and other compounds37,38,39 are available, constituting an invaluable resource for comparative studies of drugs and cell lines14. The estimation of the activity of signaling circuits from gene expression data29,30,31 provide a rich-informative type of biomarkers, thereinafter called mechanism-based biomarkers, whose direct relationship to cell functionalities can be used to suggest mechanistic explanations for the molecular basis of complex traits40.
Here we present a simple way of recoding gene expression values into mechanism-based biomarkers. Such biomarkers are subsequently used in an innovative way in the context of prediction. In particular, the method presented here is applied to predict a complex trait like drug sensitivity. Two large-scale datasets in which different cell lines are treated with a variety of drugs35,36 are used to illustrate how the predictions based on the proposed mechanism-based biomarkers are not only accurate but also provide relevant clues to understand the MoA of the drugs assayed.
Additionally, a freely available web tool for carrying out the predictions has been developed.
Results
Use of signaling circuit activation probabilities in the context of prediction
Gene expression values are recoded into signaling circuit activities (elementary components of signaling pathways) using a probabilistic model30 as described in Methods. These activity values are further used as mechanism-based biomarker features for prediction purposes. Typically predictors are built by first selecting the best informative features (mechanism-based biomarkers here) and then applying a prediction algorithm.
For each dataset, microarray gene expression data were normalized with RMA41. Normalized gene expression values were transformed into probabilities of signaling circuit activation30. Such probabilities are considered mechanism-based biomarkers and are the features used in the proposed predictive framework. Circuits whose activation statuses do not change across the conditions studied (invariant biomarkers) were initially discarded. Then, feature selection is carried out over the remaining circuits. Here, we have used Correlation-based Feature Selection (CFS). The selected biomarkers are subsequently used by the predictor algorithm. Here, we have used a regression based on Support Vector Machine (SVM)42 (SVM ɛ-regression, as implemented in the e1071 R library43) to predict the value of a continuous variable such as the IC50. Strictly speaking, in the case of SVM the previous step of most informative variables could be skipped.
The accuracy of the classification obtained was evaluated by ten-fold cross validation44, using the following parameters: total mean square error and squared correlation coefficient.
Prediction of IC50 values for cancer drugs using circuit activity values as features
Two gene expression datasets of human tumor cell lines screened for different drugs with the corresponding IC50 measurements available were used. One of them, the CGP (ArrayExpress ID: E-MTAB-783)35 was used to train the predictor and the other one, CCLE (GEO ID: GSE36139)36, to validate the predictions. Common data from both datasets were selected, resulting in a total of 317 cell lines, 12 cancers and 7 drugs. CGP data were normalized with RMA41 and normalized gene expression values were transformed into probabilities of signaling circuit activation30. Finally, a predictive model was obtained for the CGP data with a SVM ɛ-regression as explained in Methods section.
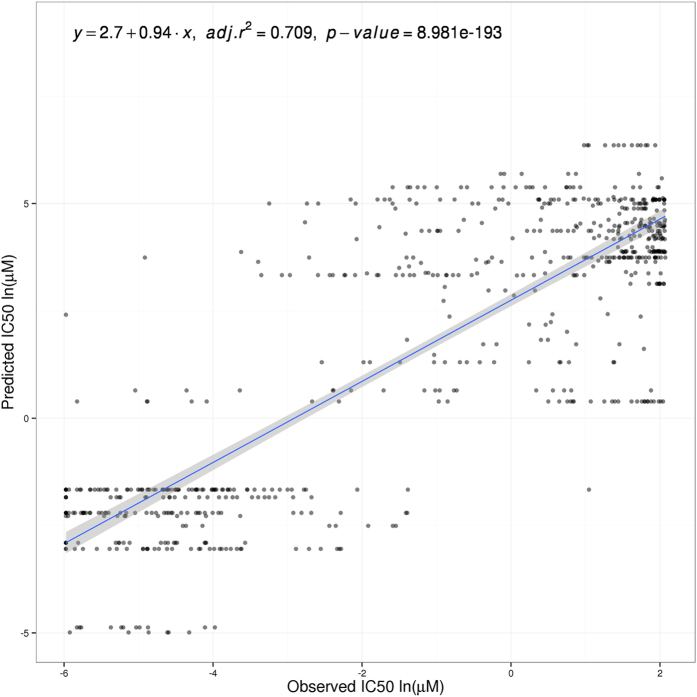
The predictor model obtained with CGP data is used to predict IC50 values in the CCLE dataset. Gene expression values were normalized as explained in Methods and used to calculate signaling circuit activation probabilities. Then, these values were used to predict an IC50 value for each cell line and drug. Figure 1 shows the agreement between predicted a real values. There is a highly significant positive correlation (r = 0.709, p = 8.98 × 10−193) between the expected, real IC50 values measured in the CCLE dataset and the values predicted by regression, based on the CGP dataset, which clearly confirms the validity of the prediction framework proposed here.
Figure 1. Predicted versus actual CCLE IC50 values using signaling circuit activities as features.
Values are represented per cell line and compound (418 cell lines, 7 compounds and 6 cancers). Data values are in natural logarithm of micro-Molar units. Linear regression is specified with intercept and slope values. Adjusted R-squared and p-value are also included.
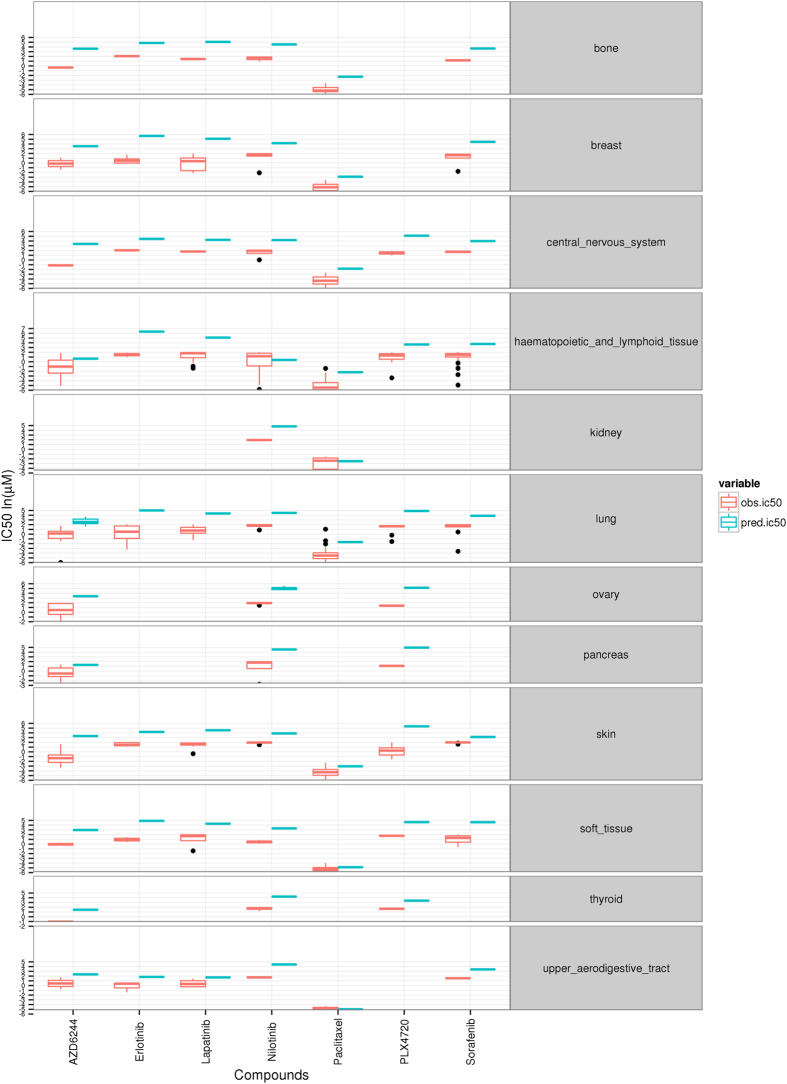
Figure 2 shows the predicted IC50 values and the corresponding real IC50 values available for the CCLE dataset averaged by tissue. Both predicted and real IC50 values were compared by estimating the root mean square error (RMSE) (Table 1). While there are some discrepancies, a global RMSE of 3.31, including all cancers and drugs, demonstrates a quite reasonable accuracy for the prediction. Specific cancers and drugs for which the prediction is especially good are: upper aerodigestive tract (RMSE = 0.52) and soft tissue (RMSE = 0.64) with Paclitaxel. The most extreme discrepancies occur in breast for Erlotinib (RMSE = 5.19) and Lapatinib (RMSE = 5.28). In general, breast tissue shows a poorer prediction than the rest of tissues. It could be due to the fact that some key pathways, such as the ERBB signaling pathway, are under-represented among the features chosen for the prediction. It can also be due to the fact that RAS signaling pathway, relevant in breast cancer, was not modeled here. Also, in skin cancer cell lines, PLX4720 (RMSE = 5.3) AZD6244 (RMSE = 4.71) seem to have selected different features to the rest of drugs which perform better in the classification (RSMEs between 1.19 and 3.22). The use of SVM, which used a combination of features for the prediction, makes difficult finding a unique explanation for the discrepancies.
Figure 2. CCLE predicted and observed IC50 values per cancer and drug.
Table 1. RMSE per cancer type and drug in the CCLE dataset.
| Cancers | Drug Paclitaxel | AZD6244 | Nilotinib | PLX4720 | Sorafenib | Erlotinib | Lapatinib |
|---|---|---|---|---|---|---|---|
| Lung | 2.99 | 3.59 | 2.77 | 3.85 | 2.82 | 5.08 | 3.87 |
| Haematopoietic and lymphoid tissue | 2.98 | 2.68 | 2.42 | 3.33 | 3.05 | 4.88 | 3.89 |
| Bone | 3.96 | 3.01 | NA | 2.5 | 2.78 | 3.61 | 3.96 |
| Skin | 1.63 | 4.71 | 2.02 | 5.3 | 1.19 | 2.66 | 3.22 |
| Ovary | NA | 3.33 | 3.18 | 3.77 | NA | NA | NA |
| Central nervous system | 2.67 | 4.51 | 2.83 | 3.66 | 2.29 | 2.41 | 2.47 |
| Pancreas | NA | 2.08 | 4.34 | 3.85 | NA | NA | NA |
| Soft tissue | 0.64 | 3.01 | 2.88 | 2.88 | 3.77 | 3.99 | 3.59 |
| Breast | 2.23 | 3.87 | 3.17 | NA | 3.67 | 5.19 | 5.28 |
| Upper aerodigestive tract | 0.52 | 2.3 | 2.73 | NA | 1.86 | 2.17 | 1.49 |
| Kidney | 1.22 | NA | 2.88 | NA | NA | NA | NA |
| Thyroid | NA | 2.51 | 2.58 | 1.71 | NA | NA | NA |
NAs appear when not enough data were available.
Comparison to predictions of IC50 values for cancer drugs derived when all the gene expression values are used as features
The potential of the full set of gene expression values as features for predicting drug sensitivity has already been proven in a recent study45. Here we have trained the predictor used in this study using directly the normalized gene expression values (instead of transforming them into signaling activities) as features. Figure 3 plots the predicted IC50 values and the corresponding real IC50 values for the CCLE dataset. The predictive power of all the genes also resulted in a highly significant positive correlation (r = 0.712, p = 1.09 × 10−114) between the expected, real IC50 values measured in the CCLE dataset and the values, predicted by regression, based on the CGP dataset.
Figure 3. Predicted versus actual CCLE IC50 values using normalized gene expression values as features.
Values are represented per cell line and compound (418 cell lines, 7 compounds and 6 cancers). Data values are in natural logarithm of micro-Molar units. Linear regression is specified with intercept and slope values. Adjusted R-squared and p-value are also included.
It must be taken into account that signaling circuit features, as defined here, will only account for the effect of the condition studied over the modeled signaling pathways. Information on any effect unrelated to signaling or related to yet unknown or undescribed signaling pathways is missing in the proposed method. In spite of this fact, the direct or indirect impact of the drugs studied on the signaling circuits is enough to produce quite reliable predictions (Fig. 1) of similar precision than the global gene expression, which potentially captures the whole reaction of the cell to the conditions studied.
MoA suggested by the mechanism-based biomarkers selected
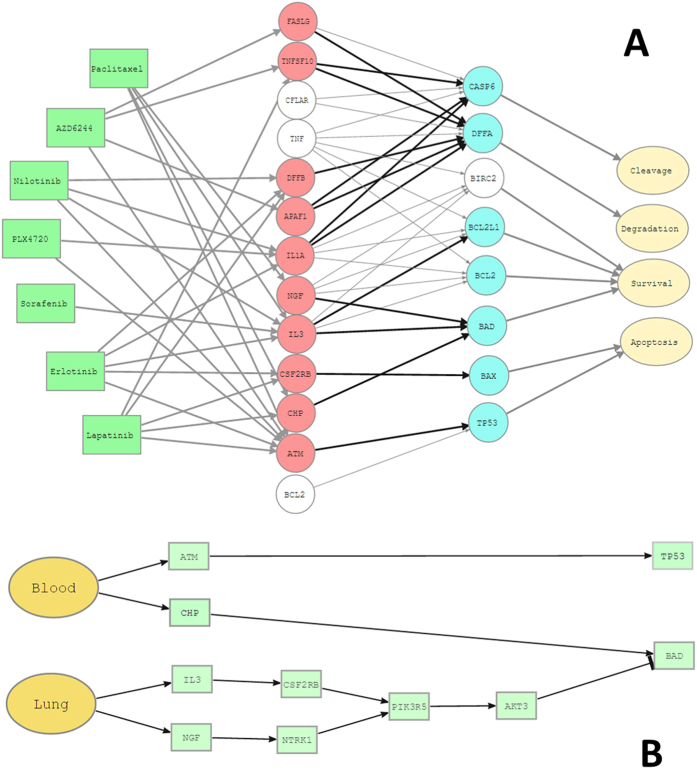
The proposed methodology selects the most predictive signaling circuits within each pathway modeled for each drug analyzed in each tissue tested. Supplementary Fig. S1 displays the signaling circuits selected for each drug/tissue combination. An interesting observation is the pervasiveness with which drugs affect to pathway activity. Almost all the pathways are affected in at least one circuit by any of the drugs in at least one of the tissues tested. There are, however, a few exceptions, such as the JAK-STAT signaling pathway, which is never affected by AZD6244 or Sorafenib; or NOTCH signaling pathway which is never affected by Nilotinib or AZD6244, along with a few more examples. The signaling circuits selected are those that exhibit the most dramatic change in activity among all the circuits affected by the drug in a particular tissue. Table 2 shows the bibliographic references that report the alteration of pathways by the studied drugs. There are numerous reports for some pathways, which are extensively affected by all the drugs, as the Apoptosis pathways, affected by Paclitaxel46, AZD624447, Nilotinib48, PLX472049, Sorafenib50, and Lapatinib51. Actually, it is interesting to see how different drugs affect the apoptosis pathway in different ways. For example, Sorafenib is the only drug that affects the pathway by exclusively inhibiting survival. It has been documented that this drug induces apoptosis by down-regulating the anti-apoptotic protein Mcl-1 via transcriptional inhibition and protein degradation50. Actually, the Mcl-1 is in the PI3K-ATK pathway and triggers survival in the Apoptosis pathway. Supplementary Fig. S1 shows how survival is inhibited by two circuits ending, respectively, in proteins BCL2 and BCL2L1. For the rest of drugs, however, the most common mechanism is the activation of the apoptosis via TP53 protein, along with a number of complementary circuits that activate functions complementary to apoptosis, such as degradation or cleavage of caspase substrate. Figure 4A shows the different signaling circuits used by the different drugs to cause cell death, which illustrate the diversity of drug MoAs, which can also differ across cell lines (Supplementary Fig. S1).
Table 2. Pathways known to be affected by different drugs with the corresponding bibliographic citation.
| Pathways | Drug Paclitaxel | AZD6244 | Nilotinib | PLX4720 | Sorafenib | Erlotinib | Lapatinib |
|---|---|---|---|---|---|---|---|
| Apoptosis pathway | 46,55 | 47 | 48 | 49 | 50 | 51 | |
| Insulin signaling pathway | 52 | 75 | 76 | 77 | |||
| mTOR signaling pathway | 78 | 79 | 80 | 81 | |||
| WNT signaling pathway | 53 | 82 | 83 | 84 | 85 | ||
| Adipocytokine signaling pathway | 86 | 86 | |||||
| Calcium signaling pathway | 54 | 87 | |||||
| ERBB signaling pathway | 88 | 89 | 89 | ||||
| Hedgehog signaling pathway | 55 | 90 | 91 | 92 | |||
| JAK-STAT signaling pathway | 56 | 93 | |||||
| p53 signaling pathway | 57 | 94 | 95 | ||||
| Chemokine signaling pathway | 58 | 96 | |||||
| PPAR signaling pathway | 59 | ||||||
| Toll-like receptor signaling pathway | 56 | ||||||
| VEGF signaling pathway | 60 | 97 | |||||
| B cell receptor signaling pathway | 98 |
Figure 4. Possible mechanism of action (MoA) of the different drugs in the apoptosis pathway as suggested by the signaling circuits selected by the predictor.
(A) Drugs (left in green) that act over the different circuits defined by the receptor (in red) and effector (in blue) proteins, respectively. Cell functionalities triggered by the circuits are labeled in pale yellow. The circuits not affected by any of the drugs are dimmed in gray. (B) Specific circuits over which the Paclitaxel drug acts in both Blood and Lung cancer cell lines. The circuits affected trigger survival and apoptosis.
There are drugs whose effect on different pathways have extensively been documented in the literature and confirm the results here presented. For example, Paclitaxel is known to affect to the Apoptosis pathway46 (see Fig. 4B), the Insulin signaling pathway52, the WNT signaling pathway53, the Calcium signaling pathway54, the Hedgehog signaling pathway55, the JAK-STAT signaling pathway56, the p53 signaling pathway57, the Chemokine signaling pathway58, the PPAR signaling pathway59, the Toll-like receptor signaling pathway56 and the VEGF signaling pathway60. Supplementary Table S1 presents detailed references that link the drug to the pathway affected in one of the cancers in which the method has selected signaling circuits from it.
Although a detailed analysis of the MoAs of each drug in each cancer would be excessively long and is beyond the scope of this paper, it is clear that the IC50 prediction is systematically based on a subset of features which have a mechanistic relevance in the action of the drug.
Experimental validation of relevant activity changes predicted
The schema used in this study encompasses the use of a training set (CGP) in which the predictors have been derived and another independent dataset (CCLE) in which the accuracy of the predictors have been demonstrated. Such demonstration involves the observation of differential activity in the signaling status of the selected circuits in the independent dataset and can be considered per se an experimental validation of the existence of such changes. However, the unlikely possibility that some of these circuits were artefactual cannot be completely ruled out because, although all the genes involved in them were actively being transcribed, they could be not translated to proteins or these could be inactive.
Although phospho-proteomic data are scarce, there is a dataset from a study of global phosphorylation changes upon Erlotinib treatment of lung adenocarcinoma cell lines61, which can be used to confirm the existence of proteins involved in the selected circuits and the change of their activation status. The study contains data on H3255, a lung adenocarcinoma cell line sensitive to EGFR-directed tyrosine kinase inhibitors, and H1975, another lung adenocarcinoma cell line resistant to first-generation reversible EGFR-TKIs, such as Erlotinib. Both cell lines are included in the CGP dataset used to generate the prediction models. Erlotinib in lung cell lines affects to a total of 23 signaling circuits across 15 pathways and include 78 nodes (see Supplementary Fig. S1). Phosphorylation data are not exhaustive and the study contains only ratio values for total of 21 proteins belonging to the circuits selected by the predictor (Table 3).
Table 3. Phosphorylation summary per protein. Mean ratios per gene were computed using its phosphosites’ phosphorylation ratios and a ratio cutoff was applied to define hyperphosphorylation (hyperP) and dephosphorylation (deP) states or no change over the threshold (NC).
| Protein | H3255 | H1975 | Pathway (KEGG ID) | Circuit | Signalling | Node Proteins |
|---|---|---|---|---|---|---|
| ADCY2 | deP | deP | Calcium signaling pathway (hsa04020) | CHRM1-ATP2A1 | Down | ADCY1 ADCY2 ADCY3 ADCY7 ADCY8 ADCY9 ADCY4 |
| SPECC1L | deP | NA | Calcium signaling pathway (hsa04020) | CHRM1-ATP2A1 | Down | CHRM1 CHRM3 CHRM5 ADORA2A SPECC1L ADORA2B ADRB1 ADRB2 ADRB3 DRD1 DRD5 HRH2 HTR4 HTR5A HTR6 HTR7 |
| ATP2A2 | NC | NA | Calcium signaling pathway (hsa04020) | CHRM1-ATP2A1 | Down | ATP2A1 LOC100510514 ATP2A2 ATP2A3 |
| ADCY9 | hyperP | NA | Calcium signaling pathway (hsa04020) | GPCR-SERCA | Down | ADCY1 ADCY2 ADCY3 ADCY7 ADCY8 ADCY9 ADCY4 |
| AKT1 | deP | hyperP | Insulin signaling pathway (hsa04910) | SKIP(INPP5K)-aPKC | Up | AKT3 AKT1 AKT2 |
| PDPK1 | deP | NA | Insulin signaling pathway (hsa04910) | SKIP(INPP5K)-aPKC | Up | PDPK1 |
| DFFA | hyperP | NA | Apoptosis (hsa04210) | DFFB-DFFA | Down | DFFA |
| ICAM1 | NA | deP | Cell adhesion molecules (CAMs) (hsa04514) | ICAM1,2,3-ITGAL | Down | ICAM1 |
| IKBKB | deP | NA | Adipocytokine signaling pathway (hsa04920) | TNF-NPY | Up | CHUK IKBKB IKBKG |
| NFKB2 | NA | NC | Adipocytokine signaling pathway (hsa04920) | TNF-NPY | Up | NFKB1 NFKB2 RELA |
| PRKAA1 | NC | NA | Adipocytokine signaling pathway (hsa04920) | TNF-NPY | Up | PRKAG2 PRKAG3 PRKAA1 PRKAA2 PRKAB1 PRKAB2 PRKAG1 |
| PRKAA2 | NC | NA | Adipocytokine signaling pathway (hsa04920) | TNF-NPY | Up | PRKAG2 PRKAG3 PRKAA1 PRKAA2 PRKAB1 PRKAB2 PRKAG1 |
| STAT3 | NA | deP | Adipocytokine signaling pathway (hsa04920) | TNF-NPY | Up | STAT3 |
| TRAF2 | hyperP | hyperP | Adipocytokine signaling pathway (hsa04920) | TNF-NPY | Up | TRAF2 |
| MAPK1 | deP | deP | Wnt signaling pathway (hsa04310) | Nkd-RhoA | Down | MAPK8 MAPK9 MAPK10 |
| MAPK1 | deP | deP | Melanogenesis (hsa04916) | SCF(KITLG)-DCT | Up | MAPK1 MAPK3 |
| MAP2K2 | deP | deP | Melanogenesis (hsa04916) | SCF(KITLG)-DCT | Up | MAP2K1 MAP2K2 |
| MAPK3 | deP | deP | Melanogenesis (hsa04916) | SCF(KITLG)-DCT | Up | MAPK1 MAPK3 |
| MITF | NA | hyperP | Melanogenesis (hsa04916) | SCF(KITLG)-DCT | Up | MITF |
| RAF1 | hyperP | NA | Melanogenesis (hsa04916) | SCF(KITLG)-DCT | Up | RAF1 |
| TP53 | hyperP | hyperP | p53 signaling pathway (hsa04115) | MDMX-Wip1(PPM1D) | Down | TP53 |
NCs indicate that the protein exist although the phosphorylation status has nos significantly changed. NAs correspond to conditions for which no data are available. For easier identification, circuits have been named according the names of the input and output nodes as they appear in KEGG. The column labelled as signaling contains indications on the change in signal activity of the circuits as predicted by the Pathiways tool30 using transcriptomic data of the untreated cell lung line vs the Erlotinib treated one. The last column contains the proteins present in the node of the protein for which phosphorylation data is available.
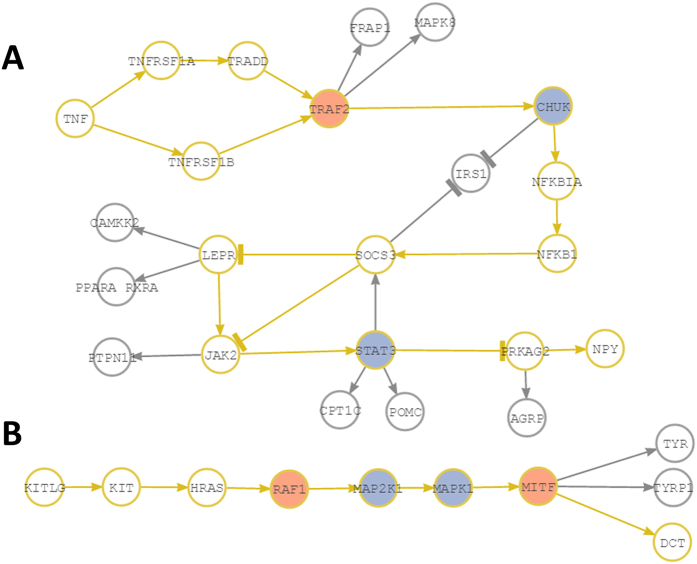
In general there is a good agreement between the change of phosphorylation status of the proteins measured that belong to a circuit selected by the predictor and the change in signaling obtained upon the application of the Pathiways tool30 for this circuit. Some apparent contradictions can easily be explained by the topology of the circuits. For example, in the case of insulin signaling pathway, PDPK1 shows a dephosphorylation and AKT1 shows inconclusive results (hyper-phosphorylation in one cell line and dephosphorylation in the other one) although Pathiways predicts an increase in signaling. However, aPKC, at the end of the circuit (according KEGG) can also be directly activated by INPP5 (SKIP node in KEGG), which explains the apparent discrepancy in the results. Another example is the circuit TNF-NPY in the Adipocytokine signaling pathway (Fig. 5A), which is predicted to be activated by Pathiways. The observed phosphorylation pattern is complex and involves hyper-phosphorylations and dephosphorylation. However, essentially what happens is that TRAF2 is activated and transmit the signal to CHUK node, where one of the three proteins (IKBKB) present in the node is dephosphorylated. There is no data on successive steps of signal transmission until the STAT protein, which is dephosphorylated. STAT is an inhibitor of PRKAG2 that transmit the last step of signaling to NPY, which allows concluding that the signal ultimately arrives to NPY, as predicted by Pathiways for this circuit selected by the predictor. Another interesting case is the circuit SCF(KITLG)-DCT from the Melanogenesis pathway (Fig. 5B). This is a lineal circuit with no bifurcations. After the hyperphosphorylation of the protein RAF1, the unique component of the node, there are two nodes that contain dephosphorylated proteins in the circuit (MAP2K1 and MAPK1). However, it seems that other proteins in the nodes managed to transmit the signal, because finally, MITF, the ultimate responsible of transmit the signal to DTC is hyperphosphorylated again. This agrees again with the Pathiways prediction of increase in the signal transmission in this circuit.
Figure 5. Two examples of circuits selected by the predictor.
The circuits are represented in yellow. Arrowheads indicate activations and “T” heads indicate inhibitions. (A) Circuit TNF-NPY from the Adipocytokine signaling pathway (hsa04920). (B) Circuit SCF(KITLG)-DCT from the Melanogenesis pathway (hsa04916). Nodes in red contain proteins found to be hyperphosphorylated when comparing treated versus untreated lung cancer cells. Nodes in blue contain proteins found to be dephosphorylated in the same comparison.
Software for the derivation of mechanism-based predictors
A web page which allows transforming gene expression values into signaling circuit activities (mechanism-based biomarkers) and further using them to calculate the best predictor of the studied experimental conditions is available at: http://pathiways.babelomics.org/.
The program inputs “.CEL” files. Gene expression values are transformed into circuit activation probabilities30. These probabilities are used to derive a predictor of the conditions studied. Additionally, a matrix of samples x probabilities can be saved to be subsequently used in other programs which can derive predictors by other algorithms (for example in Babelomics62), if desired. The program also provides estimations of the differentially activated circuits as described elsewhere30.
Discussion
Due to the inherent complexity of the cell, phenotypes cannot be understood as the result of the action of only one or a few genes12,13. Consequently, conventional approaches to study the phenotype must evolve from a gene-centric perspective towards a systems-biology-oriented view that considers the combined action of several functionally related genes63,64,65. Therefore, new approaches that transform individual gene measurements (e.g. gene expression levels) into parameters that describe functional activities of biological modules open the way to the definition of a new type of mechanism-based biomarkers which can be used to predict complex phenotypes such as disease status or drug responses66.
In particular, the identification and knowledge of drug MoAs is crucial in pharmacogenetic studies67. The fact that many drugs target signal transduction processes requires of a detailed understanding of the MoA at the signaling level, not only in the specific tissue in which the drug is aimed to act but also in other tissues that may suffer off-target effects14. Understanding such mechanisms could have an enormous impact in many aspects of drug development and personalized therapies68.
The prediction strategy proposed here introduces the use of mechanism-based biomarkers (signaling circuits), responsible for specific cell functionalities, whose anomalous activity could be the ultimate cause of a phenotype. Such biomarkers throw light on possible drug MoAs40,69, given that changes in the activity of the individual molecules is understood within the context of the system conformed by the signaling circuits64,65.
The predictions obtained are quite reasonable and even slightly better than the predictions obtained using all the genes, in agreement with previous observations70. It must be taken into account that: 1) the proposed mechanism-based biomarkers capture only the part of the effect of the drugs that either directly or indirectly affect cell signaling and 2) the predictions obtained with all the genes use much more variables (over 20,000 genes) than the corresponding ones obtained here with the signaling circuits (using only about 800 genes involved in the circuits).
Moreover, the specific circuits selected by the predictor, in the cases in which a validation has been possible using phosphorylation data, are not only transcriptionally active but also the proteins have been produced and the corresponding changes in their phosphorylation statuses are observed. Although gene expression does not necessarily imply its translation and its subsequent activation, our results strongly suggest that, when collective gene up-regulation (or down-regulation) occurs within the context of a pathway, it can be considered a reliable proxy of activation (or deactivation) of the corresponding signaling circuits.
Therefore, the use of mechanism-based biomarkers in a prediction context not only provides mechanistic explanations on the phenotypes studied but, in addition, it seems to produce comparatively better predictions.
The proposed methodology can be easily used through the software provided. Here, the methodology was focused on gene expression data obtained from microarrays because of its availability. Gene expression values can be obtained by other methodologies, such as RNA-seq71, providing the data compared are in the same scale (this is the objective of the normalization process).
Methods
Data sources
Two published large-scale datasets from the cancer genome project (CGP) (E-MTAB-783 in the ArrayExpress repository)35 and the cancer cell line encyclopedia (CCLE) (GSE36139 accession number in the Gene Expression Omnibus repository)36 were used in this study. Both datasets provide gene expression information of human tumor cell lines which have been screened for different drugs and the concentration at which the drug response reached an absolute inhibition of 50% (IC50). CGP dataset provides information about 138 drugs, 661 cell lines (including NCI-60 cell lines) which correspond with 17 cancers; on other hand, CCLE dataset includes information about 24 drugs, 493 cell lines which correspond with 24 cancers. Common data from both datasets were filtered for a total of 355 cell lines, 12 cancers and 7 drugs.
Phospho-proteomic data were obtained from a study of global phosphorylation changes upon Erlotinib treatment of lung adenocarcinoma cell lines61. Supplementary material of this study includes information of EGF stimulation (stimulated/untreated cells) and Erlotinib inhibition (treated/untreated cells) SILAC ratios.
Data preprocessing
Microarray normalization was carried out using the function RMA in the affy library41 of Bioconductor72, as implemented in Babelomics platform62.
For the phosphorylation data, mean ratios gene protein were computed using its phosphosites’ phosphorylation ratios and a ratio cutoff was applied to define hyperphosphorylation (hyperP) and dephosphorylation (deP) states. The ratio cutoff was defined as follows: > 1.1 increased, 0.9–1.1 unchanged and < 0.9 decreased. Then, hyperphosphorylation (hyperP) was defined as having a decreased ratio of stimulated/control and an increased ratio of treated/stimulated, and dephosphorylation (deP) when having an increased ratio of stimulated/control and a decreased ratio of treated/stimulated.
Derivation of mechanism-based biomarkers (signaling circuit activation probabilities) from gene expression values
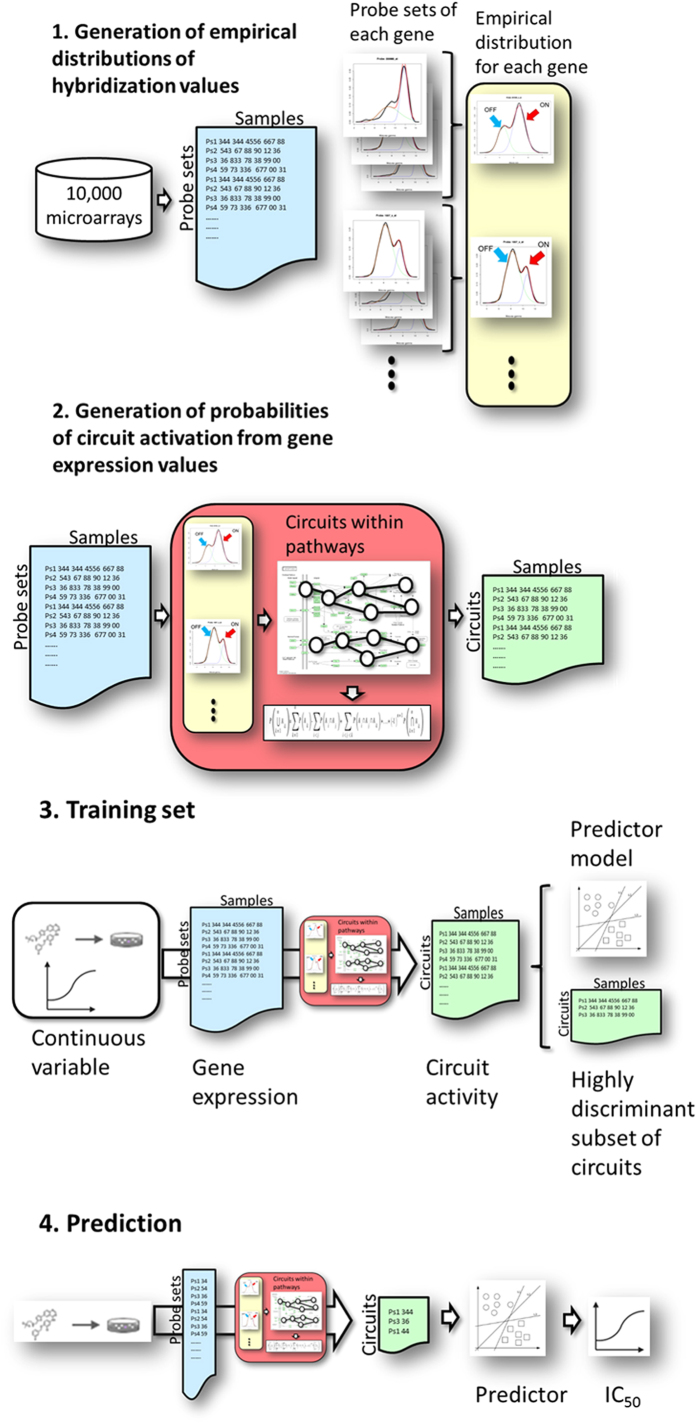
Signaling circuit activation probabilities are inferred from the estimations of gene activation probabilities corresponding to the proteins involved in the circuit. Briefly, a mixture distribution is used on a large dataset of reference microarrays to derive empirical distributions of expression values corresponding to activated and deactivated states of the probe sets in the microarrays. Probesets can be used to summarize gene expression values (Fig. 6, step 1). Within this analytic framework, gene expression is taken as a proxy of protein expression and, consequently, protein activity26,27,30,31,73.
Figure 6. Steps for the generation and use of predictors based on signaling circuit activities.
Step 1: generation of the empirical distribution of probeset values. A collection of more than 10,000 microarrays representing an enormous diversity of conditions is collected from the GEO database. For any of the probesets, an empirical distribution is derived and a mixture model is used to define the highest value peak, which corresponds to an active probe (ON), and the lowest peak that correspond to the probeset (OFF). Gene values can be obtained by summarizing the corresponding probeset values. Step 2: Given one or several microarrays, the probeset values can be contrasted with the empirical distribution values to obtain the corresponding activity probabilities which are used to derive gene activity probabilities. These, within the context of the circuits defined, are used to estimate circuit activity probabilities. Step 3: an initial training set is required to derive obtain the predictor. Gene expression values from individuals from two classes, or from different treatments (dosage, time, etc.) are obtained and transformed (as described in step 2) into the corresponding profiles of signaling activities. Then, a feature selection method obtain a sub list of highly discriminative circuits which is used to train the predictor (see below). Step 4: once the predictor is trained it can be used to predict class membership for an unknown sample (or to predict a continuous value from gene expression measurements). Gene expression values from the sample are transformed into the corresponding pattern of signaling circuit activities (see step 2) of the sample. The predictor is then used to predict the class to which most likely the sample belongs to. Identically, gene expression values of a series of conditions can be used to predict the corresponding continuous value (not shown in the figure).
The step 2 in Fig. 6 illustrates how a matrix of probeset expression values can be converted into a matrix of signaling circuit activation probabilities. Empirical distributions for probe sets previously derived are used to assign a probability of activation to probe sets in the studied microarrays30,31,73. Then, probeset activation values are combined to derive probabilities of gene activation.
On the other hand, signaling circuits are defined as the sub-pathways (within pathways taken from KEGG database18) that transmit signals from a receptor node to an effector node. Such circuits can have bi- or multi-furcations and typically consist of nodes that activate other nodes but they can also contain nodes that inhibit the activity of other nodes. Such nodes can be composed of one or several proteins. Finally, node activation probabilities are obtained by combining the probability activation values corresponding to all the genes that comprise the node (see details in30,31). Once probabilities of activation for each node in the circuit have been estimated, the probability of signal transmission can be modeled as a simple probabilistic product using the inclusion-exclusion principle30,31 (red box in the step 2 of Fig. 6 summarizes this procedure).
Probabilistic models were obtained as described above for the signaling circuits defined within a total of 26 KEGG pathways for Homo sapiens and 18 for Mus musculus. These correspond to the general categories Environmental Information Processing and Cellular Processes, which include important processes and systems such as Signal Transduction (ERBB, WNT, NOTCH, JAK-STAT, calcium, VEGF, HEDGEHOG and mTOR signaling pathways), Signaling Molecules and Interaction (neuroactive ligand-receptor interaction, cell adhesion molecules, cytokine-cytokine receptor interaction and EMC-receptor interaction), Cell Growth and Death (apoptosis and p53 signaling pathway), Cell Communication (GAP junction and tight junction), Endocrine System (insulin signaling pathway, adipocytokine signaling pathway, PPAR signaling pathway, GnRH signaling pathway and melanogenesis) and Immune System (toll-like receptor signaling pathway, B cell receptor signaling pathway, T cell receptor signaling pathway, Fc epsilon RI signaling pathway, antigen processing and presentation, and chemokine signaling pathway).
Derivation of predictors based on mechanism-based biomarkers
Prediction methods require of an initial training set to derive a trained predictor (Fig. 6, step 3) which can be further used to predict the corresponding value of a continuous variable for a new sample (Fig. 6, step 4). The step 3 in Fig. 6 summarizes how the training set is used for the training phase of the predictor. Gene expression values from different treatments are obtained and transformed as described in step 2 (Fig. 6) into the corresponding profiles of signaling activities. Since signaling circuit probabilities are used as mechanism-based biomarkers to predict drug sensitivity values, those circuits showing no variability across the treatments were initially discarded. Then, a sub list of highly discriminative circuits is obtained using Correlation-based Feature Selection (CFS) method74 as feature selection algorithm (Fig. 6, end of step 3). Drug sensitivity prediction was carried out using the highly discriminant subset of signaling circuit activities. The prediction algorithm used was Support Vector Machine (SVM)42 as implemented in the e1071 R library43. SVM ɛ-regression was performed selecting best γ and cost parameters among different values tested (10, 100 with cost values; 10−6, 10−5, 10−4, 10−3 γ values), by optimizing the mean squared error of the model with a 10-fold cross-validation.
In the particular case of the SVM algorithm, the previous feature selection step is not necessary for prediction purposes, unless a subset of relevant features is sought. K-fold cross validation (with K = 10) was used in the training step of the models. The CGP dataset35 was used to train the predictor and obtaining the prediction model (Fig. 6, end of step 3).
Once the predictor is trained it can be used to predict a drug sensitivity value from gene expression measurements in a new, unknown sample (step 4 Fig. 6).
Methods for feature selection, classifiers and performance evaluation are implemented in Babelomics platform62.
Additional Information
How to cite this article: Amadoz, A. et al. Using activation status of signaling pathways as mechanism-based biomarkers to predict drug sensitivity. Sci. Rep. 5, 18494; doi: 10.1038/srep18494 (2015).
Supplementary Material
Acknowledgments
This work is supported by grants BIO2014-57291-R from the Spanish Ministry of Economy and Competitiveness (MINECO), co-funded with European Regional Development Funds (ERDF), PROMETEOII/2014/025 from the Conselleria de Educacio of the Valencian Community, FP7-PEOPLE-2012-ITN MLPM2012 318861 from the EU FP7 and Fundació la Marató TV3 [20133134]. We also thank the support of both initiatives of the ISCIII, MINECO: the National Institute of Bioinformatics (http://www.inab.org) and the CIBER de Enfermedades Raras (CIBERER).
Footnotes
Author Contributions A.A. developed the tool and carried out the analysis; P.S.-L. developed the original algorithm and participated in the data analysis; E.V. developed the statistical aspects of the method; F.S. carried out the web implementation of the tool and J.D. conceived the work and wrote the paper.
References
- Ma Q. & Lu A. Y. Pharmacogenetics, pharmacogenomics, and individualized medicine. Pharmacol Rev 63, 437–459, 10.1124/pr.110.003533 (2011). [DOI] [PubMed] [Google Scholar]
- Staunton J. E. et al. Chemosensitivity prediction by transcriptional profiling. Proc Natl Acad Sci USA 98, 10787–10792, 10.1073/pnas.191368598 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. K. et al. A strategy for predicting the chemosensitivity of human cancers and its application to drug discovery. Proc Natl Acad Sci USA 104, 13086–13091, 10.1073/pnas.0610292104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S., Chang J. T., Andrechek E. R., Potti A. & Nevins J. R. Utilization of genomic signatures to identify phenotype-specific drugs. PLoS ONE 4, e6772, 10.1371/journal.pone.0006772 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddick G. et al. Predicting in vitro drug sensitivity using Random Forests. Bioinformatics 27, 220–224, 10.1093/bioinformatics/btq628 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eduati F. et al. Prediction of human population responses to toxic compounds by a collaborative competition. Nat Biotechnol. 10.1038/nbt.3299 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burczynski M. E. et al. Toxicogenomics-based discrimination of toxic mechanism in HepG2 human hepatoma cells. Toxicol Sci 58, 399–415 (2000). [DOI] [PubMed] [Google Scholar]
- Uehara T. et al. Prediction model of potential hepatocarcinogenicity of rat hepatocarcinogens using a large-scale toxicogenomics database. Toxicol Appl Pharmacol 255, 297–306, 10.1016/j.taap.2011.07.001 (2011). [DOI] [PubMed] [Google Scholar]
- Ioannidis J. P. et al. Repeatability of published microarray gene expression analyses. Nat Genet 41, 149–155 (2009). [DOI] [PubMed] [Google Scholar]
- Ein-Dor L., Kela I., Getz G., Givol D. & Domany E. Outcome signature genes in breast cancer: is there a unique set? Bioinformatics 21, 171–178 (2005). [DOI] [PubMed] [Google Scholar]
- Ein-Dor L., Zuk O. & Domany E. Thousands of samples are needed to generate a robust gene list for predicting outcome in cancer. Proc Natl Acad Sci USA 103, 5923–5928 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerhoff H. V. & Palsson B. O. The evolution of molecular biology into systems biology. Nat Biotechnol 22, 1249–1252 (2004). [DOI] [PubMed] [Google Scholar]
- Loscalzo J. & Barabasi A. L. Systems biology and the future of medicine. Wiley Interdiscip Rev Syst Biol Med. 10.1002/wsbm.144 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melas I. N. et al. Identification of drug-specific pathways based on gene expression data: application to drug induced lung injury. Integrative biology: quantitative biosciences from nano to macro 7, 904–920, 10.1039/c4ib00294f (2015). [DOI] [PubMed] [Google Scholar]
- Schadt E. E. et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet 37, 710–717 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Zhang Y., Marian C. & Ressom H. W. Identification of aberrant pathways and network activities from high-throughput data. Brief Bioinform 13, 406–419, 10.1093/bib/bbs001 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. J. & Ragan M. A. Understanding cellular function and disease with comparative pathway analysis. Genome Med 5, 64, 10.1186/gm468 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. et al. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 42, D199–205, 10.1093/nar/gkt1076 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft D. et al. The Reactome pathway knowledgebase. Nucleic Acids Res 42, D472–477, 10.1093/nar/gkt1102 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri P., Sirota M. & Butte A. J. Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput Biol 8, e1002375, 10.1371/journal.pcbi.1002375 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draghici S. et al. A systems biology approach for pathway level analysis. Genome Res 17, 1537–1545, 10.1101/gr.6202607 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarca A. L. et al. A novel signaling pathway impact analysis. Bioinformatics 25, 75–82 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shojaie A. & Michailidis G. Analysis of gene sets based on the underlying regulatory network. J Comput Biol 16, 407–426, 10.1089/cmb.2008.0081 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q. et al. Pathway activation strength is a novel independent prognostic biomarker for cetuximab sensitivity in colorectal cancer patients. Human Genome Variation 2, 15009, 10.1038/hgv.2015.9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y., Chen M., Wang H. & Zheng X. A network flow-based method to predict anticancer drug sensitivity. PLoS ONE 10, e0127380, 10.1371/journal.pone.0127380 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini P., Sales G., Massa M. S., Chiogna M. & Romualdi C. Along signal paths: an empirical gene set approach exploiting pathway topology. Nucleic Acids Res 41, e19, 10.1093/nar/gks866 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob L., Neuvial P. & Dudoit S. More power via graph-structured tests for differential expression of gene networks. Ann. Appl. Stat. 6, 561–600, 10.1214/11-AOAS528 (2012). [DOI] [Google Scholar]
- Chen X. et al. A sub-pathway-based approach for identifying drug response principal network. Bioinformatics 27, 649–654, 10.1093/bioinformatics/btq714 (2011). [DOI] [PubMed] [Google Scholar]
- Nam S. & Park T. Pathway-based evaluation in early onset colorectal cancer suggests focal adhesion and immunosuppression along with epithelial-mesenchymal transition. PLoS ONE 7, e31685, 10.1371/journal.pone.0031685 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian-Leon P. et al. Inferring the functional effect of gene expression changes in signaling pathways. Nucleic Acids Res 41, W213–217, 10.1093/nar/gkt451 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian-Leon P. et al. Understanding disease mechanisms with models of signaling pathway activities. BMC Syst Biol 8, 121, 10.1186/s12918-014-0121-3 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. et al. A chemical and phosphoproteomic characterization of dasatinib action in lung cancer. Nat Chem Biol 6, 291–299, 10.1038/nchembio.332 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J. The Connectivity Map: a new tool for biomedical research. Nat Rev Cancer 7, 54–60, 10.1038/nrc2044 (2007). [DOI] [PubMed] [Google Scholar]
- Lamb J. et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935, 10.1126/science.1132939 (2006). [DOI] [PubMed] [Google Scholar]
- Garnett M. J. et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 483, 570–575, 10.1038/nature11005 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J. et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607, 10.1038/nature11003 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganter B., Snyder R. D., Halbert D. N. & Lee M. D. Toxicogenomics in drug discovery and development: mechanistic analysis of compound/class-dependent effects using the DrugMatrix database. Pharmacogenomics 7, 1025–1044, 10.2217/14622416.7.7.1025 (2006). [DOI] [PubMed] [Google Scholar]
- Su Z. et al. An investigation of biomarkers derived from legacy microarray data for their utility in the RNA-seq era. Genome Biol 15, 523, 10.1186/s13059-014-0523-y (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters M. et al. CEBS—Chemical Effects in Biological Systems: a public data repository integrating study design and toxicity data with microarray and proteomics data. Nucleic Acids Res 36, D892–900, 10.1093/nar/gkm755 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryburg D. A., Song D. H., Laifenfeld D. & de Graaf D. Systems diagnostics: anticipating the next generation of diagnostic tests based on mechanistic insight into disease. Drug Discov Today 19, 108–112, 10.1016/j.drudis.2013.07.006 (2014). [DOI] [PubMed] [Google Scholar]
- Gautier L., Cope L., Bolstad B. M. & Irizarry R. A. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20, 307–315, 10.1093/bioinformatics/btg405 (2004). [DOI] [PubMed] [Google Scholar]
- Vapnik V. Statistical Learning Theory. (John Wiley and Sons, 1999). [DOI] [PubMed] [Google Scholar]
- Karatzoglou A., Meyer D. & Hornik K. Support vector machines. Journal of Statistical Software 15, 1–28 (2006). [Google Scholar]
- Shi L. et al. The MicroArray Quality Control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models. Nat Biotechnol 28, 827–838, 10.1038/nbt.1665 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papillon-Cavanagh S. et al. Comparison and validation of genomic predictors for anticancer drug sensitivity. Journal of the American Medical Informatics Association: JAMIA 20, 597–602, 10.1136/amiajnl-2012-001442 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. T. et al. Tumor suppressor BLU promotes paclitaxel antitumor activity by inducing apoptosis through the down-regulation of Bcl-2 expression in tumorigenesis. Biochem Biophys Res Commun 435, 153–159, 10.1016/j.bbrc.2013.04.061 (2013). [DOI] [PubMed] [Google Scholar]
- Holt S. V. et al. Enhanced apoptosis and tumor growth suppression elicited by combination of MEK (selumetinib) and mTOR kinase inhibitors (AZD8055). Cancer Res 72, 1804–1813, 10.1158/0008-5472.CAN-11-1780 (2012). [DOI] [PubMed] [Google Scholar]
- Shaker M. E., Ghani A., Shiha G. E., Ibrahim T. M. & Mehal W. Z. Nilotinib induces apoptosis and autophagic cell death of activated hepatic stellate cells via inhibition of histone deacetylases. Biochim Biophys Acta 1833, 1992–2003, 10.1016/j.bbamcr.2013.02.033 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F. M., Shao Y., Mayberry M. M. & Aplin A. E. Hyperactivation of MEK-ERK1/2 signaling and resistance to apoptosis induced by the oncogenic B-RAF inhibitor, PLX4720, in mutant N-RAS melanoma cells. Oncogene 30, 366–371, 10.1038/onc.2010.408 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N. K., Huang S. L., Chang T. C. & Chao C. C. Sorafenib induces endometrial carcinoma apoptosis by inhibiting Elk-1-dependent Mcl-1 transcription and inducing Akt/GSK3beta-dependent protein degradation. J Cell Biochem 114, 1819–1831, 10.1002/jcb.24530 (2013). [DOI] [PubMed] [Google Scholar]
- Long X. H. et al. Lapatinib alters the malignant phenotype of osteosarcoma cells via downregulation of the activity of the HER2-PI3K/AKT-FASN axis in vitro. Oncol Rep 31, 328–334, 10.3892/or.2013.2825 (2014). [DOI] [PubMed] [Google Scholar]
- Dziadziuszko R., Camidge D. R. & Hirsch F. R. The insulin-like growth factor pathway in lung cancer. J Thorac Oncol 3, 815–818, 10.1097/JTO.0b013e31818180f5 (2008). [DOI] [PubMed] [Google Scholar]
- Sanchez A. M. et al. Treatment with anticancer agents induces dysregulation of specific Wnt signaling pathways in human ovarian luteinized granulosa cells in vitro. Toxicol Sci 136, 183–192, 10.1093/toxsci/kft175 (2013). [DOI] [PubMed] [Google Scholar]
- Boehmerle W. et al. Paclitaxel induces calcium oscillations via an inositol 1,4,5-trisphosphate receptor and neuronal calcium sensor 1-dependent mechanism. Proc Natl Acad Sci USA 103, 18356–18361, 10.1073/pnas.0607240103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H. et al. The Hedgehog signaling pathway plays an essential role in maintaining the CD44 + CD24-/low subpopulation and the side population of breast cancer cells. Anticancer Res 29, 2147–2157 (2009). [PubMed] [Google Scholar]
- Wang T. H. et al. Paclitaxel (Taxol) upregulates expression of functional interleukin-6 in human ovarian cancer cells through multiple signaling pathways. Oncogene 25, 4857–4866, 10.1038/sj.onc.1209498 (2006). [DOI] [PubMed] [Google Scholar]
- Pasquier E. et al. Antiangiogenic activity of paclitaxel is associated with its cytostatic effect, mediated by the initiation but not completion of a mitochondrial apoptotic signaling pathway. Mol Cancer Ther 3, 1301–1310 (2004). [PubMed] [Google Scholar]
- Jackman R. W., Rhoads M. G., Cornwell E. & Kandarian S. C. Microtubule-mediated NF-kappaB activation in the TNF-alpha signaling pathway. Exp Cell Res 315, 3242–3249, 10.1016/j.yexcr.2009.08.020 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. Z. et al. PPAR signaling pathway may be an important predictor of breast cancer response to neoadjuvant chemotherapy. Cancer Chemother Pharmacol 70, 637–644, 10.1007/s00280-012-1949-0 (2012). [DOI] [PubMed] [Google Scholar]
- Cheng H. et al. In vitro sequence-dependent synergism between paclitaxel and gefitinib in human lung cancer cell lines. Cancer Chemother Pharmacol 67, 637–646, 10.1007/s00280-010-1347-4 (2011). [DOI] [PubMed] [Google Scholar]
- Zhang X. et al. Identifying novel targets of oncogenic EGF receptor signaling in lung cancer through global phosphoproteomics. Proteomics 15, 340–355, 10.1002/pmic.201400315 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso R. et al. Babelomics 5.0: functional interpretation for new generations of genomic data. Nucleic Acids Res 43, W117–121, 10.1093/nar/gkv384 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt E. E. Molecular networks as sensors and drivers of common human diseases. Nature 461, 218–223, 10.1038/nature08454 (2009). [DOI] [PubMed] [Google Scholar]
- Barabasi A. L., Gulbahce N. & Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet 12, 56–68, 10.1038/nrg2918 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M., Cusick M. E. & Barabasi A. L. Interactome networks and human disease. Cell 144, 986–998, 10.1016/j.cell.2011.02.016 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geman D., Ochs M., Price N. D., Tomasetti C. & Younes L. An argument for mechanism-based statistical inference in cancer. Hum Genet. 10.1007/s00439-014-1501-x (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S. & Iyengar R. Systems pharmacology: network analysis to identify multiscale mechanisms of drug action. Ann Rev Pharmacol Toxicol 52, 505–521, 10.1146/annurev-pharmtox-010611-134520 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopazo J. Genomics and transcriptomics in drug discovery. Drug Discov Today 19, 126–132, 10.1016/j.drudis.2013.06.003 (2014). [DOI] [PubMed] [Google Scholar]
- Dopazo J. Genomics and transcriptomics in drug discovery. Drug Discov Today. 10.1016/j.drudis.2013.06.003 (2013). [DOI] [PubMed] [Google Scholar]
- Borisov N. M. et al. Signaling pathway activation profiles make better markers of cancer than expression of individual genes. Oncotarget 5, 10198–10205 10.18632/oncotarget.2548 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Gerstein M. & Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10, 57–63 10.1038/nrg2484 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R. C. et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5, R80, 10.1186/gb-2004-5-10-r80 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni S., Schaefer C. F. & Buetow K. H. Identification of key processes underlying cancer phenotypes using biologic pathway analysis. PLoS ONE 2, e425, 10.1371/journal.pone.0000425 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanski P. & Kotthoff M. L. (2013). Package FSelector, URL http://cran.r-project.org/web/packages/FSelector/index.html.
- Gopal Y. N. et al. Basal and treatment-induced activation of AKT mediates resistance to cell death by AZD6244 (ARRY-142886) in Braf-mutant human cutaneous melanoma cells. Cancer Res 70, 8736–8747, 10.1158/0008-5472.CAN-10-0902 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou D. L. et al. Potentiating the efficacy of molecular targeted therapy for hepatocellular carcinoma by inhibiting the insulin-like growth factor pathway. PLoS ONE 8, e66589, 10.1371/journal.pone.0066589 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien N. A. et al. Activated phosphoinositide 3-kinase/AKT signaling confers resistance to trastuzumab but not lapatinib. Mol Cancer Ther 9, 1489–1502, 10.1158/1535-7163.MCT-09-1171 (2010). [DOI] [PubMed] [Google Scholar]
- Huynh H. AZD6244 (ARRY-142886) enhances the antitumor activity of rapamycin in mouse models of human hepatocellular carcinoma. Cancer 116, 1315–1325, 10.1002/cncr.24863 (2010). [DOI] [PubMed] [Google Scholar]
- Gedaly R. et al. PI-103 and sorafenib inhibit hepatocellular carcinoma cell proliferation by blocking Ras/Raf/MAPK and PI3K/AKT/mTOR pathways. Anticancer Res 30, 4951–4958 (2010). [PMC free article] [PubMed] [Google Scholar]
- Bliesath J. et al. Combined inhibition of EGFR and CK2 augments the attenuation of PI3K-Akt-mTOR signaling and the killing of cancer cells. Cancer Lett 322, 113–118, 10.1016/j.canlet.2012.02.032 (2012). [DOI] [PubMed] [Google Scholar]
- Zhu X. et al. Autophagy stimulates apoptosis in HER2-overexpressing breast cancers treated by lapatinib. J Cell Biochem 114, 2643–2653, 10.1002/jcb.24611 (2013). [DOI] [PubMed] [Google Scholar]
- Tentler J. J. et al. Identification of predictive markers of response to the MEK1/2 inhibitor selumetinib (AZD6244) in K-ras-mutated colorectal cancer. Mol Cancer Ther 9, 3351–3362, 10.1158/1535-7163.MCT-10-0376 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biechele T. L. et al. Wnt/beta-catenin signaling and AXIN1 regulate apoptosis triggered by inhibition of the mutant kinase BRAFV600E in human melanoma. Sci Signal 5, ra3, 10.1126/scisignal.2002274 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachenmayer A. et al. Wnt-pathway activation in two molecular classes of hepatocellular carcinoma and experimental modulation by sorafenib. Clin Cancer Res 18, 4997–5007, 10.1158/1078-0432.CCR-11-2322 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas-Selves M. et al. Tankyrase and the canonical Wnt pathway protect lung cancer cells from EGFR inhibition. Cancer Res 72, 4154–4164, 10.1158/0008-5472.CAN-11-2848 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena N. K. et al. Bidirectional crosstalk between leptin and insulin-like growth factor-I signaling promotes invasion and migration of breast cancer cells via transactivation of epidermal growth factor receptor. Cancer Res 68, 9712–9722, 10.1158/0008-5472.CAN-08-1952 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant J. A., Finn R. S., Slamon D. J., Cloughesy T. F. & Charles A. C. EGF activates intracellular and intercellular calcium signaling by distinct pathways in tumor cells. Cancer Biol Ther 3, 1243–1249 (2004). [DOI] [PubMed] [Google Scholar]
- Sartore-Bianchi A., Ricotta R., Cerea G., Maugeri M. R. & Siena S. Rationale and clinical results of multi-target treatments in oncology. Int J Biol Markers 22, S77–87 (2007). [DOI] [PubMed] [Google Scholar]
- Roskoski R. Jr. The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol Res 79, 34–74, 10.1016/j.phrs.2013.11.002 (2014). [DOI] [PubMed] [Google Scholar]
- Cea M. et al. Tracking molecular relapse of chronic myeloid leukemia by measuring Hedgehog signaling status. Leuk Lymphoma 54, 342–352, 10.3109/10428194.2012.708752 (2013). [DOI] [PubMed] [Google Scholar]
- Moeini A., Cornella H. & Villanueva A. Emerging signaling pathways in hepatocellular carcinoma. Liver cancer 1, 83–93, 10.1159/000342405 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad A. et al. Inhibition of Hedgehog signaling sensitizes NSCLC cells to standard therapies through modulation of EMT-regulating miRNAs. J Hematol Oncol 6, 77, 10.1186/1756-8722-6-77 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uribe P. & Gonzalez S. Epidermal growth factor receptor (EGFR) and squamous cell carcinoma of the skin: molecular bases for EGFR-targeted therapy. Pathol Res Pract 207, 337–342, 10.1016/j.prp.2011.03.002 (2011). [DOI] [PubMed] [Google Scholar]
- Dudgeon C. et al. Inhibiting oncogenic signaling by sorafenib activates PUMA via GSK3beta and NF-kappaB to suppress tumor cell growth. Oncogene 31, 4848–4858, 10.1038/onc.2011.644 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin A. R., Khuri F. R., Chen Z. G. & Shin D. M. Synergistic growth inhibition of squamous cell carcinoma of the head and neck by erlotinib and epigallocatechin-3-gallate: the role of p53-dependent inhibition of nuclear factor-kappaB. Cancer Prev Res (Phila) 2, 538–545, 10.1158/1940-6207.CAPR-09-0063 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassenmaier M. et al. CXC chemokine receptor 4 is essential for maintenance of renal cell carcinoma-initiating cells and predicts metastasis. Stem Cells 31, 1467–1476, 10.1002/stem.1407 (2013). [DOI] [PubMed] [Google Scholar]
- Shimizu S. et al. Inhibition of autophagy potentiates the antitumor effect of the multikinase inhibitor sorafenib in hepatocellular carcinoma. Int J Cancer 131, 548–557, 10.1002/ijc.26374 (2012). [DOI] [PubMed] [Google Scholar]
- Robak P. & Robak T. A targeted therapy for protein and lipid kinases in chronic lymphocytic leukemia. Curr Med Chem 19, 5294–5318 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.