Abstract
Studies investigating the association between Interleukin-23 receptor (IL-23R) gene polymorphisms and Crohn’s disease (CD) report conflicting results. Thus, a meta-analysis was carried out to assess the association between the IL-23R polymorphisms and CD. A systematic literature search was conducted to identify all relevant studies. Pooled odds ratio (ORs) with 95% confidence interval (CIs) was used to estimate the strength of association. Finally, a total of 60 case-control studies in 56 articles, involving 22,820 CD patients and 27,401 healthy controls, were included in the meta-analysis. Overall, a significant association was found between all CD and the rs7517847 polymorphism (OR = 0.699, 95% CI = 0.659 ~ 0.741, P < 0.001). Meta-analysis of the rs11209026, rs1343151, rs10489629 and rs11465804 polymorphisms indicated the same pattern as for rs7517847. Meta-analysis showed an association between the rs10889677A allele and CD (OR = 1.393, 95% CI = 1.328 ~ 1.461, P < 0.001). Similarly, meta-analysis of the rs2201840, rs1004819, rs1495965 and rs11209032 polymorphisms revealed the same pattern as that shown by meta-analysis of rs10889677. Stratification by ethnicity revealed that IL-23R gene polymorphisms were associated with CD in the Caucasian group, but not in Asians. In summary, the meta-analysis suggests a significant association between IL-23R polymorphisms and CD, especially in Caucasians.
Crohn’s disease (CD) is a chronic, inflammatory, autoimmune disease characterized by transmural inflammatory lesions that affect the entire gastrointestinal tract1. The clear mechanisms of CD pathogenesis remain to be elucidated. Studies in twins and family members indicate that CD is significantly associated with genetic susceptibility2,3. It has now been accepted that NOD2/CARD15 gene is related to innate immunity in CD4,5. Furthermore, genes of ATG16L16 and IRGM7,8 have been recognized to play pivotal roles in autophagy in CD. However, these genetic variants can not fully explain the disease onset of CD. To date, genome-wide association studies have identified approximately 80 CD-susceptibility loci, indicating a series of genes and pathogenic mechanisms, such as microbe recognition, lymphocyte activation, cytokine signaling and intestinal epithelial defense9,10,11. Thus, genetic association studies provide a foundation for the immunopathogenesis for the disease, implicating the role of innate and adaptive immunity in CD occurrence.
Interleukin-23 (IL-23), an important immuno-regulatory cytokine secreted by activated macrophages and dendritic cells, regulates the differentiation of T helper 17 cells from native CD4+ T cells12. Binding to IL-23 receptor (IL-23R) complex, IL-23 performed significantly through the Janus kinase (JAK)-signal transducer and activator of transcription (STAT) and NF-κB signaling pathways13,14. The proportion of IL-23R-expressing T cells in the periphery was 2-fold higher in ankylosing spondylitis (AS) patients than in healthy controls, specifically driven by a 3-fold increase in IL-23R-positive γ/δ T cells in AS patients15. γ/δ T cells from AS patients were skewed toward IL-17 production in response to stimulation with IL-23. In IL-23R reporter mice, γ/δ T cells responded to IL-23 during experimental autoimmune encephalomyelitis. IL-23-activated γ/δ T cells rendered αβ effector T cells refractory to the suppressive activity of regulatory T (Treg) cells and also prevented the conversion of conventional T cells into Foxp3(+) Treg cells, suggesting that IL-23/IL23R may play a potential role in autoimmune diseases16. IL-23/IL23R signaling plays critical roles in innate and adaptive inflammatory responses in the intestinal mucosa17. In mice models, IL-23/IL-23R, a key component of IL-23/Th17 pathway, is essential for intestinal inflammation of T cell-dependent colitis18,19. Furthermore, patients with CD manifested increased IL-23 secretion compared with controls20. A genome-wide association study indicated that IL-23R gene is a potential candidate for CD susceptibility21. IL-23R gene is mapped to the chromosome 1 (1p31.3)13. Recently, polymorphisms in this locus have been identified and a large number of studies indicated the association between these polymorphisms and CD risk21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76. However, previous studies in different populations reported conflicting results. The disaccord may be attributed to small sample size, various racial and ethnic backgrounds, uncorrected multiple hypothesis testing and publication bias.
Meta-analysis is a statistical method for augmenting the effective sample size through combining the results of several studies to produce a single estimate of the major effect with enhanced precision. It is considered to be a powerful tool for pooling inconsistent results from different studies77. To date, two meta-analysis52,78 of the association between the IL-23R gene polymorphisms and CD risk have been reported, but these results were still inconclusive. New studies about the role of these SNPs in the IL-23R gene in CD have been published in recent years53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70, and these studies might provide new evidence. Thus, it seems necessary to perform an updated and comprehensive meta-analysis including the latest data to investigate the association of the IL-23R gene polymorphisms and the risk of CD.
Materials and Methods
Publication research
A literature search was conducted for studies that examined association between the IL-23R gene polymorphisms and CD risk in October 13, 2015. We used PubMed database, Elsevier Science Direct, China National Knowledge Infrastructure database (CNKI) and Chinese Biomedical database (CBM) to identify articles with the following terms: “Interleukin-23 receptor, IL-23R, Crohn’s disease, Crohn disease, inflammatory bowel disease, CD and IBD”. References in the studies were also reviewed to find additional studies. The languages were limited to English and Chinese.
Inclusion and exclusion criteria
A study was included in the analysis if a) it was a case-control or cohort study; b) it included original data (independence among studies); and c) it provided sufficient data to calculate the odds ratio (OR). We excluded the followings: a) studies contained overlapping data; b) studies in which family members had been studied because of the analysis based on linkage considerations.
Data extraction
Data were collected by two independent investigators (Qi-Bing Xie and Yi Zhao). The characteristics of the selected articles were in Table 1, including first author, year of publication, study population, ethnicity, numbers of case and control, genotyping method, the concrete polymorphisms investigated in these studies. The study populations comprised Italian, German, Dutch, Hungarian, Korean, Chinese, Malaysian, African American, Australian, Canadian, Algerian and so on. The Asian subgroup included Korean, Chinese, and Malaysian populations. African American population was classified in the African subgroup and others in the Caucasian subgroup.
Table 1. Characteristics of the individual studies included in the meta-analysis.
| First Author | Year | Population (Ethnicity) | Numbers | Genotyping Methods | IL-23R polymorphisms studied |
|---|---|---|---|---|---|
| CD/Control | |||||
| Duerr21 | 2006 | Non-Jewish (C) | 547/548 | Sequencing | rs1004819, rs7517847, rs10489629, rs2201841, rs11465804, |
| Jewish (C) | 401/433 | Sequencing | rs11209026, rs1343151, rs10889677, rs11209032, rs1495965 | ||
| Baldassano22 | 2007 | American (C) | 142/281 | Sequencing | rs1004819, rs7517847, rs10489629, rs2201841, rs11465804, rs11209026, rs1343151, rs10889677, rs11209032, rs1495965 |
| Borgiani23 | 2007 | Italian (C) | 187/194 | TaqMan | rs11209026 |
| Bunning24 | 2007 | German (C) | 318/428 | PCR | rs11209026 |
| Hungarian (C) | 148/200 | PCR | rs11209026 | ||
| Dutch (C) | 157/217 | PCR | rs11209026 | ||
| Cummings25 | 2007 | British (C) | 604/1149 | Sequencing | rs1004819, rs7517847, rs10489629, rs2201841, rs11209026, rs1343151, rs11209032, rs1495965 |
| Farago26 | 2007 | Hungarian (C) | 190/220 | PCR-RFLP | rs10889677, rs2201841 |
| Glas27 | 2007 | German (C) | 833/1381 | PCR | rs1004819, rs7517847, rs10489629, rs2201841, rs11465804, rs11209026, rs1343151, rs10889677, rs11209032, rs1495965 |
| Leshinsky-Silver28 | 2007 | Israeli (C) | 282/157 | PCR | rs11209026 |
| Limbergen29 | 2007 | Scotchman (C) | 233/342 | TaqMan | rs11209026 |
| Oliver30 | 2007 | Spanish (C) | 238/342 | TaqMan | rs1004819, rs7517847, rs10489629, rs11209026, rs1343151, rs10889677, rs11209032, rs1495965 |
| Rioux31 | 2007 | American (C) | 988/1007 | Sequencing | rs7517847 |
| Roberts32 | 2007 | New Zealander (C) | 496/591 | TaqMan | rs11209026 |
| Tremelling33 | 2007 | British (C) | 1902/1345 | Sequencing | rs1004819, rs10489629, rs11465804, rs11209026, rs1343151, rs10889677, rs11209032, rs1495965 |
| Yamazaki34 | 2007 | Japanese (A) | 484/439 | TaqMan PCR-RFLP | rs1004819, rs7517847, rs10489629, rs2201841, rs11465804, rs11209026, rs1343151, rs10889677, rs11209032, rs1495965 |
| Amre35 | 2008 | Canadian (C) | 259/139 | FP-SBE | rs1004819, rs11209026, rs7517847, rs10489629, rs2201841, rs11465804, rs1343151, rs10889677, rs11209032, rs1495965 |
| Baptista36 | 2008 | Brazilian (C) | 187/255 | TaqMan | rs1004819, rs7517847, rs11209026, rs1495965, rs10889677 |
| Civitavecchia37 | 2008 | Italian (C) | 199/100 | Sequencing | rs11209026 |
| Gaj38 | 2008 | Polish (C) | 60/139 | TaqMan | rs11209026 |
| Lakatos39 | 2008 | Hungarian (C) | 266/149 | PCR | rs11209026 |
| Lappalainen40 | 2008 | Finnish (C) | 238/292 | PCR | rs1004819, rs10489629, rs2201841, rs11465804, rs11209026, rs1343151, rs10889677, rs11209032 |
| Latiano41 | 2008 | Italian (C) | 723/716 | TaqMan | rs11209026, rs7517847 |
| Lu74 | 2008 | Chinese (A) | 74/100 | PCR | rs11209026 |
| Marquez42 | 2008 | Spanish (C) | 344/547 | TaqMan | rs11209026, rs7517847 |
| Okazaki43 | 2008 | Canadian (C) | 213/310 | TaqMan | rs7517847, rs2201841, rs11209026, rs10889677, rs1495965 |
| Venegas44 | 2008 | Chilean (C) | 38/58 | PCR-RFLP | rs11209026 |
| Weersma45 | 2008 | Dutch (C) | 1684/1350 | TaqMan | rs11209026 |
| Chen71 | 2008 | Chinese (A) | 41/50 | PCR | rs11209026 |
| Dusatkova46 | 2009 | Czech (C) | 333/499 | TaqMan | rs11209026 |
| Chen72 | 2009 | Chinese (A) | 50/50 | Sequencing | rs10889677 |
| Feng73 | 2009 | Chinese (A) | 96/96 | Sequencing | rs1004819, rs7517847, rs2201841 |
| Lacher47 | 2009 | German (C) | 221/253 | TaqMan | rs11209026, rs7517847 |
| Mitrovi48 | 2009 | Slovenian (C) | 159/345 | PCR-RFLP | rs7517847 |
| Newman49 | 2009 | Canadian (C) | 443/1005 | Sequencing | rs11209026, rs7517847 |
| Weersma50 | 2009 | Dutch-Belgian (C) | 1656/1086 | Sequencing | rs11209026 |
| Yang51 | 2009 | Korean (A) | 380/380 | Sequencing | rs1004819, rs2201841, rs11209026, rs10889677, rs1495965 |
| Cotterill52 | 2009 | British (C) | 295/877 | Sequencing | rs11209026 |
| Csöngei53 | 2010 | Hungarian (C) | 315/314 | PCR-RFLP | rs1004819, rs2201841 |
| Ferguson54 | 2010 | New Zealander (C) | 339/407 | TaqMan | rs10889677, rs11209026, rs1343151, rs7517847 |
| Gazouli55 | 2010 | Greek (C) | 474/539 | PCR | rs11209026 |
| Mahurkar56 | 2010 | Indian (C) | 241/442 | Sequencing | rs11209026 |
| Sventoraityte57 | 2010 | Lithuanian (C) | 57/186 | TaqMan | rs11209026 |
| Wagner58 | 2010 | Australian (C) | 72/98 | PCR | rs11209026 |
| Lauriola59 | 2010 | Italian (C) | 19/20 | PCR | rs11209026, rs7517847 |
| Peter60 | 2011 | Ashkenazi (C) | 369/503 | TaqMan | rs11209026, rs7517847 |
| Zhao75 | 2011 | Chinese (A) | 43/134 | SNaPshot | rs1343151, rs11209032 |
| Chua61 | 2012 | Malaysian (A) | 80/100 | PCR-RFLP | rs1004819 |
| Jung62 | 2012 | Franch (C) | 798/960 | Sequencing | rs11209026 |
| Safrany63 | 2012 | Hungarian (C) | 199/253 | PCR-RFLP | rs7517847, rs2201841, rs10889677, rs11209032 |
| Szabo64 | 2012 | Hungarian (C) | 190/182 | PCR-RFLP | rs1004819, rs7517847, rs2201841, rs1343151, rs10889677 |
| Wang65 | 2012 | African American | 354/354 | TaqMan | rs7517847, rs2201841, rs11209026, rs1496965, rs10889677 |
| Mihaljevi66 | 2013 | Croatian (C) | 50/99 | PCR | rs11209026 |
| Ballester67 | 2013 | Puerto Rican (C) | 406/504 | Sequencing | rs11209026 |
| Doecke68 | 2014 | Australian (C) | 675/1255 | Sequencing | rs1004819, rs7517847, rs10489629, rs2201841, rs11465804, rs11209026, rs1343151, rs10889677, rs11209032, rs1495965 |
| New Zealander (C) | 318/533 | Sequencing | rs1004819, rs7517847, rs10489629, rs2201841, rs11209026, rs1343151 | ||
| Meddour69 | 2014 | Algerian (C) | 204/201 | TaqMan | rs11209026 |
| Zhang70 | 2014 | Chinese (A) | 420/450 | TaqMan | rs11209026, rs1004819, rs1495965 |
| Huang76 | 2015 | African American | 1088/1797 | Immunochip | rs11209026 |
CD: Crohn’s disease, C: Caucasian, A: Asian.
Statistical analysis
Allele frequencies at the IL-23R gene polymorphisms from the respective study were determined by the allele counting method. The contrast of the allelic effect of minor allele versus common allele of the IL-23R gene polymorphisms was tested. The strength of association between the IL-23R gene polymorphisms and CD susceptibility was assessed by odds ratios (OR) and 95% confidence intervals (95% CI).
The heterogeneity of between-studies was evaluated by the Chi-square test based Q-statistic79. In addition, the I2-statistic also measures the degree of inconsistency in the studies by computing what percentage of the total variation across studies was due to heterogeneity rather than by chance. A high value of I2 indicated a higher probability of the existence of heterogeneity (I2 = 0% to 25%, no heterogeneity; I2 = 25% to 50%, moderate heterogeneity; I2 = 50% to 75%, large heterogeneity; and I2 = 75% to 100%, extreme heterogeneity). If a P-value of the heterogeneity Q-statistic was more than 0.10, the pooled OR was calculated by the fixed effects model. Otherwise, a random effects model was adopted. Potential publication bias was analyzed by Egger’s linear regression test and the Funnel plot. If P-value was less than 0.05, statistically significant publication bias might exist80.
All the statistical analysis of meta-analysis was performed by STATA statistical software (version 11.0 STATA Corp LP, College Station, TX, USA).
Results
Study characteristics
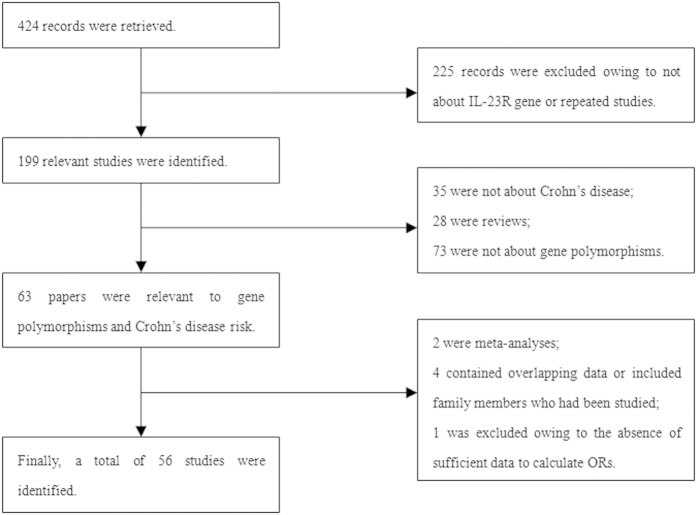
The process for selecting the studies was shown in Fig. 1. 424 potentially relevant records were first reviewed, and 56 articles met the inclusion criteria were finally included in themeta-analysis21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76. Among the 56 articles, 4 were published in Chinese and the others in English. Of the 56 articles, one article included three cohorts24, other two21,68 included two cohorts, thus, each cohort was considered as a single study. Finally, a total of 60 case-control studies in 56 articles involving 22,820 CD patients and 27,401 healthy controls were identified. There were 49 studies on rs11209026, 25 studies on rs7517847, 19 studies on rs2201841 and rs1004819 separately, 18 studies on rs10889677, 15 studies on rs1343151 and rs1495965 separately, 13 studies on rs11209032, 12 studies on rs10489629 and 8 on rs11465804. Forty-nine studies involved Caucasian populations21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,66,67,68,69, nine studies involved Asian populations34,51,61,70,71,72,73,74,75, and two study involved African populations65,76. The main characteristics of each study included in this meta-analysis were summarized in Table 1.
Figure 1. Process of selecting studies.
Evaluation of heterogeneity and publication bias
Between-study heterogeneity was found in some meta-analysis of the IL-23R gene polymorphisms in CD (Table 2). Therefore, these meta-analyses were performed in a random effects model, and the other meta-analyses were done in a fixed effects model.
Table 2. Meta-analysis of IL-23R polymorphisms in CD.
| Polymorphisms | Population | No. of studies | Test of association |
Test of heterogeneity |
Egger’s test (p) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | Z | p | Model | χ2 | p | I2(%) | ||||
| rs11209026 A versus G | Overall | 49 | 0.407(0.378 ~ 0.439) | 23.32 | <0.001 | R | 75.51 | 0.007 | 36.4 | 0.420 |
| Caucasian | 44 | 0.406(0.365 ~ 0.452) | 16.50 | <0.001 | R | 67.45 | 0.010 | 36.2 | 0.606 | |
| Asian | 3 | 0.659(0.409 ~ 1.063) | 1.71 | 0.087 | F | 0.47 | 0.791 | 0 | 0.059 | |
| African | 2 | 0.584(0.375 ~ 0.911) | 2.37 | 0.018 | F | 1.00 | 0.318 | 0 | NA | |
| rs7517847 G versus T | Overall | 25 | 0.699(0.659 ~ 0.741) | 11.93 | <0.001 | R | 46.18 | 0.004 | 48.0 | 0.303 |
| Caucasian | 22 | 0.669(0.641 ~ 0.698) | 18.68 | <0.001 | F | 26.22 | 0.198 | 19.9 | 0.545 | |
| Asian | 2 | 0.896(0.773 ~ 1.039) | 1.46 | 0.145 | F | 0.42 | 0.517 | 0 | NA | |
| African | 1 | 0.940(0.726 ~ 1.216) | 0.47 | 0.637 | NA | NA | NA | NA | NA | |
| rs10889677 A versus C | Overall | 18 | 1.393(1.328 ~ 1.461) | 13.65 | <0.001 | F | 21.98 | 0.185 | 22.7 | 0.165 |
| Caucasian | 15 | 1.438(1.366 ~ 1.513) | 13.96 | <0.001 | F | 9.32 | 0.810 | 0 | 0.196 | |
| Asian | 2 | 1.098(0.945 ~ 1.276) | 1.22 | 0.222 | F | 0.75 | 0.385 | 0 | NA | |
| African | 1 | 1.220(0.916 ~ 1.626) | 1.36 | 0.174 | NA | NA | NA | NA | NA | |
| rs1004819 T versus C | Overall | 19 | 1.352(1.264 ~ 1.446) | 8.82 | <0.001 | R | 35.77 | 0.008 | 49.7 | 0.158 |
| Caucasian | 14 | 1.441(1.373 ~ 1.514) | 14.68 | <0.001 | F | 8.73 | 0.793 | 0 | 0.943 | |
| Asian | 5 | 1.102(0.994 ~ 1.222) | 1.84 | 0.066 | F | 5.86 | 0.210 | 31.8 | 0.242 | |
| rs2201840 C versus T | Overall | 18 | 1.368(1.301 ~ 1.438) | 12.33 | <0.001 | F | 19.28 | 0.313 | 11.8 | 0.342 |
| Caucasian | 14 | 1.413(1.338 ~ 1.491) | 12.53 | <0.001 | F | 7.56 | 0.871 | 0 | 0.617 | |
| Asian | 3 | 1.088(0.943 ~ 1.255) | 1.15 | 0.250 | F | 0.53 | 0.769 | 0 | 0.996 | |
| African | 1 | 1.392(1.042 ~ 1.858) | 2.24 | 0.025 | NA | NA | NA | NA | NA | |
| rs1343151 T versus C | Overall | 15 | 0.725(0.690 ~ 0.763) | 12.49 | <0.001 | F | 19.03 | 0.164 | 26.4 | 0.576 |
| Caucasian | 13 | 0.717(0.681 ~ 0.755) | 12.73 | <0.001 | F | 11.97 | 0.448 | 0 | 0.842 | |
| Asian | 2 | 1.069(0.790 ~ 1.444) | 0.43 | 0.667 | F | 0.47 | 0.494 | 0 | NA | |
| rs1495965 G versus A | Overall | 15 | 1.197(1.103 ~ 1.299) | 4.29 | <0.001 | R | 43.42 | <0.001 | 67.8 | 0.129 |
| Caucasian | 12 | 1.218(1.111 ~ 1.335) | 4.20 | <0.001 | R | 34.76 | <0.001 | 68.4 | 0.180 | |
| Asian | 3 | 1.119(0.945 ~ 1.324) | 1.31 | 0.191 | R | 4.75 | 0.093 | 57.9 | 0.354 | |
| rs11209032 A versus G | Overall | 13 | 1.343(1.279 ~ 1.411) | 11.73 | <0.001 | F | 11.51 | 0.485 | 0 | 0.182 |
| Caucasian | 11 | 1.359(1.291 ~ 1.431) | 11.68 | <0.001 | F | 9.16 | 0.517 | 0 | 0.308 | |
| Asian | 2 | 1.181(0.995 ~ 1.403) | 1.90 | 0.057 | F | 0.02 | 0.885 | 0 | NA | |
| rs10489629 G versus A | Overall | 12 | 0.791(0.706 ~ 0.887) | 4.02 | <0.001 | R | 56.15 | <0.001 | 80.4 | 0.220 |
| Caucasian | 11 | 0.775(0.690 ~ 0.869) | 4.33 | <0.001 | R | 48.83 | <0.001 | 79.5 | 0.310 | |
| Asian | 1 | 0.998(0.815 ~ 1.222) | 0.02 | 0.986 | NA | NA | NA | NA | NA | |
| rs11465804 G versus T | Overall | 8 | 0.435(0.376 ~ 0.503) | 11.18 | <0.001 | F | 6.29 | 0.506 | 0 | 0.082 |
| Caucasian | 8 | 0.435(0.376 ~ 0.503) | 11.18 | <0.001 | F | 6.29 | 0.506 | 0 | 0.082 | |
CD: Crohn’s disease, R: random effects model, F: fixed effects model, NA: not available.
The funnel plot asymmetry and Egger’s test were performed to assess potential publication bias. If there was asymmetry, the index to determine Egger’s test will show P < 0.05. It was shown that there was no significant publication bias between all of the comparisons (all P-value > 0.05).
Meta-analysis of IL-23R gene polymorphisms in Crohn’s disease
A summary of the meta-analysis of the association between the IL-23R gene polymorphisms and CD was listed in Table 2.
IL-23R rs7517847, rs1343151, rs10489629 polymorphisms and CD
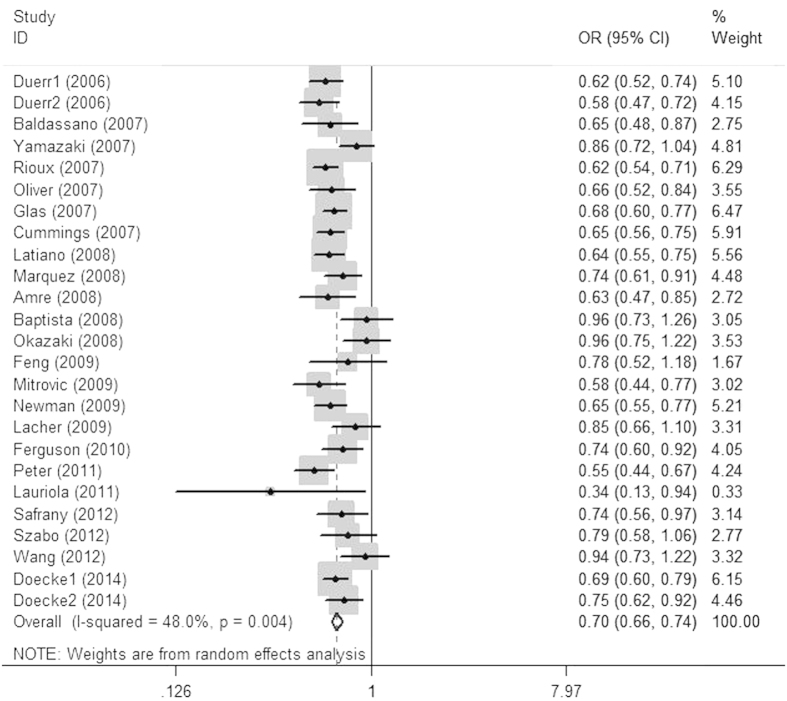
Twenty-five studies determined the relationship between the rs7517848G/T polymorphism and CD risk21,22,25,27,30,31,34,35,36,41,42,43,47,48,49,54,59,60,63,64,65,68,73. The total sample size for patients with CD and healthy controls was 9,297 and 12,643, respectively. Meta-analysis revealed an association between the rs7517847G allele and CD risk in the overall population (OR = 0.699, 95% CI = 0.659 ~ 0.741, P < 0.001; Fig. 2). Stratification by ethnicity indicated that the rs7517847G allele was significantly associated with CD risk in the Caucasian population (OR = 0.669, 95% CI = 0.641 ~ 0.698, P < 0.001), but not in the other subgroups. Meta-analysis of the rs1343151 and rs10489629 polymorphisms showed the same pattern as for rs7517847. Minor alleles of all polymorphisms might be protective alleles for CD susceptibility in Caucasians, but not in the other populations (Table 2).
Figure 2. Odds ratios and 95% confidence intervals for individual studies and pooled data for the association between the G versus T allele of the IL-23R rs7517847 polymorphism and Crohn’s disease.
IL-23R rs10889677, rs1004819, rs1495965, rs11209032 polymorphisms and CD
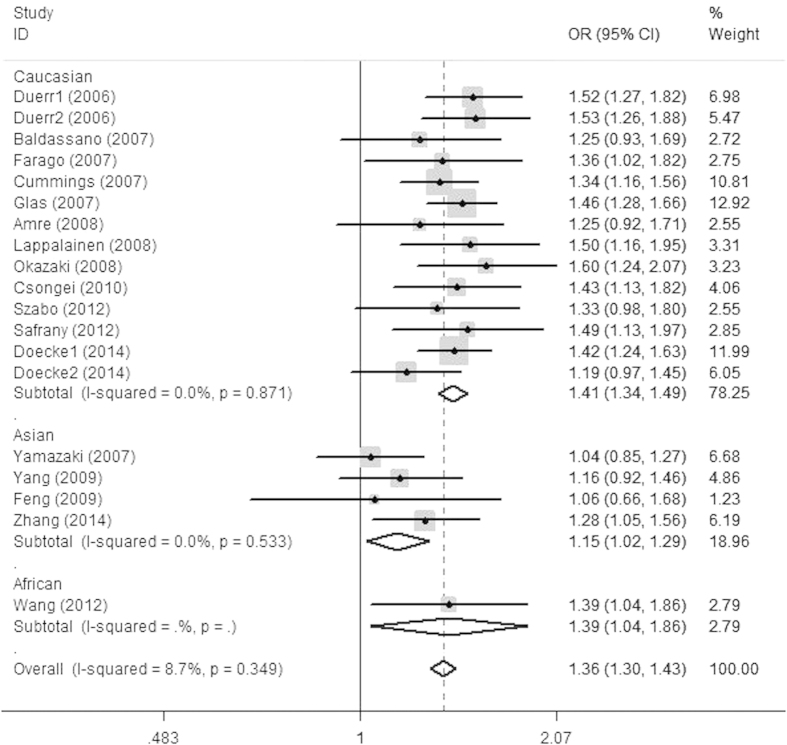
Eighteen studies containing 7,518 cases and 8,671 healthy controls examined the association of rs10889677A/C and CD21,22,26,27,30,33,34,36,40,43,51,54,63,64,65,68,72. Results indicated a significant association between the rs100889677A/C polymorphism and CD (OR = 1.393, 95% CI = 1.328 ~ 1.461, P < 0.001; Fig. 3). Stratifying by ethnicity, we found a significant association in the Caucasian population (OR = 1.438, 95% CI = 1.366 ~ 1.513, P < 0.001; Fig. 2). Results by meta-analysis of the rs1004819, rs1495965 and rs11209032 polymorphisms revealed the same pattern as for rs10889677. Minor alleles of the SNPs above might be risk alleles for CD in Caucasian subgroup. Detail results were presented in Table 2.
Figure 3. Odds ratios and 95% confidence intervals for individual studies and pooled data for the association between the A versus C allele of the IL-23R rs10889677 polymorphism and Crohn’s disease.
IL-23R rs2201841, rs11209026, rs11465804 polymorphisms and CD
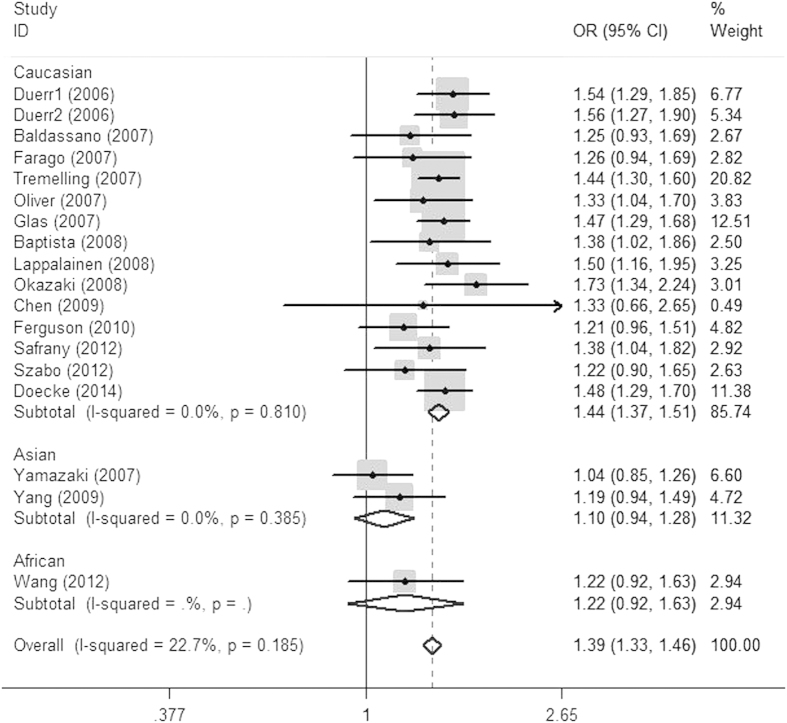
Eighteen case-control studies including 6,846 cases and 9,056 healthy controls identified a significant association between the rs2201841C/T polymorphism and susceptibility to CD21,22,25,26,27,34,35,40,43,51,53,63,64,65,68,73. The pooled OR (95% CI, P value) in the C versus T allele was 1.368 (1.301 ~ 1.438, P < 0.001). Ethnicity-specific analysis showed that the rs2201841C allele was significantly associated with CD in the Caucasian and African subjects (OR = 1.413, 95% CI = 1.338 ~ 1.491, P < 0.001; OR = 1.392, 95% CI = 1.042 ~ 1.858, P = 0.025, respectively). The forest plot was shown in Fig. 4. Meta-analysis of the rs11209026 polymorphism showed the same pattern as for rs2201841. In addition, a significant association was found between the rs11465804G allele and the risk of CD in Caucasians (OR = 0.435, 95% CI = 0.376 ~ 0.503, P < 0.001) (Table 2). However, Asians and Africans-based subgroup analysis were not performed.
Figure 4. Odds ratios and 95% confidence intervals for individual studies and pooled data for the association between the C versus T allele of the IL-23R rs2201841 polymorphism and Crohn’s disease.
Discussion
A genome-wide association study by Duerr et al.21 found strong correlation between IL-23R polymorphisms and inflammatory bowel disease (IBD). Since then, genetic variants of IL-23R have been investigated in numerous autoimmune diseases. It has been shown that the IL-23R gene plays an important role in the pathogenesis of some autoimmune diseases, such as IBD, AS, psoriasis, rheumatoid arthritis, and multiple sclerosis81. Oliver et al.30 and Baptista et al.36 found rs11209026, rs10889677 and rs1004819 polymorphisms were associated with CD disease susceptibility in Spanish. Similarly, a study on Hungarian with CD revealed that the rs1004819T carrier (TT + TC) had a 53% increasing risk when compared with the CC homozygote53. Furthermore, a study in Korean population indicated that the rs1495965G allele frequency was higher in CD cases compared to controls (OR = 1.31, 95% CI = 1.07 ~ 1.60)51. However, Okazaki et al.43 and Chua et al.61 found no significant differences in allele or genotype frequency of rs11209026 and rs1004819 polymorphisms between CD patients and controls in a Malaysian population.
Therefore, to better comprehend the association between polymorphisms in the IL-23R gene and CD susceptibility, a pooled analysis with a larger sample size, subgroup analysis performed and heterogeneity explored is needed. Overall, results of this meta-analysis suggested that IL-23R gene polymorphisms were associated with CD susceptibility (all P value < 0.001). Ethnicity-specific analysis showed that the polymorphisms in the IL-23R gene might confer susceptibility to CD in Caucasians, but not in Asians.
The diverse roles of the same gene polymorphism in ethnicity-specific analysis by ethnicity could be ascribed to the following major aspects. First, Yamazaki et al.34 showed that IL-23R gene was not a candidate gene to CD in the Japanese population. Furthermore, Lee et al.82 found that rs1004819, rs10489629, rs1343151, rs1495965, rs11209032 and rs2201841 polymorphisms revealed a different association with ankylosing spondylitis between Caucasians and Asians. Compared to the populations of Caucasian, no significant association was detected for IL-23R polymorphisms in Chinese psoriasis patients83. Similarly, IL-23R (rs1004819, rs7517847, rs10489629, rs2201841, rs1343151, rs11209032, and rs1495965) polymorphisms were not associated with systemic lupus erythematosus (SLE) in the Korean population84, while rs10889677 and rs7517847 polymorphisms were not associated with Chinese SLE patients85. In addition, CD is the most prevalent in North America and Europe, and the least prevalent among African Americans and Asians86. These findings suggested that IL-23R gene might have a susceptible nature in the Caucasians, but not in the Asians. Therefore, the inconsistent results of subgroup analysis might be attributed to ethnic differences. Second, for other variants in the IL-23R gene, Zhao et al.75 found that IL-23R rs11805303 and rs17375018 polymorphisms were not associated with CD disease susceptibility in Chinese. Similarly, a study in another Chinese population showed no significant differences in allele or genotype frequency of the rs11805303 polymorphism between CD patients and controls71. However, Chen et al.72 found that the rs11465788T allele frequency was lower in Chinese CD cases compared to controls (OR = 0.30, 95%CI = 0.15–0.60). Since few studies in Asian population focus on these polymorphisms, these results might not be reliable. Therefore, large sample size GWAS should be considered in future to confirm the role of these variants in the IL-23R gene in Asians. Third, most autoimmune diseases including CD are multifactor diseases, and these diseases were caused by an interaction of genetic and environmental factors87. Therefore, different populations lived in different environments, gene-environment interactions may partly affect the autoimmune diseases susceptibility. It has been widely accepted that genetic and environmental factors play an important role in disease initiation such as SLE, and the progression of the disease. A previous study has confirmed that the IFIH1 rs1990760 polymorphism was associated with SLE in the European population, not in the Asian population88, suggesting that interactions between different environments and genes might be different.
Although the IL-23R gene is associated with CD risk, the function of IL-23R gene variants remains unclear. Several possible mechanisms can be suggested by which polymorphisms can regulate the function of the IL-23R gene. First, the standard form of IL-23R is encoded by at least 12 exons. Zhang et al.89 demonstrated that at least six spliced isoforms of IL-23R (IL-23R1 to 6) can be generated through alternative splicing. Translation prediction revealed that spliced variants led to either premature termination to give rise to a diverse form of receptor ectodomain, or a frameshift to generate various lengths of the IL-23R endodomain89. Furthermore, the intronic polymorphisms, such as rs11805303, rs1004819, might exert their influence by regulating differential splicing. Second, the rs11209026 polymorphism, also named as Arg381Gln, is located between the transmembrane domain and the putative JAK2 binding site in the cytoplasmic portion of IL23R protein and highly conserved between species90. Replacing conserved Arg381 for Gln381 at this position probably modulates IL-17 and IL-22 expression in response to IL-23 stimulation, which may have a functional influence on IL-23R signaling pathway91,92. Third, the rs10889677 polymorphism, located in the 3′-UTR, might promote overexpression of the receptor through increasing mRNA stability and driving T cells to differentiate towards Th17, and thus, leading to inflammation by increasing release of other cytokines81. Forth, the rs10889677 variant enhanced both mRNA and protein expression of IL-23R can lead to a loss of binding capacity to the microRNAs (miRNAs) Let-7e and Let-7f. MiRNA-mediated dysregulation of IL-23R signaling correlated with a single nucleotide polymorphism in the IL-23R gene was strongly associated with CD susceptibility, suggesting that this mutation can lead to altered IL-23R signaling93.
With respect to the IL-23R haplotypes and CD, Taylor et al.94 showed that haplotype TA (rs1004819 (T) + rs790631 (A)) was 55.4% in the controls, and 64% in CD patients (P = 0.019), indicating a haplotype inferring increased CD risk. Similarly, haplotype CACCGC (rs7530511 (C) + rs7528924 (A) + rs2201841 (C) + rs10489628 (C) + rs11209026 (G) + rs1343151 (C)) was 55.8% in the controls, and 64.4% in CD patients (P = 0.013), indicating a “risk” haplotype. On the contrary, haplotype GA (rs1004819 (G) + rs790631 (A)) was 64.5% in the control group, and 54.4% in CD patients (P = 0.006), suggesting a haplotype inferring decreased CD risk. Similarly, haplotype CATTGT (rs7530511 (C) + rs7528924 (A) + rs2201841 (T) + rs10489628 (T) + rs11209026 (G)+ rs1343151 (T)) was 47.0% in the controls, and 36.6% in CD patients (P = 0.001), suggesting a “protective” haplotype. In addition, Szabo et al.64 found that haplotype GTTTAC (rs1004819 (G) + rs7517847 (T) + rs7530511 (T) + rs2201841 (T) + rs1343151 (A) + rs10889677 (C)) showed a strong protective effect in CD, and it reduced disease risk (OR = 0.37, 95% CI = 0.17 ~ 0.78, P = 0.009). Furthermore, Doecke et al.68 reported that haplotype GG (rs1004819 (G) + rs7517847 (G)) was negatively related to CD risk (OR = 0.71, 95% CI = 0.62 ~ 0.81, P < 0.001). However, haplotype AT (rs1004819 (G) + rs7517847 (G)) was positively related to CD risk (OR = 1.45, 95% CI = 1.26 ~ 1.67, P < 0.001).
Compared with the previous meta-analysis52,78, the current study involved a total of 60 case-control studies in 56 articles, which is much larger than the data of the previous meta-analysis. Moreover, we performed subgroup analysis by ethnicity to discuss the ethnic effect on the risk of CD. Thus, our meta-analysis might enhance the statistical power and draw a more reliable conclusion.
Some limitations of the present study should be considered. First, we could not analyze the potential gene-environment interactions and gene susceptibility haplotypes owing to lack of data, such as the data of environmental risk factors and genotypes. Second, our literature search was only dependent on English and Chinese, language bias might be considered. Third, potential publication bias was not found by statistical method, but it might exist because of only published articles included. Fourth, only one published studies in the African origin was included in the meta-analysis, the stratified analysis for Africans might not be reliable. Thus, the results were applicable only to the Asian and Caucasian groups. Finally, different genotyping methods and disease status might affect the data interpretation of the included studies.
In summary, the current study provides a comprehensive examination of the available evidence for the association between polymorphisms in the IL-23R gene and CD. This updated meta-analysis suggests that IL-23R gene polymorphisms are associated with CD susceptibility. However, larger sample size studies taking environmental risk factors into account and including more ethnic groups should be considered in future to confirm the results from our meta-analysis.
Additional Information
How to cite this article: Xu, W.-D. et al. Association of Interleukin-23 receptor gene polymorphisms with susceptibility to Crohn,s disease: A meta-analysis. Sci. Rep. 5, 18584; doi: 10.1038/srep18584 (2015).
Footnotes
References
- Podolsky D. K. Inflammatory bowel disease. N Engl J Med 347, 417–429 (2002). [DOI] [PubMed] [Google Scholar]
- Satsangi J., Jewell D. P. & Bell J. I. The genetics of inflammatory bowel disease. Gut 40, 572–574 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerr R. H. Update on the genetics of inflammatory bowel disease. J Clin Gastroenterol 37, 358–367 (2003). [DOI] [PubMed] [Google Scholar]
- Ogura Y. et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature 411, 603–606 (2001). [DOI] [PubMed] [Google Scholar]
- Marks D. J. et al. Defective acute inflammation in Crohn’s disease: a clinical investigation. Lancet 367, 668–678 (2006). [DOI] [PubMed] [Google Scholar]
- Hampe J. et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet 39, 207–211 (2007). [DOI] [PubMed] [Google Scholar]
- Parkes M. et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat Genet 39, 830–832 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A. et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet 42, 1118–1125 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J. H. & Brant S. R. Recent insights into the genetics of inflammatory bowel disease. Gastroenterology 140, 1704–1712 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny E. E. et al. A genome-wide scan of Ashkenazi Jewish Crohn’s disease suggests novel susceptibility loci. PLoS Genet 8, e1002559 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy C. A. et al. Divergent pro- and antiinflammatory roles for IL-23 and IL-12 in joint autoimmune inflammation. J Exp Med 198, 1951–1957 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parham C. et al. A receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rbeta1 and a novel cytokine receptor subunit, IL-23R. J Immunol 168, 5699–5708 (2002). [DOI] [PubMed] [Google Scholar]
- Cho M. L. et al. STAT3 and NF-kappaB signal pathway is required for IL-23-mediated IL-17 production in spontaneous arthritis animal model IL-1 receptor antagonist-deficient mice. J Immunol 176, 5652–5661 (2006). [DOI] [PubMed] [Google Scholar]
- Kenna T. J. et al. Enrichment of circulating interleukin-17-secreting interleukin-23 receptor-positive γ/δ T cells in patients with active ankylosing spondylitis. Arthritis Rheum 64, 1420–1429 (2012). [DOI] [PubMed] [Google Scholar]
- Petermann F. et al. γ/δ T cells enhance autoimmunity by restraining regulatory T cell responses via an interleukin-23-dependent mechanism. Immunity 33, 351–363 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier R. J. & Podolsky D. K. Unravelling the pathogenesis of inflammatory bowel disease. Nature 448, 427–434 (2007). [DOI] [PubMed] [Google Scholar]
- Yen D. et al. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J Clin Invest 116, 1310–1316 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izcue A. et al. Interleukin-23 restrains regulatory T cell activity to drive T cell-dependent colitis. Immunity 28, 559–570 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuss I. J. et al. Both IL-12p70 and IL-23 are synthesized during active Crohn’s disease and are down-regulated by treatment with anti-IL-12 p40 monoclonal antibody. Inflamm Bowel Dis 12, 9–15 (2006). [DOI] [PubMed] [Google Scholar]
- Duerr R. H. et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 314, 1461–1463 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldassano R. N. et al. Association of variants of the interleukin-23 receptor gene with susceptibility to pediatric Crohn’s disease. Clin Gastroenterol Hepatol 5, 972–976 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgiani P. et al. Interleukin-23R Arg381Gln is associated with susceptibility to Crohn’s disease but not with phenotype in an Italian population. Gastroenterology 133, 1049–51; author reply 1051-2 (2007). [DOI] [PubMed] [Google Scholar]
- Büning C. et al. Heterozygosity for IL23R p.Arg381Gln confers a protective effect not only against Crohn’s disease but also ulcerative colitis. Aliment Pharmacol Ther 26, 1025–1033 (2007). [DOI] [PubMed] [Google Scholar]
- Cummings J. R. et al. Contribution of the novel inflammatory bowel disease gene IL23R to disease susceptibility and phenotype. Inflamm Bowel Dis 13, 1063–1068 (2007). [DOI] [PubMed] [Google Scholar]
- Faragó B. et al. Functional variants of interleukin-23 receptor gene confer risk for rheumatoid arthritis but not for systemic sclerosis. Ann Rheum Dis 67, 248–250 (2008). [DOI] [PubMed] [Google Scholar]
- Glas J. et al. rs1004819 is the main disease-associated IL23R variant in German Crohn’s disease patients: combined analysis of IL23R, CARD15, and OCTN1/2 variants. PLoS One 2, e819 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshinsky-Silver E. et al. Evaluation of the interleukin-23 receptor gene coding variant R381Q in pediatric and adult Crohn disease. J Pediatr Gastroenterol Nutr 45, 405–408 (2007). [DOI] [PubMed] [Google Scholar]
- Van Limbergen J. et al. IL23R Arg381Gln is associated with childhood onset inflammatory bowel disease in Scotland. Gut 56, 1173–1174 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver J., Rueda B., López-Nevot M. A., Gómez-García M. & Martín J. Replication of an association between IL23R gene polymorphism with inflammatory bowel disease. Clin Gastroenterol Hepatol 5, 977–981, 981.e1-2 (2007). [DOI] [PubMed] [Google Scholar]
- Rioux J. D. et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet 39, 596–604 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. L. et al. IL23R R381Q and ATG16L1 T300A are strongly associated with Crohn’s disease in a study of New Zealand Caucasians with inflammatory bowel disease. Am J Gastroenterol 102, 2754–2761 (2007). [DOI] [PubMed] [Google Scholar]
- Tremelling M. et al. IL23R variation determines susceptibility but not disease phenotype in inflammatory bowel disease. Gastroenterology 132, 1657–1664 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki K. et al. Association analysis of genetic variants in IL23R, ATG16L1 and 5p13.1 loci with Crohn’s disease in Japanese patients. J Hum Genet 52, 575–583 (2007). [DOI] [PubMed] [Google Scholar]
- Amre D. K. et al. Association between genetic variants in the IL-23R gene and early-onset Crohn’s disease: results from a case-control and family-based study among Canadian children. Am J Gastroenterol 103, 615–620 (2008). [DOI] [PubMed] [Google Scholar]
- Baptista M. L. et al. CARD15 and IL23R influences Crohn’s disease susceptibility but not disease phenotype in a Brazilian population. Inflamm Bowel Dis 14, 674–679 (2008). [DOI] [PubMed] [Google Scholar]
- Civitavecchia G. et al. IL-23R determines susceptibility in Crohn’s disease in a Mediterranean area. Inflamm Bowel Dis 15, 317–318 (2009). [DOI] [PubMed] [Google Scholar]
- Gaj P., Habior A., Mikula M. & Ostrowski J. Lack of evidence for association of primary sclerosing cholangitis and primary biliary cirrhosis with risk alleles for Crohn’s disease in Polish patients. BMC Med Genet 9, 81(2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakatos P. L. et al. ATG16L1 and IL23 receptor (IL23R) genes are associated with disease susceptibility in Hungarian CD patients. Dig Liver Dis 40, 867–873 (2008). [DOI] [PubMed] [Google Scholar]
- Lappalainen M., et al. Association of IL23R, TNFRSF1A, and HLA-DRB1*0103 allele variants with inflammatory bowel disease phenotypes in the Finnish population. Inflamm Bowel Dis 14, 1118–1124 (2008). [DOI] [PubMed] [Google Scholar]
- Latiano A. et al. Replication of interleukin 23 receptor and autophagy-related 16-like 1 association in adult- and pediatric-onset inflammatory bowel disease in Italy. World J Gastroenterol 14, 4643–4651 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Márquez A. et al. IL23R and IL12B polymorphisms in Spanish IBD patients: no evidence of interaction. Inflamm Bowel Dis 14, 1192–1196 (2008). [DOI] [PubMed] [Google Scholar]
- Okazaki T. et al. Contributions of IBD5, IL23R, ATG16L1, and NOD2 to Crohn’s disease risk in a population-based case-control study: evidence of gene-gene interactions. Inflamm Bowel Dis 14, 1528–1541 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venegas M. et al. IL-23R Arg381Gln polymorphism in Chilean patients with inflammatory bowel disease. Eur Cytokine Netw 19, 190–195 (2008). [DOI] [PubMed] [Google Scholar]
- Weersma R. K. et al. Molecular prediction of disease risk and severity in a large Dutch Crohn’s disease cohort. Gut 58, 388–395 (2009). [DOI] [PubMed] [Google Scholar]
- Dusatkova P. et al. Association of IL23R p.381Gln and ATG16L1 p.197Ala with Crohn disease in the Czech population. J Pediatr Gastroenterol Nutr 49, 405–410 (2009). [DOI] [PubMed] [Google Scholar]
- Lacher M. et al. Association of the interleukin-23 receptor gene variant rs11209026 with Crohn’s disease in German children. Acta Paediatr 99, 727–733 (2010). [DOI] [PubMed] [Google Scholar]
- Mitrovič M. & Potočnik U. High Resolution Melting Curve Analysis for High-Throughput SNP Genotyping in IL23R Gene and Association of IL23R with Slovenian Inflammatory Bowel Diseases Patients. Acta Chim Slov 57, 498–505 (2010). [PubMed] [Google Scholar]
- Newman W. G., Zhang Q., Liu X., Amos C. I. & Siminovitch K. A. Genetic variants in IL-23R and ATG16L1 independently predispose to increased susceptibility to Crohn’s disease in a Canadian population. J Clin Gastroenterol 43, 444–447 (2009). [DOI] [PubMed] [Google Scholar]
- Weersma R. K. et al. Confirmation of multiple Crohn’s disease susceptibility loci in a large Dutch-Belgian cohort. Am J Gastroenterol 104, 630–638 (2009). [DOI] [PubMed] [Google Scholar]
- Yang S. K. et al. Contribution of IL23R but not ATG16L1 to Crohn’s disease susceptibility in Koreans. Inflamm Bowel Dis 15, 1385–1390 (2009). [DOI] [PubMed] [Google Scholar]
- Cotterill L. et al. Replication and meta-analysis of 13,000 cases defines the risk for interleukin-23 receptor and autophagy-related 16-like 1 variants in Crohn’s disease. Can J Gastroenterol 24, 297–302 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csöngei V. et al. Interaction of the major inflammatory bowel disease susceptibility alleles in Crohn’s disease patients. World J Gastroenterol 16, 176–183 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson L. R. et al. IL23R and IL12B SNPs and Haplotypes Strongly Associate with Crohn’s Disease Risk in a New Zealand Population. Gastroenterol Res Pract 2010, 539461 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazouli M. et al. NOD2/CARD15, ATG16L1 and IL23R gene polymorphisms and childhood-onset of Crohn’s disease. World J Gastroenterol 16, 1753–1758 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahurkar S. et al. Common variants in NOD2 and IL23R are not associated with inflammatory bowel disease in Indians. J Gastroenterol Hepatol 26, 694–699 (2011). [DOI] [PubMed] [Google Scholar]
- Sventoraityte J. et al. NOD2, IL23R and ATG16L1 polymorphisms in Lithuanian patients with inflammatory bowel disease. World J Gastroenterol 16, 359–364 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner J. et al. Interaction of Crohn’s disease susceptibility genes in an Australian paediatric cohort. PLoS One 5, e15376 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauriola M. et al. IL23R, NOD2/CARD15, ATG16L1 and PHOX2B polymorphisms in a group of patients with Crohn’s disease and correlation with sub-phenotypes. Int J Mol Med 27, 469–477 (2011). [DOI] [PubMed] [Google Scholar]
- Peter I. et al. Evaluation of 22 genetic variants with Crohn’s disease risk in the Ashkenazi Jewish population: a case-control study. BMC Med Genet 12, 63 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua K. H. et al. Association between inflammatory bowel disease gene 5 (IBD5) and interleukin-23 receptor (IL23R) genetic polymorphisms in Malaysian patients with Crohn’s disease. J Dig Dis 13, 459–465 (2012). [DOI] [PubMed] [Google Scholar]
- Jung C. et al. Genotype/phenotype analysis for 53 Crohn’s disease associated genetic polymorphisms. PLoS One 7, e52223 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safrany E. et al. Difference of interleukin-23 receptor gene haplotype variants in ulcerative colitis compared to Crohn’s disease and psoriasis. Inflamm Res 62, 195–200 (2013). [DOI] [PubMed] [Google Scholar]
- Szabo M. et al. Marked diversity of IL23R gene haplotype variants in rheumatoid arthritis comparing with Crohn’s disease and ankylosing spondylitis. Mol Biol Rep 40, 359–363 (2013). [DOI] [PubMed] [Google Scholar]
- Wang M. H. et al. Contribution of higher risk genes and European admixture to Crohn’s disease in African Americans. Inflamm Bowel Dis 18, 2277–2287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaljević S. et al. Polymorphisms of interleukin-23 receptor in patients with inflammatory bowel disease in a Croatian tertiary center. Coll Antropol 37, 1171–1177 (2013). [PubMed] [Google Scholar]
- Ballester V. et al. Association of NOD2 and IL23R with Inflammatory Bowel Disease in Puerto Rico. PLoS One 9, e108204 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doecke J. D. et al. Smoking Behaviour Modifies Il23r-Crohn’s Disease Risk in Patients with Crohn’s Disease. J Gastroenterol Hepatol 30, 299–307 (2015). [DOI] [PubMed] [Google Scholar]
- Meddour Y. et al. NOD2/CARD15 and IL23R genetic variability in 204 Algerian Crohn’s disease. Clin Res Hepatol Gastroenterol 38, 499–504 (2014). [DOI] [PubMed] [Google Scholar]
- Zhang J., Chen J., Gu J., Guo H. & Chen W. Association of IL23R and ATG16L1 with susceptibility of Crohn’s disease in Chinese population. Scand J Gastroenterol 49, 1201–1206 (2014). [DOI] [PubMed] [Google Scholar]
- Chen Z. Y. et al. Preliminary study on relationship between gene polymorphisms of interleukin-23 receptor and inflammatory bowel disease. Chin J Dig 28, 369–372 (2008). [Google Scholar]
- Bin C. et al. Contribution of rs11465788 in IL23R gene to Crohn’s disease susceptibility and phenotype in Chinese population. J Genet 88, 191–196 (2009). [DOI] [PubMed] [Google Scholar]
- Feng Y. Association analysis of IL-23R gene polymorphism in Chinese Han population with Crohn’s disease. Shanghai Traffic University Masters Dissertation (2009). [Google Scholar]
- Lu Z. K., Chen Z. R., Zheng J. J. & Shi R. H. Association study of IL-23R Arg381Gln polymorphism and inflammatory bowel disease. Chin J Dig 30, 782–784 (2010). [Google Scholar]
- Zhao X. D. et al. Association of interleukin-23 receptor gene polymorphisms with susceptibility and phenotypes of inflammatory bowel diseases in Jiangsu Han population. Zhonghua Nei Ke Za Zhi 50, 935–941 (2011). [PubMed] [Google Scholar]
- Huang C. et al. Characterization of Genetic Loci That Affect Susceptibility to Inflammatory Bowel Diseases in African Americans. Gastroenterology 10.1053/j.gastro.2015.07.065 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M. et al. Association of CTLA-4 variants with susceptibility to inflammatory bowel disease: A meta-analysis. Hum Immunol 75, 227–233 (2014). [DOI] [PubMed] [Google Scholar]
- Li Y. et al. Interleukin-23 receptor genetic polymorphisms and Crohn’s disease susceptibility: a meta-analysis. Inflamm Res 59, 607–614 (2010). [DOI] [PubMed] [Google Scholar]
- Cochran W. G. The combination of estimates from different experiments. Biometrics 10, 101–102 (1954). [Google Scholar]
- Egger M., Davey Smith G., Schneider M. & Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ 315, 629–634 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safrany E. & Melegh B. Functional variants of the interleukin-23 receptor gene in non-gastrointestinal autoimmune diseases. Curr Med Chem 16, 3766–3774 (2009). [DOI] [PubMed] [Google Scholar]
- Lee Y. H., Choi S. J., Ji J. D. & Song G. G. Associations between interleukin-23R polymorphisms and ankylosing spondylitis susceptibility: a meta-analysis. Inflamm Res 61, 143–149 (2012). [DOI] [PubMed] [Google Scholar]
- Wu Y. et al. Replication of association between interleukin-23 receptor (IL-23R) and its ligand (IL-12B) polymorphisms and psoriasis in the Chinese Han population. Hum Immunol 71, 1255–1258 (2010). [DOI] [PubMed] [Google Scholar]
- Kim H. S. et al. No association between interleukin 23 receptor gene polymorphisms and systemic lupus erythematosus. Rheumatol Int 30, 33–8 (2009). [DOI] [PubMed] [Google Scholar]
- Li Y. et al. The association between interleukin-23 receptor gene polymorphisms and systemic lupus erythematosus. DNA Cell Biol 29, 79–82 (2010). [DOI] [PubMed] [Google Scholar]
- Cho J. H. Inflammatory bowel disease: genetic and epidemiologic considerations. World J Gastroenterol 14, 338–347 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouma G. & Strober W. The immunological and genetic basis of inflammatory bowel disease. Nat Rev Immunol 3, 521–533 (2003). [DOI] [PubMed] [Google Scholar]
- Cen H. et al. Association of IFIH1 rs1990760 polymorphism with susceptibility to autoimmune diseases: a meta-analysis. Autoimmunity 46, 455–462 (2013). [DOI] [PubMed] [Google Scholar]
- Zhang X. Y. et al. Identification and expression analysis of alternatively spliced isoforms of human interleukin-23 receptor gene in normal lymphoid cells and selected tumor cells. Immunogenetics 57, 934–943 (2006). [DOI] [PubMed] [Google Scholar]
- Pidasheva S. et al. Functional studies on the IBD susceptibility gene IL23R implicate reduced receptor function in the protective genetic variant R381Q. PLoS One 6, e25038 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarin R., Wu X. & Abraham C. Inflammatory disease protective R381Q IL23 receptor polymorphism results in decreased primary CD4+ and CD8+ human T-cell functional responses. Proc Natl Acad Sci USA 108, 9560–9565 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazlett J., Stamp L. K., Merriman T., Highton J. & Hessian P. A. IL-23R rs11209026 polymorphism modulates IL-17A expression in patients with rheumatoid arthritis. Genes Immun 13, 282–287 (2012). [DOI] [PubMed] [Google Scholar]
- Zwiers A. et al. Cutting edge: a variant of the IL-23R gene associated with inflammatory bowel disease induces loss of microRNA regulation and enhanced protein production. J Immunol 188, 1573–1577 (2012). [DOI] [PubMed] [Google Scholar]
- Taylor K. D. et al. IL23R haplotypes provide a large population attributable risk for Crohn’s disease. Inflamm Bowel Dis 14, 1185–91 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]