Abstract
Enterovirus 71 (EV71), one of the major pathogens of Hand, foot and mouth disease (HFMD), results in millions of infections and hundreds of deaths each year in Southeast Asia. Biased infection and variable clinical manifestations of EV71 HFMD indicated that host genetic background played an important role in the occurrence and development of the disease. We identified the mRNA profiles of EV71 HFMD patients, which type I interferon (IFN) pathway related genes were down-regulated. Four single nucleotide polymorphisms (SNPs) of type I IFN receptor 1 (IFNAR1) were chosen to analyze their relationships to EV71 infection. We found that genotype GG of promoter variant rs2843710 was associated with the susceptibility and severity to EV71 HFMD. In addition, we assessed the regulatory effects of rs2843710 to IFN stimulated genes (ISGs), and found that the expressions of IFNAR1, OAS1 and MX1 were significantly lower in patients with rs2843710 genotype GG. And rs2843710 allele G showed weaker transcriptional activity compared with allele C. Our study indicated that rs2843710 of IFNAR1 was associated with the susceptibility and severity of EV71 HFMD in Chinese Han populations, acting as a functional polymorphism by regulating ISGs expression, such as OAS1 and MX1.
Hand, foot and mouth disease (HFMD) is a common pediatric infectious diseases, it is caused by picornaviridae family member enterovirus, mainly caused by coxsackie virus A16 (CA16) and enterovirus 71 (EV71), which characterized by fever, oral mucosa herpes, and rash on the hands, foot, and buttocks. It usually affects those less than 5 years old, in particular under 3 years1,2,3. Although most HFMD patients have good prognosis, there are some patients with severe neurological complications such as aseptic meningitis, encephalitis, brain stem encephalitis, neurogenic pulmonary edema, and hemorrhage4,5, with a high mortality6. The number of HFMD had a nearly half millions and killed 126 people in 20087,8. HFMD caused more than 1.6 million infectors and 509 deaths in 20119, most victims were infectors of EV713. Children with EV71 infection have a higher frequency of central nervous system complications. In recent 20 years, EV71 is mainly popular in Southeast Asia4,10,11, the national within the scope of the outbreak began in 2008 in China, and spread rapidly from Anhui to the other provinces7,12,13. Up to now, there are no effective vaccines and antiviral drugs to prevent or treat EV71 infection, so it is necessary to identify the susceptible factors or the warning signs to predict the disease progression.
HFMD is mainly popular in Southeast Asia, especially in China4,10,11,12, and mainly occurred in male patients6,14. Some patients may combine with severe central nervous system complications11,15,16. Even the same EV71 strain would lead to different clinical manifestations in different patients17. All together prompt that host genetic background plays an important role in the occurrence and development of EV71 HFMD. Polymorphisms of type I IFN signaling pathway genes like MX1 and OAS1 have been reported to link to the occurrence and development of HFMD18,19. Other variants of type II IFN related genes IFN-γ, IL-10 and IP-10, chemokines CCL2 and CXCL10 and eNOS also contributed to susceptibility or severity to EV71 infection20,21,22,23.
Type I IFN is the first line of host immune defense, and plays an important role in innate immune response and is an important cytokine to mediate host anti-viral response. Host recognized pathogen associated molecular patterns (PAMP) and activated type I IFN signal24. Type I IFN bound to IFNAR1/225,26, and activate a range of antiviral IFN stimulated genes (ISGs), including protein kinase R (PKR), oligoadenylate synthestase (OAS) and interferon-induced GTP-binding protein Mx (MX)27,28,29,30. Both OAS and ds RNA dependent PKR modulate virus replication, and RNAase L and MX inhibit viral transcription31,32,33. On the other hand, EV71 could be survived through down-regulating IFNAR1 and JAK1 expression34,35, indicating that IFNAR1 plays an essential role in type I IFN signaling pathway against EV71 infection. It was reported that IFNAR1 gene polymorphisms were associated with many viruses infection, including HBV, HCV and HIV-136,37,38, and variants of IFNAR1 downstream ISGs have been reported to link with the occurrence and development of HFMD18,19. However, the relationship between IFNAR1 polymorphisms and susceptibility of EV71 HFMD remains unknown.
In this study, we found that the expression of IFNAR1, IFNAR2, OAS1 and MX1 in PBMC reduced in patients with EV71 HFMD, particularly with severe symptoms. In the same time, we identified a genetic polymorphism rs2843710 C > G in the promoter of IFNAR1 gene was associated with the susceptibility and clinical phenotypes of EV71 HFMD, especially in male patients. In further, we found that rs2843710 allele G showed weaker transcriptional activity after EV71 infection. The expression of IFNAR1, OAS1 and MX1 reduced in patients with rs2843710 genotype GG compared with CC or CG. This may explain why rs2843710 allele G was associated with the susceptibility and severity of EV71 HFMD.
Results
EV71 HFMD was associated with the decrease of type I IFN related genes levels in PBMC
We performed genome-wide transcriptional analysis in PBMC isolated from healthy donors (HD, n=10), mild EV71 HFMD patients (M-EV71, n=6) and severe EV71 HFMD patients (S-EV71, n=6). Clustering analysis showed that the expression of 43 related genes was clearly different in HD, M-EV71 and S-EV71 groups, which could be divided into two gene clusters according to the different clustering model. The related genes of type I IFN signaling pathways had a similar expression patterns in the same cluster (Fig. 1).
Figure 1. The mRNA expression profiles in PBMC from different populations.
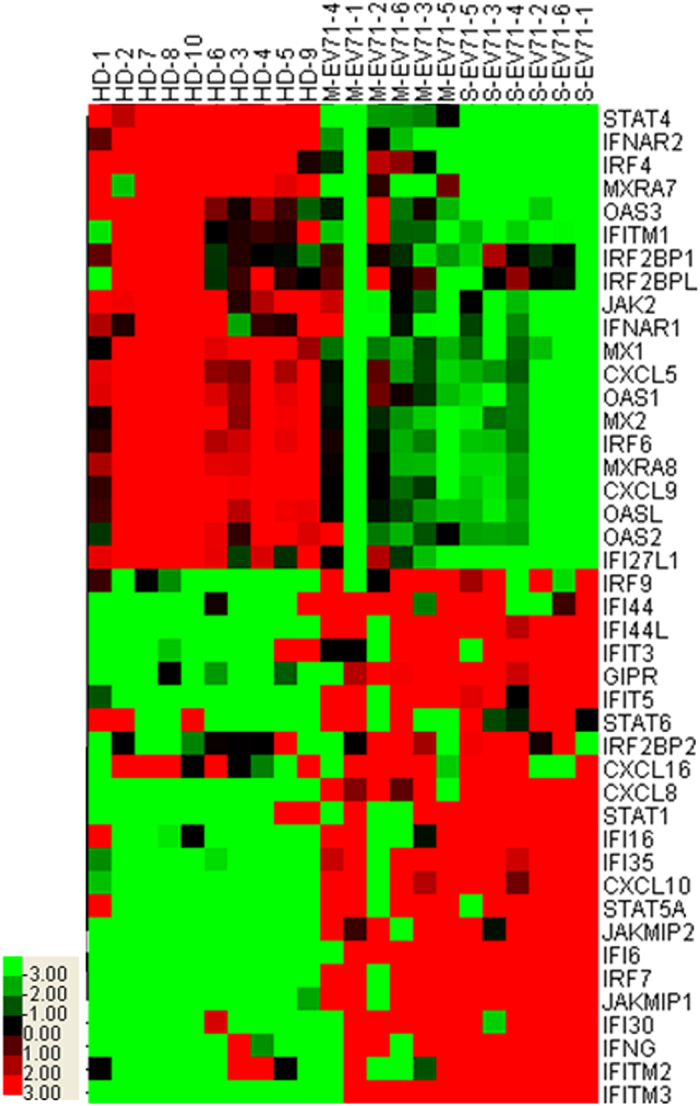
Genome-wide transcriptional analysis was performed in PBMC isolated from different groups (HD, n = 10; M-EV71, n = 6; S-EV71, n = 6). Raw data was performed using Illumina software, then delineated the false discovery rate (FDR) and normalized. P < 0.05 was regarded as statistical significant.
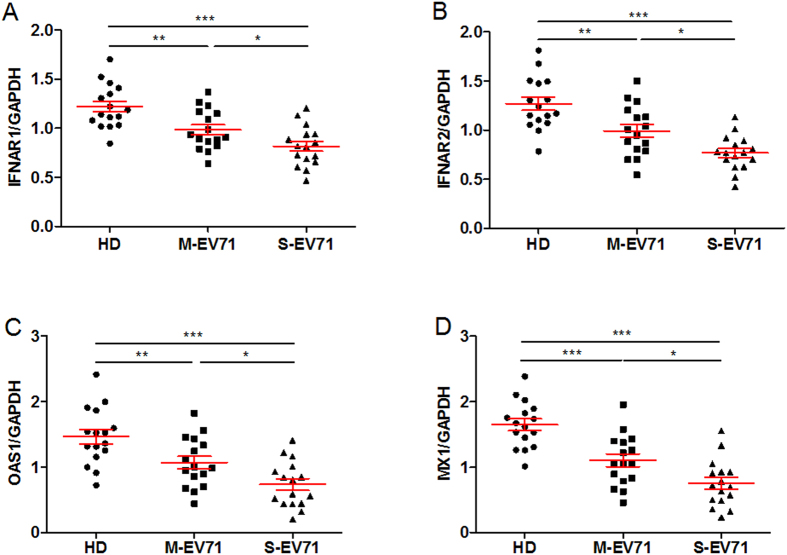
In particular, IFNAR1, IFNAR2, OAS1 and MX1 gene expression were significantly decreased in EV71 HFMD patients compared with HD. This difference was further validated by a conventional qPCR assay using another cohort (n = 16 for each group respectively, Fig. 2). Especially, the IFNAR1, IFNAR2, OAS1 and MX1 gene expression level was much lower in S-EV71 than that in M-EV71 (Fig. 2), suggesting the down-regulation of type I IFN related genes was associated with the severity of EV71 HFMD.
Figure 2. Validation of type I IFN related genes expression among different groups.
(A–D) The IFNAR1 (A) and IFNAR2 (B) and OAS1 (C) and MX1 (D) mRNA levels were verified using SYBR Green qPCR in PBMCs from healthy controls, mild patients with EV71 infection and severe patients with EV71 infection (n = 16, respectively). The expression of mRNA was 2−△△Ct relative to house keeping gene GAPDH. Differences between groups were compared with the ANOVA/Newman-Keuls multiple comparison test. *P < 0.05, **P < 0.01, ***P < 0.0001.
Rs2843710 of IFNAR1 was associated with susceptibility and severity of EV71 HFMD
We selected 4 potential functional SNPs of IFNAR1 to assess their association with EV71 HFMD. Among four SNPs of IFNAR1, rs2843710 and rs1787572 located in the promoter region, rs113181057 in exon 11, and rs1012334 in intron 3. Four SNPs genotype distribution in cases and controls were coincident with Hardy-Weinberg equilibrium (P > 0.05). As shown in Table 1, the allele frequency of rs2843710 SNP was significantly different in HD (n = 573) and EV71 HFMD (n = 1196), and allele G showed higher risk to EV71 HFMD (OR = 1.34, 95% CI, 1.15–1.55; P = 0.0001). In genotype level, the patients with rs2843710 genotype GG showed increased risk of EV71 HFMD in the recessive model (OR = 1.33, 95% CI, 1.09-1.63; P = 0.005). Genotypes CC and CG showed decreased EV71 HFMD susceptibility compared with genotype GG using an additive model (OR = 0.49, 95%CI, 0.35–0.69; P < 0.0001; and OR = 0.58, 95%CI, 0.42–0.82, P = 0.002).
Table 1. The association between IFNAR1 gene SNPs and EV71 HFMD susceptibility.
| SNP ID | Genotype | HD | EV71 | Multiplicative | Additive | Dominant | Recessive | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No (%) | No (%) | P Value | OR (95% CI) | P Value | OR (95% CI) | P Value | OR (95% CI) | P Value | OR (95% CI) | ||
| rs2843710 | CC | 251 (43.8) | 442 (37.0) | 0.0001 | 1.34(1.15–1.55) | <0.0001 | 0.49(0.35–0.69) | 0.0002 | 0.54(0.39–0.74) | 0.005 | 1.33(1.09–1.63) |
| CG | 269 (46.9) | 565 (47.2) | 0.002 | 0.58(0.42-0.82) | |||||||
| GG | 53 (9.3) | 189 (15.8) | Ref. | Ref. | |||||||
| rs1787572 | GG | 196 (34.2) | 466 (39.0) | 0.2019 | 0.91(0.79–1.05) | 0.4725 | 0.89(0.66–1.21) | 0.8764 | 1.02(0.78–1.35) | 0.0530 | 0.81(0.66–1.00) |
| GT | 290 (50.6) | 545 (45.6) | 0.4067 | 1.13(0.85–1.52) | |||||||
| TT | 87 (15.2) | 185 (15.4) | Ref. | Ref. | |||||||
| rs113181057 | TT | 573(100.0) | 1196(100.0) | — | — | — | — | — | — | — | — |
| TC | 0 (0.0) | 0 (0.0) | — | — | |||||||
| CC | 0 (0.0) | 0 (0.0) | Ref. | Ref. | |||||||
| rs1012334 | AA | 195 (34.0) | 410 (34.3) | 0.2813 | 0.92(0.80–1.07) | 0.1516 | 0.81(0.60–1.08) | 0.0607 | 0.78(0.60–1.01) | 0.9175 | 0.99(0.80–1.22) |
| AT | 268 (46.8) | 599 (50.1) | 0.0517 | 0.76(0.58–1.00) | |||||||
| TT | 110 (19.2) | 187 (15.6) | Ref. | Ref. | |||||||
Note: Hardy-Weinberg equilibrium P values of 4 SNPs (in the order of rs2843710, rs17875752, rs113181057, rs1012334) were 0.11, 0.23, 0.00, and 0.30 in HD, and were 0.71, 0.22, 0.00, and 0.19 in EV71. Abbreviations: HD, healthy donors; EV71, EV71 HFMD; CI, confidence interval; OR, odds ratio.
It was reported that EV71 HFMD mainly occurred in male patients14, so we further analyzed the association between rs2843710 and susceptibility in different sex. In male population (n = 738), the rs2843710 allele G frequency was significantly higher in cases than that in controls in multiplicative model (OR = 1.57, 95% CI, 1.29-1.90; P < 0.0001, Table 2). In genotype level, the patients with rs2843710 genotype GG increased risk of EV71 HFMD in the recessive model (OR = 1.63, 95% CI, 1.25 2.11; P = 0.0002, Table 2). In contrast, rs2843710 showed no association with susceptibility to EV71 HFMD in female population (n = 458).
Table 2. The association between the rs2843710 SNP and EV71 HFMD susceptibility stratified by sex.
| Sex | Genotype | HD | EV71 | Multiplicative | Additive | Dominant | Recessive | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No (%) | No (%) | P Value | OR (95% CI) | P Value | OR (95% CI) | P Value | OR (95% CI) | P Value | OR (95% CI) | ||
| Male | CC | 164 (47.8) | 266 (36.0) | <0.0001 | 1.57(1.29–1.90) | < 0.0001 | 0.35(0.22–0.57) | 0.0001 | 0.43(0.27–0.67) | 0.0002 | 1.63(1.25–2.11) |
| CG | 153 (44.6) | 353 (47.8) | 0.0034 | 0.50(0.32–0.80) | |||||||
| GG | 26 (7.6) | 119 (16.2) | Ref. | Ref. | |||||||
| Female | CC | 87 (37.8) | 176 (38.4) | 0.5957 | 1.07(0.84–1.34) | 0.3426 | 0.84(0.51–1.39) | 0.2075 | 0.79(0.50–1.26) | 0.8782 | 0.97(0.70–1.35) |
| CG | 116 (50.4) | 212 (46.3) | 0.1679 | 0.76(0.46–1.23) | |||||||
| GG | 27 (11.8) | 70 (15.3) | Ref. | Ref. | |||||||
Note: Hardy-Weinberg equilibrium P values of rs2843710 in males and females were 0.23, 0.99, in HD, and were 0.21, 0.54 in EV71.
Abbreviations: HD, healthy donors; EV71, EV71 HFMD; CI, confidence interval; OR, odds ratio.
To further clarify the relationship between rs2843710 and EV71 HFMD clinical manifestations, we compared different alleles’ frequencies in M-EV71 (n = 801) and S-EV71 (n = 395). The frequency of rs2843710 allele G was significantly higher in S-EV71 patients than that in M-EV71 patients, which suggested that the rs2843710 allele G was associated with the severity of EV71 HFMD (OR = 1.21, 95% CI, 1.02-1.44; P = 0.028, Table 3).
Table 3. The association between rs2843710 SNP and severity of EV71 HFMD.
| SNP ID | Genotype | HD | M-EV71 | S-EV71 | M-EV71 Vs HD | S-EV71 Vs HD | S-EV71 Vs M-EV71 | |||
|---|---|---|---|---|---|---|---|---|---|---|
| No (%) | No (%) | No (%) | P Value | OR (95% CI) | P Value | OR (95% CI) | P Value | OR (95% CI) | ||
| rs2843710 | CC | 251 (43.8) | 310 (38.7) | 132 (33.4) | 0.0053 | 1.25(1.07–1.47) | < 0.0001 | 1.52(1.26–1.84) | 0.0289 | 1.21(1.02–1.44) |
| CG | 269 (46.9) | 375 (46.8) | 190 (48.1) | |||||||
| GG | 53 (9.3) | 116 (14.5) | 73 (18.5) | |||||||
Note: Hardy-Weinberg equilibrium P values of rs2843710 were 0.11 in HD, and were 0.88 in M-EV71, and were 0.75 in S-EV71. Abbreviations: HD, healthy donors; M-EV71, mild EV71 HFMD; S-EV71, severe EV71 HFMD; CI, confidence interval; OR, odds ratio.
Rs2843710 of IFNAR1 influenced IFNAR1, OAS1 and MX1 gene expression
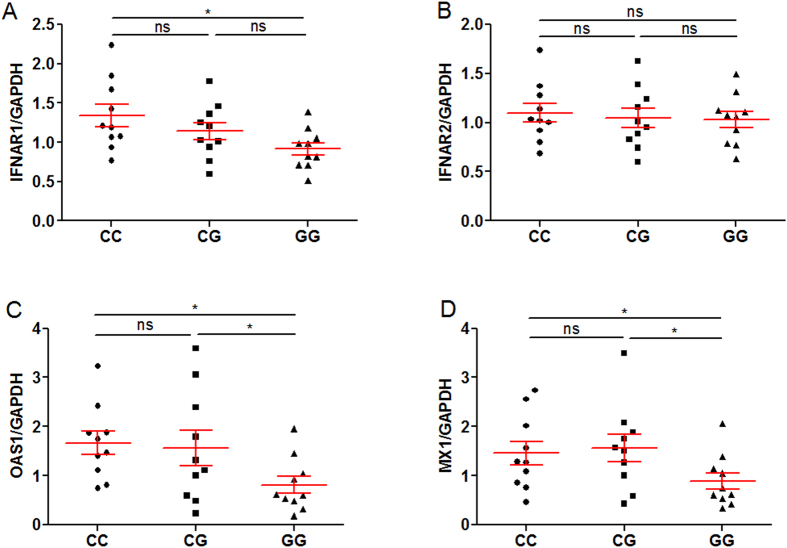
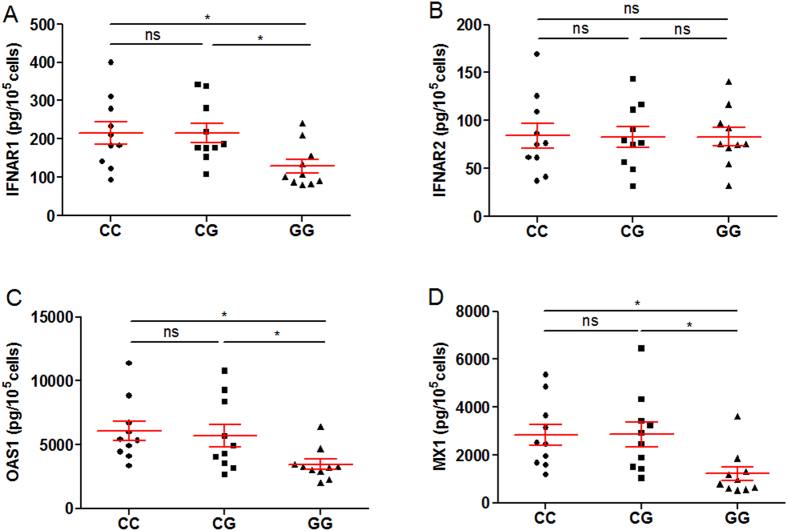
In order to explore the function of rs2843710, we detected the expression of IFNAR1, IFNAR2, OAS1 and MX1 in PBMC from patients with CC, CG and GG genotypes using qPCR and ELISA assay (n = 10, respectively). Interestingly, we found that the expression of IFNAR1, OAS1 and MX1 was much lower in genotype GG than that in genotypes CC or CG (Fig. 3A,C,D), while rs2843710 SNP did not affect the IFNAR2 gene expression (Fig. 3B). And lower protein expression of IFNAR1, OAS1 and MX1 was also observed in genotype GG carriers (Fig. 4A,C,D), while the protein expression of IFNAR2 showed no difference among all genotypes (Fig. 4B). Together, these results suggest that the rs2843710 genotype GG had specifically lower expression of IFNAR1 and down-stream genes.
Figure 3. Rs2843710 of IFNAR1 influences mRNA expression of IFNAR1, OAS1 and MX1.
(A–D) The IFNAR1 (A) and IFNAR2 (B) and OAS1 (C) and MX1 (D) mRNA levels were determined using SYBR Green qPCR in PBMCs from patients with different rs2843710 genotypes (CC, n = 10; CG, n = 10; GG, n = 10), and the expression of mRNA was 2−△△Ct relative to house keeping gene GAPDH. Differences between groups were compared with the ANOVA/Newman-Keuls multiple comparison test. *P < 0.05; ns, not significant.
Figure 4. Rs2843710 of IFNAR1 affects IFNAR1, OAS1 and MX1 expression in protein level.
(A–D) The IFNAR1 (A) and IFNAR2 (B) and OAS1 (C) and MX1 (D) protein levels were determined by ELISA assay in PBMCs from patients with different rs2843710 genotypes (CC, n = 10; CG, n = 10; GG, n = 10). Differences between groups were compared with the ANOVA/Newman-Keuls multiple comparison test. *P < 0.05; ns, not significant.
Rs2843710 of IFNAR1 affects IFNAR1 promoter transcriptional activity
To evaluate the functional significance of promoter polymorphism, transcriptional activity of IFNAR1 promoter variant rs2843710 was tested by promoter luciferase reporter assay (Fig. 5). After EV71 infection, allele G of rs2843710 showed significantly weaker transcriptional activity than allele C, while no difference was observed between both alleles without infection. These results suggest that the transcriptional activity of IFNAR1 rs2843710 allele G was attenuated after EV71 infection.
Figure 5. The rs2843710 SNP affects IFNAR1 promoter transcriptional activity.
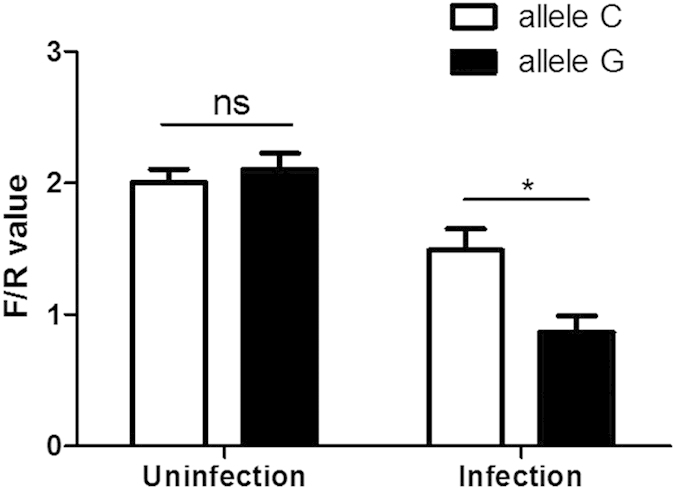
IFNAR1 promoter luciferase reporter plasmids carrying rs2843710 C or G allele were transfected into THP-1 cells, with or without EV71 infection. Luciferase activities of THP-1 cells were determined and normalized to Renillla luciferase activities. *P < 0.05; ns, not significant.
Discussion
In this study, we first illuminated the mRNA profiles of EV71 infection of HFMD by using mRNA Chip, and genes of type I IFN pathway were significantly decreased in EV71 HFMD patients. From more than 1700 volunteers and a Chinese case-control cohort, we identified a strong association between susceptibility and severity to hand, foot and mouth disease of EV71 infection and a genetic variation rs2843710 in the promoter region of IRFAR1. In contrast, other SNPs examined were not associated with disease susceptibility in the same populations. Moreover, allele G of rs2843710 showed weaker transcriptional activity compared with allele C in promoter luciferase reporter assay. The expression level of INFAR1 of rs2843710 genotype GG was lower than those with genotypes CC or CG, and coincided with the same expression pattern of INFAR1 downstream genes OAS1 and MX1, arguing that a tightly regulated type I IFN response to EV71 infection may have contributed to attenuate resistance to HFMD progression in this population.
Type I IFN comprised IFN-α, IFN-β, as well as some additional family members. It was the first line of host immune defense and directly responded to virus infection39,40. Type I IFN bound to its receptor (IFNAR), and activated a large amount of ISGs through JAK-STAT signal pathway41,42,43. Three ISGs systems present major antiviral activities, including PKR, OAS/Rnase L system and the MX GTPases27,28,29,30.
Type I IFN has protective effect against EV71 or CA16 infections. Type I IFN directly suppressed CA16 infection and controlled EV71 infection and replication44,45. IFN injection before EV71 infection could effectively protect mice, and the pre-infection effect was much stronger than post-injection45. All-trans-retinoic acid (ATRA) could protect cells apoptosis induced by EV71 infection and reduce the percentage of infected cells through enhancing the production of IFN-α46.
In contrast, EV71 could reduce the expression of IFNAR1. The 2A protein encoded by EV71 blocked type I IFN mediated STAT1, STAT2, Jak1 and Tyk2 phosphorylation through reducing IFNAR1 protein expression34. And 2A protein inhibited type I IFN responses by targeting the mitochondrial antiviral signaling (MAVS)47. The 3C protein of EV71 inhibited retinoid acid-inducible 1 gene mediated IRF3 activation and type I IFN responses48. And the 3C protein suppressed host immune response by blocking the type I IFN synthesis49. In addition, mir-146 expression increased after EV71 infection, and mir-146 promoted disease progression by inhibiting IFN production34.
HFMD is mainly popular in Southeast Asia and infects millions of children4,10,11,12, and the clinical manifestations are variable in different patients17, indicating that host genetic background played an important role in the occurrence and development of EV71 HFMD. Polymorphisms of type I IFN stimulated genes were associated with the susceptibility and clinical phenotypes of EV71 HFMD, such as −123 A and −88 T of MX1 obviously reduce the susceptibility of EV71, but individuals with the −123 A or −88 T had a higher MX1 mRNA levels in IFN-β stimulated PBMC compared with non-carriers18. A SNP rs10774671 of OAS1 contributed to the susceptibility and severity of CA16 HFMD and reduced IFN-γ expression level in severe HFMD patients19. Variants of type II IFN and other chemokines also contributed to EV71 HFMD. Interferon gamma induced proteins-10 (IP-10) gene −1596 T was the risk factor of EV71 infection20. Interleukin 10 (IL-10) and interferon gamma (IFN-γ) gene SNPs were associated with the susceptibility to EV71 HFMD encephalitis21. The −2518 G of CCL2, -201 A of CXCL10, 781 C of IL8 and 894 T of eNOS were associated with the severity of EV71 infection22,23.
We have found that IFNAR1 promoter polymorphism rs2843710 was related to susceptibility and severity of EV71 HFMD. Rs2843710, described as −568 C/G before, have been shown to be associated with susceptibility to chronic HBV infection50. And allele G of rs2843710 was associated with falciparum malaria manifestation in the endemic region51. Other variants in the promoter region of INFAR1 also related to the susceptibility of viruses infection. Carriers of 3 polymorphisms −408 T, −3 T, −77 GTL had higher risk to HBV infection50, and polymorphisms −408 C can influence the risk of developing depression of hepatitis C virus52. It was reported that allele change C > T at the −3 locus reduced the transcriptional activity of IFNAR1 promoter53.
Since rs2843710 has been predicted near the binding site of GA binding protein transcription factor (GABP), this variant may affect IFNAR1 transcription and expression53. In our study, without infection, rs2843710 seemed to have no apparent impact on the transcriptional activity of IFNAR1, which was consistent with the result Zhou et al. have reported before50. However, after EV71 infection, allele G of rs2843710 showed significantly weaker transcriptional activity compared with allele C, indicating that the difference of transcriptional activity between G and C allele might be triggered by EV71 infection. Lower transcriptional activity of rs2843710 allele G impaired IFNAR1 gene expression and resulted in down-regulation of ISGs including MX1 and OAS1. According to previous report, EV71 infection could inhibit IFNAR1 expression34. Our promoter luciferase assay results showed that EV71 infection might involve in regulating IFNAR1 transcriptional activity and influence IFNAR1 expression. The role which EV71 played in IFNAR1 transcription was needed to be further research.
Most reports showed that EV71 infection was mainly occurred in male6,14,54. In our study, the frequency of rs2843710 susceptible genotype GG was significantly higher in male patients, while this frequency showed no difference between female cases and controls. Taken together, it indicated that higher susceptible of rs2843710 genotype GG in male might be one of the reasons why EV71 HFMD had higher prevalence in male population.
In conclusion, we have identified a functional polymorphism rs2843710 in IFNAR1 gene promoter region was associated with susceptibility and severity of HFMD with EV71 infection. After EV71 infection, allele G of rs2843710 attenuated IFNAR1 transcriptional activity and expression compared with allele C, resulting in the decrease of interferon stimulated genes including MX1 and OAS1. Lower expression of MX1 and OAS1 reduced inhibition effect to virus, causing higher risk to EV71 HFMD or severer progression and complications. Higher susceptible of rs2843710 genotype GG in male might contribute to higher EV71 HFMD prevalence in male population.
Methods
Study subjects and sample
All EV71 HFMD patients were recruited from department of infectious disease or pediatric in the Third People’s Hospital of Shenzhen in 2012–2014. Healthy donors (HD, n = 573) who underwent a health examination were recruited from Shenzhen during the same period, there was no history of HFMD for HD and serum EV71 IgG test showed negative (Table 4). The criteria of HFMD diagnostics was referred to the ministry of health, 2010 version of the “hand, foot and mouth disease diagnosis and treatment guidelines”. EV71 HFMD patients were divided into mild and severe EV71 HFMD patients (M-EV71, n = 801; S-EV71, n = 395). S-EV71 presented with meningitis, encephalitis, encephalomyelitis, pulmonary edema, myocarditis, circulation problems, and ruled out merger other common infectious diseases, such as measles, cytomegalovirus infection, mumps, etc; M-EV71 didn’t get central nervous system complications and eliminated merger other common infectious diseases. We collected 3 ml peripheral blood from each volunteer using lithium heparin anticoagulant tube. 200 μl blood was cryopreserved −20 °C for genomic DNA extraction, and the remaining was for PBMC separation by density gradient centrifugation over Ficoll-Hypaque as described before55. EV71 RNA was assessed by Enterovirus type 71 nucleic acid detection kit (Da An Gene Co., Ltd., Guangzhou, China), and each patient was positive. All clinical procedures followed the protocols and were carried out in accordance with the approved guidelines of the Ethical Committee of the Third People’s Hospital of Shenzhen, under approval number TPHSZ2012001. All participants provided their written consents for the current study.
Table 4. Characteristics of patients with HFMD with EV71 infection and healthy donors.
| Cohort | Subgroup | No. | Age, months (mean ± SD) | Sex (Male:Female) |
|---|---|---|---|---|
| Healthy donors | — | 573 | 25.37 ± 11.00 | 343:230 |
| EV71 | M-EV71 | 801 | 25.01 ± 17.42 | 482:319 |
| S-EV71 | 395 | 24.71 ± 14.28 | 256:139 |
DNA and RNA Extraction
Blood genomic DNA extraction was isolated from the 200 μL whole blood using QIAamp Mini kit (Qiagen, Germany) according to the kit instructions. The quality and concentration of DNA was detected with ultramicro spectrophotometer (Agilent companies in the United States). The concentration of DNA was diluted into 5 ng/μl. RNA was extracted from 1 × 106 PBMC using RNeasy Mini Kit (Qiagen, Germany) according to the kit instructions. The quality and concentration of RNA was detected using ultramicro spectrophotometer (Agilent, USA). DNA and RNA was stored −80 °C for long-term preservation.
Microarray data analysis
RNA specimens was reverse transcribed to synthesize cDNA, then inspecting the quality of cDNA based on Illumina instructions, sample hybridization, labeling, and scanned on the iScan systems. For microarray assays, we used the HumanHT-12 v4 Expression BeadChip with 47 000 probes representing 25000 annotated genes, which coverage of well-characterized genes, gene candidates, and splice variants (Illumina, San Diego, CA, USA). Raw data generated from Illumina HT-12 was performed using the Illumina software, then delineate the false discovery rate (FDR) and normalized. P < 0.05 was regarded as Statistical significant.
Measurement of genes expression with SYBR Green qPCR assay
The mRNA expression was validated with SYBR Green qPCR assay and GAPDH was considered as a house keeping gene. RNA was reversed transcribed into cDNA using PrimeScriptTM RT reagent Kit (Takara companies, Japan). The qPCR reaction system was as follows: SYBR Premix Ex Taq II 10.0 μL, PCR Forward Primer (10 μM) 0.8 μL, PCR Reverse Primer (10 μM) 0.8 μL, ROX Reference Dye II (50×) 0.4 μL, cDNA solution 1.0 μL, RNase free H2O 7.0 μL. Reaction conditions as follows: 95 °C, 30 s; 95 °C, 5 s, 60 °C, 34 s, 40 cycles. The data were expressed as mRNA copy number relative to the house keeping gene GAPDH.
SNP Selection and Genotyping
SNPs were selected according to the SNP in HapMap database information, with focusing on their potential regulatory roles, such as transcription binding sites in the promoter region, microRNA target sites in the 3′ untranslated region (UTR), protein phosphorylation sites in the extrons and other putative regulation sites. SNPs were genotyped by time of flight mass spectrometry technology (Sequenom, San Diego. CA). The genotypes were determined by a Homogenous Mass EXTEND assay. Data analysis was performed by using MassARRAY typer software 4.0.
Measurement of IFNAR1, IFNAR2, OAS1 and MX1 expression in PBMCs with ELISA
The whole PBMCs from different genotypes patients were lysed by diluting 1:20 in hypotonic buffer as described previously56, Diluted specimens were frozen and stored at −80 °C. The levels of INFAR1 and IFNAR2 were determined using commercially available ELISA kits (PBL Assay Science, Piscataway, NJ), and the measurement of OAS1 and MX1 was also determined using a sandwich enzyme immunoassay kit BioVendor, Sweden) followed the manufacturer’s instructions.
Plasmid Constructs and Mutagenesis
The human IFNAR1 promoter (−1282 to +49) was amplified by PCR and inserted into pGL3-Basic vector (Promega, Madison, WI) upstream of the firefly luciferase coding region at Xho I and Hind III site. The primers used to PCR genomic DNA, were 5′-CATCTCGAGGAGATGGAATATAGAGATGGAATAG-3′ (forward, underlined letters showed Xho I site) and 5′-CATAAGCTTACGCGCTGCCCCTCTTAGCTCTCAC-3′ (reverse, underlined letters showed Hind III site). The pGL3-rs2843710-C vector was mutated into pGL3-rs2843710-G by site-directed mutagenesis using QuickChang II Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA). The primers used for mutation were 5’-TCTGCCCCGCTCTCGGTCTGCACACAGCAAC-3′ (forward) and 5′- GTTGCTGTGTGCAGACCGAGAGCGGGGCAGA-3′ (reverse), in which the bold letter showed the altered nucleotide.
Transient transfection and Reporter Assays
A human monocytic cell line (THP-1) was grown at 37 °C in 5% CO2 in RPMI 1640 medium with 10% FBS. To make macrophages, THP-1 cells were incubated with PMA (20 ng/ml) in 24-well plates for 24 hrs, and then washed with fresh media before transfection. A total of 0.5 ug plasmid DNA including 0.4 ug of either rs2843710-C or rs2843710-G reporter vector, and 0.1 ug of pRL-TK control vector, were co-transfected into cells using jetPEI reagents according to the manufacture’s instruction (Polyplus). 24 hrs later, the cells was infected with EV71 at a MOI of 10 for 2 hrs at 37 °C. Unbound viruses were removed by washing them with medium. The EV71 strain (GenBank: FJ607335.1) was originally isolated from the throat swabs of a HFMD patients and amplified in Rhabdomyosarcoma (RD) cell. Infected cells were then harvested and detected the Firefly and Renilla luciferase activities with Dual-Glo luciferase reporter assay system (Promega) following 48 hrs of infection. Promoter activity was measured as the ratio between Firefly and Renilla luciferase. All experiments were performed at least twice, with each transfection in triplicate.
Statistical Analysis
All SNPs investigated in this study were tested for Hardy-Weinberg equilibrium. The pearson χ2 test was used to evaluate distribution of allele frequencies in cases and controls. The unconditional logistic regression adjusted by gender and age were performed to calculate the Odd ratios (ORs), 95% confidence intervals (CIs) and corresponding P values under four alternative models (multiplicative, additive, dominant and recessive). The one-way analysis of variance (ANOVA)/Newman-Keuls multiple comparison test was used for statistical analyses to compare the differences among multiple groups. Differences were considered significant for P < 0.05. Statistical analysis of experimental data was performed using GraphPad Prism software version 5.0 (GraphPad Software, La Jolla, CA).
Additional Information
How to cite this article: Zou, R. et al. A functional polymorphism in IFNAR1 gene is associated with susceptibility and severity of HFMD with EV71 infection. Sci. Rep. 5, 18541; doi: 10.1038/srep18541 (2015).
Acknowledgments
This work was supported by the following funds: Medical Scientific Research Foundation of Guangdong Province, China (No. A2014519), Shenzhen Key Clinical Department of Emerging Infectious Diseases Foundation (No. 201161), Science and Technology Planning Project of Shenzhen, China (JCYJ20140411112047888). We thank staffs, doctors and nurses of Shenzhen Third People’s Hospital for patient management, sample collection and analysis in this work.
Footnotes
Author Contributions Y.X.L. and G.Z. conceived the project and supervised the whole study. R.Z., W.W., J.Y., YM.L. and Y.D. conducted the experiments. S.L., J.L. and Y.W. analysed the results. R.Z., S.L. and G.Z. wrote the manuscript. G.G. and B.Z. proofread the manuscript. All authors reviewed the manuscript.
References
- Zhu F. C. et al. Retrospective study of the incidence of HFMD and seroepidemiology of antibodies against EV71 and CoxA16 in prenatal women and their infants. PLoS One 7, e37206 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F. et al. Survey of enterovirus infections from hand, foot and mouth disease outbreak in China, 2009. Virol J 8, 508 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni H. et al. Epidemiological and etiological characteristics of hand, foot, and mouth disease in Ningbo, China, 2008-2011. J Clin Virol 54, 342–8 (2012). [DOI] [PubMed] [Google Scholar]
- Chan L. G. et al. Deaths of children during an outbreak of hand, foot, and mouth disease in sarawak, malaysia: clinical and pathological characteristics of the disease. For the Outbreak Study Group. Clin Infect Dis 31, 678–83 (2000). [DOI] [PubMed] [Google Scholar]
- Wang S. M. et al. Pathogenesis of enterovirus 71 brainstem encephalitis in pediatric patients: roles of cytokines and cellular immune activation in patients with pulmonary edema. J Infect Dis 188, 564–70 (2003). [DOI] [PubMed] [Google Scholar]
- Wang S. M. et al. Clinical spectrum of enterovirus 71 infection in children in southern Taiwan, with an emphasis on neurological complications. Clin Infect Dis 29, 184–90 (1999). [DOI] [PubMed] [Google Scholar]
- Tan X. et al. The persistent circulation of enterovirus 71 in People’s Republic of China: causing emerging nationwide epidemics since 2008. PLoS One 6, e25662 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J. Enterovirus 71 infection: a new threat to global public health? Lancet Neurol 7, 868–9 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Health and Family Planning Commission of the People’s Republic of China. Ministry of Health released the National Notifiable Infectious Diseases Overview of January 2012 and 2011. Epidemic information. (2012) Available at: http://www.moh.gov.cn/wsb/pyqxx/201202/54106.shtml. (Accessed: 10th February 2012).
- Ahmad K. Hand, foot, and mouth disease outbreak reported in Singapore. Lancet 356, 1338 (2000). [DOI] [PubMed] [Google Scholar]
- Ho M. et al. An epidemic of enterovirus 71 infection in Taiwan. Taiwan Enterovirus Epidemic Working Group. N Engl J Med 341, 929–35 (1999). [DOI] [PubMed] [Google Scholar]
- Yang F. et al. Enterovirus 71 outbreak in the People’s Republic of China in 2008. J Clin Microbiol 47, 2351–2 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. et al. An emerging recombinant human enterovirus 71 responsible for the 2008 outbreak of hand foot and mouth disease in Fuyang city of China. Virol J 7, 94 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W. et al. Distribution of enteroviruses in hospitalized children with hand, foot and mouth disease and relationship between pathogens and nervous system complications. Virol J 9, 8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L. Y. et al. Neurodevelopment and cognition in children after enterovirus 71 infection. N Engl J Med 356, 1226–34 (2007). [DOI] [PubMed] [Google Scholar]
- Huang C. C. et al. Neurologic complications in children with enterovirus 71 infection. N Engl J Med 341, 936–42 (1999). [DOI] [PubMed] [Google Scholar]
- Weng K. F., Chen L. L., Huang P. N. & Shih S. R. Neural pathogenesis of enterovirus 71 infection. Microbes Infect 12, 505–10 (2010). [DOI] [PubMed] [Google Scholar]
- Zhang X. et al. Association of functional polymorphisms in the MxA gene with susceptibility to enterovirus 71 infection. Hum Genet 133, 187–97 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y. et al. Association analysis of polymorphisms in OAS1 with susceptibility and severity of hand, foot and mouth disease. Int J Immunogenet 41, 384–92 (2014). [DOI] [PubMed] [Google Scholar]
- Yang J., Chen Z. Z., Lv T. G., Liu P. P. & Chen Z. B. Association of IP-10 gene polymorphism with susceptibility to Enterovirus 71 infection. Biomed Rep 1, 410–412 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. et al. Association of interleukin 10 and interferon gamma gene polymorphisms with enterovirus 71 encephalitis in patients with hand, foot and mouth disease. Scand J Infect Dis 44, 465–9 (2012). [DOI] [PubMed] [Google Scholar]
- Li J. A., Chen Z. B., Lv T. G., Han Z. L. & Liu P. P. Impact of endothelial nitric oxide synthase gene polymorphism on severity of enterovirus 71-infection in Chinese children. Clin Biochem 46, 1842–7 (2013). [DOI] [PubMed] [Google Scholar]
- Li J. A., Chen Z. B., Lv T. G. & Han Z. L. Genetic polymorphism of CCL2-2518, CXCL10-201, IL8+781 and susceptibility to severity of Enterovirus-71 infection in a Chinese population. Inflamm Res 63, 549–56 (2014). [DOI] [PubMed] [Google Scholar]
- Yoneyama M. et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol 175, 2851–8 (2005). [DOI] [PubMed] [Google Scholar]
- Cleary C. M., Donnelly R. J., Soh J., Mariano T. M. & Pestka S. Knockout and reconstitution of a functional human type I interferon receptor complex. J Biol Chem 269, 18747–9 (1994). [PubMed] [Google Scholar]
- Novick D., Cohen B. & Rubinstein M. The human interferon alpha/beta receptor: characterization and molecular cloning. Cell 77, 391–400 (1994). [DOI] [PubMed] [Google Scholar]
- Garcia M. A. et al. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol Mol Biol Rev 70, 1032–60 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B. R. PKR; a sentinel kinase for cellular stress. Oncogene 18, 6112–20 (1999). [DOI] [PubMed] [Google Scholar]
- Silverman R. H. Fascination with 2-5A-dependent RNase: a unique enzyme that functions in interferon action. J Interferon Res 14, 101–4 (1994). [DOI] [PubMed] [Google Scholar]
- Haller O. & Kochs G. Interferon-induced mx proteins: dynamin-like GTPases with antiviral activity. Traffic 3, 710–7 (2002). [DOI] [PubMed] [Google Scholar]
- Goodbourn S., Didcock L. & Randall R. E. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J Gen Virol 81, 2341–64 (2000). [DOI] [PubMed] [Google Scholar]
- George C. X. et al. Tipping the balance: antagonism of PKR kinase and ADAR1 deaminase functions by virus gene products. J Interferon Cytokine Res 29, 477–87 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth A. M., Li Z., Cattaneo R. & Samuel C. E. RNA-specific adenosine deaminase ADAR1 suppresses measles virus-induced apoptosis and activation of protein kinase PKR. J Biol Chem 284, 29350–6 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J. et al. Enterovirus 71 disrupts interferon signaling by reducing the level of interferon receptor 1. J Virol 86, 3767–76 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. et al. Enterovirus 71 inhibits cellular type I interferon signaling by downregulating JAK1 protein expression. Viral Immunol 27, 267–76 (2014). [DOI] [PubMed] [Google Scholar]
- Zhou J. et al. A non-synonymous single nucleotide polymorphism in IFNAR1 affects susceptibility to chronic hepatitis B virus infection. J Viral Hepat 16, 45–52 (2009). [DOI] [PubMed] [Google Scholar]
- He X. X. et al. Persistent effect of IFNAR-1 genetic polymorphism on the long-term pathogenesis of chronic HBV infection. Viral Immunol 23, 251–7 (2010). [DOI] [PubMed] [Google Scholar]
- Diop G. et al. Exhaustive genotyping of the interferon alpha receptor 1 (IFNAR1) gene and association of an IFNAR1 protein variant with AIDS progression or susceptibility to HIV-1 infection in a French AIDS cohort. Biomed Pharmacother 60, 569–77 (2006). [DOI] [PubMed] [Google Scholar]
- van Pesch V., Lanaya H., Renauld J. C. & Michiels T. Characterization of the murine alpha interferon gene family. J Virol 78, 8219–28 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. M., Ezashi T., Rosenfeld C. S., Ealy A. D. & Kubisch H. M. Evolution of the interferon tau genes and their promoters, and maternal-trophoblast interactions in control of their expression. Reprod Suppl 61, 239–51 (2003). [PubMed] [Google Scholar]
- Levy D. E. & Darnell J. E. Jr. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol 3, 651–62 (2002). [DOI] [PubMed] [Google Scholar]
- de Veer M. J. et al. Functional classification of interferon-stimulated genes identified using microarrays. J Leukoc Biol 69, 912–20 (2001). [PubMed] [Google Scholar]
- Der S. D., Zhou A., Williams B. R. & Silverman R. H. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci USA 95, 15623–8 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki O., Karaki T. & Imanishi J. Protective effect of interferon on infections with hand, foot, and mouth disease virus in newborn mice. J Infect Dis 153, 498–502 (1986). [DOI] [PubMed] [Google Scholar]
- Liu M. L. et al. Type I interferons protect mice against enterovirus 71 infection. J Gen Virol 86, 3263–9 (2005). [DOI] [PubMed] [Google Scholar]
- Chen S., Yang Y., Xu J., Su L. & Wang W. Effect of all-trans-retinoic acid on enterovirus 71 infection in vitro. Br J Nutr 111, 1586–93 (2014). [DOI] [PubMed] [Google Scholar]
- Wang B. et al. Enterovirus 71 protease 2Apro targets MAVS to inhibit anti-viral type I interferon responses. PLoS Pathog 9, e1003231 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei X. et al. The 3C protein of enterovirus 71 inhibits retinoid acid-inducible gene I-mediated interferon regulatory factor 3 activation and type I interferon responses. J Virol 84, 8051–61 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. P., Wang Y. F., Wang J. R., Huang S. W. & Yu C. K. Enterovirus 71 blocks selectively type I interferon production through the 3C viral protein in mice. J Med Virol 84, 1779–89 (2012). [DOI] [PubMed] [Google Scholar]
- Zhou J. et al. Polymorphisms of type I interferon receptor 1 promoter and their effects on chronic hepatitis B virus infection. J Hepatol 46, 198–205 (2007). [DOI] [PubMed] [Google Scholar]
- Kanchan K. et al. Interferon-gamma (IFNG) microsatellite repeat and single nucleotide polymorphism haplotypes of IFN-alpha receptor (IFNAR1) associated with enhanced malaria susceptibility in Indian populations. Infect Genet Evol 29, 6–14 (2015). [DOI] [PubMed] [Google Scholar]
- Rifai M. A. & Sabouni M. A. Utilizing genomic polymorphisms to personalize hepatitis C therapies. Curr Opin Organ Transplant 17, 198–203 (2012). [DOI] [PubMed] [Google Scholar]
- Zhou J. et al. Functional dissection of an IFN-alpha/beta receptor 1 promoter variant that confers higher risk to chronic hepatitis B virus infection. J Hepatol 51, 322–32 (2009). [DOI] [PubMed] [Google Scholar]
- Wang J. et al. Epidemiological analysis, detection, and comparison of space-time patterns of Beijing hand-foot-mouth disease (2008–2012). PLoS One 9, e92745 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. et al. CD4 (+) CD25 (+) FoxP3 (+) regulatory T cells suppress Mycobacterium tuberculosis immunity in patients with active disease. Clin Immunol 123, 50–9 (2007). [DOI] [PubMed] [Google Scholar]
- Vallittu A. M., Eralinna J. P., Ilonen J., Salmi A. A. & Waris M. MxA protein assay for optimal monitoring of IFN-beta bioactivity in the treatment of MS patients. Acta Neurol Scand 118, 12–7 (2008). [DOI] [PubMed] [Google Scholar]