Abstract
Dysregulated microRNAs in neurons could cause many nervous system diseases. The therapeutic manipulation of these pathogenic microRNAs necessitates novel, efficient delivery systems to facilitate microRNA modulators targeting neurons with minimal off-target effects. The study aimed to establish a lipofection protocol to upregulate expression levels of miR-21 in neurons under different conditions, including different serum-free medium, transfection conditions, and reagent concentration, by evaluating the expression levels of miR-21 and neuron injury. The expression levels of miR-21 were higher in neurons transfected by Neurobasal-A than by DMEM. Expression levels of miR-21 were already the highest at the ratio RNAiMAX:miR-21 = 3:5, but the increase of RNAiMAX's concentration had not caused the further upregulation of expression level of miR-21. Neuron injury was condition dependent and dose dependent after transfection. Compared to S-Neurobasal groups, neurons have a smaller injury in N-Neurobasal groups, and compared to ratios RNAiMAX:miR-21 = 4:5, 5:5, neuron injury was smaller at ratios of RNAiMAX:miR-21 = 1:5, 2:5, 3:5. Without the pretreatment of starvation in vitro, the lipofection protocol was that RNAiMAX/miR-21 agomir complexes were diluted in Neurobasal-A at the ratio of RNAiMAX:miR-21 = 3:5.
Introduction
Dysregulated microRNAs in neurons could cause many central nervous system (CNS) diseases. The therapeutic manipulation of these pathogenic microRNAs necessitates novel, efficient delivery systems to facilitate microRNA modulators targeting neurons with minimal off-target effects. Neuronal injury occurs in different CNS diseases, including trauma (Wee et al., 2015), tumor (Bingham et al., 2011), and ischemic (Ji et al., 2015) and neurodegenerative disorders (Li et al., 2015), and leads to many functional deficits with serious impairments in patients (Reina et al., 2013). The neurons display typical molecular and functional characteristics of mature neuronal cells in vitro and offer the opportunity to study various aspects of CNS diseases. The regulation of special gene is a crucial tool to identify the component necessary for a particular cellular process in many studies.
microRNAs play an important role in gene expression and bind to a special sequence of mRNA molecule to regulate the expression of mRNA (Choi et al., 2014). In CNS diseases, miRNAs have been reported to regulate synaptic plasticity, neuroprotection, and neurodegeneration (Saugstad, 2010). microRNA-21 (miR-21) is one of the most studied miRNAs and has been found to play an important role in most biological functions and diseases of CNS (Krichevsky and Gabriely, 2009). Growing evidence also suggests that miR-21 is detected in plasma sample in some diseases like glioma (Wang et al., 2012). Meanwhile, miR-21 may be a novel therapeutic target in human cancer (Pan et al., 2010) and miR-21 was also a therapeutic target for reversing drug resistance in cancer (Hong et al., 2013). A thorough understanding of the physiological function of miR-21 is of great importance as it can promote the development of miR-21-based biomarkers and therapeutics. However, finding an efficient transfection method that delivers miRNAs into neurons is relatively difficult. Furthermore, when transfecting neurons with high efficiency, it is important that cell damage is avoided and the original function of neurons is maintained. Another crucial limitation of transfection neurons is that neurons are very sensitive to different transfection conditions, for example, transfection solvent, transfection reagent, or pretreatment of transfection, which all influence the transfection efficiency, neuron injury, or neuronal function.
Various methods, including chemical, physical, and biological methods, have been attempted to transfect neurons (Covello et al., 2014). Physical methods, which include electrotransfection, biolistics, and injection, have been successfully used in segmentation cells but may be toxic for cells and usually have a high reagent and equipment cost and cell suspensions in vitro (Villemejane and Mir, 2009), which are not appropriate for neurons. Biological methods, which include virus and plasmid transfection, have been applied to many hard-transfected cells (Du et al., 2014), but the complexity and biosafety issues limited their widespread applications, which are not good methods for transfecting neurons. Chemical methods have widely been used to transfect, as they are relatively simple, cheap, and safe. There are many chemical methods, including calcium phosphate, liposomes, cationic lipids, cationic polymers, and cationic polysaccharides (Azzam and Domb, 2004), which are used to transfect a great number of cell lines and primary cells. In the traditional protocol, several steps are needed before transfection. Specially, the starvation pretreatment requires the cells to be cultured in only DMEM without any other substances, including fetal calf serum and antibiotics. However, the traditional protocol is also potentially harmful to cells, and transfection conditions need to be carefully optimized to improve efficiency and miRNA expression level in cells.
Cationic lipofection, a reagent of chemical methods, is the simplest and most readily available way for RNA oligo delivery. Lipofectamine RNAiMAX (RNAiMAX), a new cationic lipofection reagent, has shown higher efficiency in delivering miRNA into many stem cells and cell lines (Zhao et al., 2008; Malamas et al., 2013), although its transfection efficiency is still unclear in most primary cells, especially neurons. Here, we sought to establish a lipofection protocol to upregulate the expression level of miR-21 in neurons under different transfection conditions, including different transfection solvents (DMEM or Neurobasal), pretreatments of transfection (with starvation or not), and different reagent ratios, while maintaining viability and functional abilities of neurons. The lipofection protocol will become the basic research transfection method of miRNAs in neurons in vitro, which will provide insight into developing a novel therapeutic approach of miR-21 or other miRNA gene therapy for CNS diseases.
Materials and Methods
Neuron culture
Primary cortex fragments from the postnatal Wistar rats (<24 h), which were purchased from Experimental Animal Laboratories of the Academy of Military Medical Science and complied with NIH guidelines, were dissected and dissociated into single cells. The cell was suspended in DMEM with 10% fetal bovine serum and plated on six-well plates coated with poly-l-lysine (Sigma-Aldrich) at a density of 5 × 105 per well. After 2 h, neurons were cultured in the Neurobasal-A medium with 2% B27 and 1% glutamine (2 mL) (Gibco) for 7 days at 37°C in 95% air and 5% CO2 atmosphere.
The transfection efficiency of miR-21 and lipofection protocol
Labeling of the miR-21 agomir with a 5′-FAM oligonucleotide (FAM-miR-21; GenePharma) was done to confirm whether miR-21 oligomers could be transfected into neuronal cells or not. Immunofluorescence staining for microtubule-associated protein 2 (MAP-2) and transfection for FAM-miR-21 were performed. The neurons were cultured for 7 days, and translation was performed with RNAiMAX according to the traditional protocol with the minimum concentration transfection reagent (0.5 μL of RNAiMAX), which was used to test the transfection efficiency of miR-21. Dilute 0.5 μL RNAiMAX reagent and 2.5 μL of FAM-miR-21 agomir (20 μM) in DMEM, and the overall solution reached to 500 μL and was incubated for 20 min. Transfection solution was added to six-well plates and maintained up to 6 h. Then, the cells reacted with mouse anti-MAP-2 (1:100; BOSTER) as a neuron marker for 12 h at 4°C and reacted with TRITC-(tetra-methyl-5,6-isothiocyanate)-labeled phalloidin (1:100; BOSTER) for 1 h. DAPI was used for DNA staining. The number of MAP-2-positive cells, FAM-positive cells, and the total number of cortical neurons were counted under 200× magnification.
Neurons were cultured for 7 days and randomly divided into five groups: starvation+DMEM (S-DMEM), unstarvation+DMEM (N-DMEM), starvation+Neurobasal-A (S-Neurobasal), unstarvation+Neurobasal-A (N-Neurobasal), and the sham group (Sham). As described previously, miRNA oligomers (sequences listed in Table 1) were diluted to a final concentration of 20 mM. miRNA oligomers (2.5 μL) were then combined with RNAiMAX (0.5, 1, 1.5, 2, 2.5 μL) (Invitrogen) in an RNase-free polymerase chain reaction (PCR) tube and incubated for 20 min at room temperature, with the ratio RNAiMAX (μL):miR-21 (μL) = 1:5, 2:5, 3:5, 4:5, 5:5. The overall transfection solution reached to 500 μL (N-DMEM/Neurobasal-A) and 250 μL (S-DMEM/Neurobasal-A). Before transfection, neurons plated on six-well plates were treated with 250 μL of DMEM or Neurobasal-A for 1 h in S-DMEM/S-Neurobasal-A groups for starvation pretreatment but not in N-DMEM/N-Neurobasal-A groups. Transfection solution was added to six-well plates and maintained up to 6 h, and then the neurons were cultured in the Neurobasal-A medium with 2% B27 and 1% glutamine (2 mL). Neurons were further cultured for 48 h before analysis (Li et al., 2009). Cells of the sham group underwent the same transfection procedure without RNAiMAX. The antibiotics, such as penicillin or streptomycin, were not used in the traditional or lipofection protocol. There was no serum used in this transfection protocol.
Table 1.
The Sequences of miRNA Oligomers and Primers Used for Quantitative Real-Time RT-PCR
| miRNA oligomers | Sequences |
|---|---|
| miR-21 agomir | 5′-UAGCUUAUCAGACUGAUGUUGA-3′ |
| Agomir negative control | 5′-UUCUCCGAACGUGUCACGUTT-3′ |
| RT primers | |
| miR-21 | 5′-AATGGTTGTTCTCCACACTCTC-3′ |
| U6 | 5′-GGAACGCTTCACGAATTTG-3′ |
| PCR primers | |
| miR-21-F | 5′-ACGTTGTGTAGCTTATCAGACTG-3′ |
| miR-21-R | 5′-AATGGTTGTTCTCCACACTCTC-3′ |
| U6-F | 5′-ATTGGAACGATACAGAGAAGATT-3′ |
| U6-R | 5′-GGAACGCTTCACGAATTTG-3′ |
RT-PCR, reverse transcription polymerase chain reaction.
Isolation of total RNA and quantitative real-time PCR
After 48 h of transfection, the total RNA from neurons subjected to transfection protocols was extracted with the TRIzol reagent (Invitrogen) following the manufacturer's instructions. To get the mature miR-21, the total RNA and the Hairpin-it™ miRNAs RT-PCR Quantitation Kit/U6 snRNA RT-PCR Normalization Kit (GenePharma) were used in reverse transcription following the manufacturer's protocol. U6 small nuclear RNA (snRNA) was used as a housekeeping gene. Cycling conditions were 16°C, 30 min; 42°C, 30 min; 85°C, 10 min; and 4°C, keeping. Then, quantitative real-time (qRT)-PCR was performed with the CFX Connect™ Real-Time System (Bio-Rad Laboratories). Furthermore, the cycling conditions used were as follows: 95°C, 3 min; 40 cycles of 95°C, 12 s and 62°C, 40 s. Expression of miR-21 was calculated through a 2−ΔΔCt method (Livak and Schmittgen, 2001). Primers for amplification of miR-21 and U6 are listed in Table 1.
Western blots
After 48 h of transfection, the total protein was extracted from neurons in S/N-Neurobasal groups transfected with miR-21 agomir for immunoblotting analysis. The protein concentrations were detected with the Pierce BCA Protein Assay Kit. Equal amounts of protein were separated by SDS-PAGE and transferred onto the PVDF membrane. Then the blots were blocked by 5% nonfat milk dissolved in TBST for 2 h and incubated overnight at 4°C in the following primary antibodies: PTEN, PDCD4, and GAPDH. Subsequently, the blots were incubated with the second antibody HRP goat anti-rabbit.
TUNEL and MTT assay
To eliminate the biological effect of miR-21, miR-21-negative control (miR-21NC) was transfected into neurons according to the transfection protocol. To detect the cortical neurons and neuronal apoptosis, double immunofluorescence staining for MAP-2 and TUNEL were performed after 48 h of transfection. MAP-2 immunostaining was performed using anti-MAP-2 for 12 h and then reacted with TRITC-(tetra-methyl-5, 6-isothiocyanate)-labeled phalloidin for 1 h. Subsequently, cortical neurons were incubated with TUNEL (terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling) reacting mixture from the TUNEL Apoptosis Detection Kit (Roche) for 2 h. Finally, DAPI (4,6-diamidino-2-phenylindole) was used as a counterstain. The number of MAP-2-positive cells, TUNEL-positive cells, and the total number of neurons were counted under 200× magnification.
The cytotoxicity of the RNAiMAX was quantified by the MTT [3-(4,5-dimethylthiazol-2-yl)-2,5 diphenyl tetrazolium bromide] assay. Briefly, 15 μL of MTT (5 mg/mL) dissolved in the cell culture medium were added to each well and incubated for 4 h at 37°C after 48 h of transfection. After this period, the remaining MTT solution was removed, 150 μL of DMSO were added to each well to dissolve the formazan crystals, and then the 96-well plate was vibrated for 10 min. The absorbance was detected using a microplate reader at 490 nm.
Counting and statistics analysis
The immunostaining was analyzed with the ImageJ software. The percentage of positive cells was calculated as positive neurons/total neurons × 100%. Data are expressed as mean ± standard deviation (SD). Statistical analysis was performed with SPSS 19.0. Comparisons between any two groups of data were done using the single-factorial analysis of variance (ANOVA). When p-values were less than 0.05, statistical significance was considered.
Results
Transfection efficiency and the expression level of miR-21 in neurons
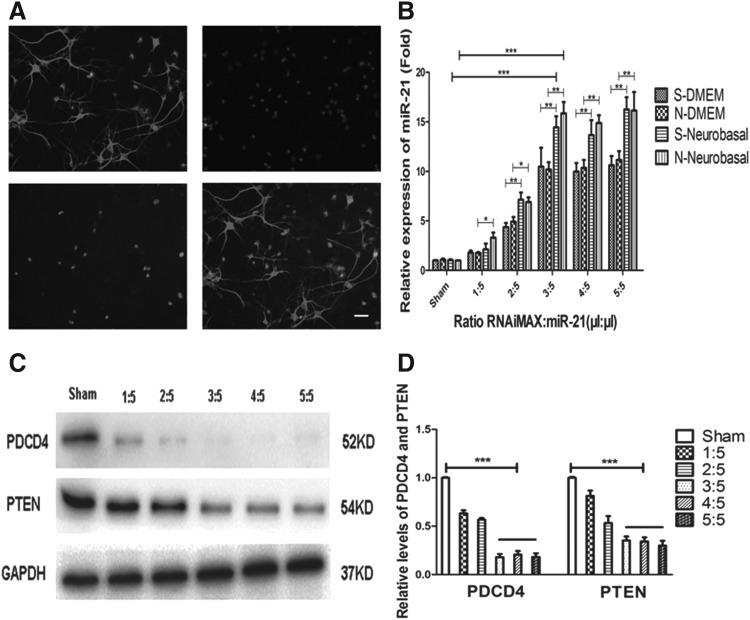
To manifest that miR-21 oligomers could be transfected into cortical neurons through RNAiMAX by the traditional protocol, we detected the transfection efficiency of FAM-miR-21 oligomers in cortical neurons. We found some intense FAM-green staining in the cytoplasm of cortical neurons, and data analyzed with the ImageJ software showed that the positive cells were 38.7% (Fig. 1A), which illustrated that the cationic lipofection reagent, RNAiMAX, could transfect miR-21 agomir into cortical neurons by the traditional protocol, but the transfection efficiency of miR-21 was not at a high level.
FIG. 1.
The transfection efficiency and expression level of miR-21 in cortical neurons. (A) Cortical neurons labeled MAP-2, DAPI, and FAM-miR-21 agomir and merged. The transfection efficiency of miR-21 oligomers was FAM-miR-21/DAPI. Scale bar: 100 μm. (B) The expression levels of miR-21 in cortical neurons. Compared to S/N-DMEM groups, the expression levels of miR-21 were higher in neurons in S/N-Neurobasal groups at ratios RNAiMAX:miR-21 = 1:5, 2:5 (*p < 0.05) and RNAiMAX:miR-21 = 3:5, 4:5, 5:5 (**p < 0.01). Compared to sham, the expression levels of miR-21 were highest at ratios RNAiMAX:miR-21 = 3:5, 4:5, 5:5 in S/N-Neurobasal (***p < 0.001) and the expression levels of miR-21 were not different at ratios RNAiMAX:miR-21 = 3:5, 4:5, 5:5 (p > 0.05). (C, D) Protein levels of PTEN and PDCD4. The protein levels of PTEN and PDCD4 decreased remarkably at ratios RNAiMAX:miR-21 = 3:5, 4:5, 5:5 (***p < 0.001). MAP-2, microtubule-associated protein 2.
To investigate the different results of lipofection protocol and traditional protocol, we detected the miR-21 expression level in cortical neurons using qRT-PCR. Although both solvents were able to mediate transfection, the expression levels of miR-21 were obviously different, and compared to S/N-DMEM groups, the expression levels of miR-21 were higher at almost all ratios in S/N-Neurobasal groups (Fig. 1B). However, the miR-21 expression level in S-DMEM groups was not different compared to N-DMEM groups and it was similar in S-Neurobasal groups and in N-Neurobasal groups (Fig. 1B). Interestingly, compared to the sham group, the highest expression levels of miR-21 was obtained at the ratio RNAiMAX:miR-21 = 3:5 and the increase of RNAiMAX's concentration had not caused the further upregulation of expression levels of miR-21 at ratios RNAiMAX:miR-21 = 4:5, 5:5 (Fig 1B). The results showed that the transfection solvent Neurobasal-A was better used to transfect miR-21 agomir into neurons than DMEM and the starvation pretreatment of transfection had no effect on the expression levels of miR-21. The above results also indicated that the expression levels of miR-21 in neurons had reached the highest level at the ratios RNAiMAX:miR-21 = 3:5, 4:5, 5:5 in S/N-Neurobasal groups in vitro.
Protein levels of PTEN and PDCD4
To further confirm the function of miR-21, we tested the protein levels of PTEN (phosphatase and tensin homolog deleted on chromosome ten) and PDCD4 (programmed cell death protein 4), which were the target genes of miR-21 (Junker et al., 2015; Xu et al., 2015). The data showed that miR-21 suppressed the protein levels of PTEN and PDCD4, and the protein levels of PTEN and PDCD4 decreased apparently at ratios RNAiMAX:miR-21 = 3:5, 4:5, 5:5 (Fig. 1C, D), which indicated that miR-21 had higher expression levels at ratios RNAiMAX:miR-21 = 3:5, 4:5, 5:5.
The expression levels of miR-9 and miR-134 in neurons
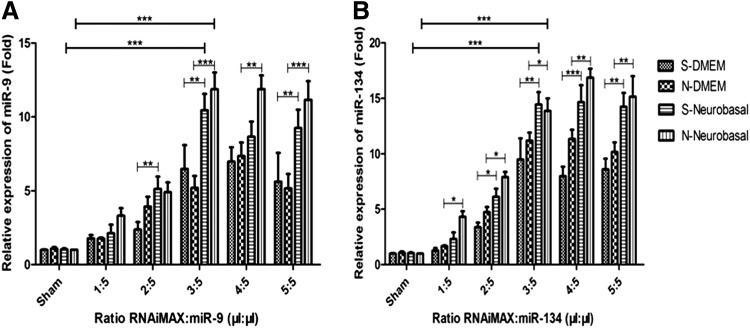
To see whether other miRNAs have similar expression levels in cortical neurons after transfection by lipofection protocol, we tested the expression levels of miR-9 and miR-134. The results showed that miR-9 and miR-134 expression levels were similar to miR-21 expression levels in cortical neurons (Fig. 2A, B). The miR-9 and miR-134 expression levels in S/N-Neurobasal groups were higher than in S/N-DMEM groups; compared to the sham group, the expression levels of miR-9 and miR-134 also achieved the highest at the ratio RNAiMAX:miR-21 = 3:5; and the increase of RNAiMAX's concentration had not caused the further upregulation of expression levels of miR-9 and miR-134 at the ratios RNAiMAX:miR-21 = 4:5, 5:5 (Fig. 2A, B). These were all similar to miR-21 expression levels in cortical neurons. The data indicated that lipofection protocol could transfect miRNAs into cortical neurons.
FIG. 2.
The expression levels of miR-9 (A) and miR-134 (B) in cortical neurons. The expression levels of miR-9 and miR-134 were similar to miR-21 in cortical neurons. Compared to S/N-DMEM groups, the expression levels of miR-9 and miR-134 were higher in S/N-Neurobasal groups at different ratios of RNAiMAX:miR-21 and not different at ratios RNAiMAX:miR-21 = 3:5, 4:5, 5:5 (p > 0.05) in cortical neurons. *p < 0.05, **p < 0.01, ***p < 0.001.
The neuron injury after transfection
We found that neuron injury was condition dependent and dose dependent after transfection (Fig. 3A–C). Compared with the sham group, the TUNEL-positive cells appeared as transfected RNAiMAX/miR-21NC complexes and were apparently increased at ratios RNAiMAX:miR-21NC = 4:5, 5:5 (Fig. 3A, B), and the cellular viability was also dramatically decreased at ratios RNAiMAX:miR-21NC = 4:5, 5:5 (Fig. 3C), which suggested that the RNAiMAX caused cortical neuron injury and the increase of RNAiMAX's concentration caused more neuron injury. Meanwhile, compared with N-Neurobasal groups, the percentage of TUNEL-positive cells was higher in the S-Neurobasal group (Fig. 3B), and the cellular viability was dramatically decreased in the S-Neurobasal group (Fig. 3C). These results indicated that cortical neurons in the N-Neurobasal group had a smaller injury than those in the S-Neurobasal group after transfection. The results above demonstrated that RNAiMAX caused injury of neurons, and compared to ratios RNAiMAX:miR-21NC = 4:5, 5:5, neuron injury was smaller at ratios RNAiMAX:miR-21NC = 1:5, 2:5, 3:5.
FIG. 3.
The apoptosis and cellular viability of cortical neurons. Cortical neurons transfected with RNAiMAX/miR-21NC complexes labeled for MAP-2, DAPI, and TUNEL and merged. Compared with RNAiMAX:miR-21NC = 1:5, 3:5, the number of apoptotic cortical neurons increased at the ratio RNAiMAX:miR-21NC = 5:5 (A). Scale bar: 100 μm. The percentage of TUNEL-positive cell: compared with S-Neurobasal groups, the percentage was lower in N-Neurobasal (*p < 0.05), and compared with ratio RNAiMAX:miR-21NC = 3:5, the percentage increased remarkably at ratios RNAiMAX:miR-21NC = 4:5, 5:5 (**p < 0.01) in N-Neurobasal groups (B). MTT for the cellular viability in cortical neurons: compared to the cortical neurons in S-Neurobasal groups, the cellular viability was higher in N-Neurobasal groups (*p < 0.05), and compared to the sham group, the cellular viability decreased dramatically at ratios RNAiMAX:miR-21 = 4:5, 5:5 in N-Neurobasal groups (**p < 0.01) (C).
Discussion
The goal of this study was to produce a parallel comparison of the traditional and lipofection protocol, including transfection conditions, solvents, and reagent concentrations, to establish the lipofection protocol for delivering active, noncoding miRNAs into cortical neurons. Some transfection reagents are currently used to introduce miRNAs into cells, but poor transfection efficiency and low expression level of miRNAs are a common problem in neurons. The cationic lipofection has been shown to perform well in many passage cells, such as osteoclasts (Liu et al., 2015), endothelial cells (Rohde et al., 2015), and human-derived epithelial cells (Le Corre et al., 2015), but the mature primary cells, like neurons, are still described as difficult-to-transfect (Morris and Labhasetwar, 2015). In some studies, the cationic lipofection reagents, like Lipofectamine 2000, were efficient in terms of delivery in primary cells (Chen et al., 2014), but there are no special protocols to maintain the functionality of the internalized RNA oligos.
In this study, by detecting the expression levels of miR-21 and injury of neurons transfected with RNAiMAX/miR-21 complexes, we successfully established the lipofection protocol of miR-21 transfecting into cortical neurons. First, we examined whether the cultured cells were cortical cells or not. The MAP-2 is a cytoskeletal protein localized in the neuronal dendritic compartment, and it is considered a marker of structural integrity because it is involved in the morphological stabilization of dendritic processes (Di Stefano et al., 2001). The cultured cells had a strong MAP-2 staining in dendrites. Meanwhile, in other primary cells, such as primary human aortic and coronary artery endothelial and vascular smooth muscle, RNAiMAX has been reported an effective gene vector (Nabzdyk et al., 2012). RNAiMAX has been used in multigene transfection and is considered an effective tool for gene delivery. As a new cationic liposome agent, RNAiMAX has a higher transfection efficiency compared with several other transfecting agents such as Oligofectamine and Lipofectamine 2000 (Zhao et al., 2008; Kamaci et al., 2011). Then, we found that RNAiMAX was effective for delivering FAM-miR-21 agomir into cortical neurons.
Second, we investigate the expression levels of miR-21 in neurons transfected with RNAiMAX/miR-21 complexes. We chose two solvents to dilute RNAiMAX/miR-21 complexes, DMEM and Neurobasal-A. DMEM is a common medium used for culturing cells and transfection (Xu et al., 2014). Neurobasal-A is a special medium used for culturing neurons (Liu et al., 2013). Both are often used as transfection solvents. By analyzing the expression levels of miR-21, we found that the expression levels of miR-21 were remarkably higher in neurons transfected by Neurobasal-A than by DMEM. Meanwhile, with different ratios of RNAiMAX:miR-21, our qRT-PCR results showed that expression levels of miR-21 were already the highest at the ratio of RNAiMAX:miR-21 = 3:5 and the increase of RNAiMAX's concentration had not caused the further upregulation of expression levels of miR-21 at ratios RNAiMAX:miR-21 = 4:5, 5:5, which may be because the saturation of transfection is reached at RNAiMAX:miR-21 = 3:5 and the expression levels of miR-21 did not have obvious differences in neurons with pretreatments of transfection. Our research certified that the expression levels of miR-21 in neurons were the highest under the conditions of RNAiMAX/miR-21 complexes diluted in Neurobasal-A and at ratios of RNAiMAX:miR-21 = 3:5, 4:5, 5:5.
Previous studies have reported that compared with untreated cells, viability of transfected cells decreased and depended on the concentration of RNAiMAX (Kamaci et al., 2011). Also, some studies reported that cationic liposome agents have been shown to cause apoptosis in human lung epithelial cell (Kongkaneramit et al., 2008). In other studies, the cell apoptosis can be induced by serum deprivation (Ferriere et al., 2013). In our research, the results demonstrated that RNAiMAX caused injury of cortical neurons and neuron injury depended on the RNAiMAX concentration. Compared to ratios RNAiMAX:miR-21 = 4:5, 5:5, the injury of primary cortical neurons was smaller at the ratio of RNAiMAX:miR-21 = 3:5, which determined that the higher RNAiMAX concentration caused more cortical neuron injury. In contrast, the pretreatment, starvation of transfection, increased neuron injury. Compared to N-Neurobasal groups, the cortical neurons' viability decreased significantly in S-Neurobasal groups and the percentage of TUNEL-positive cells increased.
Conclusion
miR-21 agomir can be transfected into neurons by RNAiMAX. Without the pretreatment of starvation in vitro, the lipofection protocol was that RNAiMAX/miR-21 agomir complexes were diluted in Neurobasal-A at the ratio of RNAiMAX:miR-21 = 3:5. The starvation pretreatment led to less apoptosis of cortical neurons, and the better solvent is Neurobasal-A, and the ratio RNAiMAX:miR-21 = 3:5 is the best transfection reagent concentration in lipofection protocol.
Acknowledgments
This work was supported by the Tianjin Research Program of Application Foundation and Advanced Technology (No. 13JCYBJC23700), the National Clinical Key Subject Construction Project of NHFPC Fund, and the National Natural Science Fund (81471252).
Disclosure Statement
No competing financial interests exist.
References
- Azzam T., and Domb A.J. (2004). Current developments in gene transfection agents. Curr Drug Deliv 1, 165–193 [DOI] [PubMed] [Google Scholar]
- Bingham D., John C.M., Panter S.S., and Jarvis G.A. (2011). Post-injury treatment with lipopolysaccharide or lipooligosaccharide protects rat neuronal and glial cell cultures. Brain Res Bull 85, 403–409 [DOI] [PubMed] [Google Scholar]
- Chen Q., Xu J., Li L., Li H., Mao S., Zhang F., et al. (2014). MicroRNA-23a/b and microRNA-27a/b suppress Apaf-1 protein and alleviate hypoxia-induced neuronal apoptosis. Cell Death Dis 5, e1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K.Y., Silvestre O.F., Huang X., Min K.H., Howard G.P., Hida N., et al. (2014). Versatile RNA interference nanoplatform for systemic delivery of RNAs. ACS Nano 8, 4559–4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covello G., Siva K., Adami V., and Denti M.A. (2014). An electroporation protocol for efficient DNA transfection in PC12 cells. Cytotechnology 66, 543–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stefano G., Casoli T., Fattoretti P., Gracciotti N., Solazzi M., and Bertoni-Freddari C. (2001). Distribution of map2 in hippocampus and cerebellum of young and old rats by quantitative immunohistochemistry. J Histochem Cytochem 49, 1065–1066 [DOI] [PubMed] [Google Scholar]
- Du J., Gao X., Deng L., Chang N., Xiong H., and Zheng Y. (2014). Transfection of the glial cell line-derived neurotrophic factor gene promotes neuronal differentiation. Neural Regen Res 9, 33–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferriere F., Habauzit D., Pakdel F., Saligaut C., and Flouriot G. (2013). Unliganded estrogen receptor alpha promotes PC12 survival during serum starvation. PLoS One 8, e69081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong L., Han Y., Zhang Y., Zhang H., Zhao Q., Wu K., et al. (2013). MicroRNA-21: a therapeutic target for reversing drug resistance in cancer. Expert Opin Ther Targets 17, 1073–1080 [DOI] [PubMed] [Google Scholar]
- Ji J., Yan H., Chen Z.Z., Zhao Z., Yang D.D., Sun X.L., et al. (2015). Iptakalim protects against ischemic injury by improving neurovascular unit function in the mouse brain. Clin Exp Pharmacol Physiol 42, 766–771 [DOI] [PubMed] [Google Scholar]
- Junker F., Chabloz A., Koch U., and Radtke F. (2015). Dicer1 imparts essential survival cues in Notch driven T-ALL via miR-21 mediated tumor suppressor Pdcd4 repression. Blood 126, 993–1004 [DOI] [PubMed] [Google Scholar]
- Kamaci N., Emnacar T., Karakas N., Arikan G., Tsutsui K., and Isik S. (2011). Selective silencing of DNA topoisomerase IIbeta in human mesenchymal stem cells by siRNAs (small interfering RNAs). Cell Biol Int Rep (2010) 18, e00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kongkaneramit L., Sarisuta N., Azad N., Lu Y., Iyer A.K., Wang L., et al. (2008). Dependence of reactive oxygen species and FLICE inhibitory protein on lipofectamine-induced apoptosis in human lung epithelial cells. J Pharmacol Exp Ther 325, 969–977 [DOI] [PubMed] [Google Scholar]
- Krichevsky A.M., and Gabriely G. (2009). miR-21: a small multi-faceted RNA. J Cell Mol Med 13, 39–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Corre S.S., Belmadi N., Berchel M., Le Gall T., Haelters J.P., Lehn P., et al. (2015). Cationic dialkylarylphosphates: a new family of bio-inspired cationic lipids for gene delivery. Org Biomol Chem 13, 1122–1132 [DOI] [PubMed] [Google Scholar]
- Li H.H., Lu F.J., Hung H.C., Liu G.Y., Lai T.J., and Lin C.L. (2015). Humic acid increases amyloid beta-induced cytotoxicity by induction of ER stress in human SK-N-MC neuronal cells. Int J Mol Sci 16, 10426–10442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Li W., Yang Y., Lu Y., He C., Hu G., et al. (2009). MicroRNA-21 targets LRRFIP1 and contributes to VM-26 resistance in glioblastoma multiforme. Brain Res 1286, 13–18 [DOI] [PubMed] [Google Scholar]
- Liu J., Dang L., Li D., Liang C., He X., Wu H., et al. (2015). A delivery system specifically approaching bone resorption surfaces to facilitate therapeutic modulation of microRNAs in osteoclasts. Biomaterials 52, 148–160 [DOI] [PubMed] [Google Scholar]
- Liu Y., Zhang Y., Lin L., Lin F., Li T., Du H., et al. (2013). Effects of bone marrow-derived mesenchymal stem cells on the axonal outgrowth through activation of PI3K/AKT signaling in primary cortical neurons followed oxygen-glucose deprivation injury. PLoS One 8, e78514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., and Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- Malamas A.S., Gujrati M., Kummitha C.M., Xu R., and Lu Z.R. (2013). Design and evaluation of new pH-sensitive amphiphilic cationic lipids for siRNA delivery. J Control Release 171, 296–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris V.B., and Labhasetwar V. (2015). Arginine-rich polyplexes for gene delivery to neuronal cells. Biomaterials 60, 151–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabzdyk C.S., Chun M., Pradhan Nabzdyk L., Yoshida S., and LoGerfo F.W. (2012). Differential susceptibility of human primary aortic and coronary artery vascular cells to RNA interference. Biochem Biophys Res Commun 425, 261–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Wang Z.X., and Wang R. (2010). MicroRNA-21: a novel therapeutic target in human cancer. Cancer Biol Ther 10, 1224–1232 [DOI] [PubMed] [Google Scholar]
- Reina C.P., Driscoll M., and Gabel C.V. (2013). Neuronal repair: Apoptotic proteins make good. Worm 2, e22285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde J.H., Weigand J.E., Suess B., and Dimmeler S. (2015). A universal aptamer chimera for the delivery of functional microRNA-126. Nucleic Acid Ther 25, 141–151 [DOI] [PubMed] [Google Scholar]
- Saugstad J.A. (2010). MicroRNAs as effectors of brain function with roles in ischemia and injury, neuroprotection, and neurodegeneration. J Cereb Blood Flow Metab 30, 1564–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villemejane J., and Mir L.M. (2009). Physical methods of nucleic acid transfer: general concepts and applications. Br J Pharmacol 157, 207–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Li P., Li A., Jiang W., Wang H., Wang J., et al. (2012). Plasma specific miRNAs as predictive biomarkers for diagnosis and prognosis of glioma. J Exp Clin Cancer Res 31, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee H.Y., Lim S.W., Chio C.C., Niu K.C., Wang C.C., and Kuo J.R. (2015). Hyperbaric oxygen effects on neuronal apoptosis associations in a traumatic brain injury rat model. J Surg Res 197, 382–389 [DOI] [PubMed] [Google Scholar]
- Xu J., Zhang W., Lv Q., and Zhu D. (2015). Overexpression of miR-21 promotes the proliferation and migration of cervical cancer cells via the inhibition of PTEN. Oncol Rep 33, 3108–3116 [DOI] [PubMed] [Google Scholar]
- Xu T., Liu W., Wang S., and Shao Z. (2014). Elucidating the role of free polycationic chains in polycation gene carriers by free chains of polyethylenimine or N,N,N-trimethyl chitosan plus a certain polyplex. Int J Nanomedicine 9, 3231–3245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M., Yang H., Jiang X., Zhou W., Zhu B., Zeng Y., et al. (2008). Lipofectamine RNAiMAX: an efficient siRNA transfection reagent in human embryonic stem cells. Mol Biotechnol 40, 19–26 [DOI] [PubMed] [Google Scholar]