Abstract
Interferons (IFNs) play a crucial role in the host's immune response and other homeostatic control actions. Three IFN types and several IFN families within the types allow for a plethora of regulatory actions. The number of distinct IFN molecules is highest among type I IFNs and, in particular, within the IFN-α family. In pigs, there are 17 IFN-α subtypes with different antiviral activities and different expression profiles; however, no data are available about biological properties other than the antiviral effector activities. Therefore, 16 porcine IFN-α genes were cloned, expressed in mammalian Chinese hamster ovary cells, and characterized for antiviral, anti-inflammatory, and MHC-modulating activities at a pre-established level of 10 IU/mL. Antiviral activity: IFN-α2, -α5, -α9, and -α10 showed the highest level of activity in a pseudorabies virus yield reduction assay. On the contrary, little, if any, activity was shown by IFN-α3, -α7, -α13, -α4, and -α15. Anti-inflammatory activity: With the exception of IFNs-α2, -α7, -α9, and -α11, all IFN-α subtypes had significant anti-inflammatory control activity in an interleukin-8 (IL-8) yield reduction assay. Gene expression analyses showed that some IFN-α subtypes can significantly downregulate the expression of IL-8, tumor necrosis factor α (TNF-α), IL-6, Toll-like receptor 4 (TLR4), βD1, and nuclear factor-κB (NF-kB) genes, while maintaining or upregulating the expression of βD4. Immunomodulation: A significant upregulation of class I and/or class II MHC was induced by all the IFNs under study, with the exception of IFNs-α11, -α15, and -α16, which instead significantly downregulated class I MHC. Our results indicate that gene duplications in the porcine IFN-α family underlie diverse effector and regulatory activities, being therefore instrumental in host survival and environmental adaptation. This role of IFN-α could be founded on fine-tuning and regulation of pro- and anti-inflammatory control actions after exposure to both infectious and noninfectious environmental stressors.
Introduction
Interferons (IFNs) are cytokines, originally discovered due to their antiviral activity (Isaacs and Lindenmann 1957), and play an important role in the host's immune response and other homeostatic control actions. In mammals, IFNs are grouped in 3 major types (I, II, and III) based on chromosomal locations, gene structures, and sequence similarity (Sang and others 2014). While there is only one type II molecular species, named IFN-γ, type I IFNs in mammalian species are a heterogeneous group consisting of at least 9 families. In humans, type I IFNs include multiple IFN-α subtypes coded on chromosome 9 and single members each of IFN-β, IFN-ω, IFN-κ, and IFN-ɛ (Takaoka and Yanai 2006). Type I IFNs also comprise other subclasses with a more limited species expression, including IFN-αω, IFN-δ, IFN-τ, and IFN-ζ (limitin) (Sang and others 2010). The same IFNs exist in mice on chromosome 4, except for IFN-ω (van Pesch and others 2004). Porcine type I IFNs comprise 39 functional genes arranged along chromosomes 1 and 10 and include 17 IFNA, 11 IFND, and 7 IFNW genes and IFNB, IFNE, IFNK, and IFNAW as single genes (Sang and others 2010, 2014). Porcine IFN-α is a multigenic family containing 17 subtypes with different antiviral activities and different expression profiles. The existence of several different subtypes, sharing high sequence similarity in the coding region, is usually the consequence of gene duplications and is often instrumental in host survival and environmental adaptation. As a result, gene duplications leading to an improvement of the host's environmental fitness often remain integrated in the genome (Kondrashov 2012). Several pieces of evidence are in agreement with this statement. In particular, IFN-α subtypes are characterized by some structural differences resulting in distinct antiviral and biological properties [antiviral, anti-inflammatory, MHC modulation, T helper regulatory, to name a few (Amadori 2007)]. Each subtype also shows a different expression with respect to tissue/cell or following different inducing stimuli, probably due to genomic differences in the promoter regions (Sang and others 2014). There is also a constitutive expression of IFNs-α and IFN-β in tissues in healthy pigs without any evidence of microbial infection (Razzuoli and others 2011b; Sang and others 2014). In addition, Razzuoli and others (2011a) demonstrated that IFN-α basal expression levels in pigs are modified after exposure to a noninfectious stressor such as early weaning. Moreover, Razzuoli and others (2010) demonstrated that a low-dose, oral IFN-α treatment can modulate the pig's inflammatory responses to the early weaning stress. Most importantly, the profile of the constitutive response observed in vivo implies the stable expression of IFN-β associated with expression of different panels of IFN-α genes (Razzuoli and others 2011b). This probably implies that IFN-α subtypes serve diverse functions, being thus selectively expressed as a result of a fine-tuning determined by physiological regulations and type of environmental stressors encountered. This makes a case for a precise characterization of swine IFN-α subtypes in terms of preferential immunoregulatory activity and homeostatic control action, underlying in vivo and in vitro expression after exposure to environmental stressors. Two previous articles by Sang and others (2010, 2014) reported the antiviral activity of 12 IFN-α subtypes against porcine reproductive and respiratory syndrome virus and vesicular stomatitis virus. On the basis of this latter study, we aimed to widen the scope of such an analysis by assessing the antiviral, immunomodulatory, and anti-inflammatory potential of each IFN-α subtype. Our working hypothesis implied that such differences could be best detected at very low cytokine concentrations. These would actually mimic the usual in vivo concentrations, underlying both autocrine and paracrine effects. The aim of this study was to highlight the preferential biological activities of IFN-α subtypes as a key to understanding the role and function of their expression as a response to infectious and noninfectious stressors.
Materials and Methods
Cells and virus
CHO.K1 (Chinese Hamster Ovary cells, IZSLER Cell Bank code BS CL 15) were grown in Ham's F12 mixture supplemented with 10% fetal bovine serum (FBS), antibiotic solution (100 U/mL penicillin and 100 μg/mL streptomycin final), and 2 mM l-glutamine, while IPEC-J2 cells (porcine intestinal epithelial cells, IZSLER Cell Bank code BS CL 205) were grown in a 1:1 mixture of Dulbecco's modified Eagle's medium and Ham's F12 mixture, enriched with 10% FBS, antibiotic solution, and 2 mM l-glutamine. MDBK cells (Madin–Darby bovine kidney, IZSLER Cell Bank code BS CL 118) were grown in complete minimum essential medium (MEM) supplemented with 10% FBS. The same medium was used for PK15cl28/BS cells (Porcine Kidney 15, clone 28, IZSLER Cell Bank code BS CL 179). Pulmonary alveolar macrophages (PAMs) were obtained from bronchoalveolar lavage on healthy pigs and frozen in liquid nitrogen until use. They were grown in RPMI 1640 medium supplemented with 10% FBS and an increased concentration of antibiotics (500 U/mL penicillin, 350 μg/mL streptomycin, 1 μg/mL amphotericin B).
Vesicular stomatitis New Jersey virus (VSV, IZSLER Cell Bank code VIR RE RSCIC 4) and pseudorabies virus Bartha BS/K61 (PRV, IZSLER, Cell Bank code VIR RE RSCIC 32) were propagated on MDBK cell monolayers.
Cloning and expression of Porcine IFN-α genes in mammalian cells
Coding regions of the porcine (Po)IFN-α2, -α8, -α12, -α13, and -α16 subtypes were amplified by polymerase chain reaction (PCR) from DNA extracted from the tonsil of a large, white landrace cross pig and the PCR products cloned into eukaryotic expression vectors pcDNA3.1/V5-His-TOPO (Life Technologies). Sequencing of the recombinant plasmids indicated that the inserts have >98% nucleotide identity to accession Nos. GQ415056, GQ415062, GQ415066, GQ415067, and GQ415070, respectively, as described elsewhere (Soule 2013). PoIFN-α3, -α4, -α5, -α6, α7, -α9, -α10, -α11, α14, -α15, and -α17 subtype genes were synthesized chemically based on the sequence of accession Nos. GQ415057, GQ415058, GQ415059, GQ415060, GQ415061, GQ415063, GQ415064, GQ415065, GQ415068, GQ415069, and GQ415071, respectively. The synthesized genes were inserted into pcDNA3.1/V5-His B (Life Technologies) and sequencing confirmed 100% nucleotide identity to expected sequences (Soule 2013). After purification by the QIAGEN Plasmid Midi Kit (Qiagen) and quantification by a spectrophotometer, a 24 μg aliquot of each recombinant plasmid was used for transfection of CHO.K1 cells. These were seeded at 5 × 105/well in 6-well plates in 2 mL of serum-free and antibiotic-free MEM and incubated at 37°C in 5% CO2 until they reached ∼90% confluence. Each plasmid was diluted in 1,500 μL of Opti-MEM I Reduced Serum Medium (Gibco, Life Technologies) and added to 1,500 μL of the same medium supplemented with 60 μL of Lipofectamine 2000 (Life Technologies). 500 microliters/well of DNA-liposome complexes were incubated for 20 min at room temperature, added to cell monolayers, and incubated at 37°C, 5% CO2. After 5 h, 2 mL of Opti-MEM I Reduced Serum Medium was added to each well and plates were incubated again under the same conditions. Cell medium containing recombinant IFN proteins was harvested after 24 h, replaced with fresh medium, and harvested again after a further 24 h. The collected medium was centrifuged at 500 × g for 10 min to remove cell debris and stocked in aliquots at −80°C. Each supernatant was identified according to the presence of the respective IFN-α subtype expressed, and supernatants were named using the su, for supernatant, acronym (eg, suIFN-α2). A pcDNA 3.1 myc-His plasmid with no insert was transfected under the same conditions to obtain a mock supernatant in the experiments described hereunder.
Titers of PoIFN-α subtypes
The titers of PoIFN-α subtypes were determined in vitro by a standard cytopathic effect (cpe) inhibition assay with VSV, as previously described (Meager 1987). Recombinant porcine IFN-α1 (rIFN-α1) was purchased from PBL Biomedical Laboratories (Cat. 17100-1) and used as the internal standard to normalize IFN titers. The units of this preparation are determined with respect to the international reference standard for human leukocyte IFN (Ga-902-530) provided by the National Institutes of Health. Positive results were expressed as the reciprocal of the dilution corresponding to 50% cpe and normalized to the IFN-α1 standard to obtain a titer in terms of IU/mL for each suIFN-α.
Purification of recombinant PoIFN-α subtypes
SuIFN-α3, -α4, and -α9 were selected for analysis of properties of purified proteins and were concentrated through Amicon Ultra-15 filter units with Ultracel-PL PLGC, 10 kDa membrane, and purified through Ni-NTA spin columns according to the manufacturer's instructions. Binding, washing, and elution buffers were 50 mM Tris–HCl, 500 mM NaCl with increasing concentrations of imidazole (Sigma-Aldrich) (15, 20, and 500 mM, respectively) to allow the specific binding of His-tagged protein to the column and its subsequent release during the elution phase. Purified eluted proteins, named with the Po prefix, were stored at −80°C until use.
Characterization of biological activities of suIFN-α and purified IFN-α subtypes
Biological activities (antiviral, anti-inflammatory, and immunomodulatory) exerted by a low IFN-α concentration (10 IU/mL based on anti-VSV results obtained on MDBK cells) of each suIFN-α and purified PoIFN-α3, -α4, -α9, and rIFN-α1 were investigated using 3 different assays. Supernatants of CHO cells treated with the mock plasmid and the above elution buffer were used as negative controls for treatments with suIFN-α and purified proteins, respectively. Except for the tests on IFN-α5, the supernatant from cells transfected with the mock plasmid was diluted 1/281 in assay media, which was the dilution factor used to obtain a 10 IU/mL concentration for the suIFN-α with the lowest titer.
All suIFN-α subtypes were screened (at a concentration of 20 IU/mL) by ELISA using the Swine IFN-α Do-It-Yourself ELISA kit according to the manufacturer's directions (KingFisher Biotech). This was performed to get an insight into the cross-reactivity of porcine IFN-α subtypes in an assay standardized with precise concentrations of recombinant IFN-α. Capture antibody was used at 1 μg/mL final concentration, while the detection antibody to porcine IFN-α1 was added to the wells at 0.1 μg/mL. Absorbance was read at 492 nm and IFN-α concentration was calculated from a standard curve created using seven 2-fold dilutions of the recombinant swine IFN-α1, provided as kit reagent, down to 0.78 ng/mL. Data were analyzed by the software, Prism 2.01 (GraphPad Software).
Yield reduction assay
The antiviral activity of each subtype was determined by measuring pseudorabies virus (PRV) genomic copies in MDBK cells treated with 10 IU/mL of each suIFN-α. MDBK cells (6 × 105 cells/well) were seeded into 12-well plates in 2 mL of MEM supplemented with 10% FBS and 10 IU/mL of suIFN-α. Cells were incubated for 18 h until confluence, washed with PBS, and infected with PRV at MOI 0.1 for 1 h. The medium containing the virus was discarded and fresh MEM supplemented with 2% FBS was added to the wells. Plates were incubated at 37°C in 5% CO2 for an additional 48 h. DNA was extracted from 200 μL of each supernatant using the QIAamp DNA Mini Kit (Qiagen) and genome copy numbers determined by a quantitative real-time PCR using a standard curve of a pJET1/blunt plasmid containing a portion of the PRV gB gene (a 195-bp amplicon).
Real-time PCR was performed using the CFX96 Touch™ Real-Time PCR System (Bio-Rad). The reaction mixture (20 μL) contained 10 μL of SsoFast EvaGreen® Supermix (Bio-Rad), 8 pmol of forward (5′-ACGGCACGGGCGTGATC-3′) and reverse (5′-GGTTCAGGGTCACCCGC-3′) primers, and 4 μL of extracted DNA. The thermocycling program was 40 cycles at 95°C for 5 s and 60°C for 30 s with an initial cycle at 95°C for 2 min. Quantity in terms of genomic copies/mL of PRV in each sample was inferred from the plasmid standard curve, ranging from 102 to 106 copies/mL. Results were expressed as log 10-transformed percentage of viral copies in IFN-α-treated cells with respect to the number of copies observed in untreated, PRV-infected control cells (100%).
Anti-inflammatory activity assay
The anti-inflammatory activity of suIFN-α subtypes was investigated on porcine IPEC-J2 cells, an established line of intestinal epithelial cells that spontaneously secretes interleukin-8 (IL-8) (Razzuoli and others 2014). IPEC-J2 cells were seeded into 12-well tissue culture plates (2 mL per well, 2 × 105/mL) and incubated at 37°C in 5% CO2 until confluence (about 24 h). In each plate, 2 wells were used as unstimulated control. The others were treated with 10 IU/mL of each suIFN-α subtype (in duplicate) for 18 h. Then, supernatants were harvested and stored at −80°C for an IL-8 ELISA, while cell monolayers were washed and conserved at −20°C for gene expression analysis. Swine IL-8 was measured by a commercial ELISA kit (R&D systems; DUOset Cat. DY535) as previously described (Razzuoli and others 2013).
Gene expression profiles
The modulation of selected genes was evaluated after treatment of IPEC-J2 cells with 10 IU/mL of suIFNs-α and incubation at 37°C in 5% CO2 for 18 h. For each 12-well plate, 2 wells were used as untreated control, while 1 well was treated with mock supernatant and 5 wells with each IFN-α subtype. Total RNA was extracted from each well of the culture plates using the RNeasy Mini Kit (Qiagen) by the Qiacube System (Qiagen) in accordance with the manufacturer's instructions. The expression of IL-1β, IL-6, IL-8, IL-4, IL-10, tumor necrosis factor α (TNF-α), IFN-β, βD1, βD2, βD3, βD4, nuclear factor κB (NF-kB), IL-18, Toll-like receptor 4 (TLR4), and suppressor of cytokine signaling 1 (SOCS1) was determined by reverse transcription (RT)-real-time PCR using primer sets described in previous studies (Amadori and others 2009; Veldhuizen and others 2009; Collado-Romero and others 2010; Ross and others 2010; Razzuoli and others 2011b). RNA was extracted from every single well of IPEC-J2 cells and tested in duplicate PCR reactions for all the cytokines under study. Porcine beta-2 microglobulin (B2M) was used as a housekeeping control gene (Table 1). cDNA synthesis was performed in the presence of random hexamers. Then, EVAGreen real-time PCR amplification was performed in a CFX96 Real-Time System as previously described (Razzuoli and others 2014). All samples were checked for DNA contamination and the relative expression of the selected genes was calculated using the formula ΔΔCt, where the Ct (cycle of threshold) value indicates the mean of 3 test replicate wells ± 1 standard deviation, ΔCt = Ct (target gene) − Ct (housekeeping), and ΔΔCt = ΔCt (IFN treatment) − ΔCt (control). Untreated cells were used as control sample, except for IL-8, TNF-α, and βD4, for which mock-treated cells were used as the control sample. Samples were scored negative when the relevant Ct was ≥40.
Table 1.
Sequences of the Primers Used in Real-Time PCR
| Gene | Protein | Primers | Accession No. NCBI |
|---|---|---|---|
| B2M | β2-Microglobulin | F:5′-CGCCCCAGATTGAAATTGATTTGC-3′ | NM_213978.1 |
| R:5′-GCTATACTGATCCACAGCGTTAGG-3′ | |||
| IL-1β | Interleukin-1β | F:5′-AATTCGAGTCTGCCCTGTACCC-3′ | NM_001005149 |
| R:5′-GCCAAGATATAACCGACTTCACCA-3′ | |||
| IL-4 | Interleukin-4 | F:5′-GGACACAAGTGCGACATCA-3′ | NM_214123.1 |
| R:5′-GCACGTGTGGTGTCTGTA-3′ | |||
| IL-6 | Interleukin-6 | F:5′-TGGCTACTGCCTTCCCTACC-3′ | NM_214399 |
| R:5′-CAGAGATTTTGCCGAGGATG-3′ | |||
| IL-8 | Interleukin-8 | F:5′-TTCGATGCCAGTGCATAAATA-3′ | M86923 |
| R:5′-CTGTACAACCTTCTGCACCCA-3′ | |||
| IL-10 | Interleukin-10 | F:5′-AGCCAGCATTAAGTCTGAGAA-3′ | HQ236499.1 |
| R:5′-CCTCTCTTGGAGCTTGCTAA-3′ | |||
| TNF-α | TNF-α | F:5′-TGCCTACTGCACTTCGAGGTTATC-3′ | NM_214022 |
| R:5′-GTGGGCGACGGGCTTATCTG-3′ | |||
| IFN-β | Interferon-β | F:5′-AGTTGCCTGGGACTCCTCAA-3′ | NM_21455 |
| R:5′-CCTCAGGGACCTCAAAGTTCAT-3′ | |||
| βD-1 | β-Defensin-1 | F:5′-CTGTTAGCTGCTTAAGGAATAAAGGC-3′ | NM_213838 |
| R:5′-TGCCACAGGTGCCGATCT-3′ | |||
| βD-2 | β-Defensin-2 | F:5′-CCAGAGGTCCGACCACTACA-3′ | NM_214442 |
| R:5′-GGTCCCTTCAATCCTGTTGAA-3′ | |||
| βD-3 | β-Defensin-3 | F:5′-CTTCCTATCCAGTCTCAGTGTTCTGC-3′ | AY460575.1 |
| R:5′-GGCTTCTGTAGACTTCAAGGAGACAT-3′ | |||
| βD-4 | β-Defensin-4 | F:5′-GTGGCTTGGATTTGAGGAGAGAGT-3′ | AY460576.1 |
| R:5′-AGTGATACACAGGCCTGGAAGGAT-3′ | |||
| IL-18 | Interleukin-18 | F:5′-CGTGTTTGAGGATATGCCTGATT-3′ | AF191088.1 |
| R:5′-TGGTTACTGCCAGACCTCTAGTGA-3′ | |||
| SOCS1 | SOCS1 | F:5′-TTCTTCGCCCTCAGTGTGAA-3′ | HM462248.1 |
| R:5′-GGCCTGGAAGTGCACGC-3′ | |||
| TLR-4 | Toll-like receptor | F:5′-TGGCAGTTTCTGAGGAGTCATG-3′ | AB188301.2 |
| R:5′-CCGCAGCAGGGACTTCTC-3′ | |||
| NF-kB1 | Nuclear factor κB | F:5′-CCCATGTAGACAGCACCACCTATGAT-3′ | NM_001048232 |
| F:5′-ACAGAGGCTCAAAGTTCTCCACCA-3′ |
Supernatants of CHO cells were collected at 24 and 48 h after transfection with mock and suIFN-α plasmids. IFN-α titers were measured in MDBK cells by a cpe inhibition assay with VSV.
CHO, Chinese hamster ovary; cpe, cytopathic effect; IFN, interferon; MDBK, Madin–Darby bovine kidney; PCR, polymerase chain reaction; SOCS1, suppressor of cytokine signaling 1; TNF, tumor necrosis factor; VSV, vesicular stomatitis New Jersey virus.
Immunomodulation assays
The immunomodulatory activity of suIFN-α subtypes was assessed by investigating the expression of (1) class I MHC (BoLA) in bovine MDBK cells, (2) class I MHC (SLA) in porcine PK15cl28 cells, and (3) class II SLA in PAMs. MDBK and PK15 cells were seeded in 12-well plates in 2 mL of MEM supplemented with 10% FBS and 10 IU/mL of suIFN-α or mock supernatant. They were incubated at 37°C in 5% CO2 for about 18 h. In each plate, 2 wells were used as unstimulated negative control. Then, cells were harvested by a cell scraper, washed in FACS buffer (PBS, 2% heat-inactivated FBS, 0.1% sodium azide), and stained with mAb W6/32, which targets a monomorphic determinant of class I MHC of both humans and cattle (IZSLER Cell Bank code BS Hy 32) and an anti-SLA-AB mAb to a monomorphic determinant of class I SLA (code BS Hy 19), respectively. The secondary antibody used was fluorescein F(ab′)2 fragment of goat anti-mouse IgG, IgM (H+L) (Life Invitrogen).
PAMs were quickly thawed and seeded in 24-well plates (2 × 106 cells in 1 mL of MEM supplemented with 10% FBS and 2× antibiotic solution). After 24 h, fresh medium supplemented with 10 IU/mL of suIFN-α (final) was added and cells were incubated at 37°C in 5% CO2 for a further 18 h. Then, cells were harvested by gently pipetting and stained with mouse anti-pig SLA class II DR: FITC (AbD Serotec) or with buffer only (control). All incubations were performed at 4°C for 30 min. Cells were washed twice in FACS buffer between each step and before being analyzed by flow cytometry analyses (Guava EasyCyte HT flow cytometer, Millipore). Cells were gated on the basis of their forward and side scatter profiles, and readings were stopped after acquisition of 10,000 events. Cells displaying fluorescence intensities above the upper limit of the negative control distribution were considered positive. For tests performed on MDBK cells and PAMs, results were reported as the number of positive cells showing fluorescence intensity higher and lower than that of untreated cells. Results of immunomodulatory activity on PK15cl28 cells were instead reported as the total number of positive and negative cells with respect to the negative control of the assay.
Statistical analysis
A D'Agostino–Pearson omnibus test was conducted to check Gaussian distributions in the data sets relative to antiviral activity after logarithmic data transformation. Then, differences between data sets of suIFN-α subtypes and mock plasmid were checked by 1-way ANOVA for repeated measures, followed by a Newman–Keuls post hoc test. Differences in immunomodulatory activity between mock plasmid and each suIFN-α subtype were verified by the chi-square test in 2 × 2 contingency tables reporting prevalence of test-positive cells. The significance threshold was set at P < 0.05 (Prism 5, GraphPad Software). Concerning gene expression, after calculation of ΔCt, data sets failing the Kolmogorov–Smirnov test were checked for significant differences by the Kruskal–Wallis test, followed by Dunn's test. This same approach was adopted for the analysis of IL-8 protein release from IPEC-J2 cells. The significance threshold was set at P < 0.05 with a correction for multiple comparisons.
Results
Expression of PoIFN-α genes in mammalian cells
Expression of PoIFN-α genes in CHO cells was confirmed by the VSV cpe inhibition assay on MDBK cells. All the transfected CHO cell cultures produced recombinant IFN proteins (Table 2). The effect was specific because it was not present in the medium collected from cells transfected with a mock plasmid. suIFN-α5 and -α11 had the lowest antiviral titers. Since no difference was observed between suIFN-α titers in the medium collected at 24 and 48 h after transfection, the 2 suspensions of each IFN-α subtype were mixed together and further processed as 1 sample only. On the basis of the cpe inhibition assay on MDBK cells, a very low concentration (10 IU/mL) was adopted for all the subsequent tests on IFN-α subtypes.
Table 2.
suIFN-α Titers in the cpe Inhibition Assay
| Titers (IU/mL) | ||
|---|---|---|
| suIFN-α | 24 H | 48 H |
| 2 | 1,111 | 1,111 |
| 3 | 10,000 | 10,000 |
| 4 | 30,000 | 30,000 |
| 5 | 94 | 94 |
| 6 | 90,000 | 90,000 |
| 7 | 1,111 | 1,111 |
| 8 | 10,000 | 10,000 |
| 9 | 3,333 | 3,333 |
| 10 | 10,000 | 10,000 |
| 11 | 281 | 281 |
| 12 | 22,781 | 22,781 |
| 13 | 22,781 | 22,781 |
| 14 | 22,781 | 22,781 |
| 15 | 7,594 | 7,594 |
| 16 | 22,781 | 22,781 |
| 17 | 22,781 | 22,781 |
| rIFN-α1 | 10,000 | 10,000 |
| MOCK | <20 | <20 |
IFN, interferon.
Purification and titration of recombinant PoIFN-α subtypes
SuIFN-α3, -α4, and -α9 were purified through Ni-NTA spin columns as described in the Materials and Methods section. The final yields of PoIFN-α detectable in the elution buffer were generally lower than expected (∼20% with respect to the initial IFN-α concentration in terms of IU/mL, data not shown).
ELISA assay
The Swine IFN-α Do-It-Yourself ELISA kit showed limited cross-reactions under our test conditions. It revealed only suIFN-α2, suIFN-α5, and suIFN-α11 under the adopted test conditions (detection of 20 IU/mL). The lower detection limit of our ELISA was 5 IU/mL of rIFN-α1 as test standard (data not shown).
Antiviral activity
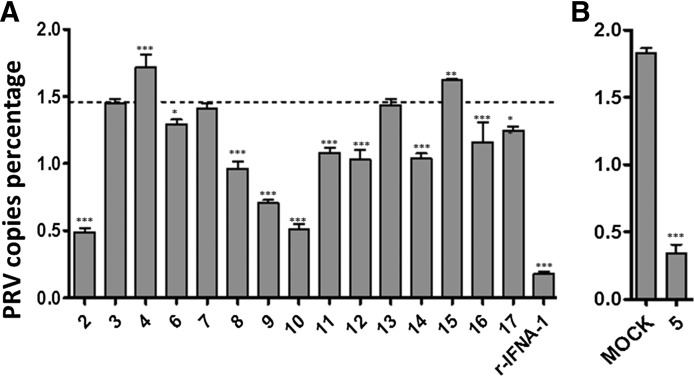
The antiviral activity of 10 IU/mL of each suIFN-α was investigated by a yield reduction assay on PRV using real-time PCR quantitation of viral copies in the supernatant of infected MDBK cells. Results are shown in Fig. 1A and B. To level out differences between different analytical sessions, all results were normalized against a control represented by the cell supernatant of PRV-infected MDBK cells not treated with IFN-α. Assuming the positive control as 100% value of PRV genome copies, all the results were expressed as a percentage of PRV copies. Numerical percentage data were log transformed and compared with results of mock supernatant-treated cells (indicated in Fig. 1A by a dashed line). Mock supernatant was used at the same dilution corresponding to 10 IU/mL of suIFN-α subtypes. At this concentration, all suIFN-α samples and mock displayed antiviral activity against PRV, some IFNs (-α2, -α5, -α9, -α10) showing a more pronounced one. Other subtypes caused instead little, if any, yield reduction with respect to the mock supernatant control. Interestingly, some suIFNs were able to reduce the PRV copy number to the same extent as mock (-α3, -α7, and -α13), while others had less antiviral activity than even the mock (-α4 and -α15, P < 0.001 and 0.01, respectively).
FIG. 1.
Antiviral activity of suIFN-α subtypes. (A, B) Antiviral activity in MDBK cells. Results are expressed as log10-transformed percentage of PRV copies (100% = 2), calculated with respect to the copy number obtained in untreated samples (PRV-infected cells in MEM without suIFN-α subtypes). Data are log10-transformed values and shown as mean ± 1 standard deviation. *P value < 0.05, **P < 0.01, and ***P < 0.001 with respect to mock supernatant. IFN, interferon; MEM, minimum essential medium; PRV, pseudorabies virus; MDBK, Madin–Darby bovine kidney.
Anti-inflammatory activity
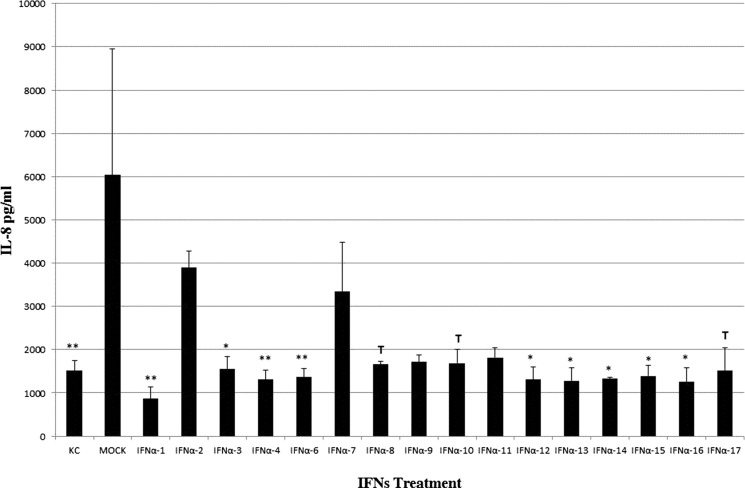
The anti-inflammatory activity of IFN-α subtypes was investigated by quantifying IL-8 release from IPEC-J2 cells treated for 18 h with 10 IU/mL of each subtype. With respect to the concentration observed in mock-treated cells (increased with respect to untreated controls), a significant reduction of IL-8 release or a tendency to significance was observed after addition of 10 IU/mL of most suIFN-α samples, with the exception of suIFNs-α2, α7, α9, and α11 (Fig. 2).
FIG. 2.
Modulation of interleukin-8 (IL-8) release after IFN-α treatments. Data are expressed as pg/mL ± 1 standard deviation and indicate the IFN-α effects on IL-8 release in IPEC-J2 cells. IPEC-J2 cells were treated with different types of IFNs at 10 IU/mL. Untreated cells (KC) and mock supernatant-treated cells were used as negative controls. After 18 h of incubation, supernatants were harvested and checked for IL-8 protein by ELISA. The asterisks indicate significant differences between mock and IFN-α treatments. *P < 0.05, **P < 0.01, and ***P < 0.001. T indicates a tendency to significance (P < 0.1).
Cytokine gene expression
Expression of cytokine genes in IPEC-J2 cells was investigated by RT real-time PCR. The following cytokines showed no significant differences in terms of gene expression after treatment with all suIFN-α subtypes: IL-6, IL-18, IL-4, SOCS1, and βD3 (Table 3).
Table 3.
Gene Expression in Terms of Ct Values in IPEC-J2 Cells After Treatments with Porcine suIFNs-α and rIFN-α1
| Cytokine expression | |||||
|---|---|---|---|---|---|
| Treatment | IL-4 | IL-6 | IL-18 | SOCS1 | bD3 |
| rIFN-α1a | 20.0 ± 0.0 | 6.7 ± 0.4 | 7.6 ± 0.1 | 17.4 ± 4.6 | 15.2 ± 0.7 |
| IFN-α2 | 20.0 ± 0.0 | 6.1 ± 0.9 | 9.7 ± 0.2 | 15.1 ± 4.3 | 17.0 ± 0.7 |
| IFN-α3 | 20.0 ± 0.0 | 4.6 ± 0.2 | 8.1 ± 0.4 | 15.5 ± 0.1 | 13.6 ± 0.7 |
| IFN-α4 | 20.0 ± 0.0 | 4.5 ± 0.4 | 7.6 ± 0.4 | 12.5 ± 0.1 | 14.4 ± 0.3 |
| IFN-α6 | 18.5 ± 2.6 | 6.2 ± 0.2 | 6.3 ± 0.1 | 18.8 ± 0.1 | 14.6 ± 0.7 |
| IFN-α7 | 18.8 ± 2.0 | 6.7 ± 0.1 | 6.0 ± 0.2 | 13.1 ± 0.1 | 15.3 ± 0.5 |
| IFN-α8 | 20.0 ± 0.0 | 6.7 ± 0.0 | 6.3 ± 0.1 | 13.1 ± 0.2 | 15.2 ± 0.6 |
| IFN-α9 | 20.0 ± 0.0 | 5.9 ± 0.7 | 7.2 ± 0.4 | 17.2 ± 5.0 | 12.9 ± 0.8 |
| IFN-α10 | 20.0 ± 0.0 | 5.2 ± 0.1 | 6.3 ± 0.0 | 10.8 ± 0.1 | 15.6 ± 3.8 |
| IFN-α11 | 20.0 ± 0.0 | 5.4 ± 0.1 | 6.0 ± 0.2 | 9.6 ± 0.9 | 17.7 ± 4.0 |
| IFN-α12 | 20.0 ± 0.0 | 4.8 ± 0.3 | 7.4 ± 0.1 | 11.8 ± 0.1 | 14.9 ± 1.0 |
| IFN-α13 | 20.0 ± 0.0 | 4.9 ± 0.2 | 7.8 ± 0.3 | 12.5 ± 1.6 | 15.3 ± 0.6 |
| IFN-α14 | 20.0 ± 0.0 | 6.0 ± 0.7 | 7.4 ± 0.2 | 11.9 ± 0.5 | 15.2 ± 0.3 |
| IFN-α15 | 20.0 ± 0.0 | 5.4 ± 0.3 | 6.8 ± 0.1 | 11.5 ± 0.3 | 14.6 ± 0.2 |
| IFN-α16 | 20.0 ± 0.0 | 5.3 ± 0.1 | 7.0 ± 0.1 | 11.4 ± 0.1 | 14.8 ± 0.6 |
| IFN-α17 | 20.0 ± 0.0 | 5.1 ± 0.2 | 7.4 ± 0.3 | 11.3 ± 0.1 | 16.4 ± 1.3 |
| Mock | 20.0 ± 0.0 | 5.5 ± 0.4 | 7.4 ± 1.9 | 11.7 ± 1.0 | 14.7 ± 1.8 |
| Untreated cells | 20.0 ± 0.0 | 5.3 ± 0.9 | 7.2 ± 7.4 | 12.2 ± 2.4 | 14.8 ± 1.9 |
Data are expressed as mean Ct ± 1 standard deviation.
Recombinant protein purchased from PBL Biomedical Laboratories.
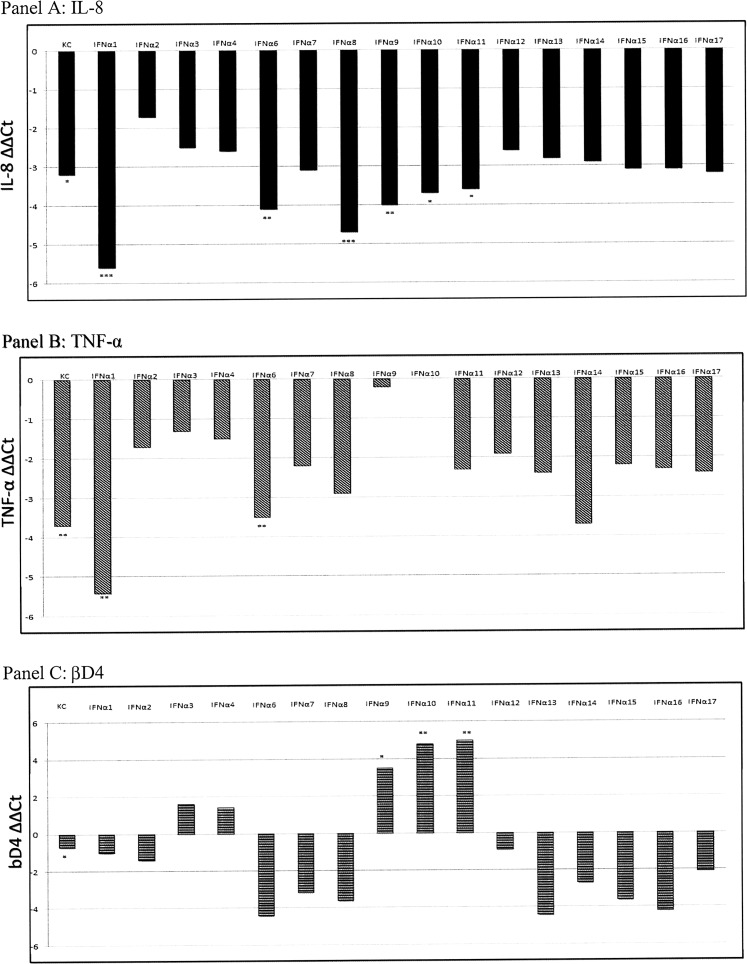
The addition of mock plasmid to cells produced an important modulation of the expression of some genes. Indeed, it caused a significant increase of βD4 (P = 0.016), TNF-α (P = 0.032), and IL-8 (P = 0.0147) gene expression with respect to untreated cells. Therefore, IL-8, TNF-α, and βD4 expression was normalized against mock-treated rather than untreated samples. Results are shown in Fig. 3. The addition of rIFN-α1 or suIFN-α6 determined a significant reduction of TNF-α (P = 0.0018 and P = 0.007, respectively) and IL-8 (P < 0.0001 and P = 0.0012, respectively) gene expression. Moreover, IL-8 was significantly downregulated by IFN-α8 (P = 0.0001), IFN-α9 (P = 0.0025), IFN-α10 (P = 0.01), and IFN-α11 (P = 0.014) with respect to the mock plasmid control. The expression of βD4 was upregulated after incubation of IPEC-J2 cells with suIFN-α9 (P = 0.0129), suIFN-α10 (P = 0.0028), or suIFN-α11 (P = 0.0028).
FIG. 3.
IL-8, TNF-α, and βD4 gene expression following suIFN-α treatments. IL-8, TNF-α, and βD4 gene expression was investigated in control untreated, suIFN-α, and mock plasmid-treated IPEC-J2 cells. Data are normalized on the values of mock plasmid-treated cells and expressed as ΔΔCt, where ΔΔCt = (ΔCt observed in mock-treated cells) − (ΔCt observed in IFN-treated cells or untreated cells). The asterisks indicate significant differences between values obtained from mock- and IFN-treated cells. *P < 0.05, **P < 0.01, and ***P < 0.001. TNF-α, tumor necrosis factor α.
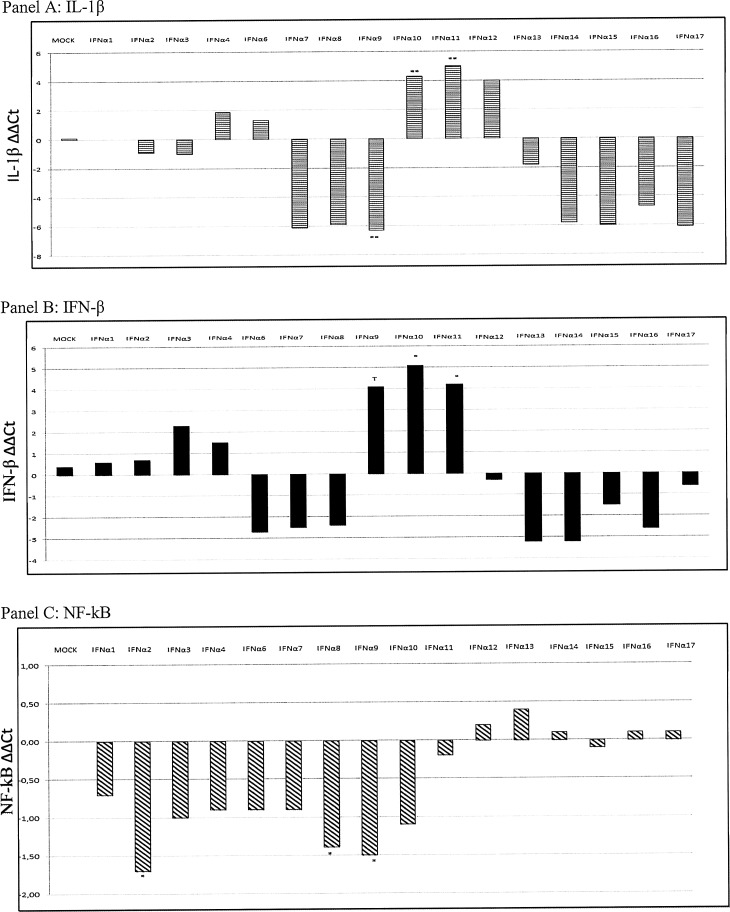
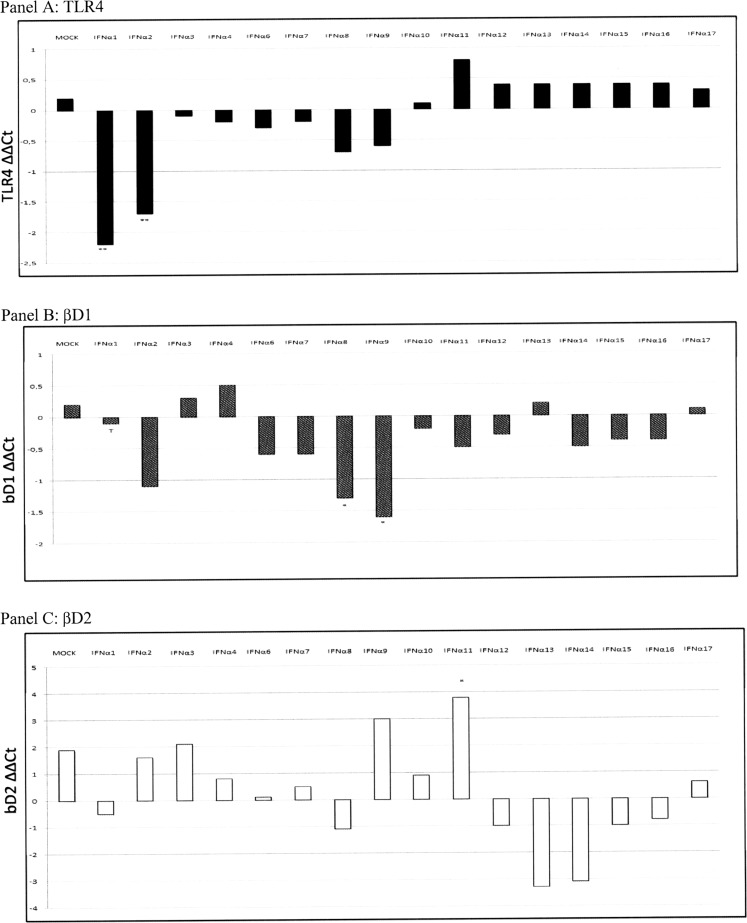
Concerning the effects of IFN-α treatments on the expression of other cytokines, we obtained the following results (Figs. 4 and 5): IFN-α1: TLR4 and βD1 gene expression was downregulated (P = 0.06 and P = 0.0123, respectively) after stimulation. IFN-α2: This IFN-α subtype significantly decreased the expression of TLR4 (P = 0.0366) and NF-kB (P = 0.02). IFN-α8: It downregulated βD1 (P = 0.043) and NF-kB (P = 0.04) gene expression. IFN-α9: Treatment with this IFN modulated cytokine gene expression, causing an increase of IFN-β (P = 0.09, tendency) and a downregulation of NF-kB (P = 0.049), IL-1β (P = 0.0018), and βD1 (P = 0.014). IFN-α10: IL-1β (P = 0.0018) and IFN-β (P = 0.012) gene expression was upregulated after IFN treatment; the same effect was observed after IFN-α11 treatment (IL-1β, P = 0.0018 and IFN-β, P = 0.046); also βD2 gene expression showed a significant increase (P = 0.014).
FIG. 4.
IL-1β, IFN-β, and NF-kB gene expression following IFN-α treatments. IL-1β, IFN-β, and NF-kB gene expression was investigated in control untreated, suIFN-α, and mock plasmid-treated IPEC-J2 cells. Data are normalized on the values of control, untreated cells, and expressed as ΔΔCt, where ΔΔCt = (ΔCt observed in untreated cells) − (ΔCt observed in IFN- or mock-treated cells). The asterisks indicate significant differences between values obtained from untreated and IFN-treated cells. *P value < 0.05, **P < 0.01, and ***P < 0.001. T indicates a tendency to significance (P < 0.1). NF-kB, nuclear factor-κB.
FIG. 5.
TLR4, βD1, and βD2 gene expression following IFN-α treatments. TLR4, βD1, and βD2 gene expression was investigated in control untreated, suIFN-α, and mock plasmid-treated IPEC-J2 cells. Data are normalized on the values of control, untreated cells, and expressed as ΔΔCt, where ΔΔCt = (ΔCt observed in untreated cells) − (ΔCt observed in IFN- or mock-treated cells). The asterisks indicate significant differences between values obtained from untreated and IFN-treated cells. *P value < 0.05, **P < 0.01, and ***P < 0.001. T indicates a tendency to significance (P < 0.1). TLR4, Toll-like receptor 4.
Immunomodulatory activity
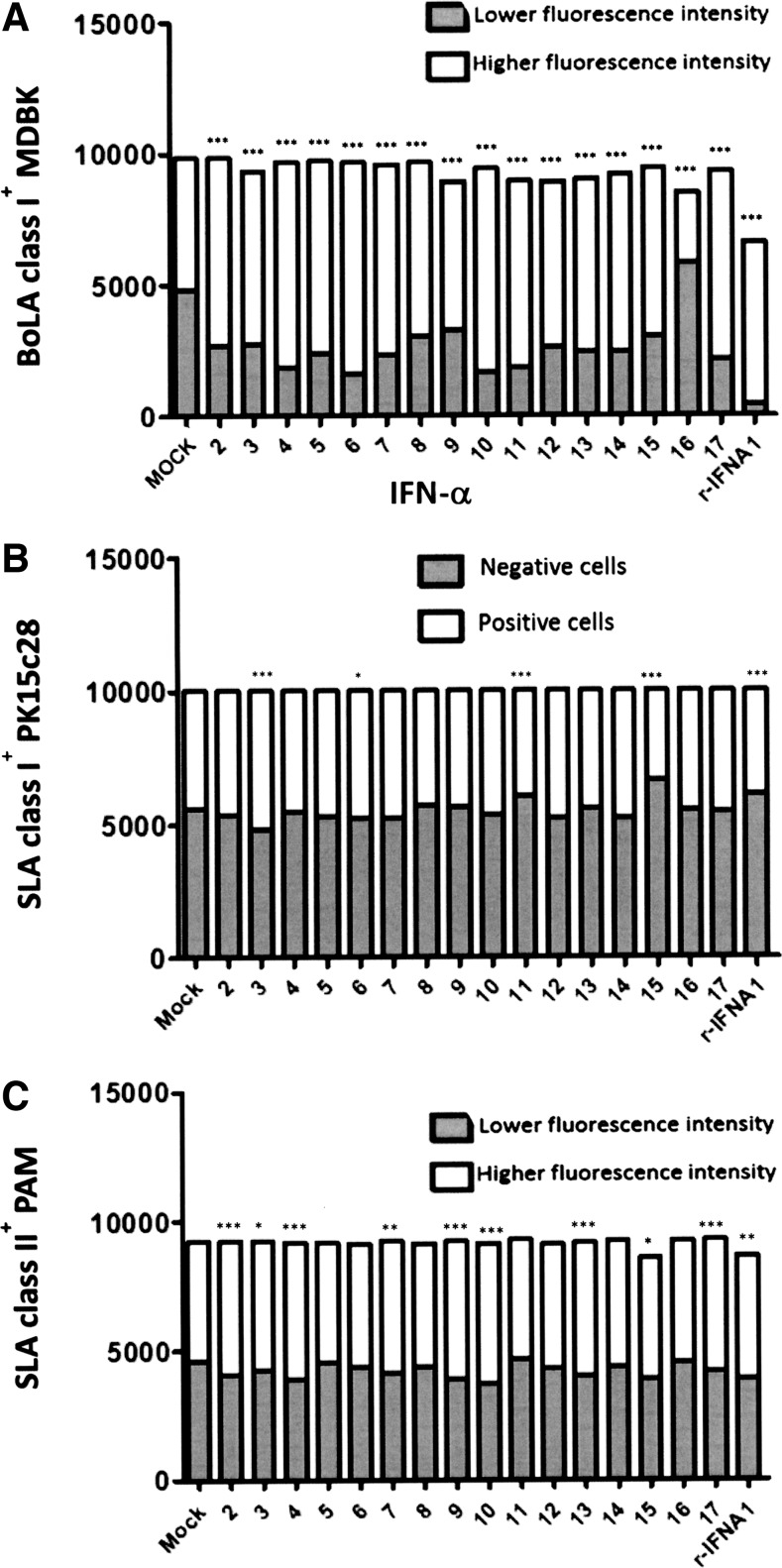
The immunoregulatory properties of the different suIFN-α subtypes under study were investigated in terms of MHC I and MHC II expression in 3 different cellular substrates. Expression of MHC I antigens was measured in MDBK and PK15c28 cells after an 18-h incubation with 10 IU/mL of IFN-α subtypes. All suIFN-α subtypes caused a significant increase in the expression of MHC class I antigens in MDBK cells compared with cells treated with mock supernatant, with little fluctuations between different subtypes. The only exception was IFN-α16, displaying an important suppression of MHC I expression with respect to the mock supernatant control level (Fig. 6A, P < 0.001).
FIG. 6.
Modulation of class I and II MHC expression by IFN-α subtypes. Expression of BoLA class I MHC in MDBK (A), SLA class I in PK15c28 cells (B), and SLA class II in PAMs (C) after treatment with suIFN-α subtypes. (A, C) Data are reported as number of positive cells, showing higher (white bars) and lower (dark bars) fluorescence intensities than those of untreated control cells. (B) Data are reported as number of SLA class I-positive (white bars) and negative cells (dark bars). Asterisks represent significant differences. *P value < 0.05, **P < 0.01, and ***P < 0.001. PAMs, pulmonary alveolar macrophages.
In contrast, modulation of class I MHC antigens in PK15cl28 cells in response to 10 IU/mL suIFN was less pronounced. Indeed, the difference in the number of MHC I-positive cells was statistically significant only for suIFN-α3 (P < 0.001), -α6 (P < 0.05), -α11, and -α15 (P < 0.001) subtypes. In particular, suIFN-α11 and -α15 reduced the expression of SLA I molecules as also rIFN-α1 did as opposed to all the other IFNs under study (Fig. 6B).
Modulation of SLA class II antigens by suIFNs was investigated in PAMs and results are reported in Fig. 6C. Most suIFNs caused a significant increase of SLA II expression (suIFN-α2, -α4, -α9, -α10, -α13, and -α17, P < 0.001; suIFN-α7, P < 0.01; suIFN-α3 and-α15, P < 0.05), as also shown for rIFN-α1 (P < 0.01).
Biological activities of purified IFN-α
Biological activities (antiviral, anti-inflammatory, and immunomodulatory) exerted by 10 IU/mL of purified PoIFN-α3, -α4, and -α9 and the respective suIFN-α subtypes were compared. These subtypes were selected as representative of the biological activities of the PoIFNs-α under study. Results are summarized in Table 4. The antiviral activity was similar for the supernatant and purified α9 subtype, while purified IFN-α4 showed a more pronounced activity against PRV infection than the nonpurified counterpart. The discrepancy about the antiviral activity of IFN-α4 was not due to a spurious effect of the imidazole-containing elution buffer of purified IFN-α (data not shown). Purified α3 had a lower ability in reducing IL-8 release from IPEC-J2 cells than the nonpurified subtype, while in contrast, purified α9 increased release. Concerning immunomodulatory properties, all purified and nonpurified subtypes caused similar changes in surface MHC expression, except for SLA class I expression on PK15cl28 cells that became undetectable after treatment with purified subtypes, as opposed to the nonpurified ones. This latter result was clearly nonspecific since it could be reproduced by the elution buffer from His-Tag columns at the same working dilution used with purified IFNs-α (data not shown).
Table 4.
Biological Activities of Nonpurified and Purified IFN-α Subtypes
| suIFN-α3 | IFN-α3 | suIFN-α4 | IFN-α4 | suIFN-α9 | IFN-α9 | |
|---|---|---|---|---|---|---|
| Antiviral activity (%)a | 28.0 | 13.7 | 52.6 | 7.4 | 5.1 | 6.5 |
| Anti-inflammatory activity (%)b | 128.8 | 100.0 | 121.2 | 119.4 | 86.1 | 113.0 |
| HLA class I expression (%)c | 70.6 | 51.8 | 80.9 | 75.8 | 62.94 | 62.4 |
| SLA class I expression (%)d | 51.5 | Neg | 45.4 | Neg | 43.37 | Neg |
| SLA class II expression (%)c | 54.1 | 50.9 | 57.3 | 44.3 | 57.47 | 46.6 |
Results are expressed as mean percentage of PRV copies with respect to the copy number measured in the untreated control sample (PRV-infected cells without any IFN-α subtype).
Results are expressed as mean percentage of IL-8 concentration (pg/mL) with respect to the level obtained in untreated control IPEC-J2 cells.
Results are expressed as mean percentage of positive cell fluorescence intensity values higher than the median of mock-treated control cells.
Results are expressed as mean percentage of SLA class I-positive cells.
PRV, pseudorabies virus.
Discussion
This study aimed to characterize porcine IFN-α subtypes by evaluating their antiviral, immunomodulatory, and anti-inflammatory activities under controlled in vitro conditions. A summary of our main findings is shown in Table 5, which refers to 16 porcine IFN-α subtypes expressed in mammalian cells and checked at a very low cytokine concentration (10 IU/mL). This was selected as a convenient, internationally recognized test parameter of type I IFNs, amenable to standardized and validated assay profiles in cell cultures of different species. Yet, the authors are aware of different antiviral activities of IFN-α subtypes on a mass basis (Zoon and others 1992), leading to different IFN protein concentrations corresponding to the same level of biological activity. This could have affected our screening in ELISA of the different IFN-α subtypes. In addition, it should be stressed that binding of mAb to different IFN-α subtypes does not imply the same final biological effect. In particular, the results of neutralization assays reveal that IFN-α mAb significantly differs in its ability to neutralize the individual IFN-α species (Viscomi and others 1999).
Table 5.
Summary Table of Porcine IFN-α Subtypes
| Cytokine gene expression | MHC expression modulation | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| IFN-α | Antiviral activity | Anti-inflammatory activity | ↑ | ↓ | BoLA class I | SLA class I | SLA class II | |||
| 2 | Very high | None | TLR4, NF-kB | 72.87 | ↑ | 46.57 | 56.20 | ↑ | ||
| 3 | Low | High | 70.57 | ↑ | 51.52 | ↑ | 54.09 | ↑ | ||
| 4 | None | Very high | 80.88 | ↑ | 45.43 | 57.35 | ↑ | |||
| 5 | Very high | Very high | 75.51 | ↑ | 47.33 | 50.40 | ||||
| 6 | High | Very high | TNF-α, IL-8 | 83.11 | ↑ | 47.81 | ↑ | 51.85 | ||
| 7 | Low | None | 75.51 | ↑ | 47.54 | 55.09 | ↑ | |||
| 8 | High | Low | IL-8, βD1, NF-kB | 68.45 | ↑ | 42.68 | 52.07 | |||
| 9 | Very high | None | βD4, IFN-β | IL-8, NF-kB, IL-1β, βD1 | 62.94 | ↑ | 43.37 | 57.47 | ↑ | |
| 10 | Very high | Low | βD4, IFN-β, IL-1β | IL-8 | 81.85 | ↑ | 46.52 | 58.94 | ↑ | |
| 11 | High | None | βD4, IFN-β, IL-1β, βD2 | IL-8 | 78.92 | ↑ | 39.17 | ↓ | 49.50 | |
| 12 | High | High | 70.41 | ↑ | 47.59 | 52.84 | ||||
| 13 | Low | High | 72.66 | ↑ | 44.01 | 56.15 | ↑ | |||
| 14 | High | High | 73.53 | ↑ | 47.42 | 52.73 | ||||
| 15 | None | High | 67.97 | ↑ | 33.6 | ↓ | 54.63 | ↑ | ||
| 16 | High | High | 31.21 | ↓ | 44.47 | 50.91 | ||||
| 17 | High | Low | 76.98 | ↑ | 45.03 | 54.89 | ↑ | |||
| r-IFNα-1 | Very high | Very high | TLR4, βD1, TNF-α, IL-8 | 93.64 | ↑ | 38.47 | ↓ | 54.93 | ↑ | |
Main differential activities of porcine IFN-α subtypes. Antiviral activity: results are expressed as arbitrary score on the basis of the yield reduction assay of pseudorabies virus. Anti-inflammatory activity: results are expressed on the basis of statistical significance in the IL-8 yield reduction assay (very high, P < 0.01; high, P < 0.05; low, P < 0.1; none, P > 0.1). Cytokine gene expression: ↑ , upregulation; ↓ downregulation (significant effects, P < 0.05). MHC expression: results are expressed in terms of % positive cells, followed by ↑ or ↓ in case of significant differences (P < 0.05).
While expression of IFN-α subtypes was successful, the purification step through His-Tag columns yielded a limited recovery of the expressed recombinant proteins, despite extensive checks and adjustments of the usual purification procedures. This despicable outcome might be due to the conformational constraints of native recombinant IFN-α proteins in this purification system based on nickel binding to histidine residues. In particular, the conformational folding of IFN-α due to its 2 intrachain S-S bonds (Viscomi 1997) might be a hindrance to an effective recognition of the histidine tag. On the whole, the purification of 3 subtypes only was performed with the main aim to reveal possible discrepancies between biological activities of purified versus nonpurified IFN-α subtypes to be considered for a meaningful comparison of test results in different laboratories.
Owing to the above, a screening was performed first on IFN-containing supernatants calibrated in terms of IU/mL. A transfection plasmid with no insert (mock plasmid) was used to discriminate between IFN-specific and nonspecific effects due to the possible pattern recognition receptor ligand properties of plasmid DNA. Supernatants from transfections with this mock plasmid had activity in the PRV yield reduction assay and caused an upregulation of IL-8 release and an increased expression of IL-8, TNF-α, and BD4 genes compared with media alone. These effects can be attributed to plasmid DNA as possible pathogen-associated molecular pattern and/or to soluble factors released by CHO cells after mock transfection.
According to our working hypothesis, we set up screening protocols based on a very low IFN-α concentration to highlight even minor differences among subtypes. The VSV/MDBK cell system had been previously characterized as a very sensitive and robust titration system for porcine IFN-α from in vivo and in vitro sources (Razzuoli and others 2011a). This is the reason why it was adopted for the quantification of the antiviral activity on the basis of an international standard. Sang and others (2010) tested in a previous study the antiviral activity of some IFN-α subtypes against porcine reproductive and respiratory syndrome arterivirus and VSV by a cpe inhibition assay. Our assay of antiviral activity differed for the adopted virus, a DNA virus as opposed to RNA, and the methodology (yield reduction evaluated by real-time PCR). Yet, despite the different analytical methods and target virus, we also found that IFN-α2 and IFN-α10 had the greatest antiviral activity among IFN-α subtypes in our PRV model (Fig. 1). Nevertheless, we cannot rule out the possibility that our results could be peculiar to the adopted virus model. Herpesviruses do not usually show a very high sensitivity to IFNs due to complex IFN evasion strategies (Mossman and Ashkar 2005), and different IFN-α subtypes might be more or less susceptible to such evasion strategies.
Our results in vitro identified different levels of anti-inflammatory control activity of porcine IFN-α subtypes in the IL-8 model. Some cytokines such as IL-8 and IL-6 showed a very high level of gene expression in IPEC-J2 cells, in agreement with our previous data (Razzuoli and others 2013, 2014). In addition, IPEC-J2 had been previously characterized for its ability to release and express IL-8 and other cytokines (Mariani and others 2009). In this respect, significant anti-inflammatory control activities or tendencies at both gene and protein levels were shown for IFN-α1, α6, α8, and α10, whereas other subtypes provided significant differences in terms of either protein or gene expression only (Figs. 2 and 3). This discrepancy might be due to the different levels of test variability in the 2 assays and/or the multiple control levels for IL-8 expression. Thus, a sustained IL-8 response can be mounted if the gene promoter is derepressed, NF-kB and JNK pathways are activated, and the resulting mRNA is stabilized through the p38 MAPK pathway (Hoffmann and others 2002). IFNs-α can antagonize this latter pathway by inducing negative regulators of mRNA stability such as tristetrapolin, targeting adenylate-uridylate-rich elements at the 3′ untranslated region of mRNA (Sauer and others 2006). Regardless of the actual targets of IFN-α-driven control actions, our findings confirm that minute concentrations of IFN-α can exert potent anti-inflammatory control actions aimed at preventing major tissue damage and waste of metabolic energy. Interestingly, the same control actions can be also triggered by IFN-α-treated lymphoid cells (Razzuoli and others 2014).
Porcine IFN-α subtypes showed a generalized stimulation of BoLA class I MHC expression with the important exception of IFN-α16 showing instead a significant inhibitory action. This was also shown for IFN-α1, α11, andα15 in porcine PK-15c28 cells. To our knowledge, this is the first report of an inhibitory control on MHC expression of type I IFN molecules. In the authors' opinion, such a peculiar feature can underlie a fine-tuning mechanism in the regulatory control actions associated with constitutive, low-titered type I IFN responses. Thus, upregulation of MHC I expression could be better regulated and adjusted vis-à-vis the actual needs of the host to promote either presentation of peptides through MHC I or NK cell activity through downregulation of MHC I (Raulet 2006). Interestingly, some IFN-α subtypes (including α16) showed no modulation of MHC class II expression (Fig. 6). The absence of an inhibitory activity on MHC II expression probably outlines a less stringent need for control over levels of expression. On the whole, the adopted methodology proved to be useful for characterizing the activity of different subtypes in vitro as both raw and purified proteins. The differences observed between nonpurified and purified IFN subtypes in terms of SLA class I expression on PK15cl28cells (Fig. 6) were shown to be caused by the imidazole-containing medium (elution buffer of the His-tag columns). Interestingly, imidazole derivatives such as imidazoquinolinamines show immunomodulatory properties and antiviral and antitumor activity through endogenous cytokine production (Dockrell and Kinghorn 2001).
As stressed in a recent review article (Davidson and others 2015), diverging activities of IFNs can be accounted for by the wide spectrum in IFN half-life and ligand affinity to both receptor subunits. Furthermore, whereas an antiviral state can be rapidly induced after binding to a low number of receptors, type I IFN immunomodulatory functions demand prolonged signaling and high receptor concentration on the cell surface.
The characterization of the main biological properties of porcine IFN-α subtypes should be offset against the observed profiles of constitutive expression in peripheral blood mononuclear cells (PBMCs) of healthy pigs. The evidence accumulated so far in the pig model indicates that constitutive expression varies in terms of both intensity and panel of involved IFN-α subtypes, in the framework of a constant expression of the IFN-β gene (Razzuoli and others 2011b), and intracellular accumulation of IFN-α molecules in a peculiar dimeric form (Amadori and others 2010) previously shown in human PBMCs too (Greenway and others 1995).
The subtypes included in this study showed widely divergent biological properties. While the standard recombinant IFN-α1 protein used cannot be properly compared because of its different expression and purification procedure, the results obtained on the other subtypes outline a balanced representation of IFNs-α endued with different prevailing properties. In this respect, constitutive expression (Razzuoli and others 2011b) would not be strictly related to antiviral defense only. This is clearly shown by the frequent involvement of IFN-α7, showing no detectable antiviral activity in our test system and also in a previous study (Sang and others 2010), as opposed instead to the potent modulation of MHC expression (Fig. 6). A prompt reaction to viral agents would be anyhow promoted by the steady-state expression of interferon regulatory factor 7 in lymphoid cells, sustained by low-level production of IFN-α proteins (Taniguchi and Takaoka 2001).
The frequent involvement in constitutive expression of IFN-β10 and α11 would be conducive to a prompt early release of βD4 (Fig. 3) and possibly other antimicrobial peptides in the framework of an effective antibacterial defense strategy. This is in line with the strong antibacterial activity of human lymphoblastoid IFN α-treated pig tonsil cells (Razzuoli and others 2012). Such a system obviously needs a tight control since antimicrobial peptides can stimulate epithelial and inflammatory cells, cell proliferation, cytokine/chemokine production, and chemotaxis (Niyonsaba and Ogawa 2005). This is probably the reason why the expression of porcine βD1 is strongly inhibited by human IFNs-α (Razzuoli and others 2014). Beyond such a caveat, our results indicate a possible prophylactic and/or therapeutic modulation of antibacterial peptides. This could be induced by selected IFN-α subtypes for either parenteral or low-dose oral treatments (Tompkins 1999), the rationale behind oral treatments being the release of second messengers from IFN-treated tonsil cells (Razzuoli and others 2014).
On the whole, our results can be of some importance in terms of both fundamental and applied research. The characterization of the porcine IFN-α subtypes involved in constitutive and stressor-driven responses could contribute to the understanding of fundamental immune mechanism of pigs. In addition, the roles of porcine type I IFNs in the response to both infectious and noninfectious stressors and in fundamental homeostatic control circuits could be conducive to new interesting advances in human diseases, having in mind the recognized advantages of pig with respect to current laboratory rodent models (Meurens and others 2012).
Acknowledgments
The authors thank C. Mantovani and D. Bonassi (IZSLER, Brescia, Italy) for their skillful technical assistance. The Italian Ministry of Health funded this study with the grant, PRC2011001. Dr. H.C. and O.S.'s contributions were funded by the DEFRA project, SE0795.
Author Disclosure Statement
No competing financial interests exist.
References
- Amadori M. 2007. The role of IFN-alpha as homeostatic agent in the inflammatory response: a balance between danger and response? J Interferon Cytokine Res 27(3):181–189 [DOI] [PubMed] [Google Scholar]
- Amadori M, Cristiano A, Ferrari M. 2010. Constitutive expression of interferons in swine leukocytes. Res Vet Sci 88(1):64–71 [DOI] [PubMed] [Google Scholar]
- Amadori M, Farinacci M, Begni B, Faita R, Podavini D, Colitti M. 2009. Effects of interferon-alpha on the inflammatory response of swine peripheral blood mononuclear cells. J Interferon Cytokine Res 29(4):241–247 [DOI] [PubMed] [Google Scholar]
- Collado-Romero M, Arce C, Ramirez-Boo M, Carvajal A, Garrido JJ. 2010. Quantitative analysis of the immune response upon salmonella typhimurium infection along the porcine intestinal gut. Vet Res 41(2):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson S, Maini MK, and Wack A. 2015. Disease-promoting effects of type I interferons in viral, bacterial, and coinfections. J Interferon Cytokine Res 35(4):252–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dockrell DH, Kinghorn GR. 2001. Imiquimod and resiquimod as novel immunomodulators. J Antimicrob Chemother 48(6):751–755 [DOI] [PubMed] [Google Scholar]
- Greenway AL, Hertzog PJ, Devenish RJ, Linnane AW. 1995. Constitutive and virus-induced interferon production by peripheral blood leukocytes. Exp Hematol 23(3):229–235 [PubMed] [Google Scholar]
- Hoffmann E, Dittrich-Breiholz O, Holtmann H, Kracht M. 2002. Multiple control of interleukin-8 gene expression. J Leukoc Biol 72(5):847–855 [PubMed] [Google Scholar]
- Isaacs A, Lindenmann J. 1957. Virus interference. I. The interferon. Proc R Soc Lond B Biol Sci 147(927):258–267 [DOI] [PubMed] [Google Scholar]
- Kondrashov FA. 2012. Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc Biol Sci 279(1749):5048–5057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani V, Palermo S, Fiorentini S, Lanubile A, Giuffra E. 2009. Gene expression study of two widely used pig intestinal epithelial cell lines: IPEC-J2 and IPI-2I. Vet Immunol Immunopathol 131(3–4):278–284 [DOI] [PubMed] [Google Scholar]
- Meager A. 1987. Quantification of interferons by anti-viral assays and their standardization. In: Clemens MJ, Morris AG, Gearing AJH, eds. Lymphokines and interferons, a practical approach. Oxford, UK: IRL Press Limited; pp 129–147 [Google Scholar]
- Meurens F, Summerfield A, Nauwynck H, Saif L, Gerdts V. 2012. The pig: a model for human infectious diseases. Trends Microbiol 20(1):50–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossman KL, Ashkar AA. 2005. Herpesviruses and the innate immune response. Viral Immunol 18(2):267–281 [DOI] [PubMed] [Google Scholar]
- Niyonsaba F, Ogawa H. 2005. Protective roles of the skin against infection: implication of naturally occurring human antimicrobial agents beta-defensins, cathelicidin LL-37 and lysozyme. J Dermatol Sci 40(3):157–168 [DOI] [PubMed] [Google Scholar]
- Raulet DH. 2006. Missing self recognition and self tolerance of natural killer (NK) cells. Semin Immunol 18(3):145–150 [DOI] [PubMed] [Google Scholar]
- Razzuoli E, Dotti S, Archetti IL, Amadori M. 2010. Clinical chemistry parameters of piglets at weaning are modulated by an oral, low-dose interferon-alpha treatment. Vet Res Commun 34 (Suppl 1):S189–S192 [DOI] [PubMed] [Google Scholar]
- Razzuoli E, Faggionato E, Dotti S, Villa R, Lombardo T, Boizza L, Ferrari M, Amadori M. 2012. Isolation and culture of pig tonsil lymphocytes. Vet Immunol Immunopathol 148(3–4):320–325 [DOI] [PubMed] [Google Scholar]
- Razzuoli E, Villa R, Amadori M. 2013. IPEC-J2 cells as reporter system of the anti-inflammatory control actions of interferon-alpha. J Interferon Cytokine Res 33(10):597–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razzuoli E, Villa R, Ferrari A, Amadori M. 2014. A pig tonsil cell culture model for evaluating oral, low-dose IFN-alpha treatments. Vet Immunol Immunopathol 160(3–4):244–254 [DOI] [PubMed] [Google Scholar]
- Razzuoli E, Villa R, Sossi E, Amadori M. 2011a. Characterization of the interferon-alpha response of pigs to the weaning stress. J Interferon Cytokine Res 31(2):237–247 [DOI] [PubMed] [Google Scholar]
- Razzuoli E, Villa R, Sossi E, Amadori M. 2011b. Reverse transcription real-time PCR for detection of porcine interferon alpha and beta genes. Scand J Immunol 74(4):412–418 [DOI] [PubMed] [Google Scholar]
- Ross JW, Ashworth MD, Mathew D, Reagan P, Ritchey JW, Hayashi K, Spencer TE, Lucy M, Geisert RD. 2010. Activation of the transcription factor, nuclear factor kappa-B, during the estrous cycle and early pregnancy in the pig. Reprod Biol Endocrinol 8:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang Y, Bergkamp J, Blecha F. 2014. Molecular evolution of the porcine type I interferon family: subtype-specific expression and antiviral activity. PLoS One 9(11):e112378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang Y, Rowland RR, Hesse RA, Blecha F. 2010. Differential expression and activity of the porcine type I interferon family. Physiol Genomics 42(2):248–258 [DOI] [PubMed] [Google Scholar]
- Sauer I, Schaljo B, Vogl C, Gattermeier I, Kolbe T, Muller M, Blackshear PJ, Kovarik P. 2006. Interferons limit inflammatory responses by induction of tristetraprolin. Blood 107(12):4790–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soule O. 2013. Immunomodulatory intervention to control classical swine fever. PhD Thesis. University of Surrey, United Kingdom [Google Scholar]
- Takaoka A, Yanai H. 2006. Interferon signalling network in innate defence. Cell Microbiol 8(6):907–922 [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Takaoka A. 2001. A weak signal for strong responses: interferon-alpha/beta revisited. Nat Rev Mol Cell Biol 2(5):378–386 [DOI] [PubMed] [Google Scholar]
- Tompkins WA. 1999. Immunomodulation and therapeutic effects of the oral use of interferon-alpha: mechanism of action. J Interferon Cytokine Res 19(8):817–828 [DOI] [PubMed] [Google Scholar]
- van Pesch V, Lanaya H, Renauld JC, Michiels T. 2004. Characterization of the murine alpha interferon gene family. J Virol 78(15):8219–8228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhuizen EJ, Koomen I, Ultee T, van Dijk A, Haagsman HP. 2009. Salmonella serovar specific upregulation of porcine defensins 1 and 2 in a jejunal epithelial cell line. Vet Microbiol 136(1–2):69–75 [DOI] [PubMed] [Google Scholar]
- Viscomi GC. 1997. Structure-activity of type I interferons. Biotherapy 10(1):59–86 [DOI] [PubMed] [Google Scholar]
- Viscomi GC, Antonelli G, Bruno C, Scapol L, Malavasi F, Funaro A, Simeoni E, Pestka S, De Pisa F, Dianzani F. 1999. Antigenic characterization of recombinant, lymphoblastoid, and leukocyte IFN-alpha by monoclonal antibodies. J Interferon Cytokine Res 19(4):319–326 [DOI] [PubMed] [Google Scholar]
- Zoon KC, Miller D, Bekisz J, zur Nedden D, Enterline JC, Nguyen NY, Hu RQ. 1992. Purification and characterization of multiple components of human lymphoblastoid interferon-alpha. J Biol Chem 267(21):15210–15216 [PubMed] [Google Scholar]