Abstract
Microbial culturomics is a new field of omics sciences that examines the bacterial diversity of human gut coupled with a taxono-genomic strategy. Using microbial culturomics, we report here for the first time a novel Gram negative, catalase- and oxidase-negative, strict anaerobic bacilli named Beduini massiliensis gen. nov., sp nov. strain GM1 (= CSUR P1440 = DSM 100188), isolated from the stools of a female nomadic Bedouin from Saudi Arabia. With a length of 2,850,586 bp, the Beduini massiliensis genome exhibits a G + C content of 35.9%, and contains 2819 genes (2744 protein-coding and 75 RNA genes including 57 tRNA and 18 rRNA genes). It is composed of 6 scaffolds (composed of 6 contigs). A total of 1859 genes (67.75%) were assigned a putative function (by COGs or by NR blast). At least 1457 (53%) orthologous proteins were not shared with the closest phylogenetic species. 274 genes (10.0%) were identified as ORFans. These results show that microbial culturomics can dramatically improve the characterization of the human microbiota repertoire, deciphering new bacterial species and new genes. Further studies will clarify the geographic specificity and the putative role of these new microbes and their related functional genetic content in health and disease. Microbial culturomics is an emerging frontier of omics systems sciences and integrative biology and thus, warrants further consideration as part of the postgenomics methodology toolbox.
Introduction
Understanding microbial diversity is at the epicenter of the postgenomics research agenda. The diversity of microbial environment is often characterized by metagenomics and transcriptomics. However, DNA and/or RNA sequencing, even using the most recent chemistry and technology, cannot fully capture the unit of the microbial environment, the microbe itself. Therefore, we have recently proposed a comprehensive approach named “microbial culturomics” aimed at isolating and obtaining pure culture of all living microbes in the microenvironment (Lagier et al., 2012a).
In the initial culturomics study, more than 200 culture conditions were tested and enabled the culture of 340 bacterial species (Lagier et al., 2012a). Subsequently, the number of culture conditions was reduced to the 18 most efficient conditions, improving the cost-effectiveness of this approach (Lagier et al., 2015).
In an effort to better characterize, understand, and intervene on the microbial universe, in addition to the progress made in isolation by culture (Lagier et al., 2015) and identification by mass spectrometry (Seng et al., 2010) and 16S rRNA sequencing, we have proposed a new system of classification and nomenclature named taxono-genomics (Fournier et al., 2015; Ramasamy et al., 2014). Indeed the combination of phylogenetic studies and genotypic characteristics, including 16S rRNA gene sequence similarity (Stackebrandt and Ebers, 2006; Tindall et al., 2010), genomic G + C content diversity, and DNA–DNA hybridization allows the current taxonomic classification of prokaryotes (Rossello-Mora, 2006; Wayne et al.,1987).
Unfortunately, a number of important flaws can be identified in these methods, mainly due to threshold values that are not applicable to all species or genera (Welker and Moore, 2011). The development of high-throughput sequencing techniques permits the availability of a high number of bacterial genome sequences in public databases (Ramasamy et al., 2014). A taxono-genomic strategy was recently developed and is based on a systematic comparison of genomic and phenotypic characteristics, especially based on the Matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) spectrum, with the phylogenetically-closest species identified in the databases (Kokcha et al., 2012; Lagier et al., 2012b; 2012c, Mishra et al., 2012a, 2012b; 2012c; 2013).
The utilization of taxono-genomics in culturomics studies improved the routine laboratory identification of current or new species or genera of bacteria in human stool (Lagier et al., 2012a). Here, we present a summary of the classification and features of B. massiliensis gen. nov., sp. nov. strain GM1 (= CSUR P1440 = DSM 100188), including a description of the complete genome sequencing and annotation.
Materials and Methods
Ethics and sample collection
This study was performed in Saudi Arabia after approval from the Ethical Committee of the King Abdulaziz University (Saudi Arabia), and the local ethic committee of the IFR48 (Marseille, France) under the numbers 014-CEGMR-2-ETH-P and 09-022, respectively. After informed consent, a stool specimen was collected on May 28, 2014 from a healthy female nomadic Bedouin donor from the Asir region in the southwestern highland of Saudi Arabia. The donor was a 42-year-old woman, who was not fasting (body mass index = 20.8), and did not have confounding health states such as pregnancy, diabetes, obesity, or gastrointestinal disorder. Rice with chicken or meat were her main food. People living in the Asir region commonly use raw milk and fermented food such as khamir (fermented bread) and laban (a type of fermented milk) without alcohol or tobacco. Fruit and vegetable consumption is very rare in the daily menu per week. Food habits and the mountainous environment have probably affected the gut microbiome of the Bedouin people. Stored at −80°C, the stool sample was sent to URMITE, Marseille, France.
Microbial culturomics
To decipher the microbial diversity of the Beduin gut microbiota by microbial culturomics, we used 6 of the 18 most efficient culture conditions summarized in Supplementary Table S1 (supplementary material is available online at www.liebertpub.com/omi) and identified in a previous work (Lagier et al., 2015). Using these six conditions, a total of 53 distinct bacterial species were identified among a total of 11,223 tested colonies (Supplementary Table S2). These bacteria are distributed in four main phyla: 72.2% Firmicutes (39/53), 16.6% Actinobacteria (9), 5.5% Proteobacteria (3), and 3.7% Bacteroidetes (2). Other 12 culture conditions are ongoing. Strain GM1 was obtained only by stool pre-incubation in 5% sheep blood broth and then 5% sheep blood agar under anaerobic conditions at 28°C (BioMerieux, Marcy l'Etoile, France) for 30 days.
Comparing our results (obtained from only six culture conditions) to previously published results from two healthy humans (Lagier et al., 2012a), we found 17 species shared by all three individuals, including Escherichia coli and several enterococci (Supplementary Table S3). Conversely, 24 species were unique to the stool analyzed in this work, including two unknown species (Supplementary Table S3). Beduini massiliensis is one of these two species, the description of the other, part of the Paenibacillus genus, is ongoing.
Matrix-assisted laser-desorption/ionization time-of-flight (MALDI-TOF MS)
MALDI-TOF MS was performed as previously reported (Seng et al., 2010). The spectrum from B. massiliensis strain GM1 colonies could not be identified in the Bruker database.
16S rRNA amplification and sequencing
The 16S rRNA PCR and sequencing were performed using a GeneAmp 2720 thermal cycler (Applied Biosystems, Bedford, MA, USA) and an ABI Prism 3130xl sequencer (Applied Biosystems), respectively, as previously described (Nkamga et al., 2015). The Chromas Pro 1.34 software (Technelysium Pty. Ltd., Tewantin, Australia) was used to correct sequences, and BLASTn searches were performed in NCBI (http://blast.ncbi.nlm.nih.gov.gate1.inist.fr/Blast.cgi). Phylogenetic inferences were analyzed following 16S rRNA sequence alignment using CLUSTALW, and using the neighbor joining method within the MEGA 6 software (Center for Evolutionary Medicine and Informatics, AZ, USA).
Growth conditions
To assess the optimum growth temperature, B. massiliensis strain GM1 was cultivated on 5% sheep blood-enriched Colombia agar (Biomerieux) in anaerobic conditions. Temperatures tested were 28°C, 37°C, 45°C, and 55°C. Both anaerobic and microaerophilic atmospheres were tested using GENbag anaer and GENbag microaer systems, respectively (BioMerieux), on 5% sheep blood-enriched Colombia agar (Biomerieux). The aerobic atmosphere was tested at 37°C, using the same medium culture without anaerobic or microarophily generators. The halophily was tested using 0%, 5%, 15% and 45% of NaCl. Three pH conditions were tested: 5, 7, and 8.5.
Biochemical, sporulation, and motility assays
Biochemical assays were performed using API strip systems: API ZYM (BioMerieux), API Rapid ID 32A (BioMerieux), and API 50CH (BioMerieux). A thermo-shock was used to perform a sporulation test. A fresh colony was observed between the blades and slats using DM1000 photonic microscope (Leica Microsystems) at 40x to assess the motility of the bacteria.
Antibiotic susceptibility
The antibiotic susceptibility of B. massiliensis was tested against 15 antibiotics including amoxicillin, amoxicillin-clavulanic acid, imipenem, doxycycline, nitrofurantoin, vancomycin, clindamycin, erythromycin, metronidazole, cotrimoxazole, amikacin, gentamicin, tobramycin, and ciprofloxacin.
Microscopy
Transmission electron microscopy of B. massiliensis strain GM1 was performed using a Tecnai G20 transmission electron microscope (FEI company, Limeil-Brevannes, France). The Gram stain was performed and observed using a DM1000 photonic microscope (Leica Microsystems).
Genome sequencing and assembly
The genomic DNA of B. massiliensis GM1 was sequenced on the MiSeq sequencer (Illumina, San Diego, CA, USA) with the Mate-Pair strategy. The gDNA was barcoded in order to be mixed with 11 other projects with the Nextera Mate-Pair sample prep kit (Illumina). All the process was performed as previously reported (Lagier et al., 2014). The reads obtained were trimmed and then assembled using CLC genomics Workbench v4.7.2 software (CLC bio, Aarhus, Denmark).
Genome annotation and comparison
Open Reading Frames (ORFs) were predicted using Prodigal with default parameters (http://prodigal.ornl.gov/). However, the predicted ORFs were excluded if they spanned a sequencing gap region. The predicted bacterial protein sequences were searched against the GenBank (Benson et al., 2010), and Clusters of Orthologous Groups (COG) databases using BLASTP. The tRNAs and rRNAs were predicted using the tRNAScan-SE and RNAmmer tools (Lagesen et al., 2007; Lowe and Eddy, 1997), respectively. Signal peptides and numbers of transmembrane helices were predicted using SignalP and TMHMM, respectively (Bendtsen et al., 2004; Krogh et al., 2001). Mobile genetic elements were predicted using PHAST and RAST (Zhou et al., 2011; Aziz et al, 2008). ORFans were identified if their BLASTP E-value was lower than 1e-03 for an alignment length greater than 80 amino acids.
If alignment lengths were smaller than 80 amino acids, we used an E-value of 1e-05. Such parameter thresholds have already been used in previous studies to define ORFans. Artemis and DNA Plotter were used for data management and visualization of genomic features, respectively (Carver et al., 2009; Rutherford et al., 2000). Mauve alignment tool (version 2.3.1) was used for multiple genomic sequence alignment (Darling et al., 2004).
To estimate the mean level of nucleotide sequence similarity at the genome level between B. massiliensis and other bacteria, we used the Average Genomic Identity Of gene Sequences (AGIOS) home-made software (Ramasamy et al., 2014). Briefly, this software combines the Proteinortho software for detecting orthologous proteins between genomes compared two by two (Lechner et al., 2011), then retrieves the corresponding genes and determines the mean percentage of nucleotide sequence identity among orthologous ORFs using the Needleman-Wunsch global alignment algorithm. The B. massiliensis strain GM1 was compared with others Firmicutes: Clostridium saccharogumia strain DSM 17460, Clostridium spiroforme strain DSM 1552, and Clostridium ramosum strain DSM 1402.
All annotation and comparaison processes were performed in the Multi-Agent Software System DAGOBAH (Gouret et al., 2011), that include Figenix (Gouret et al., 2005).
Results
Phenotypic and biochemical characterization
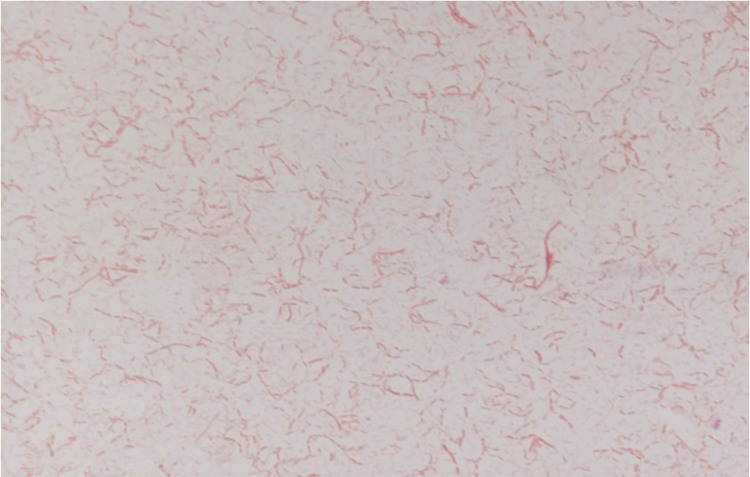
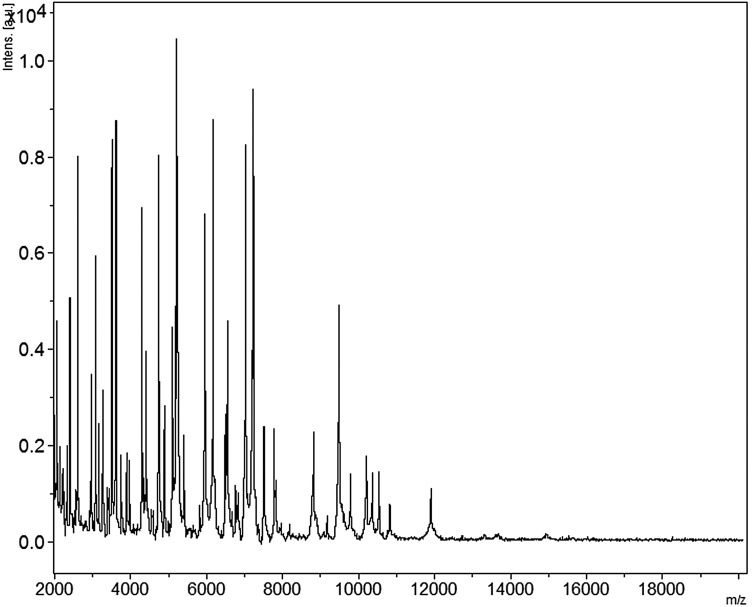
Beduini massiliensis is a Gram-negative rod that does not exhibit catalase or oxidase activities (Fig. 1). Spore forming, B. massiliensis is able to grow between 28°C and 45°C with an optimal growth temperature at 37°C, after 24 hours of incubation. The salinity range is 0%–5% (with an optimal at 0%), while the pH range is 5–8.5 (with an optimal at 7). The colonies of B. massiliensis are translucident with a diameter of 1 mm in 5% sheep blood agar. B. massiliensis strain GM1 is a anaerobic bacilli but was able to growth in a microaerophilic atmosphere. No growth of this bacterium was observed in aerobic conditions. Motile, individual cells of B. massiliensis exhibit a mean diameter of 0.5 μm and a length of 1.15 μm on electron microscopy (Fig. 2). B. massiliensis spectrum was added in the Bruker database (Fig. 3).
FIG. 1.
Gram staining of strain GM1.
FIG. 2.
Transmission electron microscopy of Beduini massiliensis strain GM1, using a Tecnai G20 (FEI company).The scale bar represents 200 nm.
FIG. 3.
Reference mass spectrum from Beduini massiliensis strain GM1. Spectra from 12 individual colonies were compared and a reference spectrum was generated.
Positive reactions were observed for arginine dihydrolase, galactosidase (α and β), β-galactosidase 6 phosphate, glucosidase (α and β), and α-fucosidase using API Rapid ID 32A strip. The same gallery shows that B. massiliensis ferments mannose and raffinose (Table 1). By contrast, reactions remain negative for urease, α-arabinosidase, β-glucuronidase, N-acetyl-β-glucosaminidase, glutamic acid decarboxylase, indole production, nitrates reduction, alkaline phosphatase, arginine arylamidase, proline arylamidase, leucyl glycine arylamidase, phenylalanine arylamidase, leucine arylamidase, pyroglutamic acid arylamidase, tyrosine arylamidase, alanine arylamidase, glycine arylamidase, histidine arylamidase, glutamyl glutamic arylamidase, and serine arylamidase.
Table 1.
Differential Characteristics of B. massiliensis strain GM1* with Clostridium saccharogumia strain DSM 17460, Clostridium spiroforme strain DSM 1552, and Clostridium ramosum strain DSM 1402
| Properties | B. massiliensis | C. saccharogumia | C. spiroforme | C. ramosum |
|---|---|---|---|---|
| Cell diameter (μm) | 0.3/1.2–1.3 | 0.9/2–3 | NA | NA |
| Oxygen requirement | Anaerobic | Anaerobic | Anaerobic | Anaerobic |
| Gram stain | − | + | + | + |
| Motility | + | − | − | − |
| Endospore formation | + | − | + | + |
| Indole | − | − | − | NA |
| Production of Alkaline phosphatase | − | − | NA | NA |
| Catalase | − | − | NA | NA |
| Oxydase | − | NA | NA | NA |
| Nitrate reductase | − | − | − | NA |
| Urease | − | − | − | − |
| β-galactosidase | + | + | NA | NA |
| N-acetyl-glucosaminidase | − | − | NA | NA |
| Acid from L-arabinose | − | − | NA | NA |
| Ribose | − | + | − | − |
| Mannose | − | + | + | + |
| Mannitol | − | NA | − | NA |
| Sucrose | − | + | + | + |
| D-glucose | + | + | + | NA |
| D-fructose | − | + | + | + |
| D-maltose | − | − | + | + |
| D-lactose | − | + | + | + |
| G + C content (%) | 35.9 | 30.1 | 28.6 | 31.4 |
| Habitat | Human gut | Human gut | Human gut | Human gut |
| References | This study | (Clavel et al., 2007) | (Kaneuchi et al., 1979; Borriello et al., 1983) | (Kaneuchi et al., 1979) |
Data from this study; NA = data not available.
Using API 50CH, starch remains the main carbohydrate substrate degraded by B. massiliensis. Result of API ZYM Gallery shows that B. massiliensis exhibited alkaline phosphatase, esterase (C4), esterase lipase (C8), lipase (C14), acid phosphatase, naphtol-AS-BI-phosphohydrolase, and α-mannosidase enzymes. API ZYM Gallery confirms also the presence of galactosidase (α and β), glucosidase (α and β), and α-fucosidase enzymes previously found using API Rapid ID 32A strip.
Antibiotic susceptibility testing shows a sensitivity of B. massiliensis for amoxicillin, doxycycline, nitrofurantoin, vancomycin, amoxicillin-clavulanic acid, clindamycin, imipenem, erythromycin, and metronidazole. By contrast, it remains resistant against cotrimoxazole, ciprofloxacin, gentamicin, tobramycin, and amikacin.
Phylogenic analysis
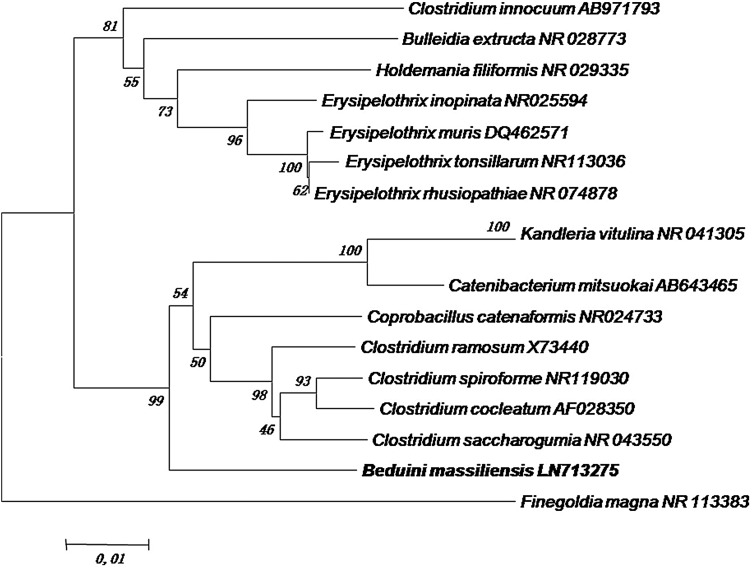
The 16S rRNA-based phylogenetic analysis showed that B. massiliensis gen. nov., sp nov., strain GM1 exhibited 91% similarity with Clostridium saccharogumia, classified in the Clostridiaceae family, created in 1933 by Pribram (Fig. 4). However, this percentage remains lower than the 95% 16S rRNA gene sequence threshold recommended by Stackebrandt and Ebers to delineate a new genus (Stackebrandt and Ebers, 2006). Therefore, this bacterium named Beduini massiliensis gen. nov., sp nov strain GM1 was classified within the new genus Beduini (Table 2). The 16S rRNA gene sequence of B. massiliensis was deposited in Genbank under the accession number LN713275.
FIG. 4.
Phylogenetic tree highlighting the position of Beduini massiliensis strain GM1 relative to other Firmicutes. Numbers at the nodes are percentages of bootstrap values obtained by repeating 1000 times the analysis to generate a majority consensus tree. The scale bar represents a 1% nucleotide sequence divergence.
Table 2.
Classification and General Features of Beduini massiliensis GM1
| Property | Term |
|---|---|
| Current classification | Domain: Bacteria |
| Phylum: Firmicutes | |
| Class: Clostridia | |
| Order: Clostridiales | |
| Family: Clostridiaceae | |
| Genus: Beduini | |
| Species: Beduini massiliensis | |
| Type strain: GM1 | |
| Gram stain | Negative |
| Cell shape | Rod |
| Motility | Motile |
| Sporulation | Yes |
| Temperature range | Mesophilic |
| Optimum temperature | 37°C |
Genome properties
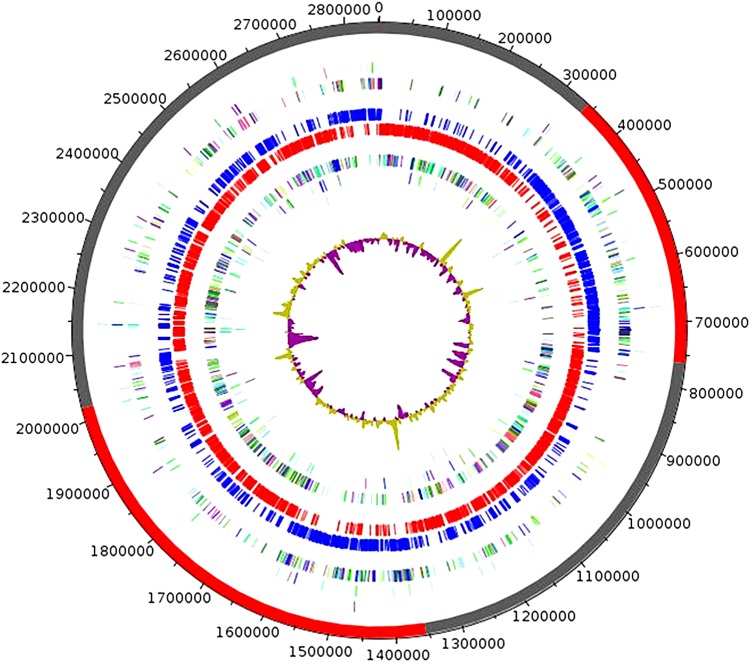
The genome is 2,850,586 bp long with 35.9 % G + C content (Table 3). It is composed of 6 scaffolds (composed of 6 contigs) (Fig. 5). Of the 2819 predicted genes, 2744 were protein-coding genes, and 75 were RNAs (6 genes are 5S rRNA, 6 genes are 16S rRNA, 6 genes are 23S rRNA, 57 genes are tRNA genes). A total of 1859 genes (67.75%) were assigned a putative function (by COGs or by NR blast). Thanks to this description, 274 genes (9.99%) completely unknown were identified as ORFans. Then, 3 genes associated with PKS or NRPS and 4 genes associated with CRISPR repeats were found. The remaining genes were annotated as hypothetical proteins (475 genes = > 17.31%). The distribution of genes into COG functional categories are presented in Table 4. The whole genome shotgun sequence of B. massiliensis strain GM1 ( = CSUR P1440 = DSM 100188) has been deposited in GenBank under accession number CDPP00000000.
Table 3.
Nucleotide Content and Gene Count Levels of the Chromosome
| Genome (total) | ||
|---|---|---|
| Attribute | Value | % of totala |
| Size (bp) | 2,850,586 | 100 |
| G + C content (%) | 1,022,794 | 35.9 |
| Coding region (bp) | 2,553,069 | 89.56 |
| Total genes | 2,819 | 100 |
| RNA genes | 75 | 2066 |
| Protein-coding genes | 2,744 | 97.33 |
| Genes with function prediction | 1,859 | 65.94 |
| Genes assigned to COGs | 1,582 | 56.11 |
| Genes with peptide signals | 248 | 8.79 |
| CRISPR repeats | 04 | 0.14 |
| ORFans genes | 274 | 9.71 |
| Genes with transmembrane helices | 683 | 24.22 |
| Genes associated with PKS or NRPS | 03 | 0.10 |
| No of antibiotic resistance genes | 0 | 0 |
The total is based on either the size of the genome in base pairs or the total number of protein- coding genes in the annotated genome.
FIG. 5.
Graphical circular map of the genome. From outside to the center: Contigs (red/gray), COG category of genes on the forward strand (three circles), genes on forward strand (blue circle), genes on the reverse strand (red circle), COG category on the reverse strand (three circles), GC content.
Table 4.
Number of Genes Associated with 25 General COG Functional Categories
| Code | Value | % of totala | Description |
|---|---|---|---|
| J | 159 | 5.79 | Translation |
| A | 0 | 0.0 | RNA processing and modification |
| K | 168 | 6.12 | Transcription |
| L | 115 | 4.19 | Replication, recombination and repair |
| B | 0 | 0.0 | Chromatin structure and dynamics |
| D | 18 | 0.66 | Cell cycle control, mitosis and meiosis |
| Y | 0 | 0.0 | Nuclear structure |
| V | 76 | 2.77 | Defense mechanisms |
| T | 52 | 1.90 | Signal transduction mechanisms |
| M | 86 | 3.13 | Cell wall/membrane biogenesis |
| N | 6 | 0.22 | Cell motility |
| Z | 0 | 0 | Cytoskeleton |
| W | 0 | 0 | Extracellular structures |
| U | 21 | 0.77 | Intracellular trafficking and secretion |
| O | 46 | 1.68 | Post-translational modification, protein turnover, chaperones |
| C | 80 | 2.92 | Energy production and conversion |
| G | 135 | 4.92 | Carbohydrate transport and metabolism |
| E | 140 | 5.10 | Amino acid transport and metabolism |
| F | 55 | 2.0 | Nucleotide transport and metabolism |
| H | 37 | 1.35 | Coenzyme transport and metabolism |
| I | 42 | 1.53 | Lipid transport and metabolism |
| P | 78 | 2.84 | Inorganic ion transport and metabolism |
| Q | 19 | 0.69 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 267 | 9.73 | General function prediction only |
| S | 136 | 4.96 | Function unknown |
| – | 277 | 9.82 | Not in COGs |
The total is based on the total number of protein coding genes in the annotated genome.
Genome comparison
The draft genome sequence of B. massiliensis GM1 (2.9 Mb) is larger than those of Clostridium spiroforme DSM 1552 (2.5 Mb) but is smaller than those of Clostridium saccharogumia strain DSM 17460 (3.4 Mb), and Clostridium ramosum DSM 1402 (3.2 Mb) (Table 5). The G + C content of B. massiliensis GM1 (35.9%) is larger than those of Clostridium saccharogumia strain DSM 17460 (30.1%), Clostridium spiroforme DSM 1552 (28.6%), and Clostridium ramosum DSM 1402 (31.4%). The gene content of B. massiliensis is larger than those of Clostridium spiroforme DSM 1552 (2533) and Clostridium saccharogumia strain DSM 17460 (2698), but smaller than that of Clostridium ramosum DSM 1402 (3,220).
Table 5.
Genome Comparison of Closely Related Species: Beduini massiliensis strain GM1
| S.NO | Name of organisms | INSDC | Size (Mb) | G + C (%) | Protein coding genes | Total genes |
|---|---|---|---|---|---|---|
| 1 | Beduini massiliensis strain GM1 | 2.85 | 35.9 | 2744 | 2819 | |
| 2 | Clostridium ramosum strain DSM 1402 | ABFX00000000.2 | 3.24 | 31.4 | 2941 | 3045 |
| 3 | Clostridium spiroforme strain DSM 1552 | ABIK00000000.2 | 2.51 | 28.6 | 2197 | 2342 |
| 4 | Clostridium saccharogumia strain DSM 17460 | JMLH00000000.1 | 3.41 | 30.1 | 2620 | 2816 |
| 5 | Coprobacillus cateniformis strain JCM 10604 | BBDT00000000.1 | 3.59 | 31.4 | 2340 | 3596 |
| 6 | Catenibacterium mitsuokai strain DSM 15897 | ACCK00000000.1 | 2.67 | 36.8 | 2412 | 2891 |
| 7 | Kandleria vitulina strain WCE2011 | JHXA00000000.1 | 2.04 | 35.0 | 1953 | 2054 |
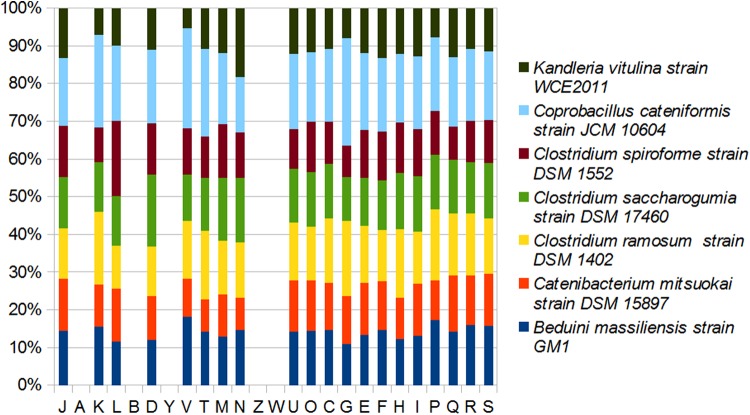
Percentage of 16S RNA similarity between between Beduini massiliensis, Clostridium saccharogumia, Clostridium spiroforme, and Clostridium ramosum remains lower than 95% (Table 6). Distribution of functional classes of predicted genes according to the clusters of orthologous groups of proteins between B. massiliensis and other Fimicutes, including Clostridium saccharogumia and Clostridium ramosum was reported (Fig. 6). The numbers of orthologous proteins shared between genomes are summarized in the Table 7, while results from Pairwise comparison are reported in Table 8.
Table 6.
Percentage of 16S RNA Similarity with Beduini massiliensis strainGM1
| S.NO | Species name | Similarity (%) | Accession no |
|---|---|---|---|
| 1 | Beduini massiliensis strain GM1 | 100 | LN713275 |
| 2 | Clostridium ramosum strain DSM 1402 | 90.38 | X73440 |
| 3 | Clostridium spiroforme strain DSM 1552 | 90.89 | NR_119030 |
| 4 | Clostridium saccharogumia strain DSM 17460 | 91.33 | NR_043550 |
| 5 | Coprobacillus cateniformis strain JCM 10604 | 90.17 | NR_024733 |
| 6 | Catenibacterium mitsuokai strain DSM 15897 | 88.25 | AB643465 |
| 7 | Kandleria vitulina strain WCE2011 | 86.59 | NR_041305 |
FIG. 6.
Distribution of functional classes of predicted genes according to the clusters of orthologous groups of proteins.
Table 7.
Numbers of Orthologous Proteins Shared Between Genomes (Upper Right)*
| B. massiliensis | C. mitsuokai | C. ramosum | C. saccharogumia | C. spiroforme | C. cateniformis | K. vitulina | |
|---|---|---|---|---|---|---|---|
| B. massiliensis | 2743 | 984 | 1286 | 1152 | 1075 | 1028 | 894 |
| C. mitsuokai | 66.31 | 2701 | 1141 | 1076 | 1005 | 909 | 969 |
| C. ramosum | 67.61 | 68.01 | 3057 | 1512 | 1396 | 1192 | 1015 |
| C. saccharogumia | 67.84 | 68.10 | 79.37 | 2822 | 1390 | 1035 | 985 |
| C. spiroforme | 67.97 | 68.66 | 78.31 | 80.76 | 2340 | 971 | 944 |
| C. cateniformis | 67.03 | 68.59 | 69.50 | 69.65 | 70.10 | 4576 | 806 |
| K. vitulina | 66.34 | 71.53 | 67.46 | 67.74 | 68.00 | 68.29 | 1983 |
Average percentage similarity of nucleotides corresponding to orthologous protein shared between genomes (lower left) and numbers of proteins per genome (bold).
Table 8.
Pairwise Comparison of Beduini massiliensis with Other Species Using GGDC, Formula 2 (DDH Estimates Based on Identities/HSP length)* Upper Right
| B. massiliensis | C. mitsuokai | C. ramosum | C. saccharogumia | C. spiroforme | C. cateniformis | K. vitulina | |
|---|---|---|---|---|---|---|---|
| B. massiliensis | 100% ± 00 | 21.7% ± 2.54 | 20.1% ± 2.56 | 20% ± 2.55 | 20.8% ± 2.56 | 22.1% ± 2.55 | 20.1% ± 2.54 |
| C. mitsuokai | 100% ± 00 | 17.9% ± 2.55 | 19.2% ± 2.54 | 18.9% ± 2.55 | 19.4% ± 2.55 | 18.2% ± 2.62 | |
| C. ramosum | 100% ± 00 | 23.2% ± 2.87 | 23.1% ± 2.83 | 21.9% ± 2.59 | 17.5% ± 2.55 | ||
| C. saccharogumia | 100% ± 00 | 24.4% ± 2.95 | 19.1% ± 2.58 | 17.9% ± 2.54 | |||
| C. spiroforme | 100% ± 00 | 20.8% ± 2.60 | 18.2% ± 2.54 | ||||
| C. cateniformis | 100% ± 00 | 18.5% ± 2.55 | |||||
| K. vitulina | 100% ± 00 |
The confidence intervals indicate the inherent uncertainty in estimating DDH values from intergenomic distances based on models derived from empirical test data sets (which are always limited in size) These results are in accordance with the 16S rRNA (Fig. 1) and phylogenomic analyses as well as the GGDC results.
Discussion
New biotechnologies and omics methodologies are intensively being developed in the postgenomics era as we traverse the complex space between structure and function, within and across species and microorganisms, in the course of integrative biology research (Adams et al., 2013; Akondi and Lakshmi, 2013; Canuel et al., 2015; Singh et al., 2013). The assessment of the human gut microbiota is one of these emerging omics frontiers and is in need of new methodological approaches. We have recently advanced a methodology for dynamic description of new bacteria, including new species and genera from the digestive flora, using the original approach of culturomics applied to samples of various geographical origins including Senegal, Niger, and Saudi Arabia.
Indeed, several new species were identified using this approach in our laboratory including Bacillus timonensis (Kokcha et al., 2012), Anaerococcus senegalensis (Lagier et al., 2012c), Alistipes senegalensis (Mishra et al., 2012a), Alistipes timonensis (Lagier et al., 2012b), Clostridium senegalensis (Mishra et al., 2012b), Peptoniphilus timonensis (Mishra et al., 2012c), and Peptoniphilus senegalensis (Mishra et al., 2013). New species from Saudi Arabia samples were also uncovered, including Clostridium jeddahense and Corynebacterium jeddahense (Edouard et al., 2014; Lagier et al., 2014).
We report here the isolation and cultivation of a new bacterial genus from a stool of a nomadic Bedouin woman from Saudi Arabia. The sustainability of this culture was confirmed by the deposit in two different collections (DSMz (Deutsche sammlung von mikroorganismen und zellkulturen) and our collection (Collection de souches de l'unité des rickettsies). The sequencing of its genome and the application of the latest taxonomic methods (Ramasamy et al., 2014) confirmed that it is indeed a new species and a new genus. This study attests to scientific value of the ‘microbial culturomics’ approach in improving the characterization of microbial communities. Since the seminal publication on this subject (Lagier et al., 2012a), we have identified 800 different species from human feces, including 124 putative new species, 48 published, and 13 officially recognized (Fournier et al., 2015).
The origin of this strain (stool of a nomadic Bedouin woman from Saudi Arabia) was supported by the very strict sampling protocols (immediate during defecation), freezing at −80°C as quickly as possible, and transport and culture in aseptic conditions (biosafety cabinet). The possibility that this species was a contaminant is very unlikely. Indeed, the contaminants are usually aerobic, including coagulase-negative Staphylococcus (Weinstein et al., 2003), Bacillus, Corynebacterium, and Micrococcus, while Beduini massiliensis is a strictly anaerobic bacterium. We cannot exclude that this bacteria is a part of the host environment known to impact the gut microbiota (Spor et al., 2011). Finally, the best proof of the digestive origin of this bacterium will be the reproducible isolation from stools from other individuals. Indeed, more than half of the new species identified by microbial culturomics from human feces in our laboratory were isolated at least a second time from another stool sample.
We note that this work does not aim to identify medical or biotechnological importance of a bacterium. The identification of a new species within a microbial community as rich and diverse as the gastrointestinal microbiota extends the human digestive microbiota repertoire (Lagier et al., 2012d). Future clinical studies (case-control studies) and experimental studies (in vitro or animal models) will clarify the geographical specificity of these new species and their role in health and disease or in biotechnology.
Conclusions
On the basis of phenotypic, phylogenetic and genomic analyses, we formally propose the creation of Beduini massiliensis gen. nov., sp. nov. that contains the strain GM1. Further features are summarized under subheadings below.
The combination of culturomics and taxono-genomics may improve the future laboratory identification of current or new bacterial species or genera and warrants further evaluation in different geographical and ecosystem settings. Microbial culturomics is an emerging frontier of omics systems sciences and integrative biology and thus, warrants further consideration as part of the postgenomics methodology toolbox.
Description of Beduini gen. nov.
Beduini (Be.dui.ni. N.L. gen. n. Beduini) is the Latin name of nomadic Bedouin population from Saudi Arabia where the stool specimen was collected. It is a Gram, catalase, and oxidase negative bacilli, spore forming with motility and strictly anaerobic individual cells. It is arginine dihydrolase, galactosidase (α and β), β-galactosidase 6 phosphate, glucosidase (α and β), α-fucosidase positive. Habitat and type species are human and Beduini massiliensis, respectively.
Description of Beduini massiliensis gen. nov., sp nov.
Beduini massiliensis (ma.si.li.en′.sis. L. fem. adj. massiliensis, of Massilia) is the Latin name of Marseille where B. massiliensis was isolated. The strain grows under temperatures ranging between 28°C and 45°C (with an optimum at 37°C). Salinity range growth is 0%–5% (with an optimum at 0%) while pH range growth is 5–8.5 (with an optimum at 7). The potential pathogenicity of the type strain GM1 (= CSUR P1440 = DSM 100188) is unknown but was isolated from a stool specimen from a nomadic Saudi Arabian Bedouin. This strain exhibited a G + C content of 35.9%. The genome and 16S rRNA sequences of B. massiliensis were deposited in Genbank under accession numbers CDPP00000000 and LN713275, respectively.
Supplementary Material
Abbreviations Used
- CSUR
Collection de Souches de l'Unité des Rickettsies
- DSM
Deutsche Sammlung von Mikroorganismen
- MALDI-TOF MS
Matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry
- TE buffer
Tris-EDTA buffer
- SDS
sodium dodecyl sulfate
- URMITE
Unité de Recherche sur les Maladies Infectieuses et Tropicales Emergentes
Acknowledgments
This project was funded by the Deanship of Scientific Research (DSR), King Abdulaziz University, Jeddah, under Grant No. (3-140-1434-HiCi). The authors, therefore, acknowledge with thanks Jaishriram Rathored for complementary data analysis and DSR for technical and financial support.
Author Disclosure Statement
The authors declare no financial conflicts of interest.
References
- Adams SH, Barnes KM, and Odle J. (2013). Comparative metabolic physiology in the ‘omics’ era: A call to arms, paws, flippers, and claws. Adv Nutr 4, 568–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akondi KB, and Lakshmi VV. (2013). Emerging trends in genomic approaches for microbial bioprospecting. OMICS 17, 61–70 [DOI] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, et al. (2008). The RAST Server: Rapid annotations using subsystems technology. BMC Genomics 9, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, and Brunak S. (2004). Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340, 783–795 [DOI] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Clark K, Lipman DJ, Ostell J, and Sayers EW. (2012). GenBank. Nucl Acids Res 40, D48–D53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borriello SP, and Carman RJ. (1983). Association of iota-like toxin and Clostridium spiroforme with both spontaneous and antibiotic-associated diarrhea and colitis in rabbits. J Clin Microbiol 17, 414–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canuel V, Rance B, Avillach P, Degoulet P, and Burgun A. (2015). Translational research platforms integrating clinical and omics data: A review of publicly available solutions. Brief Bioinform 16, 280–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver T, Thomson N, Bleasby A, Berriman M, and Parkhill J. (2009). DNAPlotter: circular and linear interactive genome visualization. Bioinformatics 25, 119–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavel T, Lippman R, Gavini F, Dore J, and Blaut M. (2007). Clostridium saccharogumia sp. nov. and Lactonifactor longoviformis gen. nov., sp. nov., two novel human faecal bacteria involved in the conversion of the dietary phytoestrogen secoisolariciresinol diglucoside. Syst Appl Microbiol 30, 16–26 [DOI] [PubMed] [Google Scholar]
- Darling AC, Mau B, Blattner FR, and Perna NT. (2004). Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14, 1394–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edouard S, Bibi F, Dhamodharan R, et al. (2014). Non-contiguous finished genome sequence and description of Corynebacterium jeddahense sp. nov. Stand Genomic Sci 9, 987–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier PE, Lagier JC, Dubourg G, and Raoult D. (2015). From culturomics to taxonomogenomics: A need to change the taxonomy of prokaryotes in clinical microbiology. Anaerobe. In press [DOI] [PubMed] [Google Scholar]
- Gouret P, Paganini J, Dainat J, Louati D, Darbo E, Pontarotti P, and Levasseur A. (2011). Integration of evolutionary biology concepts for functional annotation and automation of complex research. In: Evolution: The Multi-Agent Software System DAGOBAH, Evolutionary Biology—Concepts. Biodiversity, Macroevolution and Genome Evolution, pp. 71–87 [Google Scholar]
- Gouret P, Vitiello V, Balandraud N, Gilles A, Pontarotti P, and Danchin EG. (2005). FIGENIX: Intelligent automation of genomic annotation: Expertise integration in a new software platform. BMC Bioinformatics 6, 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneuchi C, Miyazato T, Shinjo T, and Mitsuoka T. (1979). Taxonomic study of helically coiled, sporeforming anaerobes isolated from the intestines of humans and other animals: Clostridium cocleatum sp. nov. and Clostridium spiroforme sp. nov. Int J Syst Bacteriol 29, 1–12 [Google Scholar]
- Kokcha S, Mishra AK, Lagier JC, et al. (2012). Non-contiguous-finished genome sequence and description of Bacillus timonensis sp. nov. Stand Genomic Sci 6, 346–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, and Sonnhammer EL. (2001). Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol 305, 567–580 [DOI] [PubMed] [Google Scholar]
- Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, and Ussery DW. (2007). RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucl Acids Res 35, 3100–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier JC, Armougom F, Million M, et al. (2012a). Microbial culturomics: Paradigm shift in the human gut microbiome study. Clin Microbiol Infect 18, 1185–1193 [DOI] [PubMed] [Google Scholar]
- Lagier J C, Armougom F, Mishra AK, Nguyen TT, Raoult D, and Fournier PE. (2012b). Non-contiguous-finished genome sequence and description of Alistipes timonensis sp. nov. Stand Genomic Sci 6, 315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier JC, Bibi F, Ramasany D, et al. (2014). Non contiguous-finished genome sequence and description of Clostridium jeddahense sp. nov. Stand Genomic Sci 9, 1003–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier JC, Hugon P, Khelaifia S, Fournier PE, La Scola B, and Raoult D. (2015). The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev 28, 237–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier JC, El Karkouri K, Nguyen TT, Armougom F, Raoult D, and Fournier PE. (2012c). Non-contiguous-finished genome sequence and description of Anaerococcus senegalensis sp. nov. Stand Genomic Sci 6, 116–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier JC, Million M, Hugon P, Armougom F, and Raoult D. (2012d). Human gut microbiota: Repertoire and variations. Front Cell Infect Microbiol 2, 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner M, Findeib S, Steiner L, Marz M, Stadler PF, and Prohaska SJ. (2011). Proteinortho: Detection of (Co-)orthologs in large-scale analysis. BMC Bioinformatics 12, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, and Eddy SR. (1997). tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucl Acids Res 25, 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra AK, Lagier JC, Robert C, Raoult D, and Fournier PE. (2013). Non-contiguous-finished genome sequence and description of Peptinophilus senegalensis sp. nov. Stand Genomic Sci 7, 370–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra AK, Gimenez G, Lagier JC, Robert C, Raoult D, and Fournier PE. (2012a). Non-contiguous-finished genome sequence and description of Alistipes senegalensis sp. nov. Stand Genomic Sci 6, 304–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra AK, Lagier JC, Robert C, Raoult D, and Fournier PE. (2012b). Non-contiguous-finished genome sequence and description of Clostridium senegalense sp. nov. Stand Genomic Sci 6, 386–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra AK, Lagier JC, Robert C, Raoult D, and Fournier PE. (2012c). Non-contiguous-finished genome sequence and description of Peptinophilus timonensis sp. nov. Stand Genomic Sci 7, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nkamga VD, Huynh HT, Aboudharam G, Ruimy R, and Drancourt M. (2015). Diversity of human-associated Methanobrevibacter smithii isolates revealed by multispacer sequence typing. Curr Microbiol 70, 810–815 [DOI] [PubMed] [Google Scholar]
- Pribram E. (1933). Klassification der Schizomyceten. Leipzig, pp. 1–143. Edited by Deuticke F. Deuticke, Leipzig [Google Scholar]
- Prodigal (http://prodigal.ornl.gov/). Last access: October, 2015
- Ramasamy D, Mishra AK, Lagier JC, et al. (2014). A polyphasic strategy incorporating genomic data for the taxonomic description of new bacterial species. Int J Syst Evol Microbiol 64, 384–391 [DOI] [PubMed] [Google Scholar]
- Rossello-Mora R. (2006). DNA-DNA reassociation methods applied to microbial taxonomy and their critical evaluation. In: Molecular Identification, Systematics, and Population Structure of Prokaryotes. Edited by Stackebrandt E. pp.23–50. Springer, Berlin [Google Scholar]
- Rutherford K, Parkhill J, Crook J, et al. (2000). Artemis: Sequence visualization and annotation. Bioinformatics 16, 944–945 [DOI] [PubMed] [Google Scholar]
- Seng P, Rolain JM, Fournier PE, La Scola B, Drancourt M, and Raoult D. (2010). MALDI-TOF-mass spectrometry applications in clinical microbiology. Future Microbiol 5, 1733–1754 [DOI] [PubMed] [Google Scholar]
- Singh A, Mahto KK, and Prasad R. (2013). Lipidomics and in vitro azole resistance in Candida albicans. OMICS 17, 84–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spor A, Koren O, and Ley R. (2011). Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol 9, 279–290 [DOI] [PubMed] [Google Scholar]
- Stackebrandt E, and Ebers J. (2006). Taxonomic parameters revisited: Tarnished gold standards. Microbiol Today 33, 152–155 [Google Scholar]
- Tindall BJ, Rosselló-Móra R, Busse HJ, Ludwig W, and Kämpfer P. (2010). Notes on the characterization of prokaryote strains for taxonomic purposes. Int J Syst Evol Microbiol 60, 249–266 [DOI] [PubMed] [Google Scholar]
- Wayne LG, Brenner DJ, Colwell PR, et al. (1987). Report of the ad hoc committee on reconciliation of approaches to bacterial systematic. Int J Syst Bacteriol 37, 463–464 [Google Scholar]
- Weinstein MP. (2003). Blood culture contamination: Persisting problems and partial progress. J Clin Microbiol 41, 2275–2278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker M, and Moore ER. (2011). Applications of whole-cell matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry in systematic microbiology. Syst Appl Microbiol 34, 2–11 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Liang Y, Lynch KH, Dennis JJ, and Wishart DS. (2011). PHAST: A fast phage search tool. Nucl Acids Res 39, W347–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.