Abstract
The Ross Sea, Eastern Antarctica, is considered a “pristine ecosystem” and a biodiversity “hotspot” scarcely impacted by humans. The sibling species Contracaecum osculatum sp. D and C. osculatum sp. E are anisakid parasites embedded in the natural Antarctic marine ecosystem. Aims of this study were to: identify the larvae of C. osculatum (s.l.) recovered in fish hosts during the XXVII Italian Expedition to Antarctica (2011–2012); perform a comparative analysis of the contemporary parasitic load and genetic variability estimates of C. osculatum sp. D and C. osculatum sp. E with respect to samples collected during the expedition of 1993–1994; to provide ecological data on these parasites. 200 fish specimens (Chionodraco hamatus, Trematomus bernacchii, Trematomus hansoni, Trematomus newnesi) were analysed for Contracaecum sp. larvae, identified at species level by allozyme diagnostic markers and sequences analysis of the mtDNA cox2 gene.
Statistically significant differences were found between the occurrence of C. osculatum sp. D and C. osculatum sp. E in different fish species. C. osculatum sp. E was more prevalent in T. bernacchii; while, a higher percentage of C. osculatum sp. D occurred in Ch. hamatus and T. hansoni. The two species also showed differences in the host infection site: C. osculatum sp. D showed higher percentage of infection in the fish liver. High genetic variability values at both nuclear and mitochondrial level were found in the two species in both sampling periods. The parasitic infection levels by C. osculatum sp. D and sp. E and their estimates of genetic variability showed no statistically significant variation over a temporal scale (2012 versus 1994). This suggests that the low habitat disturbance of the Antarctic region permits the maintenance of stable ecosystem trophic webs, which contributes to the maintenance of a large populations of anisakid nematodes with high genetic variability.
Keywords: Contracaecum osculatum s.l., Anisakids, Antarctic fish, Genetic variability, Allozymes, mtDNA cox2, Parasitic infections, Marine food-web
Graphical abstract
Highlights
-
•
Temporal stability of infection values of two anisakid species in Antarctic fish.
-
•
Temporal stability of genetic variability in two Antarctic anisakid parasites.
-
•
Differential distribution of two Contracaecum species in Antarctic fish.
-
•
Different host localization of the two species of Contracaecum in Antarctic fish.
1. Introduction
The Ross Sea, Eastern Antarctica, is considered a “pristine ecosystem” (Battaglia et al., 1997). It is considered to be one of the most species-rich in the Southern Ocean, and a biodiversity “hotspot” (Clarke and Johnston, 2003). The food-web of the Ross Sea continental shelf, unlike that of the slope, appears to have been largely untouched directly by humans until the last 10 years (Ainley, 2010). This provides a rare chance to investigate phenomena and factors, including complex food webs and parasites life cycles, concerning a marine ecosystem scarcely impacted by humans. The fish fauna of the Ross Sea comprises almost 80 species. The family Nothotheniidae is the dominant fish group both in terms of species richness and abundance, with 18 species (La Mesa et al., 2004). The family Channichthydae has less species richness (La Mesa et al., 2004); but it includes an euribatic species, Chionodraco hamatus, which is the most abundant channichthyid in the area (Eastman and Hubold, 1999). Fishes are an important trophic link connecting small invertebrates and top predators of the Antarctic marine ecosystem (Mintenbeck et al., 2012).
Among the parasites of pinnipeds from the Antarctic ecosystem, anisakids belonging to the Contracaecum osculatum (sensu lato) complex are the most abundant (Nascetti et al., 1993, Orecchia et al., 1994, Mattiucci et al., 2008). In the life-cycle of C. osculatum (s. l.) larval development likely occurs to the third stage (L3) inside the eggs passed out with pinniped stools (Koie and Fagerholm, 1995). Putative development from L2 to L3 in the eggs, is, however, still to be confirmed. Experimental infection trials (Koie and Fagerholm, 1995) showed that copepods could act as paratenic hosts in the life-cycle of C. osculatum (s. l.) from the Artic Boreal region. Fish act as intermediate/paratenic hosts, while various species of pinnipeds are definitive hosts (Mattiucci and Nascetti, 2008).
Previous studies using allozyme markers have demonstrated the reproductive isolation and the absence of gene flow among sympatric and allopatric populations of C. osculatum (s. l.) hosted by pinnipeds from Arctic and Antarctic regions. Those genetic markers have demonstrated the existence, within C. osculatum (s. l.) [previously considered as a cosmopolitan species and parasitic in various definitive seal hosts] of several biological species, often very similar morphologically, but reproductively isolated (sibling or cryptic species). The Arctic species are C. osculatum sp. A, C. osculatum sp. B, C. osculatum (s. s.) (see Nascetti et al., 1993, Mattiucci et al., 1998, Mattiucci et al., 2008), while the two Antarctic members are C. osculatum sp. D and C. osculatum sp. E (see Orecchia et al., 1994).
Species of the C. osculatum (s. l.) complex have been genetically characterized also on the basis of other genetic/molecular markers, such as the sequences analysis of the internal transcribed spacers of ribosomal DNA (ITS region of rDNA) (Nadler et al., 2005) and mitochondrial cox2 gene sequences analysis (Mattiucci et al., 2008). Further, the single strand conformation polymorphism (SSCP) analysis of the ITS region of rDNA was performed to screen for sequence variation within and among individuals of the C. osculatum (s. l.) species complex (Zhu et al., 2000, Hu et al., 2001).
Inter-taxon differences in SSCP profiles were detected between those Contracaecum taxa, with a reliable genetic differentiation of the sibling species from one another revealed at the ITS rDNA sequences analysis, except in the case of the two Antarctic members, i.e. C. osculatum sp. D and C. osculatum sp. E, which exhibited identical ITS of rDNA sequences and SSCP profiles at the same gene (Zhu et al., 2000). SSCP-based analyses of three mitochondrial DNA (mtDNA) regions, namely cytochrome c oxidase subunit I (cox1) and the small and the large subunit of ribosomal RNA (ssrRNA and lsrRNA, respectively), detected nucleotide differences considered diagnostic among C. osculatum sp. A, C. osculatum sp. B and C. osculatum (s. s.) in the Arctic and Antarctic members of C. osculatum (s. l.) (Hu et al., 2001). However, no differences at the same genes were detected between the two Antarctic members, i.e. C. osculatum sp. D and sp. E (Hu et al., 2001).
On the contrary, reproductive isolation and fixed alternative alleles at the multilocus allozyme electrophoresis (MAE) were found at some diagnostic loci between the two sympatric sibling species from the Antarctic Sea (Orecchia et al., 1994). In addition, more recently, sequences analysis of the mtDNA cox2 gene of specimens belonging to C. osculatum sp. D and C. osculatum sp. E, previously identified by allozymes, was able to support the existence of the two Antarctic members of C. osculatum (s. l.) as two distinct phylogenetic lineages (Mattiucci et al., 2008). Further, genetic diversity estimates at the allozyme levels were also given in the two Antarctic members, in comparison to the Arctic ones (Mattiucci and Nascetti, 2007).
The aims of this study were: 1) to identify at the species level, by means of allozymes and sequences analysis of the mtDNA cox2 gene, a high number of C. osculatum (s. l.) larvae recovered in Antarctic fish hosts, collected during the XXVIII Italian Expedition to Antarctica (2011–2012), to compare with those collected during the Expedition on 1993–1994; 2) to perform a comparative analysis of the parasitic load by C. osculatum sp. D and C. osculatum sp. E over a temporal scale (years 2011–2012 versus 1993–1994); 3) to estimate and compare at temporal scale level, the genetic variability at nuclear (allozymes) and mitochondrial level (mtDNA cox2 gene) in the two species, C. osculatum sp. D and C. osculatum sp. E; 4) to improve the knowledge about ecological aspects of those parasites in Antarctic waters, in terms of differential distribution of the two species in Antarctic fish, and their site of infection within the fish hosts.
2. Materials and methods
2.1. Sampling data
A total of 200 specimens belonging to four Antarctic fish species were sampled between the years 2011–2012, during the Antarctic summer period (December–February). Fish were caught by fishing net and by hand line in Terra Nova Bay of the Ross Sea, just off the coast where the Italian “Mario Zucchelli Station” is situated. Sampling included 50 icefish Chionodraco hamatus (Lönnberg, 1905) belonging to the Channichthydae family, 77 emerald rock cod Trematomus bernacchii Boulenger, 1902, 45 striped rock cod Trematomus hansoni Boulenger, 1902, and 28 dusty rock cod Trematomus newnesi Boulenger, 1902, belonging to the Nototheniidae family (Table 1). Some fish were examined immediately at the Mario Zucchelli Station, others were kept frozen at −30 °C and delivered, by the oceanographic ship “Italica”, to the Department of Public Health and Infectious Diseases of “Sapienza University of Rome”. Fish were measured (standard length to the nearest mm), and then subject to parasitological examination. Nematode larvae were collected from the fish body cavity, stomach wall, gonads and liver. They were formerly assigned to C. osculatum (s. l.), according to the morphological features, as suggested by Klöser and Plötz (1992) (i.e. total length of the worm with the caecum length/oesophagus length ratio), which allow to distinguish between larval C. osculatum (s. l.) and Contracaecum radiatum. They were then counted, washed in physiological saline and frozen initially at −20 °C (at the Laboratory in Antarctica), and later transferred at −50 °C at the University Department's Laboratory, until their genetic/molecular identification. The parasitological data obtained on fish collected during the present survey were compared with those obtained on the same fish species in a previous parasitological survey of the same scientific group, performed during the IX Italian Expedition to Antarctica (1993–1994).
Table 1.
Number (N) of individuals of the fish species examined for the detection of C. osculatum (s.l.) larvae during the 2011–2012 and 1993–1994 Antarctic expeditions, with the number of larvae per fish species identified by allozyme diagnostic markers and mtDNA cox2 sequences analysis.
| Sampling depth (m) | Fishing method | N | Mean standard length (min – max) (cm) | Allozymes |
mtDNA cox2 |
|||
|---|---|---|---|---|---|---|---|---|
| 1993/1994 | 2011–2012 | 1993/1994 | 2011–2012 | |||||
| Nototheniidae | ||||||||
| Trematomus bernacchii | 0–180 | Fishing nets-Hand line | 77 | 21.9 ± 2.63 (17.0–29.0) | 60 | 12 | 0 | 46 |
| T. newnesi | 30–80 | Hand line | 28 | 16.4 ± 9.80 (9.8–20.0) | 302 | 81 | 1 | 74 |
| T. hansoni | 80–180 | Fishing nets | 45 | 28.0 ± 1.34 (25.0–31.0) | 18 | 43 | 0 | 30 |
| Channichthydae | ||||||||
| Chionodraco hamatus | 80–180 | Fishing nets | 50 | 33.82 ± 3.02 (29.0–46.5) | 552 | 311 | 181 | 133 |
| TOT | 932 | 447 | 182 | 283 | ||||
2.2. Multilocus allozyme electrophoresis (MAE)
A total of 447 specimens of C. osculatum s.l. larvae collected during the 2011–2012 Antarctic survey were identified by multilocus allozyme electrophoresis (MAE), performed on those allozymes which have proven to be diagnostic between the two sibling species C. osculatum sp. D and sp. E. They are, Malate dehydrogenase-4 (Mdh-4, EC 1.1.1.37), and Adenylate kinase-2 (Adk2, EC 2.7.4.3) (see Orecchia et al., 1994). In addition, 932 larvae collected during 1993–1994 and previously stored at −80 °C (belonging to the collection of anisakid nematodes at the Section of Parasitology of Sapienza University of Rome), were also identified by MAE at these loci. In addition, a subsample of those specimens studied with MAE were also sequenced at the mitochondrial mtDNA cox2 gene, to confirm the results obtained by allozymes (see paragraph 2.3). In this aim, each single larva was divided in two parts; then, one part was used for MAE, while, the other was processed for PCR-DNA sequencing (procedures of this analysis are reported in the paragraph 2.3).
To estimate the genetic variability, at allozyme level, of the two sibling species from the two sampling periods, 200 specimens among those identified as belonging to the species C. osculatum sp. D and 200 to C. osculatum sp. E from each of two sampling periods (i.e. 2011–2012 and 1993–1994) were selected. They were studied, by allozyme markers, at the following 14 enzyme loci: Malate dehydrogenase (Mdh-1; Mdh-2; Mdh-3; Mdh-4, (EC 1.1.1.37), Isocitrate dehydrogenase (Icdh, EC1.1.1.42), 6-Phosphogluconate dehydrogenase (6Pgdh, EC 1.1.1.43), Aspartate amino transferase (Aat-2, EC 2.6.1.1), Leucine–leucine peptidase (Pep B, EC 3.4.11), Leucine-alanin peptidase (PepC-1, PepC-2, EC 3.4.11), Mannose phosphate isomerase (Mpi, EC 5.3.1.8), Glucose phosphate isomerase (Gpi, EC 5.3.1.9) and Phosphoglucomutase (Pgm-1, Pgm-2, EC 5.4.2.2). Their staining procedures are those reported in Nascetti et al. (1993). Isozymes were numbered in order of decreasing mobility from the most anodal one. Allozymes were identified by numbers indicating their mobility (in mm, standardized conditions) relative to the most common allele, designated as 100, found in the reference species (i.e. the Arctic species C. osculatum sp. A).
2.3. DNA extraction, amplification and sequencing
Among those previously identified by allozymes, a total of 283 (from 2011 to 2012) specimens of C. osculatum (s. l.) were also sequenced at the mtDNA cox2 gene. In addition, 182 specimens from the sample collected during 1993–1994 were sequenced at the mtDNA cox2 gene. Total DNA was extracted from 2 mg of tissue from each single nematode using the cetyltrithylammonium bromide method (CTAB) (Valentini et al., 2006, Mattiucci et al., 2008). The mitochondrial cytochrome C oxidase subunit II (cox2) gene was amplified using the primers 211F (5′-TTT TCT AGT TAT ATA GAT TGR TTY AT-3′) and 210R (5′-CAC CAA CTC TTA AAA TTA TC-3′) (Nadler and Hudspeth, 2000) spanning the mtDNA nucleotide position 10,639–11,248, as defined in Ascaris suum [GenBank X54253]. PCR (polymerase chain reaction) was carried out using the following conditions: 94 °C for 3 min (initial denaturation), followed by 34 cycles at 94 °C for 30 s (denaturation), 46 °C for 60 s (annealing), 72 °C for 90 s (extension), followed by post amplification at 72 °C for 10 min, as reported in Mattiucci et al. (2008).
The reference specimens and the isolated DNA samples presented in this paper are stored at the Section of Parasitology of the Department of Public Health and Infectious Diseases of “Sapienza – University of Rome”.
2.4. Statistical analysis of genetic data sets
Genetic analysis of allozyme data was performed using BIOSYS-2 software (Black, 1997). The statistical significance of departures from the Hardy–Weinberg equilibrium was estimated using the χ2 test. Genetic variability was based on 14 enzymatic loci of the two species, C. osculatum sp. D and sp. E, at the following parameters: mean number of alleles per locus (A); proportion of polymorphic loci (P) at the 0.99 criterion (a locus is considered polymorphic if the frequency of the most common allele does not exceed 0.99); and expected mean of heterozygosity per locus (He). For the genetic variability estimates, a minimum 50 of specimens belonging to the two parasite species collected on the two considered periods (2011–2012 and 1993–1994 Antarctic expeditions) was examined for each enzymatic locus.
Sequences of 519 bp of the mitochondrial cytochrome c oxidase subunit II (cox2) obtained from the specimens of the C. osculatum (s. l.), analysed in the present study, were compared with those already obtained for the same gene in previous studies (Mattiucci et al., 2008). The following sequences of all species of Contracaecum, parasites at the adult stage of pinnipeds, retrievable from GenBank (Mattiucci et al., 2008), were used for the identification of the specimens examined: C. osculatum sp. A (EU477203), C. osculatum sp. B (EU477204), C. osculatum (s. s.) (EU477206), C. baicalensis (EU477208), C. osculatum sp. D (EU477207), C. osculatum sp. E (EU477205), C. miroungae (EU477213), C. radiatum (EU477210).
Phylogenetic analysis was inferred using the Bayesian inference method, performed by MrBayes (Ronquist et al., 2012). Bayesian analysis was performed using the substitution model TrN + G (G = 0.602) as implemented in Jmodeltest (Posada, 2008), using the Akaike Information Criterion (AIC) (Posada and Buckley, 2004). For Bayesian analysis, four incrementally heated Markov Chains (using default heating values), were run for 1,000,000 generations, sampling the Markov Chains at intervals of 100 generations. Posterior probabilities were estimated and used to assess support for each branch in inferred phylogeny with probabilities where P = 95% being indicative of significant support (Reeder, 2003). The phylogenetic tree was rooted by using Pseudoterranova ceticola (DQ116435) and A. suum (X54253) as outgroups. Analysis of genetic diversity gathered from mtDNA cox2 sequences dataset were performed by using ARLEQUIN 3.5 software (Excoffier and Lischer, 2010). Mean pairwise differences among cox2 sequences of metapopulations (from each fish host species) observed at the intraspecific level, in the two species C. osculatum sp. D and C. osculatum sp. E, were estimated by using ARLEQUIN 3.5 software (Excoffier and Lischer, 2010), and by using Tamura-Nei (TrN) as the best substitution model for the sequences data set, implemented in jModeltest (Posada, 2008) using the AIC (Posada and Buckley, 2004).
2.5. Epidemiological data
The parasitic infection levels by C. osculatum (s. l.) larvae in the different fish species were calculated as the parameters of Prevalence (P, %), Mean intensity (Im) and range (min–max), following Bush et al. (1997) and Rózsa et al. (2000), using the Software Quantitative Parasitology QPweb, implemented for the web (Reiczigel and Rózsa, 2005).
The statistical significance of the differences observed in the prevalence (P) and mean intensity (Im) values of the infection by C. osculatum (s. l.) larvae, obtained by fish species at the temporal scale (two Antarctic expeditions, 2011–2012 and 1993–1994), were assessed by the Fisher's exact test and Bootstrap t-test, respectively, using the Software Quantitative Parasitology QPweb. A Bonferroni correction for multiple testing was applied to avoid inflating the Type I error rate, and the differences were considered significant when p < 0.00625.
The statistical significance of the differences observed in the relative proportions of the C. osculatum sp. D and C. osculatum sp. E identified in the different fish species and site of the infection in the host were assessed by the χ2 test (Yates corrected). Differences were considered significant when p < 0.05.
3. Results
3.1. Genetic identification of the sibling species of C. osculatum (s. l.) complex
A total of N = 1379 C. osculatum (s. l.) larvae (recovered from the sampling periods of 2011–2012 and 1993–1994) were identified genetically by allozymes markers. Among those identified, 13 C. osculatum (s. l.) larvae collected from T. bernacchii, 190 from T. newnesi, 45 from T. hansoni and 708 from Ch. hamatus were referred to the taxon indicated as C. osculatum sp. D, according to the diagnostic allozymes at the two diagnostic loci, i.e. Mdh-4104and Adk–2108. Whereas, according to the allozymes Mdh-4100 and Adk100, at those diagnostic loci, 59 larvae recovered from T. bernacchii, 193 from T. newnesi, 16 from T. hansoni and, finally, 155 larval stages collected from the icefish, Ch. hamatus, were found to correspond to the sibling species C. osculatum sp. E (Table 2).
Table 2.
Number of larval specimens of C. osculatum s.l., identified by allozymes and sequence analysis of the mtDNA cox2. The larval specimens were collected during the expeditions of 2011–2012 and 1993–1994. COSD = C. osculatum sp. D; COSE = C. osculatum sp. E.
| 2011–2012 |
1993–1994 |
|||||||
|---|---|---|---|---|---|---|---|---|
| Allozymes |
mtDNA cox2 |
Allozymes |
mtDNA cox2 |
|||||
| Fish species | COSD | COSE | COSD | COSE | COSD | COSE | COSD | COSE |
| Trematomus bernacchii | 2 | 10 | 13 | 33 | 11 | 49 | 0 | 0 |
| T. newnesi | 40 | 41 | 35 | 39 | 150 | 152 | 0 | 1 |
| T. hansoni | 33 | 10 | 8 | 22 | 12 | 6 | 0 | 0 |
| Chionodraco hamatus | 257 | 54 | 120 | 13 | 451 | 101 | 156 | 25 |
| TOT | 332 | 115 | 176 | 107 | 624 | 308 | 156 | 26 |
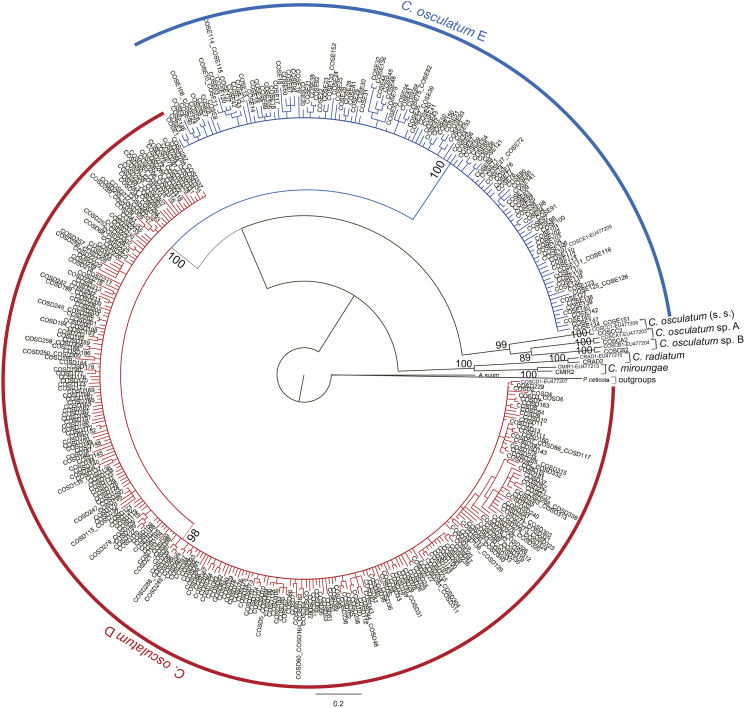
In addition, the sequence analysis of 519 bp at the mtDNA cox2 gene obtained from 272 larval specimens of C. osculatum (s. l.) – including both the specimens collected during the 2011–2012 and 1993–1994 parasitological surveys – gave a 99% or 100% match with the mtDNA cox2 sequence deposited in Genbank (EU477207) for the species named as C. osculatum sp. D. These worms include 11 larvae from T. bernacchii, 34 from T. newnesi, 7 from T. hansoni and, finally, 220 from Ch. hamatus (Table 2). Similarly, 133 larval specimens of C. osculatum (s. l.) – including specimens from both the 2011–2012 and 1993–1994 parasitological surveys – [of wich 38 collected from Ch. hamatus, 33 from T. bernacchii, 40 from T. newnesi and 22 from T. hansoni,] matched 99% the sequence of mtDNA cox2 deposited in GenBank (EU477205) under the name of C. osculatum sp. E (Table 2). The accession numbers of those sequences deposited in GenBank for C. osculatum sp. D found in Ch. hamatus, T. bernacchii T. hansoni and T. newnesi are, respectively, KT285804, KT285806, KT285808 and KT285810; the accession numbers of those sequences deposited in GenBank for C. osculatum sp. E, found in Ch. hamatus, T. bernacchii T. hansoni and T. newnesi are, respectively, KT285805, KT285807, KT285809 and KT285811. In addition, the Bayesian Inference analysis inferred from the mtDNA cox2 sequences dataset has shown that 13 larvae from T. bernacchii, 35 from T. newnesi, 8 from T. hansoni and, finally, 276 from Ch. hamatus clustered together in a well supported clade (98), including also the adult worm belonging to the species C. osculatum sp. D recovered from the pinniped host Leptonychotes weddellii, previously sequenced at the same gene and deposited in GenBank (Mattiucci et al., 2008) (Table 2, Fig. 1). Similarly, L3 individuals of C. osculatum (s. l.) collected from Ch. hamatus (N = 38), 33 from T. bernacchii, 40 from T. newnesi and 22 collected in T. hansoni, clustered in a unique clade, well supported by a high posterior probability value (100), together with the adult specimen of C. osculatum sp. E from Leptonychotes weddelli, previously sequenced by us and available from GenBank (Table 2, Fig. 1).
Fig. 1.
Circular Bayesian tree inferred from mtDNA cox-2 sequences obtained from specimens of C. osculatum sp. D and C. osculatum sp. E analysed in the present study, based on Bayesian Inference (BI) method using MrBayes v3.2.2 (Ronquist et al., 2012). Evolutionary distance was estimated using the TrN + G (G = 0.60) substitution model as implemented in jModeltest (Posada, 2008), with the AIC approach (Posada and Buckley, 2004). Posterior probability values are the result of 1.000000 of runs and are reported at the nodes. The coloured icons correspond to the two species considered in this study (red = C. osculatum sp. D and blue = C. osculatum sp. E).
3.2. Parasitic infection levels over a temporal scale
Prevalence (P) and mean intensity (Im) of C. osculatum sp. D and C. osculatum sp. E larvae recovered from 2011 to 2012 fish in comparison with those from 1993 to 1994, are reported in Table 3. Concerning the parasitic load of the fish sampled during the expeditions 2011–2012, all Ch. hamatus examined were heavily infected by C. osculatum sp. D and C. osculatum sp. E (P = 100% for both species) (Table 3). Ch. hamatus showed also the highest mean intensity with both C. osculatum sp. D larvae (A = 135.84) and C. osculatum sp. E (A = 25.86) (Table 3). Mean intensity of the infection with C. osculatum sp. D (A = 3.75) and C. osculatum sp. E (A = 4.29) was lower in T. newnesi while, T. bernacchii showed the lowest levels of prevalence and mean intensity with both C. osculatum sp. D and sp. E, i.e. P = 14.3 and A = 0.20 and P = 26.5, A = 0.57, respectively (Table 2). T. hansoni also showed low prevalence and mean intensity of infection with the two worm species (Table 3).
Table 3.
Comparison of the Prevalence (P, %), Mean intensity (Im, and min–max values) of the infection with C. osculatum sp. D and C. osculatum sp. E larvae, identified in the different fish species, over a temporal scale (2011–2012 versus 1993–1994).
| N | Fish mean length | Prevalence (%) |
Mean intensity(range) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| C. osculatum sp. D | C. osculatum sp. E | P1 | C. osculatum sp. D | C. osculatum sp. E | P2 | ||||
| T. bernacchii | 2011–2012 | 77 | 21.9 ± 2.63 | 14.3 | 26.5 | ns | 1.43 ± 0.79 | 2.15 ± 1.68 | ns |
| (17.0–29.0) | (1–3) | (1–6) | |||||||
| 1993–1994 | 75 | 22.6 ± 2.95 | 10.0 | 20.0 | ns | 1.40 ± 0.55 | 3.30 ± 2.41 | ns | |
| (15.0–30.3) | (1–2) | (1–8) | |||||||
| P3 | ns | ns | ns | ns | |||||
| T. newnesi | 2011–2012 | 28 | 16.4 ± 9.80 | 53.6 | 60.7 | ns | 7.00 ± 3.55 | 7.06 ± 4.51 | ns |
| (9.8–20.0) | (1–14) | (1–15) | |||||||
| 1993–1994 | 30 | 15.0 ± 7.25 | 90.0 | 96.7 | ns | 5.07 ± 4.66 | 4.97 ± 5.27 | ns | |
| (9.2–18.4) | (1–20) | (1–24) | |||||||
| P3 | a | a | ns | ns | |||||
| T. hansoni | 2011–2012 | 45 | 28.0 ± 1.34 | 50.0 | 33.3 | ns | 2.20 ± 1.01 | 1.40 ± 0.70 | ns |
| (25.0–31.0) | (1–4) | (1–3) | |||||||
| 1993–1994 | 40 | 26.0 ± 0.98 | 25.0 | 12.5 | ns | 1.30 ± 0.67 | 1.40 ± 0.89 | ns | |
| (24.2–29.7) | (1–3) | (1–3) | |||||||
| P3 | ns | ns | ns | ns | |||||
| Ch. hamatus | 2011–2012 | 50 | 33.82 ± 3.02 | 100 | 100 | ns | 135.84 ± 121.90 | 25.86 ± 25.15 | a |
| (29.0–46.5) | (19–550) | (1–120) | |||||||
| 1993–1994 | 50 | 30.5 ± 2.16 | 100 | 100 | ns | 102.06 ± 71.89 | 17.62 ± 13.31 | a | |
| (29.0–39.5) | (10–230) | (2–54) | |||||||
| P3 | ns | ns | ns | ns | |||||
P1 = significance level (Fisher's exact test) of differences between prevalence values of the infection by the two species of Contracaecum in each fish species; P2 = significance level of differences between mean intensity values of the infection by the two the two species of Contracaecum in each fish species (Bootstrap 2-sample t-test); P3 = significance level of differences between prevalence (Fisher's exact test) and between mean intensity values (Bootstrap 2-sample t-test) observed in the two sampling periods (2011–2012 and 1993–1994), by the two Contracaecum species in each fish species.
P < 0.0065 (Bonferroni corrected for multiple testing), ns = not significant.
Significant but slight deviation in prevalence and mean intensity values over time were observed when comparing data from 2011 to 2012 with those estimated on the same number of fish examined on 1993–1994 (Table 3). Indeed, prevalence with both Contracaecum species observed in T. newnesi showed a statistically significant differences between the two sampling expeditions (respectively, P = 53.6 in 2011–2012 versus P = 90.0 in 1993–1994 for C. osculatum sp. D, p = 0.005 calculated with Fisher's exact test), and P = 60.7 in 2011–2012 versus P = 96.7 in 1993–1994 for C. osculatum sp. E, (p = 0.0009) (Table 2).
However, considering the overall parasitic load by C. osculatum sp. D and C. osculatum sp. E in all the four fish species, no statistically significant variation (p > 0.05) was registered between the values registered in two sampling periods (2011–2012 versus 1993–1994) (Table 3).
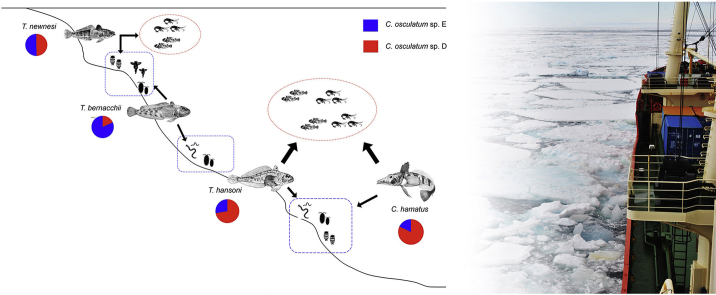
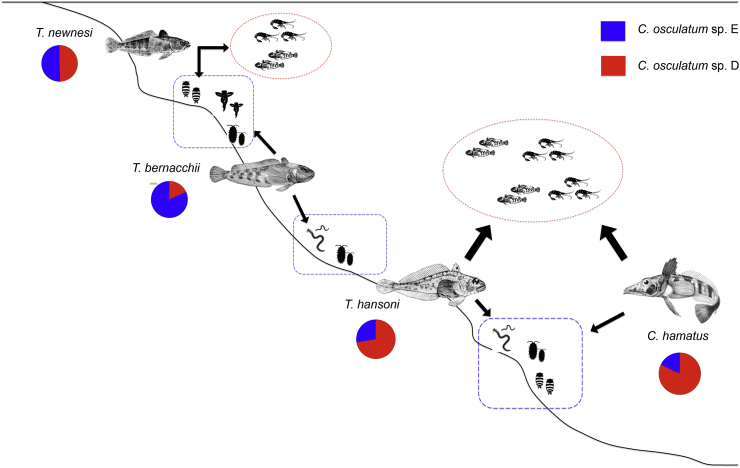
Concerning the distribution of the two worm species in the different fish species (Table 4 and Fig. 2), C. osculatum sp. D outnumbers C. osculatum sp. E in Ch. hamatus (84.8% versus 16.2%, respectively). On the contrary, C. osculatum sp. E outnumbers C. osculatum sp. D in Trematomus bernacchi (74.2% vs 25.8% of C. osculatum sp. D). T. newnesi seemed to be equally infected with C. osculatum sp. D (48.4%) and C. osculatum sp. E (51.6%). Overall, no statistically significant (p > 0.05, Yates corrected) (Table 4) difference was observed in the relative proportion of C. osculatum sp. D and C. osculatum sp. E in the four fish species understudied, between the two sampling periods (2011–2012 versus 1993–1994 expedition).
Table 4.
Relative proportions (%) of C. osculatum sp. D and C. osculatum sp. E observed in the fish species over a temporal scale (2011–2012 versus 1993–1994).
| Fish species | Parasite species |
||
|---|---|---|---|
| C. osculatum sp. D | C. osculatum sp. E | ||
| T. bernacchii | 2011–2012 | 25.8 | 74.2 |
| 1993–1994 | 18.3 | 81.7 | |
| p | ns | ||
| T. newnesi | 2011–2012 | 48.4 | 51.6 |
| 1993–1994 | 49.5 | 50.5 | |
| p | ns | ||
| T. hansoni | 2011–2012 | 56.2 | 43.8 |
| 1993–1994 | 66.7 | 33.3 | |
| p | ns | ||
| Ch. hamatus | 2011–2012 | 84.8 | 15.2 |
| 1993–1994 | 79.7 | 20.3 | |
| p | ns | ||
p = statistical significance between proportions calculated with Yates chi-squared test ***p < 0.001, *p < 0.05; ns = not significant.
Fig. 2.
Schematic distribution of the fish species examined in the present study for larval of C. osculatum sp. D and C. osculatum sp. E, along the continental shelf of the Ross Sea coastal ecosystem. Arrows indicating preferred preys and the diet preference for each fish species are reported according to the literature (La Mesa et al., 2004). The represented pelagic organisms comprise species of euphausiids and fish juveniles, benthic and epibenthic organisms are polychaetes, amphipods, decapods and gastropods. A pie chart with the relative proportions of C. osculatum sp. D and C. osculatum sp. E is given for each fish species. Squares and circles represent the hypothetical distribution of C. osculatum sp. D and C. osculatum sp. E larvae in their intermediate hosts.
3.3. Localization of C. osculatum sp. D and C. osculatum sp. E in the fish host
In order to check a differential site of infection by the two parasite species in the fish species, the infection data gathered in the two sampling periods were cumulated.
A significant difference in the occurrence of C. osculatum sp. D and C. osculatum sp. E was found with respect to the localization in the fish host (Table 5). Indeed, C. osculatum sp. D outnumbers C. osculatum sp. E in the host liver (Table 5), whereas C. osculatum sp. E was significantly (Table 5) more common in the gut of the same fish species. Indeed, in T. newnesi, characterized by an overall equal proportion of the two species, the relative frequency of C. osculatum sp. D is significantly higher in the liver (32.5% vs 13.9% of C. osculatum sp. E, p < 0.001); while C. osculatum sp. E significantly prevails in the gut (36.4% vs 17.2% of C. osculatum sp. D, p < 0.001). C. osculatum sp. D was the more frequent species observed in the liver of both T. hansoni and Ch. hamatus. In the case of T. bernacchii, C. osculatum sp. E was the predominant species in the gut of this fish host. Overall, the relative proportions of the two Contracaecum species showed always a preference of C. osculatum sp. D for the liver of the infected fish host (Table 5).
Table 5.
Relative proportions (%) of C. osculatum sp. D and C. osculatum sp. E observed by site of infection (liver versus gut) in the fish species examined (datasets from the 2011–2012 and 1993–1994 expeditions were aggregated for this analysis).
| Liver |
Gut |
p | |||
|---|---|---|---|---|---|
| C. osculatum sp. D | C. osculatum sp. E | C. osculatum sp. D | C. osculatum sp. E | ||
| T. bernacchii | – | – | 18.1 | 81.9 | *** |
| T. newnesi | 32.5 | 13.9 | 17.2 | 36.4 | *** |
| T. hansoni | 30.2 | 19.8 | 9.9 | 40.1 | *** |
| Ch. hamatus | 45.4 | 3.6 | 36.2 | 14.8 | *** |
p = probability level of the statistical significance of the comparison among number of localization of the two Contracaecum spp. from each host, obtained from χ2 test (Yates corrected). ***p < 0.0003.
3.4. Genetic variability at allozymes and mtDNA cox2, over a temporal scale
Estimates of the genetic variability parameters obtained at the considered 14 common allozyme loci, in the two species of Contracaecum, during the two Antarctic expeditions are summarized in Table 6. No significant differences were found in the genetic variability values at those considered parameters, between the two sympatric species studied in the two Antarctic expeditions. Indeed, C. osculatum sp. D showed, on average, He ≈ 0.18 in the populations of the species collected during the year 2011–2012; a similar value was observed on the samples from 1993 to 1994 estimated on 14 common allozyme loci. Similarly, the genetic variability estimate of populations of the species C. osculatum sp. E, estimated at on those allozyme loci in the present study was also close (on average, He ≈ 0.17) to that analysed from 1993 to 1994, inferred from the same loci (Table 6). Similarly, it was found that the parameter A showed high value; it was at the similar level, over time in both the two Contracaecum species (Table 6). Analogously, allele frequencies were found to be temporally stable in both worm species.
Table 6.
Estimates of genetic variability inferred from 14 allozyme loci, at the parameters of A, P99,P95 and He in the two anisakid species C. osculatum sp. D and C. osculatum sp. E, over a temporal scale (2011–2012 versus 1993–1994).
| N | n | A | P99 | He | |
|---|---|---|---|---|---|
| 2011–2012 | |||||
| C. osculatum sp. D | 200 | 14 | 2.3 (0.1) | 92.8 | 0.18 ± 0.01 |
| C. osculatum sp. E | 200 | 14 | 2.5 (0.1) | 92.8 | 0.17 ± 0.01 |
| 1993–1994 | |||||
| C. osculatum sp. D | 200 | 14 | 2.3 (0.1) | 92.8 | 0.18 ± 0.01 |
| C. osculatum sp. E | 200 | 14 | 2.4 (0.1) | 92.8 | 0.17 ± 0.02 |
N = number of larval specimens examined; n = number of allozyme loci considered in the allozyme data comparison. A = average number of alleles per locus; P99 = proportion of polymorphic loci at the 99.0% criterion; He = expected mean of heterozygosity per locus.
At the mitochondrial level, the genetic diversity estimated in the populations of C. osculatum sp. D and C. osculatum sp. E, collected during the two Antarctic expeditions, is reported in Table 7. Population structure of the two species from different fish hosts, assessed using a 519 bp portion of the cox2 mtDNA gene, showed C. osculatum sp. E had the greatest proportion of unique haplotypes (Nuh = 93%), while shared haplotypes were higher for the species C. osculatum sp. D (Nuh = 80%). Among the two species, haplotype diversity ranged from 0.995 (±0.0010) for C. osculatum sp. D to 0.999 (±0.0015) for C. osculatum sp. E with a total of 393 unique sequences recovered from 465 individual worms. Nucleotide diversity ranged from 0.017 (±0.008) to 0.025 (±0.011) and the number of polymorphic sites ranged from 51 to 144 in the two species C. osculatum sp. E and C. osculatum sp. D, respectively. At the intraspecific level, the mean pairwise sequence difference was 9.108 and 12.854 for, respectively, C. osculatum sp. E and C. osculatum sp. D.
Table 7.
Genetic variability values inferred from the mtDNA cox2 sequences analysis of C. osculatum sp. D and C. osculatum sp. E collected from the 4 fish species, T. bernacchii, T. newnesi, T. hansoni and Ch. Hamatus, during the two sampling years, i.e. 1993–1994 and 2011–2012. The number of sequences analysed (N), the nucleotide diversity (π) and relative standard deviation (sd), the haplotype diversity (Hd) and relative standard deviation (sd), the average number of nucleotide differences (K) and the number of polymorphic sites (S) are calculated by using Arlequin 3.5 software program (Excoffier and Lischer, 2010).
| N | Nh | π (sd) | Hd (sd) | K | S | |
|---|---|---|---|---|---|---|
| 1993–1994 | ||||||
| C. osculatum sp. D | 156 | 106 | 0.025(sd ± 0.011) | 0.995 (sd ± 0.001) | 12.854 | 137 |
| C. osculatum sp. E | 26 | 22 | 0.017(sd ± 0.008) | 0.988 (sd ± 0.014) | 9.108 | 51 |
| 2011–2012 | ||||||
| C. osculatum sp. D | 176 | 164 | 0.020(sd ± 0.011) | 0.999 (sd ± 0.0008) | 10.504 | 144 |
| C. osculatum sp. E | 107 | 101 | 0.019(sd ± 0.011) | 0.999 (sd ± 0.0015) | 10.278 | 124 |
4. Discussion
4.1. Genetic/molecular markers to distinguish C. osculatum sp. D and C. osculatum sp. E
The two Antarctic species C. osculatum sp. D and C. osculatum sp. E, share the same larval morphology which is also seen in L3 stages of the Arctic members of C. osculatum (s. l.) complex. Clearly, differentiation of these species by morphology at larval stage is unviable. Nevertheless, detecting and delimiting cryptic species is vital if we are to understand their responses to perturbation and variation in physiological tolerances and resilience that may determine their geographic distributions, potential host associations, and patterns of disease.
Molecular characterization of biodiversity can be useful to address scale phenomena that are critical to understanding temporal and spatial distributions in some geographic regions where cryptic biodiversity in term of species is now being revealed (Hoberg and Brooks, 2015). In this matter, the discovery of cryptic anisakid species in Antarctica has allowed an assessment of local biodiversity at both species and gene level. In the present paper, it was demonstrated that the combined use of allozymes and mtDNA cox2 sequences analysis is a valuable methodological approach in distinguishing anisakid sibling species, as it has previously been shown for other anisakid species (Mattiucci et al., 2014). The Bayesian Inference, based on the mitochondrial (mtDNA cox2) sequences dataset, supported with high posterior probability values (Fig. 1) the existence of the 2 cryptic species, i.e. C. osculatum sp. D and C. osculatum sp. E, as two independent evolutionary lineages (i.e., species); they are also shown to be sister taxa. In addition, allozymes also provided further evidence for their reproductive isolation, even in sympatric situations. Allozyme markers, by the way, provide a fast, cheap and powerful diagnostic tool for species determination and genetic variability estimates (Parker et al., 1998).
In addition, the mtDNA cox2 gene has proved to be a very polymorphic gene in the sibling species of the genus Contracaecum maturing in pinnipeds (Mattiucci et al., 2008). The same gene has also proven to be highly polymorphic in other siblings of the genus Contracaecum that mature in fish-eating birds (Mattiucci et al., 2010, Garbin et al., 2011) as well as in other species of anisakid nematodes (Mattiucci et al., 2014). Thus, in the present study, mtDNA cox2 has been confirmed to be useful in providing data sets concerning the genetic variability at the intraspecific level.
4.2. Ecological data on C. osculatum sp. D and C. osculatum sp. E
The results here presented confirm the presence of the two Antarctic species of anisakid nematodes of the genus Contracaecum (Raillet and Henry, 1912), [C. osculatum sp. D and C. osculatum sp. E (Orecchia et al., 1994)] as the most prevalent anisakid species recovered from the fish hosts captured in the Ross Sea. The two species were found in the same individual fish hosts, showing a strict simpatry and sintopy. However, a significant difference in their relative proportions in the fish species was observed. This finding could be related to the ecological and feeding habits of the fish host species. The fish species examined in the present study show trophic niche separation (Brenner et al., 2001), as demonstrated also by a study using stable carbon (C) and nitrogen (N) isotope analyses. Indeed, species of the fish genus Trematomus were differentiated in both C and N isotopic signature (Rutschmann et al., 2011), indicating respectively a different habitat use and a distinct trophic level. T. bernacchii is the most benthic species of the genus Trematomus (La Mesa et al., 2004); it has been also defined as a shallow benthic species, primarily feeding on benthic prey (Gon and Heemstra, 1990). Therefore, T. bernacchii is considered as a generalist species, which relies on a wide range of preys that are more or less associated with the sea bottom (La Mesa et al., 2004). On fish from the Ross Sea area, La Mesa et al. (2004) reported that bivalves were the main food of T. bernacchii, followed by polychaetes, and amphipods (Fig. 2). Our result showed that in T. bernacchii the species C. osculatum sp. E prevailed over C. osculatum sp. D, which was rarely seen in that fish species (Table 3, Table 4). Further, we found that small sized T. bernacchii (<25 cm) seems to be scarcely infected by Contracaecum larvae (authors personal observation). Another benthic notothenioid, T. hansoni, showed similar low infestation levels as observed in T. bernacchii (Table 3), but it had a preponderance of C. osculatum sp. D (65.0% in 1993–1994 sample, and 70.2% in 2011–2012 sample; Table 4, Fig. 2). The observed difference in the relative proportions of the two species of Contracaecum spp. could be explained by a different feeding behaviour and feeding habits of these two fish species (Fig. 2).
T. hansoni, despite sharing the benthic life style of T. bernacchii, includes more pelagic items in its diet (Eastman and Sidell, 2002). La Mesa et al., 1997, report that stomach contents of T. hansoni specimens collected in Terra Nova Bay (Ross Sea) showed a mainly piscivorous (mainly juveniles) diet, with fish eggs as secondary food items, and on several benthic organisms as polychaetes and decapods (La Mesa et al., 1997). Its food spectrum thus mainly comprises benthic organisms that are more or less associated with the substratum (La Mesa et al., 1997).
T. newnesi is considered part of cryopelagic community organism (Andriashev, 1970), and it's often associated with the under surface of the ice. The species can be defined as a frequent plankton feeder in the water column and as an occasional benthic feeder on the substratum (reviewed in Barrera-Oro, 2002); its diet in fact includes both Euphausiacea and more benthic Amphipoda, Isopoda, Polychaeta and some Mollusca (Daniels, 1982). A marked plasticity in feeding habits and diet diversity of T. newnesi has been found in areas of seasonal sea-ice coverage in the high Antarctic zone (La Mesa et al., 2000) (Fig. 2). Both C. osculatum species showed high prevalence of infection in T. newnesi, with almost all the examined fish infested (Table 3), and showing mean intensity (Im) values that were higher in T. newnesi than in the other notothenoids (Table 2). T. newnesi, with its intermediate characteristics and its peculiar cryopelagic habit, feeding both in the water column on krill, and on substratum benthic organisms (Amphipoda, Polychaeta, and Isopoda), showed a balanced mixed infection with the two Contracaecum species (48.4% C. osculatum sp. D and 51.6% C. osculatum sp. E, Table 4), feeding on both intermediate hosts for the two Contracaecum species (Fig. 2). In a study based on the stable isotope method to investigate trophic niches of Antarctic fishes, Cherel et al., 2011 reports that the intermediate δ13C value of T. newnesi is in agreement with the semipelagic life style of the species (Gon and Heemstra, 1990).
Whereas, Ch. hamatus, with a low δ13C signature, confirms its epibenthic life style, feeding mainly on pelagic prey (Gon and Heemstra, 1990). The infestation levels by both C. osculatum species in the channichtyid Ch. hamatus were remarkably higher than the levels recorded in the notothenioid fish species (Table 3, Fig. 2). A 100% prevalence (P) was recorded in samples from both Antarctic expeditions for the Contracaecum larvae of the two species, being present in all the examined Ch. hamatus specimens, and showing extremely high mean intensity values (A = 135.8 for C. osculatum sp. D and A = 25.9 for C. osculatum sp. E recorded in 2011/2012, and A = 102.0 for C. osculatum sp. D and A = 17.6 for C. osculatum sp. E recorded in 1993–1994) (Table 3). The subsample of Contracaecum spp. larvae obtained from Ch. hamatus genetically identified showed a prominent presence of C. osculatum sp. D (83.0% recorded in the subsample of 1993–1994, and 84.0% on 2011–2012) (Table 4). Piscivorous fish, such as many larger channichtyds, obviously play an important role as parathenic accumulators of Contracaecum s.l. larvae (Klöser et al., 1992). In the Ross Sea, Ch. hamatus is the most abundant and eurybathic channichtyid (Eastman and Hubold, 1999, Vacchi et al., 1999), with benthic behaviour, but also capable of vertical migration to feed on pelagic prey (Eastman and Sidell, 2002). Mintenbeck et al., 2012, defines Chionodraco spp. as demersal fish that only occasionally move in the water column. The large predatory icefish probably show a heavier helminth burden compared with the Trematomus spp. because they are likely more exposed to infection through predation on multiple intermediate hosts, such as invertebrates and smaller fish (see La Mesa et al., 2004).
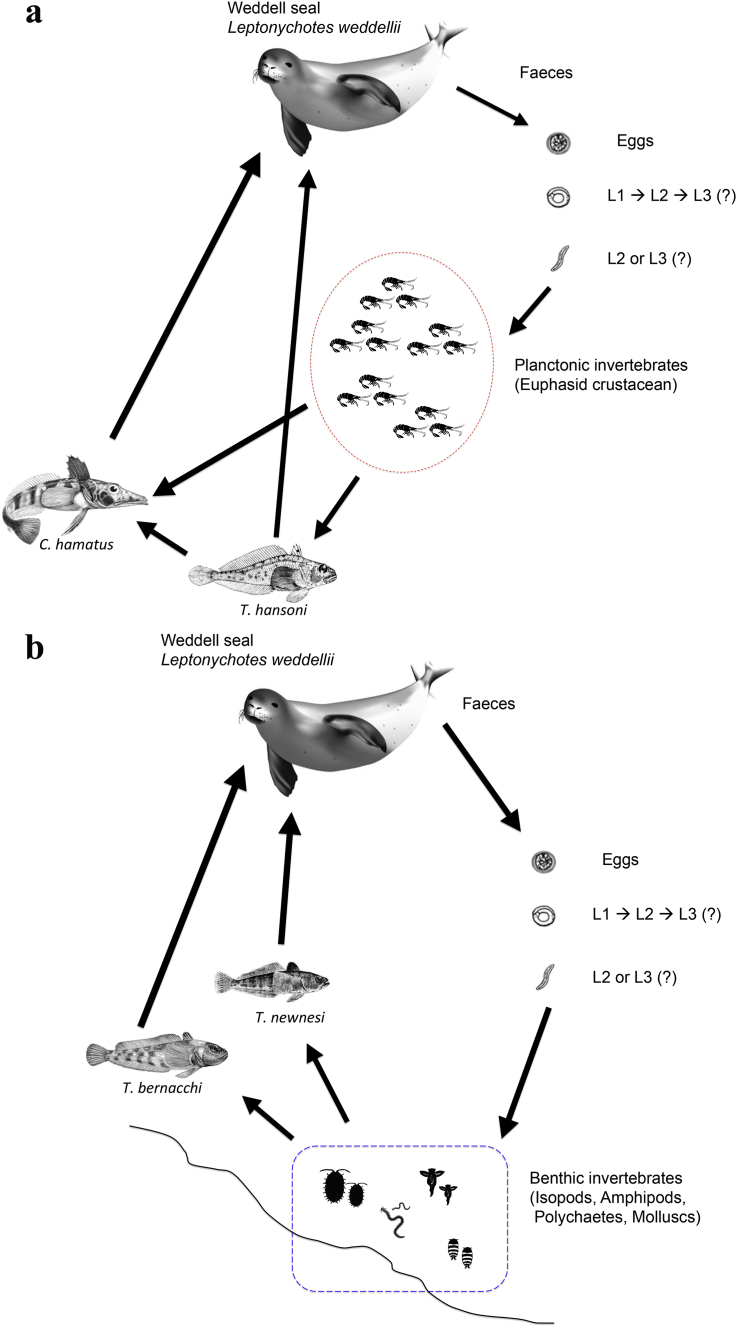
Thus, considering the relative frequencies observed of the two species of Contracaecum occurring in the different fish hosts, each one characterized by its feeding ecology and diets, some conclusion can be drawn regarding the possible life cycles of C. osculatum sp. D and C. osculatum sp. E in the Antarctic food web (Fig. 3a and b, respectively). C. osculatum sp. D seems to be associated more with fishes, such as Ch. hamatus and T. hansoni, characterized by bentho-pelagic habits, and predating on other small fishes and Antarctic krill (presumably Euphausia crystallorophias, the most frequent euphausiid present in the Ross sea). C. osculatum sp. D could include, in its life-cycle, a planktonic intermediate host, such as Euphausia crystallorophias (Fig. 3a). Instead, C. osculatum sp. E showed its higher relative proportion in T. bernacchii, which is a fish specialized in predation of strictly benthic organisms. This finding seems to suggest that a possible first intermediate host of C. osculatum sp. E could be represented by benthic organisms (Fig. 3b).
Fig. 3.
Schematic representation of the hypothetic life-cycle of C. osculatum sp. D (a) and C. osculatum sp. E (b) in the Ross Sea.
4.3. Parasitic load and genetic variability
Two complementary strategies have been suggested to examine the effects of habitat disturbance on the genetic variability of parasite populations: 1) comparison of different datasets of populations inhabiting disrupted ecosystems (spatial scale); and/or 2) comparison of particular datasets of populations through time, from the same geographical area (temporal scale) (Mattiucci and Nascetti, 2008). In this scenario, genetic diversity and parasite density (mean intensity) of anisakid populations of the genera Anisakis, Contracaecum and Pseudoterranova, have been previously compared from marine ecosystems of both Boreal and Austral regions (Mattiucci and Nascetti, 2008). Austral populations of species belonging to those genera exhibited significantly higher genetic variability values than those from the Boreal regions. Indeed, expected mean heterozygosity per locus resulted on average He ≈ 0.19 in Austral populations, while it was significantly lower (He ≈ 0.09) in Boreal anisakid populations (Mattiucci and Nascetti, 2008). Even more, a remarkable difference in genetic variability values was observed when Antarctic and sub-Antarctic populations of anisakid nematodes were compared directly with Arctic and sub-Arctic ones [on average He ≈ 0.23 in the first group versus He ≈ 0.07 in the second group] (Mattiucci and Nascetti, 2008). One conclusion could be that genetic variability values could be related to the extreme latitudes, a parameter often considered as relevant (Nevo et al., 1984). However, when comparing Antarctic and subantarctic members of the genus Contracaecum (i.e. C. osculatum sp. D and C. osculatum sp. E, C. radiatum, C. miroungae), versus those congeneric parasites of organisms of the Arctic Boreal region (i.e. C. osculatum sp. A, sp. B and C. osculatum (s. s.)), a lower level of genetic variability was observed in the second group of species (Mattiucci and Nascetti, 2008). Thus the genetic variability of anisakid parasites, at spatial scale level, was observed to be significantly different: on average, He ≈ 0.25 in the first group of species, while it resulted, on average, He ≈ 0.10 in the Arctic populations and species (Mattiucci and Nascetti, 2007, Mattiucci and Nascetti, 2008).
This study represents the first attempt to investigate the density and the genetic variability estimates of two anisakid species, i.e C. osculatum sp. D and C. osculatum sp. E, over a span of almost 20 years in the Antarctic ecosystem. This represents a long-term survey, taking into consideration the general duration of the life-cycle of anisakid nematodes, which requires some 60 days for its completion (McClelland, 2002). Indeed, no statistically significant differences have been observed in the parasite mean intensity values of C. osculatum sp. D and C. osculatum sp. E, from the considered fish species, in the two different sampling periods in the Antarctica (Table 2), with the only exception of the increasing of parasitic load found in T. hansoni collected in 2011–2012. Furthermore, no statistically significant difference was observed in the relative frequencies of the two species, C. osculatum sp. D and C. osculatum sp. E in the fish species here considered, over a temporal scale (Table 3). On the other hand, in the Antarctica, the two species, C. osculatum sp. D and C. osculatum sp. E, share the same definitive host, the weddell seal Leptonychotes weddellii, in which they occur – even syntopically – at very high parasitic burden, with several thousands of specimens collected from a single seal host (Mattiucci and Nascetti, 2008).
Thus, the high levels of infection reported for the C. osculatum (s.l.) complex in Antarctic fish species, which are prey for the Weddell seal, are consistent with the high integrity and stability of the food webs in this marine ecosystem. To support this hypothesis, there is also the finding that the Weddell seal, definitive host of those Antarctic parasites, has not suffered a decline in its population size, over a temporal scale of 30 years of study (Wickens, 1995, Kendall et al., 2003, Rotella et al., 2012, Stauffer et al., 2014). Similarly, despite there are no estimates of fish tons from the Antarctic fish species here examined and utilized as intermediate hosts by these nematodes, however, they have not been subjected to population size reduction from overfishing or any direct human impact, in such remote ecosystem. Finally, Antarctic krill (such as Euphausia superba) is estimated to have a population mass of 300–500 million tons (McBride et al., 2014).
These findings, agree with the results in the present study, and seem to support the hypothesis that the low level of habitat disturbance (pollution, overfishing, mortality by disease and hunting of seals) of the Antarctic region permits the maintenance of more stable trophic webs in this ecosystem. This same level of ecosystem health would allow definitive and intermediate/paratenic host species involved in the life-cycles of the two Antarctic species of parasites to reach higher population sizes. This will result, as a consequence, in the observation of high and stable density of parasite populations, with high and stable genetic variability values, over a temporal scale.
This is in contrast to our observations so far acquired over a temporal scale level, in other anisakid nematodes, such as Anisakis pegreffii. Indeed, despite the large effective population size and high gene flow estimates among its populations Mattiucci et al. (1997), A. pegreffii from the Mediterranean Sea, over 20 years, has shown a significant loss of genetic polymorphism and a decrease in the mean expected heterozygosity (He) value at some allozyme loci. Conversely, A. pegreffii in its Austral populations, where larger population size of their suitable definitive and intermediate/paratenic hosts are present (Shamsi, 2014) has maintained stable and similar values of genetic variability (Mattiucci and Nascetti, 2008). This finding was associated with the lower population density and geographic distribution of A. pegreffii definitive hosts (cetaceans) in the Mediterranean Sea. Interestingly, a low level of nucleotide diversity (0.007) was found at level of mtDNA cox2 sequences analysis of the Mediterranean populations of A. pegreffii (Mattiucci et al., pers. com; Blažeković et al., 2015.
5. Conclusion
Generally, in parasites there is a positive correlation between host population density and parasite abundance: as host density increases, the same should happen in the abundance of the parasite population in the community (Arneberg et al., 1998). As a consequence, variation in host dispersal and density may affect genetic diversity among their parasite populations (May, 1993, Zarlenga et al., 2014). In anisakid nematodes, the gene flow is largely determined by the different dispersal capability of hosts, and it correlates with parasite abundance. Thus, habitat disturbance of marine ecosystems and food webs can have considerable long-term effects on the genetic diversity and on demography of a parasite population, due to effects on its host's population size. Indeed, when the population size of hosts supporting the life-cycle of these parasites is reduced, the population size of their anisakid endoparasites may also be reduced. This would result in a higher probability of genetic drift in the parasite gene pools and, as a consequence, a decrease in their genetic polymorphism.
This study is a first attempt to estimate the biodiversity of anisakid parasites at both species and gene level, in an Antarctic ecosystem over an extended period. The transmission routes of the anisakid nematodes studied here follow closely the trophic relationships among their successive hosts, and thus they are parasites whose life-history stages are embedded in food webs (Mattiucci and Nascetti, 2008). The results reported here indicate the current stability of the Ross Sea Antarctic food web, as it allows the completion of two anisakid species life-cycles with high parasitic density and genetic variability values.
Clarification of the relationship between population structure and demographic history of species is critical to understanding microevolutionary processes and for predicting the resilience of species to environmental changes. Therefore, the monitoring of abundance and genetic variability, over time, of C. osculatum spp. in the Antarctic area will facilitate assessment of trophic web stability and general biodiversity of such marine ecosystem.
Conflict of interest
The authors declare that there is no conflict of interest.
Acknowledgements
The present study was granted by MIUR-PNRA (Italian Research Project in Antarctica) no. 2013AZ01/9. We are very grateful to Dr Steve C. Webb (Cawthron Institute, New Zealand) for reading the manuscript. The authors wish to thank the anonymous referees, whose suggestions were useful in improving the manuscript.
References
- Ainley D.G. A history of the exploitation of the Ross Sea, Antartica. Pol. Rec. 2010;46:233–243. [Google Scholar]
- Andriashev A.P. Cryopelagic fishes of the Arctic and Antarctic and their significance in polar ecosystems. In: Holdgate M.W., editor. Antarctic Ecology. Vol. 1. Academic; London: 1970. pp. 297–304. [Google Scholar]
- Arneberg P., Skorping A., Grenfell B., Read A.F. Host densities as determinant of abundance in parasite communities. Proc. R. Soc. Lond. 1998;265:1283–1289. [Google Scholar]
- Barrera-Oro E. The role of fish in the Antarctic marine food web: differences between inshore and offshore waters in the southern Scotia Arc and west Antarctic Peninsula. Antarct. Sci. 2002;14:293–309. [Google Scholar]
- Battaglia B., Valencia J., Walton D.W.H. Cambridge University Press; 1997. Antarctic Communities: Species, Structure and Survival. [Google Scholar]
- Black I.V.W.C. Colorado State University; Ft. Collins, CO: 1997. BIOSYS-2. A Computer Program for the Analysis of Allelic Variation in Genetics. (ftp://lamar.colostate.edu/pub/wcb4) [Google Scholar]
- Blažeković K., Pleić I.L., Duras M., Gomerčić T., Mladineo I. Three Anisakis spp. isolated from toothed whales stranded along the eastern Adriatic Sea coast. Int. J. Parasitol. 2015;45:17–31. doi: 10.1016/j.ijpara.2014.07.012. [DOI] [PubMed] [Google Scholar]
- Brenner M., Buck B.H., Cordes S., Dietrich L., Jacob U., Mintenbeck K., Schröder A., Brey T., Knust R., Arntz W. Ecological Studies in the Antarctic Sea Ice Zone. Springer; Berlin Heidelberg: 2001. The Role of Iceberg Scours in Niche Separation within the Antarctic Fish Genus Trematomus; pp. 215–220. [Google Scholar]
- Bush A.O., Lafferty K.D., Lotz J.M., Shostak A.W. Parasitology meets ecology on its own terms: Margolis et al. revisited. J. Parasitol. 1997;83:575–583. [PubMed] [Google Scholar]
- Cherel Y., Koubbi P., Giraldo C., Penot F., Tavernier E., Moteki M., Ozouf-Costaz C., Causse R., Chartier A., Hosie G. Isotopic niches of fishes in coastal, neritic and oceanic waters off Adélie land, Antarctica. Pol. Sci. 2011;5:286–297. [Google Scholar]
- Clarke A., Johnston N.A. Antarctic marine benthic diversity. In: Atkinson R.J.A., Gibson R.N., editors. vol. 41. CRC Press; 2003. pp. 47–114. (Oceanography and Marine Biology, An Annual Review). [Google Scholar]
- Daniels R.A. Feeding ecology of some fishes of the Antarctic Peninsula. Fish. Bull. 1982;80:575–588. [Google Scholar]
- Eastman J.T., Hubold G. The fish fauna of the Ross Sea, Antarctica. Antarct. Sci. 1999;11:293–304. [Google Scholar]
- Eastman J.T., Sidell B.D. Measurements of buoyancy for some Antarctic notothenioid fishes from the South Shetland Islands. Pol. Biol. 2002;25:753–760. [Google Scholar]
- Excoffier L., Lischer H.E.L. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Garbin L., Mattiucci S., Paoletti M., González-Acuña D., Nascetti G. Genetic and morphological evidences for the existence of a new species of Contracaecum (Nematoda: Anisakidae) parasite of Phalocrocorax brasilianus (Gmelin) from Chile and its genetic relationship with congeners from fish-eating bird. J. Parasitol. 2011;97:476–492. doi: 10.1645/GE-2450.1. [DOI] [PubMed] [Google Scholar]
- Gon O., Heemstra P.C. JLB Smith Institute of Ichthyology; Grahamstown, South Africa: 1990. Fishes of the Southern Ocean; pp. 1–462. [Google Scholar]
- Hoberg E.P., Brooks D.R. Evolution in action: climate change, biodiversity dynamics and emerging infectious disease. Philos. Trans. R. Soc. B. 2015;370:1665. doi: 10.1098/rstb.2013.0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M., D'Amelio S., Zhu X.Q., Paggi L., Gasser R. Mutation scanning for sequence variation in three mitochondrial DNA regions for members of the Contracaecum osculatum (Nematoda: Ascaridoidea) complex. Electrophoresis. 2001;22:1069–1075. doi: 10.1002/1522-2683()22:6<1069::AID-ELPS1069>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Kendall K.A., Ruhl H.A., Wilson R.C. Distribution and abundance of marine bird and pinniped populations within Port Forster, deception Islands, Antarctica. Deep Sea Res. Part II Top. Stud. Oceanogr. 2003;50:1873–1888. [Google Scholar]
- Klöser H., Plötz J. Morphological distinction between adult Contracaecum radiatum and Contracaecum osculatum (Nematoda, Anisakidae) from the Weddell seal (Leptonychotes weddelli) Zool. Scr. 1992;21:129–132. [Google Scholar]
- Klöser H., Plötz J., Palm H., Bartsch A., Hubold G. Adjustment of anisakid nematode life cycles to the high Antarctic food web as shown by Contracaecum radiatum and C. osculatum in the Weddell Sea. Antarct. Sci. 1992;4:171–178. [Google Scholar]
- Koie M., Fagerholm H.P. The life cycle of Contracaecum osculatum (Rudolphi, 1802) sensu strictu (Nematoda, Ascaridoidea, Anisakidae) in view of experimental infections. Parasitol. Res. 1995;81:481–489. doi: 10.1007/BF00931790. [DOI] [PubMed] [Google Scholar]
- La Mesa M., Dalù M., Vacchi M. Trophic ecology of the emerald notothen Trematomus bernacchii (Pisces, Nototheniidae) from Terra Nova Bay, Ross Sea, Antarctica. Pol. Biol. 2004;27:721–728. [Google Scholar]
- La Mesa M., Vacchi M., Castelli A., Divacco G. Feeding ecology of two nototheniid fishes, Trematomus hansoni and Trematomus loennbergii, from Terra Nova Bay, Ross Sea. Pol. Biol. 1997;17:62–68. [Google Scholar]
- La Mesa M., Vacchi M., Zunini Sertorio T. Feeding plasticity of Trematomus newnesi (Pisces, Nototheniidae) in Terra Nova Bay, Ross Sea, in relation to environmental conditions. Pol. Biol. 2000;23:38–45. [Google Scholar]
- Mattiucci S., Nascetti G. Genetic diversity and infections levels of anisakid nematodes parasitic in fish and marine mammals from Boreal and Austral Hemispheres. Vet. Parasitol. 2007;148:43–57. doi: 10.1016/j.vetpar.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Mattiucci S.G., Nascetti G. Advances and trends in the molecular systematics of anisakid nematodes, with implications for their evolutionary ecology and host-parasite co-evolutionary processes. Adv. Parasitol. 2008;66:47–148. doi: 10.1016/S0065-308X(08)00202-9. [DOI] [PubMed] [Google Scholar]
- Mattiucci S., Cipriani P., Webb S.C., Paoletti M., Marcer F., Bellisario B., Gibson D.I., Nascetti G. Genetic and morphological approaches distinguish the three sibling species of the Anisakis simplex species complex, with a species designation as Anisakis berlandi N. Sp. for A. simplex sp. C (Nematode: Anisakidae) J. Parasitol. 2014;100(2):199–214. doi: 10.1645/12-120.1. [DOI] [PubMed] [Google Scholar]
- Mattiucci S., Nascetti G., Cianchi R., Paggi L., Arduino P., Margolis L., Brattey J., Webb S., D'Amelio S., Orecchia P., Bullini L. Genetic and ecological data on the Anisakis simplex complex with evidence for a new species (Nematoda, Ascaridoidea, Anisakidae) J. Parasitol. 1997;83:401–416. [PubMed] [Google Scholar]
- Mattiucci S., Paggi L., Nascetti G., Ishikura H., Kikuchi K., Sato N., Cianchi R., Bullini L. Allozyme and morphological identification of Anisakis, Contracaecum and Pseudoterranova from Japanese waters (Nematoda: Ascaridoidea) Syst. Parasitol. 1998;40:81–92. [Google Scholar]
- Mattiucci S., Paoletti M., Solorzano A.C., Nascetti G. Contracaecum gibsoni n. sp. and C. overstreeti n. sp. (Nematoda:Anisakidae) from the Dalmatian pelican Pelicanus crispus (L.) in Greek waters: genetic and morphological evidence. Syst. Parasitol. 2010;75:207–224. doi: 10.1007/s11230-009-9220-8. [DOI] [PubMed] [Google Scholar]
- Mattiucci S., Paoletti M., Webb S.C., Sardella N., Timi J.T., Berland B., Nascetti G. Genetic relationships among species of Contracaecum Railliet and Henry, 1912 and Phocascaris Host, 1932 (Nematoda: Anisakidae) from pinnipeds based on mitochondrial cox2 sequences, and congruence with allozyme data. Parasite. 2008;15:408–419. doi: 10.1051/parasite/2008153408. [DOI] [PubMed] [Google Scholar]
- May R.M. Agriculture: resisting resistance. Nature. 1993;361:593–594. [Google Scholar]
- McBride M.M., Dalpadado P., Drinkwater K.F., Godø O.R., Hobday A.J., Hollowed A.B., Kristiansen T., Murphy E.J., Ressler P.H., Subbey S., Hofmann E.E., Loeng H. Krill, climate, and contrasting future scenarios for Arctic and Antarctic fisheries. ICES J. Mar. Sci. 2014;71:537–545. [Google Scholar]
- McClelland G. The trouble with sealworms (Pseudoterranova decipiens species complex, Nematoda): a review. Parasitology. 2002;124:183–203. doi: 10.1017/s0031182002001658. [DOI] [PubMed] [Google Scholar]
- Mintenbeck K., Barrera-Oro E., Brey T., Jacob U., Knust R., Mark F.C., Moreira E., Strobel A., Arntz W. Impact of climate change on fishes in complex Antarctic ecosystems. Adv. Ecol. Res. 2012;46:351–426. [Google Scholar]
- Nadler S.A., Hudspeth D.S.S. Phylogeny of the Ascaridoidea (Nematoda: Ascaridida) based on three genes and morphology: hypothesis of structural and sequence evolution. J. Parasitol. 2000;86:380–393. doi: 10.1645/0022-3395(2000)086[0380:POTANA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Nadler S.A., D'Amelio S., Dailey M.D., Paggi L., Siu S., Sakanari J.A. Molecular phylogenetics and diagnosis of Anisakis, Pseudoterranova, and Contracaecum from Northern Pacific marine mammals. J. Parasitol. 2005;91:1413–1429. doi: 10.1645/GE-522R.1. [DOI] [PubMed] [Google Scholar]
- Nascetti G., Cianchi R., Mattiucci S., D'Amelio S., Orecchia P., Paggi L., Brattey J., Berland B., Smith J.W., Bullini L. Three sibling species within Contracaecum osculatum (Nematoda, Ascaridida, Ascaridoidea) from the Atlantic Arctic-Boreal region: reproductive isolation and host preferences. Int. J. Parasitol. 1993;23:105–120. doi: 10.1016/0020-7519(93)90103-6. [DOI] [PubMed] [Google Scholar]
- Nevo E., Beiles A., Ben-Shlomo R. The evolutionary significance of genetic diversity: ecological, demographic and life history correlates. Evol. Dyn. Genet. Divers. 1984;53:14–213. [Google Scholar]
- Orecchia P., Mattiucci S., D'Amelio S., Paggi L., Plotz J., Cianchi R., Nascetti G., Arduino P., Bullini L. Two new members in the Contracaecum osculatum complex (Nematoda, Ascaridoidea) from the Antarctic. Int. J. Parasitol. 1994;24:367–377. doi: 10.1016/0020-7519(94)90084-1. [DOI] [PubMed] [Google Scholar]
- Parker P.G., Snow A.A., Schug M.D., Booton G.C., Fuerst P.A. What molecules can tell us about populations: choosing and using a molecular marker. Ecology. 1998;79:361–382. [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Posada D., Buckley T.R. Model selection and model averaging in phylogenetics: advantages of akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst. Biol. 2004;53:793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- Reeder T.W. A phylogeny of the Australian Sphenomorphus group (Scincidae: Squamata) and the phylogenetic placement of the crocodile skinks (Tribolonotus): Bayesian approaches to assessing congruence and obtaining confidence in maximum likelihood inferred relationships. Mol. Phylogenet. Evol. 2003;27:384–397. doi: 10.1016/s1055-7903(02)00448-7. [DOI] [PubMed] [Google Scholar]
- Reiczigel J., Rózsa L. 2005. Quantitative Parasitology 3.0.Budapest. Distributed by the authors. [Google Scholar]
- Ronquist F., Teslenko M., van der Mark P., Ayres D.L., Darling A., Höhna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotella J.J., Link W.A., Chambert T., Stauffer G.E., Garrott R.A. Evaluating the demographic buffering hypothesis with vital rates estimated for Weddel Seals from 30 years of mark-recapture data. J. An. Ecol. 2012;81:162–173. doi: 10.1111/j.1365-2656.2011.01902.x. [DOI] [PubMed] [Google Scholar]
- Rózsa L., Reiczigel J., Majoros G. Quantifying parasites in samples of hosts. J. Parasitol. 2000;86:228–232. doi: 10.1645/0022-3395(2000)086[0228:QPISOH]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Rutschmann S., Matschiner M., Damerau M., Muschick M., Lehmann M.F., Hanel R., Salzburger W. Parallel ecological diversification in Antarctic notothenioid fishes as evidence for adaptive radiation. Mol. Ecol. 2011;20:4707–4721. doi: 10.1111/j.1365-294X.2011.05279.x. [DOI] [PubMed] [Google Scholar]
- Shamsi S. Recent advances in our knowledge of Australian anisakid nematodes. Int. J. Parasitol. Parasites Wildl. 2014;3:178–187. doi: 10.1016/j.ijppaw.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauffer G.E., Rotella J.J., Garrot R.A., Kendall W.L. Environmental correlates of temporary emigration for female Weddel seals and consequences for recruitment. Ecology. 2014;95:2526–2536. [Google Scholar]
- Vacchi M., Pisano E., La Mesa G. Cold-adapted Organisms. Springer; Berlin Heidelberg: 1999. Ecological Features of Antarctic Fishes; pp. 219–238. (Springer-Verlag Berlin) [Google Scholar]
- Valentini A., Mattiucci S., Bondanelli P., Webb S.C., Mignucci-Giannone A., Colom-Llavina M.M., Nascetti G. Genetic relationships among Anisakis species (Nematoda: Anisakidae) inferred from mitochondrial cox-2 sequences and comparison with allozyme data. J. Parasitol. 2006;92:156–166. doi: 10.1645/GE-3504.1. [DOI] [PubMed] [Google Scholar]
- Wickens P.A. 1995. A Review of Operational Interactions between Pinniped and Fisheries. FAO Fish. Tech. Paper, 346. [Google Scholar]
- Zarlenga D.S., Hoberg E., Rosenthal B., Mattiucci S., Nascetti G. Anthropogenics: human influence on global and genetic homogenization of parasite populations. J. Parasitol. 2014;100:756–772. doi: 10.1645/14-622.1. [DOI] [PubMed] [Google Scholar]
- Zhu X.Q., D'Amelio S., Paggi L., Gasser R.B. Assessing sequence variation in the internal transcribed spacer of ribosomal DNA within and among members of the Contracaecum osculatum complex (Nematoda: Ascaridoidea: Anisakidae) Parasitol. Res. 2000;86:677–683. doi: 10.1007/pl00008551. [DOI] [PubMed] [Google Scholar]