Abstract
Both sterol carrier protein-2/sterol carrier protein-x (SCP-2/SCP-x) and liver fatty acid binding protein (L-FABP) have been proposed to function in hepatobiliary bile acid metabolism/accumulation. To begin to address this issue, the impact of ablating L-FABP (LKO) or SCP-2/SCP-x (DKO) individually or both together (TKO) was examined in female mice. Biliary bile acid levels were decreased in LKO, DKO, and TKO mice; however, hepatic bile acid concentration was decreased in LKO mice only. In contrast, biliary phospholipid level was decreased only in TKO mice, while biliary cholesterol levels were unaltered regardless of phenotype. The loss of either or both genes increased hepatic expression of the major bile acid synthetic enzymes (CYP7A1 and/or CYP27A1). Loss of L-FABP and/or SCP-2/SCP-x genes significantly altered the molecular composition of biliary bile acids, but not the proportion of conjugated/unconjugated bile acids or overall bile acid hydrophobicity index. These data suggested that L-FABP was more important in hepatic retention of bile acids, while SCP-2/SCP-x more broadly affected biliary bile acid and phospholipid levels.
Keywords: L-FABP, SCP-2, SCP-x, gene ablation, bile acid, sexual dimorphism
1. Introduction
Bile acids are not only biological detergents facilitating intestinal fat and fat-soluble vitamin absorption, but are also key metabolic regulators of glucose, lipid, and energy homeostasis [1]. Yet, little is known about how these relatively hydrophobic molecules are transported between the various subcellular compartments involved in their synthesis (peroxisomes, endoplasmic reticulum, mitochondria) and secretion (bile canaliculus) [2–7]. In fact, unlike non-mammalian vertebrates the mammalian liver does not contain the liver bile acid binding protein (L-BABP), which directly facilitates uptake and intracellular bile acid transport [3;4]. Instead, studies performed in vitro, with cultured hepatocytes, and with gene targeted mice suggest potential roles for three other genes encoding cytosolic proteins that bind bile acids in the mammalian liver: liver fatty acid binding protein (L-FABP), glutathione S-transferase (GST), and 3α-hydroxysteroid dehydrogenase (3α–HSD) [8;9]. Although a functional role for GST and 3α-HSD in intracellular transport of bile acids remains to be demonstrated, both ligand binding in vitro and studies in vivo with L-FABP null mice suggest potential roles for L-FABP.
L-FABP is a universal bile acid binding protein characterized by a single bile acid binding site with affinities in the 0.6–7 μM range, with highest affinities for specific bile acids with high and low, but less so intermediate, hydrophobicity indices [10–17]. L-FABP is present at very high concentration in murine (2–6% of cytosolic protein; 200–400 μM) and human (7–10% of cytosolic protein; 700–1000 μM) liver [11;18]. In addition to acting as a cytosolic bile acid transporter, L-FABP binding bile acids such as glycolithocholic acid inhibits liver microsomal sulfation of glycolithocholic acid in vitro-suggesting potential impact of L-FABP on the activities of other cytosol exposed membrane enzymes in bile acid metabolism [19].
Studies in vivo with cultured primary hepatocytes and L-FABP gene-ablated mice further support a role for L-FABP in bile acid metabolism. By binding bile acids, L-FABP reduces the toxicity of these potent detergent-like molecules [10;11;15;20]. In addition, L-FABP is thought to bind/transport bound bile acids or bile acid intermediates between sites of synthesis (peroxisomes, mitochondria, endoplasmic reticulum) and to the bile canaliculus for secretion [10;11;15;20]. Indeed, photoaffinity cross-linking studies show that L-FABP is essential for bile acid uptake and intracellular transport in rat hepatocytes [15]. Ablation of the L-FABP gene has been shown to significantly alter bile acid metabolism in male mice [21].
Finally, L-FABP, as well as the two products (SCP-2 and SCP-x, coded through alternate transcription sites) of the sterol carrier protein-2/sterol carrier protein-x gene (SCP-2/SCP-x), impact intracellular transport/targeting of cholesterol, the immediate precursor of bile acid synthesis. L-FABP [22;23] and SCP-2 [24–27] bind cholesterol with high affinity. SCP-2 stimulates liver microsomal cholesterol 7α-hydroxylase (rate limiting enzyme in hepatic bile acid synthesis) in vitro [28]. Likewise, SCP-x is the only known peroxisomal enzyme for cleaving the branched side chain of cholesterol, another key step in bile acid synthesis [28–32]. SCP-2 overexpression increases bile acid synthesis and biliary secretion in mice and in isolated rat and human hepatocytes [33;34]. Ablation of the SCP-2/SCP-x gene decreases bile acid synthesis and biliary bile acid secretion in mice [35–38]. SCP-2 and less so L-FABP also enhance intracellular transport of cholesterol to endoplasmic reticulum [39–42] and stimulate acyl-CoA cholesterol acyltransferse (ACAT) therein to form cholesteryl esters for storage/secretion [43–46]. The net effect of these opposing influences, i.e. facilitating bile acid synthesis vs potentially diverting cholesterol to storage as cholesteryl esters, on biliary bile acid levels is not clear.
Despite these advances, interpretation of studies with SCP-2/SCP-x gene-ablated mice has been complicated by concomitant upregulation [36;37;47] or downregulation [48] of liver fatty acid binding protein (L-FABP) as well as sex-differences in response. For example, mice exhibit sex-related differences in metabolism of branched-chain lipids [49], in hepatic regulation of cholesterol metabolism [50], in the hepatic lipid accumulation in mice lacking the L-FABP gene product only [22], as well as the response to a high-cholesterol diet in L-FABP gene-ablated mice [20;51] To better resolve the impact of these proteins on hepatobiliary bile acid metabolism in female mice, studies were undertaken comparing female mice singly ablated in L-FABP (LKO), singly ablated in SCP-2/SCP-x (DKO), or ablated in both L-FABP and SCP-2/SCP-x (TKO). The data herein demonstrate that L-FABP had a much greater impact on hepatic retention of bile acids while SCP-2/SCP-x more broadly affected biliary bile acid and phospholipid levels.
2. Experimental procedures
2.1. Materials
Liver homogenate protein concentration was determined using the Protein Assay Kit I (Cat # 500-0001, bovine gamma globulin) from Bio-Rad (Hercules, CA). Free cholesterol E (free cholesterol, C) and phospholipids (PL) were determined with diagnostic kits from Wako Diagnostics (Richmond, VA). Total bile acid (BA) was determined by a kit purchased from Diazyme Labs (Poway, CA). Bile acid standards (cholic acid, α-muricholic acid, β-muricholic acid, tauro-β-muricholic acid, tauro-lithocholic acid, tauro-ursodeoxycholic acid, tauro-cholic acid, tauro-chenodeoxycholic acid, and tauro-deoxycholic acid) were from Steraloids (Newport, RI). TaqMan® One-Step PCR Master Mix reagent kit and gene-specific assays for Abcg5 (Mm01226965_m1), Abcg8 (Mm00445977_m1), Cyp7a1 (Mm00484152_m1), Slc10a1 (NTCP, Mm01302718_m1), Slco1a1 (OATP1A1, Mm01267414_m1), and Slc22a7 (OATP2, Mm00460672_m1) were purchased from Applied Biosystems (Foster City, CA). Rabbit or goat polyclonal antibodies to mouse β-actin (sc-47778), BSEP (sc-17294), CYP27A1 (sc-14835), FXR (sc-13063), LXRα (sc-1201), and SHP (sc-15283) were from Santa Cruz Biotechnology (Dallas, TX). Rabbit polyclonal anti-mouse antibodies for ACAT-2 (ab66259), COX4 (ab16056), or MDR3 (ab71792) were purchased from Abcam (Cambridge, MA). Rabbit polyclonal antibody to mouse PPARα (PA1-822A) was from Pierce Antibody (Rockford, IL). Mouse monoclonal antibody against mouse GAPDH (MAB374) was from Millipore (Billerica, MA). Rabbit polyclonal antibody to 3α-HSD was purchased from US Biological (Peabody, MA). Alkaline phosphatase-conjugated goat polyclonal antibody to rabbit IgG (product # A3687) and rabbit polyclonal antibody to goat IgG (product # A4187) were from Sigma-Aldrich (St. Louis, MO). Alkaline phosphatase-conjugated rabbit polyclonal antibody against mouse IgG (product # ab6729-1) was purchased from Abcam (Cambridge, MA). All chemicals and solvents were the highest commercially available grade.
2.2. Animals
Six-week old (20–30 g) female inbred C57BL/6NCr wild-type (WT) mice were purchased from the National Cancer Institute (Frederick Cancer Research and Development Center, Frederick, MD). L-FABP gene-ablated (LKO) mice and SCP-2/SCP-x gene ablated (DKO) mice were generated as previously described [52;53] and used to create L-FABP/SCP-2/SCP-x gene ablated (TKO) mice as described [38]. All were backcrossed to the C57BL/6NCr background to ≥10 generations. Mice were maintained in a temperature-controlled facility (T = 25 °C) with a 12 h light/dark cycle and ad libitum access to food (Research Diets D11243, phytol/phytoestrogen-free, 5 g% fat, Research Diets, New Brunswick, NJ) and water. Phytol/phytoestrogen-free control diet, rather than standard rodent diet, was chosen because standard rodent diet contains variable levels of dietary phytol and phytoestrogen [37;54;55]. The major phytol metabolites (phytanic acid, pristanic acid) are highly potent naturally-occurring ligand activators of PPARα, which are known to alter transcription of L-FABP and SCP-2 as well as multiple other genes encoding proteins in cholesterol as well as fatty acid metabolism [49;56–59]. Variable levels of dietary phytoestrogens also potentially complicate hepatic lipid metabolism [54;55]. Animal protocols were Institutional Animal Care and Use Committee (Texas A&M University) approved. Mice were monitored quarterly for known rodent pathogens and determined to be pathogen-free.
2.3. Animal euthanasia and tissue collection
Mice were 12-hr fasted, weighed, and anesthetized with Avertin. Blood was collected via cardiac puncture into a polypropylene microtube, immediately processed to serum, volume of serum measured, flash-frozen on dry ice, and stored at −80 °C. While under anesthesia the mice were then euthanized by cervical dislocation, gall bladder bile collected, biliary volume determined, bile flash-frozen on dry ice, and stored at −80 °C. The liver was collected, blotted dry, weighed, and flash-frozen with phosphate buffered saline (0.5 mL, pH 7.4) on dry ice for storage at −80 °C.
2.4. Preparation of liver homogenate
Mouse liver (~0.1 g) was minced, placed in a 1.5-mL microcentrifuge tube, and homogenized in 0.5 mL of PBS (pH 7.4) on ice for 5 min with a motor-driven pestle at 2000 rpm (Tekmar Co, Cincinnati, OH). The resultant crude homogenate was sonicated on ice using a micro-tip (Sonic Dismembrator 550, Fisher Scientific, Pittsburgh, PA) set at 4, total processing time 5 min, on-time 15.0 sec, and off-time 15.0 sec. Insoluble debris was removed by centrifugation at 600 x g and 4 °C for 10 min. Protein concentration was measured in Costar 96-well assay plates (Corning, Corning, NY) using the Bradford protein micro-assay (Bio-Rad, Hercules, CA) and a BioTek Synergy 2 micro-plate reader (BioTek Instruments, Winooski, VT).
2.5. Biliary lipid and bile acid quantitation in liver and serum
Biliary free cholesterol (C) and phospholipid (PL) were quantified using Wako diagnostic kits as per the manufacturer’s instructions and a BioTek Synergy 2 micro-plate reader (BioTek Instruments, Winooski, VT). Total bile acids (BA) in liver homogenate, serum, and biliary bile were quantified using the Diazyme total bile acid diagnostic kit according to the manufacturer’s instructions (Diazyme Labs, Poway, CA) modified for use of the Bio-Tek micro-plate reader as described above. Biliary lipid ratios (C/PL, C/BA, PL/BA, PL/(PL + BA); mol/mol) and biliary cholesterol saturation index (CSI) were determined as described [60].
2.6. Western blotting of key proteins in hepatic and biliary lipid metabolism
Liver homogenate proteins were resolved by SDS-PAGE gel electrophoresis and analyzed by Western blot as described [61;62]. Control proteins (COX4, GAPDH, or β-Actin) were selected according to the protein of interest, protein molecular size, and antibody cross-reactivity. Ablating L-FABP, SCP-2/SCP-x, or both did not alter liver levels of any of the control proteins used (data not shown). Data were expressed relative to WT protein level = 1.0.
2.7. rtPCR of key proteins in hepatic and biliary lipid metabolism
Total RNA was prepared from liver using the RNeasy Mini Kit (Qiagen, Valencia, CA), purity and concentration determined with a NanoDrop 1000 Spectrophotometer (Thermo Scientific, Waltham, MA), and aliquots stored in RNA-later buffer as per the manufacturer’s instructions. Quantification of mRNA was determined using TaqMan® One Step PCR Master Mix Reagent kit and gene-specific assays for Abcg5, Abcg8, Cyp7a1, Slc10a1 (NTCP), Slco1a1 (OATP1A1) and Slc22a7 (OATP2). RNA measurements were obtained with an ABI PRISM 7000 Sequence Detection System and analyzed using ABI software in User Bulletin 2, normalized to 18S ribosomal RNA, and data presented in relative units with WT mRNA level = 1.0.
2.8. Statistics
One-way analysis of variance (ANOVA) combined with the Newman-Keuls multiple-comparisons post-test (GraphPad Prism Version 3.03, San Diego, CA) was applied to all statistical analyses. All data were examined by Bartlett’s test for equal variances, passed, and expressed as means ± standard error of the mean (n = number of mice = 8 per group). SigmaPlot 2002 for Windows Version 8.02 (SPSS, Chicago, IL) was used for all graphical representation with statistical differences (P < 0.05) indicated by labeling data with different lower-case letters.
3. Results
3.1. Ablating L-FABP (LKO), SCP-2/SCP-x (DKO) or both (TKO) differentially affected biliary, liver, and serum bile acid content
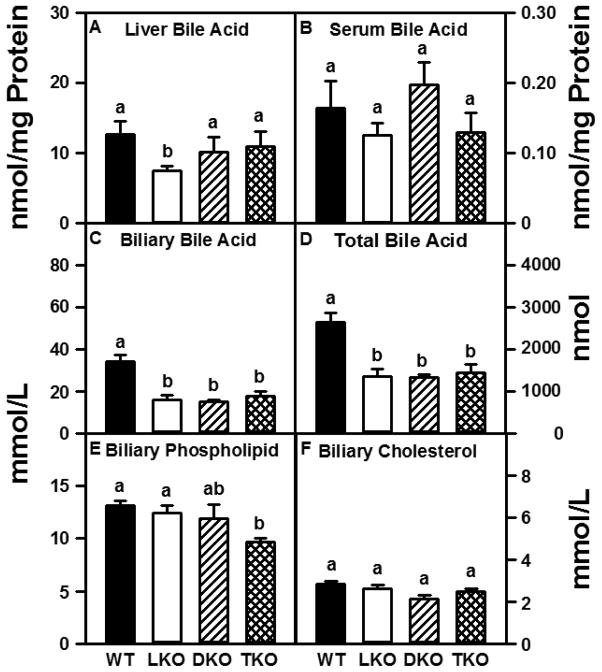
While LKO alone decreased bile acid levels in liver (Fig 1A), serum bile acid concentrations were unaffected in any gene-targeted mice (Fig 1B). In contrast, LKO, DKO and TKO each decreased biliary bile acid level by 50% (Fig 1C). Since gall bladder bile represents the largest pool of bile acids, this resulted in approximately 50% decreased total (biliary + liver + serum) bile acid levels in LKO, DKO, and TKO mice (Fig 1D).
Figure 1. Biliary total bile acid and phospholipid levels are decreased in TKO mice.
Bile acid (BA), phospholipid (PL), and cholesterol (C) concentrations were determined as described in Experimental Procedures. Liver (panel A) and serum (panel B) bile acid concentrations are expressed as nmol BA/mg liver homogenate protein. Biliary BA (panel C), PL (panel E), and C (panel F) levels are expressed as mmol/L bile. Total BA content (panel D) was determined by adding the total BA (nmol) measured in bile + liver + serum and is expressed as nmol BA. Values represent means ± SEM (n = 8). Statistically different values (P < 0.05, ANOVA) within a panel are denoted by a different lower-case letter (a, b).
Because bile acid secretion drives biliary cholesterol secretion and all gene ablations decreased biliary bile acid, the impact of LKO, DKO and TKO on biliary cholesterol and phospholipid levels was examined. The absence of both L-FABP and SCP-2/SCP-x gene products (TKO) resulted in 25% reduction in biliary phospholipid concentration (Fig 1E); however, biliary cholesterol concentration was unaffected by any gene ablation (Fig 1F).
As a result of the much larger changes in biliary bile acid (Fig 1C) than phospholipid (Fig 1E) or cholesterol (Fig 1F), several parameters reflecting ability of biliary bile to solubilize cholesterol were significantly altered. Mice lacking L-FABP (LKO, TKO) exhibited increased molar ratios of cholesterol/bile acid (C/BA), phospholipid/bile acid (PL/BA), and phospholipid/(phospholipid + bile acid) [PL/(PL + BA)] (Table 1). Mice without SCP-2/SCP-x gene products only (DKO) exhibited increased molar ratios of phospholipid/bile acid (PL/BA) and phospholipid/(phospholipid + bile acid) [PL/(PL + BA)] (Table 1). However, the biliary cholesterol saturation index (CSI) was unaffected in any of the gene-ablated mice (Table 1).
Table 1.
Biliary cholesterol saturation index and biliary lipid ratios in WT, LKO, DKO, and TKO mice.
| Component | WT mmol/L | LKO mmol/L | DKO mmol/L | TKO mmol/L |
|---|---|---|---|---|
| CSI | 6.6±0.1a | 6.6±0.1a | 6.4±0.1a | 6.4±0.1a |
| C/PL | 0.21±0.01a | 0.21±0.02a | 0.18±0.02a | 0.23±0.03a |
| C/BA | 0.11±0.01a | 0.20±0.03b | 0.14±0.01a | 0.30±0.06b |
| PL/BA | 0.39±0.04a | 1.0±0.1b | 0.8±0.1b | 0.7±0.1b |
| PL/(PL+BA) | 0.32±0.02a | 0.47±0.03b | 0.44±0.04b | 0.42±0.04b |
Biliary bile acids (BA), phospholipid (PL), and cholesterol (C), all in mmol lipid/L bile, were quantified as described in Experimental Procedures. C/PL, C/BA, PL/BA, and PL/(PL + BA) ratios (mol/mol) were calculated as described in Experimental Procedures. These values were used to determine the biliary cholesterol saturation index (CSI) according to [60]. Values represent means ± SEM (n = 8). Statistically different values (P < 0.05, ANOVA) within a row are denoted by a different lower-case letter (a, b).
3.2. Ablating L-FABP (LKO), SCP-2/SCP-x (DKO) or both (TKO) altered the species composition of biliary bile acids
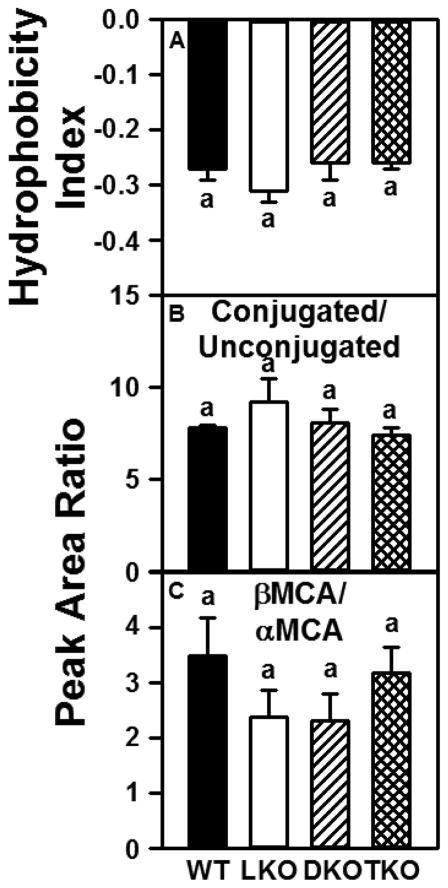
Since SCP-2/SCP-x gene products do not, but L-FABP does, bind bile acids [19;28;29], the possibility that DKO, LKO, or TKO differentially impacted the species composition of biliary bile acids was examined.
In wild-type (WT) mice, most of the biliary bile acids were conjugated (~88%), either with taurine (~80%) or less so glycine (~8%, Table 2). DKO, LKO, and TKO did not significantly alter the hydrophobicity index (HI) of the biliary bile acid population (Fig 2A), the ratio of conjugated/unconjugated biliary bile acids (Fig 2B), or the ratio of βMCA to αMCA (Fig 2C). However, there were significant differences in the levels of individual biliary bile acids as a result of differential gene ablation. The absence of L-FABP only (LKO) resulted in a 1.4-fold increase in T-MCA (Table 2); however, T-UDCA, T-CA, and G-CA levels were decreased by 25% to 30% (Table 2). In contrast to LKO mice, the loss of SCP-2/SCP-x (DKO) resulted in increased biliary levels of T-LCA (1.8-fold), T-UDCA (1.2-fold), and αMCA (1.4-fold, Table 2). Finally, the absence of both L-FABP and SCP-2/SCP-x gene products (TKO) only affected biliary levels of T-LCA (increased 1.4-fold, Table 2). Overall, the changes in biliary bile acid composition were not associated with an altered hydrophobicity index of bile acids—suggesting unaltered combined ability of these combinations of total bile acids to solubilize cholesterol.
Table 2.
Biliary bile acid composition in WT, LKO, DKO, and TKO mice.
| Bile Acid | WT % of Total | LKO % of Total | DKO % of Total | TKO % of Total |
|---|---|---|---|---|
| T-MCA | 27±2a | 37±2b | 31±5ab | 29±1a |
| T-LCA | 3.2±0.2a | 4.7±0.8a | 5.8±0.5b | 4.4±0.3b |
| T-UDCA | 4.3±0.2a | 3.2±0.3b | 5.3±0.2c | 4.0±0.1a |
| αMCA | 2.5±0.2ac | 3.8±0.6ab | 3.4±0.2b | 2.5±0.1c |
| T-CA | 43±2a | 30±2b | 36±4ab | 39±1a |
| βMCA | 7.9±0.7a | 6.8±0.6a | 8±1a | 8.6±0.3a |
| G-CA | 7.8±0.6a | 5.8±0.7b | 7.6±0.2a | 6.9±0.6ab |
| T-CDCA | 1.7±0.2a | 1.7±0.3a | 1.4±0.1a | 1.4±0.1a |
| T-DCA | 2.4±0.3a | 3.2±0.3a | 2.0±0.7a | 2.3±0.4a |
| CA | 1.0±0.6a | 0.19±0.06a | 0.25±0.07a | 0.43±0.04a |
The biliary bile composition and concentration (% of total peak area) of the following bile acids was determined as described in Experimental Procedures: cholic acid (CA), α-muricholic acid (αMCA), β-muricholic acid (βMCA), glyco-cholic acid (G-CA), tauro-cholic acid (T-CA), tauro-chenodeoxycholic acid (T-CDCA), tauro-deoxycholic acid (T-DCA), tauro-lithocholic acid (T-LCA), tauro-muricholic acid (T-MCA), tauro-ursodeoxycholic acid (T-UDCA). Values represent means ± SEM (n = 8). Statistically different values (P < 0.05, ANOVA) within a row are denoted by a different lower-case letter (a, b, c).
Figure 2. Impact of ablating SCP-2/SCP-x, L-FABP, or both on hydrophobicity index properties of biliary bile acids.
Biliary bile acid composition expressed as % of total peak area was determined as described in Experimental Procedures. This composition was then used to determine: (A) bile acid hydrophobicity index (HI) as described earlier [77]; (B) ratio of conjugated to unconjugated bile acids; (C) ratio of unconjugated βMCA/unconjugated αMCA. Values represent means ± SEM (n = 8).
3.3. DKO, LKO, and TKO differentially impacted hepatic expression of canalicular and basolateral bile salt transport proteins
The possibility that reduced biliary bile acid levels were associated with altered expression of membrane transporters of bile acids was examined.
Loss of L-FABP (LKO) resulted in a 1.4-fold increase in hepatic levels of canalicular bile salt export protein (BSEP, Table 3). In contrast, hepatic mRNA levels of the basolateral bile acid transporters OATP2 (Slc22a7) and NTCP (Slc10a1) were decreased by 60% and 30%, respectively (Table 3). Hepatic levels of OATP1A1 (Slco1a1) mRNA were unchanged in LKO mice (Table 3). Finally, loss of L-FABP had no effect on hepatic levels of the major hepatobiliary cholesterol transporter ABCG5/G8 mRNA or the hepatic phospholipid flippase protein MDR3 (Table 3). Loss of SCP-2/SCP-x gene products (DKO) also resulted in increased hepatic levels of canalicular BSEP as well as reduced levels of OATP2 and NTCP mRNA (Table 3). In contrast to LKO mice, DKO mice had reduced OATP1A1 mRNA levels (Table 3). Similar to LKO mice, the absence of SCP-2/SCP-x gene products had no effect on hepatic levels of ABCG5/G8 mRNA or MDR3 protein (Table 3). Finally, the loss of both L-FABP and SCP-2/SCP-x gene products (TKO) had a much different effect on the levels of these hepatic transporters than was observed in either LKO or DKO mice. TKO mice exhibited increased hepatic levels of ABCG5/G8, OATP2, and NTCP mRNA as well as MDR3 protein (Table 3). In contrast to LKO and DKO mice, TKO animals exhibited unchanged hepatic levels of BSEP (Table 3). OATP1A1 mRNA levels were also unchanged in TKO mice (Table 3).
Table 3.
Hepatic levels of key biliary lipid/bile acid transporters in WT, LKO, DKO, and TKO mice.
| Component | WT Relative Value |
LKO Relative Value |
DKO Relative Value |
TKO Relative Value |
|---|---|---|---|---|
| Abcg5 mRNA | 1.0±0.1a | 1.2±0.4a | 1.1±0.2a | 2.3±0.4b |
| Abcg8 mRNA | 1.0±0.2a | 1.2±0.2a | 1.1±0.2a | 2.7±0.6b |
| BSEP | 1.00±0.05a | 1.37±0.08b | 1.6±0.2b | 0.70±0.1a |
| MDR3 | 1.0±0.1a | 1.2±0.1a | 1.0±0.1a | 1.7±0.3b |
| Slco1a1/OATP1A1 mRNA | 1.0±0.1a | 0.9±0.1a | 0.6±0.2b | 1.0±0.2a |
| Slc22a7/OATP2 mRNA | 1.0±0.1a | 0.4±0.1b | 0.4±0.1b | 1.6±0.3c |
| Slc10a1/NTCP mRNA | 1.00±0.02a | 0.66±0.07b | 0.33±0.05c | 2.1±0.1d |
Liver homogenate proteins were examined by SDS-PAGE and subsequent Western blot analysis (BSEP, MDR3) as described in Experimental Procedures. Quantitative rtPCR of liver mRNA was accomplished as described in Experimental Procedures. Protein/mRNA levels are shown as relative values (WT = 1) and represent means ± SEM (n = 8). Statistically different values (P < 0.05, ANOVA) within a row are denoted by a different lower-case letter (a, b, c, d).
3.4. Key hepatic proteins involved in bile acid synthesis from cholesterol
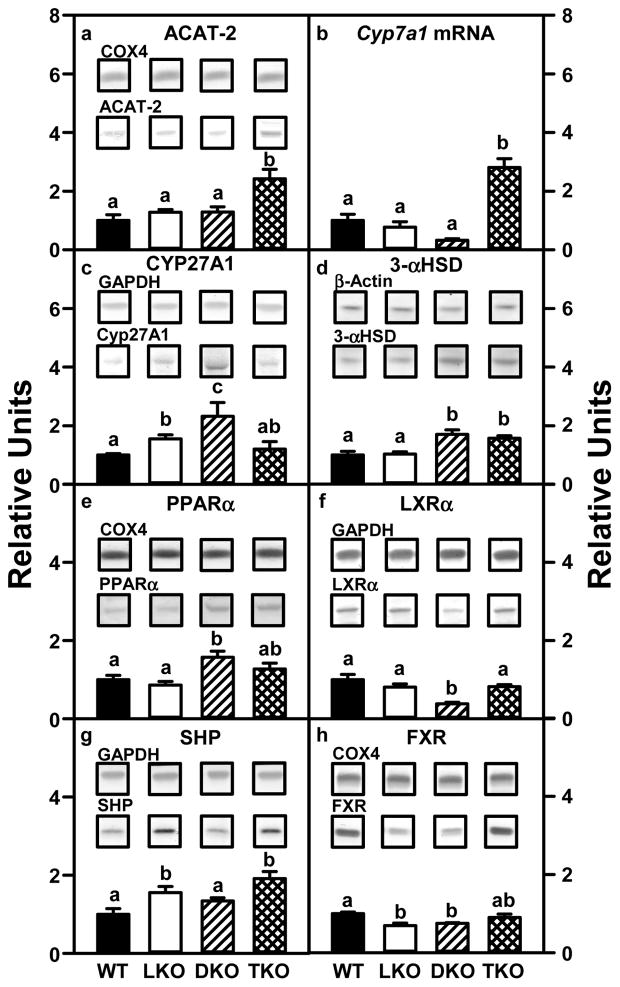
Since ablating L-FABP, SCP-2/SCP-x, or both decreased biliary bile acids, total bile acids, and in the case of LKO also hepatic bile acids, the possibility that reduced bile acid level was associated with downregulation of key enzymes in bile acid synthesis was examined.
Loss of the major mammalian bile acid binding protein L-FABP (LKO) or SCP-2/SCP-x (DKO) did not significantly alter hepatic levels of Cyp7a1 mRNA, the rate limiting enzyme in the primary bile acid synthesis pathway (Fig 3B), but increased levels of CYP27A1 protein (rate limiting enzyme in the secondary bile acid synthesis pathway) by 1.6- and 2.3-fold, respectively (Fig 3C). In contrast, neither LKO nor DKO altered levels of ACAT-2, which diverts cholesterol from bile acid synthesis to storage as cholesteryl esters, (Fig 3A); however, DKO increased levels of 3-αHSD, another bile acid binding protein potentially compensating for loss of L-FABP (Fig 3D). Although LKO and DKO altered hepatic levels of some of the major nuclear receptors regulating bile acid synthetic enzymes in a complex manner, there was no obvious pattern associated with decreased biliary bile acid and hepatic bile acid levels (Fig 3E–H). This was consistent with the fact that the activity of these nuclear receptors in bile acid synthesis is primarily regulated by ligand availability rather than altered levels of the nuclear receptor.
Figure 3. Key hepatic proteins involved in cholesterol esterification and bile acid synthesis are increased in L-FABP/SCP-2/SCP-x gene-ablated mice.
Aliquots of liver homogenate proteins were examined by SDS-PAGE and subsequent Western blot analysis to determine levels of ACAT-2 (panel A), CYP27A1 (panel C), 3-αHSD (panel D), PPARα (panel E), LXRα (panel F), SHP (panel G), and FXR (panel H) as described in Experimental Procedures. Insets show representative Western blot images of the respective protein (lower blot) and the gel-loading control protein (COX4, GAPDH, or β-Actin, upper blot). Quantitative rtPCR to determine the abundance of hepatic Cyp7a1 mRNA (panel B) was performed as described in Experimental Procedures. Relative concentration values (WT = 1) represent means ± SEM (n = 8). Statistically different values (P < 0.05, ANOVA) within a panel are denoted by a different lower-case letter (a, b, c).
Combinatorial loss of both L-FABP and SCP-2/SCP-x (TKO) did not elicit additive or synergistic effects on expression of enzymes (Fig 3B–D) or nuclear receptors in bile acid synthesis (Fig 3E–H). However, TKO did increase hepatic mRNA levels of the rate limiting enzyme in bile acid synthesis (Cyp7A1) (Fig 3B) and another potential bile acid transporter (3-αHSD, Fig 3D), while simultaneously increasing levels of ACAT-2 (Fig 3A).
Thus, the decreased biliary bile acid and total bile acid in LKO, DKO and TKO mice was not associated with reduced levels of the two rate limiting enzymes in bile acid synthesis (CYP7A1, CYP27A1), but at least in the case of TKO correlated in part with increased expression of ACAT-2, which could divert bile acid synthesis substrate cholesterol toward storage/secretion as cholesteryl esters.
4. DISCUSSION
While increasing evidence has suggested a role for L-FABP and SCP-2/SCP-x gene products in bile acid metabolism and biliary secretion, studies with individually ablating genes such SCP-2/SCP-x in mice have been complicated by concomitant upregulation of L-FABP [36;37;47]. To begin to resolve the relative contribution of L-FABP and SCP-2/SCP-x, biliary phenotype was examined in female mice singly ablated in L-FABP (LKO) or SCP-2/SCP-x (DKO) as well as upon ablating both L-FABP and SCP-2/SCP-x (TKO). The data suggest unique roles of SCP-2/SCP-x and L-FABP:
First, loss of L-FABP (LKO) alone exerted the greatest impact on biliary phenotype including: decreased (liver, bile, and total) bile acid level and increase in most parameters of biliary cholesterol saturation. These findings were consistent with the fact that L-FABP: i) is a universal bile acid binding protein with high affinity for the products and most intermediates in the bile acid synthetic pathway [10–17]; ii) binds the substrate of bile acid synthesis, i.e. cholesterol [22;23]; iii) is the most prevalent bile acid- and cholesterol-binding protein present in mammalian liver with levels as high as 2–6% of murine cytosolic protein (200–400 μM) and 7–10% of human cytosolic protein (700–1000 μM) [11;18]. L-FABP is essential for bile acid uptake and transport by rat hepatocytes [15]. Modest upregulation of canalicular salt bile export protein (BSEP) did not appear to compensate for the loss of L-FABP leading to decreased biliary bile acid. In contrast, the impact of SCP-2/SCP-x gene ablation (DKO, TKO) was less severe, decreasing only biliary bile acid (and total bile acid) with much fewer increased indices of biliary cholesterol saturation. This was consistent with the fact that DKO concomitantly upregulated two key bile acid binding proteins, i.e. L-FABP [35–38] and 3-αHSD (Fig 3D). Nevertheless, biliary bile acid secretion rates are decreased in DKO mice [36]. Finally, it is important to note that combinatorial loss of both L-FABP and SCP-2/SCP-x (TKO) did not synergistically further decrease biliary bile acid levels—suggesting that SCP-2/SCP-x played less of a role than L-FABP in transporting bile acids for secretion into bile.
Second, loss of L-FABP alone also had a greater impact on altering biliary bile acid species composition. This finding may be due to the fact that: i) L-FABP (but not SCP-2) binds multiple bile acids and intermediates in bile acid synthesis [10]; ii) L-FABP has much higher affinity for some bile acids (e.g. chenodeoxycholic acid, glycochenodeoxycholic acid, deoxycholic acid, ursodeoxycholic acid) than others (e.g. cholic acid, glycocholic acid, taurocholic acid) [10]. Unfortunately, while L-FABP increased the proportion of the most common biliary bile acid, i.e. tauro-muricholic acid (Table 2), L-FABP’s affinity for tauro-muricholic acid is not known; iii) L-FABP and SCP-2 differ markedly in intracellular level, consistent with L-FABP having a greater impact on cholesterol targeting for bile acid synthesis. L-FABP is 10–40 fold more prevalent than SCP-2 in hepatic cytoplasm, 1–10% versus 0.1% of cytosolic protein, respectively, and it is optimally localized for delivery of cholesterol and inter-organellar transport of bile acid synthetic intermediates [11;18;42]. Although neither SCP-2 nor SCP-x binds bile acids, both have roles in the synthesis of early steps in the synthesis of multiple bile acids. For example, studies in vitro show that SCP-2 stimulates the initiating step of cholesterol oxidation mediated by CYP7A1 in the endoplasmic reticulum leading to bile acid synthesis in vitro [28]. Consistent with this, SCP-2 overexpression in mice as well as in rat and human hepatocytes enhances bile acid synthesis and biliary secretion [33;34]. SCP-x is exclusively peroxisomal where it is localized for cleavage of the branched-side chain to form cholic acid from which other bile acids are derived [24;42;63]. The effect L-FABP has on the activity of endoplasmic reticulum CYP7A1 or on peroxisomal SCP-x, the key enzyme for cleaving cholesterol’s branched-side chain to form bile acids, is not known [29;32;64;65]. Despite their individual contributions, however, additive or synergistic responses were not seen in reducing biliary bile acid and total bile acid level in the combinatorial (i.e. TKO) knock out mice.
Third, there is clear sexual dimorphism in biliary phenotype expressed by mice lacking L-FABP (LKO), SCP-2/SCP-x (DKO), or both L-FABP and SCP-2/SCP-x (TKO). Since sexual dimorphism in mice has been demonstrated in the metabolism of branched-chain lipids such as phytol [49] and cholesterol [20, 51], we investigated potential sexual dimorphism in downstream bile acid metabolites of cholesterol and the biliary phenotype of female mice for comparison with an earlier study in male mice [21]. While biliary bile acid levels were decreased in female LKO, DKO, and TKO mice, as shown herein, our laboratory has previously shown that biliary bile acid levels decreased only in male DKO mice and actually increased in male LKO mice [21]. Likewise, while hepatic bile acid level decreased only in female LKO mice, as shown in the current study, both LKO and TKO decreased hepatic bile acid concentration in male mice [21]. Again, while biliary cholesterol levels were unaltered regardless of phenotype in female mice shown herein, biliary cholesterol concentration was decreased in all gene-ablated male mice (LKO, DKO, TKO) [21]. In contrast, biliary phospholipid level was decreased only in TKO female mice, while biliary phospholipid concentration was unaltered in any of the male gene targeted mice [21]. Finally, while loss of either or both genes significantly altered the molecular composition of biliary bile acids, the proportion of conjugated/unconjugated bile acids or overall bile acid hydrophobicity index remained unaltered in female mice, as shown herein. On the other hand, LKO, DKO, and TKO all increased bile acid hydrophobicity index in male mice [21]. While elucidating all the molecular bases for these sex-differences is beyond the scope of the current work, it is important to note that female WT mice express significantly more hepatic L-FABP [47–49;66] and markedly less SCP-x than their male counterparts [47;49]. L-FABP is the major bile acid binding protein in mouse and human liver [10;21]. Both in vitro and cultured cell studies have shown that SCP-x is the only known peroxisomal 3-keto-thiolase enzyme capable of oxidizing cholesterol’s branched side chain to form bile acids in the liver [28–30], sex steroids in the gonads [67], and steroid hormones in the adrenals [31;68]. Studies reporting a higher incidence of non-alcoholic fatty liver disease (NAFLD) and higher hepatic triglyceride accumulation in males than females, an increasing rate of NAFLD in postmenopausal women, and severe hepatic steatosis in mice ablated in aromatase suggest that NAFLD may be estrogen-associated [69–73]. Further, high-density lipoprotein-2 (HDL2) particles from females are larger and more capable of mediating cholesterol efflux from peripheral tissues [74;75].
Our laboratory has shown the roles played by both L-FABP and SCP-2/SCP-x in hepatic lipid accumulation in female mice [76]. The current findings with SCP-2/SCP-x, L-FABP, and SCP-2/SCP-x/L-FABP null female mice demonstrate important roles for L-FABP in hepatic bile acid accumulation and for both SCP-2/SCP-x and L-FABP in biliary phospholipid accumulation therein. Ablating each or both of these proteins all reduced biliary bile acid and total bile acid levels. Biliary indices of cholesterol saturation were increased the most upon loss of L-FABP (LKO, TKO) and less so in DKO mice, possibly due to concomitant upregulation of L-FABP in DKO mice [36;37;47]. Consequently, loss of L-FABP impacted biliary bile parameters to a greater extent than loss of SCP-2, not only by decreasing biliary bile acid levels but increasing the extent to which bile is saturated with cholesterol. Further, overall synergistic responses were not seen in biliary bile acid and total bile acid level in the combinatorial knock out mice.
Highlights.
L-FABP and/or SCP-2/SCP-x gene ablation in female mice
L-FABP gene ablation: decreased hepatic retention of bile acids
L-FABP and/or SCP-2/SCP-x gene ablation: decreased biliary bile acid levels
Significant sexual dimorphism in gene-ablated mice
Acknowledgments
This work was supported in part by the United States Public Health Service, National Institutes of Health Grants DK41402 (F.S. and A.B.K.), DK70965 (B.P.A), and HL078900 (P.N.H.). HPLC services were provided by the University of Cincinnati Mouse Metabolic Phenotyping Center (U24 DK059630).
Abbreviations
- ABCG5 or G8
ATP-binding cassette transporter G5 or G8
- ACAT-2
acyl-CoA cholesterol acyltransferase-2; B
- BA
bile acid
- C
cholesterol
- CA
cholic acid
- CE
cholesteryl ester
- CSI
cholesterol saturation index
- CYP7A1
cholesterol 7α-hydroxylase
- CYP27A1
sterol 27-hydroxylase
- DKO
SCP-2/SCP-x double null mouse
- FXR
farnesoid x receptor
- G-CA
glyco-cholic acid
- HDL
high density lipoprotein
- HI
hydrophobicity index
- 3α-HSD
3α-hydroxysteroid dehydrogenase
- LCFA
long chain fatty acid
- LDL
low density lipoprotein
- L-FABP
liver fatty acid binding protein or FABP1
- LKO
L-FABP null mouse
- LXRα
liver x receptor α
- α–MCA
α-muricholic acid
- β–MCA
β-muricholic acid
- MDR3
multidrug-resistance-3 P-glycoprotein
- OATP1 or 2
organic anion transporting polypeptide 1 or 2
- PL
phospholipid
- PPARα
−β/δ, or −γ, peroxisome proliferator-activated receptor alpha, beta/delta, or gamma
- QrtPCR
quantitative real-time polymerase chain reaction
- SCP-2
sterol carrier protein-2
- SCP-x
sterol carrier protein-x/peroxisomal thiolase 2
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SHP
short heterodimer partner
- T-CA
tauro-cholic acid
- T-CDCA
tauro-chenodeoxycholic acid
- T-DCA
tauro-deoxycholic acid
- TG
triglyceride
- TKO
L-FABP/SCP-2/SCP-x triple null mouse
- T-LCA
tauro-lithocholic acid
- T-MCA
tauro-muricholic acid
- T-UDCA
tauro-ursodeoxycholic acid
- WT
wild-type C57BL/6NCr mouse
Footnotes
Author contributions
G.G.M., planned experiments, performed experiments, analyzed data, wrote the paper; B.P.A., planned experiments, performed experiments; K.K.L., performed experiments, analyzed data; D.L., planned experiments, performed experiments, analyzed data; P.N.H., planned experiments, performed experiments, analyzed data; F.S. and A.B.K, planned experiments, wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Li T, Chiang JYL. Regulation of bile acid and cholesterol metabolism by PPARs. PPAR Research. 2009;2009:Article ID 501739–15. doi: 10.1155/2009/501739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lefebvre P, Cariou B, Lien F, Kuipers F, Staels B. Role of bile acids and bile acid receptors in metabolic regulation. Physiol Rev. 2009;89:147–191. doi: 10.1152/physrev.00010.2008. [DOI] [PubMed] [Google Scholar]
- 3.Monaco HL. The liver bile acid binding proteins. Biopolymers. 2009;91:1196–1202. doi: 10.1002/bip.21257. [DOI] [PubMed] [Google Scholar]
- 4.Guariento M, Raimondo D, Assfalg M, Zanzoni S, Pesente P, Ragona L, Tramontano A, Molinari H. Identification and functional characterization of the bile acid transport proteins in non-mammalian ileum and mammalian liver. Proteins. 2008;70:462–472. doi: 10.1002/prot.21518. [DOI] [PubMed] [Google Scholar]
- 5.Agellon LB, Torchia EC. Intracellular transport of bile acids. Biochim Biophys Acta. 2000;1486:198–209. doi: 10.1016/s1388-1981(00)00057-3. [DOI] [PubMed] [Google Scholar]
- 6.Russell DW. The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem. 2003;72:137–174. doi: 10.1146/annurev.biochem.72.121801.161712. [DOI] [PubMed] [Google Scholar]
- 7.Trauner M, Boyer JL. Bile salt transporters: Molecular characterization, function, and regulation. Physiol Rev. 2003;83:633–671. doi: 10.1152/physrev.00027.2002. [DOI] [PubMed] [Google Scholar]
- 8.Takikawa H, Kaplowitz N. Binding of bile acids, oleic acid, and organic anions by rat and human hepatic Z protein. Arch Biochem Biophys. 1986;251:385–392. doi: 10.1016/0003-9861(86)90086-x. [DOI] [PubMed] [Google Scholar]
- 9.Stolz A, Takikawa H, Ookhtens M, Kaplowitz N. The role of cytoplasmic proteins in hepatic bile acid transport. Ann Rev Physiol. 1989;51:161–176. doi: 10.1146/annurev.ph.51.030189.001113. [DOI] [PubMed] [Google Scholar]
- 10.Favretto F, Santambrogio C, D’Onofrio M, Molinari H, Grandori R, Assfalg M. Bile salt recognition by human liver fatty acid binding protein. FEBS J. 2015;282:1271–1288. doi: 10.1111/febs.13218. [DOI] [PubMed] [Google Scholar]
- 11.Favretto F, Assfalg M, Gallo M, Cicero DO, D’Onofrio M, Molinari H. Ligand binding promiscuity and human liver fatty acid binding protein: structural and dynamic insights from an interaction study with glycocholate and oleate. ChemBioChem. 2013;14:1807–1819. doi: 10.1002/cbic.201300156. [DOI] [PubMed] [Google Scholar]
- 12.Hagan RM, Worner-Gibbs J, Wilton DC. Tryptophan insertion mutagenesis of liver fatty acid binding protein. J Biol Chem. 2005;280:1782–1789. doi: 10.1074/jbc.M407131200. [DOI] [PubMed] [Google Scholar]
- 13.Di Pietro SM, Santome JA. Isolation, characterization, and binding properties of two rat liver fatty acid binding protein isoforms. Biochim Biophys Acta. 2000;1478:186–200. doi: 10.1016/s0167-4838(00)00042-x. [DOI] [PubMed] [Google Scholar]
- 14.Dietrich A, Dieminger W, Fuchte K, Stoll GH, Schlitz E, Gerok W, Kurz G. Functional signficance of interaction of hepatic FABP with sulfated and nonsulfated taurine-conjugated bile salts in rat liver. J Lipid Res. 1995;36:1745–1755. [PubMed] [Google Scholar]
- 15.Dietrich A, Dieminger W, Nelly SM, Gerok W, Kurz G. Synthesis and applicability of a photolabile 7,7-azi analogue of 3-sulfated taurine-conjugated bile acids. J Lipid Res. 1995;36:1729–1744. [PubMed] [Google Scholar]
- 16.Thumser AE, Wilton DC. The binding of cholesterol and bile salts to recombinant rat liver fatty acid-binding protein. Biochem J. 1996;320:729–733. doi: 10.1042/bj3200729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaikaus RM, Bass NM, Ockner RK. Functions of fatty acid binding proteins. Experientia. 1990;46:617–630. doi: 10.1007/BF01939701. [DOI] [PubMed] [Google Scholar]
- 18.McArthur MJ, Atshaves BP, Frolov A, Foxworth WD, Kier AB, Schroeder F. Cellular uptake and intracellular trafficking of long chain fatty acids. J Lipid Res. 1999;40:1371–1383. [PubMed] [Google Scholar]
- 19.Singer SS, Dravis D, Henkels K, Trulzsch DV. Fatty acid binding protein inhibits glycolithocholate sulfation. Biochemistry Intl. 1992;27:373–383. [PubMed] [Google Scholar]
- 20.Martin GG, Atshaves BP, McIntosh AL, Mackie JT, Kier AB, Schroeder F. Liver fatty acid binding protein (L-FABP) gene ablation alters liver bile acid metabolism in male mice. Biochem J. 2005;391:549–560. doi: 10.1042/BJ20050296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin GG, Atshaves BP, Landrock KK, Landrock D, Storey SM, Howles PN, Kier AB, Schroeder F. Ablating L-FABP in SCP-2/SCP-x null mice impairs bile acid metabolism and biliary HDL-cholesterol secretion. Am J Physiol Gastrointest and Liver Phys. 2014;307:G1130–G1143. doi: 10.1152/ajpgi.00209.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin GG, Atshaves BP, Huang H, McIntosh AL, Williams BW, Pai P-J, Russell DH, Kier AB, Schroeder F. Hepatic phenotype of liver fatty acid binding protein (L-FABP) gene ablated mice. Am J Physiol. 2009;297:G1053–G1065. doi: 10.1152/ajpgi.00116.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang H, McIntosh AL, Martin GG, Landrock KK, Landrock D, Storey SM, Gupta S, Atshaves BP, Kier AB, Schroeder F. Human L-FABP T94A variant enhances cholesterol uptake. Biochim Biophys Acta. 2015;1851:946–955. doi: 10.1016/j.bbalip.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin GG, Hostetler HA, McIntosh AL, Tichy SE, Williams BJ, Russell DH, Berg JM, Spencer TA, Ball JA, Kier AB, Schroeder F. Structure and function of the sterol carrier protein-2 (SCP-2) N-terminal pre-sequence. Biochem. 2008;47:5915–5934. doi: 10.1021/bi800251e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stolowich NJ, Frolov A, Atshaves BP, Murphy E, Jolly CA, Billheimer JT, Scott AI, Schroeder F. The sterol carrier protein-2 fatty acid binding site: an NMR, Circular Dichroic, and Fluorescence spectroscopic determination. Biochemistry. 1997;36:1719–1729. doi: 10.1021/bi962317a. [DOI] [PubMed] [Google Scholar]
- 26.Stolowich NJ, Frolov A, Petrescu AD, Scott AI, Billheimer JT, Schroeder F. Holo-sterol carrier protein-2: 13C-NMR investigation of cholesterol and fatty acid binding sites. J Biol Chem. 1999;274:35425–35433. doi: 10.1074/jbc.274.50.35425. [DOI] [PubMed] [Google Scholar]
- 27.Stolowich NJ, Petrescu AD, Huang H, Martin G, Scott AI, Schroeder F. Sterol carrier protein-2: structure reveals function. Cell Mol Life Sci. 2002;59:193–212. doi: 10.1007/s00018-002-8416-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawata S, Imai Y, Inada M, Inui M, Kakimoto H, Fukuda K, Maeda Y, Tarui S. Modulation of cholesterol 7-a hydroxylase activity by nsLTP in human liver - possibe altered regulation of its cytosolic level in patients with gallstones. Clin Chim Acta. 1991;197:201–208. doi: 10.1016/0009-8981(91)90140-8. [DOI] [PubMed] [Google Scholar]
- 29.Seedorf U, Brysch P, Engel T, Schrage K, Assmann G. Sterol carrier protein X is peroxisomal 3-oxoacyl coenzyme A thiolase with intrinsic sterol carrier and lipid transfer activity. J Biol Chem. 1994;269:21277–21283. [PubMed] [Google Scholar]
- 30.Wanders RJA, Denis S, Wouters F, Wirtz KWA, Seedorf U. Sterol carrier protein X (SCPx) is a peroxisomal branched-chain b-ketothiolase specifically reacting with 3-Oxo-pristanoyl-CoA: A new, unique role for SCPx in branched-chain fatty acid metabolism in peroxisomes. Biochem Biophys Res Commun. 1997;236:565–569. doi: 10.1006/bbrc.1997.7007. [DOI] [PubMed] [Google Scholar]
- 31.Chanderbhan RF, Kharroubi A, Pastuszyn A, Gallo LL, Scallen T. Direct evidence for sterol carrier protein-2 (SCP-2) participation in ACTH stimulated steroidogenesis in isolated adrenal cells. In: Chang TY, Freeman DA, editors. Intracellular Cholesterol Trafficking. Kluwer Academic Publishers; Boston: 1998. pp. 197–212. [Google Scholar]
- 32.Antonenkov VD, Van Veldhoven PP, Waelkens E, Mannaerts GP. Substrate specificities of 3-oxoacyl-CoA thiolase A and sterol carrier protein 2/3-oxoacyl-CoA thiolase purified from normal rat liver peroxisomes. J Biol Chem. 1997;272:26023–26031. doi: 10.1074/jbc.272.41.26023. [DOI] [PubMed] [Google Scholar]
- 33.Amigo L, Zanlungo S, Miquel JF, Glick JM, Hyogo H, Cohen DE, Rigotti A, Nervi F. Hepatic overexpression of sterol carrier protein-2 inhibits VLDL production and reciprocally enhances biliary lipid secretion. J Lipid Res. 2003;44:399–407. doi: 10.1194/jlr.M200306-JLR200. [DOI] [PubMed] [Google Scholar]
- 34.Ren S, Hylemon P, Marques D, Hall E, Redford K, Gil G, Pandak WM. Effect of increasing the expression of cholesterol transporters (StAR, MLN64, and SCP-2) on bile acid synthesis. J Lipid Res. 2004;45:2123–2131. doi: 10.1194/jlr.M400233-JLR200. [DOI] [PubMed] [Google Scholar]
- 35.Kannenberg F, Ellinghaus P, Assmann G, Seedorf U. Aberrant oxidation of the cholesterol side chain in bile acid synthesis of sterol carrier protein-2/sterol carrier protein-x knockout mice. J Biol Chem. 1999;274:35455–35460. doi: 10.1074/jbc.274.50.35455. [DOI] [PubMed] [Google Scholar]
- 36.Fuchs M, Hafer A, Muench C, Kannenberg F, Teichmann S, Scheibner J, Stange EF, Seedorf U. Disruption of the sterol carrier protein 2 gene in mice impairs biliary lipid and hepatic cholesterol metabolism. J Biol Chem. 2001;276:48058–48065. doi: 10.1074/jbc.M106732200. [DOI] [PubMed] [Google Scholar]
- 37.Seedorf U, Raabe M, Ellinghaus P, Kannenberg F, Fobker M, Engel T, Denis S, Wouters F, Wirtz KWA, Wanders RJA, Maeda N, Assmann G. Defective peroxisomal catabolism of branched fatty acyl coenzyme A in mice lacking the sterol carrier protein-2/sterol carrier protein-x gene function. Genes and Development. 1998;12:1189–1201. doi: 10.1101/gad.12.8.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Storey SM, Atshaves BP, McIntosh AL, Landrock KK, Martin GG, Huang H, Johnson JD, Macfarlane RD, Kier AB, Schroeder F. Effect of sterol carrier protein-2 gene ablation on HDL-mediated cholesterol efflux from primary cultured mouse hepatocytes. Am J Physiol. 2010;299:244–254. doi: 10.1152/ajpgi.00446.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murphy EJ, Schroeder F. Sterol carrier protein-2 mediated cholesterol esterification in transfected L-cell fibroblasts. Biochimica et Biophysica Acta. 1997;1345:283–292. doi: 10.1016/s0005-2760(97)00003-9. [DOI] [PubMed] [Google Scholar]
- 40.Jefferson JR, Slotte JP, Nemecz G, Pastuszyn A, Scallen TJ, Schroeder F. Intracellular sterol distribution in transfected mouse L-cell fibroblasts expressing rat liver fatty acid binding protein. J Biol Chem. 1991;266:5486–5496. [PubMed] [Google Scholar]
- 41.Frolov AA, Woodford JK, Murphy EJ, Billheimer JT, Schroeder F. Fibroblast membrane sterol kinetic domains: modulation by sterol carrier protein 2 and liver fatty acid binding protein. J Lipid Res. 1996;37:1862–1874. [PubMed] [Google Scholar]
- 42.Gallegos AM, Atshaves BP, Storey SM, Starodub O, Petrescu AD, Huang H, McIntosh A, Martin G, Chao H, Kier AB, Schroeder F. Gene structure, intracellular localization, and functional roles of sterol carrier protein-2. Prog Lipid Res. 2001;40:498–563. doi: 10.1016/s0163-7827(01)00015-7. [DOI] [PubMed] [Google Scholar]
- 43.Gavey KL, Noland BJ, Scallen TJ. The participation of sterol carrier protein2 in the conversion of cholesterol to cholesterol ester by rat liver microsomes. J Biol Chem. 1981;256:2993–2999. [PubMed] [Google Scholar]
- 44.Nemecz G, Schroeder F. Selective binding of cholesterol by recombinant fatty acid-binding proteins. J Biol Chem. 1991;266:17180–17186. [PubMed] [Google Scholar]
- 45.Chao H, Billheimer JT, Kier AB, Schroeder F. Microsomal long chain fatty acyl CoA transacylation: differential effect of SCP-2. Biochim Biophys Acta. 1999;1439:371–383. doi: 10.1016/s1388-1981(99)00109-2. [DOI] [PubMed] [Google Scholar]
- 46.Chao H, Zhou M, McIntosh A, Schroeder F, Kier AB. Acyl CoA binding protein and cholesterol differentially alter fatty acyl CoA utilization by microsomal acyl CoA: cholesterol transferase. J Lipid Res. 2003;44:72–83. doi: 10.1194/jlr.m200191-jlr200. [DOI] [PubMed] [Google Scholar]
- 47.Atshaves BP, McIntosh AL, Landrock D, Payne HR, Mackie J, Maeda N, Ball JM, Schroeder F, Kier AB. Effect of SCP-x gene ablation on branched-chain fatty acid metabolism. Am J Physiol. 2007;292:939–951. doi: 10.1152/ajpgi.00308.2006. [DOI] [PubMed] [Google Scholar]
- 48.Atshaves BP, McIntosh AL, Martin GG, Landrock D, Payne HR, Bhuvanendran S, Landrock K, Lyuksyutova OI, Johnson JD, Macfarlane RD, Kier AB, Schroeder F. Overexpression of sterol carrier protein-2 differentially alters hepatic cholesterol accumulation in cholesterol-fed mice. J Lipid Res. 2009;50:1429–1447. doi: 10.1194/jlr.M900020-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Atshaves BP, Payne HR, McIntosh AL, Tichy SE, Russell D, Kier AB, Schroeder F. Sexually dimorphic metabolism of branched chain lipids in C57BL/6J mice. J Lipid Res. 2004;45:812–830. doi: 10.1194/jlr.M300408-JLR200. [DOI] [PubMed] [Google Scholar]
- 50.Antoine B, Lefrancois-Martinez A-M, Guillou GL, Leturque A, Vandewalle A, Kahn A. Role of the GLUT2 glucose transporter in the response of the L-type pyruvate kinase gene to glucose in liver derived cells. J Biol Chem. 1997;272:17937–17943. doi: 10.1074/jbc.272.29.17937. [DOI] [PubMed] [Google Scholar]
- 51.Martin GG, Atshaves BP, McIntosh AL, Mackie JT, Kier AB, Schroeder F. Liver fatty acid binding protein (L-FABP) gene ablation potentiates hepatic cholesterol accumulation in cholesterol-fed female mice. Am J Physiol. 2006;290:G36–G48. doi: 10.1152/ajpgi.00510.2004. [DOI] [PubMed] [Google Scholar]
- 52.Martin GG, Danneberg H, Kumar LS, Atshaves BP, Erol E, Bader M, Schroeder F, Binas B. Decreased liver fatty acid binding capacity and altered liver lipid distribution in mice lacking the liver fatty acid binding protein (L-FABP) gene. J Biol Chem. 2003;278:21429–21438. doi: 10.1074/jbc.M300287200. [DOI] [PubMed] [Google Scholar]
- 53.Atshaves BP, McIntosh AL, Payne HR, Gallegos AM, Landrock K, Maeda N, Kier AB, Schroeder F. Sterol carrier protein-2/sterol carrier protein-x gene ablation alters lipid raft domains in primary cultured mouse hepatocytes. J Lipid Res. 2007;48:2193–2211. doi: 10.1194/jlr.M700102-JLR200. [DOI] [PubMed] [Google Scholar]
- 54.Thigpen JE, Setchell KD, Ahlmark KB, Kocklear J, Spahr T, Caviness GF, Goelz MF, Haseman JK, Newbold RR, Forsythe DB. Phytoestrogen content of purified, open- and closed-formula laboratory animal diets. Lab An Science. 1999;49:530–536. [PubMed] [Google Scholar]
- 55.Thigpen JE, Setchell KD, Goelz MF, Forsythe DB. The phytoestrogen content of rodent diets. Envron Health Persp. 1999;107:A182–A183. doi: 10.1289/ehp.107-1566530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adida A, Spener F. Intracellular lipid binding proteins and nuclear receptors inolved in branched-chain fatty acid signaling. Prost Leukot Essen Fatty Acids. 2002;67:91–98. doi: 10.1054/plef.2002.0404. [DOI] [PubMed] [Google Scholar]
- 57.Ellinghaus P, Wolfrum C, Assmann G, Spener F, Seedorf U. Phytanic acid activates the peroxisome proliferator-activated receptor alpha (PPARalpha) in sterol carrier protein-2-/sterol carrier protein x-deficient mice. J Biol Chem. 1999;274:2766–2772. doi: 10.1074/jbc.274.5.2766. [DOI] [PubMed] [Google Scholar]
- 58.Hanhoff T, Benjamin S, Borchers T, Spener F. Branched-chain fatty acids as activators of peroxisome proliferators. Eur J Lip Sci Technol. 2005;107:716–729. [Google Scholar]
- 59.Wolfrum C, Ellinghaus P, Fobker M, Seedorf U, Assmann G, Borchers T, Spener F. Phytanic acid is ligand and transcriptional activator of murine liver fatty acid binding protein. J Lipid Res. 1999;40:708–714. [PubMed] [Google Scholar]
- 60.Carey MC. Critical tables for calculating the cholesterol saturation of native bile. J Lipid Res. 1978;19:945–955. [PubMed] [Google Scholar]
- 61.Atshaves BP, McIntosh AL, Lyuksyutova OI, Zipfel WR, Webb WW, Schroeder F. Liver fatty acid binding protein gene ablation inhibits branched-chain fatty acid metabolism in cultured primary hepatocytes. J Biol Chem. 2004;279:30954–30965. doi: 10.1074/jbc.M313571200. [DOI] [PubMed] [Google Scholar]
- 62.McIntosh AL, Atshaves BP, Hostetler HA, Huang H, Davis J, Lyuksyutova OI, Landrock D, Kier AB, Schroeder F. Liver type fatty acid binding protein (L-FABP) gene ablation reduces nuclear ligand distribution and peroxisome proliferator activated receptor-alpha activity in cultured primary hepatocytes. Arch Biochem Biophys. 2009;485:160–173. doi: 10.1016/j.abb.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wouters F, Bastiaens PI, Wirtz KW, Jovin TM. FRET microscopy demonstrates molecular association of non-specific lipid transfer protein (nsL-TP) with fatty acid oxidation enzymes. EMBO J. 1998;17:7179–7189. doi: 10.1093/emboj/17.24.7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Antonenkov VD, Van Veldhoven PP, Mannaerts GP. Isolation and subunit composition of native sterol carrier protein-2/3-oxoacyl-coenzyme A thiolase from normal rat liver peroxisomes. Protein Exp Purif. 2000;18:249–256. doi: 10.1006/prep.2000.1192. [DOI] [PubMed] [Google Scholar]
- 65.Antonenkov VD, Sormunen RT, Ohlmeier S, Amery L, Fransen M. Localization of a portion of the liver isoform of fatty acid binding protein (L-FABP) to peroxisomes. Biochem J. 2006;394:475–484. doi: 10.1042/BJ20051058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Atshaves BP, McIntosh AL, Payne HR, Mackie J, Kier AB, Schroeder F. Effect of branched-chain fatty acid on lipid dynamics in mice lacking liver fatty acid binding protein gene. Am J Physiol. 2005;288:C543–C558. doi: 10.1152/ajpcell.00359.2004. [DOI] [PubMed] [Google Scholar]
- 67.Mendis-Handagama SM, Aten RF, Watkins PA, Scallen TJ, Berhman HR. Peroxisomes and sterol carrier protein-2 in luteal cell steroidogenesis: a possible role in cholesterol transport from lipid droplets to mitochondria. Tissue Cell. 1995;27:483–490. doi: 10.1016/s0040-8166(05)80056-4. [DOI] [PubMed] [Google Scholar]
- 68.Chanderbhan R, Kharroubi A, Noland BJ, Scallen TJ, Vahouny GV. Sterol carrier protein 2: Further evidence for its role in adrenol steroidogenesis. Endocrine Res. 1986;12:351–370. doi: 10.3109/07435808609035445. [DOI] [PubMed] [Google Scholar]
- 69.Farrell GC, McCullough AJ, Day CP. Non-alcoholic fatty liver disease: A practical guide. J. Wiley and Sons; Chichester, West Sussex, UK: 2013. What is non-alcoholic fatty liver disease (NAFLD), and why is it important; pp. 1–15. [Google Scholar]
- 70.Denzer C, Thiere D, Muche R, Koenig W, Mayer H, Kratzer W, Wabitsch M. Gender-specific prevalences of fatty liver in obese children and adolescents: roles of body fat distribution, sex steroids, and insulin resistance. J Clin Endocrinol Metab. 2009;94:3872–3881. doi: 10.1210/jc.2009-1125. [DOI] [PubMed] [Google Scholar]
- 71.Fan J-G, Saibara T, Chitturi S, Kim BI, Sung JY, Chutaputti A. What are risk factors and settings for NAFLD in Asia-Pacific? J Gastroent and Hepatol. 2007;22:794–800. doi: 10.1111/j.1440-1746.2007.04952.x. [DOI] [PubMed] [Google Scholar]
- 72.Vernon G, Baranova A, Younossi ZM. Systematic review: the epidemiology and natural history of NAFLD in adults. Aliment Pharmacol Ther. 2011;34:274–285. doi: 10.1111/j.1365-2036.2011.04724.x. [DOI] [PubMed] [Google Scholar]
- 73.Lonardo A, Trande P. Fatty liver, chronic hepatitis C and hepatocellular carcinoma. J Gastroent and Hepatol. 2000;15:775–782. doi: 10.1046/j.1440-1746.2000.02226.x. [DOI] [PubMed] [Google Scholar]
- 74.Catalano G, Duchene E, Julia Z, Le Goff W, Bruckert E, Chapman MJ, Guerin M. Cellular SRB1 and ABCA1 mediated cholesterol efflux are gender specific in healthy subjects. J Lipid Res. 2008;49:635–643. doi: 10.1194/jlr.M700510-JLR200. [DOI] [PubMed] [Google Scholar]
- 75.Sodre FL, Castanho VS, Castilho LN, de Barros-Mazon S, de Faria EC. HDL subfractions in normolipidemic individuals wihtout clinical atherosclerosis lipoprotein subfractions in an adult population. J Clin Lab Analysis. 2006;20:113–117. doi: 10.1002/jcla.20111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Martin GG, Atshaves BP, Landrock KK, Landrock D, Schroeder F, Kier AB. Loss of L-FABP, SCP-2/SCP-x, or both Induces Hepatic Lipid Accumulation in Female Mice. Arch Biochem Biophys. 2015;580:41–49. doi: 10.1016/j.abb.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Heuman DM. Quantitative estimation of the hydrophilic-hydrophobic balance of mixed bile salt solutions. J Lipid Res. 1989;30:719–730. [PubMed] [Google Scholar]