Abstract
Objective
Currently, depression diagnosis relies primarily on behavioral symptoms and signs, and treatment is guided by trial and error instead of evaluating associated underlying brain characteristics. Unlike past studies, we attempted to estimate accurate prediction models for late-life depression diagnosis and treatment response using multiple machine learning methods with inputs of multi-modal imaging and non-imaging whole brain and network-based features.
Methods
Late-life depression patients (medicated post-recruitment) [n=33] and elderly non-depressed individuals [n=35] were recruited. Their demographics and cognitive ability scores were recorded, and brain characteristics were acquired using multi-modal magnetic resonance imaging pre-treatment. Linear and nonlinear learning methods were tested for estimating accurate prediction models.
Results
A learning method called alternating decision trees estimated the most accurate prediction models for late-life depression diagnosis (87.27% accuracy) and treatment response (89.47% accuracy). The diagnosis model included measures of age, mini-mental state examination score, and structural imaging (e.g. whole brain atrophy and global white mater hyperintensity burden). The treatment response model included measures of structural and functional connectivity.
Conclusions
Combinations of multi-modal imaging and/or non-imaging measures may help better predict late-life depression diagnosis and treatment response. As a preliminary observation, we speculate the results may also suggest that different underlying brain characteristics defined by multi-modal imaging measures—rather than region-based differences—are associated with depression versus depression recovery since to our knowledge this is the first depression study to accurately predict both using the same approach. These findings may help better understand late-life depression and identify preliminary steps towards personalized late-life depression treatment.
Keywords: imaging, prediction, learning, late-life depression, diagnosis, treatment response
INTRODUCTION
In a given year, approximately 2 million people aged 65+ suffer from late-life depression (LLD) (Mental Health America). The current diagnosis and treatment of LLD is based on behavioral symptoms and signs. It lacks the reliability and validity that could accrue from biomarkers of underlying brain characteristics. To advance towards personalizing medicine, it is important to identify biomarkers reflecting the neural circuit abnormalities that characterize LLD.
Past studies have associated LLD diagnosis and treatment response with select few of the demographic (Blazer, 2012; Chang-Quan et al., 2010; Forlani et al., 2013; Katon et al., 2010; Luppa et al., 2012; Wild et al., 2012; Wu et al., 2012), clinical (Andreescu et al., 2008), cognition ability (Bhalla et al., 2005; Ganguli et al., 2006; Kohler et al., Apr 2010; Ribeiz et al., 2013; Wilkins et al., 2009), MR structural (Alexopoulos et al., 2008; Aizenstein et al., 2011; Change et al., 2011; Colloby et al., 2011; Crocco et al., 2010; Disabato et al., 2012; Firbank et al., 2012; Gunning et al., 2009; Gunning-Dixon et al., 2010; Kohler et al., Feb 2010; Mettenburg et al., 2012; Sexton et al., 2013; Shimony et al., 2009; Taylor et al., 2008; Taylor et al., 2011; Teodorczuk et al., 2010), and/or MR functional measures (Alalade et al., 2011; Alexopoulos et al., 2012; Andreescu et al., 2011; Andresscu et al., 2013; Bohr et al., 2012; Colloby et al., 2012; Liu et al., 2012a; Steffens et al., 2011; Wang et al., 2008; Wu et al., 2011). In this study, we use a broader spectrum of measures hoping to gain a more complete and accurate understanding of underlying brain mechanisms associated with LLD. Using a unique set of measures as features, we aimed to estimate accurate prediction models of LLD diagnosis and treatment response via machine learning; the goal being to improve the understand of LLD and take preliminary steps towards personalized treatment. Past studies have successfully done so in younger populations (Costafreda et al., 2009; Fu et al., 2008; Hahn et al., 2011; Liu et al., 2012b; Marquand et al., 2008; Mwangi et al., 2012a; Mwangi et al., 2012b; Nouretdinov et al., 2011; Zeng et al., 2012), but not for LLD.
Compared with mid-life depression, LLD has a different neural signature including gray matter (GM) and white matter (WM) structural changes (Aizenstein et al., 2014) and a more difficult treatment response (Andreescu and Reynolds, 2011). Considering the age- and disease-related complexity of brain structure and function in the elderly, we studied prediction models via generalized linear (L1 Regularized Logistic Regression (L1-LR) and Support Vector Machines with Linear Kernel (SVM-L)) and nonlinear (Alternating Decision Tree (ADTree) and Support Vector Machines with Radial Basis Function Kernel (SVM-RBF)) classification-based learning methods to accurately learn the nature of the data. SVM methods were chosen for their popularity in current literature (Costafreda et al., 2009; Fu et al., 2008; Liu et al., 2012b; Marquand et al., 2008; Mwangi et al., 2012a; Nouretdinov et al., 2011; Zeng et al., 2012), versatility in classifying data using linear and nonlinear functions, and ability to well classify data containing a large set of input features (Cortes and Vapnik, 1995). L1-LR and ADTree were chosen for their embedded feature selection abilities (i.e. inherent ability to select the most relevant features for estimating prediction models that best fit the data), easy-to-interpret resulting prediction models, and fast convergence speed (Yuan et al., 2010; Pfahringer et al., 2001). These three methods also differ in their resulting prediction models: SVMs estimate a function that optimally separates the data, L1-LR estimates a logistic function that optimally fits the data, and ADTree estimates a decision tree that optimally classifies the data.
Additionally, we focus on resting state networks (e.g. dorsal default mode network (dDMN) and anterior salience network (aSN)) associated with LLD by previous studies (Alalade et al., 2011; Alexopoulos et al., 2012; Andreescu et al., 2011; Andreescu et al., 2013; Bohr et al., 2012; Gunning et al., 2009; Steffens et al., 2011; Wu et al., 2011). Unlike past studies, which used region-based approaches (e.g. regions resulting from voxel-wise analysis (Costafreda et al., 2009; Fu et al., 2008; Hahn et al., 2011; Marquand et al., 2008; Liu et al., 2012b; Mwangi et al., 2012a; Mwangi et al., 2012b; Nouretdinov et al., 2011) or anatomical regions of interest (ROIs) (Zeng et al., 2012)), we perform a whole brain and network analyses using functional ROIs to reduce data complexity and more precisely represent the brain areas activated during a functional activity (e.g. resting state) of interest (Nieto-Castanon et al., 2003).
To our knowledge, this is the first study to estimate prediction models for LLD diagnosis and treatment response by evaluating: [1] the potential of multimodal magnetic resonance imaging (MRI) measures as biomarkers; [2] combinations and interactions between potential imaging and non-imaging predictors; [3] multiple learning methods; and [4] whole brain and network analyses using functional ROIs.
METHODS
Subject Recruitment
Non-psychotic, unipolar LLD patients (n=33) and elderly non-depressed (ND) (n=35) individuals were recruited from Pittsburgh’s Advanced Center for Intervention and Services Research for Late-Life Mood Disorders, and Alzheimer Disease Research Center’s healthy controls registry respectively. Each participant provided written informed consent after receiving a full description of the study. All participants were paid $50. Participants were evaluated using the Structured Clinical Interview for Diagnostic and Statistical Manual of Mental Disorders 4th edition (First et al., 1995). Exclusion criteria included history of Axis I disorders other than major depressive disorder and anxiety disorders, stroke, significant head injury, Alzheimer’s, Parkinson’s, and/or Huntington’s disease. Patients received 12 weeks of open-label treatment trials with duloxetine, venlafaxine, nimodipine, or escitalopram. Before treatment, participants were assessed for LLD severity using the Hamilton Depression Rating Scale (HAM-D). HAM-D score <10 after treatment defined treatment responder (Roose et al., 1994). Table 1 summarizes additional participants-related information (see Aizenstein et al. (2011) for more details). After all omissions, n>50 and n>19 participants were examined for LLD diagnosis and treatment response analyses respectively.
Table 1.
Summary of Participants-Related Information
| LLD Diagnosis Prediction | LLD Treatment Response Prediction | |||||
|---|---|---|---|---|---|---|
| Network | dDMN | aSN | Combined | dDMN | aSN | Combined |
| Number of ND | 28 (b,c) | 31 (b,c) | 28 (b,c) | n/a | n/a | n/a |
| Number of LLD | 27 (a,b,c) | 25 (a,b,c) | 22 (a,b,c) | 24 (a,b,c,d) | 22 (a,b,c,d) | 19 (a,b,c,d) |
| Number of Responders | n/a | n/a | n/a | 11 | 11 | 9 |
| Number of Non-Responders | n/a | n/a | n/a | 13 | 11 | 10 |
| Age (years) [mean (standard deviation)] | 70.20 (7.98) | 70.00 (7.85) | 69.78 (7.81) | 68.83 (7.52) | 68.36 (6.54) | 67.37 (6.45) |
| Gender (% Female) | 76.36% | 78.57% | 78.00 % | 75.00% | 81.82% | 78.95% |
| Education (years) [mean (standard deviation)] | 14.36 (2.45) | 14.43 (2.51) | 14.18 (2.40) | 14.75 (2.75) | 14.45 (2.69) | 14.37 (2.75) |
| MMSE (score) [mean (standard deviation)] | 28.40 (2.01) | 28.32 (2.08) | 28.36 (2.10) | 27.58 (2.60) | 27.41 (2.65) | 27.26 (2.83) |
| HAM-D (score) before Treatment (e) [mean (standard deviation)] | 20.33 (3.99) | 20.24 (3.84) | 20.50 (4.01) | 20.42 (4.19) | 20.32 (4.05) | 20.63 (4.26) |
| HAM-D (score) after Treatment (e) [mean (standard deviation)] | 10.08 (5.41) (f) | 9.86 (5.75) (f) | 10.11 (5.86) (f) | 10.08 (5.41) | 9.86 (5.75) | 10.11 (5.86) |
LLD participants (n=1) were omitted from the study due to excess head motion artifacts in fMRI image
ND (n=4) and LLD (n=2) participants were omitted from the study due to bad fMRI image registration to template
ND (dDMN: n=3; Combined: n=3) and LLD (dDMN: n=3; aSN: n=5; Combined: n=8) participants were omitted from the appropriate network-based analysis of this study due to bad ROIs registration to structural template
LLD (n=3) participants were omitted from the treatment response analysis of this study due to missing or partial HAM-D scores (this includes the only patient administered nimodipine)
Information regarding HAM-D scores is presented for LLD participants only.
HAM-D scores are missing for 3 participants and thus they were not included in the calculations.
Image Acquisition and Processing
Before treatment, a Siemens 3T TIM TRIO scanner and 12-channel head coil was used to acquire T1-weighted high resolution (Hi-Res), T2-weighted, T2-weighted fluid attenuated inversion recovery (FLAIR), diffusion tensor imaging (DTI) and resting state fMRI (rs-fMRI)—for which participants were asked to stay awake, think of nothing in particular and rest with eyes focused on a fixation cross—images. See Table 2 and Supplementary Table S1 for more details.
Table 2.
MRI image acquisition and processing
| MRI Image Type | T1-weighted Hi-Res | DTI | T2-weighted FLAIR | T2-weighted | rs-fMRI | |
|---|---|---|---|---|---|---|
| Image Acquisition | Slice Thickness | 1 mm | 3 mm | 3 mm | 3 mm | 3 mm |
| Resolution | 256×224 mm | 128×128 mm | 256×240 mm | 256×224 mm | 128×128 mm | |
| Field of View | 256×224 mm | 256×256 mm | 256×212 mm | 256×224 mm | 256×256 mm(a) | |
| Repetition Time | 2300 ms | 5300 ms | 9160 ms | 3000 ms | 2000 ms | |
| Echo Time | 3.43 ms | 88 ms | 88 ms | 11/101 ms | 34 ms | |
| Inversion Time | 900 ms | 2500 ms | 2500 ms | 100 ms | n/a | |
| Flip Angle | 9° | 90° | 150° | 150° | 90° | |
| Other | axial plane | axial plane | axial plane | axial plane | axial plane; integrated parallel acquisition technique = 2; gradient-echo-planar imaging sequence | |
| Image Processing (See Supplementary Table 1 for more details on feature acquisition using image processing) | Refer to Wu et al., 2006 for processing pipeline. For more details on the modified version of this pipeline used for this study, see Supplementary Table 2. | FMRIB Software Library (59) was used to preprocess images, acquire Mean Diffusivity (MD) and Fractional Anisotropy (FA) maps, and perform tractography. | Refer to Wu et al., 2006 for the processing pipeline. | Used for rs-fMRI preprocessing. | CONN toolbox (60) was used to preprocess images, extract resting state signals, and performing Fisher transformed correlations between ROIs. | |
For the rs-fMRI images, the field of view did not fully cover the cerebellum for most subjects
Feature and ROI Selection
Thirteen different feature sets (see Table 3) selected by force (i.e. a feature(s) was explicitly chosen to be removed from the full set of features) were analyzed using the machine learning methods. Forced feature selection using these specific 13 feature sets was performed to study the influence of different MRI modalities, imaging vs. non-imaging measures, and each individual feature on the prediction of LLD diagnosis and treatment response.
Table 3.
Summary of Features and Description of Feature Sets (Note: Blocks in gray indicate the features removed for each set)
| Feature Type | Feature | Feature Description | Feature Representation | Feature Sets | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | ||||
| Demographics | Age | Age | Whether younger or older old adult | |||||||||||||
| Demographics | Gender | Gender | Whether female or male | |||||||||||||
| Demographics | Education | Level of Education | Number of years formal education was received | |||||||||||||
| Cognitive Ability | MMSE | Mini-Mental State Examination score |
Whether strong or poor cognitive ability | |||||||||||||
| Functional Imaging (rs-fMRI) | Functional Connectivity Index (FCI) | Average of Fisher transformed correlation coefficients between each pair of ROIs | Degree of functional connectivity—allowing for communication between ROIs— within network | |||||||||||||
| Structural Imaging (T1-weighted Hi-Res) | Normalized whole brain tissue volume (NWBTV) | Normalized whole brain gray and white matter volume | Degree of whole brain atrophy | |||||||||||||
| Structural Imaging (T1-weighted Hi-Res) | Normalized ROIs’ tissue volume (NRTV) | Average of normalized ROIs’ gray matter volumes | Degree of network-based regional brain atrophy | |||||||||||||
| Structural Imaging (DTI) | Mean Diffusivity (MD) | Average of all ROIs’ mean gray matter MD | Amount of diffusivity within gray matter of network | |||||||||||||
| Structural Imaging (DTI) | Number of tracks | Average of the approximate number of tracks connecting each pair of ROIs | Amount of structural connectivity—allowing for communication between ROIs— within network | |||||||||||||
| Structural Imaging (DTI) | Fractional Anisotropy (FA) | Average of all weighted mean FA computed along tracks connecting each pair of ROIs | Amount of white matter integrity within network | |||||||||||||
| Structural Imaging (T2-weighted FLAIR) | Global White Matter Hyperintensities (WMHs) | WMH burden in cerebral cortex | Amount of global (i.e. whole brain) WMH burden or white matter lesions | |||||||||||||
| Structural Imaging (T2-weighted FLAIR) | Local WMHs | Average WMH burden among tracks connecting each pair of ROIs | Amount of WMH burden or white matter lesions within network | |||||||||||||
All methods were repeated for the dDMN, aSN, and combined network analyses. Network-related functional ROIs were obtained from the FIND Lab at Stanford University (Shirer et al., 2012). The regions used to compute whole network measures for the dDMN analysis include: medial prefrontal cortex, anterior and posterior cingulate cortex, left and right thalamus, left and right hippocampus, left and right angular gyrus, orbitofrontal cortex, angular gyrus, right superior frontal gyrus, precuneus, and midcingulate cortex. The regions used to compute whole network measures for the aSN analysis include: left and right middle frontal gyrus, left and right insula, anterior cingulate cortex, medial prefrontal cortex, supplementary motor area, left and right lobule VI, and crus I.
Machine Learning Methods
Each learning method was used to estimate prediction models for two expected outcome variables separately: depression and treatment response. A nested leave-one-out cross-validation method was used to perform parameter selection and determine classification accuracy of each outcome variable. Resulting average training and test set classification accuracies—along with respective specificity, sensitivity, and receiver operating characteristic (ROC) curve measures—were recorded for all tests performed.
For L1-LR, a coordinate descent method using one-dimensional Newton directions described by Yuan et al. (2010) was coded and implemented in-house using Python. To improve the results, we added an input feature of constant value equal to one for each data instance. This additional feature acts as a bias or intercept term that also affects the regularization term. To implement SVM methods, the scikit-learn Python library (Pedregosa et al., 2011) was used. To improve the results, we also implemented a feature selection filter technique using Kendall tau correlation coefficient presented by Zeng et al. (2012). The optimized version of ADTree described by Pfahringer et al. (2001) was coded and implemented in-house using Python. To improve results, we restricted the tree depth to 3 branches and minimized the number of weak hypotheses tested per boosting iteration to half the number of unique values for each feature. See Supplementary Methods for more details
RESULTS
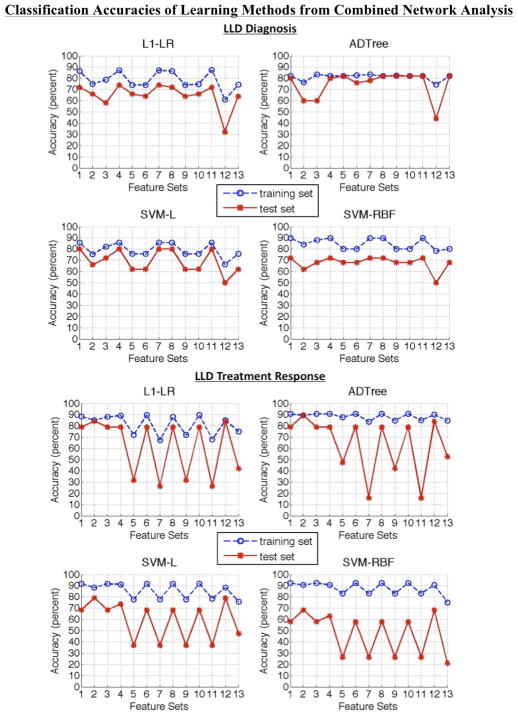
Figure 1 and Supplementary Figure S1 display a summary of all the results discussed below and in the supplemental material. Below we evaluate potential optimal biomarkers for LLD diagnosis and treatment response. First, we evaluate the performance of the different learning methods. Then, we study the prediction models of the learning method that produced the greatest classification accuracy. From the prediction models, we analyze the specific features selected and their interdependent relationship for each outcome variable. Evaluation of results across features sets to identify common results amongst all learning methods can be found in the Supplementary Results section. We were not able to appropriately evaluate results between the different networks analyses due the variance in sample sizes.
Figure 1.
Combined network analysis results—training and test classification accuracies—of the thirteen feature sets for late-life depression diagnosis and treatment response from all learning methods.
Comparing Learning Methods
To compare learning methods, we look for optimal test set classification accuracies. Also, we look for signs of overfitting or underfitting by focusing on feature set 1. A greater difference between the training and test set classification accuracies indicates a greater probability of overfitting. An improvement of classification accuracies with nonlinear compared to linear methods indicates a greater probability of underfitting by the linear methods. An improvement of classification accuracies with linear compared to nonlinear methods indicates a greater probability of overfitting by the nonlinear methods. See Supplementary Results for more details.
Diagnosis
The ADTree produced the optimal prediction model with an accuracy of 87.27% (sensitivity = 88.89%, specificity = 85.71%) using feature set 4 in dDMN analysis (see Supplementary Figure S1A) for predicting LLD diagnosis (see corresponding ROC curve in Supplementary Figure S2A). Overall, the linear classification methods showed signs of less overfitting than the nonlinear classification methods, among which ADTree overfits less. Overfitting is observed most in the aSN analysis. However, in the dDMN analysis ADTree outperforms the linear methods, suggesting a possibility of underfitting among the linear models. This may also be an indicator of ADTree being a better learning method for predicting LLD diagnosis.
Treatment Response
Again, the ADTree produced the optimal prediction model with an accuracy of 89.47% (sensitivity = 88.89%, specificity = 90.00%) using feature set 2 in combined network analysis (see Figure 1) for predicting LLD treatment response (see corresponding ROC curve in Supplementary Figure S2B). Overall, the linear classification methods showed signs of less overfitting than the nonlinear classification methods, among which ADTree overfits less. Overfitting is observed most in the dDMN analysis. Generally, all methods show signs of greater overfitting compared to LLD diagnosis—possibly due to the smaller sample size. In the dDMN and aSN analysis L1-LR outperforms the nonlinear methods, suggesting a possibility of overfitting among the nonlinear models. However, since ADTree produced the best prediction model, the problem may be a lack of relevant features in individual network analyses.
Optimal Prediction Models
We evaluate the features selected by the ADTree prediction models that produced the optimal classification accuracy for each outcome variable. These optimal models are presented below and were obtained by retaining only the most frequently splitting criterion and corresponding most frequent rules amongst all the ADTree models created during the LOOCV iterations. For interpreting these models, one must sum up the rule values associated with each splitting criterion and if the total is positive, the individual is more likely to be an LLD patient or a positive responder to treatment for LLD depending on the model’s outcome variable. See Table 3 to better understand what each feature represents in the optimal ADTrees and the interpretations provided below. The superlatives added to the measures when interpreting the models are based on the inequalities estimated by the corresponding ADTree.
Diagnosis
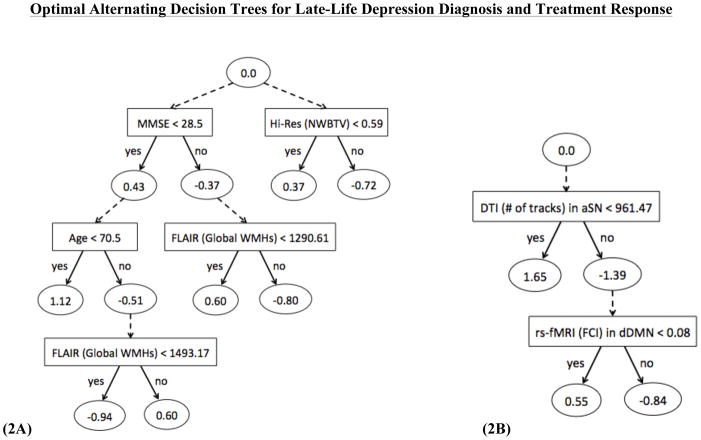
The optimal ADTree model selected mini-mental state examination (MMSE) score, age, Hi-Res whole brain atrophy, and FLAIR global white matter hyperintensity (WMH) count to be the optimal features for predicting LLD diagnosis (see Figure 2a). The selected features are not dependent on dDMN even though the optimal model was acquired during the dDMN analysis; possibly due to its larger and more evenly distributed sample of participants.
Figure 2.
Optimal prediction models in the form of alternating decision trees for predicting late-life depression (A) diagnosis and (B) treatment response [Legend: Square = Splitting Criterion; Oval = Rule]
Based on this model, an individual who is more likely to be diagnosed with LLD will have one of the following attributes: (1) poor cognitive ability + younger old adult, (2) poor cognitive ability + older old adult + high global WMH burden + high whole brain atrophy, or (3) strong cognitive ability + low global WMH burden + high whole brain atrophy.
Conversely, an individual who is not likely to diagnosed with LLD will have one of the following attributes: (1) poor cognitive ability + older old adult + low global WMH burden, (2) poor cognitive ability + older old adult + high global WMH burden + low whole brain atrophy, (3) strong cognitive ability + low global WMH burden + low whole brain atrophy, or (4) strong cognitive ability + high global WMH burden.
Treatment Response
The optimal ADTree model selected DTI number of tracks from aSN and rs-fMRI functional connectivity index (FCI) from dDMN to be the optimal features for predicting LLD treatment response (see Figure 2b).
Based on this model, an individual who is more likely to be a positive responder to treatment for LLD will have fewer structural connections—indicative of a lower WM integrity—in the aSN before treatment administration. Additionally, an individual who had a lower functional connectivity in the dDMN before treatment administration is less likely to be a negative responder to treatment for LLD.
DISCUSSION
In this study, we showed how nonlinear combinations of multimodal MRI and/or non-imaging measures can successfully estimate prediction models for diagnosis (87.27% accuracy) and treatment response (89.47% accuracy) of LLD. The optimal prediction models for both outcome variables were strikingly distinct in nature with no overlap of selected features. Additionally, the diagnosis prediction model was network-independent, while the treatment response prediction model depended on information from the dDMN and aSN. Below we evaluate our findings further using past studies for comparison.
Optimal Predictors/Biomarkers
LLD Diagnosis vs. Treatment Response Prediction Models
Non-imaging (i.e. MMSE and age) and global volume-based imaging (i.e. whole brain atrophy and global WMH burden) measures combined were found to be the optimal predictors/biomarkers of LLD diagnosis. Agreeing with past studies, poor cognitive ability (Ganguli et al., 2006; Kohler et al., Apr 2010; Wilkins et al., 2009) and/or greater whole brain atrophy (Chang et al., 2011; Ribeiz et al., 2013; Sexton et al., 2013) indicated LLD. Possibly explaining the discrepancies between past studies, age (Forlani et al., 2013; Luppa et al., 2012; Wild et al., 2012; Wu et al., 2012) and global WMH burden (Aizenstein et al., 2011; Greenwald et al., 1998; Gunning-Dixon et al., 2010; Firbank et al., 2012; Teodorczuk et al., 2010) were fully dependent on the other measures in regards to their association with LLD diagnosis. We speculate that the primary role of non-imaging measures in predicting diagnosis suggests that current neuroimaging methods cannot – yet – capture the neural complexity associated with the etiopathogenesis of LLD. The involvement of structure-related neural biomarkers (global atrophy and WM burden) in diagnosing LLD supports past studies that suggest vascular and atrophic changes trigger mood disorder in late-life (Aizenstein et al., 2014).
Contrarily, for LLD treatment response, connectivity-based imaging measures were found to be the optimal biomarkers. Specifically, lower structural connectivity—supported by the more recent of the two (Taylor et al., 2008) contradicting past findings (Alexopoulos et al., 2008; Taylor et al., 2008)—and lower functional connectivity—supporting compensation theories (Stern et al., 2003)—indicate a greater probability of treatment remission. This dependency of LLD treatment response on global network health (i.e. communication strength between network regions) may serve as a biomarker for future personalized care studies. A potential interdependence between biomarkers may explain the contradictions in results between past studies and the heterogeneity in the pathophysiology of LLD patients suggested by Taylor et al., 2008.
Overall, the mix of features predictive of diagnosis likely reflects that LLD is heterogeneous. Our observation that these particular features were not predictive of treatment response suggests that there may be a more proximal mediator of depression recovery, and perhaps the features reflecting LLD heterogeneity lead to a set of global network changes (indexed by rs-fMRI and DTI). It is intriguing that these global network biomarkers were identified as most predictive of treatment response.
Mid-Life vs. Late-life Depression Prediction Models
Unlike past studies of depression in younger populations involving prediction models, this is the first study to accurately model both diagnosis and treatment response using the same approach. While past studies have used a single imaging modality and region-based approach (Costafreda et al., 2009; Fu et al., 2008; Hahn et al., 2011; Marquand et al., 2008; Liu et al., 2012b; Mwangi et al., 2012a; Mwangi et al., 2012b; Nouretdinov et al., 2011; Zeng et al., 2012), we used a multi-modal imaging with whole brain and network-based approach that also included non-imaging measures. Our results may suggest that biomarkers of disease diagnosis and remission possibly differ on the basis of brain structure and function—i.e. the different representations of MRI modalities—as opposed to brain regions. It is possible that regional changes do not fully reflect the underlying neural vulnerabilities associated with LLD. This is supported by recent studies (Ajilore et al., 2014; Tadayonnejad et al., 2013) that describe associations of global brain networks alterations with LLD.
Past prediction model studies of mid-life depression diagnosis have shown accurate classifications can be obtained using functional (Fu et al., 2008; Hahn et al., 2011; Marquand et al., 2008; Nouretdinov et al., 2011; Zeng et al., 2012) or structural (Costafreda et al., 2009; Mwangi et al., 2012a; Mwangi et al., 2012b) imaging. Our study in LLD found structural volume-based measures in conjunction to non-imaging measures to be better predictors. We speculate that these differences in prediction factors may suggest that LLD diagnosis is primarily related to impaired structure (GM and WM), while midlife depression may stem from aberrant communication/activation of various brain regions. This hypothesis will require further testing.
Past prediction model studies of mid-life depression treatment response have primarily utilized T1-weighted Hi-Res structural imaging measures (Costafreda et al., 2009; Liu et al., 2012b; Nouretdinov et al., 2011). One study (Marquand et al., 2008) that attempted to use a task-based functional imaging measure did not achieve very high accuracy. Our study in LLD found structural and functional connectivity measures to be better predictors. Since connectivity-related imaging measures have not been tested for prediction models of mid-life depression treatment response, it is difficult to draw any conclusions.
Learning Methods
Based on our findings, modified versions of decision tree and logistic regression are potential alternative learning methods—to the traditionally used SVM—for prediction depression, particularly in late-life, diagnosis and treatment response. Modified decision tree methods with embedded feature selection capabilities, especially, may be useful for studying real-world nonlinear relationships in high-dimensional (i.e. large number of features) data.
Limitations and Future Work
Limitations to this study include: small sub-sample size for treatment response prediction (nevertheless the results were cross checked using four different learning methods), varying sample sizes for the different network analysis (this prevents us from accurately analyzing network-based effects and may be causing feature set 13 results to vary across networks despite its network-independent features), higher percentage of women (reflecting the naturalistic gender distribution in LLD (Luppa et al., 2012)), and lack of other potential cognitive ability measures and covariates (due to limited available data). Another limitation is the heterogeneous treatment. However, this may not have affected our LLD treatment response prediction results since all administered antidepressants are either selective serotonin reuptake inhibitors or serotonin-norepinephrine reuptake inhibitors, and the efficacy difference between the two is still a matter of debate (Papakostas et al., 2007; Taylor et al., 2004; Taylor et al., 2006; Thase et al., 2011). Future work includes extensive studies verifying, improving as necessary, and testing the real-world applicability of the optimal prediction models found in our study. It would also be beneficial to test other imaging and non-imaging (e.g. cognitive ability, medical comorbidities, covariates, etc.) measures as potential biomarkers. Potential clinical applications may include using machine learning and imaging to predict treatment efficacy and recommend personalized treatment for LLD. While diagnosis of LLD is not a potential application, the prediction model for LLD diagnosis can help us gain a better understanding of LLD and consequently lead to a better model for predicting treatment response.
CONCLUSIONS
Preliminary results of this study suggest that LLD diagnosis and treatment response may be better predicted using a combination of multi-modal MRI measures. The results also suggest that the incorporation of non-imaging predictors could also help improve prediction, at least for LLD diagnosis. Additionally, we speculate that whole brain and network related multi-modal MRI measures—as opposed to region-based single modality measures—may be more appropriate for comparing LLD diagnosis and treatment response in terms of associated underlying brain changes.
Supplementary Material
Acknowledgments
This research was supported by NIH grants MH076079/MH076079-04S1, MH 086686, KL2 RR024154, R21 NS060184, R37 AG025516, P30-MH52247/P30 MH71944, MH K23 086606, P30 MH90333, and Brain and Behavior Research (NARSAD) Young Investigator Award (Dr. Andreescu). It was also supported by the John A. Hartford Center of Excellence in Geriatric Psychiatry at the University of Pittsburgh.
Footnotes
Conflicts of Interest: Meenal Patel, Carmen Andreescu, Julie Price and Kathryn Edelman do not have any potential conflict of interest to acknowledge. Charles F. Reynolds III has received research support from Pfizer Inc., Eli Lilly and Co., Bristol Meyers Squibb, Forest Pharmaceuticals, and Wyeth Pharmaceuticals. Howard Aizenstein has received research support from Novartis Pharmaceuticals.
References
- 1.Mental Health America. Depression in Older Adults. http://www.mentalhealthamerica.net/conditions/depression-older-adults#2. Retrieved 2014.
- 2.Wu Z, Schimmele CM, Chappell NL. Aging and late-life depression. Journal of aging and health. 2012;24(1):3–28. doi: 10.1177/0898264311422599. [DOI] [PubMed] [Google Scholar]
- 3.Wild B, Herzog W, Schellberg D, Lechner S, Niehoff D, Brenner H, et al. Association between the prevalence of depression and age in a large representative German sample of people aged 53 to 80 years. International journal of geriatric psychiatry. 2012;27(4):375–381. doi: 10.1002/gps.2728. [DOI] [PubMed] [Google Scholar]
- 4.Forlani C, Morri M, Ferrari B, Dalmonte E, Menchetti M, De Ronchi D, et al. Prevalence and Gender Differences in Late-Life Depression: A Population-Based Study. The American journal of geriatric psychiatry : official journal of the American Association for Geriatric Psychiatry. 2013 doi: 10.1016/j.jagp.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 5.Luppa M, Sikorski C, Luck T, Ehreke L, Konnopka A, Wiese B, et al. Age- and gender-specific prevalence of depression in latest-life--systematic review and meta-analysis. Journal of affective disorders. 2012;136(3):212–221. doi: 10.1016/j.jad.2010.11.033. [DOI] [PubMed] [Google Scholar]
- 6.Chang-Quan H, Zheng-Rong W, Yong-Hong L, Yi-Zhou X, Qing-Xiu L. Education and risk for late life depression: a meta-analysis of published literature. International journal of psychiatry in medicine. 2010;40(1):109–124. doi: 10.2190/PM.40.1.i. [DOI] [PubMed] [Google Scholar]
- 7.Blazer DG. Review: antidepressants are effective for the treatment of major depressive disorder in individuals aged 55 years or older. Evidence-based mental health. 2012;15(3):72. doi: 10.1136/ebmental-2012-100629. [DOI] [PubMed] [Google Scholar]
- 8.Katon W, Unutzer J, Russo J. Major depression: the importance of clinical characteristics and treatment response to prognosis. Depression and anxiety. 2010;27(1):19–26. doi: 10.1002/da.20613. [DOI] [PubMed] [Google Scholar]
- 9.Andreescu C, Mulsant BH, Houck PR, Whyte EM, Mazumdar S, Dombrovski AY, et al. Empirically derived decision trees for the treatment of late-life depression. The American journal of psychiatry. 2008;165(7):855–862. doi: 10.1176/appi.ajp.2008.07081340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhalla RK, Butters MA, Zmuda MD, Seligman K, Mulsant BH, Pollock BG, et al. Does education moderate neuropsychological impairment in late-life depression? International journal of geriatric psychiatry. 2005;20(5):413–417. doi: 10.1002/gps.1296. [DOI] [PubMed] [Google Scholar]
- 11.Ganguli M, Du Y, Dodge HH, Ratcliff GG, Chang CC. Depressive symptoms and cognitive decline in late life: a prospective epidemiological study. Archives of general psychiatry. 2006;63(2):153–160. doi: 10.1001/archpsyc.63.2.153. [DOI] [PubMed] [Google Scholar]
- 12.Wilkins CH, Mathews J, Sheline YI. Late life depression with cognitive impairment: evaluation and treatment. Clinical interventions in aging. 2009;4:51–57. [PMC free article] [PubMed] [Google Scholar]
- 13.Kohler S, Thomas AJ, Barnett NA, O’Brien JT. The pattern and course of cognitive impairment in late-life depression. Psychological medicine. 2010;40(4):591–602. doi: 10.1017/S0033291709990833. [DOI] [PubMed] [Google Scholar]
- 14.Ribeiz SR, Duran F, Oliveira MC, Bezerra D, Castro CC, Steffens DC, et al. Structural brain changes as biomarkers and outcome predictors in patients with late-life depression: a cross-sectional and prospective study. PloS one. 2013;8(11):e80049. doi: 10.1371/journal.pone.0080049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang CC, Yu SC, McQuoid DR, Messer DF, Taylor WD, Singh K, et al. Reduction of dorsolateral prefrontal cortex gray matter in late-life depression. Psychiatry research. 2011;193(1):1–6. doi: 10.1016/j.pscychresns.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sexton CE, Mackay CE, Ebmeier KP. A systematic review and meta-analysis of magnetic resonance imaging studies in late-life depression. The American journal of geriatric psychiatry : official journal of the American Association for Geriatric Psychiatry. 2013;21(2):184–195. doi: 10.1016/j.jagp.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 17.Colloby SJ, Firbank MJ, Thomas AJ, Vasudev A, Parry SW, O’Brien JT. White matter changes in late-life depression: a diffusion tensor imaging study. Journal of affective disorders. 2011;135(1–3):216–220. doi: 10.1016/j.jad.2011.07.025. [DOI] [PubMed] [Google Scholar]
- 18.Shimony JS, Sheline YI, D’Angelo G, Epstein AA, Benzinger TL, Mintun MA, et al. Diffuse microstructural abnormalities of normal-appearing white matter in late life depression: a diffusion tensor imaging study. Biological psychiatry. 2009;66(3):245–252. doi: 10.1016/j.biopsych.2009.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mettenburg JM, Benzinger TL, Shimony JS, Snyder AZ, Sheline YI. Diminished performance on neuropsychological testing in late life depression is correlated with microstructural white matter abnormalities. NeuroImage. 2012;60(4):2182–2190. doi: 10.1016/j.neuroimage.2012.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aizenstein HJ, Andreescu C, Edelman KL, Cochran JL, Price J, Butters MA, et al. fMRI correlates of white matter hyperintensities in late-life depression. The American journal of psychiatry. 2011;168(10):1075–1082. doi: 10.1176/appi.ajp.2011.10060853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crocco EA, Castro K, Loewenstein DA. How late-life depression affects cognition: neural mechanisms. Current psychiatry reports. 2010;12(1):34–38. doi: 10.1007/s11920-009-0081-2. [DOI] [PubMed] [Google Scholar]
- 22.Teodorczuk A, Firbank MJ, Pantoni L, Poggesi A, Erkinjuntti T, Wallin A, et al. Relationship between baseline white-matter changes and development of late-life depressive symptoms: 3-year results from the LADIS study. Psychological medicine. 2010;40(4):603–610. doi: 10.1017/S0033291709990857. [DOI] [PubMed] [Google Scholar]
- 23.Firbank MJ, Teodorczuk A, van der Flier WM, Gouw AA, Wallin A, Erkinjuntti T, et al. Relationship between progression of brain white matter changes and late-life depression: 3-year results from the LADIS study. The British journal of psychiatry : the journal of mental science. 2012;201(1):40–45. doi: 10.1192/bjp.bp.111.098897. [DOI] [PubMed] [Google Scholar]
- 24.Kohler S, Thomas AJ, Lloyd A, Barber R, Almeida OP, O’Brien JT. White matter hyperintensities, cortisol levels, brain atrophy and continuing cognitive deficits in late-life depression. The British journal of psychiatry : the journal of mental science. 2010;196(2):143–149. doi: 10.1192/bjp.bp.109.071399. [DOI] [PubMed] [Google Scholar]
- 25.Disabato BM, Sheline YI. Biological basis of late life depression. Current psychiatry reports. 2012;14(4):273–279. doi: 10.1007/s11920-012-0279-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gunning-Dixon FM, Walton M, Cheng J, Acuna J, Klimstra S, Zimmerman ME, et al. MRI signal hyperintensities and treatment remission of geriatric depression. Journal of affective disorders. 2010;126(3):395–401. doi: 10.1016/j.jad.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gunning FM, Cheng J, Murphy CF, Kanellopoulos D, Acuna J, Hoptman MJ, et al. Anterior cingulate cortical volumes and treatment remission of geriatric depression. International journal of geriatric psychiatry. 2009;24(8):829–836. doi: 10.1002/gps.2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor WD, Kuchibhatla M, Payne ME, Macfall JR, Sheline YI, Krishnan KR, et al. Frontal white matter anisotropy and antidepressant remission in late-life depression. PloS one. 2008;3(9):e3267. doi: 10.1371/journal.pone.0003267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor WD, Macfall JR, Boyd B, Payne ME, Sheline YI, Krishnan RR, et al. One-year change in anterior cingulate cortex white matter microstructure: relationship with late-life depression outcomes. The American journal of geriatric psychiatry : official journal of the American Association for Geriatric Psychiatry. 2011;19(1):43–52. doi: 10.1097/JGP.0b013e3181e70cec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alexopoulos GS, Murphy CF, Gunning-Dixon FM, Latoussakis V, Kanellopoulos D, Klimstra S, et al. Microstructural white matter abnormalities and remission of geriatric depression. The American journal of psychiatry. 2008;165(2):238–244. doi: 10.1176/appi.ajp.2007.07050744. [DOI] [PubMed] [Google Scholar]
- 31.Wang L, Krishnan KR, Steffens DC, Potter GG, Dolcos F, McCarthy G. Depressive state- and disease-related alterations in neural responses to affective and executive challenges in geriatric depression. The American journal of psychiatry. 2008;165(7):863–871. doi: 10.1176/appi.ajp.2008.07101590. [DOI] [PubMed] [Google Scholar]
- 32.Liu F, Hu M, Wang S, Guo W, Zhao J, Li J, et al. Abnormal regional spontaneous neural activity in first-episode, treatment-naive patients with late-life depression: a resting-state fMRI study. Progress in neuro-psychopharmacology & biological psychiatry. 2012;39(2):326–331. doi: 10.1016/j.pnpbp.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 33.Alalade E, Denny K, Potter G, Steffens D, Wang L. Altered cerebellar-cerebral functional connectivity in geriatric depression. PloS one. 2011;6(5):e20035. doi: 10.1371/journal.pone.0020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alexopoulos GS, Hoptman MJ, Kanellopoulos D, Murphy CF, Lim KO, Gunning FM. Functional connectivity in the cognitive control network and the default mode network in late-life depression. Journal of affective disorders. 2012;139(1):56–65. doi: 10.1016/j.jad.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bohr IJ, Kenny E, Blamire A, O’Brien JT, Thomas AJ, Richardson J, et al. Resting-state functional connectivity in late-life depression: higher global connectivity and more long distance connections. Frontiers in psychiatry. 2012;3:116. doi: 10.3389/fpsyt.2012.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steffens DC, Taylor WD, Denny KL, Bergman SR, Wang L. Structural integrity of the uncinate fasciculus and resting state functional connectivity of the ventral prefrontal cortex in late life depression. PloS one. 2011;6(7):e22697. doi: 10.1371/journal.pone.0022697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andreescu C, Wu M, Butters MA, Figurski J, Reynolds CF, 3rd, Aizenstein HJ. The default mode network in late-life anxious depression. The American journal of geriatric psychiatry : official journal of the American Association for Geriatric Psychiatry. 2011;19(11):980–983. doi: 10.1097/JGP.0b013e318227f4f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Colloby SJ, Firbank MJ, He J, Thomas AJ, Vasudev A, Parry SW, et al. Regional cerebral blood flow in late-life depression: arterial spin labelling magnetic resonance study. The British journal of psychiatry : the journal of mental science. 2012;200(2):150–155. doi: 10.1192/bjp.bp.111.092387. [DOI] [PubMed] [Google Scholar]
- 39.Wu M, Andreescu C, Butters MA, Tamburo R, Reynolds CF, 3rd, Aizenstein H. Default-mode network connectivity and white matter burden in late-life depression. Psychiatry research. 2011;194(1):39–46. doi: 10.1016/j.pscychresns.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andreescu C, Tudorascu DL, Butters MA, Tamburo E, Patel M, Price J, et al. Resting state functional connectivity and treatment response in late-life depression. Psychiatry research. 2013;214(3):313–321. doi: 10.1016/j.pscychresns.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mwangi B, Ebmeier KP, Matthews K, Steele JD. Multi-centre diagnostic classification of individual structural neuroimaging scans from patients with major depressive disorder. Brain : a journal of neurology. 2012;135(Pt 5):1508–1521. doi: 10.1093/brain/aws084. [DOI] [PubMed] [Google Scholar]
- 42.Mwangi B, Matthews K, Steele JD. Prediction of illness severity in patients with major depression using structural MR brain scans. Journal of magnetic resonance imaging : JMRI. 2012;35(1):64–71. doi: 10.1002/jmri.22806. [DOI] [PubMed] [Google Scholar]
- 43.Costafreda SG, Chu C, Ashburner J, Fu CH. Prognostic and diagnostic potential of the structural neuroanatomy of depression. PloS one. 2009;4(7):e6353. doi: 10.1371/journal.pone.0006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeng LL, Shen H, Liu L, Wang L, Li B, Fang P, et al. Identifying major depression using whole-brain functional connectivity: a multivariate pattern analysis. Brain : a journal of neurology. 2012;135(Pt 5):1498–1507. doi: 10.1093/brain/aws059. [DOI] [PubMed] [Google Scholar]
- 45.Hahn T, Marquand AF, Ehlis AC, Dresler T, Kittel-Schneider S, Jarczok TA, et al. Integrating neurobiological markers of depression. Archives of general psychiatry. 2011;68(4):361–368. doi: 10.1001/archgenpsychiatry.2010.178. [DOI] [PubMed] [Google Scholar]
- 46.Fu CH, Mourao-Miranda J, Costafreda SG, Khanna A, Marquand AF, Williams SC, et al. Pattern classification of sad facial processing: toward the development of neurobiological markers in depression. Biological psychiatry. 2008;63(7):656–662. doi: 10.1016/j.biopsych.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 47.Nouretdinov I, Costafreda SG, Gammerman A, Chervonenkis A, Vovk V, Vapnik V, et al. Machine learning classification with confidence: application of transductive conformal predictors to MRI-based diagnostic and prognostic markers in depression. NeuroImage. 2011;56(2):809–813. doi: 10.1016/j.neuroimage.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 48.Marquand AF, Mourao-Miranda J, Brammer MJ, Cleare AJ, Fu CH. Neuroanatomy of verbal working memory as a diagnostic biomarker for depression. Neuroreport. 2008;19(15):1507–1511. doi: 10.1097/WNR.0b013e328310425e. [DOI] [PubMed] [Google Scholar]
- 49.Liu F, Guo W, Yu D, Gao Q, Gao K, Xue Z, et al. Classification of different therapeutic responses of major depressive disorder with multivariate pattern analysis method based on structural MR scans. PloS one. 2012;7(7):e40968. doi: 10.1371/journal.pone.0040968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aizenstein HJ, Khalaf A, Walker SE, Andreescu C. Magnetic resonance imaging predictors of treatment response in late-life depression. Journal of geriatric psychiatry and neurology. 2014;27(1):24–32. doi: 10.1177/0891988713516541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andreescu C, Reynolds CF., 3rd Late-life depression: evidence-based treatment and promising new directions for research and clinical practice. The Psychiatric clinics of North America. 2011;34(2):335–355. vii–iii. doi: 10.1016/j.psc.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cortes C, Vapnik V. Support-vector networks. Mach Learn. 1995;20(3):273–97. [Google Scholar]
- 53.Yuan G-X, Chang K-W, Hsieh C-J, Lin C-J. A Comparison of Optimization Methods and Software for Large-scale L1-regularized Linear Classification. J Mach Learn Res. 2010;11:3183–3234. [Google Scholar]
- 54.Pfahringer B, Holmes G, Kirkby R. Optimizing the Induction of Alternating Decision Trees. In: Cheung D, Williams G, Li Q, editors. Advances in Knowledge Discovery and Data Mining. Vol. 2035. Springer; Berlin Heidelberg: 2001. pp. 477–487. edn. [Google Scholar]
- 55.Nieto-Castanon A, Ghosh SS, Tourville JA, Guenther FH. Region of interest based analysis of functional imaging data. NeuroImage. 2003;(4):1303–16. doi: 10.1016/s1053-8119(03)00188-5. [DOI] [PubMed] [Google Scholar]
- 56.First M, Spitzer R, Williams J, Gibbons M. Structured clinical interview for DSM-IV-Patient version. New York: Biometrics Research Department, New York State Psychiatric Institute; 1995. [Google Scholar]
- 57.Roose SP, Glassman AH, Attia E, Woodring S. Comparative efficacy of selective serotonin reuptake inhibitors and tricyclics in the treatment of melancholia. The American journal of psychiatry. 1994;151(12):1735–1739. doi: 10.1176/ajp.151.12.1735. [DOI] [PubMed] [Google Scholar]
- 58.Wu M, Rosano C, Butters M, Whyte E, Nable M, Crooks R, et al. A fully automated method for quantifying and localizing white matter hyperintensities on MR images. Psychiatry research. 2006;148(2–3):133–142. doi: 10.1016/j.pscychresns.2006.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. Fsl. NeuroImage. 2012;62(2):782–90. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 60.Whitfield-Gabrieli S, Nieto-Castanon A. Conn: a functional connectivity toolbox for correlated and anticorrelated brain networks. Brain connectivity. 2012;2(3):125–41. doi: 10.1089/brain.2012.0073. [DOI] [PubMed] [Google Scholar]
- 61.Shirer WR, Ryali S, Rykhlevskaia E, Menon V, Greicius MD. Decoding subject-driven cognitive states with whole-brain connectivity patterns. Cerebral cortex. 2012;22(1):158–165. doi: 10.1093/cercor/bhr099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: Machine Learning in Python. J Mach Learn Res. 2011;12:2825–30. [Google Scholar]
- 63.Greenwald BS, Kramer-Ginsberg E, Krishnan KR, Ashtari M, Auerbach C, Patel M. Neuroanatomic localization of magnetic resonance imaging signal hyperintensities in geriatric depression. Stroke; a journal of cerebral circulation. 1998;29(3):613–617. doi: 10.1161/01.str.29.3.613. [DOI] [PubMed] [Google Scholar]
- 64.Stern Y. The concept of cognitive reserve: a catalyst for research. Journal of clinical and experimental neuropsychology. 2003;25(5):589–593. doi: 10.1076/jcen.25.5.589.14571. [DOI] [PubMed] [Google Scholar]
- 65.Tadayonnejad R, Ajilore O. Brain network dysfunction in late-life depression: a literature review. Journal of geriatric psychiatry and neurology. 2014;27(1):5–12. doi: 10.1177/0891988713516539. [DOI] [PubMed] [Google Scholar]
- 66.Ajilore O, Lamar M, Leow A, Zhang A, Yang S, Kumar A. Graph theory analysis of cortical-subcortical networks in late-life depression. The American journal of geriatric psychiatry : official journal of the American Association for Geriatric Psychiatry. 2014;22(2):195–206. doi: 10.1016/j.jagp.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Papakostas GI, Thase ME, Fava M, Nelson JC, Shelton RC. Are antidepressant drugs that combine serotonergic and noradrenergic mechanisms of action more effective than the selective serotonin reuptake inhibitors in treating major depressive disorder? A meta-analysis of studies of newer agents. Biological psychiatry. 2007;62(11):1217–1227. doi: 10.1016/j.biopsych.2007.03.027. [DOI] [PubMed] [Google Scholar]
- 68.Thase ME, Ninan PT, Musgnung JJ, Trivedi MH. Remission with venlafaxine extended release or selective serotonin reuptake inhibitors in depressed patients: a randomized, open-label study. The primary care companion to CNS disorders. 2011;13(1) doi: 10.4088/PCC.10m00979blu. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Taylor MJ, Freemantle N, Geddes JR, Bhagwagar Z. Early onset of selective serotonin reuptake inhibitor antidepressant action: systematic review and meta-analysis. Archives of general psychiatry. 2006;63(11):1217–1223. doi: 10.1001/archpsyc.63.11.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Taylor WD, Doraiswamy PM. A systematic review of antidepressant placebo-controlled trials for geriatric depression: limitations of current data and directions for the future. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2004;29(12):2285–2299. doi: 10.1038/sj.npp.1300550. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.