Abstract
Tc1-like transposons consist of an inverted repeat sequence flanking a transposase gene that exhibits similarity to the mobile DNA element, Tc1, of the nematode, Caenorhabditis elegans. They are widely distributed within vertebrate genomes including teleost fish; however, few active Tc1-like transposases have been discovered. In this study, 17 Tc1-like transposon sequences were isolated from 10 freshwater fish species belonging to the families Cyprinidae, Adrianichthyidae, Cichlidae, and Salmonidae. We conducted phylogenetic analyses of these sequences using previously isolated Tc1-like transposases and report that 16 of these elements comprise a new subfamily of Tc1-like transposons. In particular, we show that one transposon, Thm3 from silver carp (Hypophthalmichthys molitrix; Cyprinidae), can encode a 335-aa transposase with apparently intact domains, containing three to five copies in its genome. We then coinjected donor plasmids harboring 367 bp of the left end and 230 bp of the right end of the nonautonomous silver carp Thm1 cis-element along with capped Thm3 transposase RNA into the embryos of blunt snout bream (Megalobrama amblycephala; one- to two-cell embryos). This experiment revealed that the average integration rate could reach 50.6% in adult fish. Within the blunt snout bream genome, the TA dinucleotide direct repeat, which is the signature of Tc1-like family of transposons, was created adjacent to both ends of Thm1 at the integration sites. Our results indicate that the silver carp Thm3 transposase can mediate gene insertion by transposition within the genome of blunt snout bream genome, and that this occurs with a TA position preference.
Keywords: Tc1-like transposase, transposition, Hypophthalmichthys molitrix, Megalobrama amblycephala
Transposable elements (TEs) can move within their host genomes by changing their positions of insertion in a process called transposition (McClintock 1953; Rubin and Spradling 1982; Craig et al. 2002). Their wide distribution among all major branches of life, their diversity, and their intrinsic biological features have made TEs a considerable source of many genetic innovations during species evolution (Radice et al. 1994; Kazazian 2004; Feschotte and Pritham 2007; Janicki et al. 2011). DNA transposons constitute one of the two subclasses of Class II TEs (Craig 1995). In eukaryotic genomes, DNA transposons have been divided into at least 17 superfamilies on the basis of sequence similarities between their element-encoded transposases (Yuan and Wessler 2011). The transposition of DNA transposons follows the “cut-and-paste” mechanism and is catalyzed by functional transposase, and allows the transposon to excise itself from a donor chromosomal site and then to reinsert at a different chromosomal locus (McClintock 1953; Kidwell and Lisch 2000).
Tc1 transposon was first discovered in the nematode genome (Caenorhabditis elegans) and was shown to encode an active transposase (Emmons and Yesner 1984; Eide and Anderson 1985). Tc1-like transposons are approximately 1.3 kb long and contain a single gene without an intron encoding an ∼350-amino acid transposase. The transposase gene is flanked by inverted terminal repeats (ITRs) and is characterized by a DNA binding domain and a catalytic domain that can catalyze the DNA cleavage and TA dinucleotide target-joining steps of transposition (Plasterk et al. 1999). To date, six autonomously active Tc1-like transposons have been isolated from eukaryotes, i.e., Minos from Drosophila hydei (Franz and Savakis 1991), Mos1 from Drosophila mauritiana (Medhora et al. 1991), Famar1 from Forficula auriculata (Barry et al. 2004), Mboumar-9 from Messor bouvieri (Munoz-Lopez et al. 2008), and Passport from seawater flatfish (Pleuronectes platessa) (Clark et al. 2009). Moreover, four active Tc1-like elements have been reconstructed according to the bioinformatic consensus sequences, i.e., Sleeping Beauty (SB) from salmonid (Ivics et al. 1997), Frog Prince (FP) from Rana pipiens (Miskey et al. 2003), Himar1 from Haematibia irritans (Lampe et al. 1996), and Hsmar1 from humans (Robertson and Zumpano 1997). These works have shown the ability of Tc1-like transposases to catalyze transposition in a variety of cells and species.
DNA transposons can be developed as valuable tools for genetic analyses (Izsvak et al. 2000; Kawakami et al. 2004; Kotani and Kawakami 2008). Identification of fish-derived Tc1-like transposase will provide techniques for transgenesis and insertional mutagenesis and might provide some clues to explain the fish genome evolution (Hickman et al. 2005). In teleost fish, Tc1-like transposons are found to distribute widely within the genomes. For instance, between 6% and 10% of the genome of Atlantic salmon (Salmo salar) is occupied by Tc1-like and PiggyBac subfamily transposons (Davidson et al. 2010), and approximately 4.2% of the genome of channel catfish (Ictalurus punctatus) is composed of Tc1-like transposons (Nandi et al. 2007). Similarly, the Tc1-like transposon accounts for nearly 1.7% of the genome of common carp (Cyprinus carpio), which is second only to that of the hAT transposon superfamily (approximately 2.3%) (Wang et al. 2011). However, the majority of Tc1-like transposons that have been identified in teleost fish are defective because they contain frameshifts, insertions/deletions, and premature termination codons within the coding regions of their transposase genes during their evolution (Lohe et al. 1995).
Because multiple copies of Tc1-like transposons have accumulated in the genomes of freshwater fish during evolution (Davidson et al. 2010; Nandi et al. 2007; Wang et al. 2011), the goal of this study is to screen a few fish Tc1-like elements to see whether any copy may still retain transposition function. On the basis of 17 novel Tc1-like transposon sequences isolated from 10 freshwater fish species, we show that one of these can encode a transposase with apparently intact domains, which can mediate DNA transposition of cis-element of silver carp Thm1 in another fish species.
Materials and Methods
Experimental animals
A total of 18 freshwater fish species belonging to seven families were sampled for PCR analysis during different years from 2011 to 2014 (see Table 1 for details). The common carp, crucian carp, spotted steed, topmouth gudgeon, silver carp, grass carp, blunt snout bream, Fugu, yellowhead catfish, and Chinese longsnout catfish were collected at Qingpu Quanjie Fish Farm, Shanghai, China. The medaka, Nile Tilapia, Russian sturgeon, goldfish, common angelfish, and Denison barb were obtained from the Aquaculture Center of Shanghai Ocean University, Shanghai, China. The brown trout and naked carp were obtained at the Yadong Fish Farms, Tibet, China. The tail fin of each fish was cut and stored in 95% ethanol at −25°. All experiments were conducted following the guidelines approved by the Shanghai Ocean University Committee on the Use and Care of Animals.
Table 1. The sequence results of PCR amplifications using a single Tc1-like A primer.
| Family | Species (Sampling and PCR Analysis Year) | No. of Tc1-like Sequences | Tc1-like Elements (Designation, GenBank Acc. No.) |
|---|---|---|---|
| Cyprinidae | Common carp, Cyprinus carpio (2013) | 961 bp, 1938 bp | 1110 bp (Tcc2, KJ742725), 1473 bp (Tcc3, KJ742726), 1238 bp (Tcc4, KJ742727) |
| Crucian carp, Carassius auratus (2013) | — | 1473 bp (Tca2, KJ742724) | |
| Spotted steed, Hemibarbus maculates (2011) | 1420 bp | — | |
| Silver carp, Hypophthalmichthys molitrix (2011) | 1091 bp, 1020 bp | 1555 bp (Thm1, KJ742716), 1390 bp (Thm2, KJ742717), 1209 bp (Thm3, KJ742718) | |
| Naked carp, Gymnocypris przewalskii (2011) | 2036 bp | 961 bp (Tgpp1, KJ742728) | |
| Grass carp, Ctenopharynodon idellus (2012) | — | 1433 bp (Tci1, KJ742720), 1473 bp (Tci2, KJ742721) | |
| Blunt snout bream, Megalobrama amblycephala (2013) | — | 1329 bp (Tma2, KJ742719) | |
| Topmouth gudgeon, Pseudorasbora parva (2013) | — | — | |
| Denison barb, Puntius denisonii (2013) | 1116 bp, 1176 bp | 1345 bp (Tpd1, KJ742722), 1156 bp (Tpd2, KJ742723) | |
| Goldfish, Carassius auratus auratus (2011) | — | — | |
| Adrianichthyidae | Medaka, Oryzias latipes (2012) | 1210 bp | 1450 bp (Tmf1, KJ742729) |
| Cichlidae | Nile Tilapia, Oreochromis niloticus (2011) | 1432 bp | — |
| Common angelfish, Pterophyllums carale (2012) | 1752 bp, 1465 bp, 1528 bp | 1171 bp (Tpc1, KJ742730) | |
| Tetraodontidae | Fugu, Takifugu obscurus (2012) | 1044 bp, 907 bp, 1089 bp | — |
| Acipenseridae | Russian sturgeon, Acipenser gueldenstaedti (2012) | 1301 bp | — |
| Salmonidae | Brown trout, Salmo trutta fario (2011, 2014) | — | 1607 bp (Tbt1, JQ782179), 1473 bp (Tbt2, JQ782178) |
| Bagridae | Yellowhead catfish, Pelteobagrus fulvidraco (2012) | — | — |
| Chinese longsnout catfish, Leiocassis longirostris (2012) | — | — |
Analysis of genomic DNA
Total genomic DNA was isolated from the tail fin samples (0.1 to 0.2 g) using a standard phenol-chloroform procedure as detailed by Sambrook et al. (1989). Two microliters of genomic DNA was used as the template in PCR amplification with a single primer Tc1-like A: 5′-TACAGTTGAAGTCGGAAGTTTACATAC-3′, which was designed according to the inverted repeats of Tc1-like transposon (Izsvak et al. 1995; Liu et al. 2009; Nandi et al. 2007). PCR reaction conditions were as follows: predenaturation at 94° for 5 min; 35 cycles of denaturation at 94° for 30 sec, annealing at 51° for 30 sec, and elongation at 72° for 2 min; and a final elongation step at 72° for 10 min. PCR products were gel-purified, ligated into the T/A cloning vector pMD-19T (Takara, Dalian, China), and transformed into Escherichia coli DH5α competent cells. Positive clones were examined by PCR and direct sequencing.
Sequence analysis
The nucleotide sequences of Tc1-like transposons were analyzed using BioEdit 7.0.0.1 (Hall 1999). The complete inverted terminal repeat (ITR) sequence and the transposase open reading frame were filtered by comparing the left and right end ITR sequence and the transposase open reading frame region. The sequences of Tc1-like transposon from different fish species were obtained using the BLASTN NCBI (http://ncbi.nlm.nih.gov) search program. Alignment of the putative amino acid sequences of the Tc1-like transposases was performed with the Clustal X 1.81 program (Thompson et al. 1997). After eliminating positions with gaps, the gene genealogies were assessed by maximum likelihood using the software package PAUP*4.0b10 (Swofford 2003). For maximum likelihood, the best model and parameters were estimated by MrModeltest v2.2 (Nylander et al. 2004) based on the Akaike Information Criterion (AIC) and the following parameters: 500 bootstrap replications; ConLevel = 50; search = heuristic; brlens = yes.
Southern blot analysis
The 625-bp silver carp Thm3 probe was designed with primer pairs Thm3-f: 5′- AGATGCTGGCTGAAACTGGTAA- 3′ and Thm3-r: 5′-ATCAGGTTTGTAAGCCCTTCTC G-3′, which cover the major open reading frame (ORF) from 425 bp to 1049 bp of Thm3. The probes were digoxin-labeled in vitro using the DIG-High Prime DNA Labeling and Detection Starter Kit I (Roche, Basel, Switzerland) according to the manufacturer’s instructions. Southern blot analysis was performed as described previously by Koga et al. (2000), with the following modifications. High-molecular-weight DNA (20–30 µg for each gel slot) were digested with BglII restriction enzyme, fractionated on 0.8% agarose gels, and transferred to nylon membranes (Hybond-N; Amersham), followed by fixation via baking at 80°. The membranes were then hybridized with a digoxin-labeled silver carp Thm3 probe. Finally, stripping and reprobing DNA blots were performed after immunological detection.
Plasmid constructs
To generate the pCS2-CMV-Thm3TP construct, the ORF of the silver carp Thm3 transposase cDNA, which encodes 335 aa residues, was amplified by PCR using Pfu DNA polymerase (Stratagene, La Jolla, CA, USA) and the primers 5′-CCGCTCGAGATGGGAA AATCAAGAGAA-3′ and 5′-GCTCTAGATTAGTATTTGGTAGAATT-3′. This was then digested with XhoI and XbaI and subcloned into the XhoI-XbaI sites of the pCS2+ vector. The sequences of all plasmids were confirmed by DNA sequencing. The left (367 bp) and right (230 bp) Thm1 inverted terminal repeats (ITRs) were amplified from the silver carp using PCR primers for the left ITRs (sense, 5′-GGACTAGTTACAGTTGAAGTCGGAAG-3′; antisense, 5′-CCGCTCGAGCTCTGTGTTTGAGGTGTG-3′) and right ITRs (sense, 5′-GAAGATCTGAAGTGTATGTATACTTCTGACTT-3′; antisense, 5′-GGGGTACCTACAGTTGAAGTCGGAAGTTTACA-3′). To generate the pThm1-Mlyz2-RFP (red fluorescent protein) construct, the left ITR PCR product was digested with SpeI and XhoI, and the right ITR PCR product was digested with BglII and KpnI. These two fragments were then subcloned into the SpeI-XhoI and BglII-KpnI sites, respectively, of pTgf2-Mlyz2-RFP that contained the zebrafish myosin light chain 2 (Mlyz2) gene promoter (Guo et al. 2013).
In vitro transcription and microinjection
Capped silver carp Thm3 transposase mRNA were synthesized in vitro using the mMESSAGE mMACHINE kit (Ambion, Austin, TX) using NotI linearized pCS2-CMV-Thm3TP plasmid DNA as a template. The microinjection volume was estimated at 1 nL/embryo and contained 50 pg of circular donor plasmid pThm1-Mlyz2-RFP coinjected with 50 pg of Thm3 transposase capped mRNA. This was injected into blunt snout bream embryos at the one- to two-cell stage. The linearized donor plasmid pThm1-Mlyz2-RFP was injected without Thm3 transposase capped mRNA as a control. After injection, the embryos were placed in an embryo rearing medium and maintained at room temperature. Fluorescent expression in the embryos was analyzed using a Nikon SMZ1500 fluorescence microscope.
Transgene efficiency and insertion site analysis
The primers used for RFP PCR were: RFP-f, 5′-GCATGGAGGGCTCCGTGAACG-3′ and RFP-r, 5′-GGTGTAGTCCTCGTTGTGGG-3′. PCR amplification, cloning, and sequencing were conducted as previously described (Jiang et al. 2012). The flanking sequences of the transposon insertion sites were analyzed using the GenomeWalker Universal Kit (Clontech, California). For splinkerette PCR, 25 µg genomic DNA was digested for 12–16 hr at 37° with 80 units of Stu I and EcoR V in a 100-µL reaction volume, purified by ethanol precipitation, and 4 µL of the digestion mix was ligated with the splinkerette adaptor overnight at 16°. The linker ligation was used as a template for two rounds of PCR to amplify the transposon/genome junction. The nested primers for the 5′ flanking sequences were 5′-AGGTGTGCCTTTATATTCATCCACAGGCGT-3′ and 5′-ATCAATTAACCTATCAGAAGCTTCTT-3′. The nested primers for the 3′ flanking sequences were 5′-TAAGGAAGTGTATGTATACTTCTGACTTTGA-3′ and 5′-TCCCTCGCTCGATTCTGACATTTAACA-3′. The amplified fragments were cloned into the pMD19-T vector (TaKaRa, Dalian, China) and transformed into DH5α E. coli cells, and the positive clones were examined by PCR and direct sequencing.
Data availability
Figure S1 contains supporting figures. Further information on all plasmids and more detailed supporting data are available upon request.
Results
PCR screening and sequence analysis of Tc1-like elements from different fish species
A total of 34 specific nucleotide sequences (ranging from 907 bp to 2036 bp) were amplified from the 18 fish species using the single Tc1-like A primer complementary to ITRs (Table 1). Seventeen of these sequences exhibited structural features of the Tc1-like transposon comprising the ITRs and the transposase coding region. The remaining 17 sequences coded for other genes or were unknown sequences. The 17 Tc1-like transposons were distributed in 10 species belonging to four families (Table 1). Like most members of the Tc1-like family, the 17 cloned Tc1-like transposons were always flanked by a TA target site (data not shown). All Tc1-like transposon sequences we obtained are available in GenBank under Accession numbers KJ742716–KJ742730. Because brown trout Tbt1 and Tbt2 cloned from the samples collected in 2014 had >99% similarities to the sequences identified in 2011 (Guo et al. 2014), previous GenBank accession numbers (JQ782178–JQ782179) were used for both sequences in this study (Table 1).
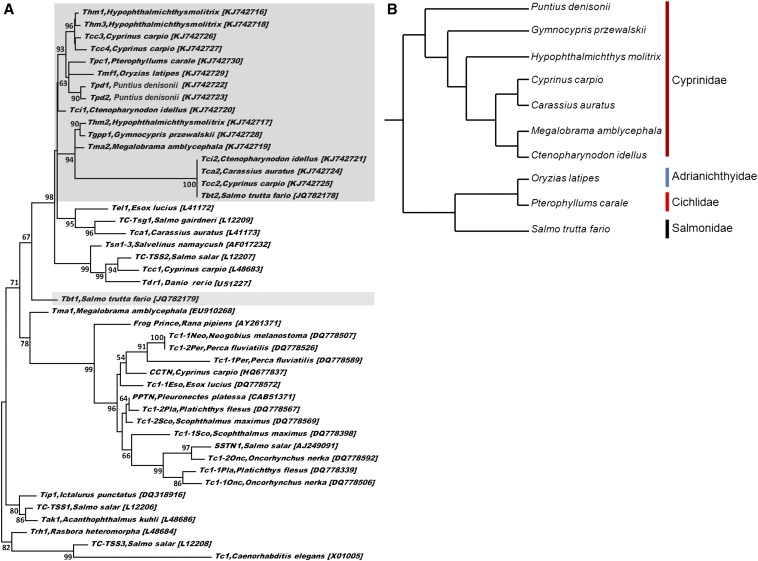
We used the 17 cloned Tc1-like transposons DNA sequence as queries for BLAST n searches of the GenBank databases; all sequences matched similarity above 50% were used for phylogenetic analysis. Our results indicate the C. elegans Tc1 does not appear as a monophyletic group, but clusters together with TC-TSS3 Salmo salar as outgroup with a high bootstrap value (Figure 1A). The isolated Tc1-like sequences seem to cluster as a distinct subfamily of the Tc1-like transposon with 98% of bootstrap, except for brown trout Tbt1. As shown in Figure 1A, the Tpd1 and Tpd2 from Denison barb do not cluster to the Tc1-like sequences from other Cyprinidae fish species, but cluster well with the medaka Tmf1 from Adrianichthyidae and the common angelfish Tpc1 from Cichlidae. Even for the isolated 13 Tc1-like sequences from seven species of the Cyprinidae family, their phylogenetic relationship is patchy clustering (Figure 1A). This means that these Cyprinidae Tc1-like sequences are not in line with the vertical evolutionary relationships (Figure 1B). By pairwise comparisons, Tc1-like transposons of the same size (1473 bp) isolated from grass carp (Tci2), common carp (Tcc2), crucian carp (Tca2), and brown trout (Tbt2) shared >99% identity to quite distantly related groups (Figure 1A; Table 1). The closely related Tc1-like transposons described here are distributed among divergent lineages of Cyprinidae and Salmonidae (diverged for >250 million years). The patchy distribution Tc1-like elements coupled with the extreme level of similarity between Tc1-like elements in the distantly divergent host species are incompatible with vertical inheritance, but are strongly indicative of multiple horizontal introductions.
Figure 1.
(A) Maximum likelihood (ML) consensus tree for Tc1-like transposases from a variety of teleost fish species. Bootstrap percentages are shown by numbers at the interior nodes. The C. elegans Tc1 sequence was used as the outgroup. The isolated 17 Tc1-like transposon sequences of this study are shown in gray in the background. (B) Phylogenetic hypotheses proposed by Nelson (2006) and Wang et al. (2007) for fish species in this study.
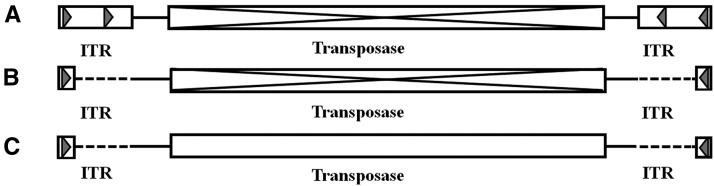
The 17 Tc1-like elements screened from different fish species could be divided into three structural patterns based on their ITRs and their transposase ORF. One pattern involved silver carp Thm1, naked carp Tgpp1, and brown trout Tbt1, and preserved complete left and right ITRs, whereas the transposase regions were interrupted by frameshifts or stop codons (Figure 2A). A second pattern involved silver carp Thm2, grass carp Tci1, Tci2, Denison barb Tpd1, Tpd2, crucian carp Tca2, common carp Tcc2, Tcc4, medaka Tmf1, common angelfish Tpc1, and brown trout Tbt2, which had incomplete ITRs as well as interrupted transposases (Figure 2B). The third pattern involved silver carp Thm3, common carp Tcc3, and blunt snout bream Tma2, which have lost some nucleotides in the ITRs, but have preserved uninterrupted transposases encoding 335 aa, 280 aa, and 238 aa Tc1-like transposases, respectively (Figure 2C).
Figure 2.
Analysis of inverted terminal repeat sequences (ITRs) and coding regions of Tc1-like transposon from different fish species. Patterns contain (A) complete left and right ITRs and an interrupted transposase, (B) incomplete left and right ITRs and an interrupted transposase, and (C) incomplete left and right ITRs and an intact transposase.
Uninterrupted Tc1-like transposase Thm3 screening from silver carp
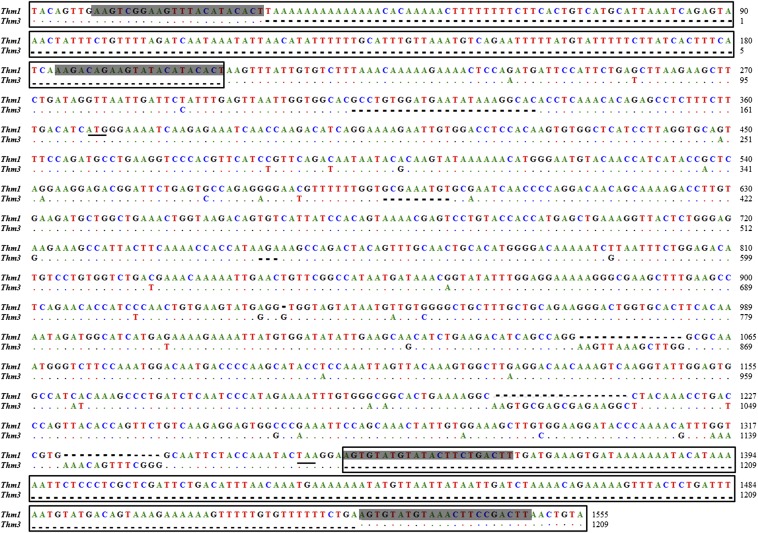
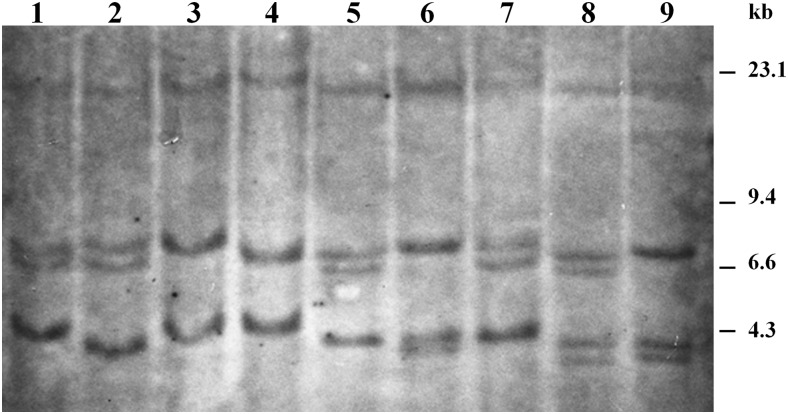
The ORF of silver carp Thm3 encodes 335 amino acids of the transposase but preserves incomplete ITRs (Figure 3). Meanwhile, the silver carp Thm1 has long ITRs of ∼210 bp with internal repeats/directed repeats (IRs/DRs), an internal structure described by Izsvak et al. (1995), but preserves interrupted transposases. The silver carp Thm3 and Thm1 elements share a high similarity (91%), except for some insertion/deletion nucleotides (Figure 3). Our further Southern blot analyses reveal that there are only three to five copies of the Thm3 transposon in the silver carp genome (Figure 4). However, the positions of these Thm3 copies are relatively constant in the silver carp genome, which indicates the silver carp Thm3 transposase may lose its autonomous transposition activity. Although this may also be attributed to the endogenous inhibition of host cell (Castaneda et al. 2011), our efforts to detect mRNAs encoded by the Thm3 element indeed failed to demonstrate the presence of the putative transposase in multiple tissues of silver carp (data not shown).
Figure 3.
Sequence alignment of Thm3 with Thm1 from silver carp. The ITRs are shown in open boxes. The gray background sequences represent internal repeats/directed repeats (IRs/DRs). Dashed lines represent missing nucleotides.
Figure 4.
Southern blot hybridization analysis of the Thm3 transposon in the silver carp genome. Genomic DNA of nine individuals was sampled from the tail fins of silver carp. Genomic DNA was digested with BglII and hybridized with a digoxin-labeled Thm3 probe. The marker sizes are indicated along the right margin.
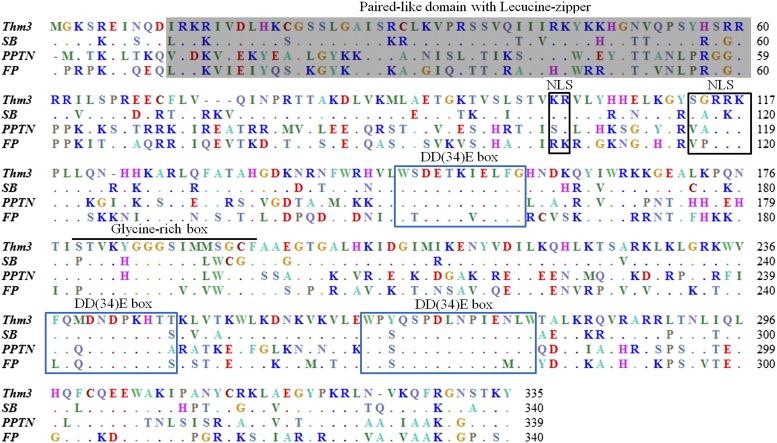
The 335-aa Thm3 transposase from silver carp exhibits greater than 80% sequence identity with reconstructed transposase SB from salmonids, but only 46% and 44% sequence identity with reconstructed transposase FP from Rana pipiens and natively active flatfish PPTN, respectively (Figure 5). As seen in salmonid SB transposase, the silver carp Thm3 transposase contains the main functional domains of Tc1-like transposases, i.e., a paired-like domain with leucine-zipper for DNA binding, a bipartite nuclear localization signal (NLS), a glycine-rich box, and complete DD(34)E domains in the C-terminal (Figure 5). The DD (34) E domain is the essential site for catalytic transposition of Tc1-like transposase. There are two Aspartates (D, D) separated by 90 amino acids, the second of which is separated from Glutamate (E) by 34 amino acid residues (Figure 5). The silver carp Thm3 transposase retains its intact functional domains, which suggests that it could be used to construct gene transfer systems in other teleost fish.
Figure 5.
Alignment of the amino acid sequences of Tc1-like transposases from the silver carp Thm3, reconstructed transposase SB from salmonid, reconstructed transposase FP from Rana pipiens, and natively active PPTN transposase from flatfish. The major functional domains are highlighted according to the structural domains of the SB transposase (Ivics et al. 1997).
Silver carp Thm3 can mediate gene transposition in blunt snout bream genome
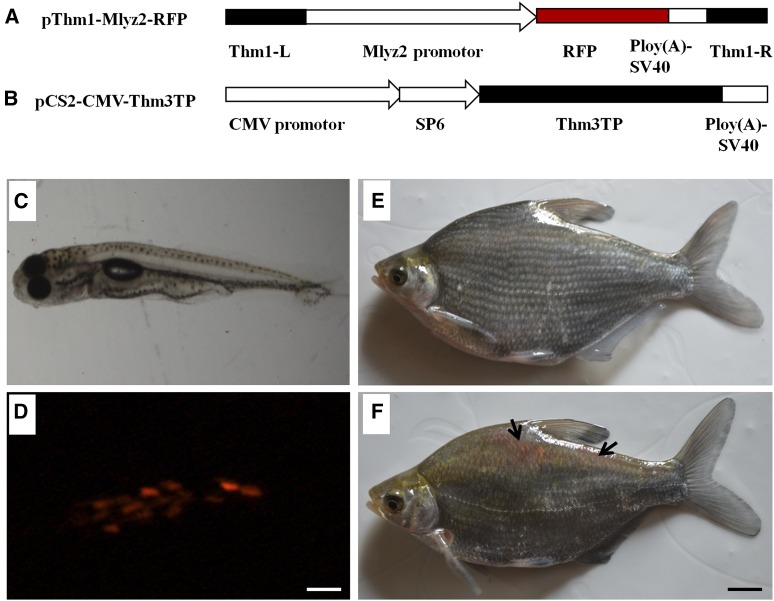
To explore whether the silver carp Thm3 transposase has transposition activity, a binary transposon vector system was developed based on Thm3 transposase and ITRs from the silver carp nonautonomous element Thm1. We constructed the donor plasmid pThm1-Mlyz2-RFP with a 367-bp left-end and 230-bp right-end of the Thm1 transposon (Figure 6A). In addition, the pCS2-CMV-Thm3TP plasmid was constructed and used to in vitro synthesize capped RNA encoding 335 aa residues Thm3 transposase (Figure 6B). We then coinjected 50 pg of donor plasmid DNA with 50 pg of capped RNA into the embryos (one- to two-cell stage) of blunt snout bream. We found that the red fluorescence expression was mainly focused in the myofibrils in blunt snout bream embryos at 72 hr postfertilization (hpf), indicative of tissue-specific expression by the zebrafish myosin light chain 2 (Mlyz2) promoter (Figure 6, C and D). In 200-d-old adult blunt snout bream, the red color could be observed in the dorsal skeletal muscles of transgenic individuals (Figure 6, E and F). PCR analysis of the transposition rate of RFP at adult blunt snout bream was performed as previously described (Jiang et al. 2012). The integration rate of RFP gene into the blunt snout bream genome when fish were 200 d old ranged from 47.0 to 59.5% (average = 50.6%, 519/1015) (Table 2). In control embryos by injected linearized donor plasmid alone, only 20.2% of embryos showed weak and high rates of the mosaic expression of RFP and few fish expressed RFP at the 200-d-old stage (data not shown). Moreover, PCR analysis showed that the average RFP-transgenic rate in control adult fish injected with the linearized donor plasmid pThm1-Mlyz2-RFP alone was 5.0% (8/184) (Table 2).
Figure 6.
Results of transposition efficiency analysis of silver carp Thm3 transposase in blunt snout bream. (A) The donor plasmid construct pThm1-Mlyz2-RFP harbors the left (367 bp) and right (230 bp) silver carp Thm1 inverted terminal repeats (ITRs). This also contains the zebrafish myosin light chain 2 (Mlyz2) promoter, the red fluorescent protein (RFP), and the SV40 poly(A) signal. (B) The transposase plasmid construct pCS2-CMV-Thm3TP. Capped silver carp Thm3 transposase mRNAs were synthesized in vitro, using linearized pCS2-CMV-Thm3TP plasmid DNA as a template. Light (C) and fluorescence (D) microscopy images of RFP expression in blunt snout bream embryos at 72 hpf after coinjection of the donor plasmid pThm1-Mlyz2-RFP and capped silver carp Thm3 transposase mRNAs in embryos at the one- to two-cell stage. Optical images of RFP expression in the negative control (E) and in transgenic positive individuals (F) in 200-d-old blunt snout bream. Arrows indicate RFP expression signals. White scale bars = 600 µm. Dark scale bars = 2 cm.
Table 2. The transposition efficiencies of silver carp Thm transposon systems in 200-d-old blunt snout bream by PCR analysis.
| Batches | No. of Survival Injection Individuals | No. of Individuals Integrated RFP | Integration Rates (%) |
|---|---|---|---|
| 1 | 66 | 31 | 47.0 |
| 2 | 147 | 64 | 43.5 |
| 3 | 121 | 72 | 59.5 |
| 4 | 87 | 41 | 47.1 |
| 5 | 212 | 107 | 50.5 |
| 6 | 187 | 96 | 51.3 |
| 7 | 195 | 108 | 55.4 |
| Average | 145 | 74 | 50.6a |
| Control 1 | 115 | 5 | 4.3 |
| Control 2 | 69 | 3 | 5.7 |
| Average | 92 | 4 | 5.0 |
Control is injected pThm1-Mlyz2-RFP plasmid only.
P < 0.01.
To precisely characterize insertions at a sequence level, we cloned the junctions of the integrated Thm1 cis-element and the surrounding genomic DNA using inverse PCR. Because the endogenous Tc1-like element Tma2 was also present in the genome of blunt snout bream (Figure 1, Table 1), its similarity with sequences of the left-end (367 bp) and right-end (230 bp) of silver carp Thm1 were 54% and 65%, respectively (supporting information, Figure S1). To avoid false positives, we chose low-similarity (<50%) regions to design the nested primers to clone the 5′ or 3′ flanking sequences (Figure S1). Among five RFP-transgenic positive individuals, there were one to four genomic integration sites in the genome of the blunt snout bream, including the specific end sequences of Thm1 DNA (Table 3; Figure S1), whereas no PCR products during splinkerette PCR were amplified in the genome of the negative control (data not shown). Moreover, a TA dinucleotide direct repeat of the target DNA at the integration site (which is the signature of Tc1-like family of transposons) was created adjacent to both ends of Thm1 at the integration sites in all individuals (Table 3). These results indicate that the silver carp Thm1 transposon insertions occurred by transposition mediated by the Thm3 transposase. It also suggests that during DNA transposition the silver carp Thm transposon system exhibits a TA insertion position preference. Furthermore, the RFP-transgenic fish were raised to maturity and crossed with the opposite sex blunt snout bream of wild-type, and F1 embryos were analyzed for RFP expression under a fluorescent microscope at 48 hpf. Among 11 RFP-transgenic positive fish that we randomly selected, 10 of 11 founders were able to express RFP in their F1 embryos from 13 to 64% (Table 4), indicating that the silver carp transposon vector inserted into the blunt snout bream genome could be transmitted to the F1 progeny.
Table 3. Exogenous Thm1 end regions and the surrounding transposon insertion site sequences in the genome of 200-d-old blunt snout bream.
| Sample No. | Copies | Sequence of Target Integration Sites |
|---|---|---|
| 1 | i | (143nt)GTACTTTATACAGTT——AACTGTATACAGCAT(11nt) |
| ii | (180nt)CAGACTTCTACAGTT——AACTGTACTGTCACT(92nt) | |
| iii | (75nt)ACATATATTACAGTT——AACTGTAAATGTGTC(138nt) | |
| iv | (85nt)CCTTGTTATACAGTT——AACTGTATTAAATGG(175nt) | |
| 2 | i | (92nt)TTATGACCTACAGTT——AACTGTATTCATATA(127nt) |
| ii | (38nt)TAAGGTTGTACAGTT——AACTGTACGCCAAAC(95nt) | |
| 3 | i | (28nt)AACTGCTCTACAGTT——AACTGTAACTGAGTA(134nt) |
| ii | (73nt)GTTGTAGTTACAGTT——AACTGTACAGTACTA(117nt) | |
| iii | (92nt)GCGTAAGTTACAGTT——AACTGTATAGTAGCG(83nt) | |
| 4 | i | (177nt)GGATAGTTTACAGTT——AACTGTATGAGTGAA(76nt) |
| 5 | i | (58nt)AGACTTGGTACAGTT——AACTGTAGACGATTT(154nt) |
| ii | (14nt)GTAGGGTTTACAGTT——AACTGTACGGTAAAT(217nt) | |
| iii | (73nt)GGTATAGGTACAGTT——AACTGTAGGTATACG(312nt) |
The TA dinucleotide direct repeats of the target DNA are marked in gray, and the end sequences of Thm1 DNA are underlined.
Table 4. RFP expression in F1 embryos of 11 RFP-transgenic positive blunt snout bream crossed with the opposite sex of wild-type.
| ID of RFP-Transgenic Positive Fisha | Gender | No. of F1 Embryos Examined | No. of RFP Positive Embryos | RFP Positive/Total F1 (%) |
|---|---|---|---|---|
| 690000116601632 | Female | 89 | 57 | 64 |
| 690020042302853 | Female | 254 | 123 | 48 |
| 690000116602112 | Male | 275 | 40 | 15 |
| 690020042302431 | Male | 212 | 27 | 13 |
| 690020042302497 | Female | 238 | 38 | 16 |
| 690020042303065 | Male | 152 | 78 | 51 |
| 690020042303056 | Male | 96 | 55 | 57 |
| 690020042302805 | Female | 321 | 0 | 0 |
| 690020042303004 | Female | 186 | 27 | 15 |
| 690000116601753 | Female | 141 | 51 | 36 |
| 690000116602026 | Male | 311 | 50 | 16 |
| Controlb | — | 215 | 0 | 0 |
The IDs of RFP-transgenic positive fish were labeled with passive integrated transponder (PIT) tags (Hongteng Barcode Corporation, Guangzhou).
The wild-type female mating with male was used as control.
Discussion
In this study, we isolated 17 Tc1-like transposons from 10 freshwater fish species, and most (16) of which clustered as a new subfamily of Tc1-like transposons. Like most members of the Tc1-like family (Eide and Anderson 1985; Emmons and Yesner 1984; Izsvak et al. 1995), the 17 cloned Tc1-like transposons were always flanked by a TA target site. Of these transposons, we have shown that the Thm3 transposon of silver carp (Hypophthalmichthys molitrix) contains the main functional domains of Tc1-like transposases and is present at relatively low copy numbers. A binary transgene vector system was developed based on the 335-amino acid Thm3 transposase and terminal inverted repeats (ITRs) from silver carp nonautonomous element Thm1. Although Thm3 is not an autonomous transposon in the silver carp, our results show that the Thm3 transposase can efficiently mediate gene insertion in the genome of one of its Cyprinidae relatives, the blunt snout bream (Megalobrama amblycephala), and can mediate transposition in the blunt snout bream germ lineage. We have shown that the silver carp Thm transposon system has a TA insertion position preference during DNA transposition in blunt snout bream.
DNA transposons transferred by the horizontal approach have been found in multiple prokaryotic and eukaryotic species (Leaver 2001; de Boer et al. 2007; Pocwierz-Kotus et al. 2007; Pace et al. 2008; Kuraku et al. 2012; Jiang et al. 2012). In the present study, the isolated Tc1-like sequences from different fish species do not conform to the vertical evolutionary relationships, and their phylogenetic relationship is patchy clustering. This means that the isolated Tc1-like transposons might be horizontally transmitted among species because of inconsistency with the accepted phylogenetic relationships of their host organisms. Also, the idea that horizontal transmission has played a role in the dissemination of cloned Tc1-like transposons in this study is supported by the presence of a complete copy in the fish genome. An almost identical 1473-bp Tc1-like transposon is found to distribute in the diverged Cyprinidae and Salmonidae, and their nucleotide sequence similarity is markedly greater than some of the most conserved protein-coding genes in vertebrates (e.g., RAG-1) (Venkatesh et al. 2001). Another convincing example is seen in the transposons Tc1-1Neo from round goby and Tc1-2Per from European perch, which have virtually identical nucleotide sequences and have been found in fish belonging to quite distantly related groups (Figure 1A and see Pocwierz-Kotus et al. 2007). Despite the lack of direct evidence, our results suggest that horizontal transmission of Tc1-like transposons seems to occur in some divergent fish lineages. Interestingly, evidence has been found that horizontal transfers of Tc1-like elements can occur between teleost fishes and lampreys and their vertebrate parasites (Gilbert et al. 2010; Kuraku et al. 2012).
Transposons are believed to have evolved via three processes: horizontal transmission, vertical inactivation, and stochastic loss (Lohe et al. 1995). An autonomous DNA transposon usually encodes an endogenous functional transposase in the host cell, which is expected to have originated relatively recently via horizontal transfer (Eide and Anderson 1985; Koga and Hori 2000; Clark et al. 2009; Jiang et al. 2012). However, most transposons, especially those in teleost fish, undergo vertical inactivation or stochastic loss to produce a large number of different copies of unautonomous transposons that have mutations in their transposase regions (Leaver 2001; Ahn et al. 2008; Wang et al. 2011; Pujolar et al. 2013). For example, in the channel catfish genome the Tip1 transposon exists at approximately 150 copies per haploid genome, whereas the Tip2 transposon exists at approximately 4000 copies per haploid genome (Nandi et al. 2007), with 32,000 copies of the nonautonomous element Tipnon present (Liu et al. 1999). However, in the silver carp genome, our results showed that low numbers of three to five copies of Thm3 transposon existed. This suggests that horizontal transmission of Thm3 transposon into the silver carp genome might not be too long, and yet vertical inactivation may not occur in the host cell. In fact, the silver carp Thm3 transposase retains its intact functional domains and it may be used to construct gene transfer systems.
The Tc1-like transposons are powerful molecular tools for transgenesis in vertebrate cells (Gallardo-Galvez et al. 2011; Garrels et al. 2014). Most Tc1-like transposons identified in fish are considered defective because they contain insertions/deletions that result in either sequence frameshifts or point mutations, which lead to a premature stop codon or a nonsense codon in the transcribed mRNA (Radice et al. 1994). Recently, several novel Tc1-like transposable elements have been found to include all the functional domains of Tc1-like transposons, e.g., CCTN in the genome of the common carp, which contains an intact 331 aa Tc1-like transposase (Wang et al. 2011), e.g., the MMTS transposon that contains motifs including DDE from mud loach (Ahn et al. 2008), and, e.g., the Tana1 transposase composed of 341 amino acids in the genome of sturgeons (Pujolar et al. 2013). However, further assays should be conducted to test if the catalytic activity of the DDE motif remains active. In this study, a binary silver carp Thm vector system was developed to enhance production of transgenic blunt snout bream as an alternative to methods of transgenesis involving the injection of donor plasmid DNA. We have shown that the silver carp Thm3 transposase can efficiently catalyze the integration of donor DNA into a TA dinucleotide site of a recipient genome. In the future, it would be good to compare the efficiency of transposition of the new element with sleeping beauty or some previously isolated Tc1-like element, although these Tc1-like elements such as SB and FP were seldom used for fish transgenesis.
Supplementary Material
Acknowledgments
We thank Dr. Jie Chen for proofreading this manuscript. This work was supported by grants from the National Science Foundation of China (No. 31572220 and 31201760 to S.M.Z. and No. 31272633 to X.Y.J.), the National High Technology Research and Development Program of China (863 Program; No. 2011AA100403 to S.M.Z.), and Shanghai Collaborative Innovation Center for Aquatic Animal Genetics and Breeding (ZF1206).
Footnotes
Supporting information is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.115.020933/-/DC1
Communicating editor: D. J. Grunwald
Literature Cited
- Ahn S. J., Kim M. S., Jang J. H., Lim S. U., Lee H. H., 2008. MMTS, a new subfamily of Tc1-like transposons. Mol. Cells 26: 387–395. [PubMed] [Google Scholar]
- Barry E. G., Witherspoon D. J., Lampe D. J., 2004. A bacterial genetic screen identifies functional coding sequences of the insect mariner transposable element Famar1 amplified from the genome of the earwig, Forficula auricularia. Genetics 166: 823–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaneda J., Genzor P., Bortvin A., 2011. piRNAs, transposon silencing, and germline genome integrity. Mutat. Res. 714: 95–104. [DOI] [PubMed] [Google Scholar]
- Clark K. J., Carlson D. F., Leaver M. J., Foster L. K., Fahrenkrug S. C., 2009. Passport, a native Tc1 transposon from flatfish, is functionally active in vertebrate cells. Nucleic Acids Res. 37: 1239–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig N. L., 1995. Unity in transposition reactions. Science 270: 253–254. [DOI] [PubMed] [Google Scholar]
- Craig N. L., Craigie R., Gellert M., Lambowitz A. M., 2002, pp. 12–23 in Mobile DNA II, ASM Press, Washington, DC. [Google Scholar]
- Davidson W. S., Koop B. F., Jones S. J., Iturra P., Vidal R., et al. , 2010. Sequencing the genome of the Atlantic salmon (Salmo salar). Genome Biol. 11: 403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer J. G., Yazawa R., Davidson W. S., Koop B. F., 2007. Bursts and horizontal evolution of DNA transposons in the speciation of pseudotetraploid salmonids. BMC Genomics 8: 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide D., Anderson P., 1985. Transposition of Tc1 in the nematode Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 82: 1756–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmons S. W., Yesner L., 1984. High-frequency excision of transposable element Tc1 in the nematode Caenorhabditis elegans is limited to somatic cells. Cell 36: 599–605. [DOI] [PubMed] [Google Scholar]
- Feschotte C., Pritham E. J., 2007. DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 41: 331–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz G., Savakis C., 1991. Minos, a new transposable element from Drosophila hydei, is a member of the Tc1-like family of transposons. Nucleic Acids Res. 19: 6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo-Galvez J. B., Mendez T., Bejar J., Alvarez M. C., 2011. Endogenous transposases affect differently Sleeping Beauty and Frog Prince transposons in fish cells. Mar. Biotechnol. (NY) 13: 695–705. [DOI] [PubMed] [Google Scholar]
- Garrels W., Holler S., Taylor U., Herrmann D., Niemann H., et al. , 2014. Assessment of fetal cell chimerism in transgenic pig lines generated by sleeping beauty transposition. PLoS One 9: e96673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C., Schaack S., Pace J. K., Brindley P. J., Feschotte C., 2010. A role for host-parasite interactions in the horizontal transfer of transposons across phyla. Nature 464: 1347–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Huang C., Shen R., Jiang X., Chen J., et al. , 2013. Insertion efficiency of Tgf2 transposon in the genome of Megalobrama amblycephala. Hereditas 35: 999–1006. [DOI] [PubMed] [Google Scholar]
- Guo X., Li F., Jiang X., Zou S., 2014. Evolution analysis of Tc1-like transposon in Salmo trutta fario genome. J. Shanghai Ocean Univ. 23: 15–21. [Google Scholar]
- Hall T.A., 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 41: 95–98. [Google Scholar]
- Hickman A. B., Perez Z. N., Zhou L., Musingarimi P., Ghirlando R., et al. , 2005. Molecular architecture of a eukaryotic DNA transposase. Nat. Struct. Mol. Biol. 12: 715–721. [DOI] [PubMed] [Google Scholar]
- Ivics Z., Hackett P. B., Plasterk R. H., Izsvak Z., 1997. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell 91: 501–510. [DOI] [PubMed] [Google Scholar]
- Izsvak Z., Ivics Z., Hackett P. B., 1995. Characterization of a Tc1-like transposable element in zebrafish (Danio rerio). Mol. Gen. Genet. 247: 312–322. [DOI] [PubMed] [Google Scholar]
- Izsvak Z., Ivics Z., Plasterk R. H., 2000. Sleeping Beauty, a wide host-range transposon vector for genetic transformation in vertebrates. J. Mol. Biol. 302: 93–102. [DOI] [PubMed] [Google Scholar]
- Janicki M., Rooke R., Yang G., 2011. Bioinformatics and genomic analysis of transposable elements in eukaryotic genomes. Chromosome Res. 19: 787–808. [DOI] [PubMed] [Google Scholar]
- Jiang X., Du X., Tian Y., Shen R., Sun C., et al. , 2012. Goldfish transposase Tgf2 presumably from recent horizontal transfer is active. FASEB J. 26: 2743–2752. [DOI] [PubMed] [Google Scholar]
- Kawakami K., Takeda H., Kawakami N., Kobayashi M., Matsuda N., et al. , 2004. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev. Cell 7: 133–144. [DOI] [PubMed] [Google Scholar]
- Kazazian H. H., 2004. Mobile elements: drivers of genome evolution. Science 303: 1626–1632. [DOI] [PubMed] [Google Scholar]
- Kidwell M. G., Lisch D. R., 2000. Transposable elements and host genome evolution. Trends Ecol. Evol. 15: 95–99. [DOI] [PubMed] [Google Scholar]
- Koga A., Hori H., 2000. Detection of de novo insertion of the medaka fish transposable element Tol2. Genetics 156: 1243–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga A., Shimada A., Shima A., Sakaizumi M., Tachida H., et al. , 2000. Evidence for recent invasion of the medaka fish genome by the Tol2 transposable element. Genetics 155: 273–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani T., Kawakami K., 2008. Misty somites, a maternal effect gene identified by transposon-mediated insertional mutagenesis in zebrafish that is essential for the somite boundary maintenance. Dev. Biol. 316: 383–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraku S., Qiu H., Meyer A., 2012. Horizontal transfers of Tc1 elements between teleost fishes and their vertebrate parasites, lampreys. Genome Biol. Evol. 4: 929–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampe D. J., Churchill M. E., Robertson H. M., 1996. A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J. 15: 5470–5479. [PMC free article] [PubMed] [Google Scholar]
- Leaver M. J., 2001. A family of Tc1-like transposons from the genomes of fishes and frogs: evidence for horizontal transmission. Gene 271: 203–214. [DOI] [PubMed] [Google Scholar]
- Liu D., You C., Liu S., Liu L., Duan W., et al. , 2009. Characterization of a novel Tc1-like transposon from bream (Cyprinidae, Megalobrama) and its genetic variation in the polyploidy progeny of bream-red crucian carp crosses. J. Mol. Evol. 69: 395–403. [DOI] [PubMed] [Google Scholar]
- Liu Z. J., Li P., Kucuktas H., Dunham R. A., 1999. Characterization of nonautonomous Tc1-like transposable elements of channel catfish (Ictalurus punctatus). Fish Physiol. Biochem. 21: 65–72. [Google Scholar]
- Lohe A. R., Moriyama E. N., Lidholm D. A., Hartl D. L., 1995. Horizontal transmission, vertical inactivation, and stochastic loss of mariner-like transposable elements. Mol. Biol. Evol. 12: 62–72. [DOI] [PubMed] [Google Scholar]
- McClintock B., 1953. Induction of instability at selected loci in Maize. Genetics 38: 579–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medhora M., Maruyama K., Hartl D. L., 1991. Molecular and functional analysis of the mariner mutator element Mos1 in Drosophila. Genetics 128: 311–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miskey C., Izsvak Z., Plasterk R. H., Ivics Z., 2003. The Frog Prince: a reconstructed transposon from Rana pipiens with high transpositional activity in vertebrate cells. Nucleic Acids Res. 31: 6873–6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Lopez M., Siddique A., Bischerour J., Lorite P., Chalmers R., et al. , 2008. Transposition of Mboumar-9: identification of a new naturally active mariner-family transposon. J. Mol. Biol. 382: 567–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi S., Peatman E., Xu P., Wang S., Li P., et al. , 2007. Repeat structure of the catfish genome: a genomic and transcriptomic assessment of Tc1-like transposon elements in channel catfish (Ictalurus punctatus). Genetica 131: 81–90. [DOI] [PubMed] [Google Scholar]
- Nelson J. S., 2006. Fishes of the World, Ed. 4th Wiley, New York. [Google Scholar]
- Nylander J. A., Ronquist F., Huelsenbeck J. P., Nieves-Aldrey J. L., 2004. Bayesian phylogenetic analysis of combined data. Syst. Biol. 53: 47–67. [DOI] [PubMed] [Google Scholar]
- Pace J. K., Gilbert C., Clark M. S., Feschotte C., 2008. Repeated horizontal transfer of a DNA transposon in mammals and other tetrapods. Proc. Natl. Acad. Sci. USA 105: 17023–17028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk R. H., Izsvak Z., Ivics Z., 1999. Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends Genet. 15: 326–332. [DOI] [PubMed] [Google Scholar]
- Pocwierz-Kotus A., Burzynski A., Wenne R., 2007. Family of Tc1-like elements from fish genomes and horizontal transfer. Gene 390: 243–251. [DOI] [PubMed] [Google Scholar]
- Pujolar J. M., Astolfi L., Boscari E., Vidotto M., Barbisan F., et al. , 2013. Tana1, a new putatively active Tc1-like transposable element in the genome of sturgeons. Mol. Phylogenet. Evol. 66: 223–232. [DOI] [PubMed] [Google Scholar]
- Radice A. D., Bugaj B., Fitch D. H., Emmons S. W., 1994. Widespread occurrence of the Tc1 transposon family: Tc1-like transposons from teleost fish. Mol. Gen. Genet. 244: 606–612. [DOI] [PubMed] [Google Scholar]
- Robertson H. M., Zumpano K. L., 1997. Molecular evolution of an ancient mariner transposon, Hsmar1, in the human genome. Gene 205: 203–217. [DOI] [PubMed] [Google Scholar]
- Rubin G. M., Spradling A. C., 1982. Genetic transformation of Drosophila with transposable element vectors. Science 218: 348–353. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T., 1989. Molecular Cloning: A Laboratory Manual, Ed. 2 New York, Cold Spring Harbor, USA. [Google Scholar]
- Swofford, D., 2003 PAUP: Phylogenetic analysis using parsimony and other methods, Version 4, Handbook and Software. Sunderland, M. A., Sinauer Associates, USA. [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G., 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh B., Erdmann M. V., Brenner S., 2001. Molecular synapomorphies resolve evolutionary relationships of extant jawed vertebrates. Proc. Natl. Acad. Sci. USA 98: 11382–11387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Ji P., Xu P., Sun X., 2011. Identification and characterization of a novel Tc1-like transposon in the Cyprinus carpio genome. J. Fish. Sci. China 18: 1392–1398. [Google Scholar]
- Wang X., Li J., He S., 2007. Molecular evidence for the monophyly of East Asian groups of Cyprinidae (Teleostei: Cypriniformes) derived from the nuclear recombination activating gene 2 sequences. Mol. Phylogenet. Evol. 42: 157–170. [DOI] [PubMed] [Google Scholar]
- Yuan Y. W., Wessler S. R., 2011. The catalytic domain of all eukaryotic cut-and-paste transposase superfamilies. Proc. Natl. Acad. Sci. USA 108: 7884–7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Figure S1 contains supporting figures. Further information on all plasmids and more detailed supporting data are available upon request.