Abstract
Sex chromosomes and the sex-determining (SD) gene are variable in vertebrates. In particular, medaka fishes in the genus Oryzias show an extremely large diversity in sex chromosomes and the SD gene, providing a good model to study the evolutionary process by which they turnover. Here, we investigated the sex determination system and sex chromosomes in six celebensis group species. Our sex-linkage analysis demonstrated that all species had an XX-XY sex determination system, and that the Oryzias marmoratus and O. profundicola sex chromosomes were homologous to O. latipes linkage group (LG) 10, while those of the other four species, O. celebensis, O. matanensis, O. wolasi, and O. woworae, were homologous to O. latipes LG 24. The phylogenetic relationship suggested a turnover of the sex chromosomes from O. latipes LG 24 to LG 10 within this group. Six sex-linkage maps showed that the former two and the latter four species shared a common SD locus, respectively, suggesting that the LG 24 acquired the SD function in a common ancestor of the celebensis group, and that the LG 10 SD function appeared in a common ancestor of O. marmoratus and O. profundicola after the divergence of O. matanensis. Additionally, fine mapping and association analysis in the former two species revealed that Sox3 on the Y chromosome is a prime candidate for the SD gene, and that the Y-specific 430-bp insertion might be involved in its SD function.
Keywords: sex-determining gene, convergent evolution, sex chromosome, Sox, medaka, genetics of sex
Sex determination mechanisms in animals are extremely diverse. Most vertebrates have genetic sex determination with sex chromosomes, while their origins differ among taxa. Such turnover of sex chromosomes is associated with the substitution or translocation of master sex-determining (SD) genes. To date, nine SD genes or candidates have been identified on different sex chromosomes in mammals, birds, frogs, and fishes (Gubbay et al. 1990; Sinclair et al. 1990; Matsuda et al. 2002; Yoshimoto et al. 2008; Smith et al. 2009; Hattori et al. 2012; Kamiya et al. 2012; Myosho et al. 2012; Yano et al. 2012; Takehana et al. 2014), confirming that different SD genes have contributed to the development of new sex chromosomes during vertebrate evolution. These studies also suggest that several groups of genes tend to be recruited as the master SD signals, because five of them belong to the transcription factor genes (Sox3-related or Dmrt1-related) and other three are involved in the TGF-beta signaling pathway (Amh, Amhr2, and Gsdf). To understand the limited diversity of SD genes, we must first elucidate a common mechanism of SD gene emergence by identifying the molecular mechanism by which each gene arose. However, most of the above vertebrate species whose SD genes have been identified are distantly related to each other, which makes tracing the evolutionary process of SD gene turnover by comparing them difficult. However, it may be easier in closely related species that have common genes and pathways involved in sex determination because they share recent common ancestors. In some fish groups, such as salmonids and sticklebacks, high rates of sex chromosome turnover have been documented among closely related species (Ross et al. 2009; Woram et al. 2003). Among them, the SD genes have not been uncovered in sticklebacks, and the majority of salmonids share the same SD gene, sdY, which has potentially jumped into different ancestral autosomes (Yano et al. 2013).
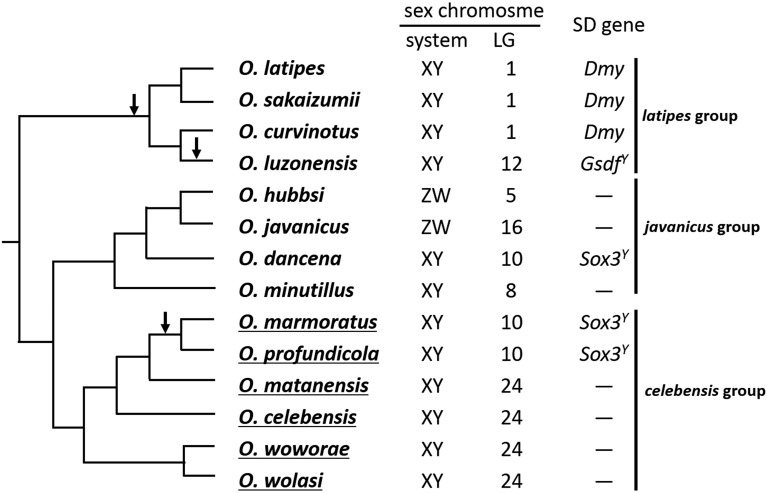
Medaka fishes in the genus Oryzias show an amazingly large diversity in their sex determination systems and sex chromosomes, providing an excellent model group for investigating the molecular mechanisms underlying the rapid turnover of sex chromosomes. They possess both XY and ZW systems, and their sex chromosomes differ from one species to another (Tanaka et al. 2007; Takehana et al. 2007a, 2007b, 2008; Nagai et al. 2008). Among the eight species analyzed to date, only three (Oryzias latipes, O. sakaizumii, and O. curvinotus) share the same sex determination mechanism (Figure 1; Matsuda et al. 2002, 2003). In these species, Dmy on the homologous Y chromosome is the SD gene, but this gene has not been found in other congeneric species. Instead, different SD genes have been isolated; GsdfY in O. luzonensis (Myosho et al. 2012) and Sox3Y in O. dancena (Takehana et al. 2014), suggesting that different sex chromosomes have evolved through the acquisition of new SD genes. However, all of these species are members of the latipes and javanicus groups in Oryzias, and no information is available on the members of the remaining celebensis group.
Figure 1.
Phylogenetic relationships and sex determination in Oryzias fishes. The phylogenetic information was taken from Takehana et al. (2005) and Mokodongan and Yamahira (2015). Arrows indicate nodes at which the sex chromosome turnovers occurred. Species examined in this study are underlined.
All species in the celebensis group are endemic to Sulawesi Island, Indonesia. To date, 14 described species—approximately half of the species in the genus—are endemic to this island, suggesting that Sulawesi is a diversity hotspot for the medaka fishes. Previous molecular phylogenetic analysis has revealed that the celebensis group is monophyletic, and genetic divergences among the species are relatively lower than those in other groups (Takehana et al. 2005). In this study, we isolated sex-linked DNA markers and identified the sex chromosomes of six species in the celebensis group. All species had an XY sex determination system, but their sex chromosomes differed: those of O. marmoratus and O. profundicola were homologous to O. latipes LG 10, while those of O. celebensis, O. matanensis, O. wolasi, and O. woworae were homologous to O. latipes LG 24. We also identified possible SD gene candidates on these sex chromosomes. In particular, the O. marmoratus and O. profundicola SD loci were tightly linked to the Sox3 gene, which is also the SD gene in O. dancena from the javanicus group, suggesting that it has been independently recruited as the SD gene in different lineages of this genus.
Materials and Methods
Fish
O. celebensis, O. marmoratus, O. matanensis, and O. profundicola were supplied by Niigata University, a subcenter of the National BioResource Project (Medaka) (http://www.shigen.nig.ac.jp/medaka/). These species were originally collected at Ujung Pandang, Lake Towuti, Lake Matano, and Lake Towuti in Sulawesi, Indonesia, respectively. O. wolasi and O. woworae were obtained commercially from aquarium shops in Japan. Their collection localities are unknown, but their original distributions are southeastern Sulawesi (Parenti and Hadiaty 2010; Parenti et al. 2013). All fish were raised and maintained at 27 ± 2° under a 14:10 h light:dark cycle.
Genetic crosses, sexing, and DNA extraction
In each species, one female and one male were crossed to generate their F1 progeny. Single pairs of the progeny were subsequently intercrossed to obtain F2 and F3 generations. We obtained 361 progeny (72 F1, 153 F2, and 136 F3) in O. celebensis, 181 (54 F1 and 127 F2) in O. marmoratus, 83 F1 in O. matanensis, 78 F1 in O. profundicola, 201 (32 F1 and 169 F2) in O. wolasi, and 66 F1 in O. woworae. Phenotypic sex was determined by adult fish secondary sex characteristics, namely, the shapes of dorsal and anal fins. Total DNA was extracted from the caudal fin by proteinase K digestion, phenol-chloroform extraction, and isopropanol precipitation (Shinomiya et al. 1999). DNA samples were dissolved in TE buffer.
Isolation of sex linked markers and linkage analysis
To identify polymorphisms between the male and female parents, we screened expressed sequence tag (EST) markers developed in O. latipes (http://mbase.nig.ac.jp/mbase/medaka_top.html) (Naruse et al. 2004). As for the markers on LG 10 and LG 24, we designed new STS primers on conserved exons across introns based on the medaka and stickleback genomic sequences (http://asia.ensembl.org/index.html) (Supporting Information, Table S1). ESTs and STSs were amplified under the following touchdown PCR conditions: 5 min at 95°; 12 cycles of 30 s at 95°, 30 s at 56–65° (−3° for every three cycles), 2 min at 72°; 22 cycles of 30 s at 95°, 30 s at 53°, 2 min at 72°; and 2 min at 72°. Polymorphisms in each amplified marker were analyzed by polyacrylamide gel electrophoresis to detect insertion, deletion, heteroduplex, and PCR-RFLPs. Sex-linkage maps were constructed for the polymorphic markers in all six species.
SD region sequencing in O. marmoratus and O. profundicola
We designed original primers based on the consensus regions between O. latipes (http://asia.ensembl.org/index.html) and O. dancena (accession numbers: AB909496 and AB909497) LG 10 sequences, and determined the DNA sequences of O. marmoratus by PCR direct sequencing and sequencing after cloning of the PCR products. We used an XX female and an XY male to determine the X and Y chromosome sequences. The X and Y sequences were deposited in GenBank under the accession numbers LC037173–LC037176. We also determined partial O. profundicola X and Y chromosome sequences to check 55 Y-specific mutation sites found in the O. marmoratus SD region.
Association analysis
We obtained wild O. marmoratus (N = 68) and O. profundicola (N = 34) stocks from Shinshu University and the National Institute for Basic Biology. We genotyped 10 Y-specific mutation regions conserved between the O. marmoratus and O. profundicola Y chromosomes by PCR-direct sequencing. Primer sequences for genotyping were as follows: the #1 and #2 common Y-specific mutations, CGGATTGAATGTCGTTTTCC and TGACCTCATTCCCGGAAAGGA; #3, TTTTTGATTGAAAAGAAGAGAGTTT and ACATGCATTTCACCCCTCTC; #4 and #5, TCTGTTGAAGGTCTGGGTCTC and TTTACGCACGCTCTGTTGTC; #6–9, TTCCGAGTTCAAAACTCTTGC and CCAAACGTTGGCTTCAAATC; #10, GTACCCCCACATCCTGTCAC and TTATTACCAGTGGGGCAACG.
Results
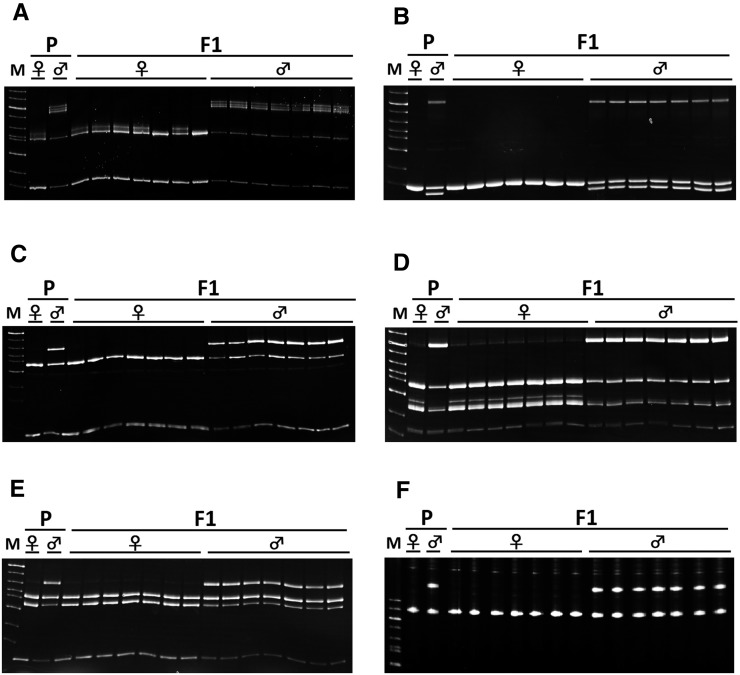
To isolate sex-linked markers in six species, O. celebensis, O. marmoratus, O. matanensis, O. profundicola, O. wolasi, and O. woworae, we screened over 250 ESTs established in O. latipes. We detected polymorphic patterns in the EST markers between the parents, and assessed whether or not the polymorphisms cosegregated with the phenotypic sex. We also designed and genotyped new STS markers on the same chromosome as the ESTs to obtain additional sex-linked markers. As a result, we successfully isolated sex-linked markers from all six species, namely five markers from O. celebensis, nine from O. marmoratus, six from O. matanensis, five from O. profundicola, five from O. wolasi, and four from O. woworae. For the completely sex-linked markers, all male progeny had the paternal genotype, while all female progeny had the maternal genotype, indicating that all of these species had an XX-XY sex determination system (Figure 2).
Figure 2.
Male heterogametic inheritance of sex-linked markers in celebensis group Oryzias fishes. Electrophoretic patterns of SOX3 PCR product digested with MseI in O. marmoratus (A), MF01SSA075C05 PCR product in O. profundicola (B), PCMT1 digested with BsmAI in O. celebensis (C), HNRPU digested with HindIII in O. matanensis (D), PGM digested with HphI in O. wolasi (E), and SOX7 in O. woworae (F). Note that the male-specific bands in the parents segregated perfectly with F1 males. F1, F1 progeny from the parents; M, marker; P, parents.
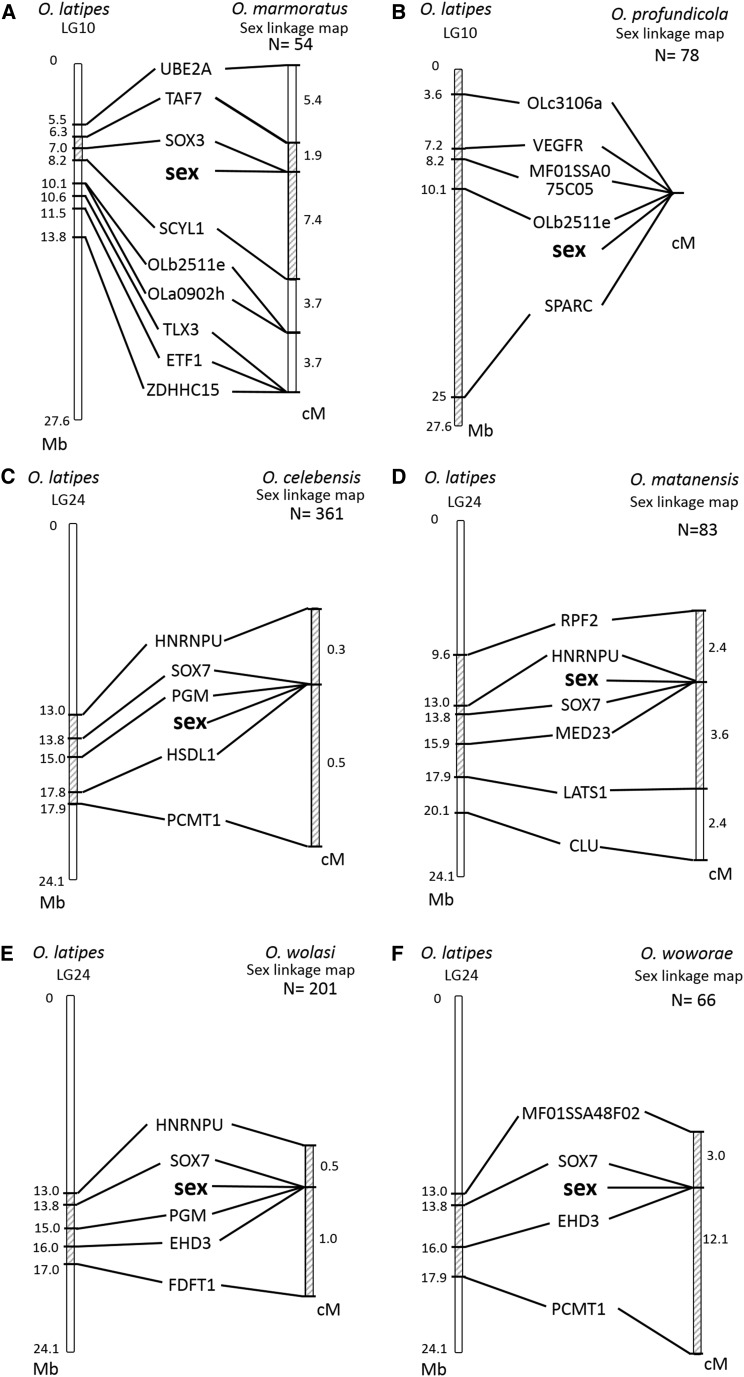
We constructed sex-linkage maps using the markers, and identified the SD loci in all six species (Figure 3). All of the O. marmoratus and O. profundicola sex-linked markers were located on O. latipes LG 10, while those of O. celebensis, O. matanensis, O. wolasi, and O. woworae were on O. latipes LG 24. Gene orders were conserved between the sex-linkage maps and the physical O. latipes maps, indicating conserved syntenies around the SD locus between the species. These results suggest that the sex O. marmoratus and O. profundicola chromosomes are homologous to O. latipes LG 10, and those of O. celebensis, O. matanensis, O. wolasi, and O. woworae to O. latipes LG 24. Furthermore, the mapped SD loci positions were consistent between O. marmoratus and O. profundicola and among O. celebensis, O. matanensis, O. wolasi, and O. woworae, suggesting single origins of both types of sex chromosome.
Figure 3.
Sex-linkage maps of DNA markers in O. marmoratus (A), O. profundicola (B), O. celebensis (C), O. matanensis (D), O. wolasi (E), and O. woworae (F). Comparison of the gene orders between the sex-linkage map in six species and the physical O. latipes LG 10 and LG 24 maps are also shown. Lines between the compared chromosomes connect the positions of orthologous gene pairs. The distances between flanking markers are shown as either physical length or map distance. The shaded area represents the sex-determining (SD) regions on the genetic maps and the corresponding regions on the physical maps.
To elucidate the exact location of the SD locus in O. marmoratus, we performed further linkage analysis by adding 127 F2 progeny (in total N = 181). This analysis pinpointed the SD locus within a 1.4 cM region between two STS markers: SOX3-up and SOX3-down. This region was approximately 34.4 kb in O. latipes, and contained only one gene, Sox3, based on its draft genome sequence data (Figure 3A). We then determined entire nucleotide sequences for the O. marmoratus SD region, and found that the X and Y sequences were 29.8 kb and 30.2 kb in length, respectively. Both SD regions contained only the Sox3 gene, suggesting this as a prime candidate for the SD gene in O. marmoratus.
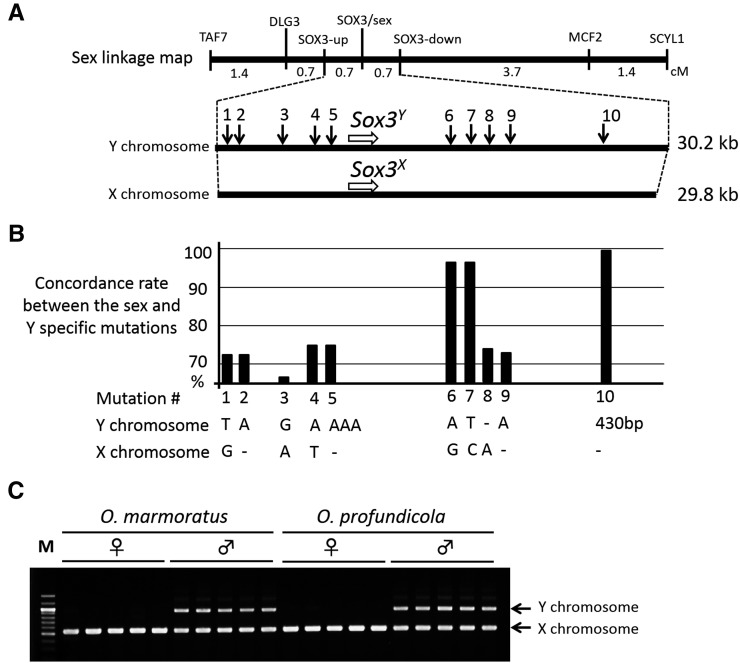
No mutations in the Sox3 gene coding sequences suggested that its regulatory mutations might contribute to the sex-determining function, as in the case of Sox3Y in O. dancena (Takehana et al. 2014). To identify the responsible sequences in the SD region, we compared the sequences between the O. marmoratus X and Y chromosomes, and detected 55 Y-specific mutations, comprising 37 SNPs and 18 indels. To find conserved mutations between species, we checked these mutation sites on the O. profundicola X and Y chromosomes, because this species was assumed to have the same SD locus. By comparing X and Y chromosome sequences between the two species, we successfully identified ten common Y-specific mutations, five SNPs and five indels (Figure 4A). Further association analysis using wild O. marmoratus (N = 68) and O. profundicola (N = 34) stocks revealed that they only had a single 430-bp insertion Y chromosome-specific mutation in common (Figure 4, B and C). This insertion was not homologous to other sequences deposited in the public database, and was not found in the O. dancena SD region, suggesting that it might contribute to the Sox3 SD function in O. marmoratus and O. profundicola.
Figure 4.
Positional cloning and association analysis of the SD locus on the sex chromosomes shared between O. marmoratus and O. profundicola. (A) A high resolution genetic map around the O. marmoratus SD region using 181 progeny (top), and physical maps of the region on the Y and X chromosomes (bottom). The SD locus is mapped within a genetic interval (1.4 cM) between two markers (SOX3-up and SOX3-down), and completely links to the marker SOX3. Arrows indicate Y-specific mutations common to O. marmoratus and O. profundicola. (B) Association analysis between the phenotypic sex and Y-specific mutations in O. marmoratus and O. profundicola. Only the 430-bp insertion perfectly matches phenotypic sex in O. marmoratus (N = 68) and O. profundicola (N = 34). (C) Electrophoretic pattern of PCR product including the 430-bp insertion in O. marmoratus and O. profundicola. Upper bands indicate the Y chromosome alleles having the insertion. Lower bands indicate the X chromosome alleles without the insertion.
Discussion
The present study clearly demonstrated the sex determination system, sex chromosomes, and SD loci in six celebensis group medaka fishes. All sex-linked markers isolated showed male-heterogametic inheritances, indicating that all six species have an XX-XY sex determination system. Previous studies have revealed that all four species, in the latipes group (Matsuda et al. 2003; Hamaguchi et al. 2004), and two species, O. dancena and O. minutillus, in the javanicus group have an XX-XY sex determination system (Takehana et al. 2007a; Nagai et al. 2008). However, two species, O. hubbsi and O. javanicus, in the javanicus group have a ZZ-ZW system (Takehana et al. 2007b, 2008). Therefore, 12 of the 14 Oryzias species have an XX-XY sex determination system, suggesting that the XX-XY system is dominant in this genus.
Sex LGs have been identified in eight Oryzias species; namely O. latipes and O. sakaizumii (Naruse et al. 2000), O. curvinotus (Matsuda et al. 2003), O. luzonensis (Tanaka et al. 2007), O. minutillus (Nagai et al. 2008), O. dancena (Takehana et al. 2007a), O. hubbsi (Takehana et al. 2007b), and O. javanicus (Takehana et al. 2008). The sex chromosomes differed from species to species, with the exception of O. latipes, O. sakaizumii, and O. curvinotus, which have the same SD gene Dmy (see Figure 1). In the present study, we successfully identified the sex LG in the six celebensis group species. The O. marmoratus and O. profundicola sex chromosomes were homologous to O. latipes LG 10, while those of O. celebensis, O. matanensis, O. wolasi, and O. woworae were homologous to O. latipes LG 24. Sex chromosomes of LG 10 and 24 were considered to share the same SD genes, respectively, because the positions of the SD loci were conserved between the species.
Although O. marmoratus and O. profundicola shared the same sex chromosomes, their sex-linkage maps differed in terms of mapping distance. In the O. marmoratus sex-linkage map, we identified a small SD region because of frequent recombination. In contrast, no recombination (0/78) was observed in the O. profundicola sex chromosome pair, suggesting that it might be suppressed along these chromosomes in this species. Similar situations were also observed in the O. celebensis, O. matanensis, O. wolasi, and O. woworae sex-linkage maps. Their sex-linked markers clustered around the SD loci, 0/361 recombination in O. celebensis, 0/83 in O. matanensis, 0/200 in O. wolasi, and 0/66 in O. woworae. This recombination suppression is likely associated with chromosome rearrangements, such as an inversion around the SD locus. The absence of recombination associated with chromosome rearrangements has been reported in other Oryzias species (Takehana et al. 2007b) and sticklebacks (Peichel et al. 2004; Ross and Peichel 2008). However, Matsuda et al. (1999) reported that maleness rather than heterogametic sex chromosome suppresses recombination in O. latipes. To test two possibilities, we need to examine the sex chromosome structure by FISH analysis and the recombination frequency in sex-reversed females.
We have successfully identified Sox3 as a prime candidate for the SD gene in O. marmoratus and probably in O. profundicola. This is the sole gene in the O. marmoratus SD region, and the 430-bp Y-specific insertion is associated with phenotypic sex in wild stocks of these species, suggesting that the insertion might contribute to its male-determining function. In another Oryzias species, O. dancena, it has been shown that the Sox3 gene on the Y chromosome is required and sufficient in male determination (Takehana et al. 2014). However, the 430-bp insertion was not found in the O. dancena SD region, suggesting that these Sox3-dependent sex determination mechanisms have evolved independently. Furthermore, the mammalian SD gene, Sry, is considered to have evolved from Sox3 as an allelic variant (Foster and Graves 1994; Sutton et al. 2011). Thus, the independent recruitments of Sox3 to male determination appears to have occurred at least three times in vertebrates.
In O. celebensis, O. matanensis, O. wolasi, and O. woworae, we identified the common SD loci on their sex chromosomes, which were homologous to O. latipes LG 24. Using the Ensemble genome browser, we identified 125 protein-coding genes within the region ranging from 13.0 to 17.0 Mb of O. latipes LG 24, which corresponded to their SD loci. There is no well-known sex-related gene among these genes, except for Sox7. Our linkage analysis showed that this gene was tightly linked to phenotypic sex in all four species. More than 20 Sox family genes have been identified so far, and classified into nine groups based on their sequence homology (Bowles et al. 2000). Of these, seven genes (Sry, Sox3, Sox8, Sox9, Sox10, Sox15, and Sox17) are involved in either mammalian sex determination or differentiation, suggesting that these Sox genes potentially have an SD function (Koopman et al. 1991; Sutton et al. 2011; Barrionuevo et al. 2009; Chaboissier et al. 2004; Polanco et al. 2010; Sarraj et al. 2003; Kanai et al. 1996). Further analysis of the SD function of Sox7 in the Oryzias species is required.
In the celebensis group, the basal lineages shared the same SD loci, suggesting that the common ancestor of this group had sex chromosomes homologous to O. latipes LG 24. Thus, a transition of the different sex determination mechanisms likely occurred in the common ancestor of the sister species pair, O. marmoratus and O. profundicola, after the divergence of O. matanensis, because they share the common SD loci on the sex chromosomes that were homologous to O. latipes LG 10. A recent molecular phylogenetic study reconfirmed the close genetic relationship between O. marmoratus, O. profundicola, and O. matanensis, and estimated their divergence time at less than 2.5 million years ago (Mokodongan and Yamahira 2015). Additionally, these three species inhabit in the same Malili Lake System in central Sulawesi, the former two species in Lake Towuti and the latter in Lake Matano (Kottelat 1990). Thus, these findings suggest that the turnover of the sex chromosomes occurred very recently in these species, likely during the speciation process in in the different lakes.
Supplementary Material
Acknowledgments
We thank T. Sato for mating O. woworae, and N. Shibata for O. marmoratus and O. profundicola DNA sampling at Shinshu University. This work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan to M.S. (25440178). The authors declare no conflict of interest.
Footnotes
Supporting information is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.115.021543/-/DC1
Sequence data from this article have been deposited with the DDBJ data library under accession numbers LC037173–LC037176.
Communicating editor: J. A. Birchler
Literature Cited
- Barrionuevo F., Georg I., Scherthan H., Lecureuil C., Guillou F., et al. , 2009. Testis cord differentiation after the sex determination stage is independent of Sox9 but fails in the combined absence of Sox9 and Sox8. Dev. Biol. 327: 301–312. [DOI] [PubMed] [Google Scholar]
- Bowles J., Schepers G., Koopman P., 2000. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev. Biol. 227: 239–255. [DOI] [PubMed] [Google Scholar]
- Chaboissier M. C., Kobayashi A., Vidal V. I., Lutzkendorf S., van de Kant H. J., et al. , 2004. Functional analysis of Sox8 and Sox9 during sex determination in the mouse. Development 131: 1891–1901. [DOI] [PubMed] [Google Scholar]
- Foster J. W., Graves J. A., 1994. An SRY-related sequence on the marsupial X chromosome: implications for the evolution of the mammalian testis-determining gene. Proc. Natl. Acad. Sci. USA 91: 1927–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubbay J., Collignon J., Koopman P., Capel B., Economou A., et al. , 1990. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature 346: 245–260. [DOI] [PubMed] [Google Scholar]
- Hamaguchi S., Toyazaki Y., Shinomiya A., Sakaizumi M., 2004. The XX–XY sex-determination system in Oryzias luzonensis and O. mekongensis revealed by the sex ratio of the progeny of sex-reversed fish. Zoolog. Sci. 21: 1015–1018. [DOI] [PubMed] [Google Scholar]
- Hattori R. S., Murai Y., Oura M., Masuda S., Majhi S. K., et al. , 2012. A Y-linked anti-Mullerian hormone duplication takes over a critical role in sex determination. Proc. Natl. Acad. Sci. USA 109: 2955–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya T., Kai W., Tasumi S., Oka A., Matsunaga T., et al. , 2012. A trans-species missense SNP in Amhr2 is associated with sex determination in the tiger pufferfish, Takifugu rubripes (fugu). PLoS Genet. 8: e1002798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y., Kanai-Azuma M., Noce T., Saido T. C., Shiroishi T., et al. , 1996. Identification of two Sox17 messenger RNA isoforms, with and without the high mobility group box region, and their differential expression in mouse spermatogenesis. J. Cell Biol. 133: 667–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman P., Gubbay J., Vilain N., Goodfellow P., Lovell-Badge R., 1991. Male development of chromosomally female mice transgenic for Sry. Nature 351: 117–121. [DOI] [PubMed] [Google Scholar]
- Kottelat M., 1990. The ricefishes (Oryziidae) of the Malili Lakes, Sulawesi, Indonesia, with description of a new species. Ichthyol. Explor. Freshwaters. Ichthyol. Explor. Freshwat. 1: 151–166. [Google Scholar]
- Matsuda M., Sotoyama S., Hamaguchi S., Sakaizumi M., 1999. Male-specific restriction of recombination frequency in the sex chromosomes of the medaka, Oryzias latipes. Genet. Res. 73: 225–231. [Google Scholar]
- Matsuda M., Nagahama Y., Shinomiya A., Sato T., Matsuda C., et al. , 2002. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417: 559–563. [DOI] [PubMed] [Google Scholar]
- Matsuda M., Sato T., Toyazaki Y., Nagahama Y., Hamaguchi S., et al. , 2003. Oryzias curvinotous has DMY, a gene that is required for male development in the Medaka, O. latipes. Zoolog. Sci. 20: 159–161. [DOI] [PubMed] [Google Scholar]
- Mokodongan D. F., Yamahira K., 2015. Origin and intra-island diversification of Sulawesi endemic Adrianichthyidae. Mol. Phylogenet. Evol. 93: 150–160. [DOI] [PubMed] [Google Scholar]
- Myosho T., Otake H., Masuyama H., Matsuda M., Kuroki Y., et al. , 2012. Tracing the emergence of a novel sex-determining gene in medaka, Oryzias luzonensis. Genetics 191: 163–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T., Takehana Y., Hamaguchi S., Sakaizumi M., 2008. Identification of the sex-determining locus in the Thai medaka, Oryzias minutillus. Cytogenet. Genome Res. 121: 137–142. [DOI] [PubMed] [Google Scholar]
- Naruse K., Fukamachi S., Mitani H., Kondo M., Matsuoka T., et al. , 2000. A detailed linkage map of medaka, Oryzias latipes: comparative genomics and genome evolution. Genetics 154: 1773–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naruse K., Tanaka M., Mita K., Shima A., Postlethwait J., et al. , 2004. A medaka gene map: the trace of ancestral vertebrate proto-chromosomes revealed by comparative gene mapping. Genome Res. 14: 820–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenti L. R., Hadiaty R. K., 2010. A new, remarkably colorful, small ricefish of the genus Oryzias (Beloniformes, Adrianichthyidae) from Sulawesi, Indonesia. Copeia 2: 268–273. [Google Scholar]
- Parenti L. R., Hadiaty R. K., Lumbantobing D., Herder F., 2013. Two new ricefishes of the genus Oryzias (Atherinomorpha: Beloniformes: Adrianichthyidae) augment the endemic freshwater fish fauna of southeastern Sulawesi, Indonesia. Copeia 3: 403–414. [Google Scholar]
- Peichel C. L., Ross J. A., Matson C. K., Dickson M., Grimwood J., et al. , 2004. The master sex-determination locus in threespine sticklebacks is on a nascent Y chromosome. Curr. Biol. 14: 1416–1424. [DOI] [PubMed] [Google Scholar]
- Polanco J. C., Wilhelm D., Davidson T. L., Knight D., Koopman P., 2010. Sox10 gain-of-function causes XX sex reversal in mice: implications for human 22q-linked disorders of sex development. Hum. Mol. Genet. 19: 506–516. [DOI] [PubMed] [Google Scholar]
- Ross J. A., Peichel C. L., 2008. Molecular cytogenetic evidence of rearrangements on the Y chromosome of the threespine stickleback fish. Genetics 179: 2173–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J. A., Urton J. R., Boland J., Shapiro M. D., Peichel C. L., 2009. Turnover of sex chromosomes in the stickleback fishes (Gasterosteidae). PLoS Genet. 5: e1000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarraj M. A., Wilmore H. P., McClive P. J., Sinclair A. H., 2003. Sox15 is up regulated in the embryonic mouse testis. Gene Expr. Patterns 3: 413–417. [DOI] [PubMed] [Google Scholar]
- Shinomiya A., Matsuda M., Hamaguchi S., Sakaizumi M., 1999. Identification of genetic sex of the medaka, Oryzias latipes, by PCR. Fish Biol. J. Medaka 10: 31–32. [Google Scholar]
- Sinclair A., Berta P., Palmer M. S., Hawkins J. R., Griffiths B. L., et al. , 1990. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 346: 240–244. [DOI] [PubMed] [Google Scholar]
- Smith C. A., Roeszler K. N., Ohnesorg T., Cummins D. M., Farlie P. G., et al. , 2009. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature 461: 267–271. [DOI] [PubMed] [Google Scholar]
- Sutton E., Hughes J., Sekido R., Tan J., Arboleda V., et al. , 2011. Identification of SOX3 as an XX male sex reversal gene in mice and humans. J. Clin. Invest. 121: 328–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takehana Y., Naruse K., Sakaizumi M., 2005. Molecular phylogeny of the medaka fishes genus Oryzias (Beloniformes: Adrianichthyidae) based on nuclear and mitochondrial DNA sequences. Mol. Phylogenet. Evol. 36: 417–428. [DOI] [PubMed] [Google Scholar]
- Takehana Y., Demiyah D., Naruse K., Hamaguchi S., Sakaizumi M., 2007a Evolution of different Y chromosomes in two medaka species, Oryzias dancena and O. latipes. Genetics 175: 1335–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takehana Y., Naruse K., Hamaguchi S., Sakaizumi M., 2007b Evolution of ZZ/ZW and XX/XY sex-determination systems in the closely related medaka species, Oryzias hubbsi and O. dancena. Chromosoma 116: 463–470. [DOI] [PubMed] [Google Scholar]
- Takehana Y., Hamaguchi S., Sakaizumi M., 2008. Different origins of ZZ/ZW sex chromosomes in closely related medaka fishes, Oryzias javanicus and O. hubbsi. Chromosome Res. 16: 801–811. [DOI] [PubMed] [Google Scholar]
- Takehana Y., Matsuda M., Myosho T., Suster M. L., Kawakami K., et al. , 2014. Co-option of Sox3 as the male-determining factor on the Y chromosome in the fish Oryzias dancena. Nat. Commun. 5: 4157. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Takehana Y., Naruse K., Hamaguchi S., Sakaizumi M., 2007. Evidence for different origins of sex chromosomes in closely related Oryzias fishes: substitution of the master sex-determining gene. Genetics 177: 2075–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woram R. A., Gharbi K., Sakamoto T., Hoyheim B., Holm L. E., et al. , 2003. Comparative genome analysis of the primary sex-determining locus in salmonid fishes. Genome Res. 13: 272–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano A., Guyomard R., Nicol B., Jouanno E., Quillet E., et al. , 2012. An immune-related gene evolved into the master sex-determining gene in rainbow trout, Oncorhynchus mykiss. Curr. Biol. 22: 1423–1428. [DOI] [PubMed] [Google Scholar]
- Yano A., Nicol B., Jouanno E., Quillet E., Fostier A., et al. , 2013. The sexually dimorphic on the Y-chromosome gene (sdY) is a conserved male-specific Y-chromosome sequence in many salmonids. Evol. Appl. 6: 486–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto S., Okada E., Umemoto H., Tamura K., Uno Y., et al. , 2008. A W-linked DM-domain gene, DM-W, participates in primary ovary development in Xenopus laevis. Proc. Natl. Acad. Sci. USA 105: 2469–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.