Abstract
Symbiotic interactions between microbes and their multicellular hosts have manifold biological consequences. To better understand how bacteria maintain symbiotic associations with animal hosts, we analyzed genome-wide gene expression for the endosymbiotic α-proteobacteria Wolbachia pipientis across the entire life cycle of Drosophila melanogaster. We found that the majority of Wolbachia genes are expressed stably across the D. melanogaster life cycle, but that 7.8% of Wolbachia genes exhibit robust stage- or sex-specific expression differences when studied in the whole-organism context. Differentially-expressed Wolbachia genes are typically up-regulated after Drosophila embryogenesis and include many bacterial membrane, secretion system, and ankyrin repeat-containing proteins. Sex-biased genes are often organized as small operons of uncharacterized genes and are mainly up-regulated in adult Drosophila males in an age-dependent manner. We also systematically investigated expression levels of previously-reported candidate genes thought to be involved in host-microbe interaction, including those in the WO-A and WO-B prophages and in the Octomom region, which has been implicated in regulating bacterial titer and pathogenicity. Our work provides comprehensive insight into the developmental dynamics of gene expression for a widespread endosymbiont in its natural host context, and shows that public gene expression data harbor rich resources to probe the functional basis of the Wolbachia-Drosophila symbiosis and annotate the transcriptional outputs of the Wolbachia genome.
Keywords: Wolbachia, Drosophila, symbiosis, development, cytoplasmic incompatibility
Intracellular bacterial symbioses provide powerful systems to investigate the diverse consequences of coevolution between microbes and their hosts. Some bacterial endosymbiotic interactions are beneficial to both organisms, resulting in coadapations that generate “obligate” dependency. Obligate symbioses are often characterized by ancient phylogenetic associations, restriction of microbes to specialized host cells, provision of essential nutrients from microbe to host, and extreme microbial genome reduction (reviewed in Dale and Moran 2006; Moran et al. 2008). Other microbial endosymbiotic interactions are obligate for the microbe, but are nonessential (“facultative”) from the standpoint of the host. Facultative endosymbionts are of particular interest since some may represent a transitional state between free-living bacteria and obligate mutualists, thus offering insights into both the early evolutionary stages of mutualism and the propagation of invasive pathogens (Dale and Moran 2006; Moran and Degnan 2006).
Efforts to identify microbial genes that maintain infections of facultative endosymbionts are hampered by the inability to culture and manipulate these species in a free-living state. Likewise, the lack of extreme genome reduction in facultative endosymbionts does not allow the mere existence of a gene to provide prima facie evidence of its importance in a particular host context, as it does in mutualist species with highly-reduced genomes (Moran and Degnan 2006). Therefore, candidate genes in facultative endosymbionts that might mediate interaction with their hosts have been primarily identified using comparative genomic approaches. For example, initial sequencing of the Wolbachia pipientis genome from the arthropod Drosophila melanogaster revealed an unusually large number of ankyrin repeat domain (ANK) encoding genes relative to other bacteria (Wu et al. 2004). Large numbers of ANK-containing genes are also observed in the genomes of other Wolbachia strains that form facultative associations with arthropod hosts (Iturbe-Ormaetxe et al. 2005; Duron et al. 2007; Siozios et al. 2013), while few ANK-containing genes are found in the obligate Wolbachia endosymbionts of nematodes (Foster et al. 2005; Darby et al. 2012). Comparative genomic analysis of more closely-related strains of Wolbachia has also been used to identify candidate genes involved in host-symbiont interaction (Iturbe-Ormaetxe et al. 2005; Sinkins et al. 2005; Duron et al. 2007; Chrostek et al. 2013; Woolfit et al. 2013). For example, a cluster of eight genes (called the Octomom region), identified as being specifically duplicated in the pathogenic “Popcorn” (wMelPop) strain of Wolbachia from D. melanogaster (Chrostek et al. 2013; Woolfit et al. 2013), was recently shown to cause the high bacterial titers and virulence associated with this strain (Chrostek and Teixeira 2015).
Genome-wide gene expression profiling offers another promising approach to identify candidate genes involved in host-symbiont interactions. Both transcriptomics and proteomics have been used successfully to study how bacterial gene regulation changes in native host tissues for obligate endosymbionts (Wilcox et al. 2003; Moran et al. 2005; Reymond et al. 2006; Bennuru et al. 2011; Darby et al. 2012; Rao et al. 2012; Luck et al. 2014). However, genome-wide expression profiling has not yet been used extensively to study gene expression dynamics for facultative endosymbionts in their native host context (Slatko et al. 2014). Recently, Darby et al. (2014) conducted transcriptomic and proteomic analysis of a Wolbachia strain from D. melanogaster (wMelPop-CLA) and Baldridge et al. (2014) profiled the proteome of Wolbachia wStr from the planthopper Laodelphax striatellus. Both of these studies used stably transinfected nonnative host cell lines from the mosquito Aedes albopictus. Likewise, two recent studies have also used wMelPop-CLA transfected in nonnative Aedes cell lines to identify small noncoding RNAs (ncRNAs) by high-throughput sequencing (Mayoral et al. 2014; Woolfit et al. 2015). Woolfit et al. (2015) also generated transcriptomic data from two Wolbachia strains (wMelPop and wMelCS) in native host tissues (heads of D. melanogaster), but did not attempt to identify differentially expressed genes that may be involved in host-microbe interactions.
Here, we report global gene expression dynamics for a facultative endosymbiont across the life cycle of a native arthropod host. The rationale for this analysis is to identify bacterial genes involved in maintaining facultative endosymbiotic associations on the basis of their differential expression across host life-cycle stages. Our work takes advantage of a previously uncharacterized Wolbachia infection in the ISO1 reference strain that was used for the D. melanogaster genome project (Brizuela et al. 1994; Adams et al. 2000). We show that the D. melanogaster ISO1 strain was originally infected with Wolbachia prior to being donated to the Drosophila stock center, whereafter it was used by the modENCODE project to generate deep total RNA-seq data from 30 time points across the D. melanogaster life cycle including embryos, larvae, pupae, adult males, and adult females (Graveley et al. 2011; Brown et al. 2014; Duff et al. 2015). Using this rich transcriptomic resource, we show that the majority of Wolbachia genes are expressed across the life cycle, but that most Wolbachia genes show stable expression across different host stages and sexes when studied at the whole-fly level. We identify a set of 80 genes that show reproducible changes in expression levels in at least one life-cycle stage, the majority of which are up-regulated after embryonic development with peaks of expression in early larval, late pupal or adult stages. We also identify 41 genes that show expression differences between adult males and females, with the majority of these sex-biased genes being up-regulated in males and showing age-dependent effects. Genes with stage- or sex-specific expression differences include chaperones, ANK-containing genes, and genes with predicted membrane or secretion system function, but most have no known function. Our results provide general insight into the dynamics of gene expression in a facultative endosymbiont across different life cycle stages and sexes of an arthropod host, and provide a rich set of resources to further explore the functional basis of the Wolbachia-Drosophila symbiosis.
Materials and Methods
D. melanogaster strains and husbandry
Substrains of the D. melanogaster ISO1 strain originally described in Brizuela et al. (1994) were obtained from several sources: (i) the Bloomington Drosophila Stock Center (BDSC ISO1, stock #2057); (ii) Jim Kennison (National Institute of Child Health and Human Development); (iii) Todd Laverty (Howard Hughes Medical Institute Janelia Farms); and (iv) Sue Celniker (Lawrence Berkeley National Laboratory). The construction of the isogenic line carrying Wolbachia variant wMel in a DrosDel w1118 background (Ryder et al. 2004) is described in Chrostek et al. (2013). D. melanogaster lines were maintained on a standard cornmeal diet at a constant temperature of 25°.
We generated versions of all ISO1 substrains cured of any potential Wolbachia infection by treating with tetracycline for two generations. Adults were allowed to lay eggs for 5 d on Formula 4-24 food (Carolina, cat #173210) mixed with equal part water containing 0.25 mg/ml tetracycline. Offspring from the first generation were collected at 10 d and transferred to new food containing 0.25 mg/ml tetracycline and allowed to lay eggs for 5 d. Offspring from the second generation were collected at 10 d and transferred onto standard cornmeal-agar food to establish Wolbachia-free stocks.
Wolbachia infection status
DNA for polymerase chain reaction (PCR) screening of Wolbachia infection status was prepared from single flies by placing individual males in a standard fly squish buffer (50 μl of 1M Tris pH 8.0, 0.5M EDTA, 5M NaCl) plus 1 μl of 10 mg/ml Proteinase K. Flies were then placed in a thermocycler at 37° for 30 min, 95° for 2 min followed by a 4° hold. PCR was performed using 4 μl of fly squish product in a total volume of 50 μl. The presence of Wolbachia was confirmed by PCR using two sets of primers: (i) Wolbachia_F2 (5′-TGGCTCACATAGATGCTGGT- 3′) and Wolbachia_R2 (5′-GTCCCATTTCTCACGCATTT-3′); and (ii) Wolbachia_F3 (5′-ATCCTGCAAATTGGCGTACT-3′) and Wolbachia_R3 (5′-ATAACGCACACCTGGCAAAT-3′). To ensure DNA preparation was sufficient for PCR amplification, control primers were used from the D. melanogaster genome: rDNA-F (5′-AAACTAGGATTAGATACCCTATTAT-3′) and rDNA-R (5′-AAGAGCGACGGGCGATGTGT-3′). PCR was performed with Kappa HiFi polymerase (KAPA Biosystems, KK2502) using the following reaction conditions: 30 cycles of 95° for 20 sec, 60° for 15 sec, and 72° for 90 sec.
Genome sequencing and data analysis
Genomic DNA for the BDSC ISO1 strain was prepared from 10 starved, adult males using the Qiagen DNeasy Blood and Tissue Kit (Qiagen, 69504). A total of 1 μg of DNA was fragmented using a Covaris S220 sonicator (Covaris Inc.) to 250 bp fragments by adjusting the treatment time to 85 sec. Following the manufacturer’s directions, short fragment libraries were made using KAPA Library Preparation Kits (KAPA Biosystems, KK8201) and Bioo Scientific NEXTflex DNA Barcodes (Bioo Scientific, 514104). The resulting libraries were purified using the Agencourt AMPure XP system (Beckman Coulter, A63880), then quantified using a Bioanalyzer (Agilent Technologies) and a Qubit Fluorometer (Life Technologies). Libraries were pooled with other strains, requantified, and run for 100 cycles in paired-end high output mode over multiple lanes on an Illumina HiSeq 2000 instrument using HiSeq Control Software v1.5.15.1 and Real-Time Analysis v1.13.48.0. CASAVA v1.8.2 was run to demultiplex reads and generate fastq files.
Fastq sequences were mapped against a “holo-genome” consisting of the Release 5 version of the D. melanogaster genome (Ensembl Genomes Release 24, Drosophila_melanogaster.BDGP5.24.dna.toplevel.fa) and the Wolbachia wMel reference genome (Ensembl Genomes Release 24, Wolbachia_endosymbiont_of_drosophila_melanogaster.GCA_000008025.1.24) (Cunningham et al. 2015; Kersey et al. 2014). Holo-genome reference mapping was performed using bwa mem v0.7.5a (Li 2013) with default parameters in paired-end mode. Mapped reads for all runs from the same sample were merged, sorted and converted to BAM format using samtools v0.1.19 (Li et al. 2009). BAM files were then used to create BCF and fastq consensus sequence files using samtools mpileup v0.1.19 (options -d 100000). Fastq consensus sequence files were converted to fasta using seqtk v1.0-r76-dirty (https://github.com/lh3/seqtk) and concatenated with consensus sequences of Wolbachia-type strains from Chrostek et al. (2013). Maximum-likelihood phylogenetic analysis on resulting multiple alignments was performed using raxmlHPC-PTHREADS v8.1.16 (options -T 6 -f a -x 12345 -p 12345 -N 100 -m GTRGAMMA) (Stamatakis 2014).
RNA-seq data analysis
We analyzed total RNA-seq libraries from the modENCODE developmental time course, which samples 30 time points from the BDSC ISO1 substrain across the D. melanogaster life cycle including embryos, larvae, pupae, adult males, and adult females. Information about the RNA sample collection (Graveley et al. 2011; Brown et al. 2014) and RNA-seq library construction and sequencing (Duff et al. 2015) for these datasets has been described previously. Total RNA-seq libraries analyzed are 100 bp read length, rRNA-depleted, paired-end, and stranded, with two biological replicates available for 24 of the 30 time points. All nonadult samples are from mixed sex organisms in unknown ratios; adult female samples are from mated and virgin flies in unknown ratios.
Total RNA-seq fastq sequences from SRP001696 were downloaded and mapped against the holo-genome described above in paired-end mode using bwa mem v0.7.5a with default parameters (accession numbers for samples used in this study are given in Supporting Information, Table S1). Resulting mapped reads were sorted and converted to BAM format using samtools v0.1.19. Counts for both forward and reverse reads together were used to summarize numbers of reads mapping to the Wolbachia and D. melanogaster genomes. Forward reads from each read-pair (which correspond to the antisense orientation in the Illumina TruSeq Stranded Total RNA kit used) were converted to the opposite strand and combined with reverse reads to generate wiggle plots of strand-specific RNA-seq coverage.
Sorted BAM files were used to count reads overlapping protein-coding genes on the sense orientation by one or more bp using BEDtools v2.22.0 (Quinlan and Hall 2010) with the Ensembl Genomes Release 24 version of the Wolbachia genome annotation (Wolbachia_endosymbiont_of_drosophila_melanogaster.GCA_000008025.1.24.gtf). We did not attempt to estimate expression levels for annotated ncRNA genes because bacterial rRNA transcripts were not targeted for depletion by modENCODE and thus Wolbachia rRNA expression levels are high and variable among samples, which affects normalization of all protein-coding genes. Since current gene models in Wolbachia correspond only to coding regions and not full-length transcripts, we chose a read counting strategy that allowed RNA-seq reads to extend beyond currently-annotated gene model limits. Only counts for the reverse read from each read-pair (which corresponds to the sense orientation in the Illumina TruSeq Stranded Total RNA kit) were used for expression level estimates, differential expression analysis, and clustering.
We performed differential expression analysis using edgeR v3.6.8 (Robinson et al. 2010) with p-values adjusted using the method of Benjamini and Hochberg (1995) to correct for multiple testing. Read counts were normalized using the trimmed mean of M-values method (Robinson and Oshlack 2010) and models were fitted using tagwise dispersion (Robinson and Smyth 2007). To identify Wolbachia genes that change in any stage across the life cycle, we performed a single analysis using an ANOVA-like generalized linear model approach (McCarthy et al. 2012) using all stages of the ISO1 developmental time course that had replicates (24 time points) with an adjusted p-value cutoff of 0.05. To identify Wolbachia genes that change between pairs of samples we used an exact test approach (Robinson and Smyth 2008) with adjusted p-value 0.01 and twofold change cutoffs.
Probabilistic clustering on all samples from the ISO1 time course (both with and without biological replicates) was performed with MBcluster.seq v.1.0 (Si et al. 2014) using the Poisson model with two clusters and the expectation maximization method. Because MBcluster.seq is a probabilistic method, we performed 1000 runs of the clustering analysis. We matched cluster identifiers from different runs using the fact that the majority of genes are stably expressed across the life cycle, and defined the cluster with the majority of genes as “cluster 1” and the remaining genes as “cluster 2”. Genes assigned to cluster 2 were further classified into subclusters 2a and 2b on the basis of the number of runs in which a gene was assigned to cluster 2.
Within-sample normalized read counts in units of transcripts per million (TPM) (Li and Dewey 2011) were also used to generate between-sample correlation and gene-by-sample heatmaps. Effective gene length in TPM normalization was set to be to account for reads that extend beyond annotated gene models. Normalization by library size in TPM and differential expression analyses removes the ability to detect global up- or down-regulation of all Wolbachia genes that might occur from differences in Wolbachia titer among samples, but does not affect the ability to detect difference among samples due to differential expression of individual genes. Differential expression, clustering, and visualization were performed using R software v3.1.1 (R Development Core Team 2012).
Reverse transcription real-time quantitative PCR
RNA for reverse transcription real-time quantitative PCR (RT-qPCR) was obtained from embryos, adult males, and adult virgin females for the BDSC ISO1 and DrosDel w1118 strains. Two independent collections, each with five biological replicates per stage, were performed for each D. melanogaster line. For embryo collection, flies laid eggs for 2 hr on agar plates supplemented with 1:1 yeast/water paste. After 16 hr at 25° the embryos were collected, treated with 2% sodium hypochlorite, and washed with sterile water before RNA extraction. A total of 500 embryos were used per sample. For adult collection, males and females were separated immediately after eclosion and maintained on a standard diet for 24 hr before RNA extraction. Ten adult flies were used per sample. Samples were homogenized with a plastic pestle in 1 ml of Trizol Reagent (Ambion, 15596-018). RNA was extracted according to the manufacturer’s protocol and resuspended in 50 μl of diethylpyrocarbonate-treated water (Ambion, AM9915G). RNA concentrations were determined using a NanoDrop ND-1000 Spectrophotometer. cDNA was prepared from 4 μg of total RNA using random primers (Promega, C1181) and Moloney Murine Leukemia Virus reverse transcriptase (Promega, M1705). Primers were allowed to bind to the template RNA at 70° for 5 min and the reaction proceeded at 25° for 10 min, 37° for 60 min, and 85° for 10 min.
RT-qPCR reactions were carried out in a CFX384 Real-Time PCR Detection System (Bio-Rad). Reactions were carried out in 384-well plates (Bio-Rad, HSP3805) using iTaq Universal SYBR Green Supermix (Bio-Rad, 172-5125), 0.15 μM of each primer, and 5 μl of cDNA diluted 1:50 in water. Each complete, independent collection of each D. melanogaster line was analyzed in one plate. Each plate contained two technical replicates of every sample for each set of primers. Sequences of the primers used for RT-qPCR can be found in Table S4. Amplification conditions were set up as follows: 50° for 2 min, 95° for 10 min, followed by 40 cycles of 95° for 30 sec, 57° for 1 min, and 72° for 30 sec. Melting curves were analyzed to confirm the specificity of amplified products and Ct values were obtained using Bio-Rad CFX Manager default threshold settings. Relative transcript expression levels were calculated by the method of Pfaffl (2001). Gene expression was normalized using as reference genes the three stably-expressed Wolbachia genes WD1043, WD1063, and WD1071, which were selected because they exhibit low fold-change and low coefficient of variation across the ISO1 life cycle time course (de Jonge et al. 2007). Expression values were calculated relative to embryonic expression levels.
Relative gene expression values were analyzed using R software v3.1.1 (R Development Core Team 2012) by fitting a linear mixed-effect model to the data of each gene using the lmer package (v2.0-20), comparing the effect of stage (embryo, adult male, and adult female) with a Tukey’s all-pair comparison using the glht package (v1.3-9). The data of the two genotypes were analyzed separately and together. For the linear mixed-model, the stage and genotype (in the joint analysis) were considered fixed effects, while independent collection was considered a random effect. No correction was applied to p-values, and thus α-levels for significance were set at 0.001 to account for multiple testing.
Functional and comparative annotation of Wolbachia genes
We generated functional annotations for Wolbachia genes using three sources: (i) by querying wMel open reading frames against the Genbank nucleotide (nt) database (April 2012, 15,938,872 sequences; 40,783,330,152 letters) using TBLASTN v2.2.25+ (Altschul et al. 1997) with default options; (ii) by querying wMel open reading frames against the Pfam-A.hmm database (v26.0) (Finn et al. 2014) using hmmscan v3.0 with default options (http://hmmer.org); and (iii) by using the original functional annotations generated by TIGR.
We identified homologs of wMel genes by conducting an all-vs.-all search of genes from the following complete Wolbachia genomes using BLASTP 2.2.27+ with default options: wRi (supergroup A strain from D. simulans, NC_012416), wPip-Pel (supergroup B strain from Culex quinquefasciatus, NC_010981), and wBm (supergroup D strain from Brugia malayi, NC_006833). The best hit to a gene in genome A was defined as the gene in genome B that had the highest bit score. Homology groups were defined such that a member had to have a reciprocal best hit to at least one other member of the group (single linkage), which permits paralogs to be included in a group.
Data availability
DNA-seq fastq reads for BDSC ISO1 were submitted to European Nucleotide Archive as experiment ERX645969. Table S1 contains a summary of the number of mapped RNA-seq reads for D. melanogaster and Wolbachia, number of expressed genes (defined as genes with nonzero TPM or genes with ≥2 mapped reads per gene), mean TPM for the sample (same for all samples, inverse of gene number times one million), and standard deviation of TPM for the sample for each total RNA sample in SRP001696. Table S2 contains gene IDs, coordinates, number of reads (from read 2 of paired-end data) mapping to the sense strand in each sample (_r1 = replicate 1, _r2 = replicate 2), estimated TPM for each sample, number of runs found in cluster 2, cluster assignment, adjusted p-value in life cycle GLM, log2 fold-change in life cycle GLM vs. embryo 0-2 hr, maximum fold change between any two stages in life cycle GLM, adjusted p-values and log2 fold change for pairwise exact tests between male and female samples at 1, 5, and 30 d, gene name, annotated gene product, effective number of codons, GC content, and number of homologs in wMel, wRi, wPip-Pel, and wBm genomes. Table S3 contains RT-qPCR GLM analysis results for ISO1 and DrosDel w1118. The p-values reported in Table S3 are not adjusted for multiple testing, and thus α-levels for significance were set at 0.001. Table S4 contains sequences of PCR primers used for RT-qPCR experiments.
Results and Discussion
The D. melanogaster ISO1 reference strain is infected with Wolbachia
As a control for another project, we obtained the ISO1 reference strain (Brizuela et al. 1994) used for the D. melanogaster genome project from the Bloomington Drosophila Stock Center (BDSC) and sequenced its genome. We discovered that the BDSC ISO1 sample contained a large number of Wolbachia sequences (4.5 million reads, 2.5% of total) when mapped against a “holo-genome” comprised of the D. melanogaster plus W. pipientis wMel reference genomes (Adams et al. 2000; Wu et al. 2004). The observation of Wolbachia sequences in the ISO1 stock was unexpected, since at no point since its original sequencing by the Berkeley Drosophila Genome Project (BDGP) and Celera Genomics in 2000 had Wolbachia sequences been reported in this strain (Adams et al. 2000; Celniker et al. 2002; Hoskins et al. 2015). In fact, direct searches of assembled or unassembled ISO1 genomic sequences from the BDGP failed to detect any evidence of Wolbachia (Wu et al. 2004; Salzberg et al. 2005). By investigating the provenance and conducting PCR-based assays of Wolbachia infection status of a panel of ISO1 substrains (see details in File S1), we confirmed that the BDSC ISO1 substrain is indeed infected with Wolbachia and established that loss of the Wolbachia infection occurred on the lineage leading to the BDGP ISO1 substrain.
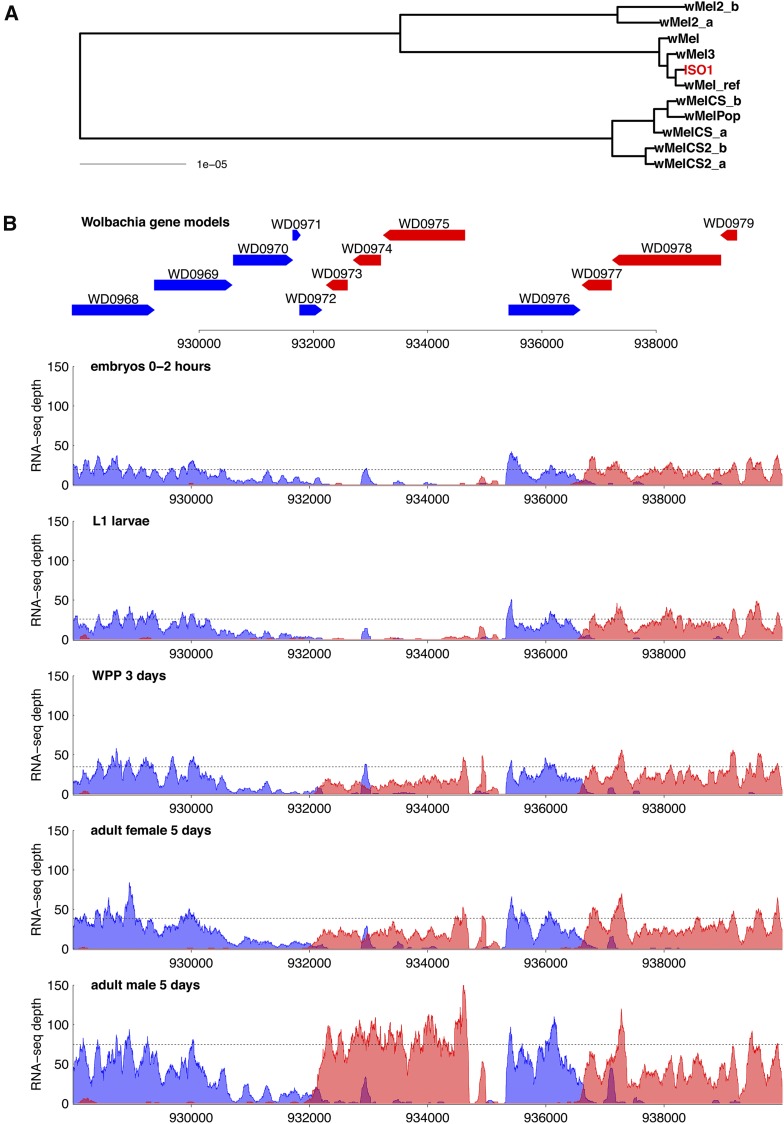
We next addressed which of the major variants of Wolbachia that are known to exist in D. melanogaster infects the ISO1 reference strain (Richardson et al. 2012; Chrostek et al. 2013; Woolfit et al. 2013). To do this, we assembled a consensus sequence from BDSC ISO1 reads that mapped to the wMel reference, then generated a whole-genome phylogeny jointly with the wMel reference genome (Wu et al. 2004) and genomes from known Wolbachia genotypes (Chrostek et al. 2013). This analysis showed that the Wolbachia infection in the BDSC ISO1 substrain is from a wMel-like genotype that is very closely related to both the wMel reference genome sequence (Wu et al. 2004) and the wMel-type strain recently reported by Chrostek et al. (2013) (Figure 1A). The very high sequence similarity between the wMel genotype in ISO1 and the wMel reference genome allows functional genomic data collected in ISO1 to be easily and accurately mapped to the reference genome sequence, and implies that wMel reference genome annotations closely reflect the content of the ISO1 Wolbachia genome.
Figure 1.
Phylogeny and expression landscape of Wolbachia in D. melanogaster. (A) Phylogenetic tree of Wolbachia strains based on whole genome sequences from this study (ISO1, red), Wu et al. (2004) (wMel_ref), and Chrostek et al. (2013) (all others). The scale bar for branch lengths is units of substitutions per site. The Wolbachia variant in ISO1 is very closely related to the wMel reference genome and to the wMel variant from Chrostek et al. (2013). (B) Gene models and RNA-seq coverage plots for a 12-gene window of the Wolbachia genome showing gene expression levels in representative stages of the D. melanogaster modENCODE RNA-seq life cycle time course. Gene models (pointed rectangles) and RNA-seq coverage (strand-specific wiggle plots of number of reads mapped to each base pair) are shown on the forward and reverse strands in blue and red, respectively. RNA-seq plots are shown on the same absolute y-axis scale. To provide an internal normalization factor for comparison across samples, mean coverage of the stably-expressed Wsp/WD1063 gene (not shown in this interval) divided by 20 is depicted by the dashed line in each panel. This example shows a set of three consecutive genes (WD0973, WD0974 and WD0975) that are cotranscribed as a single operon and specifically up-regulated in males in comparison to neighboring genes, as well as an unannotated noncoding RNA transcript that is expressed antisense to the 3′-end of WD0974.
The modENCODE developmental time course reveals a small subset of Wolbachia genes with dynamic expression across the D. melanogaster life cycle
Establishing that the BDSC ISO1 substrain is infected with Wolbachia is important since this strain is widely used in Drosophila genomics, including being one of the strains used by modENCODE to profile the transcriptome of D. melanogaster (Graveley et al. 2011; Brown et al. 2014; Duff et al. 2015). In particular, modENCODE-generated total RNA-seq libraries from BDSC ISO1 that span 30 time points across the D. melanogaster life cycle including multiple stages from embryos, larvae, pupae, and both adult sexes, with two biological replicates being available for 24 of the 30 time points (Graveley et al. 2011; Brown et al. 2014; Duff et al. 2015). We tested whether the Wolbachia infection in ISO1 could be detected in modENCODE total RNA-seq libraries by mapping reads to the combined D. melanogaster plus W. pipientis holo-genome reference. We found that the modENCODE total RNA-seq libraries do contain large numbers of Wolbachia sequences, with a median of 1.7 million reads per sample (range: 0.1–7 million) mapping to the Wolbachia genome, corresponding to a median of 1.6% (range: 0.3%–8.5%) of the total number of RNA-seq reads mapped in each sample (Table S1). As shown in Figure 1B, coverage and strand-specificity of the modENCODE RNA-seq dataset is high enough to show clear correspondence with the boundaries of most annotated Wolbachia gene models, given their presumed operonic structure and lack of annotated untranslated regions. Analysis of modENCODE RNA-seq libraries showed that the majority of Wolbachia genes were expressed in each sample, and that expression levels of genes were highly correlated among biological replicates (Figure S1 and File S1). These results further confirm that the BDSC ISO1 substrain is indeed infected with Wolbachia, and allow the modENCODE RNA-seq developmental time course to be analyzed in the context of the Wolbachia-Drosophila symbiosis.
We exploited our observation that modENCODE time course contains an essentially-complete Wolbachia transcriptome to study how Wolbachia expression varies across the D. melanogaster life cycle. Globally, we saw high correlations in expression levels across all stages (Figure S1 and File S1), with two weakly-differentiated, partially-overlapping clusters spanning embryonic to white prepupal (WPP) stages, and late larval to adult stages, respectively. In addition to the larger embryonic/pupal and pupal/adult clusters, stage-specific clusters could be observed for embryonic 10–12 hr, larval L1, larval L2, and larval L3 samples. Genome-wide expression changes at these particular stages suggest a potential link to pulses of ecdysone, a steroid hormone that regulates many aspects of arthropod development, which has been implicated in mediating the phenotypic effects of Wolbachia on its hosts (Negri 2012).
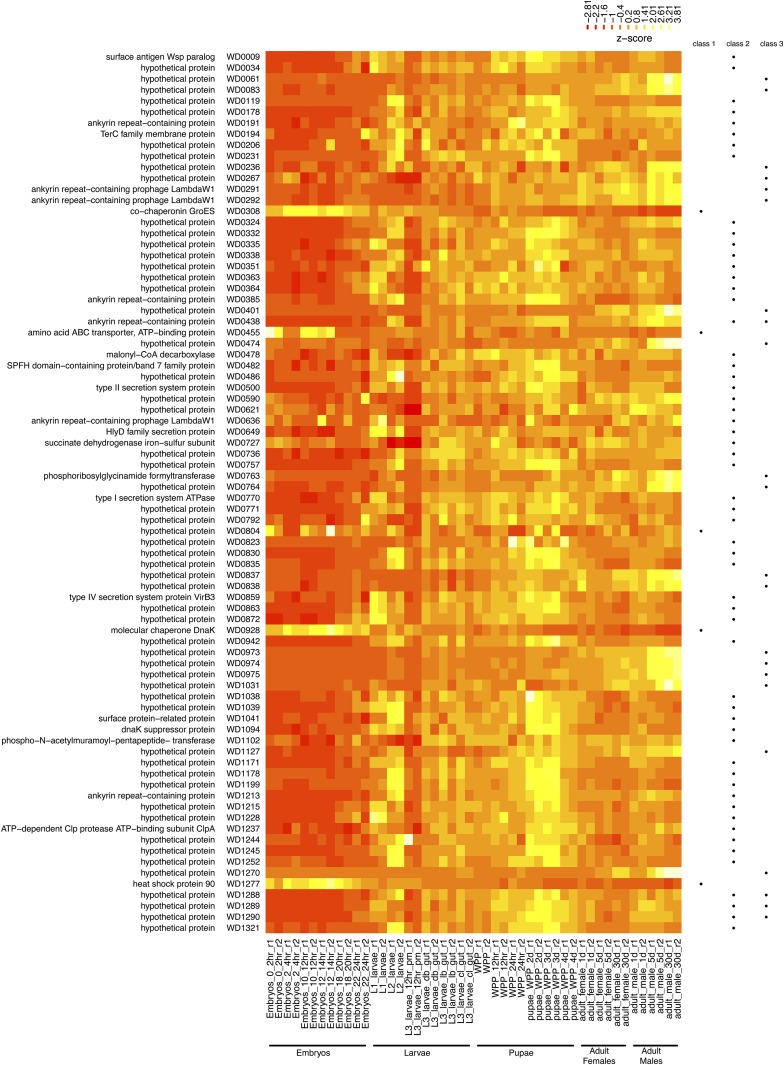
To identify specific genes whose expression levels vary reproducibly across life cycle stages, we performed differential expression analysis across all 24 stages that had biological replicates using an ANOVA-like GLM approach (McCarthy et al. 2012). We chose to perform an omnibus test of changes in expression across life cycle stages simultaneously because of the large number of life cycle stages and their complex developmental dependencies. This analysis revealed a small subset of Wolbachia genes (80/1195, 6.7%) that were differentially expressed in one or more life cycle stage at an adjusted p-value of less than 0.05 (Figure 2 and Table S2). All 80 genes had a greater than twofold change between at least one pair of stages. The vast majority of Wolbachia genes identified as differentially expressed across the D. melanogaster life cycle in the modENCODE time course showed a common pattern of being expressed at lower relative levels in embryos (75/80, 93.8%), with higher expression in either larval, pupal, and/or adult stages, and a transient decrease in expression at larval L3 (12 hr). However, five genes show the opposite pattern of having higher relative expression in embryos with down-regulation later in the life cycle (GroES/WD0308, ABC transporter/WD0455, WD0804, DnaK/WD0928, and Hsp90/WD1277). The dynamics of up-regulated and down-regulated genes show nearly complementary transitions at the end of embryogenesis, suggesting a response to common signals or possible cross-talk between these gene sets. We note that both up- and down-regulated genes exhibit a wide range of absolute expression levels, and many show quantitative shifts rather than dramatic qualitative changes in expression level.
Figure 2.
A small subset of Wolbachia genes show differential expression across the D. melanogaster life cycle. Row-normalized expression levels are visualized as a heatmap where each row represents a gene (ordered top-to-bottom by its position in the genome) and each cell represents the relative expression level for a particular sample in terms of Z-scores [observed transcripts per million (TPM) minus row mean TPM, divided by the standard deviation of TPMs for that row]. Values higher than row means are represented by yellow, and values lower than row means are represented by red. Gene names and identifiers are shown on the left. Membership in dynamically-expressed gene classes is shown by dots on the right. Class 1 includes genes that show down-regulation after embryogenesis. Class 2 includes genes that show up-regulation after embryogenesis, with peaks of expression in larval and pupal stages. Class 3 includes genes that show up-regulation after embryogenesis, with peaks of expression in adults. Classification of gene sets is not mutually exclusive. Stages that lack biological replicates in the modENCODE total RNA-seq time course were not used in this analysis and are not shown here.
To support conclusions about the global pattern of Wolbachia gene expression dynamics across the D. melanogaster life cycle based on differential expression analysis, we performed RT-qPCR for a sample of 10 genes in ISO1 and a second D. melanogaster strain (DrosDel w1118) carrying a wMel infection (Figure S2 and File S1). RT-qPCR results validated the main Wolbachia gene expression dynamics inferred from whole-organism RNA-seq in D. melanogaster, and indicated that gene expression information from the ISO1 RNA-seq time course can likely be extrapolated to other strains carrying wMel-like Wolbachia infections. We also performed probabilistic clustering on the entire modENCODE RNA-seq time course (including stages without replicates) (Si et al. 2014) (Figure S3 and File S1). This analysis identified two main clusters that could be matched across independent clustering runs (Figure S3A and Table S2). The first cluster contained the majority of Wolbachia genes (1033, 86.4%) and showed a pattern of relatively stable expression levels across the life cycle (Figure S3B). The second cluster contained the remaining 162 genes (13.6%), which generally showed up-regulation after embryogenesis and included the vast majority of genes identified as differentially expressed in the life cycle GLM (74/80, 92.5%). Overall, clustering analysis supported the main conclusions of the differential expression analysis that only a small proportion of Wolbachia genes show robust differences in expression across the modENCODE life cycle time course at the level of the whole organism, and that the majority of dynamically-expressed Wolbachia genes show up-regulation after embryogenesis. However, the greater number of genes identified as being dynamically expressed by clustering relative to the life cycle GLM suggests that our differential expression analysis may have detected only a conservative subset of Wolbachia with the strongest expression differences across D. melanogaster development.
Dynamically-expressed Wolbachia genes are predicted to be involved in stress response and host-microbe interactions
The 80 Wolbachia genes that exhibited dynamic expression across the modENCODE D. melanogaster life cycle time course fall into three broad classes (Figure 2). The first is a small class of five genes that show high relative expression in embryos with down-regulation later in the life cycle. Three of these genes are involved in chaperone function (GroES/WD0308, DnaK/WD0928, and Hsp90/WD1277). The chaperone GroEL/WD0307, which putatively forms a complex with GroES/WD0308, is cotranscribed with GroES/WD0308 and shows similar down-regulation at later stages of the life cycle, but does not pass the significance threshold in the life cycle GLM (p = 0.15). Both GroES/WD0308 and GroEL/WD0307 were in the top 15 most abundant transcripts based on average TPM across all stages, confirming that chaperones are among the most highly expressed genes in Wolbachia (Bennuru et al. 2011; Darby et al. 2012, 2014). High basal expression of GroEL or other chaperone proteins has been suggested to be a compensatory mechanism for the accumulation of slightly deleterious nonsynonymous mutations in endosymbionts that arise because of their small population size and lack of recombination (Moran 1996; Fares et al. 2002). The differential expression of Wolbachia chaperones during the D. melanogaster life cycle that we have observed may result from different exposure to external sources of stress or different requirements for protein folding/stability between eggs and larvae vs. pupae and adults.
The second class, comprising the majority of up-regulated genes detected (57/80), shows increases in relative expression starting with the larval L1 or L2 stages carrying on into adulthood, with decreases at the larval L3 (12 hr) stage and increases at the white prepupal 2 and 3 d stages. Genes in this class are mostly unannotated, but include eight genes that code for proteins with membrane or secretion system function (WspB/WD0009, TerC/WD0194, SPFH domain/WD0482, type II secretion/WD0500, HlyD/WD0649, type I secretion/WD0770, VirB3/WD0859, Rhoptry surface protein related/WD1041) and four ANK-containing genes (WD0191, WD0385, WD0438, WD1213). ANK-containing genes from several bacterial species have been shown to be type IV secretion system effector molecules that have diverse effects on eukaryotic cells (Caturegli et al. 2000; Sisko et al. 2006; Lin Et al., 2007; Pan Et al., 2008; O’Brien et al. 2015). Thus, secretion of ANK-containing genes into the host cell may be enriched during early larval and mid-to-late pupal stages of D. melanogaster development. Up-regulation of components for secretion systems (type III) has been observed in pupal stages of other arthropod endosymbionts (Dale et al. 2002), suggesting that metamorphosis may be a general period that is enriched for up-regulation of secreted symbiont effector proteins involved in host interaction. Wolbachia genes up-regulated during pupal stages could play roles in bacterial proliferation or tissue-specific migration, since Wolbachia in D. melanogaster have previously been shown to increase in numbers during pupal stages of testis development (Clark et al. 2002). The presence of a homolog for the Escherichia coli transcriptional regulator DksA/WD1094 in this class also provides a potential mechanism to understand the common differential regulation of these genes (Paul et al. 2005; Costanzo et al. 2008).
A third class of 22 Wolbachia genes show up-regulation primarily in D. melanogaster adults, with higher expression in adult males relative to adult females at the same age (see more below). Most of the genes in this class also have no known function. However, three are ANK-containing genes (WD0291, WD0292, WD0438). Our observation of sex-biased expression of ANK-containing genes based on global gene expression profiles of Wolbachia in D. melanogaster extends results from targeted RT-PCR analysis showing sex-biased expression of ANK-containing genes in Wolbachia strains from other insects (Sinkins et al. 2005; Duron et al. 2007; Klasson et al. 2009; Papafotiou et al. 2011; Wang et al. 2014). Finally, we note that our qualitative classification of up-regulated genes in classes 2 and 3 is not mutually exclusive, and the existence of four genes (WD0438, WD1288, WD1289, and WD1290) with sex-biased expression that also show differential expression at larval or pupal stages suggests possible shared regulation of these classes.
Wolbachia genes with sex-biased expression show age-dependent effects
Wolbachia is known to cause a variety of sex-specific phenotypes in its hosts (Werren et al. 2008), including a form of embryonic mortality arising from matings between infected males and uninfected females known as cytoplasmic incompatibility (CI). The wMel Wolbachia variant from D. melanogaster induces CI in the laboratory. However, this effect is partial and transient (Hoffmann et al. 1994; Yamada et al. 2007), and absent in field conditions (Hoffmann et al. 1998). To identify Wolbachia genes with sex-biased expression that might play a role in CI, we performed a more in-depth analysis of Wolbachia expression between males and females at matched ages. For this analysis, we used an exact testing framework (Robinson and Smyth 2008) because the GLM-based approach used for the complete life cycle is not the optimal method to use in a pairwise context. We identified a total of 41 genes that exhibited greater than 1.5-fold difference at an adjusted p-value cutoff of 0.01 in pairwise tests between male and female samples at either 1 d, 5 d, or 30 d post eclosion, respectively (Figure 3 and Table S2). Most sex-biased genes in this analysis were identified in one or both of the up-regulated classes in the life cycle GLM above (28/41, 68.3%), indicating that these complementary approaches identify a similar set of Wolbachia genes with detectable sex-biased expression in the modENCODE data set. Likewise, sex-biased genes comprise over one-third of differentially-expressed genes identified in the life cycle GLM (28/80, 35%), suggesting that sex-biased expression is a dominant component of the major differences in Wolbachia gene expression that can be observed across the D. melanogaster life cycle. Neither the GLM nor pairwise analysis revealed sex-biased expression in D. melanogaster for homologs of Wolbachia genes from Culex pipiens (WD0631, WD0632; WD0254, WD0255, WD0508, WD0622, WD0623, WD0626), which have recently been suggested to play a role in CI (Beckmann and Fallon 2013; Pinto et al. 2013).
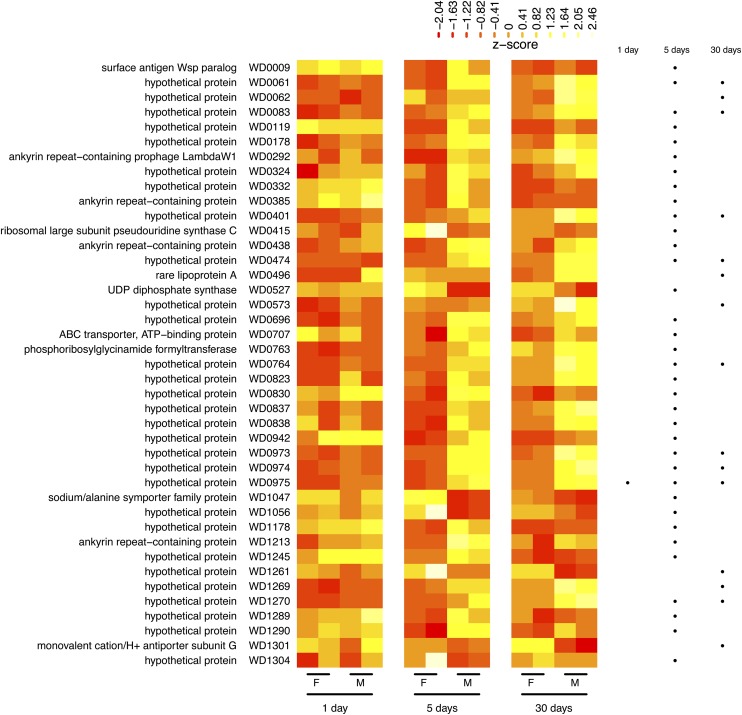
Figure 3.
Wolbachia genes show age-dependent sex-biased expression. Row-normalized expression levels are visualized as a heatmap where each row represents a gene (ordered top-to-bottom by its position in the genome), and each cell represents the relative expression level for a particular sample in terms of Z-scores [observed transcripts per million (TPM) minus row mean TPM, divided by the standard deviation of TPMs for that row]. For each stage, two biological replicates are shown for each female (F) and male (M) sample as distinct columns. Values higher than row means are represented by yellow, and values lower than row means are represented by red. Gene names and identifiers are shown on the left. Dots on the right indicate if a gene is differentially expressed between males and females at 1, 5, or 30 d post eclosion, respectively. All 41 genes identified as differentially expressed in any of the three pairwise comparisons between males and females in ISO1 are shown here.
The majority of sex-biased genes in the pairwise male-vs.-female analyses showed higher expression in males relative to females at matched stages, with only seven genes (rluC/WD0415, uppS/WD0527, sodium/alanine symporter/WD1047, WD1056, WD1261, cation antiporter subunit G/WD1301, WD1304) showing relatively higher expression in females at one or more time points. Many Wolbachia genes with male-biased expression are found in operons (Figure S4 and File S1). Additionally, most genes with sex-biased expression were identified at 5 d post eclosion (35/41), many of which maintained sex-biased expression until 30 d post eclosion. At 5 d post eclosion, whole-organism RNA-seq correctly predicted the presence (3/3) or lack (13/14) of sex-biased expression differences for 16/17 ANK-containing genes in a wMel strain previously classified by RT-qPCR to have over a 1.5-fold difference in expression level between testes and ovaries of 2-d-old flies (Papafotiou et al. 2011) (the only exception being that WD0292 shows sex-biased expression in the RNA-seq data at 5 d that is not observed in the RT-qPCR at 2 d). The general lack of sex-biased expression at 1 d post eclosion inferred from RNA-seq is also supported by RT-qPCR results (Figure S2): the five up-regulated genes we tested are all sex-biased at 5 d but not 1 d post eclosion in the RNA-seq data (Figure 3), and none of these genes show sex-biased expression at 1 d post eclosion in our RT-qPCR data.
Our finding that Wolbachia genes with sex-biased expression are typically up-regulated at 5 d post eclosion is puzzling considering previous work showing a decline in the strength of CI in D. melanogaster males at 1 vs. 5 d post eclosion (Reynolds and Hoffmann 2002; Reynolds et al. 2003; Yamada et al. 2007). Given that the CI phenotype can vary in D. melanogaster (Hoffmann et al. 1998), it is possible that CI was not expressed in the ISO1 samples used for the modENCODE time course and, thus, Wolbachia genes that are up-regulated as males age have nothing to do with CI. If so, these results may imply that Wolbachia responds to or affects other sexually dimorphic host phenotypes that vary with age. If these genes are in fact involved in CI, however, the observed pattern of sex-biased genes being up-regulated in older males would be compatible with these Wolbachia genes playing a role in attenuating the modification of D. melanogaster sperm that leads to embryonic lethality in incompatible crosses (Poinsot et al. 2003). Alternatively, if the host is responsible for reducing the effects of Wolbachia on the sperm of older males, up-regulation of Wolbachia genes in older males could represent an attempt by Wolbachia to compensate against host attenuation and hence indicate these genes play a role in promoting CI.
wMel genes with dynamic expression in D. melanogaster are conserved in other Wolbachia strains
To understand if candidate genes identified on the basis of differential expression in D. melanogaster might interact more broadly with other hosts, we asked whether wMel genes that show stage- and sex-specific expression are conserved in other divergent Wolbachia strains. For this analysis, we used complete Wolbachia genome sequences from wRi (an arthropod supergroup A strain from D. simulans), wPip-Pel (an arthropod supergroup B strain from Culex quinquefasciatus) and wBm (a nematode supergroup D strain from Brugia malayi) (Klasson et al. 2008, 2009; Foster et al. 2005). We identified and clustered homologs in all genomes analyzed, and reconstructed homology groups that included wMel homologs for 86 of 93 genes that show either stage- or sex-specific expression (seven dynamically-expressed wMel genes were too small to pass BLAST filtering cutoffs). Only three of the 86 dynamically-expressed genes in homology groups (3.5%) were restricted to the wMel genome, whereas 30 genes (34.9%) had homologs in Wolbachia genomes from other arthropods, and a further 53 genes (61.6%) also had homologs in Wolbachia genomes from nematodes. The phylogenetic distribution of dynamically-expressed genes does not differ from genome-wide expectations (χ2 = 4.82, p = 0.09, d.f. = 2). These results indicate that the majority of genes identified as dynamically expressed in D. melanogaster are core components of the Wolbachia gene repertoire and are not unusual in their degree of conservation. Nevertheless, many dynamically-expressed candidate genes are arthropod-specific but only a few are D. melanogaster-specific, as might be expected for candidate host-interaction genes in a facultative endosymbiont that can switch arthropod hosts by horizontal transfer. Arthropod-specific dynamically-expressed genes include several ANK-containing genes (WD0191, WD0636, WD1213) and membrane/secretion system genes (ABC transporter/WD0455, SPFH domain/WD0482, type II secretion/WD0500, sodium/alanine symporter/WD1047, ClpA/WD1237), emphasizing the importance of intercellular communication in explaining how Wolbachia forms facultative symbioses with its arthropod hosts.
Expression of Wolbachia genes previously implicated in host-microbe interaction
In addition to identifying new candidates for mediating host-microbe interaction on the basis of their stage- or sex-specific differential expression, we also investigated expression levels of Wolbachia genes previously suggested to be candidates for mediating interaction with D. melanogaster. The most widely hypothesized set of candidates for host-microbe interaction are the 23 ANK-containing genes that are possible type IV secretion system effectors in Wolbachia (Wu et al. 2004; Iturbe-Ormaetxe et al. 2005; Papafotiou et al. 2011; Siozios et al. 2013), which the modENCODE data show are expressed at widely different levels in D. melanogaster (Figure 4A). The five most weakly-expressed ANK-containing genes (WD0285, WD0286, WD0514, WD0636, WD0637) are found in the Octomom and prophage regions, and are the same five genes that Papafotiou et al. (2011) found were too weakly expressed to obtain reliable RT-qPCR data in adult gonads. Thirteen ANK-containing genes are highly expressed (WD0191, WD0291, WD0292, WD0294, WD0385, WD0438, WD0441, WD0498, WD0550, WD0633, WD0754, WD0766, WD1213), which include the majority of differentially expressed ANK-containing genes identified in this study or by Papafotiou et al. (2011) (WD0191, WD0291, WD0292, WD0294, WD0385, WD0438, WD0550, WD0636, WD1213). The nine genes that make a complete type IV secretion system in wMel are all highly expressed in all D. melanogaster life cycle stages, including the virB8 paralog (WD0817), which is not a part of the two genomic clusters that contain the remaining eight type IV secretion system genes. These results together support a model where a functionally competent type IV Wolbachia secretion system is expressed throughout the D. melanogaster life cycle, with both constitutive and regulated secretion of subsets of ANK-containing effectors.
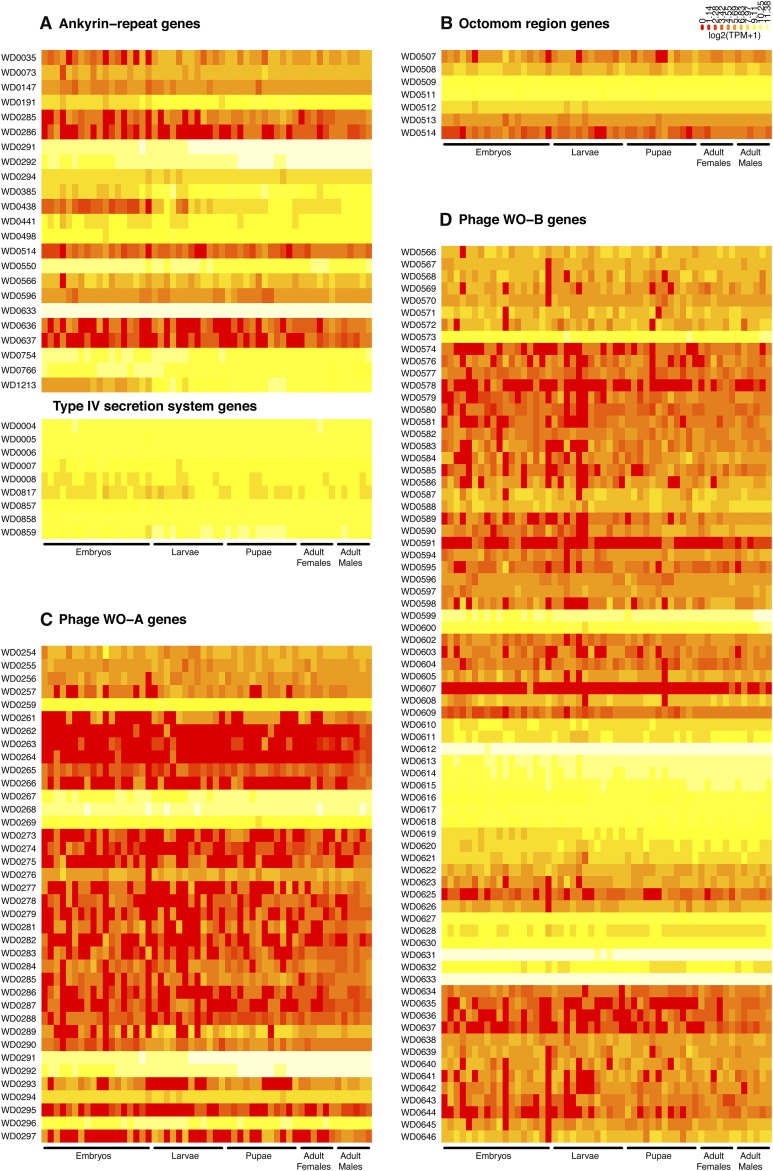
Figure 4.
Expression profiles of Wolbachia genes previously implicated in host-microbe interactions. (A) ANK-containing and type IV secretion system genes. (B) Octomom genes. (C) Prophage WO-A genes. (D) Prophage WO-B genes. The 23 ANK-containing genes in panel (A) are distributed throughout the Wolbachia genome. The nine type IV secretion system genes are found in three different genomic intervals. The Octomom, prophage WO-A, and prophage WO-B regions are each from single intervals in the Wolbachia genome. The Octomom region only contains seven genes, since WD0510 is not included in the Ensembl annotation for Wolbachia wMel. Phage coordinates are from Metcalf et al. (2014). Expression levels are not row-normalized and are visualized as a heatmap where each row represents a gene (ordered top-to-bottom by its position in the genome) and each cell represents expression in units of observed transcripts per million (TPM). A pseudocount of one was added to each gene’s TPM before transforming to log2 scale. Values with higher levels of expression are represented by yellow, and values with lower levels of expression are represented by red. All panels are on the same heatmap color scale. Gene names and identifiers are shown on the left. All stages including those that lack biological replicates in the modENCODE time course are shown here.
The Octomom region is part of the accessory genome of Wolbachia (Iturbe-Ormaetxe et al. 2005; Chrostek et al. 2013) and contains eight genes whose copy number controls Wolbachia growth and pathogenesis (Chrostek and Teixeira 2015). In the wMel variant, where Octomom is present in one copy, all genes in this region (WD0507–WD0514) are expressed at relatively constant levels across the life cycle (Figure 4B). None of these genes show a significant change in gene expression during host development in the life cycle GLM. However, different Octomom genes do vary considerably in their expression levels relative to each other, with the most highly expressed genes being found in the middle of this interval (WD0509–WD0512). Given that two Octomom genes with possible regulatory (the helix-turn-helix-containing gene WD0508) or effector (the ANK-containing gene WD0514) function are relatively weakly expressed in the nonpathogenic wMel variant, it is possible that overexpression of one or both of these genes may be responsible for the pathogenic phenotype when Octomom is amplified in wMelPop (Chrostek and Teixeira 2015).
Unlike obligate endosymbionts with streamlined genomes, prophages are often present in Wolbachia from arthropod hosts and have been suggested to directly or indirectly influence Wolbachia-host interactions (Kent and Bordenstein 2010; Metcalf et al. 2014). Two major prophage regions are present in the wMel genome, called WO-A and WO-B, both of which have undergone degeneration and rearrangement since insertion (Wu et al. 2004; Kent et al. 2011). There is no clear evidence that the WO-A and WO-B prophages from wMel can enter a lytic phase as they can in other arthropods (Masui et al. 2001; Fujii et al. 2004; Bordenstein et al. 2006; Sanogo and Dobson 2006). However, phage-like particles have been reported in extracts of D. melanogaster strains infected with wMel (Gavotte et al. 2004). Consistent with previous results from wMelPop-CLA (Darby et al. 2014) and prophages in Salmonella enterica (Perkins et al. 2009), expression levels of most genes in the WO-A and WO-B prophage regions are typically very low across the entire D. melanogaster life cycle (Figure 4, C and D), and define the largest segments of the wMel genome with consecutive lowly-expressed genes. The most conspicuous exception to this pattern is the 21 gene interval in WO-B (WD0611–WD0634) that contains genes laterally-transferred between Wolbachia and the Rickettsia endosymbiont of the tick Ixodes scapularis (WD0612–WD0621) (Ishmael et al. 2009; Gillespie et al. 2012), a region not typically present in WO prophage from other Wolbachia strains (Kent et al. 2011). In addition, two very abundant currently-unannotated antisense ncRNA transcripts can be detected overlapping the major capsid genes of both WO-A (WD0274) and WO-B (WD0604) (Figure S5), which may play a role in the regulation of prophage genes.
Most prophage-encoded structural genes are expressed at low levels, with only genes in the tail (WD0567–WD0574) and base-plate (WD0638–WD0644) regions of WO-B being expressed at appreciable levels. Likewise, most nonstructural prophage-encoded genes previously suggested to be candidates for host interaction (VrlC.2/WD0579, VrlC.1/WD0580, Patatin/WD0565, DNA methylases WD0263 and WD0594) (Kent and Bordenstein 2010) are expressed at low levels. Intriguingly, each prophage region contains a highly-expressed operon (WD0267–WD0269 in WO-A and WD0599–WD0600 in WO-B) that encodes homologs of the E. coli RelE toxin (WD0269 and WD0600) (Figure 4, C and D). RelE is a stress-inducible cytotoxic translational repressor that is counteracted by the antitoxin RelB, a small protein, the gene of which is cotranscribed in the same operon as RelE (Christensen et al. 2001; Pedersen et al. 2003; Yamaguchi et al. 2011). The genes adjacent to the RelE homologs in WO-A and WO-B (WD0267, WD0268 and WD0599) are also cotranscribed and encode small peptides, and thus could be acting as antitoxins. In fact, WD0268–WD0269 and WD0599–WD0600 have been computationally predicted to be toxin-antitoxin pairs (Shao et al. 2011), and toxin-antitoxin pairs have been previously reported in other cryptic prophages (Van Melderen and Saavedra De Bast 2009). RelE-containing gene clusters are also found in similar positions (between the terminase and portal genes that form the phage head) in divergent prophages from D. simulans (WOriA) and Nasonia vitripinnis (WOVitA2) (Kent et al. 2011), further indicating that they may play some conserved functional role such as stabilizing the Wolbachia prophage genomic regions by preventing large-scale deletions (Van Melderen and Saavedra De Bast 2009). Low expression levels of phage structural genes together with highly-expressed putative toxin-antitoxin pairs suggests that prophages in wMel are maintained in the lysogenic state by self-preservation.
Conclusions and Future Directions
We have shown that the ISO1 reference strain used by the modENCODE project is infected with Wolbachia, and used this fortuitous observation to study the global expression dynamics of a facultative endosymbiont over the life cycle of the model insect species D. melanogaster. Our work represents the most comprehensive gene expression profiling of an endosymbiotic bacteria in its native host context to date. We have established that most Wolbachia genes are expressed in all D. melanogaster life cycle stages, but that major changes in expression levels of Wolbachia genes are rare when studied simultaneously across all D. melanogaster tissues. It is important to emphasize, however, that the modENCODE total RNA-seq libraries were made from whole animals, and thus any tissue-specific Wolbachia gene expression differences that may exist cannot be detected in these data, nor can sex-specific differences in nonadult stages. Nevertheless, we identify a set of 93 Wolbachia wMel genes that show robust stage- or sex-specific differential expression at the whole-fly level, many of which share common expression dynamics and therefore may be coregulated. These genes provide many new candidate genes for understanding, and possibly manipulating, the genetic basis of how Wolbachia interacts with arthropod hosts. Importantly, we also provide the first detailed insight into the developmental dynamics of Wolbachia gene expression in an insect host, which suggests that the larval and pupal stages [where Wolbachia have been detected cytologically (Clark et al. 2002, 2003, 2005)] merit further study to understand how Wolbachia manipulates host biology to maintain persistent infections and affect transmission.
Future studies can leverage our finding that the modENCODE total RNA-seq dataset contains a nearly-complete Wolbachia transcriptome to functionally annotate the transcriptional landscape of the Wolbachia genome. Currently, only protein-coding regions and a small number of ncRNAs are included in the wMel genome annotation (Wu et al. 2004), and recent work has identified a handful of additional Wolbachia ncRNAs (Mayoral et al. 2014; Woolfit et al. 2015). The strand-specific total RNA-seq data from modENCODE can now be used to generate high quality transcript models to annotate 5′- and 3′-untranslated regions of protein-coding genes, delimit operons, and identify new ncRNA genes in Wolbachia (see examples in Figure S4 and Figure S5). The possibility of a more comprehensive annotation of ncRNAs in Wolbachia is particularly exciting given recent work suggesting that ncRNAs provide an important layer of posttranscriptional regulation to modulate protein expression levels in the Buchnera endosymbiont of aphids (Hansen and Degnan 2014). Together with other recently published transcriptomic data (Darby et al. 2014; Mayoral et al. 2014; Woolfit et al. 2015), the necessary materials are now available to undertake a systematic reannotation of the Wolbachia wMel genome in order to support basic and applied research on this important model organism.
Supplementary Material
Acknowledgments
We thank Jim Kennison, Todd Laverty, and Sue Celniker for providing stocks; Jim Kennison, Gerry Rubin, Todd Laverty, and Roger Hoskins for information on the provenance of ISO1 substrains; Brent Graveley, Peter Cherbas, and Robert Eisman for information about how modENCODE RNA-seq samples were generated; Sarah Bordenstein and Seth Bordenstein for information about prophage coordinates; Raquel Linheiro for R programming guidance; Github for providing free private repositories that enabled this collaboration; and Douda Bensasson, Sarah Bordenstein, Seth Bordenstein, Tim Karr, and members of the Bergman Lab for comments on the manuscript. We are especially grateful to Roger Hoskins for his suggestion to look in the modENCODE RNA-seq data to confirm our observation that the BDSC substrain of ISO1 was infected with Wolbachia, which led to the results presented here. This work was supported by a National Environment Research Council Ph.D. studentship (F.G.), Fundação para a Ciência e a Tecnologia postdoctoral fellowship SFRH/BPD/73420/2010 (C.R.C), an award from the Stowers Institute for Medical Research (R.S.H.), National Science Foundation grant NSF-IOS-1456545 (I.G.L.N.), Human Frontier Science Program grant RGY0093/2012 (C.M.B.), and Biotechnology and Biological Sciences Research Council grant BB/L002817/1 (C.M.B.).
Footnotes
Supporting information is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.115.021931/-/DC1
Sequence data from this article have been deposited with ENA under accession no. ERX645969.
Communicating editor: J. M. Comeron
Literature Cited
- Adams M. D., Celniker S. E., Holt R. A., Evans C. A., Gocayne J. D., et al. , 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195. [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Madden T. L., Schffer A. A., Zhang J., Zhang Z., et al. , 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldridge G. D., Baldridge A. S., Witthuhn B. A., Higgins L., Markowski T. W., et al. , 2014. Proteomic profiling of a robust Wolbachia infection in an Aedes albopictus mosquito cell line. Mol. Microbiol. 94: 537–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann J. F., Fallon A. M., 2013. Detection of the Wolbachia protein WPIP0282 in mosquito spermathecae: implications for cytoplasmic incompatibility. Insect Biochem. Mol. Biol. 43: 867–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57: 289–300. [Google Scholar]
- Bennuru S., Meng Z., Ribeiro J. M. C., Semnani R. T., Ghedin E., et al. , 2011. Stage-specific proteomic expression patterns of the human filarial parasite Brugia malayi and its endosymbiont Wolbachia. Proc. Natl. Acad. Sci. USA 108: 9649–9654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordenstein S. R., Marshall M. L., Fry A. J., Kim U., Wernegreen J. J., 2006. The tripartite associations between bacteriophage, Wolbachia, and arthropods. PLoS Pathog. 2: e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brizuela B. J., Elfring L., Ballard J., Tamkun J. W., Kennison J. A., 1994. Genetic analysis of the brahma gene of Drosophila melanogaster and polytene chromosome subdivisions 72ab. Genetics 137: 803–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. B., Boley N., Eisman R., May G. E., Stoiber M. H., et al. , 2014. Diversity and dynamics of the Drosophila transcriptome. Nature 512: 393–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caturegli P., Asanovich K. M., Walls J. J., Bakken J. S., Madigan J. E., et al. , 2000. ankA: an Ehrlichia phagocytophila group gene encoding a cytoplasmic protein antigen with ankyrin repeats. Infect. Immun. 68: 5277–5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker S. E., Wheeler D. A., Kronmiller B., Carlson J. W., Halpern A., et al. , 2002. Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 3: 0079.1–0079.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen S. K., Mikkelsen M., Pedersen K., Gerdes K., 2001. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc. Natl. Acad. Sci. USA 98: 14328–14333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrostek E., Teixeira L., 2015. Mutualism breakdown by amplification of Wolbachia genes. PLoS Biol. 13: e1002065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrostek E., Marialva M. S. P., Esteves S. S., Weinert L. A., Martinez J., et al. , 2013. Wolbachia variants induce differential protection to viruses in Drosophila melanogaster: a phenotypic and phylogenomic analysis. PLoS Genet. 9: e1003896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M. E., Veneti Z., Bourtzis K., Karr T. L., 2002. The distribution and proliferation of the intracellular bacteria Wolbachia during spermatogenesis in Drosophila. Mech. Dev. 111: 3–15. [DOI] [PubMed] [Google Scholar]
- Clark M. E., Veneti Z., Bourtzis K., Karr T. L., 2003. Wolbachia distribution and cytoplasmic incompatibility during sperm development: the cyst as the basic cellular unit of CI expression. Mech. Dev. 120: 185–198. [DOI] [PubMed] [Google Scholar]
- Clark M. E., Anderson C. L., Cande J., Karr T. L., 2005. Widespread prevalence of Wolbachia in laboratory stocks and the implications for Drosophila research. Genetics 170: 1667–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo A., Nicoloff H., Barchinger S. E., Banta A. B., Gourse R. L., et al. , 2008. ppGpp and DksA likely regulate the activity of the extracytoplasmic stress factor E in Escherichia coli by both direct and indirect mechanisms. Mol. Microbiol. 67: 619–632. [DOI] [PubMed] [Google Scholar]
- Cunningham F., Amode M. R., Barrell D., Beal K., Billis K., et al. , 2015. Ensembl 2015. Nucleic Acids Res. 43: D662–D669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale C., Moran N. A., 2006. Molecular interactions between bacterial symbionts and their hosts. Cell 126: 453–465. [DOI] [PubMed] [Google Scholar]
- Dale C., Plague G. R., Wang B., Ochman H., Moran N. A., 2002. Type III secretion systems and the evolution of mutualistic endosymbiosis. Proc. Natl. Acad. Sci. USA 99: 12397–12402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby A. C., Armstrong S. D., Bah G. S., Kaur G., Hughes M. A., et al. , 2012. Analysis of gene expression from the Wolbachia genome of a filarial nematode supports both metabolic and defensive roles within the symbiosis. Genome Res. 22: 2467–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby A. C., Christina Gill A., Armstrong S. D., Hartley C. S., Xia D., et al. , 2014. Integrated transcriptomic and proteomic analysis of the global response of Wolbachia to doxycycline-induced stress. ISME J. 8: 925–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge H. J. M., Fehrmann R. S. N., de Bont E. S. J. M., Hofstra R. M. W., Gerbens F., et al. , 2007. Evidence based selection of housekeeping genes. PLoS One 2: e898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duff M. O., Olson S., Wei X., Garrett S. C., Osman A., et al. , 2015. Genome-wide identification of zero nucleotide recursive splicing in Drosophila. Nature 521: 376–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duron O., Boureux A., Echaubard P., Berthomieu A., Berticat C., et al. , 2007. Variability and expression of ankyrin domain genes in Wolbachia variants infecting the mosquito Culex pipiens. J. Bacteriol. 189: 4442–4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares M. A., Barrio E., Sabater-Muoz B., Moya A., 2002. The Evolution of the Heat-Shock Protein GroEL from Buchnera, the Primary Endosymbiont of Aphids, Is Governed by Positive Selection. Mol. Biol. Evol. 19: 1162–1170. [DOI] [PubMed] [Google Scholar]
- Finn R. D., Bateman A., Clements J., Coggill P., Eberhardt R. Y., et al. , 2014. Pfam: the protein families database. Nucleic Acids Res. 42: D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J., Ganatra M., Kamal I., Ware J., Makarova K., et al. , 2005. The Wolbachia genome of Brugia malayi: endosymbiont evolution within a human pathogenic nematode. PLoS Biol. 3: e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii Y., Kubo T., Ishikawa H., Sasaki T., 2004. Isolation and characterization of the bacteriophage WO from Wolbachia, an arthropod endosymbiont. Biochem. Biophys. Res. Commun. 317: 1183–1188. [DOI] [PubMed] [Google Scholar]
- Gavotte L., Vavre F., Henri H., Ravallec M., Stouthamer R., et al. , 2004. Diversity, distribution and specificity of WO phage infection in Wolbachia of four insect species. Insect Mol. Biol. 13: 147–153. [DOI] [PubMed] [Google Scholar]
- Gillespie J. J., Joardar V., Williams K. P., Driscoll T., Hostetler J. B., et al. , 2012. A Rickettsia genome overrun by mobile genetic elements provides insight into the acquisition of genes characteristic of an obligate intracellular lifestyle. J. Bacteriol. 194: 376–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley B. R., Brooks A. N., Carlson J. W., Duff M. O., Landolin J. M., et al. , 2011. The developmental transcriptome of Drosophila melanogaster. Nature 471: 473–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen A. K., Degnan P. H., 2014. Widespread expression of conserved small RNAs in small symbiont genomes. ISME J. 8: 2490–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A. A., Clancy D. J., Merton E., 1994. Cytoplasmic incompatibility in Australian populations of Drosophila melanogaster. Genetics 136: 993–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A. A., Hercus M., Dagher H., 1998. Population dynamics of the Wolbachia infection causing cytoplasmic incompatibility in Drosophila melanogaster. Genetics 148: 221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins R. A., Carlson J. W., Wan K. H., Park S., Mendez I., et al. , 2015. The Release 6 reference sequence of the Drosophila melanogaster genome. Genome Res. 25: 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishmael N., Dunning Hotopp J. C., Ioannidis P., Biber S., Sakamoto J., et al. , 2009. Extensive genomic diversity of closely related Wolbachia strains. Microbiology 155: 2211–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturbe-Ormaetxe I., Burke G. R., Riegler M., O’Neill S. L., 2005. Distribution, expression, and motif variability of ankyrin domain genes in Wolbachia pipientis. J. Bacteriol. 187: 5136–5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent B. N., Bordenstein S. R., 2010. Phage WO of Wolbachia: lambda of the endosymbiont world. Trends Microbiol. 18: 173–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent B. N., Funkhouser L. J., Setia S., Bordenstein S. R., 2011. Evolutionary genomics of a temperate bacteriophage in an obligate intracellular bacteria (Wolbachia). PLoS One 6: e24984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey P. J., Allen J. E., Christensen M., Davis P., Falin L. J., et al. , 2014. Ensembl Genomes 2013: scaling up access to genome-wide data. Nucleic Acids Res. 42: D546–D552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasson L., Walker T., Sebaihia M., Sanders M. J., Quail M. A., et al. , 2008. Genome evolution of Wolbachia strain wPip from the Culex pipiens group. Mol. Biol. Evol. 25: 1877–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasson L., Westberg J., Sapountzis P., Nslund K., Lutnaes Y., et al. , 2009. The mosaic genome structure of the Wolbachia wRi strain infecting Drosophila simulans. Proc. Natl. Acad. Sci. USA 106: 5725–5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C. N., 2011. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., 2013 Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv: 1303.3997. Available at: http://arxiv.org/abs/1303.3997. Accessed: 9 Nov 2015 [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M., den Dulk-Ras A., Hooykaas P. J. J., Rikihisa Y., 2007. Anaplasma phagocytophilum AnkA secreted by type IV secretion system is tyrosine phosphorylated by Abl-1 to facilitate infection. Cell. Microbiol. 9: 2644–2657. [DOI] [PubMed] [Google Scholar]
- Luck A. N., Evans C. C., Riggs M. D., Foster J. M., Moorhead A. R., et al. , 2014. Concurrent transcriptional profiling of Dirofilaria immitis and its Wolbachia endosymbiont throughout the nematode life cycle reveals coordinated gene expression. BMC Genomics 15: 1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui S., Kuroiwa H., Sasaki T., Inui M., Kuroiwa T., et al. , 2001. Bacteriophage WO and virus-like particles in Wolbachia, an endosymbiont of arthropods. Biochem. Biophys. Res. Commun. 283: 1099–1104. [DOI] [PubMed] [Google Scholar]
- Mayoral J. G., Hussain M., Joubert D. A., Iturbe-Ormaetxe I., O’Neill S. L., et al. , 2014. Wolbachia small noncoding RNAs and their role in cross-kingdom communications. Proc. Natl. Acad. Sci. USA 111: 18721–18726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy D. J., Chen Y., Smyth G. K., 2012. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40: 4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf J. A., Jo M., Bordenstein S. R., Jaenike J., Bordenstein S. R., 2014. Recent genome reduction of Wolbachia in Drosophila recens targets phage WO and narrows candidates for reproductive parasitism. PeerJ 2: e529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran N. A., 1996. Accelerated evolution and Muller’s rachet in endosymbiotic bacteria. Proc. Natl. Acad. Sci. USA 93: 2873–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran N. A., Degnan P. H., 2006. Functional genomics of Buchnera and the ecology of aphid hosts. Mol. Ecol. 15: 1251–1261. [DOI] [PubMed] [Google Scholar]
- Moran N. A., Dunbar H. E., Wilcox J. L., 2005. Regulation of transcription in a reduced bacterial genome: nutrient-provisioning genes of the obligate symbiont Buchnera aphidicola. J. Bacteriol. 187: 4229–4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran N. A., McCutcheon J. P., Nakabachi A., 2008. Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 42: 165–190. [DOI] [PubMed] [Google Scholar]
- Negri I., 2012. Wolbachia as an infectious extrinsic factor manipulating host signaling pathways. Wolbachia 2: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien K. M., Lindsay E. L., Starai V. J., 2015. The Legionella pneumophila Effector Protein, LegC7, Alters Yeast Endosomal Trafficking. PLoS One 10: e0116824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Lhrmann A., Satoh A., Laskowski-Arce M. A., Roy C. R., 2008. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science 320: 1651–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papafotiou G., Oehler S., Savakis C., Bourtzis K., 2011. Regulation of Wolbachia ankyrin domain encoding genes in Drosophila gonads. Res. Microbiol. 162: 764–772. [DOI] [PubMed] [Google Scholar]
- Paul B. J., Berkmen M. B., Gourse R. L., 2005. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc. Natl. Acad. Sci. USA 102: 7823–7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen K., Zavialov A. V., Pavlov M. Y., Elf J., Gerdes K., et al. , 2003. The Bacterial Toxin RelE Displays Codon-Specific Cleavage of mRNAs in the Ribosomal A Site. Cell 112: 131–140. [DOI] [PubMed] [Google Scholar]
- Perkins T. T., Kingsley R. A., Fookes M. C., Gardner P. P., James K. D., et al. , 2009. A strand-specific RNAseq analysis of the transcriptome of the typhoid Bacillus salmonella typhi. PLoS Genet. 5: e1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W., 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto S. B., Stainton K., Harris S., Kambris Z., Sutton E. R., et al. , 2013. Transcriptional regulation of Culex pipiens mosquitoes by Wolbachia influences cytoplasmic incompatibility. PLoS Pathog. 9: e1003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poinsot D., Charlat S., Merot H., 2003. On the mechanism of Wolbachia-induced cytoplasmic incompatibility: confronting the models with the facts. BioEssays 25: 259–265. [DOI] [PubMed] [Google Scholar]
- Quinlan A. R., Hall I. M., 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team , 2012. R: A Language and Environment for Statistical Computing, Vienna, Austria. [Google Scholar]
- Rao R. U., Huang Y., Abubucker S., Heinz M., Crosby S. D., et al. , 2012. Effects of doxycycline on gene expression in Wolbachia and Brugia malayi adult female worms in vivo. J. Biomed. Sci. 19: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond N., Calevro F., Viuelas J., Morin N., Rahb Y., et al. , 2006. Different levels of transcriptional regulation due to trophic constraints in the reduced genome of Buchnera aphidicola APS. Appl. Environ. Microbiol. 72: 7760–7766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds K. T., Hoffmann A. A., 2002. Male age, host effects and the weak expression or non-expression of cytoplasmic incompatibility in Drosophila strains infected by maternally transmitted Wolbachia. Genet. Res. 80: 79–87. [DOI] [PubMed] [Google Scholar]
- Reynolds K. T., Thomson L. J., Hoffmann A. A., 2003. The effects of host age, host nuclear background and temperature on phenotypic effects of the virulent Wolbachia strain popcorn in Drosophila melanogaster. Genetics 164: 1027–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson M. F., Weinert L. A., Welch J. J., Linheiro R. S., Magwire M. M., et al. , 2012. Population genomics of the Wolbachia endosymbiont in Drosophila melanogaster. PLoS Genet. 8: e1003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., Oshlack A., 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., Smyth G. K., 2007. Moderated statistical tests for assessing differences in tag abundance. Bioinformatics 23: 2881–2887. [DOI] [PubMed] [Google Scholar]
- Robinson M. D., Smyth G. K., 2008. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics 9: 321–332. [DOI] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K., 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder E., Blows F., Ashburner M., Bautista-Llacer R., Coulson D., et al. , 2004. The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics 167: 797–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg S. L., Hotopp J. C. D., Delcher A. L., Pop M., Smith D. R., et al. , 2005. Serendipitous discovery of Wolbachia genomes in multiple Drosophila species. Genome Biol. 6: R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanogo Y. O., Dobson S. L., 2006. WO bacteriophage transcription in Wolbachia-infected Culex pipiens. Insect Biochem. Mol. Biol. 36: 80–85. [DOI] [PubMed] [Google Scholar]
- Shao Y., Harrison E. M., Bi D., Tai C., He X., et al. , 2011. TADB: a web-based resource for Type 2 toxin-antitoxin loci in bacteria and archaea. Nucleic Acids Res. 39: D606–D611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si Y., Liu P., Li P., Brutnell T. P., 2014. Model-based clustering for RNA-seq data. Bioinformatics 30: 197–205. [DOI] [PubMed] [Google Scholar]
- Sinkins S. P., Walker T., Lynd A. R., Steven A. R., Makepeace B. L., et al. , 2005. Wolbachia variability and host effects on crossing type in Culex mosquitoes. Nature 436: 257–260. [DOI] [PubMed] [Google Scholar]
- Siozios S., Ioannidis P., Klasson L., Andersson S. G. E., Braig H. R., et al. , 2013. The diversity and evolution of Wolbachia ankyrin repeat domain genes. PLoS One 8: e55390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisko J. L., Spaeth K., Kumar Y., Valdivia R. H., 2006. Multifunctional analysis of Chlamydia-specific genes in a yeast expression system. Mol. Microbiol. 60: 51–66. [DOI] [PubMed] [Google Scholar]
- Slatko B. E., Luck A. N., Dobson S. L., Foster J. M., 2014. Wolbachia endosymbionts and human disease control. Mol. Biochem. Parasitol. 195: 88–95. [DOI] [PubMed] [Google Scholar]
- Stamatakis A., 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Melderen L., Saavedra De Bast M., 2009. Bacterial toxin-antitoxin systems: more than selfish entities? PLoS Genet. 5: e1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.-H., Niu L.-M., Ma G.-C., Xiao J.-H., Huang D.-W., 2014. Large proportion of genes in one cryptic WO prophage genome are actively and sex-specifically transcribed in a fig wasp species. BMC Genomics 15: 893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren J. H., Baldo L., Clark M. E., 2008. Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6: 741–751. [DOI] [PubMed] [Google Scholar]
- Wilcox J. L., Dunbar H. E., Wolfinger R. D., Moran N. A., 2003. Consequences of reductive evolution for gene expression in an obligate endosymbiont. Mol. Microbiol. 48: 1491–1500. [DOI] [PubMed] [Google Scholar]
- Woolfit M., Iturbe-Ormaetxe I., Brownlie J. C., Walker T., Riegler M., et al. , 2013. Genomic evolution of the pathogenic Wolbachia strain, wMelPop. Genome Biol. Evol. 5: 2189–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfit M., Algama M., Keith J. M., McGraw E. A., Popovici J., 2015. Discovery of putative small non-coding RNAs from the obligate intracellular bacterium Wolbachia pipientis. PLoS One 10: e0118595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Sun L. V., Vamathevan J., Riegler M., Deboy R., et al. , 2004. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2: E69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada R., Floate K. D., Riegler M., O’Neill S. L., 2007. Male development time influences the strength of Wolbachia-induced cytoplasmic incompatibility expression in Drosophila melanogaster. Genetics 177: 801–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., Park J.-H., Inouye M., 2011. Toxin-antitoxin systems in bacteria and archaea. Annu. Rev. Genet. 45: 61–79. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA-seq fastq reads for BDSC ISO1 were submitted to European Nucleotide Archive as experiment ERX645969. Table S1 contains a summary of the number of mapped RNA-seq reads for D. melanogaster and Wolbachia, number of expressed genes (defined as genes with nonzero TPM or genes with ≥2 mapped reads per gene), mean TPM for the sample (same for all samples, inverse of gene number times one million), and standard deviation of TPM for the sample for each total RNA sample in SRP001696. Table S2 contains gene IDs, coordinates, number of reads (from read 2 of paired-end data) mapping to the sense strand in each sample (_r1 = replicate 1, _r2 = replicate 2), estimated TPM for each sample, number of runs found in cluster 2, cluster assignment, adjusted p-value in life cycle GLM, log2 fold-change in life cycle GLM vs. embryo 0-2 hr, maximum fold change between any two stages in life cycle GLM, adjusted p-values and log2 fold change for pairwise exact tests between male and female samples at 1, 5, and 30 d, gene name, annotated gene product, effective number of codons, GC content, and number of homologs in wMel, wRi, wPip-Pel, and wBm genomes. Table S3 contains RT-qPCR GLM analysis results for ISO1 and DrosDel w1118. The p-values reported in Table S3 are not adjusted for multiple testing, and thus α-levels for significance were set at 0.001. Table S4 contains sequences of PCR primers used for RT-qPCR experiments.