Abstract
Background
Some isoquinoline alkaloids from Macleaya cordata (Willd). R. Br. (Bo Luo Hui) exhibited antibacterial, antiparasitic, antitumor, and analgesic effects. The targets of these isoquinoline alkaloids are undefined. This study aims to investigate the compound–target interaction network and potential pharmacological actions of isoquinoline alkaloids of M. cordata by reverse pharmacophore database screening.
Methods
The targets of 26 isoquinoline alkaloids identified from M. cordata were predicted by a pharmacophore-based target fishing approach. Discovery Studio 3.5 and two pharmacophore databases (PharmaDB and HypoDB) were employed for the target profiling. A compound–target interaction network of M. cordata was constructed and analyzed by Cytoscape 3.0.
Results
Thirteen of the 65 predicted targets identified by PharmaDB were confirmed as targets by HypoDB screening. The targets in the interaction network of M. cordata were involved in cancer (31 targets), microorganisms (12 targets), neurodegeneration (10 targets), inflammation and autoimmunity (8 targets), parasitosis (5 targets), injury (4 targets), and pain (3 targets). Dihydrochelerythrine (C6) was found to hit 23 fitting targets. Macrophage migration inhibitory factor (MIF) hits 15 alkaloids (C1–2, C11–16, C19–25) was the most promising target related to cancer.
Conclusion
Through in silico target fishing, the anticancer, anti-inflammatory, and analgesic effects of M. cordata were the most significant among many possible activities. The possible anticancer effects were mainly contributed by the isoquinoline alkaloids as active components.
Background
Macleaya cordata (Willd). R. Br. (Bo Luo Hui) (Fig. 1) has been used for the treatment of cancer [1], insect bites [2], and ringworm infection [3] in Mainland China, North America, and Europe. Phytochemical and pharmacological studies demonstrated that the isoquinoline alkaloids derived from M. cordata are its major active components [4]. Thirty isoquinoline alkaloids have been isolated from M. cordata (Fig. 2), including chelerythrine (C12), sanguinarine (C15), sanguidimerine (C17), chelidimerine (C18), berberine (C21), coptisine (C23), allocryptopine (C24, C25), and protopine (C26). These alkaloids exhibited a broad spectrum of biological activities, such as antitumor [5–8], anti-inflammatory [9–11], antimicrobial [12–14], analgesic [15], and antioxidant [16] activities.
Fig. 1.

The original plant of Macleaya cordata
Fig. 2.
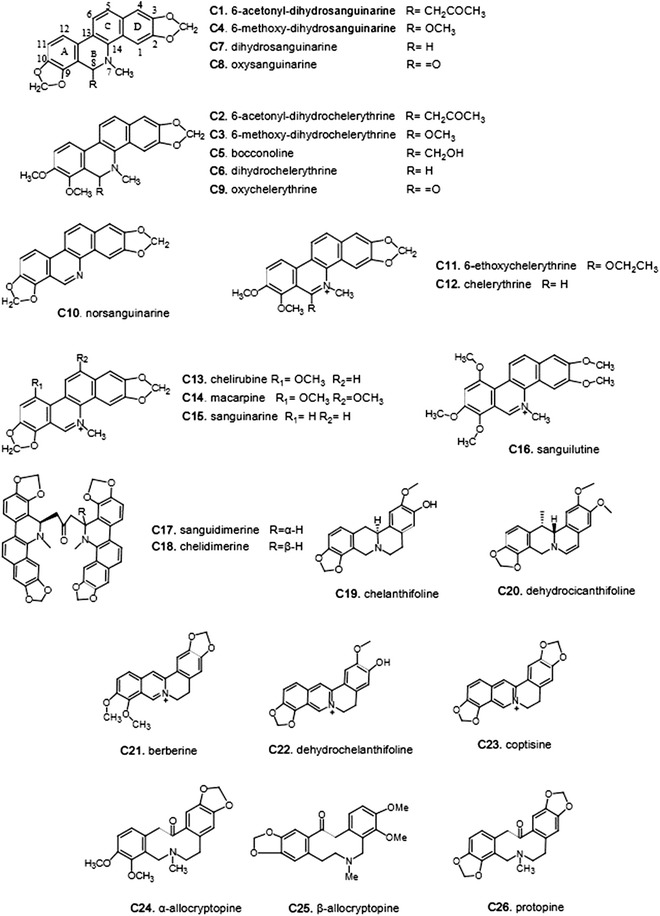
The isoquinoline alkaloids of Macleaya cordata
In our previous study [17], we found that M. cordata could be counted not only as one of the richest resources in Mainland China among all species of the tribe Chelidonieae, but also as one of the most promising natural resources for drug discovery. M. cordata has gained the attention of pharmacognosists since early 1990s (Fig. 3). However, its obscure molecular actions have hindered its use in drug development.
Fig. 3.
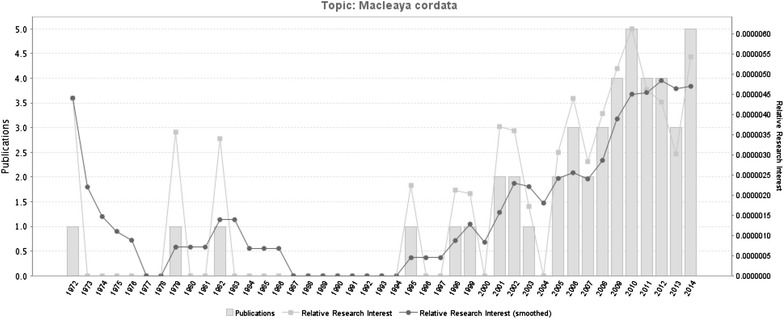
The statistics of Pubmed publications on Macleaya cordata between 1972 and 2014
Although protein–ligand docking techniques have been available in virtual drug screening for specific targets, such as tumor necrosis factor α-converting enzyme (TACE) [18], inducible nitric oxide synthase (iNOS) [19], and Janus-activated kinase 2 (JAK2) [20], these docking approaches to virtual screening are often too computationally expensive [21].
This study aims to investigate the compound-target interaction network of isoquinoline alkaloids of M. cordata by reverse pharmacophore database screening technology, and outline its potential action mechanisms.
Methods
Workflow
Figure 4 shows the workflow of this study. The structures and bioactivities of the isoquinoline alkaloids of M. cordata were collected by literature review [17]. The alkaloids were then applied to target fishing with two pharmacophore and target databases, PharmaDB and HypoDB. The hit pharmacophore models were picked out according to the threshold of a predetermined fit value. The results from PharmaDB screening were compared with those from HypoDB screening. After analysis of the hit targets and their associated pathways and diseases, as well as the interactions between the alkaloids and the targets, an action network of M. cordata was constructed. Literature retrieval was simultaneously carried out to verify the findings.
Fig. 4.
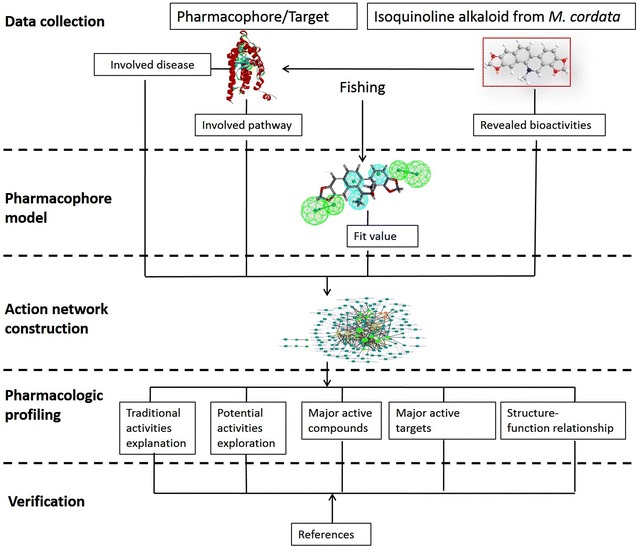
The workflow of this study
Compound collection
The active components of M. cordata were collected from our own database [17] and the literature. All 26 isoquinoline alkaloids of M. cordata and their bioactivities are listed in Table 1. As shown in Fig. 2, the alkaloids were divided into three classes: benzo[c]phenanthridines (Ben, C1–C18), protoberberines (Ber, C19–C23), and protopines (Pro, C24–C26). Based on the replacement of the C-ring, C1–C9 belong to the dihydro-benzo[c]phenanthridines, C10 is a N-demethyl subtype, and C11–C16 are quaternary ammonium bases that share an iminium moiety (C=N+). The remaining two bisbenzo[c]phenanthridines (BisBen, C17–C18) are epimers to one another.
Table 1.
Basic information of the isoquinoline alkaloids in M. cordata
| No. | Compounds | Bioactivities | Virtual hitting targets |
|---|---|---|---|
| 1 | 6-Acetonyl-dihydrosanguinarine | Anti-bacteria Insecticidal |
MIF; TTR; NQO1 |
| 2 | 6-Acetonyl-dihydrochelerythrine | Anti-oxidant Anti-HIV |
HSD1; MIF; PDE4D; NQO1; PLA2s; nAChR 7α; AknH; TtgR |
| 3 | 6-Methoxy-dihydrochelerythrine | Anti-cancer Anti-parasitic |
CAR/RXR; MR; ERα; JNK3; SHBG; AR; 15S-LOX; MMP12; PPARγ; SARS M(pro); Scy D; MAO-A |
| 4 | 6-Methoxy-dihydrosanguinarine | Anti-bacteria Anti-cancer Anti-platelet aggregation |
MR; ERα; FNR; MAO-A |
| 5 | Bocconoline | Anti-bacteria Anti-fungal |
Opsin 2; HSD1; CAR/RXR; MD; ERα; JNK3; SHBG; Chk1; AR; 15S-LOX; CDK2; CAMKII; Aurora A; PIM1; MMP12; Tankyrase 2; SARS M(pro); PfENR; FabZ; DHODH; CDPKs; FNR; ENR; Scy D; MAO-A |
| 6 | Dihydrochelerythrine | Anti-bacteria Anti-fungal |
CAR/RXR; MR; ERα; PPO; TTR; JNK3; SHBG; NQO1; RBP4; 15S-LOX; CK2; PIM1; FabZ; DHODH; SnoaL; FNR; ENR; Scy D; MAO-A; MAO-B; AchE; HIV-1 RT; OSBP |
| 7 | Dihydrosanguinarine | Anti-bacteria Anti-fungal |
MR; ERα; PPO; SHBG; 15S-LOX; CDK2; CK2; MAO-A; AchE |
| 8 | Oxysanguinarine | Anti-platelet aggregation | PIM1; CK2 |
| 9 | Oxychelerythrine | Cytotoxic | CAR/RXR; TTR; JNK3; SHBG; 15S-LOX; CLK1; CK2; PIM1; MMP12; MAPK p38; COMP; FabZ; SonaL; FNR; ENR; MAO-A; MAO-B; AchE; OSBP |
| 10 | Norsanguinarine | Anti-fungal | CK2; NmrA |
| 11 | 6-ethoxychelerythrine | Anti-bacteria Anti-fungal |
MIF; TTR; JNK3; GAPDH; nAChR 7α; FabZ; CAT; LmrR; HS5B Pol |
| 12 | Chelerythrine | Anti-bacteria Anti-fungal Anti-parasitic Anti-cancer |
MIF; TTR; FabZ; HS5B Pol |
| 13 | Chelirubine | Anti-proliferative | MIF; NQO1; GR; ZipA-FtsZ; AknH; opdA |
| 14 | Macarpine | Cytotoxic Anti-proliferative |
PDE4B; PDE 4B; MIF; TTR; NQO1; PIM1; MAPK p38; GR; ZipA-FtsZ; AknH |
| 15 | Sanguinarine | Anti-bacteria Anti-fungal Anti-parasitic Anti-cancer Anti-oxidant Hepatotoxicity |
MIF; nAChR 7α |
| 16 | Sanguilutine | Anti-proliferative | HSD1; MIF; PDE4D; PLA2s; FabZ |
| 17 | Sanguidimerine | Unreported | ATTP |
| 18 | Chelidimerine | Unreported | MDR HIV-1 Protease |
| 19 | Chelanthifoline | Anti-malarial | ALR; ERα; ERβ; MIF; PDK-1; CK2; PIM1; Pi3 Kγ; GR; nAChR 7α; TEM-1; ActR; MAO-B; HIV-1 RT; OSBP |
| 20 | Dehydrocicanthifoline | Unreported | HSD1; MR; PDE4B; PDE4D; PPO; MIF; TTR; JNK3; CRBP-2; MAPK p38; AR; PIM1; ZipA-FtsZ; HS5B Pol; HIV-1 RT |
| 21 | Berberine | Anti-fungal Anti-malarial Anti-cancer Cytotoxic Anti-inflammatory Anti-Alzheimer’s Anti-fertility Anti-diabetes |
MIF; FabZ; Scy D; AchE |
| 22 | Dehydrochelanthifoline | Anti-virus | ERα; ERβ; MIF; GSK-3β; TTR; CDK2; PLA2s; MAO-B |
| 23 | Coptisine | Cytotoxic Anti-diabetes CYP2D6 inhibition Anti-oxidative Anti-spasmodic |
MIF |
| 24 | α-Allocryptopine | Anti-funga Anti-arrhythmic |
HSD1; MIF; HS5B Pol; Scy D; BACE1 |
| 25 | β-Allocryptopine | Anti-parasitic Anti-hepatic fibrosis |
HSD1; MIF; HS5B POl; Scy D; BACE1; CRALBP; PPO; TTR; nAChR 7α |
| 26 | Protopine | Anti-malarial Anti-parasitic Anti-fertility Anti-spasmodic |
NQO1; PfENR; TtgR |
Conformation analysis
The structures of all 26 alkaloid candidates were prepared in MOL format, and converted from 2D drawings to 3D models. Their energies were minimized by the software Discovery Studio (DS, v3.5) developed by BIVIA (USA) with the CHARMM force field. A Monte Carlo-based conformational analysis (FAST mode) was performed to generate conformers from the initial conformations. The maximal 255 conformers were allowed with an energy interval of 20 kcal/mol. These alkaloid molecules were rigid, and the number of conformers for each compound was much fewer than 255. Hence, a total of 135 conformers were generated for the 26 isoquinoline alkaloids.
Ligand profiling
A pharmacophore model represented a series of common features of a set of ligands with a special pharmacological target. The features of a pharmacophore model reflected the target–ligand interaction mode. Pharmacophore-based virtual screening was an alternative to docking. By fitting a compound against a panel of pharmacophore models derived from multiple pharmacological targets, the potential targets of the compound can be outlined.
Automated ligand profiling was available in DS 3.5 as the so-called “Ligand Profiler” protocol. The software offered automated pharmacophore-based activity profiling and reporting [22]. In this study, the default parameters of DS 3.5 were used. For each candidate ligand, three or more features were mapped.
Pharmacophore databases
DS 3.5 was equipped with two available pharmacophore databases, i.e., HypoDB [23] and PharmaDB [24]. HypoDB contained about 2500 pharmacophore models derived from protein–ligand 3D complex structures as well as structural data on small bioactive organic molecules. PharmaDB was created from the sc-PDB, a well-accepted data source in structure-based profiling protocols. The sc-PDB was a collection of 3D structures of binding sites found in the Protein Data Bank (PDB). The binding sites were extracted from crystal structures in which a complex between a protein cavity and a small molecule ligand could be identified. PharmaDB consisted of about 68,000 pharmacophores derived from 8000 protein–ligand complexes from the sc-PDB dataset. PharmaDB is a new and updated pharmacophore database developed in collaboration with Prof. Didier Rognan [25, 26]. The target and pharmacophore models from PharmaDB and HypoDB were not entirely consistent. PharmaDB had a larger quantity of targets, while the models in the HypoDB were fewer and described as being experimentally validated. Therefore, in this study, PharmaDB was employed in the target fishing, and HypoDB was used to validate the results.
Regarding PharmaDB, multiple pharmacophores with shape or excluded volume constraints were generated for each protein target. For the pharmacophores with shape constraints, the suffix “-s” was added to the name. In addition, a numerical suffix referred to the ranking of selectivity evaluated by a default algorithm in DS v3.5. In this study, only the best models with “−1” in their names were employed in the ligand profiling [23]. For each pharmacophore database, a classification tree was available, from which the individual models could be selected.
Parameters
In the profiling with PharmaDB, all the pharmacophore models with the shape of the binding pocket were selected for the virtual screening with default settings. The RIGID mode was used as the molecular mapping algorithm. No molecular features were allowed to be missed while mapping these ligands to the pharmacophore models to increase selectivity. The minimal inter-feature distance was set at 0.5 Å. Parallel screening technology for one or more compounds against a multitude of pharmacophore models was available as a Pipeline Pilot protocol. The number of parallel processing procedures was set at 4. The whole calculation was carried on a T5500 workstation (DELL inc., USA).
Binding mode refinement
All the poses of the ligands mapped into the pharmacophore were preserved. A series of target-ligand pairs were selected as emphasis for further examinations. The selection was based upon compatibility with the reported pharmacological activities, as well as traditional usage of M.cordata. A further refinement was carried out in Molecular Operating Environment (MOE) developed by CCG (Canada) to identify the protein–ligand binding modes. Energy minimization was carried out by conjugated gradient minimization with the MMFF94x force field, until an RMSD of 0.1 kcal mol−1 Ǻ−1 was reached.
Network construction
An interaction table between alkaloids and targets was presented as the ligand profiling results. For each target, the name and pathway information were collected from the PDB and KEGG. The diseases related to the targets were collected from the Therapeutic Target Database (TTD; http://bidd.nus.edu.sg/group/cjttd/) [27] and DrugBank (http://www.drugbank.ca/) [28] databases. Compound-Target-Pathway networks were generated by Cytoscape 3.0 (Cytoscape Consortium, USA) [29]. In the networks, nodes represented the compounds, targets, and biological pathways. The edges linking the compound-target and target-pathway represented their relationships and were marked with different types of lines. After the network was built, the basic parameters of the network were computed and analyzed.
Results and discussion
The profiling results are presented in two HTML tables, designated MoleculeFits and PharmacophoreFits. Two descriptors, fit value and shape similarity, were used to measure the fitness of the ligand and pharmacophore. A fit value equal to or greater than 0.3 was used as a heuristic threshold to select targets from the activity profiler. For each pharmacophore model, the classification information of the target can be indicated in a HTML table created by DS 3.5 called as Pharmacophores. Finally, 98 pharmacophore models were mapped. The models belonged to 65 protein targets, and were involved in 60 pathways. A complete list of the 241 target-ligand pairs is shown in Table 2. The name and indication information of the targets are shown in Table 3. The 13 targets verified by HypoDB screening are marked with an asterisk in Table 3.
Table 2.
The results of ligand profiling
| Class | CMD-ID | ph4 | Target short name | Gene | Uniprot-AC | Fit value | Shape similarity |
|---|---|---|---|---|---|---|---|
| Ben | 1 | 3cfn | TTR | TTHY_HUMAN | P02766 | 0.750635 | 0.508475 |
| Ben | 1 | 1h69 | NQO1 | NQO1_HUMAN | P15559 | 0.923086 | 0.536437 |
| Ben | 2 | 3kba | Progesterone receptor | PRGR_HUMAN | P06401 | 0.334698 | 0.506897 |
| Ben | 2 | 1xom | PDE4D | PDE4D_HUMAN | Q08499 | 0.346437 | 0.527574 |
| Ben | 2 | 2wnj | nAChR 7α | Q8WSF8_APLCA | Q8WSF8 | 0.43985 | 0.505495 |
| Ben | 2 | 1h69 | NQO1 | NQO1_HUMAN | P15559 | 0.928518 | 0.504604 |
| Ben | 3 | 2oz7 | AR | ANDR_HUMAN | P10275 | 0.360685 | 0.500849 |
| Ben | 3 | 2a3i | MR | MCR_HUMAN | P08235 | 0.375601 | 0.528195 |
| Ben | 3 | 5std | ScyD | SCYD_MAGGR | P56221 | 0.418672 | 0.543119 |
| Ben | 3 | 1l2i | ERα | ESR1_HUMAN | P03372 | 0.420147 | 0.542969 |
| Ben | 3 | 1xvp | CAR/RXR | NR1I3_HUMAN | Q14994 | 0.460385 | 0.534672 |
| Ben | 3 | 3lmp | PPARγ | PPARG_HUMAN | P37231 | 0.526039 | 0.500787 |
| Ben | 3 | 1d2s | SHBG | SHBG_HUMAN | P04278 | 0.558685 | 0.563525 |
| Ben | 3 | 2gz7 | SARS M(pro) | R1AB_CVHSA | P0C6X7 | 0.559512 | 0.547348 |
| Ben | 3 | 2p0m | 15S-LOX | LOX15_RABIT | P12530 | 0.639897 | 0.537344 |
| Ben | 3 | 3f15 | MMP12 | MMP12_HUMAN | P39900 | 0.725254 | 0.50503 |
| Ben | 3 | 2bxr | MAO-A | AOFA_HUMAN | P21397 | 0.799156 | 0.521008 |
| Ben | 3 | 2o2u | JNK3 | MK10_HUMAN | P53779 | 0.835637 | 0.577825 |
| Ben | 4 | 2bgi | FNR | Q9L6V3_RHOCA | Q9L6V3 | 0.427793 | 0.516878 |
| Ben | 4 | 1l2i | ERα | ESR1_HUMAN | P03372 | 0.452535 | 0.593291 |
| Ben | 4 | 2bxr | MAO-A | AOFA_HUMAN | P21397 | 0.793632 | 0.533917 |
| Ben | 5 | 2ol4 | PfENR | Q9BH77_PLAFA | Q9BH77 | 0.315455 | 0.518182 |
| Ben | 5 | 3g0u | DHODH | PYRD_HUMAN | Q02127 | 0.369491 | 0.508604 |
| Ben | 5 | 2bxr | MAO-A | AOFA_HUMAN | P21397 | 0.387312 | 0.544118 |
| Ben | 5 | 2a3i | MR | MCR_HUMAN | P08235 | 0.387489 | 0.563771 |
| Ben | 5 | 7std | ScyD | SCYD_MAGGR | P56221 | 0.422146 | 0.504744 |
| Ben | 5 | 2uue | CDK2 | CDK2_HUMAN | P24941 | 0.424268 | 0.567108 |
| Ben | 5 | 2oz7 | AR | ANDR_HUMAN | P10275 | 0.426337 | 0.510961 |
| Ben | 5 | 3coh | Aurora-A | STK6_HUMAN | O14965 | 0.4511 | 0.531532 |
| Ben | 5 | 3kr8 | Tankyrase 2 | TNKS2_HUMAN | Q9H2K2 | 0.464801 | 0.548729 |
| Ben | 5 | 1d2s | SHBG | SHBG_HUMAN | P04278 | 0.480784 | 0.571721 |
| Ben | 5 | 1l2i | ERα | ESR1_HUMAN | P03372 | 0.493882 | 0.57529 |
| Ben | 5 | 2wel | CAMKII | KCC2D_HUMAN | Q13557 | 0.493929 | 0.516729 |
| Ben | 5 | 3fne | ENR | INHA_MYCTU | P0A5Y6 | 0.50498 | 0.546169 |
| Ben | 5 | 1xvp | CAR/RXR | NR1I3_HUMAN | Q14994 | 0.508683 | 0.576427 |
| Ben | 5 | 5std | ScyD | SCYD_MAGGR | P56221 | 0.522671 | 0.566972 |
| Ben | 5 | 2brg | Chk1 | CHK1_HUMAN | O14757 | 0.541427 | 0.51711 |
| Ben | 5 | 3doz | FabZ | Q5G940_HELPY | Q5G940 | 0.546379 | 0.507843 |
| Ben | 5 | 2bgi | FNR | Q9L6V3_RHOCA | Q9L6V3 | 0.553334 | 0.549296 |
| Ben | 5 | 3fnf | ENR | INHA_MYCTU | P0A5Y6 | 0.562321 | 0.511494 |
| Ben | 5 | 3fnh | ENR | INHA_MYCTU | P0A5Y6 | 0.614823 | 0.507547 |
| Ben | 5 | 2p0m | 15S-LOX | LOX15_RABIT | P12530 | 0.673148 | 0.541414 |
| Ben | 5 | 3dp1 | FabZ | Q5G940_HELPY | Q5G940 | 0.687924 | 0.53816 |
| Ben | 5 | 2gz7 | SARS M(pro) | R1AB_CVHSA | P0C6X7 | 0.694125 | 0.551789 |
| Ben | 5 | 2o2u | JNK3 | MK10_HUMAN | P53779 | 0.805153 | 0.553719 |
| Ben | 5 | 3f15 | MMP12 | MMP12_HUMAN | P39900 | 0.893862 | 0.507187 |
| Ben | 6 | 3fj6 | DHODH | PYRD_HUMAN | Q02127 | 0.341464 | 0.566038 |
| Ben | 6 | 5std | ScyD | SCYD_MAGGR | P56221 | 0.364451 | 0.555556 |
| Ben | 6 | 1d2s | SHBG | SHBG_HUMAN | P04278 | 0.367805 | 0.529289 |
| Ben | 6 | 2a3i | MR | MCR_HUMAN | P08235 | 0.368001 | 0.559289 |
| Ben | 6 | 1xvp | CAR/RXR | NR1I3_HUMAN | Q14994 | 0.423023 | 0.572534 |
| Ben | 6 | 2v60 | MAO-B | AOFB_HUMAN | P27338 | 0.436774 | 0.511294 |
| Ben | 6 | 1l2i | ERα | ESR1_HUMAN | P03372 | 0.451108 | 0.529175 |
| Ben | 6 | 1rbp | RBP4 | RET4_HUMAN | P02753 | 0.486949 | 0.516378 |
| Ben | 6 | 2nsd | ENR | INHA_MYCTU | P0A5Y6 | 0.509088 | 0.530738 |
| Ben | 6 | 1kgj | TTR | TTHY_RAT | P02767 | 0.522255 | 0.529412 |
| Ben | 6 | 1tv6 | HIV-1 TR | POL_HV1B1 | P03366 | 0.564494 | 0.529981 |
| Ben | 6 | 2bgi | FNR | Q9L6V3_RHOCA | Q9L6V3 | 0.636419 | 0.536325 |
| Ben | 6 | 2p0m | 15S-LOX | LOX15_RABIT | P12530 | 0.663082 | 0.545045 |
| Ben | 6 | 3imu | TTR | TTHY_HUMAN | P02766 | 0.690803 | 0.577011 |
| Ben | 6 | 3dp1 | FabZ | Q5G940_HELPY | Q5G940 | 0.705916 | 0.565401 |
| Ben | 6 | 2o2u | JNK3 | MK10_HUMAN | P53779 | 0.705945 | 0.507463 |
| Ben | 6 | 1h69 | NQO1 | NQO1_HUMAN | P15559 | 0.755827 | 0.508911 |
| Ben | 6 | 2bxr | MAO-A | AOFA_HUMAN | P21397 | 0.795152 | 0.541573 |
| Ben | 6 | 1sjw | SnoaL | Q9RN59_STRNO | Q9RN59 | 0.904111 | 0.661572 |
| Ben | 6 | 2j3q | AChE | ACES_TORCA | P04058 | 0.992134 | 0.661327 |
| Ben | 7 | 2x1n | CDK2 | CDK2_HUMAN | P24941 | 0.329358 | 0.521253 |
| Ben | 7 | 1d2s | SHBG | SHBG_HUMAN | P04278 | 0.340019 | 0.542857 |
| Ben | 7 | 1l2i | ERα | ESR1_HUMAN | P03372 | 0.465563 | 0.553846 |
| Ben | 7 | 2j3q | AChE | ACES_TORCA | P04058 | 0.470546 | 0.67 |
| Ben | 7 | 2p0m | 15S-LOX | LOX15_RABIT | P12530 | 0.639874 | 0.545254 |
| Ben | 7 | 2bxr | MAO-A | AOFA_HUMAN | P21397 | 0.822509 | 0.548694 |
| Ben | 8 | 3bgp | PIM-1 | PIM1_HUMAN | P11309 | 0.659102 | 0.52193 |
| Ben | 9 | 2wu7 | CLK1 | CLK3_HUMAN | P49761 | 0.333353 | 0.541053 |
| Ben | 9 | 1d2s | SHBG | SHBG_HUMAN | P04278 | 0.40988 | 0.526096 |
| Ben | 9 | 2nsd | ENR | INHA_MYCTU | P0A5Y6 | 0.417703 | 0.522727 |
| Ben | 9 | 1tha | TTR | TTHY_HUMAN | P02766 | 0.42188 | 0.505071 |
| Ben | 9 | 1xvp | CAR/RXR | NR1I3_HUMAN | Q14994 | 0.459386 | 0.600775 |
| Ben | 9 | 1fbm | COMP | COMP_RAT | P35444 | 0.540756 | 0.509542 |
| Ben | 9 | 2p0m | 15S-LOX | LOX15_RABIT | P12530 | 0.609094 | 0.548596 |
| Ben | 9 | 2bxr | MAO-A | AOFA_HUMAN | P21397 | 0.636415 | 0.524336 |
| Ben | 9 | 3iw7 | MAPK p38 | MK14_HUMAN | Q16539 | 0.671331 | 0.532803 |
| Ben | 9 | 1sjw | SnoaL | Q9RN59_STRNO | Q9RN59 | 0.679871 | 0.665953 |
| Ben | 9 | 2bgi | FNR | Q9L6V3_RHOCA | Q9L6V3 | 0.68446 | 0.509554 |
| Ben | 9 | 3dp1 | FabZ | Q5G940_HELPY | Q5G940 | 0.723083 | 0.601732 |
| Ben | 9 | 2v60 | MAO-B | AOFB_HUMAN | P27338 | 0.818911 | 0.501006 |
| Ben | 9 | 2o2u | JNK3 | MK10_HUMAN | P53779 | 0.878824 | 0.532609 |
| Ben | 9 | 2j3q | AChE | ACES_TORCA | P04058 | 0.992667 | 0.679157 |
| Ben | 10 | 2wmd | NmrA | NMRL1_HUMAN | Q9HBL8 | 0.635677 | 0.601671 |
| Ben | 11 | 3doz | FabZ | Q5G940_HELPY | Q5G940 | 0.380298 | 0.514677 |
| Ben | 11 | 3kvx | JNK3 | MK10_HUMAN | P53779 | 0.408174 | 0.518987 |
| Ben | 11 | 2wnj | nAChR 7α | Q8WSF8_APLCA | Q8WSF8 | 0.408648 | 0.512476 |
| Ben | 11 | 3doy | FabZ | Q5G940_HELPY | Q5G940 | 0.456816 | 0.515444 |
| Ben | 11 | 1k3t | GAPDH | G3PG_TRYCR | P22513 | 0.56209 | 0.500931 |
| Ben | 11 | 3lmp | PPARγ | PPARG_HUMAN | P37231 | 0.648243 | 0.508527 |
| Ben | 11 | 1qca | CAT | CAT3_ECOLX | P00484 | 0.780585 | 0.505747 |
| Ben | 11 | 3f8f | LmrR | A2RI36_LACLM | A2RI36 | 0.817481 | 0.51932 |
| Ben | 13 | 1xan | GR | GSHR_HUMAN | P00390 | 0.573697 | 0.520833 |
| Ben | 13 | 3kba | Progesterone receptor | PRGR_HUMAN | P06401 | 0.651629 | 0.522059 |
| Ben | 13 | 1h69 | NQO1 | NQO1_HUMAN | P15559 | 0.811055 | 0.503055 |
| Ben | 13 | 3a3w | opdA | Q93LD7_RHIRD | Q93LD7 | 0.833715 | 0.510158 |
| Ben | 14 | 3huc | MAPK p38 | MK14_HUMAN | Q16539 | 0.345074 | 0.534091 |
| Ben | 14 | 1xan | GR | GSHR_HUMAN | P00390 | 0.517848 | 0.510823 |
| Ben | 14 | 1h69 | NQO1 | NQO1_HUMAN | P15559 | 0.723867 | 0.515504 |
| Ben | 14 | 1xom | PDE4D | PDE4D_HUMAN | Q08499 | 0.745933 | 0.503704 |
| Ben | 14 | 1xlx | PDE4B | PDE4B_HUMAN | Q07343 | 0.796555 | 0.52037 |
| Ben | 15 | 2wnj | nAChR 7α | Q8WSF8_APLCA | Q8WSF8 | 0.495424 | 0.509356 |
| Ben | 16 | 3kba | Progesterone receptor | PRGR_HUMAN | P06401 | 0.342606 | 0.537671 |
| Ben | 16 | 1xom | PDE4D | PDE4D_HUMAN | Q08499 | 0.816692 | 0.539427 |
| BisBen | 17 | 1r5 l | ATTP | TTPA_HUMAN | P49638 | 0.32356 | 0.514156 |
| BisBen | 18 | 1rq9 | MDR HIV-1 Protease | Q5RTL1_9HIV | Q5RTL1 | 0.621229 | 0.507743 |
| Ber | 19 | 1u3s | ERβ | ESR2_HUMAN | Q92731 | 0.408794 | 0.535377 |
| Ber | 19 | 2j3q | AChE | ACES_TORCA | P04058 | 0.485705 | 0.596737 |
| Ber | 19 | 3l54 | Pi3 Kγ | PK3CG_HUMAN | P48736 | 0.498919 | 0.59589 |
| Ber | 19 | 1pzo | TEM-1 | BLAT_ECOLX | P62593 | 0.523404 | 0.526667 |
| Ber | 19 | 2ikg | ALR | ALDR_HUMAN | P15121 | 0.561704 | 0.507109 |
| Ber | 19 | 1c1c | HIV-1 TR | POL_HV1H2 | P04585 | 0.577074 | 0.542373 |
| Ber | 19 | 2r7b | PDK-1 | PDPK1_HUMAN | O15530 | 0.587223 | 0.533049 |
| Ber | 19 | 1yye | ERβ | ESR2_HUMAN | Q92731 | 0.679767 | 0.56691 |
| Ber | 19 | 1qkt | ERα | ESR1_HUMAN | P03372 | 0.681913 | 0.56351 |
| Ber | 19 | 1xan | GR | GSHR_HUMAN | P00390 | 0.715506 | 0.548544 |
| Ber | 19 | 2wnj | nAChR 7α | Q8WSF8_APLCA | Q8WSF8 | 0.87937 | 0.501031 |
| Ber | 19 | 3b6c | ActR | Q53901_STRCO | Q53901 | 0.880577 | 0.597561 |
| Ber | 19 | 1x78 | ERβ | ESR2_HUMAN | Q92731 | 0.909059 | 0.522565 |
| Ber | 20 | 1xm4 | PDE4B | PDE4B_HUMAN | Q07343 | 0.402388 | 0.569138 |
| Ber | 20 | 1tha | TTR | TTHY_HUMAN | P02766 | 0.45615 | 0.514286 |
| Ber | 20 | 1tv6 | HIV-1 TR | POL_HV1B1 | P03366 | 0.459002 | 0.521154 |
| Ber | 20 | 2nw4 | AR | ANDR_RAT | P15207 | 0.463505 | 0.541203 |
| Ber | 20 | 1opb | CRBP2 | RET2_RAT | P06768 | 0.485837 | 0.534653 |
| Ber | 20 | 2waj | JNK3 | MK10_HUMAN | P53779 | 0.572085 | 0.603104 |
| Ber | 20 | 1kgj | TTR | TTHY_RAT | P02767 | 0.740087 | 0.56531 |
| Ber | 20 | 1xom | PDE4D | PDE4D_HUMAN | Q08499 | 0.811727 | 0.542406 |
| Ber | 20 | 1xlx | PDE4B | PDE4B_HUMAN | Q07343 | 0.859016 | 0.51341 |
| Ber | 20 | 3i6d | PPO | PPOX_BACSU | P32397 | 0.97618 | 0.570499 |
| Ber | 21 | 5std | ScyD | SCYD_MAGGR | P56221 | 0.324086 | 0.505682 |
| Ber | 21 | 2j3q | AChE | ACES_TORCA | P04058 | 0.992907 | 0.672209 |
| Ber | 22 | 1di8 | CDK2 | CDK2_HUMAN | P24941 | 0.406566 | 0.501094 |
| Ber | 22 | 1u3s | ERβ | ESR2_HUMAN | Q92731 | 0.661777 | 0.509434 |
| Pro | 24 | 5std | ScyD | SCYD_MAGGR | P56221 | 0.538905 | 0.543636 |
| Pro | 24 | 3ine | BACE1 | BACE1_HUMAN | P56817 | 0.54561 | 0.522968 |
| Pro | 25 | 1tyr | TTR | TTHY_HUMAN | P02766 | 0.356425 | 0.526412 |
| Pro | 25 | 3inf | BACE1 | BACE1_HUMAN | P56817 | 0.37118 | 0.504303 |
| Pro | 25 | 2wnj | nAChR 7α | Q8WSF8_APLCA | Q8WSF8 | 0.553029 | 0.533461 |
| Pro | 25 | 3ine | BACE1 | BACE1_HUMAN | P56817 | 0.597712 | 0.51259 |
| Pro | 25 | 3hx3 | CRALBP | RLBP1_HUMAN | P12271 | 0.604625 | 0.513158 |
| Pro | 25 | 5std | ScyD | SCYD_MAGGR | P56221 | 0.763034 | 0.522523 |
| Pro | 26 | 2ow2 | PfENR | MMP9_HUMAN | P14780 | 0.302479 | 0.507865 |
| Pro | 26 | 2f1o | NQO1 | NQO1_HUMAN | P15559 | 0.432407 | 0.52183 |
Table 3.
The targets identified
| Targets | Short name | Type | Pathway | Diseases |
|---|---|---|---|---|
| Retinaldehyde-binding protein | CRALBP | Research | Retinaldehyde metabolism | Retinitis pigmentosa |
| Rhodopsin | Opsin 2 | Research | Retina metabolism | Retinitis pigmentosa |
| 11-Beta-hydroxysteroid dehydrogenase | HSD1 | Successful | Glucocorticoid concentration | Diabetes Osteoporosis Hepatotoxicity |
| CAR/RXR heterodimer | CAR/RXR | Research | Triglyceride metabolism | Diabetes Hepatitis |
| Aldose reductase | ALR | Successful | Glucolipid metabolism | Diabetes Pain |
| Mineralocorticoid receptors | MR | Successful | Na+/K+ equilibrium | Inflammatory, autoimmune disease Injury |
| Phosphodiesterase 4B | PDE4B | Successful | AKT/mTOR pathway | Cancer Obesity |
| Phosphodiesterase 4D | PDE4D | Successful | Intracellular cAMP//CREB signaling | Cancer Alzheimer’s |
| Protoporphyrinogen oxidase | PPO | Research | Heme biosynthesis | Cancer Parasitosis |
| Transthyretin | TTR | Clinic Trial | Thyroxine carrier | Cancer Alzheimer’s |
| Mitogen-activated protein kinase 10 | JNK3 | Research | GbRH/ErbB/MAPK/insulin signaling pathway | Cancer Alzheimer’s |
| Sex hormone-binding globulin | SHBG | Research | Sex steroids biosynthesis | Cancer |
| NAD(P)H:quinone oxidoreductase | NQO1 | Research | Quinones metabolism | Cancer |
| Cellular retinol binding protein II | CRBP2 | Research | Retinol metabolism | Cancer |
| Estrogen receptor alphaa | ERαa | Successful | Estrogen metabolism Insulin-like growth factor pathway |
Cancer Alzheimer’s Injury Osteoporosis |
| Alpha-tocopherol (alpha-T) transfer protein | ATTP | Research | α-Tocopherol metabolism | Cancer |
| Human serum retinol binding protein 4 | RBP4 | Research | Retinol metabolism | Cancer |
| Estrogen receptor betaa | ERβa | Successful | Estrogen metabolism MAPK, PI3K signaling |
Cancer Alzheimer’s Injury |
| Checkpoint kinase 1a | Chk1a | Research | DNA damage response | Cancer |
| Androgen receptor | AR | Successful | Hormone metabolism | Cancer |
| Reticulocyte 15S-lipoxygenase | 15S-LOX | Research | Arachidonic acid metabolism | Cancer |
| 3-Phosphoinositide-dependent kinase-1a | PDK-1a | Research | Phosphatidylinositol 3 kinase (PI3K) signaling | Cancer |
| Casein kinase 2a | CK2a | Research | Ser/Thr pathway | Cancer |
| Cyclin dependent kinase 2a | CDK2a | Research | Cell cycle | Cancer |
| Calcium/calmodulin dependent protein kinase II delta | CAMKII | Research | NF-κB-mediated inflammatory response Ca2+-linked signaling |
Cancer Inflammatory, autoimmune disease |
| Dual-specificity protein kinase 1 | CLK1 | Research | Nuclear redistribution of SR proteins | Cancer |
| Proto-oncogene serine threonine kinasea | PIM-1a | Research | Cell cycle regulation JAK/STAT pathway | Cancer |
| Aurora kinase A | Aurora-A | Clinical trial | Cell cycle arrest | Cancer |
| Matrix metalloproteinases | MMP12 | Research | Cell invasion, metastasis | Cancer Inflammatory, autoimmune disease |
| Phospholipase A2 | PLA2s | Successful | VEGF/MAPK/GnRH signaling | Cancer Inflammatory, autoimmune disease |
| Mitogen-Activated Protein Kinases p38 | MAPK p38 | Clinical trial | MAPK signaling | Cancer Pain Inflammatory, autoimmune disease Dermatosis |
| Tankyrase 2 | Tankyrase 2 | Research | Canonical Wnt signaling | Cancer |
| Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | Pi3Kγ | Research | Cancer migration, invasion Inositol phosphate metabolism |
Cancer Inflammatory, autoimmune disease |
| PPARgamma-LBDa | PPARγa | Research | LPS-induced iNOS expression | Cancer Inflammatory, autoimmune disease Osteoporosis |
| Cartilage oligomeric matrix protein | COMP | Research | Bone regeneration | Autoimmune disease Injury |
| Severe acute respiratory syndrome coronavirus (SARS-CoV) main protease (M(pro)) | SARS M(pro) | Research | Virus maturation | Virus infection |
| Glycosomal glyceraldehyde-3-Phosphate Dehydrogenase | GAPDH | Successful | Glyceraldehydes metabolism | Parasitosis |
| Glutathione disulfide oxidoreductase | GR | Research | Glutathione metabolism | Parasitosis |
| Acyl carrier protein reductasea | PfENRa | Successful | Fatty acid biosynthesis | Parasitosis |
| Acetylcholine binding protein alpha7 | nAChR 7α | Successful | Calcium signaling pathway | Alzheimer’s Pain |
| 3R-hydroxyacyl-acyl carrier protein dehydratase | FabZ | Research | Fatty acid biosynthesis | Parasitosis |
| Dihydroorotate dehydrogenase | DHODH | Successful | Pyrimidine metabolism | Parasitosis |
| TEM-1 Beta-Lactamasea | TEM-1a | Successful | Cefotaxime metabolism | Bacterial infection |
| Chloramphenicol acetyltransferase | CAT | Research | Chloramphenicol metabolism | Bacterial infection |
| Polyketide cyclase SnoaL | SnoaL | Research | Nogalamycin biosynthesis | Bacterial infection |
| ZipA attaches FtsZ protein | ZipA-FtsZ | Research | Cell division | Bacterial infection |
| Ferredoxin-NADP+ reductase | FNR | Successful | Redox metabolism | Bacterial infection |
| Polyketide cyclase AknH | AknH | Research | Aclacinomycin biosynthesis | Bacterial infection |
| Enoyl-acyl carrier protein reductase | ENR | Successful | Fatty acid biosynthesis | Bacterial infection |
| Multidrug binding protein TtgR | TtgR | Research | Active extrusion of drug | Bacterial infection |
| NmrA-like family domain | NmrA | Research | Transcriptional repress | Fungal infection |
| Bacterial phosphotriesterase | opdA | Research | Organophosphate metabolism | Bacterial infection |
| Streptomyces coelicolor TetR family protein ActRa | ActRa | Research | Transcriptional repress | Bacterial infection |
| Multidrug binding transcriptional regulator LmrR | LmrR | Research | Autoregulatory mechanism | Bacterial infection |
| Scytalone Dehydratase | ScyD | Research | Fungicide | Fungal infection |
| Human monoamine oxidase A | MAO-A | Successful | Monoamines metabolism | Depression |
| Acetylcholin esterase | AChE | Successful | Glycerophospholipid metabolism | Alzheimer’s Parkinson’s |
| β-Site amyloid precursor protein cleaving enzyme | BACE1 | Clinical trial | Neuregulin processing | Alzheimer’s |
| Multidrug-resistant HIV-1 proteasea | MDR HIV-1 proteasea | Successful | Self-activation | AIDs |
| HIV-1 reverse transcriptase | HIV-1 TR | Successful | ATP-dependent excision, pyrophosphorolysis | AIDs |
| Oxysterol binding protein | OSBP | Research | Intracellular lipid homeostasis Signal conduction |
Virus infection Cancer |
| Rhodopsin | Opsin 2 | Research | Rod photoreceptor | Retinitis pigmentosa |
| Macrophage migration inhibitory factor | MIF | Clinical trial | Phenylalanine, tyrosine metabolism | Cancer Inflammatory, autoimmune disease |
| Glycogen synthase kinase-3 beta | GSK-3β | Research | Glycogen biosynthesis | Cancer Alzheimer’s Diabetes |
| Hepatitis C virus (HCV) polymerase | HS5B Pol | Successful | DNA biosynthesis | Virus infection |
aThe targets verified by HypoDB screening
Analysis of the interaction network
A topological analysis of the interaction network offered insights into the biologically relevant connectivity patterns, and highly influential compounds or targets. Some Chinese medicines had been investigated by interaction network analysis [30–32].
The pharmacological network of M. cordata had three types of nodes (Fig. 5). The 26 alkaloid nodes formed the core of the network, and were surrounded by 65 target nodes. Each target was linked to at least one pathway. A total of 60 pathway nodes constituted the outer layer of the network. Each alkaloid was the center of a star-shaped action net except for the two bisbenzo[c]phenanthridines (BisBen), which were only linked to one target and one pathway, respectively. The alkaloids and targets were strongly interconnected in many-to-many relationships.
Fig. 5.
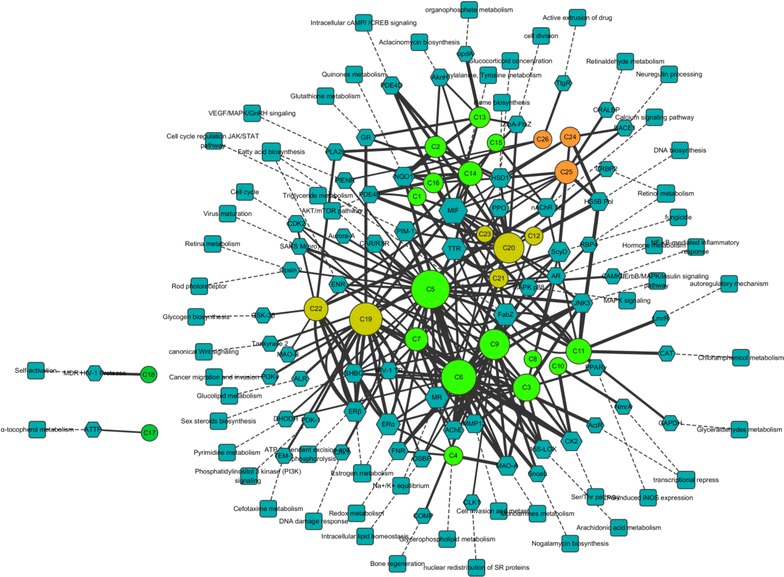
The pharmacological network of Macleaya cordata. Hexagon, targets; Rectangle, biopathway; Ellipse, alkaloids (bright green Ben, dark green BisBen, breen Ber, orange Pro)
A general overview of the global topological properties of the network was obtained from the statistical data by the Network Analyzer of Cytoscape. The diameter of the network was 8.0, the centralization was 0.14, and the density was 0.024. The node degree indicated the number of edges linking to other nodes. The highly connected nodes were referred to as the hubs of the network. The degrees of all the alkaloids (Fig. 6a) and important targets (Fig. 6b) were investigated. The compounds with higher degree values, such as C5, C6, C9, C19, and C20, that might participate in more interactions than the other components were the hubs in the network. The target degree values mostly ranged between 2 and 7. The targets with the highest degree values included MIF (16), TTR (11), FabZ* (11), ERα* (10), and MR (10). The targets with higher degree values might be involved in the pharmacological actions of M. cordata.
Fig. 6.
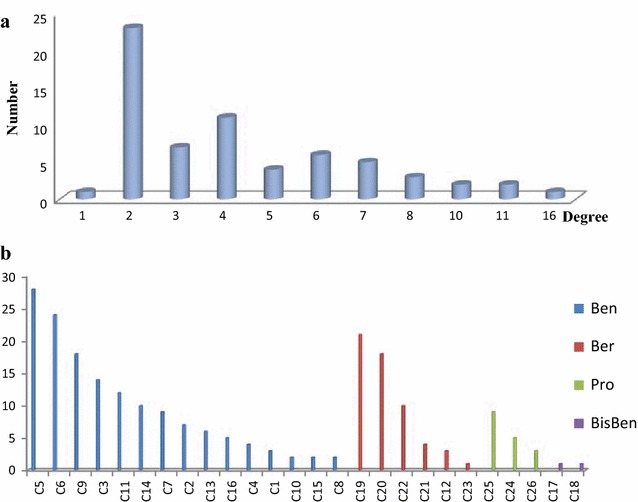
Degree distribution in the network. a alkaloids, b targets
Interpreting the pharmacological actions
By mining the PubMed and TTD, the targets of M. cordata in the PharmaDB profiling results were annotated with biological functions and clinical indications (Table 3). Furthermore, the targets were classified according to the reported pharmacological activities of M. cordata as follows: microorganism (including bacterial, fungal, and viral) infection (12 targets, with 3 targets verified by HypoDB screening), parasitic disease (5 targets, with 2 targets validated by HypoDB screening), pain (3 targets), cancer (31 targets, with 8 targets confirmed by HypoDB screening), inflammation (8 targets, with 1 target verified by HypoDB screening), and injury (4 targets, with 2 targets fished by HypoDB screening).
Antibacterial activity
The extracts and their purified alkaloids from M. cordata exhibited notable activities against Staphylococcus aureus, Pseudomonas aeruginosa, Escherichia coli, Bacillus subtilis, Tetracoccus spp., and methicillin-resistant Staphylococcus aureus (MRSA) [12, 33]. In this study, 12 proposed targets were closely related to microorganisms, and seven of them exhibited antibacterial activities (Fig. 7). the key types of alkaloids with antibacterial activity were dihydro-benzo[c]phenanthridine alkaloids and protoberberines.
Fig. 7.
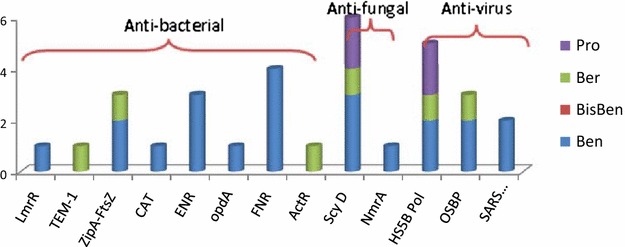
The compounds mapping of microorganism related targets
Five targets (LmrR, TEM-1*, CAT, FNR, and ActR) were related to multidrug-resistant bacterial strains. LmrR, a multidrug binding transcriptional regulator and the predicted target of C11, was a PadR-related transcriptional repressor that regulated the production of LmrCD, a major multidrug ABC transporter in Lactococcus lactis [34, 35]. TEM-1* (TEM-1 beta-lactamase) fished by C19 was one of the antibiotic-resistance determinants for penicillins, early cephalosporins, and novel drugs from their derivatives [36]. A new drug, Avibactam™, innovated by AstraZeneca is a TEM-1 inhibitor that has already entered phase III clinical development [37]. In addition, chloramphenicol acetyltransferase (CAT), an antibiotic-inactivating enzyme predicted by C11, catalyzed the acetyl-S-CoA-dependent acetylation of chloramphenicol at the 3-hydroxyl group and resulted in chloramphenicol-resistance in bacteria [38]. Ferredoxin-NADP+ reductase (FNR), targeted in silico by C4, C5, C6, and C9, participated in numerous electron transfer reactions, had no homologous enzyme in humans, and was a target for the accumulation of multidrug-resistant microbial strains [39]. The Streptomyces coelicolor TetR family protein ActR* was found by C19. ActR* may mediate timely self-resistance to an endogenously-produced antibiotic. TetR-mediated antibiotic-resistance might have been acquired from an antibiotic-producer organism [40].
Two targets indicating other pathways were involved in the antibacterial activity. The ZipA-FtsZ complex was fished by C13, C14, and C20 (Fig. 8). ZipA was a membrane-anchored protein in E. coli that interacted with FtsZ-mediated bacterial cell division, and was considered a potential target for antibacterial agents [41]. The target ENR catalyzed an essential step in fatty acid biosynthesis. ENR was a target for narrow-spectrum antibacterial drug discovery because of its essential role in metabolism and its sequence conservation across many bacterial species [42].
Fig. 8.
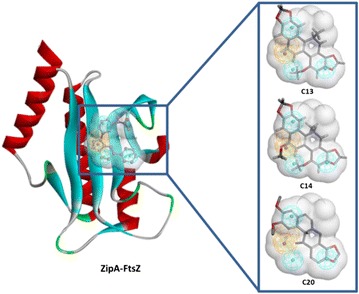
Three alkaloids mapped to ZipA-FtsZ. Left the crystal structure and pharmacophore of target, right the alkaloids fit to the pharmacophore
Antiparasitic activity
M. cordata showed remarkable effects against Ichthyophthirius multifiliis in grass carp [43] and richadsin [44], as well as against Dactylogyrus intermedius in Carassius auratus [45]. The total alkaloids of M. cordata were able to kill gastrointestinal parasites [46].
In this study, five targets involved in parasitic diseases were predicted. Because of the lack of reported protein–ligand crystal structures for parasitosis, these five targets were not related to the above parasitosis in either humans or other animals. However, the findings suggested the potential of M.cordata to treat other parasitosis, such as malaria, Chagas disease, and Kala-azar. The enoyl-acyl carrier reductase PfENR* fished by two alkaloids (C5 and C26) and the (3R)-hydroxymyristoyl acyl carrier protein dehydratase FabZ* in silico targeted by six alkaloids (C5, C6, C9, C11, C12, and C16) were involved in the fatty acid biosynthesis of Plasmodium falciparum. The antioxidant enzyme GR fished by C13, C14, and C19 was a target for antimalarial drug development [47]. The target glycosomal glyceraldehyde-3-phosphate dehydrogenase (GAPDH) found by C11 was a target for the development of novel chemotherapeutic agents for the treatment of Chagas disease [48]. Dihydroorotate dehydrogenase (DHODH) retrieved by C5 and C6 was related to both Leishmania infection and Trypanosoma infection [49].
Analgesic activity
A mixture of the isoquinoline alkaloids from M. cordata exhibited strong analgesic activity towards the pain caused by inflammatory cytokines and direct peripheral nerve stimulation [50]. In this study, three targets related to pain were identified. nAChR7α was abundantly expressed in the central and peripheral nervous systems, and involved in subchronic pain and inflammation [51]. In the profiling results, nAChR7α was picked out by five alkaloids (C2, C11, C15, C19, and C25). MAPK p38 fished by C9, C14, and C20 was involved in the development and maintenance of inflammatory pain [52, 53]. The reductase ALR fished by C19 was a specific target of painful diabetic neuropathy [54, 55]. Inhibitors of ALR relieved pain and improved somatic and autonomic nerve function [56]. In addition, based on the action network, berberines (Ber) such as C19 and C20 may also be involved in the analgesic activity of M. cordata.
Anti-inflammatory activity
Eight targets related to inflammation were identified in this study. Phosphatidylinositol-4, 5-bisphosphate 3-kinase catalytic subunit gamma isoform (PI3 Kγ) fished by C19 recruited leukocytes [57]. The proteinase MMP12, also known as macrophage metalloelastase (MME) or macrophage elastase (ME), was identified with three fitted compounds (C3, C5, and C9) in this study. MMP12 mediated neutrophil and macrophage recruitment and T cell polarization [58], and was a potential therapeutic target for asthma [59]. PPARγ* fished by C3 was another inflammation-related target. Some early findings demonstrated the anti-inflammatory effects of PPARγ by activating human or murine monocytes/macrophages and monocyte/macrophage cell lines [60].
MAPK p38 was involved in a signaling cascade controlling cellular responses to inflammatory cytokines, and it was verified for this pathway in murine macrophage RAW264.7 cells that the M. cordata extract increased both the mRNA and protein levels of cytoprotective enzymes including heme oxygenase-1 (HO-1) and thioredoxin 1 via activation of the p38 MAPK/Nrf2 pathway [16]. The kinase calcium/calmodulin-dependent protein kinase II (CAMKII) was a regulator of intracellular Ca2+ levels, which triggered activation of the transcription factor nuclear factor-kappa B (NF-κB) after T-cell receptor stimulation. An inhibitory effect of CAMKII on NF-κB was confirmed [61]. Phospholipase A2 (PLA2s) was a key enzyme in prostaglandin (PG) biosynthesis for discharging arachidonic acid. Selective inhibitors of PLA2s were implicated in inflammation and connected to diverse diseases, such as cancer, ischemia, atherosclerosis, and schizophrenia [62].
The target mineralocorticoid receptor (MR) fished by five compounds (C3, C4, C6, C7, and C20) was activated by mineralocorticoids, such as aldosterone and deoxycorticosterone, as well as by glucocorticoids, like cortisol. Antagonists of MR had cardioprotective and anti-inflammatory effects in vivo via aldosterone-independent mechanisms [63]. Macrophage migration inhibitory factor (MIF) was involved in both innate and adaptive immune responses. Inhibitors of MIF were potential anti-inflammatory agents [64].
Seven of the eight predicted targets were also related to cancer. These dual correlative targets were PI3Kγ, MMP12, PPARγ*, MAPK p38, CAMKII, PLA2s, and MIF. Their matching compounds are shown in Fig. 9, and the benzo[c]phenanthridine (Ben) alkaloids and berberine (Ber) alkaloids were involved in the anti-inflammatory activity.
Fig. 9.
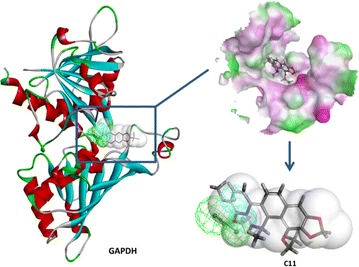
Alkaloid C11 mapped to GAPDH. Left the crystal structure and pharmacophore of GAPDH, upper right the alkaloid C11 docked into the target, lower right C11 fitting into the pharmacophore and the shape of the pocket
Injury healing activity
In this study, four predicted targets (ERα*, ERβ*, MR, and COMP) were involved in injury repair. Among them, ERα*, ERβ*, and MR were linked with internal injuries, such as brain injury [65], vascular injury [66], and neuronal injury [67]. The other target, cartilage oligomeric matrix protein (COMP), found by C9 was a non-collagenous extracellular matrix protein found predominantly in cartilage, but also in tendon, ligament, and meniscus [68]. COMP was a marker for joint destruction associated with osteoarthritis, rheumatoid arthritis, trauma, and intense activity [69].
Antitumor activity
Both the mixed and single alkaloids of M. cordata strongly inhibited proliferation and induced apoptosis of cancer cells [6, 70]. The anticancer drug Ukrain™ is an isoquinoline type. The major components of Ukrain™ are chelidonine, sanguinarine, chelerythrine, protopine, and allocryptopine. Ukrain™ exerted cytotoxic effects in cancer cells without negative effects on normal cells [71], and had radiosensitization effects on cancer cells, while exerting radioprotective effects on normal cells [72].
In the pharmacological profiling results, almost half of the predicted targets (31 of 65 targets) had a close relationship with cancer, and ten of them (Table 3) successfully entered into clinical trial observations. In total, nine targets related to cancer were fished by more than five compounds. The results revealed promising prospects for M. cordata in antitumor drug research and development. Based on the action network (Fig. 5), possible antitumor molecular mechanisms of M.cordata were analyzed as follows: (1) most possible effective targets and (2) most likely contributing compounds.
The MIF column was particularly tall (Fig. 10) because it was fished by 15 compounds, including all quaternary benzo[c]phenanthridine (Ben) alkaloids (C11–C16), two other benzo[c]phenanthridine (Ben) alkaloids, five protoberberine (Ber) alkaloids, and two protopine (Pro) alkaloids. The discovered pathways of these 15 compounds mainly included NF-κB and ERK signaling pathways [73, 74], Bax/Bcl and caspase-dependent pathway [75], ROS-mediated mitochondrial pathway [76], p38 MAPK/Nrf2 pathway [77], and VEGF-induced Akt phosphorylation pathway [78]. All of these pathways were linked closely with MIF [79–84]. However, there have been no experimental reports on to the interactions between MIF and these alkaloids.
Fig. 10.
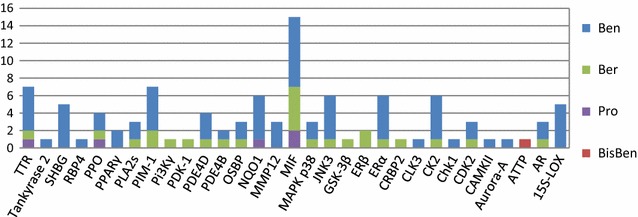
The alkaloids mapping of cancer related targets
Both transthyretin (TTR) and proto-oncogene serine threonine kinase* (PIM-1) were found by seven compounds. TTR was a biomarker for lung cancer [85] and pancreatic ductal adenocarcinoma [86], but has not yet been confirmed as a therapeutic target. PIM-1* fished by C5, C6, C8, C9, C14, C19, and C20, and also verified by HypoDB screening, was responsible for cell cycle regulation, antiapoptotic activity, mediation of homing, and migration of receptor tyrosine kinases via the JAK/STAT pathway. PIM-1 was upregulated in many hematological malignancies and solid tumors. Although PIM kinases were described as weak oncogenes, they were heavily targeted for anticancer drug discovery [87]. C12 was partially involved in the JAK/STAT pathway [88].
The benzo[c]phenanthridine (Ben) alkaloids of M. cordata hit cancer-related targets a total of 75 times, compared with 25 times for protoberberines (Ber), five times for protopines (Pro), and one time for bis-benzo[c]phenanthridines (BisBen) (Fig. 11). According to the quantitative determination of alkaloids from M. cordata, the quaternary benzo[c]phenanthridine alkaloids C12, C13, and C15 were the main active components [89]. However, the dihydro-benzo[c]phenanthridines such as C5, C6, and C9 rarely reached the limit of detection (LOD), and hit more targets than the main alkaloids. As the quaternary and dihydro-benzo[c]phenanthridines can be transformed into one another, the dihydro-benzo[c]phenanthridines could be active compounds in vivo. The metabolism of C15 was examined in pig liver microsomes and cytosol by electrospray ionization hybrid ion trap/time-of-flight mass spectrometry, and C7 was one of the main metabolites in liver microsomes and the only metabolite in cytosol [90]. Hence, the issue of whether the dihydro-benzo[c]phenanthridines were the main compounds combining with the targets in vivo requires further investigation.
Fig. 11.
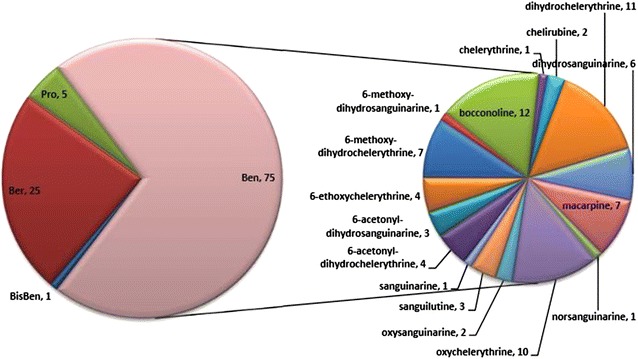
The hit number of the alkaloids to cancer related targets
Among the 31 cancer-related targets, at least seven (including MIF, PPARγ*, CAMKII, and Pi3Kγ) were involved in the immune system. These immune-associated targets might be crucial to for oncotherapy with M. cordata.
Potential pharmacological activities
According to the pharmacological profiling, some unreported pharmacological performances of M.cordata emerged. In this study, 10 targets linked with neurodegeneration were fished, among which AChE and MAO-B were crucial therapeutic targets in Alzheimer’s disease and Parkinson’s disease [91–94].
In addition, antiviral activities, especially anti-HIV, anti-SARS coronavirus, and antifungal activities, were kinds of extensions of the antibacterial function of M. cordata. The possible anti-HIV activity was notable, because HIV-1 reverse transcriptase and multidrug-resistant HIV-1 protease* were particularly related to AIDS [95–99]. Meanwhile, the anti-HIV activity was partly confirmed by HypoDB screening. The protein SARS-CoV M(pro) predicted by C3 and C5 was an attractive target for structure-based drug design of anti-SARS drugs owing to its indispensability for the maturation of severe acute respiratory syndrome coronavirus (SARS-CoV) [100]. Another target, HS5B Pol, fished by five alkaloids was a target for anti-HCV therapeutic advances [101]. Inhibitors of HS5B Pol would be a principal option for the treatment of HCV [102]. Meanwhile, scytalone dehydratase and negative transcriptional regulator NmrA were suggested to be physiological targets of new fungicides and the subjects of inhibitor design and optimization [103–105].
In this paper, we proposed a very wide range of the promising targets for the isoquinoline alkaloids of M. cordata. Most of the hits are not yet proven by pharmacological experiment.
Conclusion
Through in silicotarget fishing, the anticancer, anti-inflammatory, and analgesic effects of M. cordata were the most significant among many possible activities. The possible anticancer effects were mainly contributed by the isoquinoline alkaloids as active components.
Authors’ contributions
HBL, QFL, PGX and YP conceived and designed the study. HBL, QFL and PGX performed the experiments. HBL, QFL and YP wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
The authors would like to thank Prof. Jun Xu (Sun Yat-sen University, China) and Prof. Yanze Liu (IMPLAD, China) for assistance in preparing the manuscript and Dr. Rong Zhao (National Yang-Ming University, Taiwan) for assistance in analyzing the pathway. This work was supported by National Natural Science Foundation of China (Grant No. 81072995), and Peking Union Medical College Youth Fund (Grant No. 3332013079).
Competing interests
The authors declare that they have no competing interests.
Abbreviations
- CHARMM
chemistry at Harvard Macromolecular Mechanics
- MOE
molecular operating environment
- RMSD
root mean square deviation
- MMFF
Merck molecular force field
- PDB
Protein Data Bank
- KEGG
Kyoto Encyclopedia of genes and genomes
- TTD
Therapeutic Target Database
- HTML
hypertext markup language
Contributor Information
Qifang Lei, Email: leiqifang66@163.com.
Haibo Liu, Email: hbliu@implad.ac.cn.
Yong Peng, Email: ypeng@implad.ac.cn.
Peigen Xiao, Email: xiaopg@public.bta.net.cn.
References
- 1.Xu GJ, Wang Q, Yu BM. Color illustrations of antitumor traditional Chinese medicine. Fuzhou: Fujian Sci Technol Publ House; 1997. p. 759.
- 2.Grieve M. A modern herbal. Middlesex: Penguin Books; 1984.
- 3.Duke JA, Ayenus ES. Medicinal plants of China. Inc. Algonac: Reference Publications; 1984.
- 4.Psotova J, Vecera R, Zdarilova A, Anzenbacherova E, Kosina P, Svobodova A, Hrbac J. Safety assessment of sanguiritrin, alkaloid fraction of Macleaya cordata, in rats. Vet Med-Czech. 2006;51(4):145–155. [Google Scholar]
- 5.Pang JX, Ma RQ, Liu LM, Jiang YP, Sun LS. In vitro cytotoxic effect on Hep3B cells and in vivo antitumor effect in mice. J First Mil Med Univ. 2005;25(3):325–328. [PubMed] [Google Scholar]
- 6.Yang S, Liu Y, Yang QF, Xiang JF, Tang YL, Xu GZ. Antitumor effect of Macleaya cordata and its molecular mechanism on inducement of human telomeric DNA to form G-quadruplex. Chin Trad Herb Drugs. 2011;42(4):738–742. [Google Scholar]
- 7.Pang FG. Study on the Anticancer Constituents of Macleaya cordata (Willd) R.Br. Master Thesis. Shenyang Pharmaceutical University; 2005.
- 8.Kemeny-Beke A, Aradi J, Damjanovich J, Beck Z, Facsko A, Berta A, Bodnar A. Apoptotic response of uveal melanoma cells upon treatment with chelidonine, sanguinarine and chelerythrine. Cancer Lett. 2006;237(1):67–75. doi: 10.1016/j.canlet.2005.05.037. [DOI] [PubMed] [Google Scholar]
- 9.Lenfeld J, Kroutil M, Marsálek E, Slavik J, Preininger V, Simánek V. Antiinflammatory activity of quaternary benzophenanthridine alkaloids from Chelidonium majus. Planta Med. 1981;43(2):161–165. doi: 10.1055/s-2007-971493. [DOI] [PubMed] [Google Scholar]
- 10.Park JE, Cuong TD, Hung TM, Lee I, Na M, Kim JC, Ryoo S, Lee JH, Choi JS, Woo MH. Alkaloids from Chelidonium majus and their inhibitory effects on LPS-induced NO production in RAW264.7 cells. Bioorg Med Chem Lett. 2011;21:6960–6963. doi: 10.1016/j.bmcl.2011.09.128. [DOI] [PubMed] [Google Scholar]
- 11.Xiao L, Yi J, Zhao J, Xu L, Liu BY, Liu DM, Zeng JG. Protective effect of Macleaya cordata extract on alcohol-induced acute hepatic injury in rats. Cent South Pharm. 2011;9(7):485–489. [Google Scholar]
- 12.Kosina P, Gregorova J, Gruz J, Vacek J, Kolar M, Vogel M, Roos W, Naumann K, Simanek V, Ulrichova J. Phytochemical and antimicrobial characterization of Macleaya cordata herb. Fitoterapia. 2010;81:1006–1012. doi: 10.1016/j.fitote.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 13.Cheng RB, Chen X, Liu SJ, Zhang GH. Effect of Chelerythrine on glucosyltransferase and water-insoluble glucan of Streptococcus mutans. Shanghai J Stomatol. 2007;16(3):324–327. [PubMed] [Google Scholar]
- 14.Yu JP, Zhao DL, Meng XB, Zhou XQ. The antibacterial effect of the alkaloids from Macleaya cordata on eight kinds of fungi. J Mount Agri Biol. 2006;25(1):89–91. [Google Scholar]
- 15.Hiller KO, Ghorbani M, Schilcher H. Antispasmodic and relaxant activity of chelidonine, protopine, coptisine, and Chelidonium majus extracts on isolated guinea-pig ileum. Planta Med. 1998;64(8):758–760. doi: 10.1055/s-2006-957576. [DOI] [PubMed] [Google Scholar]
- 16.Vrba J, Orolinova E, Ulrichova J. Induction of heme oxygenase-1 by Macleaya cordata extract and its constituent sanguinarine in RAW264.7 cells. Fitoterapia. 2012;83(2):329–335. doi: 10.1016/j.fitote.2011.11.022. [DOI] [PubMed] [Google Scholar]
- 17.Lei QF, Zhao XL, Xu LJ, Peng Y, Xiao PG. Chemical constituents of plants from tribe Chelidonieae and their bioactivities. Chin Herb Med. 2014;6(1):1–21. doi: 10.1016/S1674-6384(14)60001-0. [DOI] [Google Scholar]
- 18.Liu LJ, Leung KH, Lin S, Chan DS, Susanti D, Rao W, Chan PW, Ma DL, Leung CH. Pharmacophore modeling for the identification of small-molecule inhibitors of TACE. Methods. 2015;71(1):92–97. doi: 10.1016/j.ymeth.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 19.Zhong HJ, Liu LJ, Chong CM, Lu L, Wang M, Chan DS, Chan PW, Lee SM, Ma DL, Leung CH. Discovery of a natural product-like iNOS inhibitor by molecular docking with potential neuroprotective effects in vivo. PLoS One. 2014;9(4):e92905. doi: 10.1371/journal.pone.0092905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma DL, Chan DS, Wei G, Zhong HJ, Yang H, Leung LT, Gullen EA, Chiu P, Cheng YC, Leung CH. Virtual screening and optimization of Type II inhibitors of JAK2 from a natural product library. Chem Commun (Camb) 2014;50(90):13885–13888. doi: 10.1039/C4CC04498C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Gao Z, Kang L, Zhang H, Yang K, Yu K, Luo X, Zhu W, Chen K, Shen J. TarFisDock: a web server for identifying drug targets with docking approach. Nucleic Acids Res. 2006;34(suppl 2):W219–W224. doi: 10.1093/nar/gkl114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schuster D. 3D pharmacophores as tools for activity profiling. Drug Discov Today Tech. 2010;7(4):e205–e211. doi: 10.1016/j.ddtec.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 23.Steindl TM, Schuster D, Laggner C, Langer T. Parallel screening: a novel concept in pharmacophore modelling and virtual screening. J Chem Inf Model. 2006;46(5):2146–2157. doi: 10.1021/ci6002043. [DOI] [PubMed] [Google Scholar]
- 24.Steindl TM, Schuster D, Wolber G, Laggner C, Langer T. High throughput structure-based pharmacophore modeling as a basis for successful parallel virtual screening. J Comput Aided Mol Des. 2006;20(12):703–715. doi: 10.1007/s10822-006-9066-y. [DOI] [PubMed] [Google Scholar]
- 25.Meslamani J, Li J, Sutter J, Stevens A, Bertrand HO, Rognan D. Protein–ligand-based pharmacophores: generation and utility assessment in computational ligand profiling. J Chem Inf Model. 2012;52(4):943–955. doi: 10.1021/ci300083r. [DOI] [PubMed] [Google Scholar]
- 26.Meslamani J, Rognan D, Kellenberger E. sc-PDB: a database for identifying variations and multiplicity of ‘druggable’ binding sites in proteins. Bioinformatics. 2011;27(9):1324–1326. doi: 10.1093/bioinformatics/btr120. [DOI] [PubMed] [Google Scholar]
- 27.Zhu F, Shi Z, Qin C, Tao L, Liu X, Xu F, Zhang L, Song Y, Liu X, Zhang J. Therapeutic target database update 2012: a resource for facilitating target-oriented drug discovery. Nucleic Acids Res. 2012;40(D1):D1128–D1136. doi: 10.1093/nar/gkr797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knox C, Law V, Jewison T, Liu P, Ly S, Frolkis A, Pon A, Banco K, Mak C, Neveu V. DrugBank 3.0: a comprehensive resource for ‘omics’ research on drugs. Nucleic Acids Res. 2011;39(suppl 1):D1035–D1041. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27(3):431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ehrman TM, Barlow DJ, Hylands PJ. In silico search for multi-target anti-inflammatories in Chinese herbs and formulas. Bioorg Med Chem. 2010;18(6):2204–2218. doi: 10.1016/j.bmc.2010.01.070. [DOI] [PubMed] [Google Scholar]
- 31.Li B, Xu X, Wang X, Yu H, Li X, Tao W, Wang Y, Yang L. A systems biology approach to understanding the mechanisms of action of Chinese herbs for treatment of cardiovascular disease. Int J Mol Sci. 2012;13(10):13501–13520. doi: 10.3390/ijms131013501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X, Xu X, Wang J, Yu H, Wang X, Yang H, Xu H, Tang S, Li Y, Yang L. A system-level investigation into the mechanisms of Chinese Traditional Medicine: compound Danshen formula for cardiovascular disease treatment. PLoS One. 2012;7(9):e43918. doi: 10.1371/journal.pone.0043918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao DL, Yu JP, Zhou XQ. Antibacterial effect of the Sanguinarine Hydrochloride and Bocconoline from Macleaya cordata. Food Sci. 2005;26(1):45–47. [Google Scholar]
- 34.Fibriansah G, Kovacs AT, Pool TJ, Boonstra M, Kuipers OP, Thunnissen AM. Crystal structures of two transcriptional regulators from Bacillus cereus define the conserved structural features of a PadR subfamily. PLoS One. 2012;7(11):e48015. doi: 10.1371/journal.pone.0048015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agustiandari H, Peeters E, de Wit JG, Charlier D, Driessen AJ. LmrR-mediated gene regulation of multidrug resistance in Lactococcus lactis. Microbiology. 2011;157(Pt 5):1519–1530. doi: 10.1099/mic.0.048025-0. [DOI] [PubMed] [Google Scholar]
- 36.Salverda ML, De Visser JA, Barlow M. Natural evolution of TEM-1 beta-lactamase: experimental reconstruction and clinical relevance. FEMS Microbiol Rev. 2010;34(6):1015–1036. doi: 10.1111/j.1574-6976.2010.00222.x. [DOI] [PubMed] [Google Scholar]
- 37.Ehmann DE, Jahic H, Ross PL, Gu RF, Hu J, Kern G, Walkup GK, Fisher SL. Avibactam is a covalent, reversible, non-beta-lactam beta-lactamase inhibitor. Proc Natl Acad Sci USA. 2012;109(29):11663–11668. doi: 10.1073/pnas.1205073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shaw WV. Chloramphenicol acetyltransferase: enzymology and molecular biology. CRC Crit Rev Biochem. 1983;14(1):1–46. doi: 10.3109/10409238309102789. [DOI] [PubMed] [Google Scholar]
- 39.Catalano-Dupuy DL, Lopez-Rivero A, Soldano A, Ceccarelli EA. Redox proteins as targets for drugs development against pathogens. Curr Pharm Des. 2013;19(14):2594–2605. doi: 10.2174/1381612811319140009. [DOI] [PubMed] [Google Scholar]
- 40.Willems AR, Tahlan K, Taguchi T, Zhang K, Lee ZZ, Ichinose K, Junop MS, Nodwell JR. Crystal structures of the Streptomyces coelicolorTetR-like protein ActR alone and in complex with actinorhodin or the actinorhodin biosynthetic precursor (S)-DNPA. J Mol Biol. 2008;376(5):1377–1387. doi: 10.1016/j.jmb.2007.12.061. [DOI] [PubMed] [Google Scholar]
- 41.Tsao DH, Sutherland AG, Jennings LD, Li Y, Rush TR, Alvarez JC, Ding W, Dushin EG, Dushin RG, Haney SA. Discovery of novel inhibitors of the ZipA/FtsZ complex by NMR fragment screening coupled with structure-based design. Bioorg Med Chem. 2006;14(23):7953–7961. doi: 10.1016/j.bmc.2006.07.050. [DOI] [PubMed] [Google Scholar]
- 42.Ling LL, Xian J, Ali S, Geng B, Fan J, Mills DM, Arvanites AC, Orgueira H, Ashwell MA, Carmel G. Identification and characterization of inhibitors of bacterial enoyl-acyl carrier protein reductase. Antimicrob Agents Chemother. 2004;48(5):1541–1547. doi: 10.1128/AAC.48.5.1541-1547.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yao JY, Shen JY, Li XL, Xu Y, Hao GJ, Pan XY, Wang GX, Yin WL. Effect of sanguinarine from the leaves of Macleaya cordata against Ichthyophthirius multifiliis in grass carp (Ctenopharyngodon idella) Parasitol Res. 2010;107(5):1035–1042. doi: 10.1007/s00436-010-1966-z. [DOI] [PubMed] [Google Scholar]
- 44.Yao JY, Zhou ZM, Li XL, Yin WL, Ru HS, Pan XY, Hao GJ, Xu Y, Shen JY. Antiparasitic efficacy of dihydrosanguinarine and dihydrochelerythrine from Macleaya microcarpa against Ichthyophthirius multifiliis in richadsin (Squaliobarbus curriculus) Vet Parasitol. 2011;183(1–2):8–13. doi: 10.1016/j.vetpar.2011.07.021. [DOI] [PubMed] [Google Scholar]
- 45.Wang GX, Zhou Z, Jiang DX, Han J, Wang JF, Zhao LW, Li J. In vivo anthelmintic activity of five alkaloids from Macleaya microcarpa (Maxim) Fedde against Dactylogyru sintermedius in Carassiusauratus. Vet Parasitol. 2010;171(3–4):305–313. doi: 10.1016/j.vetpar.2010.03.032. [DOI] [PubMed] [Google Scholar]
- 46.Juskiewicz J, Gruzauskas R, Zdunczyk Z, Semaskaite A, Jankowski J, Totilas Z, Jarule V, Sasyte V, Zdunczyk P. Raceviciute-StupelieneA: effects of dietary addition of Macleaya cordata alkaloid extract on growth performance, caecal indices and breast meat fatty acids profile in male broilers. J Anim Physiol Anim Nutr (Berl) 2011;95(2):171–178. doi: 10.1111/j.1439-0396.2010.01037.x. [DOI] [PubMed] [Google Scholar]
- 47.Sarma GN, Savvides SN, Becker K, Schirmer M, Schirmer RH, Karplus PA. Glutathione reductase of the malarial parasite Plasmodium falciparum: crystal structure and inhibitor development. J Mol Biol. 2003;328(4):893–907. doi: 10.1016/S0022-2836(03)00347-4. [DOI] [PubMed] [Google Scholar]
- 48.Leitão A, Andricopulo AD, Oliva G, Pupo MT, de Marchi AA, Vieira PC, da Silva MF, Ferreira VF, de Souza MC, Sá MM, Moraes VR, Montanari CA. Structure–activity relationships of novel inhibitors of glyceraldehyde-3-phosphate dehydrogenase. Bioorg Med Chem Lett. 2004;14(9):2199–2204. doi: 10.1016/j.bmcl.2004.02.025. [DOI] [PubMed] [Google Scholar]
- 49.Pinheiro MP, Emery FS, Nonato MC. Target sites for the design of anti-trypanosomatid drugs based on the structure of dihydroorotatedehydrogenase. Curr Pharm Des. 2013;19(14):2615–2627. doi: 10.2174/1381612811319140011. [DOI] [PubMed] [Google Scholar]
- 50.Cai YT, Ju CY, Yan HR, Sun LQ. Experimental research on analgesic effect of the total alkaloid from Celandine. J Mudanjiang Med Univ. 2011;32(6):29–30. [Google Scholar]
- 51.Freitas K, Carroll FI, Damaj MI. The antinociceptive effects of nicotinic receptors alpha7-positive allosteric modulators in murine acute and tonic pain models. J Pharmacol Exp Ther. 2013;344(1):264–275. doi: 10.1124/jpet.112.197871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Medicherla S, Reddy M, Ying J, Navas TA, Li L, Nguyen AN, Kerr I, Hanjarappa N, Protter AA, Higgins LS. p38alpha-selective MAP kinase inhibitor reduces tumor growth in mouse xenograft models of multiple myeloma. Anticancer Res. 2008;28(6A):3827–3833. [PubMed] [Google Scholar]
- 53.Laufer S, Lehmann F. Investigations of SCIO-469-like compounds for the inhibition of p38 MAP kinase. Bioorg Med Chem Lett. 2009;19(5):1461–1464. doi: 10.1016/j.bmcl.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 54.Steuber H, Heine A, Klebe G. Structural and thermodynamic study on aldose reductase: nitro-substituted inhibitors with strong enthalpic binding contribution. J Mol Biol. 2007;368(3):618–638. doi: 10.1016/j.jmb.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 55.Suryanarayana P, Kumar PA, Saraswat M, Petrash JM, Reddy GB. Inhibition of aldose reductase by tannoid principles of Emblica officinalis: implications for the prevention of sugar cataract. Mol Vis. 2004;10:148–154. [PubMed] [Google Scholar]
- 56.Young RJ, Ewing DJ, Clarke BF. A controlled trial of sorbinil, an aldose reductase inhibitor, in chronic painful diabetic neuropathy. Diabetes. 1983;32(10):938–942. doi: 10.2337/diab.32.10.938. [DOI] [PubMed] [Google Scholar]
- 57.Wymann MP, Solinas G. Inhibition of phosphoinositide 3-kinase gamma attenuates inflammation, obesity, and cardiovascular risk factors. Ann N Y Acad Sci. 2013;1280:44–47. doi: 10.1111/nyas.12037. [DOI] [PubMed] [Google Scholar]
- 58.Dufour A, Overall CM. Missing the target: matrix metalloproteinase antitargets in inflammation and cancer. Trends Pharmacol Sci. 2013;34(4):233–242. doi: 10.1016/j.tips.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 59.Mukhopadhyay S, Sypek J, Tavendale R, Gartner U, Winter J, Li W, Page K, Fleming M, Brady J, O’Toole M. Matrix metalloproteinase-12 is a therapeutic target for asthma in children and young adults. J Allergy Clin Immunol. 2010;126(1):70–76. doi: 10.1016/j.jaci.2010.03.027. [DOI] [PubMed] [Google Scholar]
- 60.Clark RB. The role of PPARs in inflammation and immunity. J Leukoc Biol. 2002;71(3):388–400. [PubMed] [Google Scholar]
- 61.Maubach G, Sokolova O, Wolfien M, Rothkotter HJ, Naumann M. Ca/calmodulin-dependent kinase II contributes to inhibitor of nuclear factor-kappa B kinase complex activation in Helicobacter pylori infection. Int J Cancer. 2013;133(6):1507–1512. doi: 10.1002/ijc.28148. [DOI] [PubMed] [Google Scholar]
- 62.Mahalka AK, Kinnunen PK. Class specific peptide inhibitors for secretory phospholipases A2. Biochem Biophys Res Commun. 2013;436(2):349–353. doi: 10.1016/j.bbrc.2013.05.110. [DOI] [PubMed] [Google Scholar]
- 63.Usher MG, Duan SZ, Ivaschenko CY, Frieler RA, Berger S, Schutz G, Lumeng CN, Mortensen RM. Myeloid mineralocorticoid receptor controls macrophage polarization and cardiovascular hypertrophy and remodeling in mice. J Clin Invest. 2010;120(9):3350–3364. doi: 10.1172/JCI41080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu L, Li Y, Sun H, Zhen X, Qiao C, Tian S, Hou T. Current developments of macrophage migration inhibitory factor (MIF) inhibitors. Drug Discov Today. 2013;18(11–12):592–600. doi: 10.1016/j.drudis.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 65.Dubal DB, Zhu H, Yu J, Rau SW, Shughrue PJ, Merchenthaler I, Kindy MS, Wise PM. Estrogen receptor alpha, not beta, is a critical link in estradiol-mediated protection against brain injury. Proc Natl Acad Sci USA. 2001;98(4):1952–1957. doi: 10.1073/pnas.041483198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Karas RH, Hodgin JB, Kwoun M, Krege JH, Aronovitz M, Mackey W, Gustafsson JA, Korach KS, Smithies O, Mendelsohn ME. Estrogen inhibits the vascular injury response in estrogen receptor beta-deficient female mice. Proc Natl Acad Sci USA. 1999;96(26):15133–15136. doi: 10.1073/pnas.96.26.15133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Macleod MR, Johansson IM, Soderstrom I, Lai M, Gido G, Wieloch T, Seckl JR, Olsson T. Mineralocorticoid receptor expression and increased survival following neuronal injury. Eur J Neurosci. 2003;17(8):1549–1555. doi: 10.1046/j.1460-9568.2003.02587.x. [DOI] [PubMed] [Google Scholar]
- 68.Smith RK, Heinegard D. Cartilage oligomeric matrix protein (COMP) levels in digital sheath synovial fluid and serum with tendon injury. Equine Vet J. 2000;32(1):52–58. doi: 10.2746/042516400777612053. [DOI] [PubMed] [Google Scholar]
- 69.Posey KL, Hecht JT. The role of cartilage oligomeric matrix protein (COMP) in skeletal disease. Curr Drug Targets. 2008;9(10):869–877. doi: 10.2174/138945008785909293. [DOI] [PubMed] [Google Scholar]
- 70.Fan SL, Jiao F, Zhang Y, An CX, Fu JM. Study on effect of total alkaloids of Macleaya cordata to animal’s transplanted tumor. Shanxi Onco Med. 2000;8(3):174–175. [Google Scholar]
- 71.Hohenwarter O, Strutzenberger K, Katinger H, Liepins A, Nowicky JW. Selective inhibition of in vitro cell growth by the anti-tumour drug Ukrain. Drugs Exp Clin Res. 1992;18:S1–S4. [PubMed] [Google Scholar]
- 72.Cordes N, Plasswilm L, Bamberg M, Rodemann HP. Ukrain, an alkaloid thiophosphoric acid derivative of Chelidonium majus L. protects human fibroblasts but not human tumour cells in vitro against ionizing radiation. Int J Radiat Biol. 2002;78(1):17–27. doi: 10.1080/09553000110089991. [DOI] [PubMed] [Google Scholar]
- 73.Li H, Zhai Z, Liu G, Tang T, Lin Z, Zheng M, Qin A, Dai K. Sanguinarine inhibits osteoclast formation and bone resorption via suppressing RANKL-induced activation of NF-kappaB and ERK signaling pathways. Biochem Biophys Res Commun. 2013;430(3):951–956. doi: 10.1016/j.bbrc.2012.12.051. [DOI] [PubMed] [Google Scholar]
- 74.Ansari KM, Das M. Skin tumor promotion by argemone oil/alkaloid in mice: evidence for enhanced cell proliferation, ornithine decarboxylase, cyclooxygenase-2 and activation of MAPK/NF-kappaB pathway. Food Chem Toxicol. 2010;48(1):132–138. doi: 10.1016/j.fct.2009.09.029. [DOI] [PubMed] [Google Scholar]
- 75.Lee JS, Jung WK, Jeong MH, Yoon TR, Kim HK. Sanguinarine induces apoptosis of HT-29 human colon cancer cells via the regulation of Bax/Bcl-2 ratio and caspase-9-dependent pathway. Int J Toxicol. 2012;31(1):70–77. doi: 10.1177/1091581811423845. [DOI] [PubMed] [Google Scholar]
- 76.Choi WY, Kim GY, Lee WH, Choi YH. Sanguinarine, a benzophenanthridine alkaloid, induces apoptosis in MDA-MB-231 human breast carcinoma cells through a reactive oxygen species-mediated mitochondrial pathway. Chemotherapy. 2008;54(4):279–287. doi: 10.1159/000149719. [DOI] [PubMed] [Google Scholar]
- 77.Vrba J, Orolinova E, Ulrichova J. Induction of heme oxygenase-1 by Macleaya cordata extract and its constituent sanguinarine in RAW264.7 cells. Fitoterapia. 2012;83(2):329–335. doi: 10.1016/j.fitote.2011.11.022. [DOI] [PubMed] [Google Scholar]
- 78.Basini G, Santini SE, Bussolati S, Grasselli F. Sanguinarine inhibits VEGF-induced Akt phosphorylation. Ann N Y Acad Sci. 2007;1095:371–376. doi: 10.1196/annals.1397.040. [DOI] [PubMed] [Google Scholar]
- 79.Hussain F, Freissmuth M, Volkel D, Thiele M, Douillard P, Antoine G, Thurner P, Ehrlich H, Schwarz HP, Scheiflinger F. Human anti-macrophage migration inhibitory factor (MIF) antibodies inhibit growth of human prostate cancer cells in vitro and in vivo. Mol Cancer Ther. 2013;12(7):1223–1234. doi: 10.1158/1535-7163.MCT-12-0988. [DOI] [PubMed] [Google Scholar]
- 80.Guo Y, Hou J, Luo Y, Wang D. Functional disruption of macrophage migration inhibitory factor (MIF) suppresses proliferation of human H460 lung cancer cells by caspase-dependent apoptosis. Cancer Cell Int. 2013;13(1):28–36. doi: 10.1186/1475-2867-13-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wadgaonkar R, Somnay K, Garcia JG. Thrombin induced secretion of macrophage migration inhibitory factor (MIF) and its effect on nuclear signaling in endothelium. J Cell Biochem. 2008;105(5):1279–1288. doi: 10.1002/jcb.21928. [DOI] [PubMed] [Google Scholar]
- 82.Sun B, Nishihira J, Suzuki M, Fukushima N, Ishibashi T, Kondo M, Sato Y, Todo S. Induction of macrophage migration inhibitory factor by lysophosphatidic acid: relevance to tumor growth and angiogenesis. Int J Mol Med. 2003;12(4):633–641. [PubMed] [Google Scholar]
- 83.Chuang YC, Su WH, Lei HY, Lin YS, Liu HS, Chang CP, Yeh TM. Macrophage migration inhibitory factor induces autophagy via reactive oxygen species generation. PLoS One. 2012;7(5):e37613. doi: 10.1371/journal.pone.0037613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mathew B, Jacobson JR, Siegler JH, Moitra J, Blasco M, Xie L, Unzueta C, Zhou T, Evenoski C, Al-Sakka M. Role of migratory inhibition factor in age-related susceptibility to radiation lung injury via NF-E2-related factor-2 and antioxidant regulation. Am J Respir Cell Mol Biol. 2013;49(2):269–278. doi: 10.1165/rcmb.2012-0291OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu L, Sun S, Liu J, Wu S, Dai S, Wang X, Huang L, Xiao X, He D. A new serum biomarker for lung cancer—transthyretin. Zhongguo Fei Ai Za Zhi. 2009;12(4):300–305. doi: 10.3779/j.issn.1009-3419.2009.04.08. [DOI] [PubMed] [Google Scholar]
- 86.Chen J, Chen LJ, Xia YL, Zhou HC, Yang RB, Wu W, Lu Y, Hu LW, Zhao Y. Identification and verification of transthyretin as a potential biomarker for pancreatic ductal adenocarcinoma. J Cancer Res Clin Oncol. 2013;139(7):1117–1127. doi: 10.1007/s00432-013-1422-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Blanco-Aparicio C, Carnero A. Pim kinases in cancer: diagnostic, prognostic and treatment opportunities. Biochem Pharmacol. 2013;85(5):629–643. doi: 10.1016/j.bcp.2012.09.018. [DOI] [PubMed] [Google Scholar]
- 88.Pan J, Fukuda K, Saito M, Matsuzaki J, Kodama H, Sano M, Takahashi T, Kato T, Ogawa S. Mechanical stretch activates the JAK/STAT pathway in rat cardiomyocytes. Circ Res. 1999;84(10):1127–1136. doi: 10.1161/01.RES.84.10.1127. [DOI] [PubMed] [Google Scholar]
- 89.Pencikova K, Urbanova J, Musil P, Taborska E, Gregorova J. Seasonal variation of bioactive alkaloid contents in Macleaya microcarpa (Maxim.) Fedde. Molecules. 2011;16(4):3391–3401. doi: 10.3390/molecules16043391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang HH, Wu Y, Sun ZL, Liu ZY. Identification of sanguinarine metabolites in pig liver preparations by accurate mass measurements using electrospray ionization hybrid ion trap/time-of-flight mass spectrometry. Rapid Commun Mass Spectrum. 2013;27(9):979–984. doi: 10.1002/rcm.6538. [DOI] [PubMed] [Google Scholar]
- 91.Fang L, Gou S, Fang X, Cheng L, Fleck C. Current progresses of novel natural products and their derivatives/analogs as anti-Alzheimer candidates: an update. Mini Rev Med Chem. 2013;13(6):870–887. doi: 10.2174/1389557511313060009. [DOI] [PubMed] [Google Scholar]
- 92.Greenfield S, Vaux DJ. Parkinson’s disease, Alzheimer’s disease and motor neurone disease: identifying a common mechanism. Neurosci. 2002;113(3):485–492. doi: 10.1016/S0306-4522(02)00194-X. [DOI] [PubMed] [Google Scholar]
- 93.Huang L, Lu C, Sun Y, Mao F, Luo Z, Su T, Jiang H, Shan W, Li X. Multitarget-directed benzylideneindanone derivatives: anti-beta-amyloid (Abeta) aggregation, antioxidant, metal chelation, and monoamine oxidase B (MAO-B) inhibition properties against Alzheimer’s disease. J Med Chem. 2012;55(19):8483–8492. doi: 10.1021/jm300978h. [DOI] [PubMed] [Google Scholar]
- 94.DeMarcaida JA, Schwid SR, White WB, Blindauer K, Fahn S, Kieburtz K, Stern M, Shoulson I. Effects of tyramine administration in Parkinson’s disease patients treated with selective MAO-B inhibitor rasagiline. Mov Disord. 2006;21(10):1716–1721. doi: 10.1002/mds.21048. [DOI] [PubMed] [Google Scholar]
- 95.Boyer PL, Clark PK, Hughes SH. HIV-1 and HIV-2 reverse transcriptases: different mechanisms of resistance to nucleoside reverse transcriptase inhibitors. J Virol. 2012;86(10):5885–5894. doi: 10.1128/JVI.06597-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Klarmann GJ, Hawkins ME, Le Grice SF. Uncovering the complexities of retroviral ribonuclease H reveals its potential as a therapeutic target. AIDS Rev. 2002;4(4):183–194. [PubMed] [Google Scholar]
- 97.Ambrose Z, Herman BD, Sheen CW, Zelina S, Moore KL, Tachedjian G, Nissley DV, Sluis-Cremer N. The human immunodeficiency virus type 1 nonnucleoside reverse transcriptase inhibitor resistance mutation I132M confers hypersensitivity to nucleoside analogs. J Virol. 2009;83(8):3826–3833. doi: 10.1128/JVI.01968-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sadiq SK, Noe F, De Fabritiis G. Kinetic characterization of the critical step in HIV-1 protease maturation. Proc Natl Acad Sci USA. 2012;109(50):20449–20454. doi: 10.1073/pnas.1210983109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Martin P, Vickrey JF, Proteasa G, Jimenez YL, Wawrzak Z, Winters MA, Merigan TC, Kovari LC. “Wide-open” 1.3 A structure of a multidrug-resistant HIV-1 protease as a drug target. Structure. 2005;13(12):1887–1895. doi: 10.1016/j.str.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 100.Lu IL, Mahindroo N, Liang PH, Peng YH, Kuo CJ, Tsai KC, Hsieh HP, Chao YS, Wu SY. Structure-based drug design and structural biology study of novel nonpeptide inhibitors of severe acute respiratory syndrome coronavirus main protease. J Med Chem. 2006;49(17):5154–5161. doi: 10.1021/jm060207o. [DOI] [PubMed] [Google Scholar]
- 101.Patel BA, Krishnan R, Khadtare N, Gurukumar KR, Basu A, Arora P, Bhatt A, Patel MR, Dana D, Kumar S. Design and synthesis of ʟ- and d-phenylalanine derived rhodanines with novel C5-arylidenes as inhibitors of HCV NS5B polymerase. Bioorg Med Chem. 2013;21(11):3262–3671. doi: 10.1016/j.bmc.2013.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Varshney J, Sharma PK, Sharma A. A review on an update of NS5B polymerase hepatitis C virus inhibitors. Eur Rev Med Pharmacol Sci. 2012;16(5):667–671. [PubMed] [Google Scholar]
- 103.Jordan DB, Basarab GS, Steffens JJ, Schwartz RS, Doughty JG. Tight binding inhibitors of scytalonedehydratase: effects of site-directed mutations. Biochemistry-Us. 2000;39(29):8593–8602. doi: 10.1021/bi000467m. [DOI] [PubMed] [Google Scholar]
- 104.Stammers DK, Ren J, Leslie K, Nichols CE, Lamb HK, Cocklin S, Dodds A, Hawkins AR. The structure of the negative transcriptional regulator NmrA reveals a structural superfamily which includes the short-chain dehydrogenase/reductases. EMBO J. 2001;20(23):6619–6626. doi: 10.1093/emboj/20.23.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhao X, Hume SL, Johnson C, Thompson P, Huang J, Gray J, Lamb HK, Hawkins AR. The transcription repressor NmrA is subject to proteolysis by three Aspergillus nidulans proteases. Protein Sci. 2010;19(7):1405–1419. doi: 10.1002/pro.421. [DOI] [PMC free article] [PubMed] [Google Scholar]


