Abstract
Background
Mounting evidence indicates that the indigenous gut microbiota exerts long-lasting programming effects on brain function and behaviour.
Objective
In this study, we used the germ-free (GF) mouse model, devoid of any microbiota throughout development, to assess the influence of the indigenous microbiota on social preference and repetitive behaviours (e.g. self-grooming).
Methods and results
Using the three-chambered social approach task, we demonstrate that when adult GF mice were given a choice to spend time with a novel mouse or object, they spent significantly more time sniffing and interacting with the stimulus mouse compared to conventionally raised mice (specific pathogen-free, SPF). Time spent in repetitive self-grooming behaviour, however, did not differ between GF and SPF mice. Real-time PCR–based gene expression analysis of the amygdala, a key region that is part of the social brain network, revealed a significant reduction in the mRNA levels of total brain-derived neurotrophic factor (BDNF), BDNF exon I-, IV-, VI-, IX-containing transcripts, and NGFI-A (a signalling molecule downstream of BDNF) in GF mice compared to SPF mice.
Conclusion
These results suggest that differential regulation of BDNF exon transcripts in the amygdala by the indigenous microbes may contribute to the altered social development of GF mice.
Keywords: brain development, germ-free mice, gene expression, synaptic plasticity genes, amygdala, BDNF
It is now increasingly recognised that indigenous gut microbiota exerts an influence on brain development and behaviour. A series of recent studies have demonstrated that mice raised under germ-free (GF) conditions display decreased anxiety-like behaviour compared to conventionally raised mice, suggesting a role of the gut microbiota in emotional behaviour (1–3). Other studies have demonstrated increased motor activity (2) and memory impairments in GF mice (4). In addition, GF mice display altered physiology (e.g. enhanced hypothalamic-pituitary-adrenal axis response to stress), neurochemistry (e.g. increased monoamine neurotransmission in striatum and hippocampus), and gene expression (e.g. genes involved in neurotransmission, synaptic plasticity, and metabolism), among others (1–5). The molecular mechanisms underlying the effects of the gut microbiota on brain development and function are not fully understood. One potential neuronal target that may mediate central effects of the gut microbiota is brain-derived neurotrophic factor (BDNF), a member of the neurotrophin family. BDNF plays an important role in the formation and modulation of brain circuits through its influence on neuronal survival, differentiation, synapse formation, and plasticity (6, 7). In the adult brain, BDNF regulates synaptic transmission and plasticity, and therefore is fundamental to normal brain function. Moreover, a number of studies have shown that the expression of BDNF is highly sensitive to perturbations of the gut microbiota (1–3, 5, 8). Furthermore, several neurodevelopmental and psychiatric disorders (e.g. schizophrenia and mood disorders) that often co-occur with gastrointestinal (GI) problems have been linked to abnormalities in the BDNF signalling pathway (9, 10). Interestingly, microbial influence on brain activity in response to emotional challenge has also been reported in healthy humans (11). Clinical reports demonstrating alteration in the composition of the gut microbiota in neurodevelopmental and psychiatric disorders have lent further support to the theory that intestinal bacteria may play a role in the pathophysiology of human brain disorders (12). Autism spectrum disorder (ASD) is one such disorder; it has been associated with GI problems and an abnormal composition of the gut microbiota (13–15). The main core symptoms of ASD are impaired social communication and the presence of a restricted, repetitive pattern of behaviour or interest. Recently, Desbonnet et al. (16) reported impaired social behaviour and increased repetitive grooming in GF mice, thus suggesting a link between ASD symptoms and gut microbiota.
In the present study, we assessed the influence of gut microbiota on social approach behaviour using the three-chamber social approach task for mice, which has been widely used to characterise animal models of ASD (17). This task takes advantage of the fact that mice are highly social animals and have a natural tendency to approach and interact with unfamiliar conspecifics. In addition, we evaluated exploratory behaviour and repetitive self-grooming. Given that multiple promoters have been shown to regulate the rodent BDNF gene, we also examined the influence of gut microbiota on the levels of BDNF exon-specific transcripts in the amygdala, a key brain region involved in anxiety-like behaviour and processing of social stimuli.
Material and methods
Animals
Adult male GF and specific pathogen-free (SPF) Swiss-Webster mice (3 months old) were obtained from Taconic Farms, Inc. (Germantown, NY, USA). In order to avoid litter effects, it was requested that the animals originate from different mothers. All animals were shipped in guaranteed Taconic Germfree Shippers with free access to food and water. Upon arrival, GF mice were transferred to special sterile isolators and allowed to rest for 1 week before testing. Conventionally raised mice were also housed in similar sterile isolators to ensure similar environmental conditions for both GF and SPF mice. Within the isolators, animals were kept in standard sterilised plastic cages under controlled temperature, humidity, and light (12:12 h light–dark cycle) conditions, with free access to autoclaved water and food. All experiments were conducted in accordance with European Directive 86/609/EEC and Recommendation 2007/526/65/EC and were approved by the Animal Experimentation Ethics Committee, Stockholm North.
Behavioural studies
Testing took place between 9:00 and 16:00 h under low illumination to reduce stress. On the day of testing, animals were brought in sterile filtered cages to the testing room and allowed to rest for at least 1 h before testing. Test chambers were cleaned first with disinfectant and then with 70% ethanol and water after each animal.
Three-chamber social approach task
Social preference was evaluated in a three-chambered apparatus as previously described (18). The rectangular three-chambered box was made of clear Plexiglas (64 cm long, 43.7 cm wide, and 35 cm high) and was divided into three equally sized chambers (43.7 cm long, 20 cm wide, and 35 cm high). The walls dividing the chambers contained a small rectangular opening (10 cm wide and 6.5 cm high) to allow the test mouse access to the adjacent chambers. Clear rectangular Plexiglas doors were used to close the openings when required. The test session began with a 10-min habituation in the centre chamber without access to the side chambers, followed by another 10-min habituation session to the entire empty box, with access to all three chambers. The test mouse was then briefly confined to the centre chamber while the experimenter placed the stimuli. To test for social approach behaviours towards a live unfamiliar mouse (i.e. same age and sex, but different strain), a novel DBA/2NCrl mouse previously habituated to a grid enclosure (Noldus, Wageningen, The Netherlands) was placed in one of the side chambers. A control novel object, an identical empty grid enclosure devoid of social odours, was placed in the other side chamber. The location of the novel object and the novel mouse alternated between the left and right side chambers across subjects. After both stimuli were positioned in the side chambers, the two doorways were simultaneously opened and the test mouse was allowed access to all three chambers for 10 min. All sessions were recorded from the front of the three-chambered apparatus using a Samsung (Seoul, South Korea) HMX-H100P high-definition camcorder. The time the test mouse spent interacting with either the stimulus mouse or empty enclosure was analysed by three observers with stopwatches, as well as the time spent grooming all body regions. The social index (SI) was calculated as follows, as previously described (19): SI=(time exploring novel mouse)/(time exploring novel mouse+time exploring novel object).
Open-field test
Animals were placed individually in the centre of an open-field box (48 cm×48 cm; Acti-Mot detection system, TSE Systems, Bad Homburg, Germany), and their spontaneous motor activity was recorded as described (20). The computer programme automatically recorded the following parameters: distance travelled, a count of rearing activity (vertical infrared photo beam breaks), and time spent in the centre.
Quantitative real-time polymerase chain reaction
Total RNA from the amygdala (n=6 per group) was isolated using RNeasy® Mini Kits (QIAGEN AB, Sollentuna, Sweden) according to the manufacturer's instructions and quantified by spectrophotometry using a NanoDrop® ND-2000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). First-strand cDNA synthesis was carried out with equal amounts of total RNA (1 µg/reaction) using the iScript cDNA synthesis kit (Bio-Rad, Sundbyberg, Sweden) according to the manufacturer's instructions and stored at −20°C until used.
Quantitative real-time polymerase chain reaction (qRT-PCR) was carried out using the CFX384 Touch Real-Time PCR Detection System (Bio-Rad). Briefly, each PCR reaction contained 30 ng cDNA, 0.5 µM each of primer and nuclease-free water, and 5 µl iQ™ SYBR® Green Supermix (SYBR® Green I dye: Bio-Rad), 50 U/ml iTaq™ DNA polymerase (Bio-Rad), Deoxynucleotide triphosphates (dNTPs), 6 mM MgCl2, 100 mM KCl, 20 nM fluorescein including stabilisers, and 40 mM Tris–HCl (pH 8.4) in a 10-µl reaction. The housekeeping gene, glyceraldehyde three-phosphate dehydrogenase (GAPDH; gene ascension number NM_008084.2), was used for normalisation. All samples were performed in triplicate. The cycling programme was set as follows: Step 1, 98°C for 30 sec; Step 2, 40 cycles of 98°C for 5 sec, followed by 50–60°C for 15 sec; Step 3, 90 cycles for 5 sec each, beginning at 50°C and increasing by 0.5°C with each subsequent cycle. Subsequent to the amplification procedure, a melting curve analysis was performed (set point, 50°C) in order to confirm amplification specificity. The specificity of the gene products was determined via melting curve analyses. Primer sequences and annealing temperatures used for the total BDNF, BDNF exon-containing transcripts (I–IX), nerve growth factor-inducible clone A (NGFI-A), and GAPDH are listed in Table 1. The data analysis was based on the 2−ΔΔCt method (21). The normalised ΔCt for each gene of interest (GOI) was calculated by deducting the Ct of the reference gene GAPDH (as this gene showed no expression difference among the different RNA samples) from the Ct of each GOI. Then, the double delta Ct (ΔΔCt) for each GOI was calculated by deducting the average ΔCt of GOI in the SPF group from the ΔCt of each GOI in the GF group. The fold changes of each GOI compared with the SPF group were calculated as 2−ΔΔ Ct.
Table 1.
PCR primers used to assay gene expression
| Gene | Primer | Sequence | Accession number | Tm (°C) | Product size (bp) |
|---|---|---|---|---|---|
| BDNF | Forward | 5′-TGG CTG ACA CTT TTG AGC AC-3′ | NM_001048142 | 55 | 188 |
| Reverse | 5′-GTT TGC GGC ATC CAG GTA AT-3′ | ||||
| Exon I | Forward | 5′-TGA GAG TTG AAG CTT TGC GG-3′ | EF125669.1 | 55 | 189 |
| Reverse | 5′-ATT GTG GCT TTG CTG TCC TG-3′ | ||||
| Exon IIC | Forward | 5′-TTT GGT CCC CTC ATT GAG CT-3′ | EF125672.1 | 55 | 159 |
| Reverse | 5′-TTC TTT GCG GCT TAC ACC AC-3′ | ||||
| Exon III | Forward | 5′-TCT ATC ATC CCT CCC CGA GA-3′ | EF125681.1 | 55 | 123 |
| Reverse | 5′-AAC TGG GCT CAA GGA AGC AT-3′ | ||||
| Exon IV | Forward | 5′-AGC ATG AAA TCT CCC AGC CT-3′ | EF125673.1 | 55 | 213 |
| Reverse | 5′-CGG TCC CCA AGG TTC TAG AC-3′ | ||||
| Exon V | Forward | 5′-TAG CTT TGT GGT GCG GGA AG-3′ | EF125682.1 | 57 | 146 |
| Reverse | 5′-AAG TTG CCT TGT CCG TGG A-3′ | ||||
| Exon VI | Forward | 5′-GGG CTT GGA GAA GGA AAC CG-3′ | EF125674.1 | 55 | 188 |
| Reverse | 5′-GGT CCA CAC AAA GCT CTC GG-3′ | ||||
| Exon VII | Forward | 5′-CTG TCA CCT GCT CTC TAG GG-3′ | EF125683.1 | 55 | 118 |
| Reverse | 5′-AGT TCC GCA GAC CCT TTC AG-3′ | ||||
| Exon VIII | Forward | 5′-CAA CTG GAT GTG TGG AAC CA-3′ | EF125684.1 | 55 | 128 |
| Reverse | 5′-AGT GTG TGG GTA GAT GCC AA-3′ | ||||
| Exon IXA | Forward | 5′-ATT TGT GTC CCC TGC AGC T-3′ | EF125685.1 | 55 | 151 |
| Reverse | 5′-GTG GGA AGG AAG CAG AGA CA-3′ | ||||
| GAPDH | Forward | 5′-TCC ATG ACA ACT TTG GCA TT-3′ | NM_008084.2 | 55 | 377 |
| Reverse | 5′-GTT GCT GTT GAA GTC GCA GG-3′ |
BDNF, brain-derived neurotrophic factor; GAPDH, glyceraldehyde three-phosphate dehydrogenase.
Statistical analysis
Statistical analyses were performed using the STATVIEW computer software (version 4.0). All behavioural experiments were analysed using either repeated-measures analysis of variance (ANOVA) or factorial ANOVA when appropriate. All post hoc comparisons were made using a Bonferroni/Dunn test when significant ANOVA effects were found. Data from gene expression studies were analysed using an unpaired t-test (two-tailed). The threshold for statistical significance was set as P≤0.05. All data are presented as the mean±SEM.
Results
Three-chamber social approach task
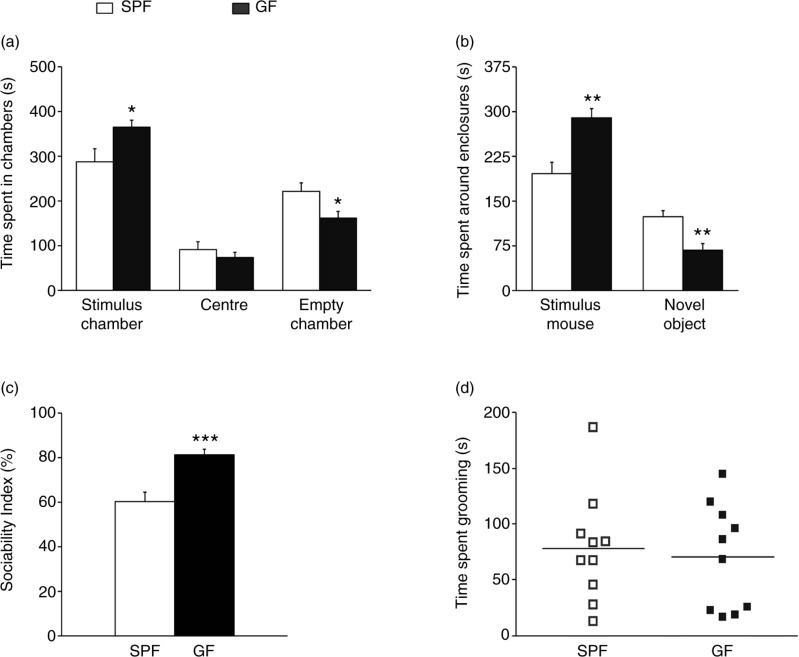
Figure 1 illustrates the results of the social approach session. Repeated-measures ANOVA showed a significant phenotype effect on time spent in the different chambers (F 1, 18=6.0; p<0.05), as well as a significant phenotype by chamber interaction (F 2, 36=4.7; p<0.05). Post hoc Bonferroni/Dunn analyses showed that GF mice spent significantly more time in the chamber containing the novel mouse than the chamber with the novel object, compared to SPF mice (p<0.05; Fig. 1a). No differences were found in time spent in the centre (p>0.1). GF mice also spent significantly more time around the novel mouse (e.g. sniffing and/or interacting with the stimulus mouse) than SPF mice (p<0.01; Fig. 1b), indicating high sociability. Indeed, the sociability index of GF mice was significantly higher than that of SPF mice (p<0.001; Fig. 1c). Total time engaged in repetitive self-grooming behaviour did not differ between GF and SPF mice (p>0.1; Fig. 1d).
Fig. 1.
Germ-free (GF) mice display increased sociability. (a) Bars show time (seconds) spent in the different chambers during the social approach session by specific pathogen-free (SPF) and GF mice. (b) Bars show time (seconds) spent interacting with the stimulus mouse or in close proximity to the novel object. (c) Bars show sociability indexes (percentage) of SPF and GF mice. (d) Bars show time (seconds) spent in self-grooming by SPF and GF mice. All data (a–d) are presented as means (±SEM; n=10 per group). *p<0.05, *p<0.01, ***p<0.001 compared with SPF mice.
Open-field test
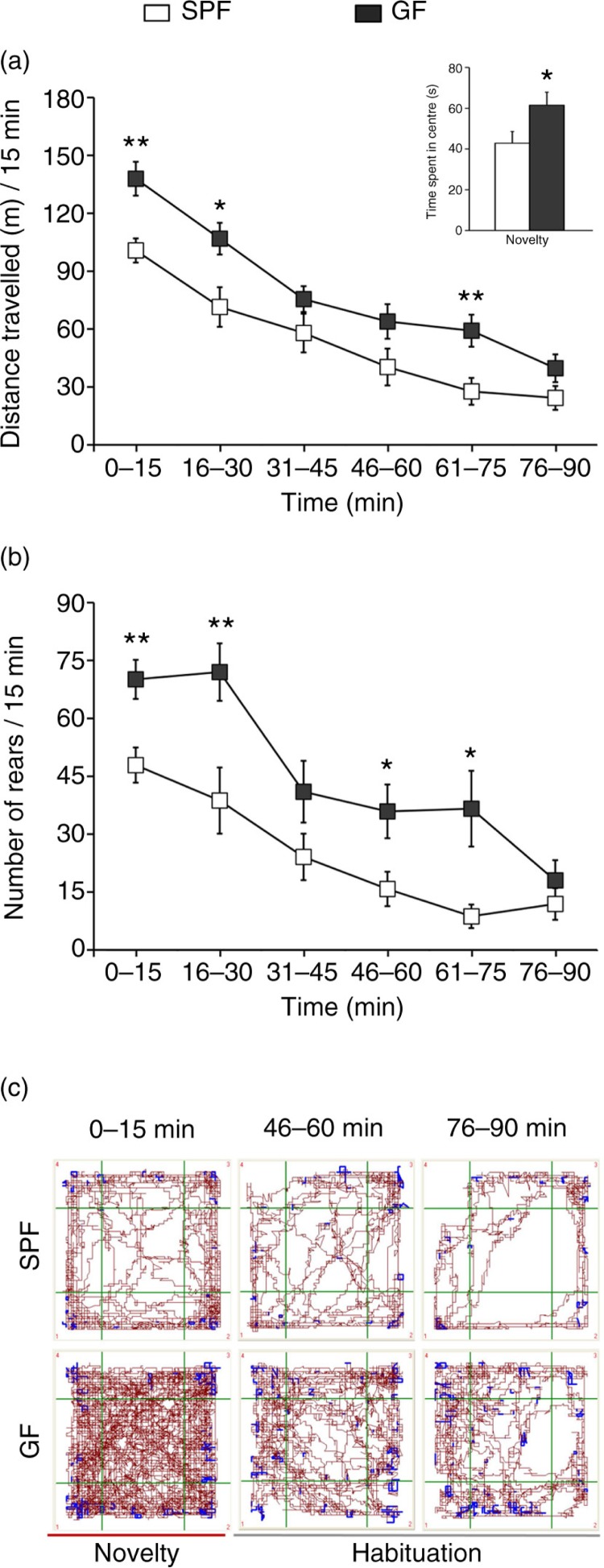
Figure 2 illustrates the results from the open-field test. Repeated-measures ANOVA showed a significant phenotype effect on distance travelled (F1,18=9.8; p<0.01). Post hoc Bonferroni/Dunn analyses showed that GF mice travelled significantly longer distances during both the initial first 15 min of testing (p<0.01; see Fig. 2a) and the habituation phase (i.e. 16–30 and 61–75 min time intervals; p<0.05 and p<0.01, respectively; see Fig. 2a) compared to SPF mice. In addition, GF mice spent more time in the centre of the open field during the novelty period (p<0.05; see inset in Fig. 2a), thus indicating reduced anxiety-like behaviour. Repeated-measures ANOVA also showed a significant phenotype effect on number of rears (F1, 18=13.4; p<0.01, see Fig. 2b). Further analysis revealed that GF mice exhibited a higher number of rears during the 0–15 min, 16–30 min, 46–60 min, and 61–75 min time intervals (p<0.01, p<0.01, p<0.05, and p<0.05, respectively; see Fig. 2b).
Fig. 2.
GF mice display increased spontaneous motor activity. (a) Average distance travelled (metres) measured in 15-min time bins across a 90-min session in an open-field box. (Inset) Bars show time (seconds) spent in the centre during the initial 15 min of testing. (b) Average number of rears measured in 15-min bins across a 90-min session in an open-field box. (c) Representative tracks of movement patterns of SPF and GF mice at the 0–15, 46–60, and 76–90-min intervals of the 90-min open-field test session; distance travelled and rearing activity are shown in dark red and blue, respectively. All data (a–b) are presented as means (±SEM; n=10 per group). *p<0.05, **p<0.01 compared with SPF mice.
Gene expression studies
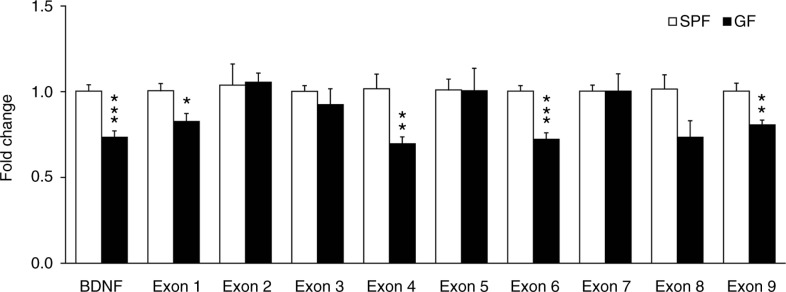
We investigated the expression of the common BDNF-encoding transcript and BDNF exon transcript variants in the amygdala of a new set of naïve GF and SPF mice, by means of quantitative real-time polymerase chain reaction (qRT-PCR). The level of the common BDNF-encoding transcript was significantly reduced in GF mice (t (10)=5.08, p<0.001; Fig. 3). Among the nine different BDNF exon transcript variants examined (I–IX), only the levels of BDNF exon I, IV, VI and IX transcript variants were significantly reduced in GF mice compared to SPF mice [(t (10)=2.87, p<0.05), (t (10)=3.42, p<0.01), (t (10)=5.64, p<0.001) and (t (10)=3.64, p<0.01), respectively; Fig. 3].
Fig. 3.
Differential promoter control of the BDNF gene by gut microbiota in the amygdala. Quantitative real-time polymerase chain reaction was used to examine expression levels of the common and exon-specific BDNF transcripts in the amygdala of GF and SPF mice. Expression level of each transcript examined was normalised to heat shock protein 90 (Hsp90) levels and expressed relative to the SPF group. Data are presented as means (±SEM; n=6 per group). *p<0.05, **p<0.01, ***p<0.001 compared with SPF mice.
In our previous study with the NMRI strain, we found that the expression of the immediate-early gene NGFI-A (also known as egr1and zif268) was greatly reduced in the absence of gut microbiota in several brain regions, including the amygdala (2). We therefore studied the expression of this gene in the amygdala of Swiss-Webster GF and SPF mice for comparisons with our previous study. In GF mice, the mRNA levels of NGFI-A were found to be significantly reduced compared to SPF mice (GF vs. SPF: 0.64±0.03 vs. 1.00±0.07; t (10)=5.13, p<0.001).
Discussion
The main finding of this study is that the indigenous microbiota alters the development of social behaviour in mice, which is in agreement with the study by Desbonnet et al. (16). In contradiction to their findings, however, we found that adult GF Swiss-Webster mice display significantly higher levels of sociability compared to SPF mice, as indicated by a stronger preference for time spent close to the novel mouse versus the novel object (i.e. empty enclosure). Moreover, time spent in ‘stereotypic behaviours’ (e.g. repetitive self-grooming) during the choice session did not differ between GF and SPF mice. The reasons for the discrepancies between our studies are unclear, but they could be related, in part, to differences in experimental design. For example, we used the DBA/2NCrl strain as stimulus mice, whereas in their study Desbonnet et al. used the NIH Swiss strain. Importantly, our animals were older than those in the aforementioned study. It is well established that social brain networks continue to undergo dynamic structural and functional changes throughout adolescence and into late adulthood, and therefore the behavioural phenotype of GF mice may change with age. Although the same strain of mice was used in these two studies, another aspect that must be considered with regard to the influence of the gut microbiota on brain development and behaviour is the genetic background of the host. For example, unlike in the NMRI and Swiss-Webster strains, the absence of gut microbiota actually exacerbates anxiety-like behaviour in genetically anxiety-prone strains (e.g. BALB/c mice, F344 rats) (22, 23). Moreover, GF rats of the stress-sensitive F344 strain spent less time sniffing an unknown partner during the initial first 2 min of a 10-min social interaction test compared to their conventionally raised counterparts. However, GF F344 rats spent the same amount of time in social contacts and self-grooming for the remaining testing period, indicating that the initial lack of social interaction was mediated by increased anxiety-like behaviour in response to a novel situation rather than deficits in social behaviour per se (23). Taken together, these results highlight the importance of the genetic background and age of the host on gut microbiome–brain interactions.
In the present study, we also compared the exploratory activity and habituation profile of adult GF Swiss-Webster mice after exposure to a novel open field. Typically, mice exposed to a novel environment display initial high levels of exploratory behaviour (i.e. novelty period), followed by a progressive reduction in exploratory activity that occurs as the novel environment becomes familiar (i.e. habituation phase). Our results showed that adult GF Swiss-Webster mice display increased levels of motor activity both during the novelty period and habituation phase. These findings are in agreement with our previous observations showing that adult GF NMRI mice display higher motor activity during the habituation phase (2), as well as other findings reporting elevated home-cage activity counts in GF C57BL/6J mice (24). Interestingly, oral administration of antibiotics to conventionally raised mice also increases exploratory behaviour (8). This effect was not observed in GF mice treated with antibiotics, thus supporting the crucial role of the gut microbiota. Another study by Neufeld et al. (3) failed to detect significant differences in the locomotor activity of naïve adult female GF Swiss-Webster mice. These discrepancies are likely due to the fact that these authors used shorter testing periods than ours (i.e. 30 min vs. 90 min used in the present study). Indeed, careful examination of their open-field data shows that GF mice travelled a greater mean distance than SPF mice towards the end of testing period (i.e. 25–30 min intervals; see Fig. 1 in the aforementioned study). Alternatively, there might be potential sex differences in the modulatory central effects of the gut microbiota on motor control, since these authors used female Swiss-Webster mice. Consistent with this idea, the study by Clarke et al. (1) demonstrated that some of the neurochemical changes found in the hippocampus of GF mice are only observed in males.
Regulation of synaptic plasticity-related genes by gut microbiota
Several recent studies of adult male GF mice (NMRI, Swiss-Webster, and BALB/c strains) have demonstrated that BDNF mRNA levels or protein concentration are decreased in several brain regions, including the hippocampus (1, 2, 5), prefrontal cortex (2), and amygdala (2) when compared to conventionally raised mice. In contrast, another study by Neufeld et al. (3) found an increase in BDNF mRNA expression in the dentate gyrus of female GF Swiss-Webster mice, suggesting potential sex differences in the modulatory central effects of the gut microbiota. These previous studies, however, did not explore whether microbes differentially regulate mRNA levels of specific BDNF transcripts. Studies over the last few years have revealed that BDNF has a complex genomic structure and transcriptional regulation (10). Structurally, the rodent BDNF gene consists of at least eight 5′ noncoding exons and a 3′ coding exon (IX) (25). Although the various transcripts encode the same BDNF protein, they are differentially expressed in distinct brain regions, cell types, and even in different cell compartments (e.g. soma vs. dendrites). In addition, specific signalling pathways and transcription factors have been linked to the transcription of the different BDNF transcripts (10). In this study, we examined the influence of the gut microbiota on expression levels of the common BDNF transcript and BDNF exon-containing transcripts (I–IX) in the amygdala, a brain region readily activated in the three-chambered approach paradigm (26) and a central hub in the social brain network (27, 28). We found that total BDNF and BDNF exon-containing transcripts I-, IV-, VI-, and IX-mRNA levels are significantly decreased in the amygdala of adult male GF Swiss-Webster mice. The present findings are consistent with our previous study showing decreased expression levels of BDNF in the basolateral amygdala of adult NMRI GF mice using an in situ hybridisation approach that did not distinguish between the various BDNF transcripts (2). In a recent study, however, Stilling et al. (26) found an upregulation of the BNDF exon IV transcript in the amygdala of adult GF Swiss-Webster mice using a next-generation whole transcriptome sequencing (RNA-Seq) approach. The reasons for this discrepancy are not clear, since we used the same strain and sex of mice. Nevertheless, these observations suggest that expression of BDNF in the brain is strongly influenced by the presence of indigenous microbes. The transcription factors involved in the regulation of the distinct exon-specific BDNF transcripts are not fully understood. However, several studies have implicated the Cyclic adenosine monophosphate (cAMP) response element binding protein (CREB) in the regulation of BDNF in the amygdala and other brain regions (10, 29). These studies have shown that the mouse BDNF exon transcript IV is upregulated via CREB binding to a cAMP/Ca2+-response element. These observations are in line with our genome-wide profiling study showing that the cAMP-mediated signalling pathway is one of four key canonical pathways strongly regulated by the gut microbiota (2).
More recent studies have also demonstrated that the expression of BDNF is strongly regulated by a variety of epigenetic mechanisms, such as DNA methylation and posttranslational modifications of histones (30–32). In this regard, short-chain fatty acids (SCFA) such as butyrate and propionate (the end products of fermentation of dietary fibres by the gut microbiota) are of particular interest since they are capable of inhibiting histone deacetylases (33, 34). Indeed, it has been recently demonstrated that systemic administration of butyrate increases expression of BDNF transcript IV in the hippocampus (35). In addition, recent studies have demonstrated that SCFA (i.e. butyrate and propionate) are capable of modulating the expression of synapse-related genes (36). Moreover, SCFA have been shown to activate the CREB-dependent pathways in both an animal model of ASD (37, 38) and PC12 cells (36). However, SCFA can also activate cells via G-protein-coupled receptors (GPRs), such as GPR41 and GPR43 (39). Deciphering the details of how gut microbiota regulates central BDNF expression and function will be an important step in understanding how indigenous microbes shape the development of neural circuits, brain functions, and behaviour.
Acknowledgements
This work was supported by the Olle Engkvist Byggmästare Foundation, Swedish Brain Foundation, Swedish Research Council (K2015-62X-22745-01-4), and Strategic Neuroscience Programme at Karolinska Institutet.
Authors' contributions
TA, HF, and RDH designed the study. TA, HR, and YQ performed the experiments and analysed the data. TA, HR, HF, and RDH wrote the paper with input from YQ.
Conflict of interest and funding
The authors report no financial interests or conflicts of interest.
References
- 1.Clarke G, Grenham S, Scully P, Fitzgerald P, Moloney RD, Shanahan F, et al. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol Psychiatry. 2013;18:666–73. doi: 10.1038/mp.2012.77. [DOI] [PubMed] [Google Scholar]
- 2.Diaz Heijtz R, Wang S, Anuar F, Qian Y, Bjorkholm B, Samuelsson A, et al. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci USA. 2011;108:3047–52. doi: 10.1073/pnas.1010529108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neufeld KM, Kang N, Bienenstock J, Foster JA. Reduced anxiety-like behavior and central neurochemical change in germ-free mice. Neurogastroenterol Motil. 2011;23:255–64. doi: 10.1111/j.1365-2982.2010.01620.x. e119. [DOI] [PubMed] [Google Scholar]
- 4.Gareau MG, Wine E, Rodrigues DM, Cho JH, Whary MT, Philpott DJ, et al. Bacterial infection causes stress-induced memory dysfunction in mice. Gut. 2011;60:307–17. doi: 10.1136/gut.2009.202515. [DOI] [PubMed] [Google Scholar]
- 5.Sudo N, Chida Y, Aiba Y, Sonoda J, Oyama N, Yu XN, et al. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. J Physiol. 2004;558(Pt 1):263–75. doi: 10.1113/jphysiol.2004.063388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vicario-Abejon C, Owens D, McKay R, Segal M. Role of neurotrophins in central synapse formation and stabilization. Nat Rev Neurosci. 2002;3:965–74. doi: 10.1038/nrn988. [DOI] [PubMed] [Google Scholar]
- 7.Benarroch EE. Brain-derived neurotrophic factor: regulation, effects, and potential clinical relevance. Neurology. 2015;84:1693–704. doi: 10.1212/WNL.0000000000001507. [DOI] [PubMed] [Google Scholar]
- 8.Bercik P, Denou E, Collins J, Jackson W, Lu J, Jury J, et al. The intestinal microbiota affect central levels of brain-derived neurotropic factor and behavior in mice. Gastroenterology. 2011;141:599–609. doi: 10.1053/j.gastro.2011.04.052. e1–3. [DOI] [PubMed] [Google Scholar]
- 9.Castrén E. Neurotrophins and psychiatric disorders. In: Lewin GR, Carter BD, editors. Neurotrophic factors. Handbook of experimental pharmacology. Vol. 220. Berlin: Springer; 2014. pp. 461–79. [DOI] [PubMed] [Google Scholar]
- 10.Adachi N, Numakawa T, Richards M, Nakajima S, Kunugi H. New insight in expression, transport, and secretion of brain-derived neurotrophic factor: implications in brain-related diseases. World J Biol Chem. 2014;5:409–28. doi: 10.4331/wjbc.v5.i4.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tillisch K, Labus J, Kilpatrick L, Jiang Z, Stains J, Ebrat B, et al. Consumption of fermented milk product with probiotic modulates brain activity. Gastroenterology. 2013;144:1394–401. doi: 10.1053/j.gastro.2013.02.043. 1401.e1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13:701–12. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- 13.Finegold SM, Dowd SE, Gontcharova V, Liu C, Henley KE, Wolcott RD, et al. Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe. 2010;16:444–53. doi: 10.1016/j.anaerobe.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 14.Williams BL, Hornig M, Buie T, Bauman ML, Cho Paik M, Wick I, et al. Impaired carbohydrate digestion and transport and mucosal dysbiosis in the intestines of children with autism and gastrointestinal disturbances. PLoS One. 2011;6:e24585. doi: 10.1371/journal.pone.0024585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krajmalnik-Brown R, Lozupone C, Kang D-W, Adams JB. Gut bacteria in children with autism spectrum disorders: challenges and promise of studying how a complex community influences a complex disease. Microb Ecol Health Dis. 2015;26:26914. doi: 10.3402/mehd.v26.26914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desbonnet L, Clarke G, Shanahan F, Dinan TG, Cryan JF. Microbiota is essential for social development in the mouse. Mol Psychiatry. 2014;19:146–8. doi: 10.1038/mp.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silverman JL, Yang M, Lord C, Crawley JN. Behavioural phenotyping assays for mouse models of autism. Nat Rev Neurosci. 2010;11:490–502. doi: 10.1038/nrn2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang M, Silverman JL, Crawley JN. Curr Protoc Neurosci. 2011. Automated three-chambered social approach task for mice. Chapter 8: Unit 8.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pietropaolo S, Guilleminot A, Martin B, D'Amato FR, Crusio WE. Genetic-background modulation of core and variable autistic-like symptoms in Fmr1 knock-out mice. PLoS One. 2011;6:e17073. doi: 10.1371/journal.pone.0017073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diaz Heijtz R, Scott L, Forssberg H. Alteration of dopamine D1 receptor-mediated motor inhibition and stimulation during development in rats is associated with distinct patterns of c-fos mRNA expression in the frontal-striatal circuitry. Eur J Neurosci. 2004;19:945–56. doi: 10.1111/j.0953-816x.2004.03154.x. [DOI] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Nishino R, Mikami K, Takahashi H, Tomonaga S, Furuse M, Hiramoto T, et al. Commensal microbiota modulate murine behaviors in a strictly contamination-free environment confirmed by culture-based methods. Neurogastroenterol Motil. 2013;25:521–8. doi: 10.1111/nmo.12110. [DOI] [PubMed] [Google Scholar]
- 23.Crumeyrolle-Arias M, Jaglin M, Bruneau A, Vancassel S, Cardona A, Daugé V, et al. Absence of the gut microbiota enhances anxiety-like behavior and neuroendocrine response to acute stress in rats. Psychoneuroendocrinology. 2014;42:207–17. doi: 10.1016/j.psyneuen.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 24.Backhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci USA. 2007;104:979–84. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aid T, Kazantseva A, Piirsoo M, Palm K, Timmusk T. Mouse and rat BDNF gene structure and expression revisited. J Neurosci Res. 2007;85:525–35. doi: 10.1002/jnr.21139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stilling RM, Ryan FJ, Hoban AE, Shanahan F, Clarke G, Claesson MJ, et al. Microbes & neurodevelopment – absence of microbiota during early life increases activity-related transcriptional pathways in the amygdala. Brain Behav Immun. 2015;50:209–20. doi: 10.1016/j.bbi.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 27.Amaral DG. The amygdala, social behavior, and danger detection. Ann N Y Acad Sci. 2003;1000:337–47. doi: 10.1196/annals.1280.015. [DOI] [PubMed] [Google Scholar]
- 28.Bickart KC, Dickerson BC, Barrett LF. The amygdala as a hub in brain networks that support social life. Neuropsychologia. 2014;63:235–48. doi: 10.1016/j.neuropsychologia.2014.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng F, Zhou X, Luo Y, Xiao H, Wayman G, Wang H. Regulation of brain-derived neurotrophic factor exon IV transcription through calcium responsive elements in cortical neurons. PLoS One. 2011;6:e28441. doi: 10.1371/journal.pone.0028441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boulle F, van den Hove DLA, Jakob SB, Rutten BP, Hamon M, van Os J, et al. Epigenetic regulation of the BDNF gene: implications for psychiatric disorders. Mol Psychiatry. 2012;17:584–96. doi: 10.1038/mp.2011.107. [DOI] [PubMed] [Google Scholar]
- 31.Karpova NN. Role of BDNF epigenetics in activity-dependent neuronal plasticity. Neuropharmacology. 2014;76(Pt C):709–18. doi: 10.1016/j.neuropharm.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 32.Sun H, Kennedy PJ, Nestler EJ. Epigenetics of the depressed brain: role of histone acetylation and methylation. Neuropsychopharmacology. 2013;38:124–37. doi: 10.1038/npp.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sealy L, Chalkley R. The effect of sodium butyrate on histone modification. Cell. 1978;14:115–21. doi: 10.1016/0092-8674(78)90306-9. [DOI] [PubMed] [Google Scholar]
- 34.Kiefer J, Beyer-Sehlmeyer G, Pool-Zobel BL. Mixtures of SCFA, composed according to physiologically available concentrations in the gut lumen, modulate histone acetylation in human HT29 colon cancer cells. Br J Nutr. 2006;96:803–10. doi: 10.1017/bjn20061948. [DOI] [PubMed] [Google Scholar]
- 35.Intlekofer KA, Berchtold NC, Malvaez M, Carlos AJ, McQuown SC, Cunningham MJ, et al. Exercise and sodium butyrate transform a subthreshold learning event into long-term memory via a brain-derived neurotrophic factor-dependent mechanism. Neuropsychopharmacology. 2013;38:2027–34. doi: 10.1038/npp.2013.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nankova BB, Agarwal R, MacFabe DF, La Gamma EF. Enteric bacterial metabolites propionic and butyric acid modulate gene expression, including CREB-dependent catecholaminergic neurotransmission, in PC12 cells – possible relevance to autism spectrum disorders. PLoS One. 2014;9:e103740. doi: 10.1371/journal.pone.0103740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacFabe DF, Cain DP, Rodriguez-Capote K, Franklin AE, Hoffman JE, Boon F, et al. Neurobiological effects of intraventricular propionic acid in rats: possible role of short chain fatty acids on the pathogenesis and characteristics of autism spectrum disorders. Behav Brain Res. 2007;176:149–69. doi: 10.1016/j.bbr.2006.07.025. [DOI] [PubMed] [Google Scholar]
- 38.MacFabe DF. Enteric short-chain fatty acids: microbial messengers of metabolism, mitochondria, and mind: implications in autism spectrum disorders. Microb Ecol Health Dis. 2015;26 doi: 10.3402/mehd.v26.28177. 28177, doi: http://dx.doi.org/10.3402/mehd.v26.28177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Natarajan N, Pluznick JL. From microbe to man: the role of microbial short chain fatty acid metabolites in host cell biology. Am J Physiol Cell Physiol. 2014;307:C979–85. doi: 10.1152/ajpcell.00228.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]