Abstract
The current study aimed to isolate and characterize a chromate-resistant bacterium from tannery effluent, able to reduce Cr(VI) aerobically at high pH and salinity. Environmental contamination by hexavalent chromium, Cr(VI), presents a serious public health problem. Enrichment led to the isolation of 12 bacteria displaying different degrees of chromate reduction. Phenotypic characterization and phylogenetic analysis based on 16S rDNA sequence comparison indicated that the most potent strain belonged to the genus Halomonas. The new strain designated as Halomonas sp. M-Cr was able to reduce 82% of 50 mg L−1 Cr(VI) in 48 h, concomitant with discolouring of yellow colour of the medium and formation of white insoluble precipitate of Cr(III). It exhibited growth up to 3500 mg L−1 Cr(VI), 20% NaCl and showed strong Cr(VI) reduction under alkaline condition, pH 10. Scanning electron microscopy revealed precipitation of chromium hydroxide on bacterial cell surfaces, which showed characteristic peak of chromium in energy-dispersive X-ray analysis. Plackett–Burman design was used to evaluate the influence of related parameters for enhancing Cr(VI) reduction. Glucose, yeast extract and KH2PO4 were confirmed as significant variables in the medium. Data suggest Halomonas sp. M-Cr as a promising candidate for bioremediation of Cr(VI) contaminated effluents particularly in saline and alkaline environments. Up to our knowledge, this is the first report on isolation of haloalkaliphilic Halomonas sp. from tannery effluent.
Keywords: bioremediation, Cr(VI) reduction, Halomonas sp. M-Cr, haloalkaliphilic, scanning electron microscopy, Plackett–Burman design
Introduction
Chromium (Cr) is a toxic heavy metal extensively used in a variety of industrial processes, owing to its impressive corrosion resistance.[1] Cr(VI) containing wastewater has become a well-recognized hazard in water pollution control. Soluble Cr(VI) is extremely toxic and shows carcinogenic and mutagenic effect on biological systems due to its strong oxidizing nature.[2] In contrast, Cr(III) being sparingly soluble, less toxic and bioavailable in comparison to Cr(VI), readily forms insoluble oxides and hydroxides above pH 5.[3] Thus, biotransformation of Cr(VI) to less toxic Cr(III) is an effective strategy for the remediation of Cr(VI) pollution worldwide.[4] The process has been demonstrated in several bacterial species under both aerobic and anaerobic conditions.[5–7]
Leather tanning is an environmentally challenging process and is one of the leading foreign exchanges earning industries in Egypt. Million litres of wastewater from tanneries containing a high amount of Cr(VI) are discharged into the sewage drains and ponds without any treatment. Therefore, in this paper we report the isolation and characterization of a chromate-resistant bacterium from tannery effluent able to reduce Cr(VI) aerobically at high pH and salinity. Statistical optimization of process parameters that enhance reduction of Cr(VI) was also performed. Up to our knowledge this is the first report about chromate (VI) reduction by haloalkaliphilic Halomonas sp. isolated from tannery effluent.
Materials and methods
Sampling
Samples were collected from different stages of the tanning process (El-Halafawy Leather Tanning Company, Damanhour, EL-Bahera, Egypt) using screw capped sterilized glass bottles, maintained at 4 °C and immediately transported to the laboratory.
Enrichment and isolation of chromate detoxifying alkaliphilic bacteria
One mL from each sample was enriched in 250 mL Erlenmeyer flasks containing 50 mL of Luria-Bertani (LB) medium (g L−1) tryptone 10, yeast extract 5, NaCl 5, supplemented with 50 mg L−1 Cr(VI) in the form of K2CrO4 and adjusted to pH 10 with sodium carbonate. The inoculated flasks were incubated at 30 °C for 72 h in a rotary shaker at 120 rpm, serving as the initial enrichment culture. Subsequent enrichment transfer cultures were established using 5 mL as inoculum. From flasks showing turbidity and colour change from yellow to turbid white,[8] 100 μL aliquots were spread on LB agar plates amended with the same Cr(VI) concentration and incubated at 30 °C for 48 h. Bacterial colonies showing distinct morphologies were selected, purified and preserved at 4 °C or in 30% (V/V%) sterile glycerol.
Identification of the bacterial strain
The basic biochemical and physiological properties of M-Cr isolate were analysed according to Bergey's Manual of Determinative Bacteriology.[9] Cell morphology was examined by scanning electron microscope (JEOL JEM-5300).
Molecular characterization
Molecular characterization of the isolate was done by 16S rDNA sequence analysis. DNA was isolated from M-Cr cells using standard procedures.[10] The purity of the isolated DNA was confirmed by gel electrophoresis. Amplification of 16S rDNA gene was performed as previously reported [11] using F 5′AGAGTTTGATCMTGGCTCAG3′ and R 5′TACGGYTACCTTGTTACGACTT3′ as forward and reverse primers. The polymerase chain reaction (PCR) amplification products were analysed by electrophoresis on a 1% agarose gel and purified. An amplified product of 16S rDNA was sequenced using an ABI PRISM 377 DNA Sequencer and ABI PRISM Big Dye Terminator Cycle Sequencing (Perkin Elmer). The 16S rDNA sequence was uploaded to NCBI database using BLASTN program (http://www.ncbi.nlm.nih.gov/blast/; version 2.0) and compared with sequences available in the GenBank database. Sequences of most close members were aligned using CLUSTALW program (http://www.ebi.ac.uk/clustalw). A phylogenetic tree was constructed, using the phylogeny inference package (PHYLIP; version 3.6).
Chromate reduction by Halomonas sp. M-Cr
A seed culture was prepared by transferring a loopfull of 48 h old slant into 25 mL LB media without chromate, pH 10 and incubated aerobically at 30 °C, by shaking at 120 rpm until O.D.600 of 1.0. Reduction of chromium by Halomonas sp. M-Cr was examined by inoculating 25 mL/100 mL flask with 0.5 mL of seed culture. Sterile inoculated broth without Cr(VI) served as the biotic control, and the uninoculated broth with Cr(VI) served as the abiotic control. The biotic control was used to compare the growth of bacteria with or without Cr(VI) and the abiotic control was used to test if any change in Cr(VI) appeared as a result from the presence of the media components. All the cultures including blanks were incubated at 30 °C with continuous shaking (120 rpm). Samples were collected under sterile conditions at regular time intervals to monitor Cr(VI) reduction as well as growth. All experiments were performed in duplicate and mean values were recorded.
Quantification of growth
Growth of Halomonas sp. M-Cr was determined according to Ibrahim et al. [5] by measuring absorbance at 600 nm against distilled water as blank.
Estimation of hexavalent chromium
The concentration of residual hexavalent chromium was determined spectrophotometrically in the culture supernatant after centrifugation at 10,000 rpm for 10 min at 4 °C to remove any suspended biomass, and assayed at 540 nm using 1,5-diphenylcarbazide (DPC) method.[12]
Effect of pH and salinity on Cr(VI) reduction
The influence of pH on bacterial growth and chromate reduction was examined by adjusting pH of medium to values ranging from 6 to 11 with predetermined amounts of filter-sterilized (0.22 μm) 1M Na2CO3 or 1M HCl. The effect of salt concentration was examined by adding different concentrations (50–200 g L−1) of NaCl.
Scanning electron microscopy (SEM) and SEM-EDX analysis of Halomonas sp. M-Cr cells
Bacterial cells grown in liquid media with and without Cr(VI) were harvested by centrifugation at 10 000 rpm for 10 min at 4 °C. Cells were fixed, dehydrated and dried using the critical point method.[13] Elemental analysis of reduction product was carried out with the help of a computer controlled field emission scanning electron microscopy (JEOL JEM-5300) equipped with an energy-dispersive X-ray (EDX) probe to detect Cr and its precipitate compound distribution on and around the cell surface.
Selection of significant variables by Plackett–Burman
A total of nine independent variables: glucose, (NH4)2SO4, yeast extract, tryptone, KH2PO4, NaCl, MgSO4·7H2O, Cr(VI) and inoculum size were used. For the selection of significant variables affecting chromate reduction by Halomonas sp. M-Cr, a variety of variables were tested and identified via the Plackett–Burman design experiment.[14] Based on this design, each variable was examined at two levels: −1 for low level and +1 for high level, and a centre point was run to evaluate the linear and curvature effects of the variables. The experimental design with nine factors under investigation with the name, symbol code and actual level of the variables is shown in Table 1. Plackett–Burman experimental design is based on the first-order polynomial model:
Table 1.
Plackett–Burman experimental design matrix for evaluation of nine components with the actual and coded levels and the design response for Cr(VI) reduction by Halomonas sp. M-Cr.
| Variables/Levels |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Glucose (g L−1) | (NH4)2 SO4 (g L−1) | Yeast extract (g L−1) | Tryptone (g L−1) | KH2PO4 (g L−1) | NaCl (g L−1) | Inoculum size (%) | MgSO4.7H2O (g L−1) | Cr(VI) (mg L−1) | Chromate reduction (%) | |
| Trials | G | A | Y | Tr | K | N | Is | M | Cr | |
| 1 | +1(15) | +1(1.5) | −1(0.25) | +1(5) | +1(0.75) | +1(100) | −1(1) | −1(0.05) | −1(30) | 81 |
| 2 | +1(15) | −1(0.5) | +1(0.75) | +1(5) | +1(0.75) | −1(50) | −1(1) | −1(0.05) | +1(70) | 74 |
| 3 | −1(5) | +1(1.5) | +1(0.75) | +1(5) | −1(0.25) | −1(50) | −1(1) | +1(0.15) | −1(30) | 60 |
| 4 | +1(15) | +1(1.5) | +1(0.75) | −1(1) | −1(0.25) | −1(50) | +1(3) | −1(0.05) | +1(70) | 55 |
| 5 | +1(15) | +1(1.5) | −1(0.25) | −1(1) | −1(0.25) | +1(100) | −1(1) | +1(0.15) | +1(70) | 47 |
| 6 | +1(15) | −1(0.5) | −1(0.25) | −1(1) | +1(0.75) | −1(50) | +1(3) | +1(0.15) | −1(30) | 78 |
| 7 | −1(5) | −1(0.5) | −1(0.25) | +1(5) | −1(0.25) | +1(100) | +1(3) | −1(0.05) | +1(70) | 55 |
| 8 | −1(5) | −1(0.5) | +1(0.75) | −1(1) | +1(0.75) | +1(100) | −1(1) | +1(0.15) | +1(70) | 39 |
| 9 | −1(5) | +1(1.5) | −1(0.25) | +1(5) | +1(0.75) | −1(50) | +1(3) | +1(0.15) | +1(70) | 67 |
| 10 | +1(15) | −1(0.5) | +1(0.75) | +1(5) | −1(0.25) | +1(100) | +1(3) | +1(0.15) | −1(30) | 63 |
| 11 | −1(5) | +1(1.5) | +1(0.75) | −1(1) | +1(0.75) | +1(100) | +1(3) | −1(0.05) | −1(30) | 54 |
| 12 | −1(5) | −1(0.5) | −1(0.25) | −1(1) | −1(0.25) | −1(50) | −1(1) | −1(0.05) | −1(30) | 43 |
| 13 | 0(10) | 0(1) | 0(0.5) | 0(3) | 0(0.5) | 0 (70) | 0(2) | 0(0.1) | 0(50) | 60 |
| 14 | 0(10) | 0(1) | 0(0.5) | 0(3) | 0(0.5) | 0 (70) | 0(2) | 0(0.1) | 0(50) | 61 |
where Y is the response (chromate reduction), β0 is the model intercept and βi is the linear coefficient, and xi is the level of the independent variable. In the present work, 9 assigned variables were screened in 12 experimental runs in addition to 2 runs at their centre point. All trials were performed in duplicate and the averages of chromate reduction results after 24 h were treated as the responses Y (Table 1). The results were analysed by the STATISTICA (version 6.0, StatSoft, USA (including parameters estimation and analyses of variance (ANOVA). From the regression analysis of the variables, the factors with significant levels greater than 90% (P-value < 0.1) were considered to have a significant effect on chromate reduction.
Results and discussion
Screening for chromate reducing alkaliphilic bacteria
While many reports of microbial Cr(VI) reduction are in circulation, very few [15,16] have demonstrated Cr(VI) reduction under alkaline conditions. In addition, high concentration of salts in wastewater treatment systems can be a major problem for conventional biological treatments. Halophilic and alkaliphilic microorganisms produce unique biocatalysts that function under harsh conditions in which their mesophilic counterparts could not survive, permitting the development of additional industrial and bioremediation processes.[5] In this study, 30 unique colonies were selected according to their colony morphology and growth on LB plate amended with 50 mg L−1Cr(VI). Preliminary selection of chromate reducing bacteria was estimated qualitatively and quantitatively. A strain designated as M-Cr showed the highest reduction efficiency and was subsequently chosen for further study.
Characterization of M-Cr
Strain M-Cr formed cream, circular, smooth, bright, mucoid, convex colonies on alkaline LB agar with entire opaque margin, 2–3 mm in diameter. Cells were motile, gram-negative, non-sporulating straight rods, as observed by SEM, with a length of 0.75–1.37 μm and a width of 0.65–1.0 μm (Figure 6A). In addition, the strain M-Cr was oxidase and catalase positive, and best grown in a medium containing up to 20% of NaCl with an optimum at 5% NaCl. The pH growth ranged from 7.0 to 11 with an optimum at pH 9–10.0. According to its growth characteristics, the strain was described as an alkaliphilic, moderate halophilic bacterium.
Figure 6.
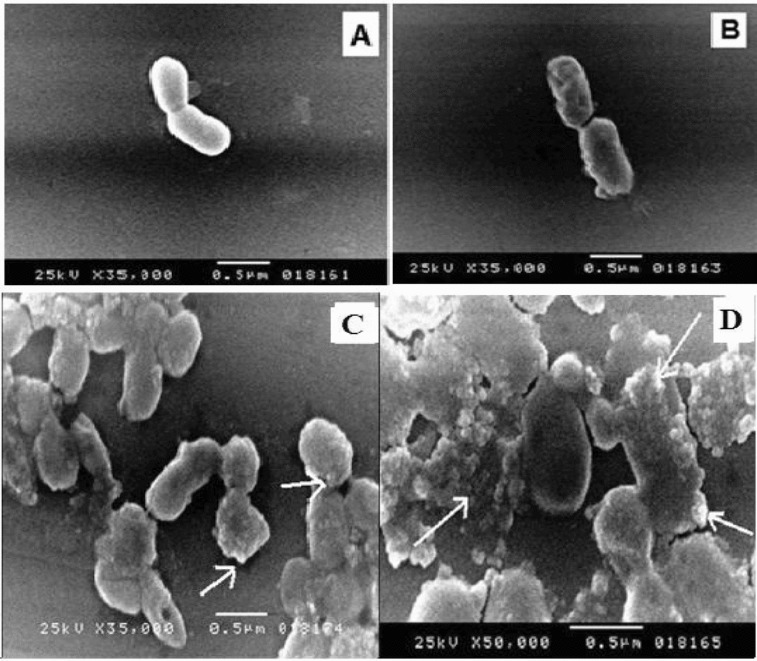
SEM micrographs of Halomonas sp. M-Cr cells grown in: (A) LB medium without Cr(VI) (control); (B) LB medium amended with 50 mg L−1 Cr(VI) for 24 h; (C) Cr(III) precipitates found as discrete particles bound to the cell surface (arrows); (D) Amorphous Cr(III) hydroxide precipitates not attached to cells are also present (arrows), the largest precipitates were slightly rounded.
The 16S rDNA sequence analysis revealed that strain M-Cr shared 91%–99% sequence homology to the species of the genus Halomonas, which was included in Phyllum Proteobacteria subgroup Gamma, in the family Halomonadaceae. The 16S rDNA gene sequence was submitted to NCBI GenBank database as Halomonas sp. M-Cr with accession number JX025759. The constructed phylogenetic tree (Figure 1) shows that Halomonas sp. M-Cr is tightly clustered with Halomonas campisalis strain 4A (GenBank accession No. NR028702), isolated from saline lakes in Washington State (USA).[17] Phylogenetic analysis revealed that Halomonas sp. M-Cr is included in a group of alkaliphilic, chromate reducers halomonads comprehending Halomonas mongoliensis Z-7009 (similarity level 99%), Halomonas kenyensis AIR-2 (96%) [18] and Halomonas campaniensis 5AG (96%).[19] The identified strain exhibited a high degree of Cr(VI) resistance, being able to grow in the presence of 3500 mg L−1 Cr(VI). This was much higher than those of other Cr(VI)-resistant strains: Halomonas elongate ATCC33173, Halomonas subglaciescola UQM2926 and Halomonas sp. TA-04, with values of 50, 125 and 200 mg L−1, respectively,[20,21] while it was comparable to other bacterial species, such as Serratia sp. Cr-10 (1500 mg L−1 Cr(VI)) [22] and Bacillus sp. MDS05 (2500 mg L−1 Cr(VI)).[23]
Figure 1.

Phylogenetic tree based on 16S rDNA gene sequence, and reference sequences extracted from the GenBank Database, showing the phylogenetic relationship of Halomonas sp. M-Cr within representative species of the genus Halomonas. Numbers in bracket represents GenBank accession numbers.
Time course of Cr (VI) reduction by Halomonas sp. M-Cr
The growth pattern of Halomonas sp. M-Cr in alkaline LB medium (pH 10) containing 50 mg L−1 Cr(VI), followed the same growth pattern as without Cr(VI), but with slight inhibition (Figure 2). Reduction was found to be growth associated. No reduction was detected in cell-free medium from the initial to the final stage of the experiment, this indicated no evidence of spontaneous Cr(VI) reduction and the major mechanism of reduction was attributable to microbial metabolism.[24] Also the formation of precipitates only in cultures supplemented with Cr(VI) illustrated the transformation of this soluble oxyanion to Cr(III) that forms insoluble hydroxide Cr(OH)3 as a white precipitate (Figure 3). Comparing chromate reduction between different studies can be difficult, given the wide range of culturing conditions used and the effect such conditions can have on reduction process.[25] Under alkaline conditions, H. chromatireducens AGD 8-3 was able to reduce 80% of 5 mg Cr(VI)/l, while H. campisalis Z-7398, H. desiderata FB2, H. kenyensis AIR-2, H. natronophila Z-7009 and H. campaniensis 5AG reduced approximately 50%, 35%, 25%, 25% and 5% of chromate, respectively.[26] Halomonas sp. MV-2007, reduced about 75% of the 5 mg L−1 Cr(VI) after 25 days.[16]
Figure 2.
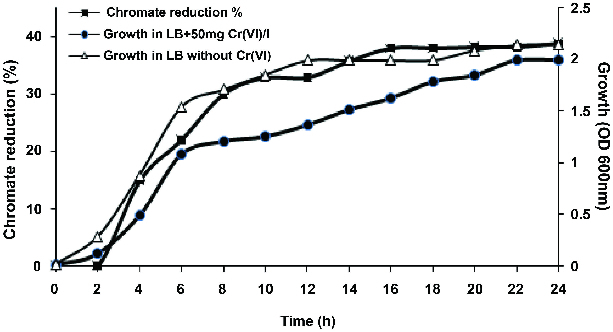
Growth and Cr(VI) reduction efficiency of Halomonas sp. M-Cr grown in LB broth, pH 10, in the absence and presence of 50 mg L−1 Cr(VI) and incubated at 30 °C under shaking at 120 rpm.
Figure 3.
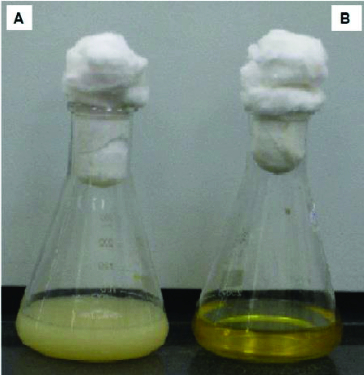
Chromate reduction by Halomonas sp. M-Cr. Cells were grown in alkaline LB medium (pH 10) amended with 50 mg L−1 Cr(VI), and incubated at 30 °C with shaking at 120 rpm. Complete Cr(VI) reduction was achieved within 120 h and white-precipitate was visible at the bottom of the flask (A). Cell-free control was used to monitor any abiotic reduction of Cr(VI) (B).
Effect of pH and salinity on Halomonas sp. M-Cr growth and Cr (VI) reduction
Data in Figure 4 confirm that Halomonas sp. M-Cr cells favour the reduction under alkaline conditions compared to neutral or acidic conditions. The first report on Cr(VI) reduction under alkaline conditions by Halomonads was reported by Van Engelen et al. [16] for H. sp. MV-2007, isolated from Soap Lake, a chemical stratified alkaline lake located in central Washington State, USA, followed by Shapovalova et al. [26] for H. chromatireducens AGD 8-3.
Figure 4.

Effect of pH on growth and chromate reduction by Halomonas sp. M-Cr after incubation for 48 h with 50 mg L−1 Cr(VI), NaCl 0.5%, and agitation of 120 rpm, 30 °C.
As depicted in Figure 5, maximum growth and chromate reduction (82.22%) were observed in the presence of 5% NaCl after 48 h incubation. Regarding the genus Halomonas, only H. chromatireducens, isolated from soda solonchak soils of the Kulunda steppe (Russia) and H. sp. TA-04, isolated from polluted marine sediments near a stainless steel plant in Southern Italy have been described as a Cr(VI) reducer under high salinity.[21,26]
Figure 5.
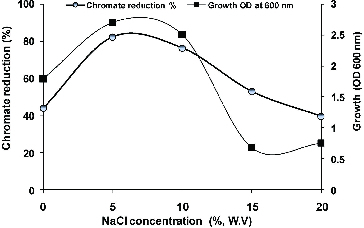
Effect of different concentrations of NaCl on growth and chromate reduction by Halomonas sp. M-Cr growing in LB medium of pH 10 with initial Cr(VI) concentration of 50 mg L−1 Cr(VI) after incubation for 48 h at 30 °C.
SEM and EDX analysis of Halomonas sp. M-Cr cells
Trivalent form of chromium is known to readily precipitate as chromium hydroxide [Cr(OH)3] above pH of 5.0.[27] Cell morphology of Halomonas sp. M-Cr was observed with SEM after cultivation of bacteria for 48 h with and without Cr(VI). In the presence of Cr(VI), elongation of cells (2.0–3.0 μm) with appearance of wrinkles on the surface was observed (Figure 6B). Similar effects on cellular morphology and surface topology has been observed in other gram-negative bacteria like: Acinetobacter haemolyticus and Serratia sp. Cr-10.[22,28] Also SEM analysis revealed some bacterial cells encrusted with amorphous precipitates or the precipitate was formed at random sites (Figure 6C, Figure 6D). The largest precipitates were slightly rounded. Similar results were obtained for Shewanella oneidensis and Acinetobacter haemolyticus [29,30] in which various sizes of Cr(III) precipitates were bound to the cell and restricted to the outer surface after chromate reduction. The precipitate was assumed to be in the Cr(III) form, Cr(OH)3, due to the inability of chromate anions to bind with electronegative surface functional groups (e.g. carboxyl, phosphoryl and hydroxyl) commonly found on gram-negative envelopes.[28,31]
Elemental analysis of the amorphous precipitate by EDX revealed that the peak corresponding to chromium was higher than for other elements (Figure 7), indicating that Cr was the major element comprising 46.3% of the total weight of the precipitate. The SEM image (Figure 7, inset A) shows the presence of Cr(OH)3 precipitates adhered to the surface of the rods shaped cells.
Figure 7.
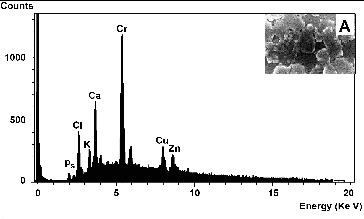
EDX spectrum analysis of amorphous precipitates that surrounded Halomonas sp. M-Cr cell surfaces during Cr(VI) reduction (A). An EDX spectrum from the dense particles generated a large Cr peak, indicating that it is most likely an amorphous Cr(III) hydroxide. Inset: SEM images of Halomonas sp. M-Cr cells and precipitates. The whole area was analysed with EDX.
Screening of significant variables using Plackett–Burman design
To the best of our knowledge, very few studies applied statistical design to optimize bioreduction of Cr(VI).[32–35] Plackett–Burman design, an efficient technique for medium component optimization,[35,36] was employed to identify significant variables that enhance chromate reduction by Halomonas sp. M-Cr and to find out their probable optimal levels in a limited number of experiments. Nine variables were analysed with regard to their effects on chromate reduction using a Plackett–Burman design. The responses in Table 1 show a wide variation in Cr(VI) reduction efficiency, ranging from 39% to 81% corresponding to the combined effect of the nine parameters in their specific ranges.
Analysis of variance (ANOVA) for the results of Plackett–Burman design is shown at Table 2. The model determination coefficient of the regression model (R 2 = 0.9669) indicates that 96.69% of the variability in the response could be explained by the model. Adjusted determination coefficient (Adj R 2 = 0.8179) was also high in order to support a high significance of the model.
Table 2.
Statistical analysis of Plackett–Burman design results.
| Variables | Coefficient | Effect | t-value | P-value | Confidence level (%) |
|---|---|---|---|---|---|
| Intercept | 59.71 | – | – | – | – |
| Glucose | 6.71 | 13.42 | 4.02 | 0.06 | 94 |
| (NH4)2SO4 | 1.04 | 2.08 | 0.62 | 0.6 | 40 |
| Yeast extract | −2.18 | −4.36 | −1.31 | 0.32 | 68 |
| Tryptone | 7.015 | 14.03 | 4.21 | 0.05 | 95 |
| KH2PO4 | 5.85 | 11.7 | 3.51 | 0.07 | 93 |
| MgSO4·7H2O | −0.71 | −1.42 | −0.42 | 0.71 | 29 |
| NaCl | −3.15 | −6.3 | −1.89 | 0. 2 | 80 |
| K2CrO4 | −3.52 | −7.04 | −2.11 | 0.17 | 83 |
| Inoculum size | 2.32 | 4.64 | 1.39 | 0.3 | 70 |
Note: R 2 = 0.9669; Adj R 2 = 0.8179.
Main effect analysis revealed that, five out of the nine variables (glucose, tryptone, KH2PO4, (NH4)2SO4 and inoculum size) included in this study were found to have a positive influence on Cr(VI) reduction, indicating that the higher concentrations of these variables are ideal for enhancing Cr(VI) reduction, whereas chromate, yeast extract, NaCl and MgSO4·7H2O had negative effect towards reduction process, indicating that lower concentrations of these factors in experimental range were favourable for increasing Cr(VI) reduction (Figure 8). Variables with the confidence levels greater than 90% were considered as significant. Tryptone was considered the most significant factor (95% confidence level), followed by glucose at 94%, and KH2PO4 at 93%. The confidence levels of other variables were below 90%; hence, their individual effects were negligible. If the variables which were insignificant were to be neglected, the model equation for chromate reduction efficiency can be written as
 |
Figure 8.
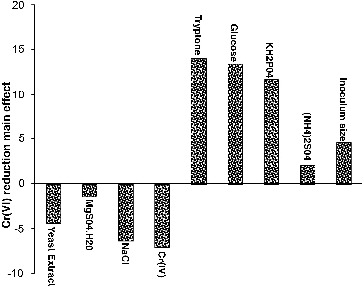
Positive and negative influence of different variables on Cr(VI) reduction by Halomonas sp. M-Cr based on the result of Plackett–Burman design.
The positive correlation between tryptone concentration and chromate reduction implies that a higher concentration is more effective in increasing reduction efficiency in the chosen experimental limits. This indicates nutrient requirement for optimum chromium reduction which depends on the nature of microbial employed species.[34] The positive significance effect of glucose on reduction process by Halomonas sp. M-Cr was probably due to the increase in metabolic activity. It has been found that chromium transport into the bacterial cell depends on energy. Therefore, it is a glucose dependent process.[37] These results are in consistence with other reports indicating requirement of glucose as electron donor for Cr(VI) reduction.[5,21,35,38] Phosphate sources play a crucial role in cellular respiration and metabolism of the microbes which induces the microbe to uptake the metal ions.[39]
Verification of the predicted optimum variables
The Plackett–Burman design predicted that the higher chromate reduction efficiency was achieved in a medium containing (g L−1): glucose 15, (NH4)2SO4 1.5, yeast extract 0.25, tryptone 5, KH2PO4 0.75, MgSO4·7H2O 0.05, NaCl 50 and pH 10.0. In order to evaluate the accuracy of the applied Plackett–Burman design, a verification experiment was carried out in triplicate. The average of reduction of predicted near optimum levels of independent variables were examined and compared to the centre condition settings. Optimization improved the Cr(VI) reduction efficiency to 100% of 50 mg L−1 Cr(VI) in 12 h compared with 60% removal in 24 h before optimization, which represented an increase in reduction efficiency of 40%. These results confirm the validity of the optimized medium.
Conclusions
This study reports the isolation of a potent Cr(VI) reducing moderate halophilic and alkaliphilic Halomonas sp. M-Cr from tannery effluent. Potential reduction of chromium by Halomonas sp. M-Cr was optimized by employing Plackett–Burman design. This design helped in locating the optimum levels of the most significant parameters which contribute to the maximum Cr(VI) reduction. Plackett–Burman not only demonstrated the increase in chromate reduction by Halomonas sp. M-Cr at the optimized conditions but also proved to be simple, efficient and time and material saving. The ability of the strain to reduce Cr(VI) in the presence of high salinity as well as at high pH expands the opportunities for bioremediation of marine polluted environments or wastewaters containing high salt concentrations. To the best of our knowledge, this is the only report available in the literature on chromium reduction by Halomonas sp. isolated from tannery effluents.
References
- Sarangi A, Krishnan C. Comparison of in vitro Cr(VI) reduction by CFEs of chromate resistant bacteria isolated from chromate contaminated soil. Bioresour Technol. 2008;99:4130–4137. doi: 10.1016/j.biortech.2007.08.059. [DOI] [PubMed] [Google Scholar]
- Xu L, Luo M, Jiang C, Wei X, Kong P, Liang X, Zhao J, Yang L, Liu H. In vitro reduction of hexavalent chromium by cytoplasmic fractions of Pannonibacter hragmitetus LSSE-09 under aerobic and anaerobic conditions. Appl Biochem Biotech. 2012;166:933–941. doi: 10.1007/s12010-011-9481-y. [DOI] [PubMed] [Google Scholar]
- Park CH, Keyhan M, Wielinga B, Fendrof S, Matin A. Purification to homogeneity and characterization of a novel Pseudomonas putida chromate reductase. Appl Environ Microbiol. 2000;66:1788–1795. doi: 10.1128/aem.66.5.1788-1795.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz MIR, Perez CD, Vargas E, Riveros-Rosas H, Campos Garcia J, Cervantes C. Mechanisms of bacterial resistance to chromium compounds. Biometals. 2008;21:321–332. doi: 10.1007/s10534-007-9121-8. [DOI] [PubMed] [Google Scholar]
- Ibrahim ASS, El-Tayeb AM, Elbadawi BY, Al-Salamah AA, Antranikian G. Hexavalent chromate reduction by alkaliphilic Amphibacillus sp. KSUCr3 is mediated by copper-dependent membrane-associated Cr(VI) Extremophiles. 2012;16:659–668. doi: 10.1007/s00792-012-0464-x. [DOI] [PubMed] [Google Scholar]
- Liu Z, Wu Y, Lei C, Liu P, Gao M. Chromate reduction by a chromate-resistant bacterium Microbacterium sp. World J Microbiol Biotechnol. 2012;28:1585–1592. doi: 10.1007/s11274-011-0962-5. [DOI] [PubMed] [Google Scholar]
- Soni SK, Singh R, Awasthi A, Singh M, Kalra A. In vitro Cr(VI) reduction by cell-free extracts of chromate-reducing bacteria isolated from tannery effluent irrigated soil. Environ Sci Pollut Res. 2013;20:1661–1674. doi: 10.1007/s11356-012-1178-4. [DOI] [PubMed] [Google Scholar]
- He M, Xiangyang L, Liu H, Miller SJ, Wang G, Rensing C. Characterization and genomic analysis of a chromate resistant and reducing bacterial strain Lysinibacillus fusiformis ZC1. J Hazard Mater. 2011;185:682–688. doi: 10.1016/j.jhazmat.2010.09.072. [DOI] [PubMed] [Google Scholar]
- Holt JG, Krieg NR, Sneath PHA, Staley JT, Williams ST. Bergey's manual of determinative bacteriology. 9th ed. Baltimore (MD): Williams and Wilkins; 1994. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning A: Laboratory manual. New York (NY): Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Ausuble FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Short protocols in molecular biology. New York (NY): Wiley; 1999. [Google Scholar]
- American Public Health Association (APHA) Standard methods for the examination of water and waste water. 21st ed. Washington D C, USA: 2005. [Google Scholar]
- Mary MS, Vincent S, Janarthanan S, Rao TS, Tata BVR. Bioreduction of Cr(VI) by alkaliphilic Bacillus subtilis and interaction of the membrane groups. Saudi J Biol Sci. 2011;18:157–167. doi: 10.1016/j.sjbs.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plackett RL, Burman JP. The design of optimum multifactorial experiments. Biometrica. 1946;33:305–325. [Google Scholar]
- Ye Q, Roh Y, Carroll SL, Blair B, Zhou J, Zhang CL, Fields MW. Alkaline anaerobic respiration isolation and characterization of a novel alkaliphilic and metal-reducing bacteria. Appl Environ Microbiol. 2004;70:5595–5602. doi: 10.1128/AEM.70.9.5595-5602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Engelen MR, Peyton BM, Mormile MR, Pinkart HC. Fe(III), Cr(VI) and Fe(III) mediated Cr(VI) reduction in alkaline media using a Halomonas isolate from Soap Lake, Washington. Biodegradation. 2008;19:841–850. doi: 10.1007/s10532-008-9187-1. [DOI] [PubMed] [Google Scholar]
- Mormile MR, Romine MF, Garcia MT, Ventosa A, Bailey TJ, Peyton BM. Halomonas campisalis sp.nov., a denitrifying, moderately haloalkaliphilic bacterium. Syst Appl Microbiol. 1999;22:551–558. doi: 10.1016/S0723-2020(99)80008-3. [DOI] [PubMed] [Google Scholar]
- Boltyanskaya YV, Kevbrin VV, Lysenko AM, Kolganova TV, Tourova TP, Osipov GA, Zhilina TN. Halomonas mongoliensis sp. nov. and Halomonas kenyensis sp. nov., New haloalkaliphilic denitrifiers capable of N2O reduction, isolated from Soda Lakes. Microbiology. 2007;76:739–747. [PubMed] [Google Scholar]
- Romano I, Giordano A, Lama L, Nicolaus B, Gambacorta A. Halomonas campaniensis sp.nov., haloalkaliphilic bacterium isolated from a mineral pool of Campania Region, Italy. Syst Appl Microbiol. 2005;28:610–618. doi: 10.1016/j.syapm.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Nieto JJ, Fernandez-Castillo R, Marquez MC, Ventosa A, Quesada E, Ruiz, Berraquero F. Survey of metal tolerance in moderately halophilic eubacteria. Appl Environ Microbiol. 1989;55:2385–2390. doi: 10.1128/aem.55.9.2385-2390.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focardi S, Pepi M, Landi G, Gasperini S, Ruta M, Biasio PD, Focardi SE. Hexavalent chromium reduction by whole cells and cell free extract of the moderate halophilic bacterial strain Halomonas sp. TA-04. Int J Biodeterioration Biodegradation. 2012;66:63–70. [Google Scholar]
- Zhang K, Li F. Isolation and characterization of a chromium-resistant bacterium Serratia sp. Cr-10 from a chromate-contaminated site. Appl Microbiol Biotechnol. 2011;90:1163–1169. doi: 10.1007/s00253-011-3120-y. [DOI] [PubMed] [Google Scholar]
- Cheng G, Li X. Bioreduction of chromium (VI) by Bacillus sp. isolated from soils of iron mineral area. Eur J Soil Biol. 2009;45:483–487. [Google Scholar]
- Faisal M, Abdul H, Hasnain Sh. Chromium-resistant bacteria and cyanobacteria: impact on Cr(VI) reduction potential and plant growth. J Ind Microbiol Biotechnol. 2005;32:615–621. doi: 10.1007/s10295-005-0241-2. [DOI] [PubMed] [Google Scholar]
- Guha H, Jayachandran K, Maurrasse F. Kinetics of chromium(VI) reduction by a type strain Shewanella alga under different growth conditions. Environ Pollut. 2001;115:209–218. doi: 10.1016/s0269-7491(01)00108-7. [DOI] [PubMed] [Google Scholar]
- Shapovalova AA, Khijniak TV, Tourova TP, Sorokin D Yu. Halomonas chromatireducens sp. nov., a new denitrifying facultatively haloalkaliphilic bacterium from solonchak soil capable of aerobic chromate reduction. Microbiology. 2009;78:102–111. [PubMed] [Google Scholar]
- Rai D, Sass BM, Moore DA. Chromium (III) hydrolysis constants and solubility of chromium (III) hydroxide. Inorg Chem. 1987;26:345–349. [Google Scholar]
- Pei QH, Shahir S, Raj ASS, Zakaria ZA, Ahmad WA. Chromium (VI) resistance and removal by Acinetobacter haemolyticus . World J Microbiol Biotechnol. 2009;25:1085–1093. [Google Scholar]
- Daulton TL, Little BJ, Jones-Meehan J, Blom DA, Allard LF. Microbial reduction of chromium from the hexavalent to divalent state. Geochim Cosmochim Acta. 2007;71:556–565. [Google Scholar]
- Zakaria ZA, Zakaria Z, Surif S, Ahmad WA. Hexavalent chromium reduction by Acinetobacter haemolyticus isolated from heavy-metal contaminated wastewater. J Hazard Mater. 2007;146:30–38. doi: 10.1016/j.jhazmat.2006.11.052. [DOI] [PubMed] [Google Scholar]
- Mclean SJ, Beveridge TJ. Chromate reduction by a Pseudomonad isolated from a site contaminated with chromated copper arsenate. Appl Environ Microbiol. 2001;67:1076–1084. doi: 10.1128/AEM.67.3.1076-1084.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazouki M, Keyanpour-Rad M, Shafie Sh, Shahhoseini Sh. Efficiency of Penicillium chrysogenum PTCC5037 in reducing low concentration of chromium hexavalent in a chromium electroplating plant wastewater. Bioresour Technol. 2007;98:2116–2122. doi: 10.1016/j.biortech.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Mabrouk MEM. Statistical optimization of medium components for chromate reduction by halophilic Streptomyces sp. MS-2. Afr J Microbiol Res. 2008;2:103–109. [Google Scholar]
- Venil CK, Mohan V, Lakshmanaperumalsamy P, Yerima MB. Optimization of chromium removal by the indigenous bacterium Bacillus spp. REP02 using the response surface methodology. ISRN Microbiology. 2011;2011:1–9. doi: 10.5402/2011/951694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulimi M, Subika J, Jastin S, Natarajan C, Amitava M. Enhancing the hexavalent chromium bioremediation potential of Acinetobacter junii VITSUKMW2 using statistical design experiments. J Microbiol Biotechnol. 2012;22:1767–1775. doi: 10.4014/jmb.1203.03063. [DOI] [PubMed] [Google Scholar]
- Mabrouk MEM, ElAhwany AMD, Beliah MMB, Sabry SA. Xanthan production by a novel mutant strain of Xanthomonas campestris: Application of statistical design for optimization of process parameters. Life Sci J. 2013;10:1660–1667. [Google Scholar]
- Silva B, Figueiredo H, Neves IC, Tavares T. The role of pH on Cr(VI) reduction and removal by Arthrobacter viscosus . Int J Chem Biol Eng. 2009;2:100–103. [Google Scholar]
- Kathiravan MN, Karthick R, Muthu N, Muthukumar K, Velan M. Sonoassisted microbial reduction of chromium. Appl Biochem Biotechnol. 2010;160:2000–2013. doi: 10.1007/s12010-009-8716-7. [DOI] [PubMed] [Google Scholar]
- Nasseri S, Assadi MM, Sepehr NM, Rostami KH, Sharit M, Nadafi K. Chromium removal from tanning effluent using biomass of Aspergillus oryzae . Pakistan J Biol Sci. 2002;5:1056–1059. [Google Scholar]


