Abstract
This article elucidates that strain Pseudomonas aeruginosa (IES-Ps-1) is a versatile toxic organic compound degrader. With the degradation of malathion and cypermethrin (studied by other researchers previously), this strain was able to degrade phenol. Two other indigenous soil flora (i.e., Pseudomonas sp. (IES-S) and Bacillus subtilis (IES-B)) were also found to be potential phenol degraders.
Phenol was degraded with Monod kinetics during growth in nutrient broth and mineral salts medium. Before entering into the growth inhibition phase, strains IES-Ps-1, IES-S and IES-B could tolerate up to 400, 700 and 500 mg/L phenol, respectively, when contained in nutrient broth. However, according to the Luong–Levenspiel model, the growth of strains IES-Ps-1, IES-S and IES-B would cease at 2000, 2174 and 2190 mg/L phenol, respectively. Strain IES-Ps-1 degraded 700, 900 and 1050 mg/L phenol contained in mineral salts medium with the specific rates of 0.034, 0.075 and 0.021 h−1, respectively. All these strains grew by making clusters when exposed to phenol in order to prevent damages due to high substrate concentration. These strains transformed phenol into catechol, which was then degraded via ortho-cleavage pathway.
Keywords: biodegradation, phenol, Pseudomonas, Bacillus, kinetics
Introduction
Being the basic structural units for a variety of organic compounds, phenol and its derivatives are used in oil refineries, manufacturing phenol and its derivatives, paints, pharmaceuticals, industries of resins, textile dyes, disinfectants, petrochemicals and paper mills, hence found in the effluents of these industries.[1–4]
Phenol is highly toxic to all life forms in all concentrations (5–2000 mg/L) and considered as a priority pollutant.[5,6] The daily average amount of phenol to human exposure should not exceed 20 mg.[1] Also, phenol can pose lethal effects in marine species at concentrations of 5–25 ppm.[4] Therefore, it is extremely important to develop efficient techniques to remove these pollutants from the environment to save natural ecosystems for sustainable development.[7]
A variety of methods are available for the removal of phenols from industrial effluents. The efficiency of these methods is mainly dependent on the time taken for the complete removal and the initial concentrations of phenols to be degraded. Some processes can be very costly and this is a large determinant when choosing a system. Adsorption of phenol on activated carbon,[8] solvent extraction [9] and chemical processes (i.e., chlorination, ozonation, benzol extraction and oxidation) can give up to 90% removal efficiency.[10,11] However, these processes usually produce a variety of intermediates and by-products, which are usually more toxic than the original substrate and lead to secondary effluent problems.[5,6,12] Toxic emissions from such processes can also be produced and these can be more damaging than the reactants.
Biological degradation is the most efficient and low-cost method for the removal of phenols from wastewaters [12–14] as it leads to complete mineralization of phenols and environmentally acceptable and less expensive.
Studies have shown that phenol can be aerobically degraded by a wide variety of microorganisms of genera Pseudomonas,[15–22] Acinetobacter,[23] Alcaligenes,[24,25] Bacillus,[26] Burkholderia,[27,28] Nocardia,[29] Nocardioides,[30] Ralstonia [31] and Rhodococcus.[32] In this study, we have presented the pathway and degradation kinetics of phenol by Pseudomonas and Bacillus species.
Materials and methods
Culture medium and growth conditions
Bacterial strains were grown aerobically at 35 °C under shake flask conditions. Growth medium (mineral salts medium, MM) contained per litre: 5.4 g of Na2HPO4⋅12H2O, 1.4 g of KH2PO4, 0.5 g of (NH4)2SO4, 0.2 g of MgSO4⋅7H2O, supplemented with trace elements solution (5 mL L−1) containing per litre 780 mg of Ca(NO3)2⋅4H2O, 200 mg of FeSO4⋅7H2O, 20 mg of Na2SeO4⋅10H2O, 10 mg of ZnSO4⋅7H2O, 10 mg of H3BO3, 10 mg of CoCl2⋅6H2O, 10 mg of CuSO4⋅5H2O, 4 mg of MnSO4⋅H2O, 3 mg of Na2MoO4⋅2H2O, 2 mg of NiCl2⋅6H2O.
Culture used
Three bacterial strains were used in this study. One of them was Pseudomonas sp. IES-Ps-1, which was initially isolated and identified in 1995 from soil using enrichment technique with malathion (an organophosphate pesticide) as the sole source of carbon and energy.[33] Later in 2004, strain IES-Ps-1 was used to study the degradation of cypermethrin (a common synthetic pyrethroid insecticide).[34] Since IES-Ps-1 showed high tolerance against hazardous chemicals, it was then used to degrade phenol in this study. The culture is characterized as motile, gram-negative and thin short rod shaped.
Isolation of new bacterial strains able to degrade phenol
Two different species of phenol degraders were isolated and characterized by enrichment technique. For this, samples were collected from activated sludge of the treatment plant of a steel mill, which is situated at Karachi, Pakistan. Three test tubes containing 5 mL sterile nutrient broth (NB) were incubated with 1 g (wet weight) activated sludge and incubated under non-shaking conditions at 35 °C. Growth of microbial strains was observed after 24 h and samples of the enrichment culture were spread on nutrient agar (NA) plates. Morphologically different colonies from NA plates were separately incubated into 120 mL flasks containing 50 mL MM supplemented with 100 mg/L phenol. After every 7–8 days, cultures were inoculated into series of flasks containing 50 mL MM with increasing concentration of phenol (200–2000 mg/L). Phenol tolerant bacterial strains were isolated by streaking the loopful of MM on NA plates. Morphologically different strains were separately streaked on NA plates and preserved on NA slants, temporarily. The cultures were sub-cultured after every two–three months. Purity of the cultures was regularly checked during preservation and study period.
Identification and characterization of isolated strains
Strain IES-Ps-1 was previously identified up to the genus level.[33] In this study, the specie type of this strain was identified. The genera and/or species of other isolates were identified by performing gram staining, motility, morphological characteristics and biochemical tests with some modifications described by Bergey's manual and Sivaraj et al.[35,36] Gram reactions and morphological characteristics of the isolated strains were observed under the oil immersion objectives of the microscope. True motility of the bacterial isolates was observed under high microscope magnification (1000X) using hanging drop method.
Growth and degradation kinetics under batch conditions
For studying growth and degradation kinetics of isolated strains, pure cultures of strain IES-Ps-1, IES-S and IES-B were separately inoculated into a series of 500 mL Erlenmeyer flasks containing 250 mL NB and MM supplemented with 100–1200 mg/L of phenol. Flasks were incubated at 35 °C in a shaking water bath (Grant SS30, UK) at 125 rpm. Samples were drawn every hour from NB and after every day from the MM flasks. Growth kinetics of the strains was calculated by monitoring the optical densities at 600 nm and degradation kinetics were studied by measuring the residual concentration of phenol in the medium.
Growth kinetics
For studying growth kinetics of Pseudomonas and Bacillus strains with phenol as the sole source of carbon and energy, mathematical modelling was performed. For this, we used the Monod equation:[37]
| (1) |
where µ is the specific growth rate (h−1), µmax the maximum specific growth rate (h−1), S the substrate concentration (mmol/L) at time t, and Ks is the half saturation coefficient (mmol/L).
For studying the growth inhibition of bacterial strains at high phenol concentrations, the Haldane–Andrew model [38,39] was used, which is given by the following equation:
 |
(2) |
where Ki is the substrate inhibition constant (mmol/L).
Luong [40] and Levenspiel [41] extended the Monod type model to describe the growth inhibition at high substrate concentrations:
| (3) |
where Sm is the critical inhibitor concentration above which growth stops and n is an empirical constant.
Growth and degradation rates of Pseudomonas and Bacillus strains on phenol were calculated by measuring the slopes of decreasing optical densities at 600 nm and phenol concentrations at different time points.
Removal of phenol from the bioreactor
Degradation of phenol was studied by inoculating the respective bacterial strains in a bioreactor containing 4 litres of domestic wastewater collected from University of Karachi, Pakistan campus under non-sterile conditions. Physical conditions of the reactor were maintained (i.e., 35 °C, 350 rpm and normal pH) with constant supply of air in order to maintain 5 mg/L dissolved oxygen.
Determination of cell concentration and electron microscopy of microbial strains
Cell concentrations of bacterial strains were determined from optical density measurements at 600 nm using a UV/visible spectrophotometer (Shimadzu, Japan) with the NB or MM as blank.
For visualization of bacterial strains, samples were prepared by taking 5 mL tryptic soy broth in corning tube. The desired culture was inoculated in the sterilized medium and incubated at 37 °C. After 24 h, 2 mL of 20% glucose solution was added in the culture medium and further incubated. All materials were discarded from corning tube after 24 h. Five millilitres of 99% methanol were added in the tube and left for 15 minutes, then discarded. Finally, 0.1% crystal violet was added and left for 20 minutes, then washed with sterilized distilled water. Air dried samples were cut into miniature pieces and placed under analytical scanning electron microscope (SEM).
The samples were plated with gold in JEOL quick auto coater JFC-1500 Ion Sputtering Device and observed with SEM (JEOL JSM – 6380, MP- 41080, Japan).
Analytical methods
Residual concentrations of phenol were estimated with spectrophotometer (Shimadzu UV-Vis 1201, Japan) by 4-aminoantipyrene method.[22,42] and by reverse phase high pressure liquid chromatography (HPLC) using a Novapak C18 column (250 × 4.6 mm, 5 µm particle size). The mobile phase used was 100% methanol. An injection volume of 20 µL was used. The flow rate was 0.8 mL/min and detection at 254 nm was with a UV-Vis absorbance detector.
Results and discussion
Isolation of phenol degraders
Bacterial strains of different genera were found as potential degraders of phenol. These strains were separately streaked on agar plates containing varying concentrations of phenol. The concentration of phenol was gradually increased to check their tolerance. The highest concentration of phenol was 2000 mg/L. Finally, one gram positive Bacillus and two gram negative short rods were selected for studying the degradation of phenol.
Identification and characterization of phenol degraders
The identification and characterization of bacterial isolates were performed using morphological, cultural and biochemical tests (Table 1) as described in Bergey's manual.[36] The identification of strain IES-Ps-1 was confirmed to be Pseudomonas aeruginosa. The other two new isolates were identified as Pseudomonas sp. and Bacillus subtilis and designated as IES-S and IES-B, respectively.
Table 1.
Morphological, cultural and biochemical characteristics of isolated strains able to degrade phenol.
| Test |
IES-Ps-1 |
IES-S |
IES-B |
| Description of colonies | Large, flat, greenish pigment | Pale coloured mucoid colonies on MacConkey | Large dry cream coloured irregular colonies on agar |
| Shape | Irregular | Regular | Irregular |
| Elevation | Umbonate | Convex | Flat |
| Gram reaction | Gram-negative short rods | Gram-negative short rods | Gram-positive short rods |
| Motility | Motile | Motile | Motile |
| O2 need | Strict aerobe | Strict aerobe | Strict aerobe |
| Optimal growth temperature (C) | 37 | 37 | 37 |
| Acid from glucose | + | − | + |
| Oxidase | + | + | − |
| Catalase | + | + | + |
| Indole | − | − | − |
| Citrate | + | + | − |
| H2S production | − | − | − |
| Nitrate reduction | + | − | + |
| Urea hydrolysis | + | + | − |
| Gelatin hydrolysis | + | + | + |
| Lactose fermentation | − | − | − |
| Sucrose | − | − | + |
| Fructose | + | − | − |
| Mannose | + | − | − |
| Ribose | + | − | − |
| Xylose | + | − | − |
| Cetramide agar base (CAB) | Fluorescent green colonies | − | − |
Kinetics of phenol degradation in NB
In order to study the kinetic properties of Pseudomonas sp. strain IES-Ps-1, B. subtilis sp. strain IES-B and Pseudomonas sp. strain IES-S, we performed a number of growth experiments in batch cultures. Cells of these strains were separately incubated in NB containing increasing concentrations (50–1200 mg/L) of phenol (data not shown). A comparison in growth kinetics of strain IES-Ps-1 in two different media (enriched and artificial) was studied by incubating cells in separate flasks containing NB and MM supplemented with 100–1050 mg/L phenol. Cell growth with respect to time was measured in each batch culture with a certain phenol concentration. The specific growth rates at different phenol concentrations were calculated by measuring the slopes of increasing optical densities at 600 nm. From the results, it is obvious that the specific growth rate increases with an increase in substrate concentration until a maximum value is reached. With strain IES-Ps-1, IES-S and IES-B, the maximum specific growth rates were 0.26, 0.43 and 0.78 h−1, respectively (Figure 1). However, specific growth rates started to decrease when phenol concentrations were above 220, 310 and 320 mg/L in cultures of IES-Ps-1, IES-S and IES-B, respectively. This decline indicates the inhibition in growth, which can be caused due to cell damage or disruption of membrane integrity at higher phenol concentrations.[43,44]
Figure 1.
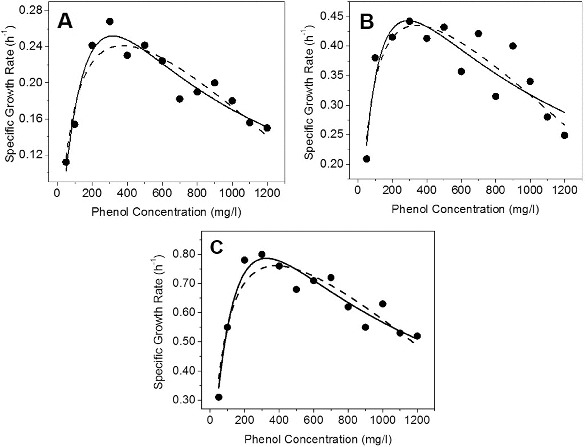
Growth kinetics of (A) IES-Ps-1, (B) IES-S and (C) IES-B in nutrient broth under shake flask conditions supplemented with phenol. Experimental data points are shown with dots. Haldane–Andrew regression curves are mentioned with regular lines and Luong–Levenspiel regression curves are indicated with dashed line.
In order to determine the growth parameters, the Monod model was used. The Haldane–Andrew and Luong–Levenspiel models were used to determine the effects of substrate inhibition on growth rates. Growth kinetic data were fitted by non-linear regression and the parameters of the kinetic constants were calculated with Microcal Origin 8.0. The parameters estimated by using these models are summarized in Table 2.
Table 2.
Kinetic parameters obtained using Monod, Luong–Levenspiel and Haldane-Andrew models in batch mode with pure culture of Pseudomonas (IES-Ps-1), Pseudomonas (IES-S) and Basillus (IES-B) growing in NB supplemented with phenol.
| Strain IES-Ps-1 |
Strain IES-S |
Strain IES-B |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Kinetic parameter | Monod model | Luong–Levenspiel model | Haldane–Andrew model | Monod model | Luong–Levenspiel model | Haldane–Andrew model | Monod model | Luong–Levenspiel model | Haldane–Andrew model |
| mmax (h−1) | 0.30 | 0.38 | 0.60 | 0.51 | 0.63 | 0.8 | 0.9 | 1.2 | 1.8 |
| Ks (mg/l) | 98 | 111 | 234 | 71 | 77 | 121 | 92 | 102 | 206 |
| Ki (mg/l) | – | 394 | – | – | 696 | – | – | 520 | |
| Sm (mg/l) | 2000 | – | – | 2174 | – | – | 2190 | – | |
| n | 1 | – | – | 1 | – | – | 1 | – | |
The data of specific growth rate was fitted to Monod's model when the concentrations of phenol were lower than 400, 700 and 500 mg/L in strains IES-Ps-1, IES-S and IES-B, respectively. Haldane-Andrew and Luong–Levenspiel models were used when inhibition in growth started due to increase in phenol concentrations. The Ks values in Monod's model indicate the capability of the isolates to grow at lower substrate concentrations. The values of Ki in Haldane–Andrew model indicate the inhibition in growth of microbial strains that starts at a particular substrate concentration over which the specific growth and degradation rate decline. The higher Ki (i.e., 696 mg/L) of strain IES-S and 520 mg/L of strain IES-B compared to lower value (i.e., 394 mg/L) of strain IES-Ps-1 indicated a higher tolerance level in growth.
In Luong–Levenspiel model, the value of maximum substrate concentration (Sm) is used, at which the microbes cease to grow. The highest Sm value was obtained in strain IES-B, indicating that this strain can grow with up to 2190 mg/L of phenol. Similarly, strain IES-Ps-1 and strain IES-S can survive and grow with up to 2000 and 2174 mg/L of phenol, respectively (Table 2).
Kinetics of phenol degradation in mineral salts medium
In order to study the growth kinetics of IES-Ps-1 and degradation kinetics of phenol in MM, we performed a number of batch experiments in shake flask conditions. Strain IES-Ps-1 grew exponentially between 16 and 93, 0 and 72 and 33 and 140 h with specific growth rates of 0.034, 0.075 and 0.021 h−1 when 700 (Figure 2A), 900 (Figure 2B) and 1050 mg/L (Figure 2C) of phenol was incubated in MM, respectively.
Figure 2.
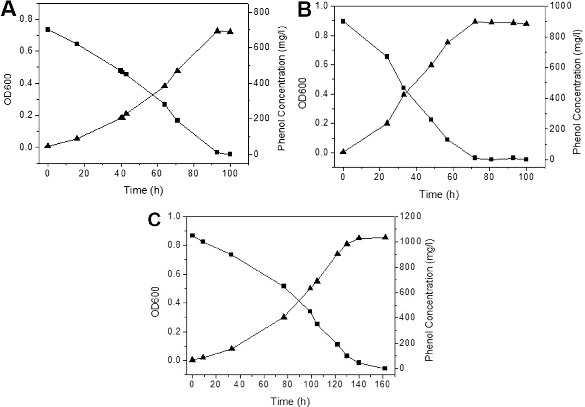
Growth kinetics of phenol degradation by Pseudomonas IES-Ps-1 in MM medium supplemented with phenol. (A) 700 mg/L, (B) 900 mg/L and (C) 1050 mg/L.
Electron microscopy of phenol degraders
The cultural morphology of bacterial isolates was studied by electron microscopy. The SEM images reveal that the isolated strains are rod-shaped cells with approximate 1 1 μm mu;m in width and 2–4 4 μm mu;m in length (Figure 3). It was also observed that the cells growing in plain NB were scattered. However, cells growing in NB containing phenol formed clusters in order to prevent damages due to high phenol concentration and the metabolites, which are formed intermediately. This behaviour of isolated strains can be explained with the fact that cluster formation is a type of defence mechanism in several microbial strains. Proteins, which are required to mineralize a substrate are mainly expressed under normal growth conditions. However, several other proteins (i.e., chaperonin, succinyl-CoA synthetase-β-subunit, thioredoxin reductase and superoxide dismutase) are also expressed that help cells growing under stress (i.e., high substrate concentration in this study).[45–47]
Figure 3.
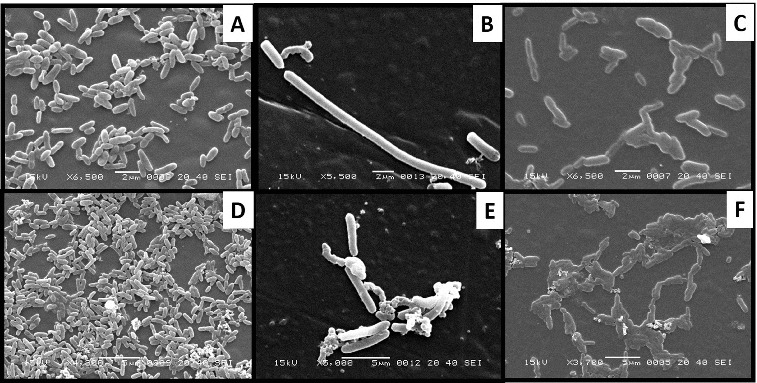
Cellular morphology of (A) Pseudomonas sp. IES-Ps-1 at 6500X magnification, (B) Bacillus sp. IES-B at 5500X magnification and (C) Pseudomonas sp. IES-S at 6500X magnification when grown in nutrient broth and the respective cells of IES-Ps-1, (D) IES-B (E) and IES-S (F) grown in mineral medium supplemented with phenol.
Degradation pathway of phenol
Degradation of phenol in MM by Pseudomonas sp. strain IES-Ps-1 in shake flask conditions was very slow during the first 24 h (Figure 4(A)). During this period, a peak of catechol with increasing concentration was observed through HPLC. After 24 h, quick degradation of phenol occurred and catechol disappeared. This indicates that the enzyme required for the degradation of phenol (i.e., phenol hydroxylase) was not as active as the enzyme that transforms catechol (i.e., catechol 1, 2-dioxygenase). Later, trace of cis, cis-muconate was observed when over 95% phenol and 99% catechol were degraded.
Figure 4.
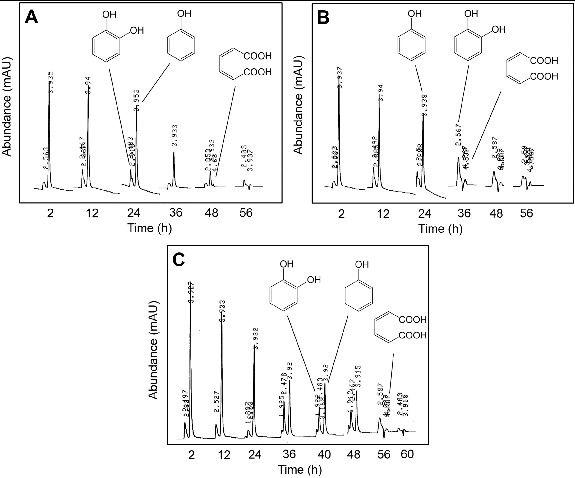
HPLC chromatograms showing the degradation of phenol in a bioreactor under non-sterile conditions supplemented with 700 mg/L of phenol when the bioreactor was inoculated with the cells of (A) Pseudomonas sp. strain IES-Ps-1, (B) Bacillus sp. strain IES-B and (C) Pseudomonas sp. strain IES-S.
Similar behaviour in Bacillus sp. strain IES-B was observed during degradation of phenol. A very slow degradation of phenol in the first 24 h was observed with the accumulation of trace amount of catechol (Figure 4(B)). A rapid degradation of phenol and catechol was observed in subsequent hours. Complete degradation of phenol occurred after 36 h and trace of cis,cis-muconate was monitored.
Pseudomonas sp. strain IES-S degraded phenol very slowly in the first 24 h (Figure 4C). Later, the concentration of phenol decreased quickly and became constant for 12 h with the accumulation of catechol. Phenol was completely degraded after 56 h with the accumulation of traces of catechol and muconic acid, which disappeared after 60 h.
Degradation studies of phenol by various Pseudomonas species have been conducted by many researchers under aerobic and anaerobic conditions.[19,20,48–53] The findings of this study suggest that phenol is aerobically degraded in MM by the isolated strains through ortho-cleavage pathway. The degradation is initiated with the formation of catechol, in which phenol hydroxylase (a monooxygenase) attached a hydroxyl group at ortho position with the benzene ring. Catechol is the main intermediate formed during biodegradation of phenol by different microbial strains, [54] which is degraded either via meta-cleavage by catechol 2,3-dioxygenase [13,55] or ortho-cleavage by catechol 1,2-dioxygenase.[56] The transformation of catechol via any route occurs very quickly by dioxygenases that results in quick disappearance of the immediate metabolites (i.e., cis, cis-muconate through ortho-cleavage or 2-hydroxymuconic semialdehyde through meta-cleavage).[19,48,49] In this study, we observed the formation of muconic acid with all three isolates (Figure 4), which indicates that phenol degradation occurs through an ortho-cleavage pathway. Later, the metabolites enter into the tricarboxylic acid cycle for complete degradation of phenol (Figure 5).
Figure 5.
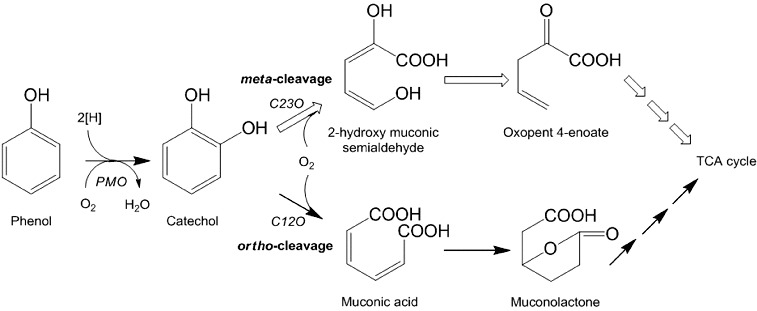
Phenol degradation pathway. The enzymes involved in first two steps of degradation are, PMO = phenol monooxygenase, C12O = catechol 1,2-dioxygenase and C23O = catechol 2,3-dioxygenase.
Conclusion
The results obtained through this study indicate that the isolated strains IES-Ps-1, IES-S and IES-B degraded phenol through an ortho-cleavage pathway. These strains possess high tolerance to phenol toxicity and are capable to degrade up to 400, 700 and 500 mg/L phenol, respectively, without any significant inhibition in batch conditions, hence could be utilized as capable candidates for bioremediation of phenolic wastewaters.
Acknowledgements
The authors would like to thank Mr Aamir Alamgir for proof reading the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Funding
The authors acknowledge the financial support from office of the dean faculty of science, University of Karachi through project grant invoice [NO. DFS/2007-162].
References
- ATSDR 2003 http://www.atsdr.cdc.gov/MHM1/mmg115.html Agency for toxic substances and disease registry. Medical management guidelines for phenol. Available from.
- Bandhyopadhyay K. Das D. Bhattacharyya P. Maiti BR. Reaction engineering studies on biodegradation of phenol by Pseudomonas putida MTCC 1194 immobilized on calcium alginate. Biochem Eng J. 2001;8:179–186. [Google Scholar]
- Bandyopadhyay K. Das D. Maiti BR. Kinetics of phenol degradation using Pseudomonas putida MTCC 1194. Bioprocess Eng. 1998;18:373–377. [Google Scholar]
- Mollaei M. Abdollahpoura S. Atashgahi S. Abbasi H. Masoomi F. Rada I. Lotfi AS. Zahiri HS. Vali H. Noghabi KA. Enhanced phenol degradation by Pseudomonas sp. Sa01: gaining insight into the novel single and hybrid immobilizations. J Hazard Mater. 2010;175:284–292. doi: 10.1016/j.jhazmat.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Kasikara-Pazarlioglu N. Telefoncu A. Biodegradation of phenol by Pseudomonas putida immobilized on activated pumice particles. Process Biochemistry. 2005;40:1807–1814. [Google Scholar]
- Han Y. Quan X. Chen S. Zhao H. Cui C. Zhao Y. Electrochemically enhanced adsorption of phenol on activated carbon fibers in basic aqueous solution. J Colloid Interface Sci. 2006;299:766–771. doi: 10.1016/j.jcis.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Puerto-Tello A. Moreno-piraján JC. Guzmán AM. Escudero ME. Velázquez L. Giraldo L. Sapag K. Decomposition of phenol by Pseudomonas aeruginosa immobilized on activated carbons. J Environ Eng Manag. 2009;19:73–78. [Google Scholar]
- Nakhla GF. Suidan MT. Pfeffer JT. Control of anaerobic GAC reactors treating inhibitory waste eaters. J Water Pollut Control Fed. 1990;62:65–72. [Google Scholar]
- Patterson JW. Wastewater treatment technology. Ann: Arbor, MI: Ann Arbor Science Publishers; 1975. [Google Scholar]
- Bond RG. Straub CP. Handbook of Environmental Control. Vol. IV, Wastewater: treatment and disposal. Cleveland, OH: CRC Press; 1974. p; p. 905. [Google Scholar]
- Wang YT. Effect of chemical oxidation on anaerobic biodegradation of model phenolic compounds. Water Environ Res. 1992;64:268–273. [Google Scholar]
- Hamed TA. Bayraktar E, Mehmetoğlu U, Mehmetoğlu T. The biodegradation of benzene, toluene and phenol in a two-phase system. Biochem Eng J. 2004;19:137–146. [Google Scholar]
- Gibson DT. Subramanian V. Microbial degradation of aromatic hydrocarbons. Microbial degradation of organic compounds. Dekker Inc; 1984. pp. 181–252. [Google Scholar]
- Wang SJ. Loh KC. Modeling the role of metabolic intermediates in kinetics of phenol biodegradation. Enzym Microb Technol. 1999;25:177–184. [Google Scholar]
- Kumar A. Kumar S. Kumar S. Biodegradation kinetics of phenol and catechol using Pseudomonas putida MTCC 1194. Biochem Eng J. 2004;22:151–159. [Google Scholar]
- Fava F. Armenante PM. Kafkewitz D. Marchetti L. Influence of organic and inorganic growth supplements on the aerobic biodegradation of chlorobenzoic acid. Appl Microbiol Biotechnol. 1995;43:171–177. doi: 10.1007/BF00170640. [DOI] [PubMed] [Google Scholar]
- Bettman H. Rehm HJ. Degradation of phenol by polymer entrapped microorganisms. Appl Microbiol Biotechnol. 1984;20:285–290. [Google Scholar]
- Feist C. Hegeman GD. Phenol and benzoate metabolism by Pseudomonas putida: regulation of tangential pathways. J Bacteriol. 1969;100:869–877. doi: 10.1128/jb.100.2.869-877.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill GA. Robinson CW. Substrate inhibition kinetics: phenol degradation by Pseudomonas putida . Biotechnol Bioeng. 1975;17:599–615. [Google Scholar]
- Hinteregger C. Leitner R. Loidl M. Ferschl A. Streichsbier F. Degradation of phenol and phenolic compounds by Pseudomonas putida EKII. Appl Microbiol Biotechnol. 1992;37:252–259. doi: 10.1007/BF00178180. [DOI] [PubMed] [Google Scholar]
- Sokol W. Dynamics of continuous stirred-tank biochemical reactor utilizing inhibitory substrate. Biotechnol Bioeng. 1988;31:198–202. doi: 10.1002/bit.260310303. [DOI] [PubMed] [Google Scholar]
- Yang RD. Humphery AE. Dynamic and steady sate studies of phenol biodegradation in pure and mixed cultures. Biotechnol Bioeng. 1975;17:1211–1235. doi: 10.1002/bit.260170809. [DOI] [PubMed] [Google Scholar]
- Paller G. Hommel RK. Kleber HP. Phenol degradation by Acinetobacter calcoaceticus NCIB 8250. J Basic Microbiol. 1995;35:325–335. doi: 10.1002/jobm.3620350508. [DOI] [PubMed] [Google Scholar]
- Hughes EJ. Bayly RC. Skurray RA. Evidence for isofunctional enzymes in the degradation of phenol, m- and p-toluate, and p-cresol via catechol meta-cleavage pathways in Alcaligenes eutrophus . J Bacteriol. 1984;158:79–83. doi: 10.1128/jb.158.1.79-83.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard D. Lindley ND. Carbon and energy flux constraints in continuous cultures of Alcaligenes eutrophus grown on phenol. Microbiology. 1998;144:241–248. doi: 10.1099/00221287-144-1-241. [DOI] [PubMed] [Google Scholar]
- Buswell JA. Metabolism of phenol and cresols by Bacillus stearothermophilus . J Bacteriol. 1975;124:1077–1083. doi: 10.1128/jb.124.3.1077-1083.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder M. Muller C. Posten C. Deckwer W-D. Hecht V. Inhibition kinetics of phenol degradation from unstable steady state data. Biotechnol Bioeng. 1997;54:567–576. doi: 10.1002/(SICI)1097-0290(19970620)54:6<567::AID-BIT7>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Solomon BO. Posten C. Harder MPF. Hecht V. Deckwer W-D. Energetics of Pseudomonas cepacia growth in a chemostat with phenol limitation. J Chem Technol Biotechnol. 1994;60:275–282. [Google Scholar]
- Rizzuti L. Augugliaro V. Torregrossa V. Savarino A. Kinetics of phenol removal by Nocardia species. Eur J Appl Microbiol Biotechnol. 1979;8:113–118. [Google Scholar]
- Cho Y. Rhee S. Lee S. Influence of phenol on biodegradation of p-nitrophenol by freely suspended and immobilized Nocardioides sp. NSP.41. Biodegradation. 2000;11:21–28. doi: 10.1023/a:1026512922238. [DOI] [PubMed] [Google Scholar]
- Leonard D. Lindly NA. Growth of Ralstonia eutropha on inhibitory concentration of phenol: diminished growth can be attributed to hydrophilic perturbation of phenol hydroxylase activity. Enzym Microb Technol. 1999;25:271–277. [Google Scholar]
- Hensel J., Straube G. Kinetic studies of phenol degradation by Rhodococcus sp. P1.II. Continuous cultivation. Antonievan Leeuwenhoek. 1990;57:33–36. doi: 10.1007/BF00400333. [DOI] [PubMed] [Google Scholar]
- Hashmi I. Microbial transformations of hazardous waste during biological waste treatment. 2000 www.eprints.hec.gov.pk dissertation] University of Karachi.
- Jilani S. Biodegradation of hazardous waste during biological treatment process. 2004 www.eprints.hec.gov.pk University of Karachi.
- Sivaraj R. Dorthy CAM. Veneckatesh R. Isolation, characterization, and growth kinetics of bacteria metabolizing textile effluent. J Biosci Technol. 2011;2:324–330. [Google Scholar]
- John GH. Sneath PH. Krieg NR. Holt JG. Holt JG. Bergey's manual of determinative bacteriology. Lippincott Williams and Wilkins. New York: 2000. 9th ed. [Google Scholar]
- Monod J. The growth of bacterial cultures. Annu Rev Microbiol. 1979;3:371–394. [Google Scholar]
- Haldane JSB. Enzymes, longmans, green, UK. Cambridge, MA: MIT Press; 1930. 1965. [Google Scholar]
- Andrew JF. A mathematical model for continuous culture of microorganisms utilizing inhibitory substrates. Biotechnol Bioeng. 1968;10:707–723. [Google Scholar]
- Luong JHT. Generalization of Monod kinetics for analysis of growth data with substrate inhibition. Biotechnol Bioeng. 1987;29:242–248. doi: 10.1002/bit.260290215. [DOI] [PubMed] [Google Scholar]
- Levenspiel O. The Monod equation. a revisit and a generalization to product inhibition situations. Biotechnol Bioeng. 1980;22:1671–1687. [Google Scholar]
- APHA (American Public Health Association) Standard methods for the examination of water and wastewater. 18th ed. Washington: , DC: American Public Health Association; 1992. [Google Scholar]
- Sikkema J. de Bont FA. Poolman B. Mechanisms of membrane toxicity of hydrocarbons. Microbiol Rev. 1995;59:201–222. doi: 10.1128/mr.59.2.201-222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber FJ. de Bont JA. Adaptation mechanisms of microorganisms to the toxic effects of organic solvents on membranes. Biochimica et Biophysica Acta. 1996;1286:225–245. doi: 10.1016/s0304-4157(96)00010-x. [DOI] [PubMed] [Google Scholar]
- Cabiscol E. Tamarit J. Ros J. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int Microbiol. 2000;3:3–8. [PubMed] [Google Scholar]
- Feder ME. Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol. 1999;61:243–282. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- Scott MD. Meshnick SR. Eaton JW. Superoxide dismutase-rich bacteria. Paradoxical increase in oxidant toxicity. J Biol Chem. 1987;262:3640–3645. [PubMed] [Google Scholar]
- Kotturi G. Robinson CW. Inniss WE. Phenol degradation by a psychrotrophic strain of Pseudomonas putida . Appl Microb Biotechnol. 1991;34:539–543. [Google Scholar]
- Nikakhtari H. Hill GA. Continuous bioremediation of phenol-polluted air in an external loop airlift bioreactor with a packed bed. J Chem Technol Biotechnol. 2006;81:1029–1038. [Google Scholar]
- Molin G. Nilsson I. Degradation of phenol by Pseudomonas putida ATCC 11172 in continuous culture at different ratios of biofilm surface to culture volume. Appl Environ Microbiol. 1985;50:946–950. doi: 10.1128/aem.50.4.946-950.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed AM. Nakhla FG. Farooq S. Phenol degradation by Pseudomonas aeruginosa . J Environ Sci Health A: Environ Sci Eng Toxicol. 1994;30:99–107. [Google Scholar]
- Allsop PJ. Chisti Y. Moo-Young M. Sullivan GR. Dynamics of phenol degradation by Pseudomonas putida . Biotechnol Bioeng. 1993;41:572–580. doi: 10.1002/bit.260410510. [DOI] [PubMed] [Google Scholar]
- Sá CSA. Boaventura RAR. Biodegradation of phenol by Pseudomonas putida . Biochem Eng J. 2001;9:211–219. doi: 10.1016/s1369-703x(00)00072-3. [DOI] [PubMed] [Google Scholar]
- Agarry SE. Solomon BO. Kinetics of batch microbial degradation of phenols by indigenous Pseudomonas fluorescence . Int J Environ Sci Technol. 2008;5:223–232. [Google Scholar]
- Fujita M. Ike M. Kamiya T. Accelerated phenol removal by amplifying the gene expression with a recombinant plasmid encoding catechol 2,3-oxygenase. Water Res. 1993;27:9–13. [Google Scholar]
- Tuah PM. Rashid NAA. Salleh MM. Degradation pathway of phenol through ortho-cleavage by Candida tropicalis RETL-Cr1. Borneo Sci. 2009;24:1–8. [Google Scholar]


