We prospectively showed that when anaplastic lymphoma kinase (ALK) immunohistochemistry (IHC) is sensitive enough and appropriately interpreted, it can be a stand-alone diagnostic for ALK inhibitor therapies. Our data will further spread and promote IHC for the screening of patients who will benefit from ALK inhibitor therapy, and cut the time and money costs for that purpose.
Keywords: immunohistochemistry, ALK, FISH, companion diagnostics, iScore, alectinib
Abstract
Background
Anaplastic lymphoma kinase (ALK) fusions need to be accurately and efficiently detected for ALK inhibitor therapy. Fluorescence in situ hybridization (FISH) remains the reference test. Although increasing data are supporting that ALK immunohistochemistry (IHC) is highly concordant with FISH, IHC screening needed to be clinically and prospectively validated.
Patients and methods
In the AF-001JP trial for alectinib, 436 patients were screened for ALK fusions through IHC (n = 384) confirmed with FISH (n = 181), multiplex RT-PCR (n = 68), or both (n = 16). IHC results were scored with iScore.
Result
ALK fusion was positive in 137 patients and negative in 250 patients. Since the presence of cancer cells in the samples for RT-PCR was not confirmed, ALK fusion negativity could not be ascertained in 49 patients. IHC interpreted with iScore showed a 99.4% (173/174) concordance with FISH. All 41 patients who had iScore 3 and were enrolled in phase II showed at least 30% tumor reduction with 92.7% overall response rate. Two IHC-positive patients with an atypical FISH pattern responded to ALK inhibitor therapy. The reduction rate was not correlated with IHC staining intensity.
Conclusions
Our study showed (i) that when sufficiently sensitive and appropriately interpreted, IHC can be a stand-alone diagnostic for ALK inhibitor therapies; (ii) that when atypical FISH patterns are accompanied by IHC positivity, the patients should be considered as candidates for ALK inhibitor therapies, and (iii) that the expression level of ALK fusion is not related to the level of response to ALK inhibitors and is thus not required for patient selection.
Registration number
JapicCTI-101264 (This study is registered with the Japan Pharmaceutical Information Center).
introduction
Patient selection is a key factor for molecular-targeted therapy. A molecular-targeted drug does not work appropriately, or rather may be harmful, if used for the patients without its molecular target. Good molecular targets are those that are not widely expressed in normal tissues and on which cancers are heavily ‘addicted’ for growth and survival. Accordingly, anaplastic lymphoma kinase (ALK) is one of the most desirable molecular targets. Only 4 years after the discovery of an oncogenic fusion, EML4-ALK [1], an ALK inhibitor, crizotinib, with an overall response rate (ORR) of 57%, was approved for therapy of ALK-positive lung cancer (ALK+ LC) [2].
ALK fusions are found in only 4%–6% of lung adenocarcinomas [3, 4]. There are three major conventional diagnostics for ALK fusions: fluorescence in situ hybridization (FISH), RT-PCR, and immunohistochemistry (IHC). As of September 2015, the Vysis ALK Break Apart FISH Probe Kit (Vysis FISH) (Abbott) is the only approved companion diagnostic test for prescribing crizotinib in Japan. In the United States, a high-sensitive IHC, VENTANA ALK (D5F3) CDx Assay, was also approved, resulting in two companion diagnostics. In Europe, crizotinib is approved for patients with ALK+ LC, but the type of test used for ALK fusion detection is not specified.
Alectinib is a second-generation ALK inhibitor with potent in vitro activity against both wild-type and mutated ALK, including mutations that confer resistance to crizotinib [5]. The results of the phase I/II study, AF-001JP, showed that alectinib is highly effective against ALK+ LC [ORR, 93.5%; 95% confidence interval (CI) 82.1–98.6] and well tolerated [6]. Two ALK inhibitors, crizotinib and alectinib, are currently approved in Japan. A highly sensitive immunohistochemical companion diagnostic, Histofine ALK iAEP kit (Nichirei Bioscience), which is based on the intercalated antibody-enhanced polymer (iAEP) method [4] was also approved as a companion diagnostic for alectinib. Although the increase in therapeutic options benefits clinical practice, this evokes some confusion about their companion diagnostics.
Here, we describe the details of our screening process, which contributed to the high ORR in AF-001JP. Additionally, several practical problems related to companion diagnostics are discussed: that is, how unexpected test results are addressed and how strictly the principle of companion diagnostics should be adhered to.
methods
screening for ALK fusions
In the trial, 436 patients were examined for ALK fusions between 10 September 2010, and 18 April 2012. The patients were screened with IHC (n = 384), multiplex RT-PCR (n = 68), or both (n = 16). Formalin-fixed paraffin-embedded (FFPE) specimens were used for IHC screening with the ALK Detection Kit (now renamed as Histofine ALK iAEP kit), and unfixed specimens (pleural effusion, bronchoalveolar lavage, sputum, or frozen tumor tissues) were used for multiplex RT-PCR screening [3]. In the samples for RT-PCR, the presence of cancer cells was not always examined by morphology.
ALK IHC results were classified into four categories according to the rate of positively stained tumor cells: positive (positive tumor cells >80%), probably positive (80%≥ positive tumor cells >50%), probably negative (50%≥ positive tumor cells >0%), and negative (0%). After patient selection for the trial, these four categories were renamed as iScore 3, 2, 1, and 0, respectively [7], and therefore, we use the term ‘iScore’ hereafter. For patients with iScore >0, FISH was subsequently performed for confirmation using in-house probes, which were confirmed to be sufficiently concordant with Vysis FISH. An experienced board-certified pathologist (KT) judged IHC and FISH results. The intensity of IHC staining was evaluated with Ariol (Leica Biosystems). Multiplex RT-PCR was performed and judged by a commercial clinical laboratory (SRL, Tokyo).
results
screening for ALK fusions
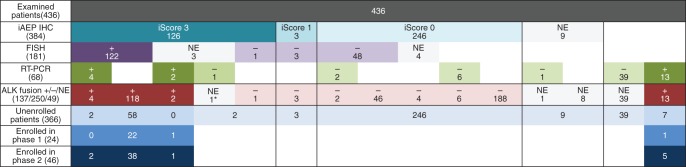
The results of ALK screening are summarized in Figure 1. Among the 436 patients examined, 137 and 250 patients were diagnosed as positive and negative for ALK fusion, respectively. Since the presence of cancer cells in the samples for RT-PCR was not always examined, ALK fusion negativity could not be ascertained in 49 patients.
Figure 1.
Demographics of the patients examined for ALK fusions. One patient with iScore 3 had negative results for RT-PCR and the FISH analysis failed. However, the patient had positive FISH results outside this trial and was enrolled in another trial for crizotinib. In this study, this patient was assigned ‘not evaluable’ for the presence of ALK fusions (*), because the presence was confirmed outside the trial.
Among the 384 patients screened with IHC, 9 patients could not be scored because no cancer cells were observed in their specimens. FISH was initially performed for the 129 patients with iScore >0, and after the inclusion period, for the 52 patients who had iScore 0 and provided consent for the additional FISH study. Since 7 patients failed in FISH, results of both IHC and FISH were evaluable in 174 patients. When iScore 3 was defined as IHC-positive and others as IHC-negative, sensitivity, specificity, positive predictive value, and negative predictive value of IHC compared with those of FISH were 100% (122/122), 98.1% (51/52), 99.2% (122/123), and 100% (51/51), respectively. The concordance rate was 99.4% (173/174).
The patient who showed iScore 3 and a negative FISH result had a large-cell neuroendocrine carcinoma. Although the proportion of positively stained cancer cells was slightly >80%, IHC staining intensity of each cancer cell was highly variable (a checker-board pattern) (supplementary Figure S1, available at Annals of Oncology online). In ALK fusion gene-positive samples stained with ALK iAEP IHC, almost all cancer cells stain positive, while heterogeneous staining patterns including a checker-board pattern indicated the expression of wild-type ALK [7, 8]. Therefore, after the trial, the original scoring criteria were slightly amended to exclude cases with a checker-board pattern from iScore 3 to iScore 2. The current version of iScore is given in Table 1.
Table 1.
Current version of iScore
| ALK iScore 3 | ALK iScore 2 | ALK iScore 1 | ALK iScore 0 | |
|---|---|---|---|---|
| Inclusion criteria | Proportion of positive tumor cells in the tested sample >80% | a. 80%≥ proportion of positive tumor cells in the tested sample >50% | 50%≥ proportion of positive tumor cells in the tested sample >0% | Absence of positive tumor cells |
| (cases with a checker-board pattern are classified as iScore 2b) | b. Cases with a checker-board pattern (positive and negative cells are frequently adjacent to each other) | |||
| Interpretation | An adenocarcinoma is ALK fusion-positive. | Despite the negativity of the ALK fusion gene, some or all cells may undergo neuroendocrine differentiation. Or, in other cases, the ALK fusion gene is positive, but the amount of ALK fusion protein decreases at sites (e.g. where cells undergo squamous differentiation). | The ALK fusion gene is negative, but some or all cells may undergo neuroendocrine differentiation. | ALK fusion gene is negative. |
| A large-cell neuroendocrine carcinoma may be ALK fusion-positive or express the full-length ALK protein. | ||||
| A small-cell carcinoma expresses the full-length ALK protein. | ||||
| Instruction | In overt adenocarcinomas, further confirmation using methods such as FISH is not required. Further confirmation using methods such as FISH is necessary for other histological subtypes. | Further confirmation for the absence (or presence) of the ALK fusion gene using methods such as FISH is preferable. | Further confirmation for the absence of the ALK fusion gene using methods such as FISH is preferable. | Further confirmation using methods such as FISH is not required. However, in cases that are highly likely to be ALK fusion-positive, further verifications are preferable. |
In cells containing a large amount of mucus, such as signet-ring cells, the cytoplasm is small; and therefore, such cells are less likely to be stained even if the ALK fusion gene is positive. If cells containing a large amount of mucus are negative in a sample containing positive cells, do not include the mucous-containing negative cells when calculating the rate of positive cells.
RT-PCR was performed in 68 patients including 16 patients who underwent both IHC and RT-PCR screening. Two patients who had iScore 3 but failed in FISH turned positive in RT-PCR. One patient showed a discordant result (IHC-positive, RT-PCR-negative, and failed in FISH). Outside the trial, however, the patient had a positive FISH result and was enrolled in another trial for crizotinib.
ALK fusion-positive lung cancer patients: age and sex distributions and response to alectinib
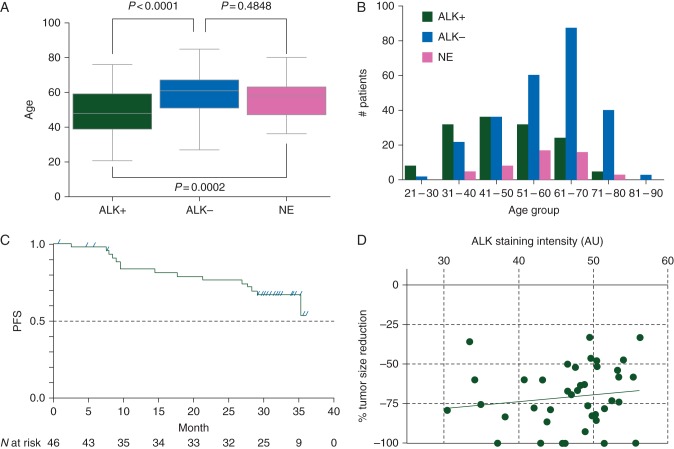
From 387 patients judged either positive or negative for ALK fusions in the trial, 208 were men and 179 were women; the age range was 21–85 years (median age, 57 years). ALK-positive patients were significantly younger (median: 48 versus 61 years. P < 0.0001; one-way ANOVA) (Figure 2A and B) and showed a female predilection [54% (74/137) versus 42% (105/250), P = 0.026; Fisher's exact test].
Figure 2.
ALK-positive patients: age distribution and response to alectinib. In comparison with ALK-negative patients, ALK-positive patients were significantly younger (median: 48 versus 61 years. P < 0.0001; one-way ANOVA). Patients who were not evaluable for the presence of ALK fusions (NE) were not significantly different from ALK-negative patients in age distribution (median: 58 versus 61 years. P = 0.4848; one-way ANOVA), suggesting that most of them were negative for ALK fusions (A and B). Median PFS was not yet reached at the time of data cutoff, but was estimated to be longer than 29 months. Fifteen of 46 (32.6%) of patients had a PFS event and the 2-year event-free duration rate was 0.76 (95% CI 0.60–0.87). Progressive disease was confirmed in 12 patients (26.1%) (C). Tumor size reduction rate and ALK IHC intensity of each of the 41 patients who had iScore 3 and were enrolled in phase II were plotted. Among the 41 patients, 9, 29, 1, and 2 patients had CR, PR, SD, and UN, respectively (ORR: 92.7%). The correlation was not significant (R2 = 0.01868) (D). AU, arbitrary unit.
As of 31 October 2014, all 46 patients in phase II showed at least 30% reduction in tumor size, and 28 were on treatment with alectinib [9]. Nine and 34 patients achieved complete response (CR) and partial response (PR), respectively. One patient remained in stable disease (SD), and 2 had an unknown response (UN) due to early withdrawal. The ORR was 93.5%. The median follow-up duration reached 30 months (range: 1–36), but PFS events were confirmed in only 15 patients (32.6%; 12 PD and 3 deaths) (Figure 2C). Therefore, the median PFS will be more than 29 months. ALK IHC specimens of the 41 patients were examined for staining intensity. The maximum staining intensity was not correlated with the rate of tumor size reduction (Figure 2D).
iScore 3 cases with atypical FISH patterns
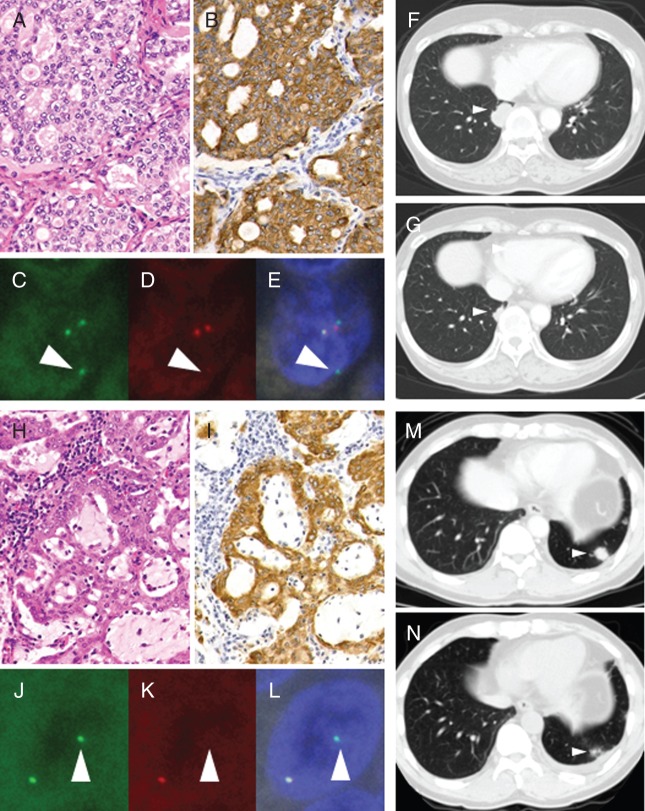
In 122 patients who had iScore 3 and were judged positive for FISH, two showed an atypical FISH pattern, the isolated 5′-side signal pattern. We performed FISH again with Vysis FISH, and obtained the same results (Figure 3C–E and J–L). This pattern is currently defined as negative in the criteria for Vysis FISH. However, we judged these cases positive for ALK fusion because (i) all the cancer cells clearly showed ALK expression by IHC (iScore 3; Figure 3B and I); (ii) both cases showed a mucinous cribriform pattern on histopathological analysis, which is the characteristic of ALK+ LC [10] (Figure 3A and H); and (iii) the ALK locus was indeed rearranged although the observed FISH pattern was atypical. One patient (IHCF8007) was enrolled in phase II and showed PR with a maximum tumor reduction of 48.1% (Figure 3F and G). The other patient (IHCF12028) was not enrolled for alectinib therapy but was treated with crizotinib outside the trial, and showed good response (Figure 3M and N). After the therapy, we examined these cases for ALK fusion partners by 5′-RACE optimized for FFPE specimens [11]. In IHCF8007, the variant 3 of the EML4-ALK transcript (E6;E20) was identified. We failed to identify the fusion partner in IHCF12028, probably due to severe mRNA degeneration.
Figure 3.
Patients with an atypical ALK FISH pattern. Two patients with iScore 3 showed an atypical FISH pattern (IHCF8007, A–G; IHCF12028, H–N). Sample from both patients showed a mucinous cribriform pattern on morphological analysis (A and H), iScore 3 in IHC (B and I), and the isolated 5′-side signal pattern in FISH (5′-side signal, C and J; 3′-side signal, D and K; merged, E and L). Figures of computed tomography before (F) and 63 days after (G) the first intake of alectinib and before (M) and 39 days after (N) the first intake of crizotinib.
discussion
Among the 126 patients with iScore 3, 124 were confirmed positive for ALK fusions in the trial, and 1 was outside of the trial. All the 41 patients who continued into phase II showed at least 30% tumor reduction with an ORR of 92.7%. This rate indicated that ALK iAEP IHC judged by iScore has sufficient specificity. The trial might have some limitations with respect to analyzing sensitivity. FISH was initially performed only for patients with >iScore 0 and was done retrospectively only for 52 additional patients with iScore 0. In addition, for the analysis of retrospectively performed FISH, the knowledge of IHC results might introduce a potential bias. However, the results of two other studies compensate for these limitations. In these two studies, in which the concordance of IHC and FISH was examined, ALK iAEP (ALK Detection Kit results judged by iScore) and Vysis FISH were blindly performed without any knowledge of the other test results. One study was previously reported [7], and the data of the other study have been submitted to the Ministry of Health, Labour, and Welfare for approval of the ALK iAEP kit. Both studies showed high concordance rates. As a total of the three studies including the present one, the sensitivity, specificity, and positive and negative predictive values of IHC to FISH were 99.5% (182/183), 99.9% (594/595), 99.5% (182/183), and 99.9% (594/595), respectively (Table 2).
Table 2.
Three studies to compare between ALK iAEP IHC and FISH
| IHC+ | IHC− |
|||
|---|---|---|---|---|
| iScore 3 | iScore 2 | iScore 1 | iScore 0 | |
| Takamochi et al. [7] | ||||
| FISH+ | 10 | 0 | 0 | 0 |
| FISH− | 0 | 1 | 2 | 347 |
| Data submitted to the Ministrya | ||||
| FISH+ | 50 | 0 | 0 | 1b |
| FISH− | 0 | 1 | 1 | 191 |
| AF-001JP | ||||
| FISH+ | 122 | 0 | 0 | 0 |
| FISH− | 1 | 0 | 3 | 48 |
| Total | ||||
| FISH+ | 182 | 0 | 0 | 1 |
| FISH− | 1 | 2 | 6 | 586 |
Sensitivity: 99.5% (182/183)
Specificity: 99.9% (594/595)
Concordance: 99.7% (776/778)
aHistofine ALK iAEP Kit package insert.
bAlthough in the examined specimen (∼250 mm2), the rate of cancer cells with positive FISH signals was 5.0%, the rate was as high as 44% in only a small area (∼0.15 mm2). The commercial laboratory judged the case as FISH-positive by consulting with the FISH probe manufacture, Abbott.
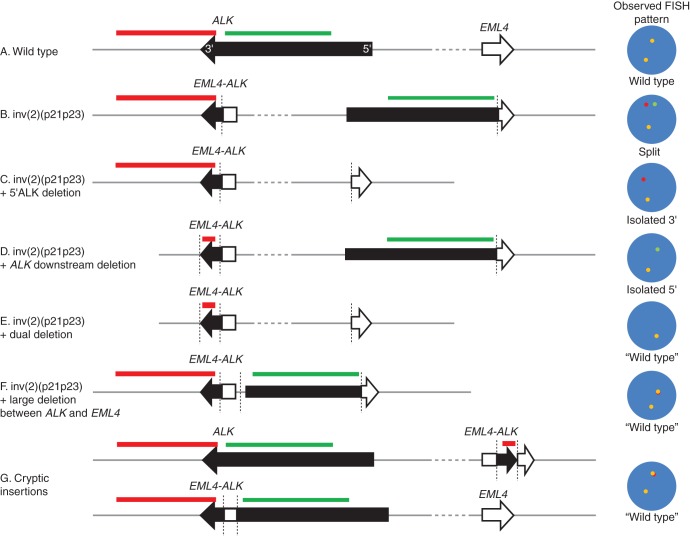
We encountered two cases of iScore 3 that showed an atypical FISH pattern, that is, an isolated 5′-side signal pattern. Given that an EML4-ALK transcript was identified in one of these cases and that both cases responded to ALK inhibitor therapy, the ALK kinase domain was preserved in the rearranged allele, although the 3′-side FISH signal was not observed. The length of the ALK kinase domain is only 30 kb, that is, 5–20 times shorter than typical FISH probes; thus, most of the 3′-side ALK FISH probe covers the downstream region outside ALK (Figure 4A–C). In cases with an isolated 5′-side signal pattern, the ALK downstream region is highly likely to be largely deleted, and the remaining region including the ALK kinase domain, ∼30 kb, is too short to be clearly observed with FISH on FFPE specimens. This results in an isolated 5′-side signal pattern (Figure 4D). There are several reports of cases showing an isolated 5′-side signal pattern accompanied by IHC positivity [12, 13] or negativity [14]. The latter IHC-negative cases harbored KRAS mutations indicating that ALK was not functionally rearranged [14], because KRAS mutations are usually mutually exclusive to ALK fusions. One of the IHC-positive patients was treated with, and responded to, an ALK inhibitor [13], like our two patients. In addition, some patients did not show isolated signals, but showed only the merged signal(s), known as ‘wild-type pattern’, although their IHC and RT-PCR results were positive [15]. In such cases, ALK fusion formation may be accompanied by some very rare events, for example, simultaneous deletions of 5′ and downstream regions, a large deletion between ALK and EML4, and small insertions (Figure 4E–G). Among the patients with atypical FISH patterns, which are regarded as negative in the current criteria, some had positive results for IHC, RT-PCR, or both and responded to ALK inhibitor therapy [13, 16]. Therefore, when atypical FISH patterns are accompanied by positivity in other methods, most often IHC, it is reasonable to recognize such patterns as functional ALK rearrangements. The molecular target of ALK inhibitors is ALK protein, not the ALK gene.
Figure 4.
Chromosomal rearrangement patterns for EML4-ALK and corresponding FISH patterns. ALK (729-kb length) and EML4 (163-kb length) are on 2p23 and 2p21, respectively, and separated from each other by 12.3 Mb. FISH probes are usually about 150∼500 kb long (A). EML4-ALK is produced thorough several patterns of chromosomal rearrangements involving 2p21–23. Most commonly, EML4-ALK is produced as a result of simple inv(2)(p21p23), and, in this case, a pair of 5′- and 3′-signals and a fused (normal) signal in the ALK split FISH assay were seen (B). About 40% of the cases with EML4-ALK show an isolated 3′-side signal pattern, and in this case, the 5′-region of ALK is likely to be deleted during rearrangement (C). Some cases (<1%) show an isolated 5′-side signal pattern. Such a patient responds to ALK inhibitor therapy when he or she obtains positive results for anti-ALK immunohistochemistry. In such a case, ALK kinase domain is preserved and the downstream region of ALK is deleted (D). Some patients very occasionally show the ‘wild-type pattern’ although their IHC and RT-PCR results are positive. In such cases, ALK fusion formation may be accompanied by some very rare events, including, for example, simultaneous deletions of 5′ and downstream regions, a large deletion between ALK and EML4, or small insertions (E–G).
After the trial, alectinib was approved on 4 July 2014 and became the second approved ALK inhibitor in Japan. Meanwhile, Histofine ALK iAEP kit (ALK iAEP) was approved as a companion diagnostic for alectinib, but not for crizotinib. Alectinib and ALK iAEP were listed on the National Health Insurance reimbursement list on 2 September 2014. Currently, in Japan, Vysis FISH is the only companion diagnostic for crizotinib (approved in 2012), and can also be used for alectinib because it was confirmed to be equivalent to the in-house ALK FISH probes used in the present trial. ALK iAEP, Vysis FISH, and the in-house probes were proven equivalent to each other for the detection of ALK fusions (Table 2). However, according to the package insert of alectinib, both Vysis FISH and ALK iAEP have to be performed before patients can start alectinib therapy, although FISH was performed in the trial only for confirmation of IHC screening results and the two tests were almost concordant with each other. In contrast, Vysis FISH results alone are sufficient for initiating crizotinib therapy.
On 9 September 2014, the Japan Lung Cancer Society (JLCS) made a statement concerning such a complicated situation: ‘… The concept of companion diagnostics is based on the principle that the same diagnostics used in the trial should also be used in the practice to reproduce the efficacy and safety of the drug proven in the trial … However, it is self-evident that what should be companioned with a molecular-targeted drug is a target, not a diagnostic. If the regulatory agency strictly adheres to the principle and instructs physicians to comply with the regulations, then scientifically illogical situations might occur. For example, patients might not be reimbursed for ALK iAEP if it is used for crizotinib, or the patients diagnosed with Vysis FISH and treated with crizotinib might have to further undergo ALK iAEP screening to receive second-line alectinib therapy …’ (authors' translation). Crizotinib and alectinib share the same targeted molecule and disease, but their companion diagnostic test requirements are different. This situation is leading to concerns in clinical practice in Japan, such as the following: there will be several molecular-targeted drugs to the same molecular target X; there will be several diagnostics for the same molecular target X; however, each drug to X will be companioned only with its specific diagnostic for X, and thus patients will have to take another diagnostic for X when they take another drug to X. Such a ‘tragicomedy’ should be scientifically considered and avoided for the benefit of patients. A more scientifically down-to-earth principle is recommended; when the response of a drug is proved to be predicted reliably with a biomarker and the biomarker is proved to be detected reliably with a diagnostic, the diagnostic should be companioned with the drug even if it is not used in the trial.
In conclusion, our study showed (i) that when ALK IHC is sufficiently sensitive and appropriately interpreted, it can be a stand-alone diagnostic for ALK inhibitor therapies; (ii) that when atypical FISH patterns are accompanied by IHC positivity, the patients should be considered as candidates for ALK inhibitor therapies; and (ii) that the expression level of ALK fusion is not related to the level of response to ALK inhibitors and is thus not required at least for patient selection. With the diversification of diagnostics for molecular targets, the interpretation of and the regulation policy for companion diagnostics are expected to evolve further.
funding
AF-001JP was funded by the study sponsor, Chugai Pharmaceutical Co. Ltd. The sponsor entered a contract with two central laboratories for screening (JFCR and SRL). For the detection of ALK fusion partners, this study was supported, in part, by The Ministry of Education, Culture, Sports, Science and Technology, the Japan Society for the Promotion of Science KAKENHI (25250020, 25871073) and the Japan Agency for Medical Research and Development.
disclosure
KT Honoraria: Nichirei, Chugai Pharma, Pfizer; Consulting or Advisory Role: Chugai Pharma, Nichirei; Research Funding: Chugai Pharma; Patents, Royalties, Other Intellectual Property: Nichirei. YK and TF are employed in Chugai Pharma. HY Honoraria: Chugai Pharma, Pfizer; Research Funding: Chugai Pharma. AI Honoraria: Chugai Pharma, Pfizer; Research Funding: Chugai Pharma. HK is employed in Chugai Pharma. KK Honoraria: Chugai Pharma, Pfizer, Astellas; Research Funding: Chugai Pharma, Pfizer, Astellas, Novartis. KN Honoraria: Chugai Pharma, Pfizer Japan; Research Funding: Chugai Pharma, Pfizer Japan. TS Honoraria: Pfizer Japan, Chugai Pharma; Research Funding: Chugai Pharma, Astellas Pharma, Novartis, Pfizer Japan. MM Honoraria: Chugai Pharma, Pfizer, Novartis; Research Funding Company: Chugai Pharma, Novartis. TH Honoraria: Chugai, Pfizer; Research Funding: Pfizer, Inc., Chugai Pharma, Novartis. MH Honoraria Chugai Pharma, Pfizer. YO Employment: Chugai (An Immediate Family Member); Honoraria: Chugai Pharma, Pfizer; Consulting or Advisory Role: Chugai Pharma, Novartis; Research Funding: Chugai Pharma, Pfizer, Novartis. NN Honoraria: Chugai Pharma, Pfizer. NY Honoraria: Chugai Pharma, Pfizer; Research Funding: Chugai Pharma. MN Honoraria: Nichirei, Chugai Pharma, Pfizer, Novartis; Consulting or Advisory Role: Chugai Pharma, Pfizer, Novartis; Research Funding: Chugai Pharma, Pfizer, Novartis, Astellas Pharma. TT Honoraria: Chugai Pharma, Novartis, Astellas Pharma.
Supplementary Material
acknowledgements
We thank the patients, their families, all of the investigators who participated in the study, and the central laboratory, SRL, which did the ALK rearrangement testing by RT-PCR method. We thank Satoko Baba for the technical assistance in IHC and FISH. This manuscript was reviewed and commented on by all authors. The corresponding author had full access to the study data and takes full responsibility for the final decision to submit the manuscript.
references
- 1.Soda M, Choi YL, Enomoto M et al. . Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007; 448: 561–566. [DOI] [PubMed] [Google Scholar]
- 2.Kwak EL, Bang YJ, Camidge DR et al. . Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 2010; 363: 1693–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takeuchi K, Choi YL, Soda M et al. . Multiplex reverse transcription-PCR screening for EML4-ALK fusion transcripts. Clin Cancer Res 2008; 14: 6618–6624. [DOI] [PubMed] [Google Scholar]
- 4.Takeuchi K, Choi YL, Togashi Y et al. . KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin Cancer Res 2009; 15: 3143–3149. [DOI] [PubMed] [Google Scholar]
- 5.Kodama T, Tsukaguchi T, Yoshida M et al. . Selective ALK inhibitor alectinib with potent antitumor activity in models of crizotinib resistance. Cancer Lett 2014; 351: 215–221. [DOI] [PubMed] [Google Scholar]
- 6.Seto T, Kiura K, Nishio M et al. . CH5424802 (RO5424802) for patients with ALK-rearranged advanced non-small-cell lung cancer (AF-001JP study): a single-arm, open-label, phase 1–2 study. Lancet Oncol 2013; 14: 590–598. [DOI] [PubMed] [Google Scholar]
- 7.Takamochi K, Takeuchi K, Hayashi T et al. . A rational diagnostic algorithm for the identification of ALK rearrangement in lung cancer: a comprehensive study of surgically treated Japanese patients. PLoS One 2013; 8: e69794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takeuchi K. Interpretation of anti-ALK immunohistochemistry results. J Thorac Oncol 2013; 8: e67–e68. [DOI] [PubMed] [Google Scholar]
- 9.Ohe Y, Nishio M, Kiura K et al. . A phase I/II study with a CNS-penetrant, selective ALK inhibitor alectinib in ALK-rearranged non-small cell lung cancer (ALK+NSCLC) patients (pts): updates on progression free survival (PFS) and safety results from AF-001JP. J Clin Oncol 2015; 33(suppl: abstr 8061). [Google Scholar]
- 10.Takeuchi K, Soda M, Togashi Y et al. . RET, ROS1 and ALK fusions in lung cancer. Nat Med 2012; 18: 378–381. [DOI] [PubMed] [Google Scholar]
- 11.Togashi Y, Soda M, Sakata S et al. . KLC1-ALK: a novel fusion in lung cancer identified using a formalin-fixed paraffin-embedded tissue only. PLoS One 2012; 7: e31323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshida A, Tsuta K, Nitta H et al. . Bright-field dual-color chromogenic in situ hybridization for diagnosing echinoderm microtubule-associated protein-like 4-anaplastic lymphoma kinase-positive lung adenocarcinomas. J Thorac Oncol 2011; 6: 1677–1686. [DOI] [PubMed] [Google Scholar]
- 13.Ren S, Hirsch FR, Varella-Garcia M et al. . Atypical negative ALK break-apart FISH harboring a crizotinib-responsive ALK rearrangement in non-small-cell lung cancer. J Thorac Oncol 2014; 9: e21–e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gainor JF, Varghese AM, Ou SH et al. . ALK rearrangements are mutually exclusive with mutations in EGFR or KRAS: an analysis of 1,683 patients with non-small cell lung cancer. Clin Cancer Res 2013; 19: 4273–4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murakami Y, Mitsudomi T, Yatabe Y. A screening method for the ALK fusion gene in NSCLC. Front Oncol 2012; 2: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peled N, Palmer G, Hirsch FR et al. . Next-generation sequencing identifies and immunohistochemistry confirms a novel crizotinib-sensitive ALK rearrangement in a patient with metastatic non-small-cell lung cancer. J Thorac Oncol 2012; 7: e14–e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.