Abstract
Transcription errors occur in all living cells; however, it is unknown how these errors affect cellular health. To answer this question, we monitored yeast cells that were genetically engineered to display error-prone transcription. We discovered that these cells suffer from a profound loss in proteostasis, which sensitizes them to the expression of genes that are associated with protein-folding diseases in humans; thus, transcription errors represent a new molecular mechanism by which cells can acquire disease. We further found that the error rate of transcription increases as cells age, suggesting that transcription errors affect proteostasis particularly in aging cells. Accordingly, transcription errors accelerate the aggregation of a peptide that is implicated in Alzheimer’s disease, and shorten the lifespan of cells. These experiments reveal a novel, basic biological process that directly affects cellular health and aging.
Keywords: Transcriptional fidelity, mutagenesis, proteotoxic stress, heat-shock proteins, aging
Introduction
DNA replication, transcription and translation provide the foundation for life itself. Together, these three processes ensure the inheritance of our genome and the faithful expression of our genetic code. To preserve the integrity of this code, it is essential that these processes are carried out with remarkable precision. Sporadic errors are unavoidable though, and these errors reveal how important biological fidelity is for organismal health. For example, errors that occur during DNA replication contribute to carcinogenesis(31), and errors that occur during translation contribute to amyloidosis(32). How transcription errors affect cellular health is less well understood. Several experiments have now demonstrated though, that transcription errors have the potential to profoundly affect human health. For example, transcription errors in the gene that encodes the β-amyloid precursor protein generate toxic versions of the Aβ protein in patients with non-familial Alzheimer’s disease(6, 7). Similar transcription errors can generate toxic versions of the ubiquitin-B protein in patients with Down’s syndrome(6, 7), or activate oncogenic pathways in quiescent cells(8). These transcription errors, which occur at disease-related loci could directly contribute to human disease; however, underlying these specific, disease-related errors, is a much larger population of errors that occur randomly throughout the genome. How these random errors affect cellular health is completely unknown. Recently though, several alleles have been identified in the budding yeast Saccharomyces cerevisiae that result in error prone transcription(9, 10). These alleles provide the first opportunity to fill this gap in our knowledge. Here, we monitored the health of two cell lines that exhibit error prone transcription and demonstrate that random transcription errors profoundly affect cellular proteostasis, as well as the rate at which yeast cells age; thus, transcription errors represent a new molecular mechanism by which cells can acquire disease.
Results
Cells that exhibit error prone transcription display increased levels of molecular chaperones
To determine how transcription errors affect cellular health, we monitored two cell lines that exhibit error prone transcription. The first cell line carries a point mutation in the gene that encodes Rpb1 (rpb1-E1103G, MVY0002), a core catalytic subunit of the RNA polymerase II complex(9) (RNAPII). The second cell line carries a deletion of the gene that encodes Rpb9 (rpb9Δ, MVY0003), a non-essential subunit of the RNAPII complex(10). These alleles display a 3–9 fold increase in the error rate of transcription in vivo and in vitro, and the mechanisms that are responsible for this increase are well established(9, 10, see Methods for information on the genetic background of these strains).
Interestingly, both of these cell lines exhibit a growth defect (fig. 1a, b), suggesting that transcription errors have the potential to affect the overall health of a cell. To understand the mechanism that is responsible for this observation, we used an unbiased mass spectrometry approach to identify proteins that were significantly upregulated in rpb1-E1103G and rpb9Δ cells compared to WT cells (MVY0001). In total we detected 390 proteins. Out of these 390 proteins, we found that 22 proteins were significantly upregulated ≥1.5 fold in the rpb1-E1103G cells, while 32 proteins were significantly upregulated ≥1.5 fold in the rpb9Δ cells (table S1). Fifteen of these proteins were shared among the error prone cell lines, and further analysis indicated that 7 of these play an important role in protein folding and protein quality control (PQC, table 1A). In addition, two chaperones were exclusively detected in the error prone cells and not in the WT cells (table 1B), which prohibited a quantitative comparison, while two additional chaperones were significantly upregulated only in the rpb9Δ cells (table 1C). The upregulation of multiple chaperones in the error prone cells suggests that they suffer from increased levels of proteotoxic stress.
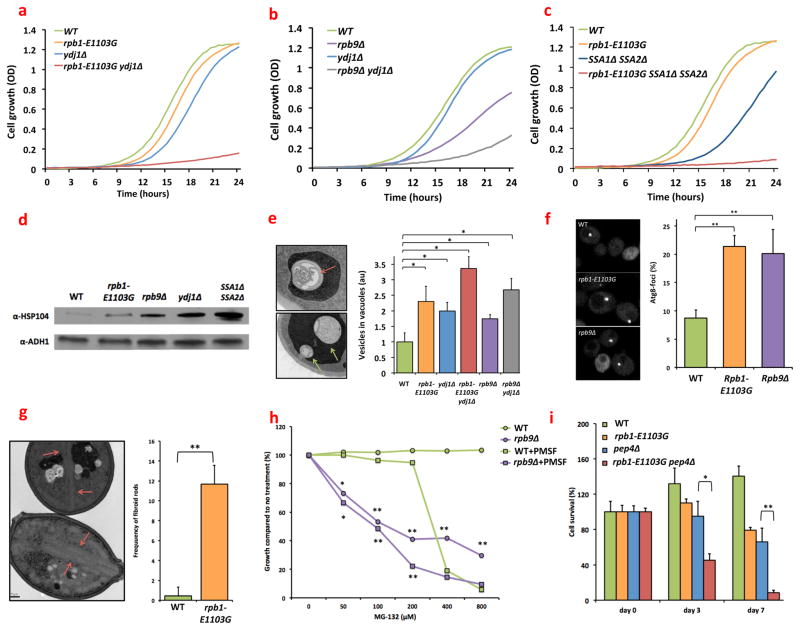
Fig. 1.
Genetic, biochemical and ultrastructural data suggest that cells that exhibit error prone transcription experience proteotoxic stress. (a) In the absence of YDJ1, rpb1-E1103G (MVY0001-4) and cells grow very slowly. (b) In the absence of YDJ1, rpb9Δ cells grow very slowly (MVY0001, 3, 4, 6). Each cell line was diluted to OD 0.001, and aliquoted in triplicate into a 96-well plate. Cell strains were then allowed to grow uninterrupted for 24 hours at 30°C while an OD measurement was made every 15 minutes. (c) In the absence of SSA1 and SSA2, rpb1-E1103G cells grow very slowly (MVY0001, 2, 7, 8). The protocol described in figure legend 1a and b was used for this experiment as well. (d) Similar to cells that have lost YDJ1 or SSA1 and SSA2, rpb1-E1103G cells and rpb9Δ cells display increased expression of Hsp104 (MVY0001-4, 7). Cells were grown into log phase, lysed, and analyzed by Western blotting with an anti-body against Hsp104. (e) Autophagic remnants (top panel, red arrows) and inclusion bodies (middle panel, green arrows) are more frequently present in the vacuoles of rpb1-E1103G and rpb9Δ cells compared to WT cells (MVY0001-6). The prevalence of these vesicles can be exacerbated further by YDJ1 deletion. (f) Cells were transformed with a GFP-tagged copy of ATG8. Foci indicate locations where autophagosome formation occurs. The number of foci present per cell is an indication of autophagy activity (MVY0001-3). (g) Fibroid, crystalline protein aggregates (red arrows) are present in rpb1-E1103G cells that were chronologically aged for 96 hours in a 30°C incubator (MVY0001-2). (h) Rpb9Δ cells are more sensitive to increasing concentrations of MG-132 than WT cells. The sensitivity of Rpb9 cells to MG-132 can be enhanced further by simultaneously inhibiting proteases present in the vacuole by 200uM PMSF (MVY0001, 3). (i) Deletion of PEP4 shortens the chronological lifespan of rpb1-E cells (MVY0001, 2, 13, 14). Cells were allowed to grow into stationary phase for 48 hours (day 0) and the number of colony forming units (CFUs) determined. After 3 and 7 days of incubation and constant shaking at 30°C, the number of CFUs were determined again and compared to day 0. At least 3 biological replicates were used for each genotype for each experiment presented above. Images and western blots were quantified with ImageJ software.
*= P<0.05, **= P<0.01
Table 1.
Increased expression of molecular chaperones in cells that display error prone transcription. (a) List of all proteins that were significantly upregulated ≥1.5-fold in both rpb1-E1103G cells and rpb9Δ cells when compared to WT cells (MVY0001-3). (b) Molecular chaperones that were detected in the error prone cells, but not in WT cells, so that a precise fold-increase could not be calculated (MVY0001-3). (c) Molecular chaperones that were significantly upregulated in rpb9Δ cells, but not rpb1-E1103G cells. For comparison, in the same experiment, we found that Hsp104 was upregulated 3.5 fold in ydj1Δ cells. Proteins involved in protein quality control are labeled in red (MVY0001-3).
| rpb1-E1103G | rpb9Δ | |||||
|---|---|---|---|---|---|---|
| Gene | Function | Fold-increase | P-value | Fold-increase | P-value | |
| A | SSC1 | Mitochondrial heat shock protein of the HSP70 family | 1.8 | 0.021 | 1.7 | 0.016 |
| PMA1 | Plasma membrane ATP-ase | 1.9 | 0.009 | 1.7 | 0.009 | |
| HSC82 | Cytosolic chaperone of the HSP90 family | 2 | 0.001 | 1.5 | 0.015 | |
| SSB2 | Ribosome associated chaperone, functions with Zuo1 | 2.2 | 0.001 | 2.4 | 0.001 | |
| SSA1 | Heat shock protein of the HSP70 family | 2.2 | 0.001 | 4.5 | 0.001 | |
| DUG1 | Metallopeptidase that degrades gluthathione | 2.3 | 0.03 | 2.3 | 0.023 | |
| CDC60 | Leucyl tRNA synthetase | 2.4 | 0.015 | 3.3 | 0.001 | |
| CDC48 | Involved in ER-associated protein degradation | 2.4 | 0.006 | 2.2 | 0.006 | |
| ACO1 | Required for the tricarboxylic acid cyle | 2.4 | 0.003 | 2.9 | 0.001 | |
| PFK1 | Involved in glycolysis | 2.5 | 0.007 | 2.3 | 0.007 | |
| PFK2 | Involved in glycolysis | 2.6 | 0.011 | 4.2 | 0.001 | |
| SSE1 | ATP-ase component of HSP90 chaperone complex | 2.7 | 0.001 | 2.1 | 0.001 | |
| HXK2 | Hexokinase that the catalyzes phosphorylation of glucose | 3.5 | 0.005 | 4.3 | 0.001 | |
| STI1 | HSP90 co-chaperone, interacts with SSA chaperones | 4 | 0.001 | 2.9 | 0.006 | |
| TDH2 | Involved in glycolysis and gluconeogenesis | 4.1 | 0.003 | 3.4 | 0.01 | |
| B | SIS1 | HSP40 co-chaperone | detected | N/A | detected | N/A |
| HSP82 | Cytosolic chaperone of the HSP90 family | detected | N/A | detected | N/A | |
| C | HSP60 | Mitochondrial chaperone | 1.6 | 0.15 | 2.4 | 0.001 |
| HSP104 | Disaggregase of protein aggregates | 1.6 | 0.28 | 2.8 | 0.003 | |
The health of the error prone cells depends on multiple molecular chaperones
To investigate this idea further, we used genetics, biochemistry, fluorescence microscopy, and electron microscopy to find additional evidence for proteotoxic stress in the error prone cells. First, we used a genetic approach to test whether molecular chaperones are indeed critical for the health of the error prone cells. To this end, we introduced a YDJ1 deletion into the error prone cells using a standard mating and sporulation approach. YDJ1 encodes an Hsp40 co-chaperone that contributes to the folding process of nascent proteins, and helps refold proteins that were previously misfolded(11). In the absence of YDJ1, we found that rpb1-E1103G (MVY0005) and rpb9Δ cells (MVY0006) produce exceptionally small colonies (fig. S1a), grow at a slow rate (fig. 1a, b), and exhibit a swollen appearance (fig. S1b). Taken together, these results indicate that YDJ1 is critical for the overall health of cells that display error prone transcription.
To determine whether this finding was unique to YDJ1, or whether it could be extended to other chaperones, we also deleted SSA1 and SSA2 in rpb1-E1103G cells. Ssa1 and Ssa2 are two chaperones that are in the Hsp70 family(11) and are 98% identical. Accordingly, they need to be deleted simultaneously in WT cells to affect cellular function (MVY0007). We found that the growth rate of rpb1-E1103G ssa1Δ ssa2Δ cells (MVY0008) closely resembles the growth rate of rpb1-E1103G ydj1Δ cells (fig. 1c), and that the rpb1-E1103G ssa1Δ ssa2Δ cells exhibit a similar swollen appearance. Rpb9Δ cells, and to a lesser degree rpb1-E1103G cells, were also more sensitive to inhibition of Hsp82 with radicicol(12) than WT cells (fig. S2). Hsp82 is a key molecular chaperone that is upregulated in response to heat and stress(13) and contributes to the folding process of a specific set of proteins that is particularly difficult to fold(14). Thus, multiple chaperones in addition to Ydj1 are required to maintain the health of the error prone cells, supporting the hypothesis that these cells suffer from proteotoxic stress.
The error prone cells suffer from proteotoxic stress
To find additional evidence for the presence of proteotoxic stress in the error prone cells, we monitored the expression of Hsp104 (fig. 1d), a protein that disassembles protein aggregates, and whose upregulation is a common response to proteotoxic stress and protein aggregation(15). We found that rpb1-E1103G and rpb9Δ cells indeed exhibit increased expression of Hsp104 compared to WT cells, similar to cells that are known to experience proteotoxic stress due to loss of YDJ1 (MVY0004) or SSA1 and SSA2(15) (MVY0007, fig. 1d). This observation was confirmed in vivo using fluorescent imaging of a GFP tagged copy of HSP104, which was chromosomally integrated into WT (MVY0009), rpb1-E1103G (MVY0010) and rpb9Δ cells (MVY0011, fig. S3). The upregulation of Hsp104 indicates that despite the efforts of multiple molecular chaperones, protein aggregation may be occurring to a greater degree in the error prone cell lines. Because protein aggregates and other forms of proteotoxic stress are frequently cleared from cells by autophagy, we used electron microscopy to detect increased levels of autophagy as well as the protein aggregates themselves(16). We found that rpb1-E1103G cells and rpb9Δ cells indeed carry increased amounts of autophagic bodies inside their vacuoles compared to WT cells (fig. 1e), which contained condensed multilammelar remnants of autophagosomes (fig. 1e, top). Autophagic bodies are normally rapidly degraded inside the vacuole, but they can accumulate under stressful conditions that increase autophagic activity, such as proteotoxic stress(17). Accordingly, we observed more autophagic bodies inside the vacuoles of the error prone cells when YDJ1 was deleted (MVY0005, fig. 1e) and less after ATG1 was deleted (MVY0012), a gene that is important for phagophore formation (fig. S4). Together, these two observations indicate increased autophagic activity in the error prone cells. This conclusion was supported by the observation that more Atg8-GFP foci are present in rpb1-E1103G and rpb9Δ cells compared to the WT cells when these cells are transformed with a plasmid that contains a GFP tagged copy of Atg8 (Atg8-GFP foci indicate locations where autophagosome formation occurs, fig. 1f). In addition to detecting increased levels of autophagy, we found that chronologically aged rpb1-E1103G cells accumulated fibroid, crystalline protein aggregates inside their cytoplasm that could span the length of an entire cell (fig. 1g).
Cells can degrade autophagic debris and toxic proteins with proteases that are located in the proteasome and the vacuole. To test the importance of these proteases for the health of the error prone cells, we treated WT cells and error prone cells with MG-132, a drug that inhibits the proteasome (18), and PMSF, a drug that inhibits proteases present in the vacuole(19). We found that rpb9Δ cells exhibit a greatly increased sensitivity to MG-132 (fig. 1h) and PMSF compared to WT cells, and a combination of these two drugs was the most effective (fig. 1g). Although these drugs did not significantly reduce the growth rate of the rpb1-E1103G cells, we found that deletion of PEP4 (which encodes a key protease inside the vacuole(20))in the rpb1-E1103G cells, greatly diminished the lifespan of rpb1-E1103G cells (MVY0014, fig. 1i), indicating that protease activity is also important for the health of rpb1-E1103G cells, although not enough to affect the rate at which the cells grow. The combined weight of all of these experiments strongly suggests that the error prone cells suffer from increased levels of proteotoxic stress compared to WT cells. The enhanced growth defect of rpb9Δ cells, in combination with their elevated sensitivity to PMSF, MG-132 and Hsp82 inhibition, as well as their enhanced induction of Hsp104, further suggest that rpb9Δ cells suffer from greater amounts of proteotoxic stress than rpb1-E1103G cells. It is important to note though, that in contrast to the rpb1-E1103G allele, deletion of rpb9 results in a measurable effect on transcription itself. It cannot be excluded therefore that this perturbation contributes to the enhanced sensitivity of rpb9Δ cells as well.
Transcription errors sensitize cells to genes associated with protein-folding diseases
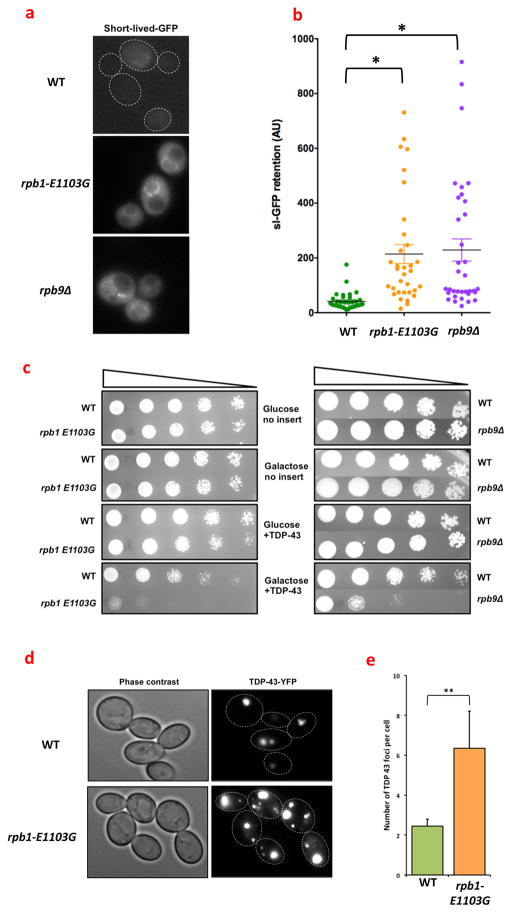
Recent evidence indicates that the capacity of cells to combat proteotoxic stress is limited. For example, the expression of an aggregation prone version of the Huntingtin protein (a human protein that causes a neuromuscular disorder(21)) in C. elegans, overwhelms the capacity of the PQC machinery to such an extent, that regular maintenance tasks (such as assisting with the folding process of endogenous proteins) are neglected(22). To test whether error prone transcription could overwhelm the PQC machinery in a similar manner, we monitored the status of the PQC machinery with a fluorescent reporter protein. This reporter protein consists of a short-lived GFP molecule (sl-GFP) that is a target for destruction by the proteasome(23). Retention of sl-GFP inside a cell is therefore a barometer for the workload of the PQC machinery(23). Notably, rpb1-E1103G and rpb9Δ cells retain more sl-GFP than WT cells (fig. 2a, b), which reinforces the idea that rpb1-E1103G cells suffer from proteotoxic stress and indicates that the PQC machinery is indeed overextended. To explore this idea further we expressed a YFP tagged copy of TDP-43 in the error prone cells(24), a toxic protein that causes Amyotrophic Lateral Sclerosis. Although TDP-43 is only slightly toxic in WT cells, we noticed that the viability of rpb1-E1103G cells and rpb9Δ cells was greatly diminished by TDP-43 (fig. 2c). We observed a similar phenomenon when we overexpressed a toxic repeat of 17 alanines in the rpb1-E1103G cells, while a non-toxic control containing a repeat 8 alanines did not affect cellular viability (fig. S5a). The increased toxicity of TDP-43 was also apparent at the molecular level, as a fluorescently tagged copy of TDP-43 aggregated into larger and more numerous foci in rpb1-E1103G cells compared to WT cells (fig. 2d, e). These results indicate that WT cells degrade TDP-43 more efficiently than error prone cells, mirroring our results with sl-GFP (fig. 2a).
Fig. 2.
Transcription errors overextend the protein quality control machinery. (a) WT cells and rpb1-E1103G cells were transformed with a short-lived-GFP molecule, which is an indicator of the strain placed upon cellular protein quality control pathways. rpb1-E1103G and rpb9Δ cells display a brighter GFP signal than WT cells, indicating that sl-GFP is degraded less efficiently in the error prone cells compared to the WT cells and that PQC is overextended in the error prone cells. Dashed lines outline cells that are difficult to see due to low fluorescence (MVY0001-3). (b) Quantification of the retention of sl-GFP in WT cells, rpb1-E1103G cells and rpb9Δ cells (MVY0001-3). (c) Cells were either transformed with an empty vector, or a plasmid that contained a YFP-tagged copy of TDP-43, which was placed under the control of a GAL4 promoter. The transformed cells were then spotted in 4-fold dilutions on plates that contained gluocose (no expression) or galactose (expression). Expression of TDP-43 inhibited the growth of rpb1-E1103G and rpb9Δ cells to a greater degree than WT cells, as shown by reduced growth of the error prone cells at multiple dilutions (MVY0001-3). (d) Cells were transformed with a YFP-tagged copy of TDP-43, grown into log-phase and monitored under a fluorescence microscope. Despite identical expression patterns, rpb1-E1103G cells contain more protein aggregates than WT cells, indicating that they have greater difficulty destroying the TDP-43 protein. The aggregates in rpb1-E1103G cells are also larger in size compared to WT cells (MVY0001-2). (e) Quantification of the number of TDP-43 foci in WT cells and rpb1-E1103G cells. At least 3 biological replicates were used for each genotype for each experiment presented above. Images were quantified with ImageJ software (MVY0001-2).
**= P<0.01
To determine the breadth of these observations, we expressed two additional proteins in the error prone cells: the toxic glutamine repeat region of the Huntingtin protein, and the yeast prion Rnq1(25). These proteins form amyloid-like aggregates that are structurally distinct from the amorphous aggregates of TDP-43. Despite this mechanistic difference though, we found that Rnq1 and a toxic version of the Huntingtin protein (Htt103Q) greatly reduced the viability of rpb9Δ cells compared to WT cells, whereas a benign version of Huntingtin (Htt25Q) did not (fig. S5b). However, Htt103Q and Rnq1 did not affect rpb1-E1103G cells more than WT cells, which demonstrates that rpb9Δ cells have a wider sensitivity to toxic proteins than rpb1-E1103G cells, consistent with the observation that rpb9Δ cells suffer from a greater amount of proteotoxic stress. Taken together, these results indicate that transcription errors can overextend the PQC machinery, which allows toxic proteins that are associated with disease to escape degradation, thereby enhancing their toxicity and rate of aggregation.
Transcription errors increase with age
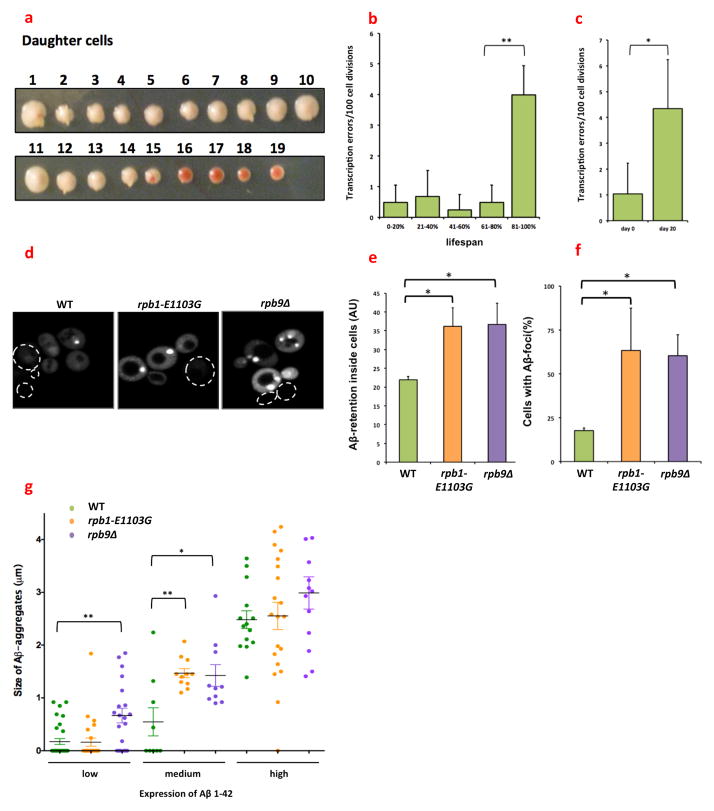
Proteotoxic stress is a universal hallmark of aging cells(26). However, the molecular mechanisms that cause proteotoxic stress in aging cells remain unknown. Since transcription errors induce proteotoxic stress, we wondered whether transcription errors could represent one of these mechanisms. To test this possibility, we measured the transcriptional error rate of WT cells as a function of age. For these measurements, we used an assay that records transcription errors on a track of 10 cytosines (9) as red sectors in growing colonies (fig. 3a). This cytosine track places the gene that encodes Cre-recombinase out of frame. If RNAPII slips on this track and reinstates the proper reading frame, active proteins are generated. Cre-recombinase then excises a target elsewhere in the genome, which results in the accumulation of red pigment inside the cells. As a result, a temporary transcription error is converted into a tractable, genetic mutation. To determine if aging affects the transcriptional error rate, we aged these reporter cells (a cross between GRY3337 and GRY3724) replicatively and chronologically: the replicative lifespan of yeast cells is measured by the number of daughter cells that are born from a single mother cell and is thought to be a model for the lifespan of mitotic cells, whereas the chronological lifespan is measured by the time a cell can stay alive in a non-dividing state, which is thought to be a model for the lifespan of post-mitotic cells(27). We discovered that the error rate of transcription increases ≈8-fold in the final stages of the replicative lifespan of the cells (fig. 3b), and 4–10 fold during their chronological lifespan (fig. 3c). We confirmed that these events were the result of transcription errors and not genetic mutations in two ways. First, we sequenced the Cre-locus to search for genetic mutations, but found none in the 48 clones we tested. Second, we sporulated cells that were derived from the red colonies and mated them to a second reporter. If a genetic error had reactivated Cre-recombinase, these cells would continuously express an active version of Cre-recombinase and activate the second reporter immediately. However, no Cre-activity was detected in any of the 48 clones tested. As a result of the increased number of transcription errors in aging cells, we conclude that transcription errors place an increasing burden on the PQC of the aging cells, which suggests that transcription errors are partially responsible for the increased levels of proteotoxic stress seen in aging cells. In humans, this decline in proteostasis predisposes cells to age-related pathology that is mediated by toxic proteins, as seen in Alzheimer’s and Parkinson’s disease (26). To specifically test whether transcription errors are capable of modulating the degradation of proteins that are associated with age-related diseases, we expressed a GFP tagged copy of Aβ1-42 in our cell lines. Aβ1-42 is an aggregation-prone, short peptide, which forms large amyloid plaques in Alzheimer’s patients and is a powerful target for cellular PQC. Similar to TDP-43, we found that Aβ1-42 is destroyed less efficiently in rpb1-E1103G and rpb9Δ cells compared to WT cells (fig. 3d, e), and aggregates into larger, more numerous foci in the error prone cells (fig. 3f). The aggregation of Aβ1-42 strongly depends on the level of expression, and we found that aggregation of Aβ1-42 is initiated at lower levels of expression in the error prone cells compared to the WT cells (fig. 3g). Taken together, these experiments indicate that transcription errors increase with age in yeast, and that these errors can affect the degradation and aggregation of proteins that are associated with age-related diseases in humans.
Fig. 3.
The fidelity of transcription changes as a function of age. (a) Example of a replicative assay for one mother cell. Daughter cells were collected from an aging mother cell, and displayed in a row across a plate. In the image shown, each consecutive daughter is numbered. These daughter cells were grown into a colony, which allowed us to score transcription errors by monitoring red sectoring in the growing colonies. A transcription error could occur in either the mother cell, or one of the daughter cells. If a transcription error occurred in the mother cell, all the subsequent daughter cells born after this event would grow into red colonies (cell 16–19). If a transcription error occurred in the daughter cell though, then only that colony would be affected. For instance, while daughter 15 was growing into a colony, one of its progeny experienced a transcription error, resulting in a clearly visible red sector inside the colony (diploid cross between GRY3337 and GRY3724). (b) The error rate of transcription increases approximately 8-fold as a function of the replicative age of the cell. Virgin cells containing the reporter construct were selected and the transcriptional error rate of each cell was determined as a function of their age. For example, if a cell gave birth to a total of 20 daughters, and a transcription error occurred at daughter 17, then the error occurred at 85% of its lifespan(diploid cross between GRY3337 and GRY3724). (c) The error rate of transcription increases approximately 4-fold as a function of the chronological age of the cell. Cells were grown into stationary phase for 48 hours in YAPD (day 0) and aged for extended periods of time while the number of CFUs was determined every 3 days (diploid cross between GRY3337 and GRY3724). (d) Aβ1-42 aggregates into more numerous foci in rpb1-E1103G and rpb9Δ cells than in WT cells. Cells were transformed with a plasmid that contains a GFP-tagged copy of Aβ1-42, which was placed under the control of a PGK1 promoter and monitored with a fluorescence microscope. Dashed outlines outline cells that are difficult to see due to low fluorescence (MVY0001-3). (e) More Aβ1-42 is present inside rpb1-E1103G compared to WT cells despite equal expression levels; however, rpb9Δ cells have a 20% lower expression of Aβ1-42 compared to WT cells. We therefore normalized Aβ1-42 expression in rpb9Δ cells to GFP expression. With or without this correction, more Aβ1-42 is retained inside both of the error prone cells compared to WT cells. Approximately 120 cells were analyzed for each genotype (MVY0001-3). (f) A greater number of Aβ1-42 foci appear in rpb1-E1103G and rpb9Δ cells (MVY0001-3). (g) Aβ1-42 starts to aggregate at lower levels of expression in rpb1-E1103G and rpb9Δ cells compared. This threshold is reached first in rpb9Δ cells, and then in the rpb1-E1103G cells. At least 3 biological replicates were used for each genotype for each experiment presented above. Images were quantified with ImageJ software (MVY0001-3).
*= P<0.05, **= P<0.01
Transcription errors decrease the lifespan of cells, which can be rescued by dietary restriction
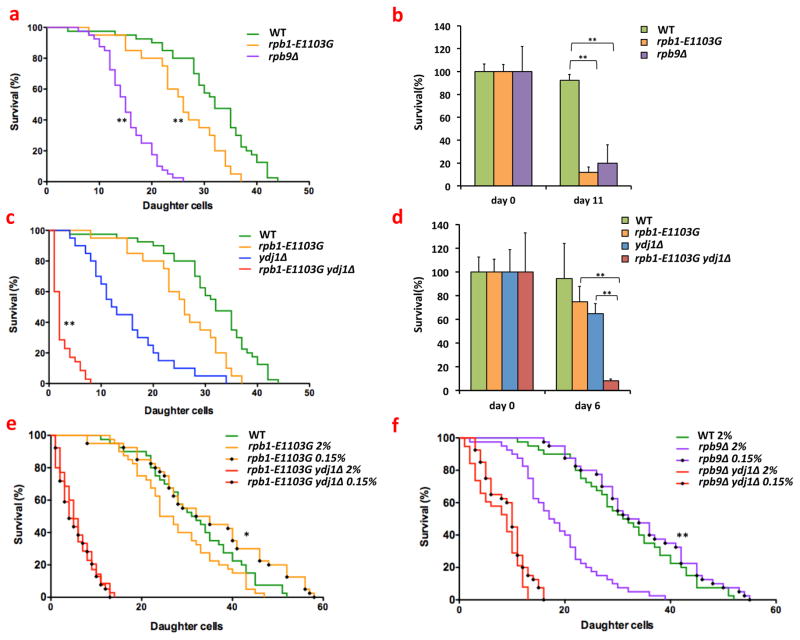
In addition to age-related pathology, proteotoxicity also contributes to the mortality of aging cells(28). It is therefore possible that age-related transcription errors also contribute to cellular aging by shortening cellular lifespan. To investigate this possibility, we monitored the replicative and chronological lifespan of rpb1-E1103G and rpb9Δ cells, and found that these cell lines exhibit a shorter replicative and chronological lifespan compared to WT cells (fig. 4a, b). This lifespan could be shortened further by exacerbating the levels of proteotoxic stress through YDJ1 or SSA1 and SSA2 deletion (fig. 4c, d, S6a, b). Transcription errors had the strongest effect on the chronological lifespan of the cells though, which is consistent with the observation that proteotoxic stress primarily affects post-mitotic cells. After determining how transcription errors affect aging cells, we decided to investigate whether these effects are permanent, or reversible. Because dietary restriction delays the onset of age-related pathology and extends the lifespan of cells(29), we tested whether dietary restriction could also reverse the deleterious effects of transcription errors on the lifespan of aging cells. To do this, we monitored the replicative lifespan of error prone cells that were grown on normal (2%) or restricted concentrations of glucose. Importantly, restricting glucose to 0.05%, 0.1% or 0.15% rescued the lifespan of both rpb1-E1103G and rpb9Δ cells (fig. 4e, f, S7a, b), which demonstrates that the deleterious effects of transcription errors on the lifespan of aging cells can indeed be reversed. Furthermore, the lifespan extension of the error prone cells depended on the presence of YDJ1 and SSA1 and SSA2 (fig. 4e, f, S7c), indicating that this extension is orchestrated by the manipulation of PQC pathways, a well-known effect of dietary restriction(30).
Fig. 4.
Error prone cells have a shortened lifespan. (a) rpb1-E1103G and rpb9Δ cells live shorter replicative lifespans than WT cells. For example, the average lifespan of WT cells is ≈30 divisions, while the average lifespan of rpb9Δ cells is ≈18 divisions. In all replicative lifespan assays, the lifespan of 20–40 mother cells was measured per sample (MVY0001-3). (b) rpb1-E1103G and rpb9Δ cells live shorter chronological lifespans than WT cells. For all chronological lifespan assays, the lifespan of 3–5 replicates was measured per sample (MVY0001-3). (c) Deletion of YDJ1 results in a profound reduction of the replicative lifespan of rpb1-E1103G cells (MVY0001, 2, 4, 5). (d) Deletion of YDJ1 results in a profound reduction of the chronological lifespan of rpb1-E1103G cells (MVY0001, 2, 4, 5). (e) Dietary restriction rescues the shortened lifespan of rpb1-E1103G cells, but not rpb1-E1103G ydj1Δ cells (MVY0001, 2, 5). (f) Dietary restriction rescues the shortened lifespan of rpb9Δ cells, but not rpb9Δ ydj1Δ cells (MVY0001, 3, 6). At least 3 biological replicates were used for each genotype for each experiment presented above. Kaplan Meier curves and analysis was performed with Prism Graphpad software.
*= P<0.05, **= P<0.01
Discussion
Basic biological consequences of transcription errors
The genome provides a precise, biological blueprint of life. To implement this blueprint correctly, the genome must be read with great precision; however, due to the constraints of biological fidelity, it is impossible for this process to be completely error-free. As a result, transcription errors can occur at any time, in any transcript, and how these random errors affect cellular health is unknown. Here, we answer this question by studying cells that express an error prone version of RNAPII, and we demonstrate that transcription errors affect cellular health by inducing proteotoxic stress. Most likely, this increase in proteotoxic stress is caused by transcription errors that change structurally important amino acids in polypeptide chains. For example, a transcription error that replaces a hydrophobic amino acid with a hydrophilic amino acid could substantially distort the folding process of a protein. Similar distortions are caused by genetic mutations(33) and translation errors(32). As a result, our experiments expose a surprising new role for the PQC machinery in living cells: to counteract the deleterious effects of random transcription errors.
Impact of transcription errors on disease-related proteins
We further found that transcription errors have the potential to affect cells in a medically relevant manner by encumbering the PQC machinery, which allows disease related proteins to escape degradation, thereby increasing their toxicity; thus, randomly occurring transcription errors represent a new molecular mechanism by which cells can acquire disease. We also discovered that the error rate of transcription increases with age in yeast, which contributes to the decline in proteostasis seen in aging cells. As a result, transcription errors allow toxic proteins to escape the PQC machinery particularly in aging cells. In humans, an age-related increase in the error rate of transcription could allow proteins associated with age-related diseases such as Alzheimer’s and Parkinson’s disease to accumulate in aging neurons, which would provide valuable new insight into the molecular etiology of numerous age-related diseases. To test this possibility, it will be important to produce animal models and human cell lines that display error prone transcription.
Transcription errors, DNA damage, and predictions from our experiments
The potential link between neurons and transcription errors is especially intriguing because oxidative damage is a powerful source of transcription errors. As oxidative damage accumulates in aging neurons, it could raise the transcriptional error rate and facilitate the onset of age-related diseases. Accordingly, transcription errors may represent a molecular link between aging and age-related diseases. In theory though, any form of DNA damage could contribute to the transcriptional error rate, including most of the mutagens we are currently aware of. If so, we may be substantially underestimating the harmful effects mutagens have on cellular health. By taking advantage of recently developed next-gen sequencing technology, it may be possible to test this hypothesis. DNA-damaging reagents are especially dangerous, because they can cause repeated transcription errors at a single base(4, 8, 34). These errors can generate a large number of proteins with a single, potentially harmful mutation. For example, transcription errors that occur repeatedly in the gene that encodes the amyloid precursor protein generate toxic versions of the Aβ protein in Alzheimer’s patients(6, 7). A remarkable prediction from our observations is therefore that an age-related increase in transcription errors may not only result in highly specific, disease-related proteins, but also provide the very conditions that allow these proteins to escape the surveillance of the PQC machinery.
Methods
Cell strains and plasmids
All the strains that were used for our experiments were backcrossed 15 times into the BY4742/BY4741 background, directly pulled from the BY4742 MATα knock-out collection, or sporulated from the BY4743 heterozygous knock-out collection to create haploid cells in the BY4742/BY4741 background. Unless otherwise noted, WT cells, rpb1-E1103 cells or rpb9Δ cells refer to MVY0001, MVY0002 or MVY0003 respectively. All other strains are specifically referred to with genotype and strain number in the text. In addition, each figure legend contains information about the exact strains that were used to acquire the data that is depicted. Double mutants were generated using standard mating and sporulation protocols using the BY4742 MATα knock-out collection from Open Biosystems as a source for the deletions of key genes. For instance, to delete a single gene in a WT or error prone background, we crossed rpb1-E1103G or rpb9Δ cell lines into the appropriate strain from the MATα knock-out collection. The diploid cells were then sporulated, microdissected, and genotyped so that each strain was known to posses the desired auxotrophy markers (see strain list). All the plasmids used for our experiments were previously generated, and no modifications were made. All plasmid transformations were performed using standard lithium acetate protocols.
Cell lines
| MVY0001: | ura3Δ0, leu2Δ0, trp1-Δ1, met15Δ0, his3-Δ1, MATa |
| MVY0002: | rpb1-E1103G, ura3Δ0, leu2Δ0, trp1-Δ1, met15Δ0, his3-Δ1 MATa |
| MVY0003: | rpb9::kanMX, ura3Δ0, leu2Δ0, trp1-Δ1, met15Δ0, his3-Δ1 MATa |
| MVY0004: | ydj1::kanMX, ura3Δ0, leu2Δ0, trp1-Δ1, met15Δ0, his3-Δ1 MATa |
| MVY0005: | rpb1-E1103G, ydj1::kanMX, ura3Δ0, leu2Δ0, trp1-Δ1, met15Δ0, his3-Δ1 MATa |
| MVY0006: | rpb9::kanMX ydj1::kanMX, ura3Δ0, leu2Δ0, trp1-Δ1, met15Δ0, his3-Δ1 MATa |
| MVY0007: | ssa1::kanMX, ssa2::kanMX, ura3Δ0, leu2Δ0, trp1-Δ1, met15Δ0, his3-Δ1 MATa |
| MVY0008: | rpb1-E1103G ssa1::kanMX, ssa2::kanMX, ura3Δ0, leu2Δ0, trp1-Δ1, met15Δ0, his3-Δ1, MATa |
| MVY0009: | HSP104-GFP-His3MX6, ura3Δ0, leu2Δ0, trp1-Δ1, met15Δ0, his3-Δ1 MATa |
| MVY0010: | rpb1-E1103G, HSP104-GFP-His3MX6, ura3Δ0, leu2Δ0, trp1-Δ1, met15Δ0, MATa |
| MVY0011: | rpb9::kanMX, HSP104 -GFP-His3MX6, ura3Δ0, leu2Δ0, trp1-Δ1, met15Δ0, MATa |
| MVY0012: | rpb1-E1103G, atg1::kanMX, ura3Δ0, leu2Δ0, trp1-Δ1, met15Δ0, his3-Δ1, MATa |
| MVY0013: | pep4::kanMX, ura3Δ0, leu2Δ0, trp1-Δ1, met15Δ0, his3-Δ1, MATa |
| MVY0014: | rpb1-E1103G, pep4::kanMX, ura3Δ0, leu2Δ0, trp1-Δ1, met15Δ0, his3-Δ1, MATa |
| GRY3337: | ade2Δ, ade6-AI2floxkanMX, his3-Δ1, leu2Δ0, met15Δ0, trp1-Δ1, ura3Δ0, rpb1::kanMX, MATa [pJS725 URA3 RPB1] |
| GRY3724: | ade2Δ, ade6Δ0::zeoMX, his3-Δ1, leu2::PHIS3-malE-C10EB-cre:hygMX, ura3Δ0, MATα |
Plasmids
| pRNQ1: | pRS416, Gal1-Rnq1-YFP (URA3, CEN, Summers, Cyr, JBC, 2009) |
| pHTT25: | pYES, Gal1-HTT25Q-GFP (URA3, 2μ, Wolfe, Cyr, PLOSone, 2014) |
| pHtt103Q: | pYES, Gal1-HTT103Q-GFP (URA3, 2μ, Wolfe, Cyr, PLOSone, 2014) |
| pTDP43: | pRS416, Gal1-TDP-43-YFP (URA3, CEN, Johnson, Gitler, PNAS 2008) |
| pYFP: | pRS416, Gal1-YFP (URA3, CEN, Summers, Cyr, JBC, 2009) |
| pslGFP: | pRS426, ADH1-slGFP (URA3, 2μ, Summers, Cyr, JBC, 2009) |
| pGFP: | pRS416, PGK1- GFP (URA3, CEN, Caine, Macreadie, FEMS 2007) |
| pAβ1-42: | pRS416, PGK1-Aβ1-42-GFP (URA3, CEN, Caine, Macreadie, FEMS 2007) |
| pATG-8: | pRS426, ATG8-GFP (URA3, 2μ, Lang, Duncan, JBC 2014) |
| pJS725: | RPB1 (URA3, 2μ, Strathern, Kashlev, JBC 2013) |
| pRG1361: | pRS416, Gal1-Pab18A-GFP (URA3, CEN, Konopka, Gardner, MBC, 2011) |
| pRG1365: | pRS416, Gal1-Pab117A-GFP (URA3, CEN, Konopka, Gardner, MBC, 2011) |
Lifespan experiments
Replicative lifespan assays were performed as previously reported. Briefly, single colonies were streaked out on fresh YAPD plates, grown overnight and re-streaked onto a new YAPD plate in the morning. After 6 hours of growth, a small amount of cells was moved onto a fresh, experimental plate, and 20–60 mother cells were displayed for each genotype. The daughters, or the grand daughters of these cells were used for our lifespan assays, so that each cell was a virgin at the start of the experiment. Chronological aging assays were performed by inoculating single colonies grown on YAPD plates into 3ml of YAPD medium. These cultures were grown overnight, and diluted ten-fold into fresh YAPD medium the next morning. This culture was then allowed to grow uninterrupted for 48 hours to drive the cells into stationary phase. After 48 hours, the number of colony forming units present in the culture was determined by diluting the cells in water, and plating them on freshly made YAPD plates. Three days, seven days, ten days and fourteen days later, the number of colony forming units was assessed again and compared to the 48-hour time point to calculate survival. The survival of each genotype was monitored in triplicate or quadruplicate, starting from a unique, single colony.
Western blotting
Single colonies were inoculated in YAPD, grown overnight, and then diluted in fresh YAPD the next morning. The cells were then cultured until they reached log-phase, and 2ODs from each cell line were collected and suspended in 100μl of SDS loading buffer adapted for cellular lysis, supplemented with 100μl of acid washed glass beads. The cells were then boiled immediately for 30 seconds, and subsequently vortexed for 30 seconds to lyse them. These boiling and vortexing steps were repeated 4 more times to fully lyse the cells. Then, 50μl of loading buffer was added to the lysates, which was spun down to pellet large cell debris. After centrifugation, the protein lysate was immediately loaded onto a 4–12% gel and blotted for the appropriate antibody after transfer onto a nitrocellulose membrane. The membrane was scanned using Typhoon imager system. The rabbit HSP104 antibody was purchased from Abcam, the ADH1 antibody was purchased from Abcam and the goat anti rabbit secondary antibody, conjugated to Alexafluor 488 was purchased from Invitrogen and Molecular Probes.
Electron microscopy
Yeast cells were prepared for transmission electron microscopy using a modification of methods published previously (Wright, 2000). Specifically, cells were fixed with 2% glutaraldehyde in 0.1M PIPES buffer, pH 6.8, containing 1mM MgCl2, 1mM CaCl2, and 0.1M sorbitol for overnight at 4°C. Following cell wall permeabilization with 1% sodium metaperiodate, the cell pellets were post-fixed for 1 hour with 1% osmium tetroxide/1.25% potassium ferrocyanide/0.1M PIPES buffer, pH 6.8, and stained en bloc in 2% aqueous uranyl acetate for 20 minutes. The pellets were dehydrated using an increasing ethanol series followed by propylene oxide and embedment in Spurr’s epoxy resin (Electron Microscopy Sciences, Hatfield, PA). Ultrathin sections (70nm) were cut and mounted on 200mesh copper grids and stained with Reynolds’ lead citrate (Reynolds, 1963). The sections were observed at 80kV using a LEO EM910 transmission electron microscope (Carl Zeiss Microscopy, LLC, Thornwood, NY), and digital images were taken using a Gatan Orius SC1000 digital camera with Digital Micrograph 3.11.0 software (Gatan, Inc., Pleasanton, CA).
Protein digestion for mass spectrometry
WT cells, rpb1-E1103G cells, rpb9Δ cells, were pulverized and frozen (~−80 °C) until further use. Each sample contained three biological replicates. Lysed lyophilized cells were suspended in 50 mM ammonium bicarbonate with protease inhibitors (Roche cOmplete Protease Inhibitor Cocktail) prepared to manufacturer’s recommendations. Protein concentrations were determined via a Coomassie Plus™ (Bradford) Assay Kit (Thermo Scientific, 23236) with BSA standard. Samples for digestion were diluted with 50 mM ammonium bicarbonate to provide equivalent protein concentration in all samples.
A standard in-solution tryptic digestion protocol (http://www.genome.duke.edu/cores/proteomics/sample-preparation/) was followed for shotgun digestion of all samples. In short, Rapigest SF acid-labile surfactant (Waters Corporation, Milford, MA) was added to 50 μg of protein to a final concentration of 0.1%. Samples were vortexed, heated at 40 °C for 10 minutes and centrifuged to condensate. Dithiothreitol (Sigma-Aldrich Co., St.Louis, MO) was added to a final concentration of 10 mM to reduce disulfide bonds. Samples were vortexed and heated to 80 °C for 15 minutes. All samples were then cooled, centrifuged to condensate, and iodoacetamide (Sigma-Aldrich Co.) was added to a final concentration of 20 mM to alkylate disulfide bonds. The samples were incubated in the dark for 30 minutes before trypsin (TPCK-modified, Sigma-Aldrich Co.) was added in a 1:50 trypsin to protein ratio. Digestion occurred overnight at 37°C, followed by acidification with trifluoroacetic acid (Sigma-Aldrich Co.) to 1.0 % by volume. Samples were then centrifuged (15,000 rpm) for 5 min and the supernatant was transferred into total recovery vials. For quantification, bovine serum albumin digest (Waters Corporation, 186002329) was added as an internal standard at a concentration of 25 fmol/μL.
Liquid Chromatography and mass spectrometry analysis
Each sample was injected (2 μL) onto a RPLC-MS/MS Waters nanoAcquity/SynaptG2 system. Samples were initially trapped on a 5 mm × 180 um trap column packed with 5 um Symmetry C18 particles (Waters) at a flow rate of 5 ul/min for 6 minutes. Analytical separations were performed on a 75 um × 250 mm column packed with 1.8 μm HSS T3 C18 particles (Waters Corporation). Mobile phase A was water with 0.1% formic acid, and mobile phase B was acetonitrile with 0.1% formic acid (Fisher). Water and acetonitrile were Optima LC-MS grade, and all other chemicals were ACS reagent grade or higher. At a flow rate of 400 nL/min, a 90 min gradient from 5 to 40 % B was used to separate the peptides. The outlet of the column was connected to a silica electrospray emitter (New Objective, Inc) operated at +2.6 kV. The Synapt G2 mass spectrometer analyzer operated at a resolution of ~20,000 in ion-mobility data-independent acquisition (IMS-MSE) mode. Parent ions were scanned from 50 to 2000 m/z over 0.6 s at 4eV. MSE scans to collect fragment ion data were performed by ramping collision energy from 27 to 50 eV over 0.6s.
Peptide data processing
Peptide data was initially processed using ProteinLynx Global Server 2.5.2 (Waters Corporation) and deconvoluted MS/MS spectra were searched against a database of known yeast proteins from Uni-Prot (www.uniprot.org) protein knowledgebase with a 1X reversed sequence appended and 100% false discovery rate. These results were then imported into Scaffold 4.2.0 (Proteome Software, Portland, OR) with a 4.9% protein false discovery rate and a minimum of 3 peptides for protein identification. Peptides matching multiple proteins were exclusively assigned to the protein with the most evidence. The spectral counts for each peptide assigned to a protein were summed to give the quantitative value of the protein. The value was normalized by multiplying the average total number of spectra, for each biological sample, divided by the total number of spectra for each analytical replicate. Bovine serum albumin was used as the standard to covert the average intensity of the top 3 ‘best-flier’ peptides for each proteins to moles using the method of Silva(35). The quantified amounts (fmol) for proteins identified in 2 of the 3 biological replicates were averaged and a Fisher’s exact test was performed between WT cells and cells that display error prone transcription. Proteins that contained a p-value <0.050 between the two samples and a fold change >1.5 were considered differentially expressed.
Spotting assay
Single colonies were picked up from glucose plates and inoculated into medium containing 3% raffinose. Cells were grown overnight, and diluted ten-fold in fresh medium containing 3% raffinose. After 4.5 hours of growth, the cells were diluted to OD 0.2, and spotted on plates containing either galactose or glucose.
Fluorescence microscopy
Single colonies were inoculated in YAPD, grown overnight, and then diluted in fresh YAPD the next morning. Cells were grown into log phase, collected and imaged using a Nikon Lumencore microscope at 250x magnification in synthetic complete buffer using an LED lamp for illumination and standard GFP and RFP filters for data collection. All images were collected with Metamorph and Zen software, and analyzed in ImageJ.
Growth assays
Single colonies were inoculated in YAPD, grown overnight, and then diluted in fresh YAPD the next morning. Cells were grown into log phase, collected, and diluted to OD 0.001. These cells were then aliquoted into a 96 well plate, and grown for 24 hours at 30°C in a 96-well bioananalyzer that recorded the OD in each well every 10 minutes. The growth rate of each genotype was monitored in triplicate or quadruplicate, starting from a unique, single colony.
Transcription fidelity measurements
Transcription fidelity assays were performed as previously reported(36). Briefly, transcriptional errors on a tract of 10 cytosines result in accurate synthesis of the Cre-recombinase gene, which excises a drug resistance gene inserted into an artificial intron in the ADE6 gene, ade6-AI2floxkanMX. The kanMX gene is flanked by two lox sites. Cre recombinase excises the kanMX gene generating the functional ADE6-AI2lox allele. In yeast that carry an ade2 mutation, the Cre-mediated switch from ade6-AI2flox::kanMX to ADE6-AI2lox results in a colony color change from white to red.
Supplementary Material
Significance Statement.
Transcription is required for every biological process inside a cell. Although most transcripts are generated faithfully from their DNA template, errors do occur from time to time. How these errors affect cellular function is unknown. We discovered that these errors cause proteotoxic stress, and allow proteins associated with neurodegenerative diseases to escape the surveillance of the protein quality control machinery. These observations uncover the basic biological consequences of transcription errors, and suggest that transcription errors are novel modifiers of numerous neurodegenerative diseases.
Acknowledgments
We thank Kerry Bloom and Jolien Verdaasdonk for extensive help with fluorescent microscopy. We thank the lab of Douglas Wallace, Liming Pei, Gary Pielak and Kevin Weeks for use of their equipment and Dr. Richard Gardner for plasmids carrying toxic alanine repeats. We thank Eeva-Liisa Eskelinen, Greg Odorizzi, Daniel Klionsky and his laboratory for help interpreting the electron microscopy images.
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Wanagat J, Cao Z, Pathare P, Aiken JM. Mitochondrial DNA deletion mutations colocalize with segmental electron transport system abnormalities, muscle fiber atrophy, fiber splitting, and oxidative damage in sarcopenia. Faseb J. 2001;15(2):322–332. doi: 10.1096/fj.00-0320com. [DOI] [PubMed] [Google Scholar]
- 3.Kraytsberg Y, et al. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat Genet. 2006;38(5):518–520. doi: 10.1038/ng1778. [DOI] [PubMed] [Google Scholar]
- 4.Viswanathan A, You HJ, Doetsch PW. Phenotypic change caused by transcriptional bypass of uracil in nondividing cells. Science. 1999;284(5411):159–162. doi: 10.1126/science.284.5411.159. [DOI] [PubMed] [Google Scholar]
- 5.Schimmel P. Development of tRNA synthetases and connection to genetic code and disease. Protein Sci. 2008;17(10):1643–1652. doi: 10.1110/ps.037242.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Leeuwen FW, Burbach JP, Hol EM. Mutations in RNA: a first example of molecular misreading in Alzheimer’s disease. Trends Neurosci. 1998;21(8):331–335. doi: 10.1016/s0166-2236(98)01280-6. [DOI] [PubMed] [Google Scholar]
- 7.van Leeuwen FW, et al. Frameshift mutants of beta amyloid precursor protein and ubiquitin-B in Alzheimer’s and Down patients. Science. 1998;279(5348):242–247. doi: 10.1126/science.279.5348.242. [DOI] [PubMed] [Google Scholar]
- 8.Saxowsky TT, Meadows KL, Klungland A, Doetsch PW. 8-Oxoguanine-mediated transcriptional mutagenesis causes Ras activation in mammalian cells. Proc Natl Acad Sci U S A. 2008;105(48):18877–18882. doi: 10.1073/pnas.0806464105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strathern J, et al. The fidelity of transcription: RPB1 (RPO21) mutations that increase transcriptional slippage in S. cerevisiae. The Journal of biological chemistry. 2013;288(4):2689–2699. doi: 10.1074/jbc.M112.429506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walmacq C, et al. Rpb9 subunit controls transcription fidelity by delaying NTP sequestration in RNA polymerase II. The Journal of biological chemistry. 2009;284(29):19601–19612. doi: 10.1074/jbc.M109.006908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475(7356):324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 12.Jarosz DF, Lindquist S. Hsp90 and environmental stress transform the adaptive value of natural genetic variation. Science. 2010;330(6012):1820–1824. doi: 10.1126/science.1195487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borkovich KA, Farrelly FW, Finkelstein DB, Taulien J, Lindquist S. hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Molecular and cellular biology. 1989;9(9):3919–3930. doi: 10.1128/mcb.9.9.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nathan DF, Vos MH, Lindquist S. In vivo functions of the Saccharomyces cerevisiae Hsp90 chaperone. Proc Natl Acad Sci U S A. 1997;94(24):12949–12956. doi: 10.1073/pnas.94.24.12949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bosl B, Grimminger V, Walter S. The molecular chaperone Hsp104--a molecular machine for protein disaggregation. Journal of structural biology. 2006;156(1):139–148. doi: 10.1016/j.jsb.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 16.Eskelinen EL, Reggiori F, Baba M, Kovacs AL, Seglen PO. Seeing is believing: the impact of electron microscopy on autophagy research. Autophagy. 2011;7(9):935–956. doi: 10.4161/auto.7.9.15760. [DOI] [PubMed] [Google Scholar]
- 17.Baba M. Electron microscopy in yeast. Methods in enzymology. 2008;451:133–149. doi: 10.1016/S0076-6879(08)03210-2. [DOI] [PubMed] [Google Scholar]
- 18.Tsubuki S, Saito Y, Tomioka M, Ito H, Kawashima S. Differential inhibition of calpain and proteasome activities by peptidyl aldehydes of di-leucine and tri-leucine. Journal of biochemistry. 1996;119(3):572–576. doi: 10.1093/oxfordjournals.jbchem.a021280. [DOI] [PubMed] [Google Scholar]
- 19.Powers JC, Asgian JL, Ekici OD, James KE. Irreversible inhibitors of serine, cysteine, and threonine proteases. Chemical reviews. 2002;102(12):4639–4750. doi: 10.1021/cr010182v. [DOI] [PubMed] [Google Scholar]
- 20.Van Den Hazel HB, Kielland-Brandt MC, Winther JR. Review: biosynthesis and function of yeast vacuolar proteases. Yeast. 1996;12(1):1–16. doi: 10.1002/(sici)1097-0061(199601)12:1<1::aid-yea902>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 21.Walker FO. Huntington’s disease. Lancet. 2007;369(9557):218–228. doi: 10.1016/S0140-6736(07)60111-1. [DOI] [PubMed] [Google Scholar]
- 22.Gidalevitz T, Ben-Zvi A, Ho KH, Brignull HR, Morimoto RI. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 2006;311(5766):1471–1474. doi: 10.1126/science.1124514. [DOI] [PubMed] [Google Scholar]
- 23.Summers DW, Wolfe KJ, Ren HY, Cyr DM. The Type II Hsp40 Sis1 cooperates with Hsp70 and the E3 ligase Ubr1 to promote degradation of terminally misfolded cytosolic protein. PLoS One. 2013;8(1):e52099. doi: 10.1371/journal.pone.0052099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson BS, McCaffery JM, Lindquist S, Gitler AD. A yeast TDP-43 proteinopathy model: Exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proc Natl Acad Sci U S A. 2008;105(17):6439–6444. doi: 10.1073/pnas.0802082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sondheimer N, Lindquist S. Rnq1: an epigenetic modifier of protein function in yeast. Molecular cell. 2000;5(1):163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 26.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaeberlein M. Lessons on longevity from budding yeast. Nature. 464:7288, 513–519. doi: 10.1038/nature08981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes & development. 2008;22(11):1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fontana L, Partridge L, Longo VD. Extending healthy life span--from yeast to humans. Science. 328(5976):321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kennedy BK, Steffen KK, Kaeberlein M. Ruminations on dietary restriction and aging. Cell Mol Life Sci. 2007;64(11):1323–1328. doi: 10.1007/s00018-007-6470-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bielas JH, Loeb LA. Mutator phenotype in cancer: timing and perspectives. Environ Mol Mutagen. 2005;45(2–3):206–213. doi: 10.1002/em.20111. [DOI] [PubMed] [Google Scholar]
- 32.Lee JW, et al. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443(7107):50–55. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- 33.Guo HH, Choe J, Loeb LA. Protein tolerance to random amino acid change. Proc Natl Acad Sci U S A. 2004;101(25):9205–9210. doi: 10.1073/pnas.0403255101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bregeon D, Doddridge ZA, You HJ, Weiss B, Doetsch PW. Transcriptional mutagenesis induced by uracil and 8-oxoguanine in Escherichia coli. Molecular cell. 2003;12(4):959–970. doi: 10.1016/s1097-2765(03)00360-5. [DOI] [PubMed] [Google Scholar]
- 35.Silva JC, Gorenstein MV, Li GZ, Vissers JP, Geromanos SJ. Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Molecular & cellular proteomics: MCP. 2006;5(1):144–156. doi: 10.1074/mcp.M500230-MCP200. [DOI] [PubMed] [Google Scholar]
- 36.Strathern JN, Jin DJ, Court DL, Kashlev M. Isolation and characterization of transcription fidelity mutants. Biochim Biophys Acta. 2012;1819(7):694–699. doi: 10.1016/j.bbagrm.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.