Abstract
Reconstitution of CMV-specific immunity following transplant remains a primary clinical objective to prevent CMV disease, and adoptive immunotherapy of CMV-specific T cells can be an effective therapeutic approach. Due to viral persistence, most CMV-specific CD8pos T cells become terminally differentiated effector cells (TEFF). A minor subset retains a memory-like phenotype (TM), but it is unknown whether these cells retain memory function or persist over time. Interestingly, recent studies suggest that CMV-specific CD8pos T cells with different phenotypes have different abilities to reconstitute sustained immunity following transfer. The immunology of human CMV (HCMV) infections is reflected in the murine model (MCMV). We found that HCMV- and MCMV-specific T cells displayed shared genetic programs, validating the MCMV model for studies of CMV-specific T cells in vivo. The MCMV-specific TM population was stable over time and retained a proliferative capacity that was vastly superior to TEFF cells. Strikingly, after transfer, TM cells established sustained and diverse T cell populations even after multiple challenges. Although both TEFF and TM cells could protect Rag−/− mice, only TM cells persisted after transfer into immune replete, latently-infected recipients and responded if recipient immunity was lost. Interestingly, transferred TM cells did not expand until recipient immunity was lost, supporting that competition limits the antigen stimulation of TM cells. Ultimately, these data show that CMV-specific TM cells retain memory function during MCMV infection and can reestablish CMV immunity when necessary. Thus, TM cells may be a critical component for consistent, long-term adoptive immunotherapy success.
Introduction
Latent Cytomegalovirus (CMV) is present within a large percentage of the population but is effectively controlled by the immune system (1-6). However, in transplant patients, immune suppression can allow CMV reactivations to progress to disease and increase mortality. Despite the advancements of anti-viral medications, long-term prevention of CMV disease is dependent on the reconstitution of CMV-specific immunity, which can be achieved through adoptive immunotherapy (5-18).
In adoptive immunotherapy, healthy CMV-seropositive donors provide CMV-specific T cells to an immune suppressed recipient. Due to the persistent nature of CMV infection, CMV-seropositive donors accumulate large numbers of CMV-specific CD8pos T cells (approximately 5-10% of the total CD8pos T cells), a process termed “memory inflation,” (19-28). Studies in humans and the well-characterized mouse model (MCMV) have shown that the majority of inflationary populations are composed of terminally differentiated effector phenotype (TEFF) T cells that presumably develop as a result of repeated antigen stimulation and may not possess the proliferative or survival capacity necessary for long-term maintenance of CMV immunity (22, 27, 29-34). Interestingly however, a fraction of these inflationary T cells retain a memory-like (TM) phenotype, despite sharing epitope specificity and T cell receptor sequences with the TEFF subset (23, 25, 35-37). Studies with other infection models have shown that such a memory phenotype can identify cells that have “stem-cell like” characteristics (38, 39). If this model holds true for CMV immunity, the CMV-specific TM cells would be ideal to use in an adoptive immunotherapy setting. Recent evidence supports this hypothesis. In a non-human primate model, CMV-specific effector T cells that were expanded in vitro from sorted TM cell had a superior ability to survive after adoptive transfer (40). Moreover, a human study showed a positive correlation between the presence of CMV-specific TM cells in a donor transfer and the long-term maintenance of donor derived cells (41).
The goal of our study was to utilize the mouse model (MCMV) to directly address the capacity of the CMV-specific TM population to restore long-term CMV-specific immunity after transfer. Importantly, we found that the MCMV-specific TM cells share a common genetic program with their human CMV-specific counterparts and that these cells could repeatedly restore long-term CMV-specific immunity under a spectrum of transfer scenarios. Our data suggest that adoptive immunotherapy with CMV-specific TM cells will improve consistency and clinical outcomes in patients at-risk for developing CMV disease.
Materials and Methods
Mice
Unless otherwise indicated, C57BL/6 mice, CD45.1 mice (B6.SJL-Ptprca Pepcb/BoyJ), Thy1.1 mice (B6.PL-Thy1a/CyJ) and Rag−/− mice (B6.129S7-Rag1<tm1Mom>J) were purchased from Jackson Laboratory. OT-I transgenic mice (C57BL/6-Tg(TcraTcrb)1100Mjb/J) also purchased from Jackson, were bred with CD45.1 mice to produce double positive (CD45.2pos/CD45.1pos) OT-I mice. All protocols were approved by the Thomas Jefferson University Institutional Animal Care and Use Committee.
Infections
Unless otherwise indicated, mice were infected intraperitoneally (i.p.) with 2 × 105 pfu of MCMV strain MW97.01 (42). Mice were considered latently-infected at 8 weeks post-infection. Rag−/− mice were infected with 5 × 104 pfu of MCMV-TK virus (43). OT-I T cell transfer recipients were challenged with 2 × 105 pfu MCMV-SL8, which expresses the SIINFEKL peptide (44, 45).
Tetramer Staining, Antibodies and FACS Analysis
MHC-tetramers were provided by the NIH Tetramer Core Facility (http://tetramer.yerkes.emory.edu/) and have been described previously (27). Staining was performed as described previously (27) with tetramers and the following antibodies: [CD8(53-6.7); CD44(IM7); CD27(LG.3A10); CD127(A7R34); KLRG1(2F1); CD62L(MEL-14); CD45.1(A20); CD45.1(104); Thy1.1(OX-7); Thy1.2(30-H12); IFN-γ(XMG1.2); TNF−α(MP6-XT22); CD107a(1D4B)]. In all cases, samples were collected on an LSR II and analyzed with FlowJo software (Tree Star). The gating strategy for phenotypic characterization of tetramerpos CD8pos T cells involved first gating lymphocytes and then singlets. CD8pos cells were gated as frequency of singlets. Tetramerpos cells were identified as a frequency of CD8pos cells. A B8R tetramer (specific for the B8R peptide from Vaccinia), was used as a negative tetramer control. Tetramerpos cells were phenotypically defined by they expression of KLRG1, CD27, CD127 or CD62L.
Adoptive Transfers
CD8pos splenocytes from latently-infected donors were enriched using EasySep Biotin selection kit (StemCell Technologies) and biotinylated antibodies against RBCs(Ter-119), CD4(GK1.5) and CD19(6D5) according to the recommended protocol. Enriched cells were stained to determine the frequency of tetramerpos cells within the enriched fraction and then sorted on either a MoFlo (Dako Cytomation) or an ARIA II (BD Biosciences) cell sorter. Sorted cells were counted, and 5 × 104 cells were transferred via the retro-orbital sinus. Sort purity was analyzed on an LSR II. The number of transferred tetramer-binding CD8pos T cells was estimated using the tetramer frequency within the enriched CD8pos population and the post-sort purity analysis. Fold change was calculated as the number of tetramer-binding T cells in the spleen 7 days post-challenge over the total number of tetramerpos cells transferred (assuming 100% engraftment). The gating strategy for analyzing donor cells in the recipients was identical to that described above with antibodies specific for the relevant congenic marker (CD45.1 or Thy1.2).
For OT-I adoptive transfers, splenocytes from naive mice containing 600 OT-I T cells were transferred. Recipients were challenged with MCMV-SL8. To establish secondary and tertiary populations, OT-I TM CD8pos T cells were FACS sorted and transferred as described above. Following challenge with MCMV-SL8, the frequencies of donor OT-Is were determined in the blood of recipients using the strategy described above except that singlets were not identified and OT-I donors were identified by expression CD45.1 and Vα2.
Intracellular Stimulation (ICS)
ICS and staining was performed as previously described (27, 45), with minor modifications. Specifically, cells were incubated with 1 μg/mL peptide (Genemed Synthesis), 1 μg/mL brefeldin A (GolgiPlug, BD Biosciences) and CD107a-specific antibody for 3 hours.
CD70 Blocking Antibody Treatment
CD70 antibody blockade was performed as previously described (46), with minor modifications. Briefly, mice received either 150 μg of anti-CD70(FR70) or control rat IgG2b (both from BioXCell) via the i.p. route. Injections were administered at days -1, 0 and 3 post-infection.
Antibody Depletions
Antibody depletions were performed with Thy1.1(19E12), CD4(GK1.5) and NK1.1(PK136) antibodies. 300 μg of each antibody were administered i.p. in PBS. Three subsequent injections of 100 μg of each antibody were given at 7 day intervals.
Microarray
Splenocytes from latently-infected mice were co-stained with tetramers loaded with the antigenic peptides from M38, m139 and IE3 (25) and sorted on a MoFlo (Dako Cytomation) cell sorter. MCMV-specific T cells were identified as CD8pos, CD44hi and tetramer binding and then further segregated into TM and TEFF cells subsets by expression of KLRG1 and CD127. Naïve CD8pos cells were CD44lo. Total RNA was isolated using the Qiagen RNAeasy Plus Kit (Qiagen), quantified on a NanoDrop 2000c Spectrophotometer (Thermo Scientific) and processed at the Microarray Core Facility at Thomas Jefferson University. Briefly, 2.5 μg fragmented and biotinylated cDNA was hybridized to Mouse gene 1.0 ST array (Affymetrix). Chips were scanned on an Affymetrix Gene Chip Scanner 3000 and data were analyzed using the R programming language and various packages from Bioconductor (47). The oligo package (48) was used to extract expression data from the Affymterix CEL files and perform background and RMA normalization (49). Annotation information was added using the mogene10sttranscriptcluster.db (50) package. Probes without valid annotations (7,196 of 35,556 probes) were removed before differential expression analysis using the limma package's (51) linear modeling and Bayes methods (52). Genes showing up- or down-regulation of at least twofold and p-value < 0.05 in each of three contrasts (TEFF vs. Naïve, TM vs. Naïve, and TEFF vs. TM) were considered for gene set enrichment analysis (GSEA). Microarray data have been deposited in the Gene Expression Omnibus (GEO) database (53) (accession number: GSE61927 http://www.ncbi.nlm.nih.gov/genbank)
Gene Set Enrichment Analysis
Human data for series GSE24151 (54) was retrieved from NCBI's GEO database (53), extracted using Partek® Genomics Suite® software, version 6.6 (Partek Inc., St. Louis, MO) and curated for input into GSEA software (55) (http://www.broadinstitute.org/gsea/). Since the data for GSE24151 have been deposited in GEO as log10 ratios of the reference pool to sample, each value was inverted by multiplying by -1. Gene names in the six mouse gene lists (up- or down-regulated in each of the three contrasts) were converted to human names using data from NCBI's Homologene database, Release 68 (http://www.ncbi.nlm.nih.gov/homologene). The converted gene lists along with genes specific to the liver and the TCR receptor pathway from the Molecular Signature Database (MSigDB) (55) were analyzed for enrichment in the human data using recommended settings for the GSEA command-line interface.
Results
MCMV-specific inflationary TM populations are stable and share a common transcriptional program with HCMV-specific CD8pos T cells in humans
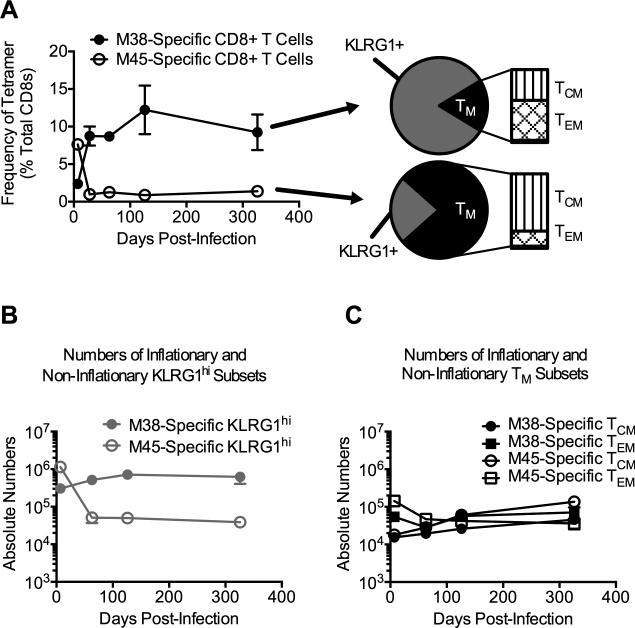
In the mouse model, MCMV infection of C57BL/6 (B6) mice results in inflation of select MCMV-specific CD8pos T cells specific for peptides from the viral proteins M38, m139 and IE3 (Fig. 1A and (25, 27, 28)). As in humans infected with human CMV (HCMV), the majority of MCMV-specific inflationary T cells express a TEFF phenotype (often defined as TEMRA in humans), while only a small fraction express a TM-like phenotype, defined here as KLRG1lo/CD27hi and further sub-divided into central memory (TCM – CD127hi/CD62Lhi) and effector memory (TEM – CD127hi/CD62Llo) subsets (Fig. 1A and (22, 23, 27, 29-33, 56)). In contrast, “non-inflationary” MCMV-specific CD8pos T cell responses, represented by the response against the viral protein M45, contract after acute infection and are thought to be maintained by homeostatic mechanisms thereafter (Fig. 1A and (25, 27, 57)). As expected, noninflators express a predominately memory (TM) phenotype, which also includes both TCM and TEM subsets (Fig. 1A and (23, 27)).
Figure 1. The non-inflationary and inflationary CD8pos T cell populations retain similar numbers of TM phenotype cells.
Cohorts of age-matched B6 female mice were infected with MCMV and sacrificed at the indicated time points (n = 4 per time point). Tetramer staining and phenotypic analyses were performed on blood and splenocytes. (A) Frequency of tetramer-binding CD8pos T cells in the blood at indicated time points. The phenotypic analysis shown was performed at d326 post-infection. TM cells were identified as CD27hi/KLRG1lo. TCM and TEM were further identified as CD127hi and either CD62Lhi or CD62Llo, respectively. (B) Absolute numbers of KLRG1hi tetramer-binding CD8pos T cells in the spleen. (C) Absolute numbers of TCM and TEM tetramer-binding CD8pos T cells. Data are displayed as mean ± SEM and represent two independent experiments.
It remains unknown whether the constant immune stimulation needed to maintain memory inflation causes a decline of the TM subset within inflationary populations over time. Using infection-matched cohorts, we found that the numbers of TM cells that were specific for inflationary antigens were stable over time and remarkably similar to the numbers of non-inflationary TM cells, despite great differences between the numbers of inflationary and non-inflationary TEFF cells (Fig. 1B, 1C). Thus, although continuous antigen stimulation maintains memory inflation, the inflationary TM population remains stable.
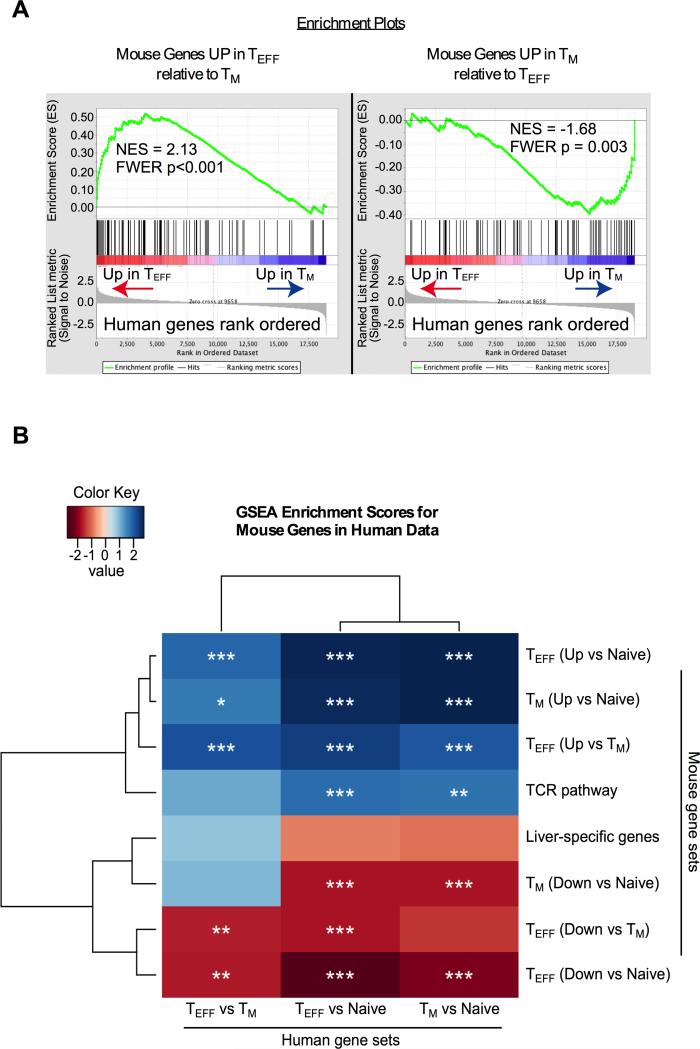
The MCMV model is well characterized and the T cell responses clearly recapitulate those seen in HCMV-infected people. To determine whether MCMV-specific TM and TEFF cells share a common transcriptional program with their human counterparts, we sorted MCMV-specific TM (CD44hi/CD127hi/KLRG1lo) and TEFF (CD44hi/CD127lo/KLRG1hi) cells specific for the M38, m139 and IE3 antigens. Microarray analyses were performed on these cells. Genes that were significantly up- or down-regulated in TM and TEFF subsets relative to each other or to naïve (CD44low) T cells, were mapped to the corresponding human genes and compared with the profiles of HCMV-specific T cells, previously defined by the van Lier group as CD27hi/CD45RAlo (TM) or CD27lo/CD45RAhi (TEFF) (54). The CD27 and CD127 (IL-7Rα) molecules both mark CMV-specific T cells with a memory phenotype in mice and humans (27, 29, 32, 58, 59) and nearly all MCMV-specific KLRG1lo/CD27hi cells (TM) co-expressed CD127 (either TCM or TEM, Figure 1A). Gene set enrichment analyses (GSEA) were used to measure the overall correlation between the mouse and human gene expression data. As shown in Fig. 2A, genes that distinguished mouse TEFF and TM cells from each other were highly enriched within the corresponding human data set. That is: genes up-regulated specifically in mouse TM cells relative to mouse TEFF cells were highly enriched within the genes that distinguish human TM cells from human TEFF and vice versa. Moreover, relative to naive T cells, mouse genes that were up and down-regulated by TEFF or TM cells were highly enriched within genes that distinguished their human counterparts from human naive T cells (Fig. 2B). The analyzed mouse genes and the core enrichment profiles of each comparison are listed in Supplemental Table 1. Importantly, several of these genes corresponded to our sorting parameters and the known phenotypes of TM and TEFF populations. As controls, identical analyses were performed with genes associated with the T cell receptor signaling pathway or liver and the data exhibited expected patterns (Fig. 2B).
Figure 2. Gene Set Enrichment Analyses reveal significant overlap between the transcriptional profile of CMV-specific T cells in humans and mice.
(A) Gene set enrichment was performed as described in the Methods. Shown are the enrichment plots for mouse genes that differed in a TEFF vs. TM comparison, plotted relative to human TEFF and TM cells. Values represent the normalized enrichment score (NES) and Family Wise Error Rate (FWER), which estimates the probability of a false positive NES. (B) Lists of significantly altered mouse genes (2-fold up or down and P<.05) were generated for TEFF and TM cells relative to each other and relative to naive (CD44low) T cells. GSEA analyses were performed with these mouse gene sets relative to each of the indicated human data sets, rank ordered by expression (see methods). Stars indicate FWER corrected significance to control for multiple testing (* P<.05, ** P<.01, *** P<.001).
Overall, these data show that MCMV-specific and HCMV-specific T cells share a common genetic program, validating the use of the MCMV model to investigate the function of HCMV-specific T cells. To our knowledge, this is the first direct comparison of mouse and human CMV-specific T cell gene expression profiles.
The inflationary TM population retains proliferative capacity
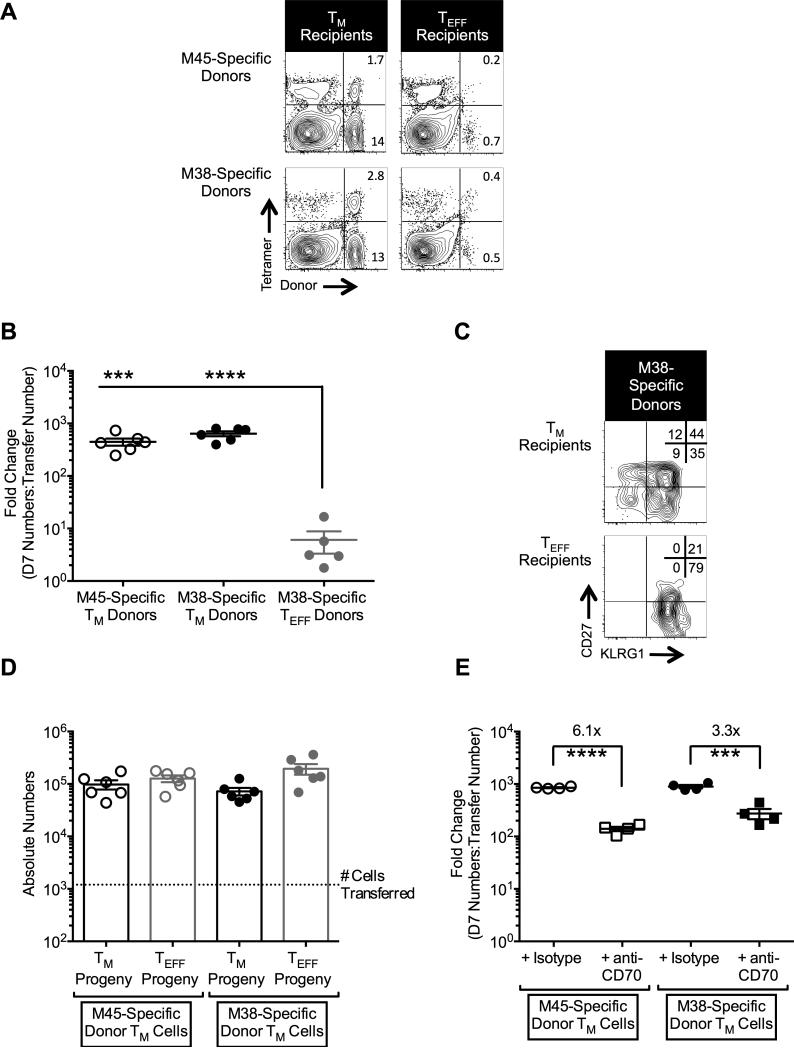
To test the proliferative capacity of the TM and TEFF cells, both populations were sorted from spleens of latently-infected B6 mice (>3 months post-infection) using their differential expression of KLRG1 and CD27. Sorted cells were transferred into naive congenic recipients and re-challenged. The M45- and M38-specific TM cells proliferated robustly within 7 days after challenge, each expanding almost 1000-fold in the spleen alone, assuming 100% engraftment of the donor cells (Fig. 3A, 3B). In contrast, the M38-specific TEFF population expanded less than 10-fold in the same time period. Importantly, while the TEFF donor cells remained exclusively KLRG1hi, the TM donor cells produced large numbers of both TEFF and TM progeny (Fig. 3C). In fact, donor M45- and M38-specific TM phenotype cells were present in the spleen 7 days after challenge at numbers that were approximately 50 to 100-fold higher than had been transferred (dotted line, Fig. 3D), indicating expansion of this subset without terminal differentiation. These data show that MCMV-specific TM cells retain robust proliferative capacity and can produce phenotypically diverse progeny including new TM-phenotype cells.
Figure 3. TM cells dramatically expand 7 days post-challenge and produce both TM and TEFF progeny.
Age matched B6 mice received either TM or TEFF cells and were challenged with MCMV as described in the Methods. Spleens were collected 7 days later for analysis. (A) Representative FACS plots of tetramerpos donors in the spleen 7 days post-challenge. Frequencies in the corner are relative to total CD8pos cells. (B) Fold change of donor cells in the spleen, calculated as described in the Methods, 7 days after challenge. As antigen-specific T cells were not sorted, approximately equal numbers of M38- and M45-specific TM cells were transferred but ~10-fold more M38-specific TEFF cells were transferred compared with the TM cells. Due to the extremely low number of M45-specific TEFF transferred and the minimal expansion at day 7, it was not possible to calculate a comparable fold change value for the M45-specific TEFF population. Data were collected from two independent experiments (TM: n = 6 total; TEFF: n = 5 total) are shown. Statistical significance was determined by a Student's t-test (*** P<.001; **** P<.0001). (C) Representative FACS plots of M38-specific CD8pos T cell progeny from either TM or TEFF donors in the spleen at 7 days post-challenge. Frequencies in the corner are relative to M38-specific CD8pos cells. (D) Absolute number of TM and TEFF phenotypic progeny that were produced from TM donors. Data are from the same experiments described in (B). (E) Fold change of donor cells in the spleen following treatment with either isotype control or anti-CD70 antibody. Data were collected 7 days post-challenge and represent two independent experiments (n = 6 total). Statistical significance was determined by a Student's t-test (*** P<.001; **** P<.0001). All graphical data are displayed as mean ± SEM.
Recent work has shown that interaction between CD27 and its ligand CD70 plays a functional role in the proliferation of MCMV-specific inflationary T cells (46). To test the contribution of this interaction specifically within the TM population, we sorted and transferred TM cells as above and blocked the CD27-CD70 interaction as described in the Methods. Blocking the CD27-CD70 interaction significantly decreased the expansion of the M38- and M45-specific TM cells 7 days post-challenge by approximately 4- to 6-fold (Fig. 3E), which is in line with the impact of CD70 blockade on unsorted (i.e. combined TM and TEFF populations) inflationary T cells (46). These data further suggest that the majority of proliferative potential of inflationary T cells is contained within the minor TM subset. It should be noted that even in the presence of CD70 blockade, the TM population retained a proliferative capacity that was greater than the TEFF population, suggesting that additional pathways contribute to the total proliferative potential of these cells (Fig. 3B, 3E and unpublished observations).
The inflationary TM population persists and can repeatedly recapitulate memory inflation
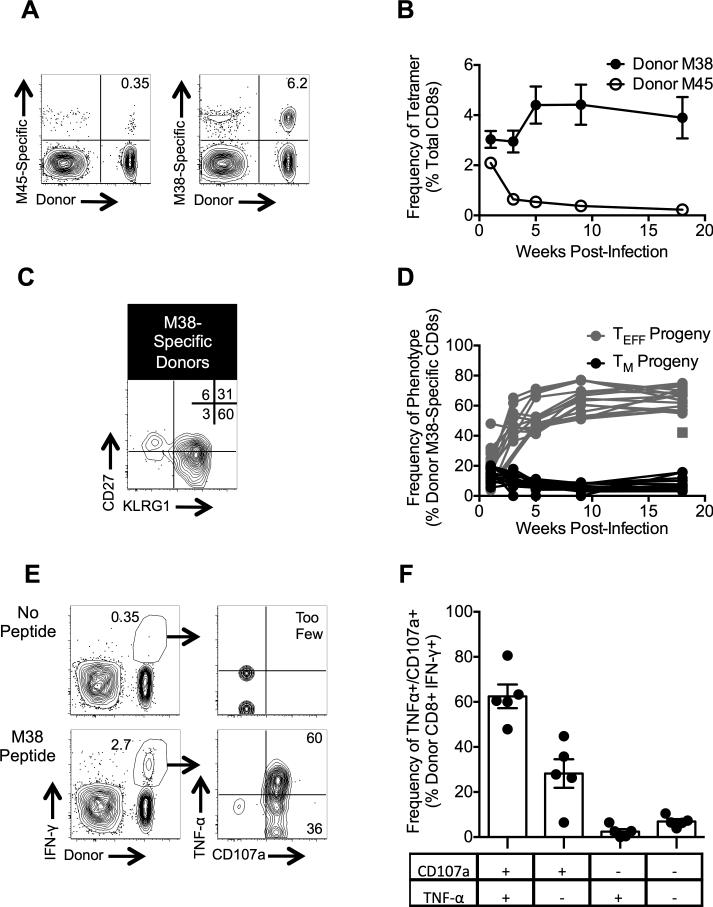
To determine the ability of the TM donor cells to persist long-term, we tracked the progeny from TM donor cells in the blood after re-challenge. M38-specific T cells from TM-sorted donors persisted at high frequencies in recipients, while the M45-specific donor cells contracted after their initial expansion in the same mice (Fig. 4A, 4B). Despite their initial TM phenotype, the donor M38-specific T cells largely expressed a TEFF phenotype after challenge (Fig. 4C, 4D), consistent with a typical “inflationary” population. The population as a whole retained its ability produce IFN-γ, TNF-α and expose CD107a (Fig. 4E, 4F). Importantly, a small portion of donor T cells retained their TM phenotype even after this secondary challenge (Fig. 4C, 4D).
Figure 4. TM cells reinflate following re-challenge and retain function.
Age matched B6 mice received TM cells and were challenged with MCMV as in the Methods. (A) Representative FACS plots of donor-derived T cells in the blood 126 days post-challenge. (B) Frequencies of tetramer-binding T cells in the blood over time. Data were collected from three independent experiments (n = 17 total). (C) Representative FACS plot of the phenotype of donor-derived M38-specific T cells in the blood 126 days post-challenge. (D) Frequencies of donor-derived, M38-specific TM and TEFF cells in the blood over time. Data are from the same experiments described in (B). Each line represents an individual mouse. The square datum point represents a mouse that appeared to lose the donor T cells after day 7 post-challenge, but effectors appeared ~20 weeks after challenge. (E-F) Intracellular cytokine staining was performed on splenocytes 221 days post-challenge. Shown are representative FACS plots of stimulated (with M38 peptide) and unstimulated cells (E) and the frequencies of IFN-γ positive cells that also express TNF-α and/or CD107a (F). Data were collected from a single experiment (n = 5) described above. All graphical data are displayed as mean ± SEM.
To understand whether these persistent TM phenotype donors continued to be functional, we turned to the OT-I transgenic system to facilitate sorting and avoid the possible selection of different T cell clones (Fig. 5A). As shown previously, transferred naive OT-Is undergo inflation and produce both TM and TEFF progeny after primary challenge with SIINFEKL-expressing MCMV-SL8 (45). We sorted the TM phenotype OT-I cells that formed after primary challenge, transferred these cells and challenged the recipients to establish secondary populations (Supplemental Fig. 1A). As with non-transgenic T cells (Fig. 4), secondary challenge of TM OT-Is induced inflation and TEFF formation as well as a persistent KLRG1lo population (Supplemental Fig. 1B). These secondary TM cells were again sorted (Supplemental Fig. 1C), transferred into a 3rd set of naïve recipients and re-challenged. Incredibly, the donor secondary TM population inflated and produced both KLRG1hi and KLRG1lo progeny following this tertiary challenge (Fig. 5B-5E).
Figure 5. TM cells can reinflate following multiple re-challenges.
(A) Schematic of experimental design. To establish primary OT-I inflationary populations, 600 naïve OT-I T cells expressing CD45.1 were transferred into naïve B6 (CD45.2) recipients followed by infection with MCMV-SL8 (i.e. primary challenge). Thirteen weeks later, 6,000 TM phenotype primary OT-Is, isolated by FACS sorting, were transferred into new B6 recipients followed by MCMV-SL8 challenge (i.e. secondary challenge). This process was repeated a third time, transferring 3,500 TM OT-Is into naive mice and challenging with MCMV-SL8 (i.e. tertiary challenge). (B-C) Representative FACS plot of the donor stain 91 days post tertiary challenge (B) and frequencies of donor OT-Is (relative to total CD8s) in the blood at the indicated time points after tertiary challenge (C). Data were collected from two independent experiments (n = 12 total). Each line represents an individual mouse. (D-E) Phenotypic analyses of the mice described in (B-C). Representative FACS plot of the donor stain 30 weeks post-challenge Frequencies are relative to donor CD8s. (F-G) Intracellular cytokine staining was performed on splenocytes approximately 20 weeks after the tertiary challenge. Shown are representative FACS plots of stimulated (with SIINFEKL peptide) and unstimulated cells (F) and the frequencies of IFN-γ positive cells that also express TNF-α and/or CD107a (G). Data were collected from two independent experiments (n = 12). Data are displayed as mean ± SEM.
Repeated acute viral challenges of small numbers of T cells in naive mice drives TEFF differentiation (60-63) and indeed the overall frequency of tertiary inflationary cells that retained a TM phenotype was reduced (Fig. 5E and Supplemental Fig. 1B). However, these tertiary stimulated OT-Is remained functional, producing both IFN-γ and TNF-α, as well as exposing CD107a (Fig. 5F, 5G). These data show that TM phenotype T cells specific for inflationary antigens can repeatedly recapitulate memory inflation upon viral challenge and produce functional TEFF and TM progeny.
Memory and Effector Subsets protect Rag−/− mice
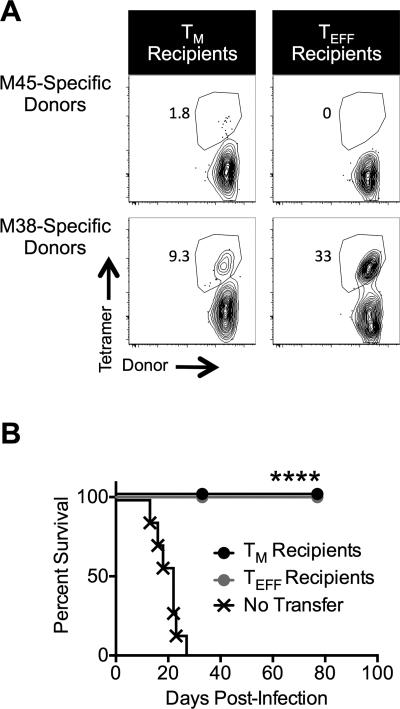
To test the ability of transferred TM cells to protect against a lethal MCMV challenge, TM and TEFF populations were sorted from latently-infected B6 mice as above and transferred into Rag−/− recipients. One day later, the Rag−/− recipients were challenged with MCMV-TK, which lacks the m157 gene and is therefore resistant to NK-mediated control (43). Both transferred TM or TEFF cells expanded following the challenge and were sufficient to protect the recipients (Fig. 6A, 6B). In contrast, Rag−/− mice that received no T cell therapy became moribund in 2-4 weeks and had to be sacrificed (Fig. 6B). Notably, the TEFF population, which proliferated very poorly in immune replete mice (Fig. 3), expanded and persisted in immune deficient hosts for at least 11 weeks post-challenge (Fig. 6A, 6B). However, the TEFF response in 5 out of 6 recipients M45-specific, non-inflationary T cells (Fig. 6A). These data show that MCMV-specific TM cells are capable of protecting immune deficient mice and producing immune responses with broad specificities.
Figure 6. TM and TEFF cells protect Rag−/− mice following an acute MCMV challenge.
Age matched Rag−/− mice received either TM or TEFF cells and were challenged with MCMV-TK as described in the Methods. Mice were monitored daily for signs of morbidity (lethargy, raised hair and shaking) and sacrificed if they displayed clear signs of morbidity. Data were collected from two independent experiments. One experiment was carried out until 77 days post-challenge. A second experiment was censored at 33 days post-challenge. (A) Representative tetramer staining of T cells in Rag−/− that received either TM or TEFF transfers. Data were collected 11 weeks post-challenge. Frequencies are relative to total CD8s. (B) Survival curve (n = 7 for control group; n = 7 for TM group; n = 6 for TEFF group). Statistical significance was determined by a log-rank (Mantel-Cox) test (**** P<.0001).
The TM population can persist long-term and respond when necessary
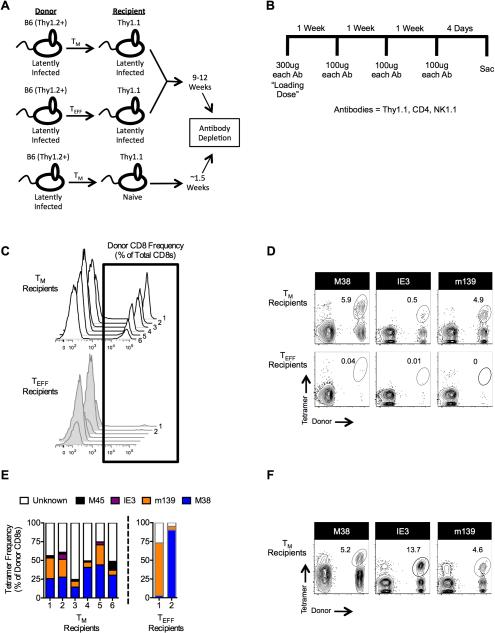
Patients undergoing hematopoietic stem cell transplantation (HSCT) are most susceptible to late-onset (>100 days) reactivating CMV, as opposed to an acute CMV infection ((12, 64-66) and reviewed in (67)). Furthermore, transferred CMV-specific T cells will need to compete with host immunity. Therefore, we developed a model to test whether TM and TEFF subsets could respond to viral reactivation after a long delay. To this end, TM and TEFF cells were sorted from latently-infected mice (> 3 months post-infection) and transferred into immune replete, infection matched or naive, congenic recipients differing at the Thy1 locus (Fig. 7A). Following the transfer, the latently-infected recipients were rested as described in the figure legend. Donor T cells did not expand dramatically in any animal following transfer (Supplemental Fig. 2A) supporting our previous conclusion that competition between T cells dictates MCMV-specific T cell expansion (45). Recipient T cells and NK cells were then eliminated in all mice using a cocktail of depletion antibodies that targeted the host cells (Thy1.1pos), but left the donor cells (Thy1.2pos) intact (Fig. 7B and Supplemental Fig. 2B). This depletion protocol did not induce detectible viral transcription in any animal as assessed by nested RT-PCR (unpublished observations), likely due to the presence of anti-viral antibodies (68). Despite the 9-12 week rest period, MCMV-specific donor TM cells responded robustly in all infected recipients after host depletion (Fig. 7C and 7D). Importantly, donor TM cells did not expand to detectible levels in depleted naive recipients (Supplemental Fig. 2C). However, viral challenge of naïve mice that received TM donor cells 12 weeks previously induced a robust donor response in 3 of the 4 animals, indicating that the TM cells persisted in these mice, even without any antigen (Supplemental Fig. 2C). Thus, antigen rather than homeostatic mechanisms accounts for the donor TM response in infected recipients.
Figure 7. TM cells persist in latently-infected, immune replete mice and expand when host immunity is lost.
(A) Schematic of experimental design. Age matched B6 and Thy1.1 mice were infected with 1×106 pfu MCMV-Smith. Following the establishment of viral latency (>8 weeks post-infection), either TM or TEFF cells from the B6 donors were transferred, as described in the Methods, into the latently-infected Thy1.1 recipients or into naïve Thy1.1 mice. Latently-infected recipients were rested for 9-12 weeks, while the naïve recipients were rested for approximately 1.5 weeks. (B) Antibody depletion schedule. (C-E) The presence of tetramerpos donors was analyzed by flow cytometry immediately following the depletion schedule. Data were collected from two independent experiments (n = 6 total). Three mice from each group were depleted 9 weeks after the transfer; three mice from each group were depleted 12 weeks after transfer. (C) Histograms of donor T cells within each individual recipient. (D) Representative FACS plots of tetramerpos donors immediately following the depletion regimen. (E) Frequency within each individual recipient of each analyzed tetramer as a percent of total donor CD8+ cells. TEFF recipients 3-6 are excluded because they did not have a donor population. (F) Tetramer staining was performed 11 weeks after depletion in one experiment described above (n = 3).
In marked contrast, after depletion, donor T cells were only detectible in 2 animals that had received TEFF cells and then only at very low frequencies (Fig. 7C, 7D). Control experiments (Supplemental Fig. 3A-3C) supported previous work (69) suggesting that the KLRG1-specific antibody did not induce depletion of the transferred TEFF subset. Thus, the failure of TEFF cells to expand in this setting is not a sorting artifact, but rather the inability to persist and/or expand in response to low amounts of viral antigen.
After expansion, all infected mice that received TM cells had a donor population specific for multiple epitopes and the progeny had differentiated to form new TEFF populations (Fig. 7E and unpublished observations). Furthermore, the four tetramers used only stained ~60% of the total donor population in each animal (Fig. 7E), suggesting that the remaining 40% of each donor population contained cells specific for additional MCMV antigens. In contrast, in the two animals in which TEFF donors expanded to detectible levels, each was skewed substantially towards a single inflationary epitope (Fig. 7E). Since these sorted TEFF populations included large numbers of T cells specific for M38, m139 and IE3, this “hit-or-miss” expansion of donor T cells with select specificities implies that a very small number of non-TEFF cells may have contaminated the transfer.
In the mice that received TM donor cells, their diverse progeny persisted in recipients for more than 11 weeks after termination of the depletion regimen, even though host immunity had returned (Fig. 7F). These data suggest that TM cells with inflationary specificities are capable of surviving in an environment with very little or no antigen stimulation and then responding as needed during a period in which the host is immune compromised and viral antigen becomes available.
In total, these data show that protective MCMV-specific TM cells persist throughout infection, retain superior proliferative function, and can respond to viral antigen as needed, in contrast to the numerically dominant TEFF cells. Since MCMV-specific TM cells share a transcriptional program with HCMV-specific TM cells, our data suggest that TM cells may be ideal candidates to restore functional immune surveillance in patients at risk for CMV reactivation.
Discussion
Adoptive immunotherapy using CMV-specific CD8pos T cells can be a successful therapeutic strategy for combating CMV reactivations (5-18). However, the majority of CMV-specific CD8pos T cells isolated from healthy donors will express an effector-differentiated phenotype (CD27lo/CD127lo/CD45RAhi/KLRG-1hi/CD57hi - reviewed in (70)), and in vitro expansion of CMV-specific T cells drives their differentiation towards an effector phenotype (40). We used the MCMV model to show that the ability to restore MCMV-immunity is contained almost entirely within the minor TM subset that retains CD27. Although both TM and TEFF cells protected Rag−/− mice (Fig. 6), humans are unlikely to remain completely immune depleted like Rag−/− mice, and bolus CMV infections are of lesser concern than reactivation following transplantation. The inability of the TEFF population to consistently expand after immune depletion in latently-infected hosts, suggests that these cells will only be protective under limited conditions. These data support a previous study in humans that correlated the transfer of CD27hi CMV-specific T cells with an increased likelihood of T cell persistence and expansion (41).
To validate the use of the MCMV model, we compared human and mouse MCMV-specific T cells and show for the first time that TM and TEFF populations in mice and humans share a common transcriptional profile. The power of the GSEA analysis used for this comparison is that it identifies significant correlations across the entire transcriptional profile, rather than comparing individual genes. Nevertheless, we expect that future studies examining conserved and divergent genetic pathways will reveal significant and relevant information about CMV-specific immunity in mouse and man. These results highlight the usefulness of the MCMV model to: 1) perform CMV-specific CD8pos T cell functional studies that are difficult or impossible to perform in humans and 2) provide translational insights into novel or improved therapeutic strategies.
Understanding how CMV-specific T cell immunity is maintained is critical for the improvement of CMV adoptive immunotherapy. Persistent antigen stimulation from CMV reactivations results in the majority of inflationary CD8pos T cells developing a TEFF phenotype and function. However, our previous work showed that unsorted inflationary CD8pos T cells, containing primarily TEFF cells, declined after transfer into congenic, latently-infected recipients (27). These data suggest that MCMV-specific TEFF cells are unable to sustain themselves in an immune replete environment, even in the presence of antigen. It has been proposed that the accumulation of TEFF cells is the result of continual antigen stimulation of the TM population. Our data show that a small, stable MCMV-specific TM population has strong functional similarities to classical memory T cells that develop following acute infections and can recapitulate memory inflation. For example, the ability to proliferate in response to antigen without terminal differentiation is hallmark of functional memory T cells (71). In addition to producing differentiated progeny that accumulated after MCMV challenge (Figure 4D), donor TM cells also produced TM phenotype progeny that outnumbered the cells transferred (Fig. 3C and D) and persisted throughout our observation period (Fig. 4C and D). These data suggest that MCMV-specific TM cells have the ability to replace themselves even while producing differentiated progeny in response to antigen. Importantly, this was true through at least three rounds of stimulation using sorted splenic TM cells (Figure 5). Thus, MCMV-specific TM cells have the capacity to respond repeatedly to viral antigen during this persistent infection.
It is interesting that transferred TM cells failed to expand in immune-replete, latently-infected hosts. Indeed, detectible numbers of donor T cells were only evident in one out of six mice prior to immune depletion (Supplemental Fig. 2A). In this case, the donors were not positive for any of the tetramers used in the analyses and made up less than 1% of the total CD8pos population. However, loss of the host T cell populations led to rapid and robust expansion of donor T cells with diverse specificities and phenotypes in all TM cell recipients (Figure 7). The failure of transferred TM cells to expand in the presence of host MCMV-specific immunity may reflect the relative lack of available antigen during the latent phase of MCMV infection. Indeed, viral reactivations occur in only a fraction of latently-infected cells at any given time, and only rarely produce infectious viral particles (72, 73). Moreover, we have found that competition between T cells for access to this limited antigen regulates the expansion of individual T cell clones (45). Thus, the combination of low antigen and large numbers of MCMV-specific T cells in the recipients may have “shielded” the majority of the donor TM cells from the ongoing infection – an idea we have proposed previously (45, 74). Importantly, MCMV antigen is not required for MCMV-specific TM cell survival. We have previously shown that MCMV-specific TM cells divide at a consistent rate with or without antigen (28) and our new data (Supplemental Fig. 2C) show that inflationary TM cells can survive in naïve mice without any antigen. Thus, homeostatic mechanisms can support the inflationary TM population when it does not have access to antigen, which may partially explain the preservation of memory function within the TM subset. Taken together, these data suggest that the highly functional TM population, which can persist without access to antigen, proliferates robustly and produces new TM cells as well as more differentiated progeny upon antigen stimulation.
Overall, our data further support the model that the burden of maintaining memory inflation falls on the functional TM population, which can provide a stable and consistent source of new TEFF progeny whenever needed, over for prolonged periods of time. However, T cell competition for limited antigen appears to prevent the continuous stimulation of most TM cells. Nonetheless, the TM population is capable of robustly responding if T cell competition is lost - a conclusion with important clinical implications for adoptive immunotherapy. Variations in transplant protocols, patients and anti-viral therapy responses make it difficult to predict and standardize CMV prevention therapies. Our data suggest that the plasticity of the TM population, transferred before any disease develops, may offer a “personalized” therapy, where the treatment adapts to the conditions of the patient and responds if and when antigen becomes available. Future studies will be needed to explore whether the addition of homeostatic cytokines (e.g. IL-15) or pharmacotherapeutics (e.g. rapamycin (75)) preserves the TM phenotype either in vivo or during in vitro expansion.
Supplementary Material
Acknowledgments
We thank the Kimmel Cancer Center Flow Cytometry Facility and Animal Facility at Thomas Jefferson University.
This work was supported by a faculty start-up package from Thomas Jefferson University and grants from the National Institutes of Health (K22-AI081866 and RO1AI106810 grants awarded to CMS).
Abbreviations
- CMV
Cytomegalovirus
- HCMV
Human Cytomegalovirus
- MCMV
Murine Cytomegalovirus
- TM
memory phenotype CD8+ T cells
- TEFF
terminal differentiated effector phenotype CD8+ T cells
- TCM
central memory CD8+ T cells
- TEM
effector memory CD8+ T cells
- TEMRA
terminal differentiated CD8+ T cell phenotype in humans
- i.p.
intraperitoneal route
- GSEA
gene set enrichment analysis
- GEO
gene expression omnibus
- SEM
standard error of the mean
Footnotes
Conflict-of-Interest
The authors declare no conflict-of-interests.
Experiments conceived and designed by MQ and CMS. Experiments conducted by MQ, HT, BD and TM. Data analyzed by MQ, HT, MT and CMS. Manuscript written by MQ and CMS.
References
- 1.Staras SA, Dollard SC, Radford KW, Flanders WD, Pass RF, Cannon MJ. Seroprevalence of cytomegalovirus infection in the United States, 1988-1994. Clin. Infect. Dis. 2006;43:1143–1151. doi: 10.1086/508173. [DOI] [PubMed] [Google Scholar]
- 2.Polić B, Hengel H, Krmpotić A, Trgovcich J, Pavić I, Lučin P, Jonjić S, Koszinowski UH. Hierarchical and redundant lymphocyte subset control precludes cytomegalovirus replication during latent infection. J. Exp. Med. 1998;188:1047–1054. doi: 10.1084/jem.188.6.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simon CO, Holtappels R, Tervo H-M, Böhm V, Däubner T, Oehrlein-Karpi SA, Kühnapfel B, Renzaho A, Strand D, Podlech J. CD8 T cells control cytomegalovirus latency by epitope-specific sensing of transcriptional reactivation. J. Virol. 2006;80:10436–10456. doi: 10.1128/JVI.01248-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holtappels R, Bohm V, Podlech J, Reddehase MJ. CD8 T-cell-based immunotherapy of cytomegalovirus infection: “proof of concept” provided by the murine model. Med. Microbiol. Immunol. 2008;197:125–134. doi: 10.1007/s00430-008-0093-2. [DOI] [PubMed] [Google Scholar]
- 5.Walter EA, Greenberg PD, Gilbert MJ, Finch RJ, Watanabe KS, Thomas ED, Riddell SR. Reconstitution of cellular immunity against cytomegalovirus in recipients of allogeneic bone marrow by transfer of T-cell clones from the donor. N. Engl. J. Med. 1995;333:1038–1044. doi: 10.1056/NEJM199510193331603. [DOI] [PubMed] [Google Scholar]
- 6.Riddell SR, Watanabe KS, Goodrich JM, Li CR, Agha ME, Greenberg PD. Restoration of viral immunity in immunodeficient humans by the adoptive transfer of T cell clones. Science. 1992;257:238–241. doi: 10.1126/science.1352912. [DOI] [PubMed] [Google Scholar]
- 7.Barron MA, Gao D, Springer KL, Patterson JA, Brunvand MW, McSweeney PA, Chang Z, Barón AE, Weinberg A. Relationship of reconstituted adaptive and innate cytomegalovirus (CMV)-specific immune responses with CMV viremia in hematopoietic stem cell transplant recipients. Clin. Infect. Dis. 2009;49:1777–1783. doi: 10.1086/648423. [DOI] [PubMed] [Google Scholar]
- 8.Feuchtinger T, Opherk K, Bethge WA, Topp MS, Schuster FR, Weissinger EM, Mohty M, Or R, Maschan M, Schumm M, Hamprecht K, Handgretinger R, Lang P, Einsele H. Adoptive transfer of pp65-specific T cells for the treatment of chemorefractory cytomegalovirus disease or reactivation after haploidentical and matched unrelated stem cell transplantation. Blood. 2010;116:4360–4367. doi: 10.1182/blood-2010-01-262089. [DOI] [PubMed] [Google Scholar]
- 9.Reusser P, Riddell SR, Meyers JD, Greenberg PD. Cytotoxic T-lymphocyte response to cytomegalovirus after human allogeneic bone marrow transplantation: pattern of recovery and correlation with cytomegalovirus infection and disease. Blood. 1991;78:1373–1380. [PubMed] [Google Scholar]
- 10.Li CR, Greenberg PD, Gilbert MJ, Goodrich JM, Riddell SR. Recovery of HLA-restricted cytomegalovirus (CMV)-specific T-cell responses after allogeneic bone marrow transplant: correlation with CMV disease and effect of ganciclovir prophylaxis. Blood. 1994;83:1971–1979. [PubMed] [Google Scholar]
- 11.Gratama JW, Boeckh M, Nakamura R, Cornelissen JJ, Brooimans RA, Zaia JA, Forman SJ, Gaal K, Bray KR, Gasior GH, Boyce CS, Sullivan LA, Southwick PC. Immune monitoring with iTAg MHC Tetramers for prediction of recurrent or persistent cytomegalovirus infection or disease in allogeneic hematopoietic stem cell transplant recipients: a prospective multicenter study. Blood. 2010;116:1655–1662. doi: 10.1182/blood-2010-03-273508. [DOI] [PubMed] [Google Scholar]
- 12.Boeckh M, Leisenring W, Riddell SR, Bowden RA, Huang ML, Myerson D, Stevens-Ayers T, Flowers ME, Cunningham T, Corey L. Late cytomegalovirus disease and mortality in recipients of allogeneic hematopoietic stem cell transplants: importance of viral load and T-cell immunity. Blood. 2003;101:407–414. doi: 10.1182/blood-2002-03-0993. [DOI] [PubMed] [Google Scholar]
- 13.Peggs KS, Verfuerth S, Pizzey A, Chow SL, Thomson K, Mackinnon S. Cytomegalovirus-specific T cell immunotherapy promotes restoration of durable functional antiviral immunity following allogeneic stem cell transplantation. Clin. Infect. Dis. 2009;49:1851–1860. doi: 10.1086/648422. [DOI] [PubMed] [Google Scholar]
- 14.Peggs KS, Thomson K, Samuel E, Dyer G, Armoogum J, Chakraverty R, Pang K, Mackinnon S, Lowdell MW. Directly selected cytomegalovirus-reactive donor T cells confer rapid and safe systemic reconstitution of virus-specific immunity following stem cell transplantation. Clin. Infect. Dis. 2011;52:49–57. doi: 10.1093/cid/ciq042. [DOI] [PubMed] [Google Scholar]
- 15.Schmitt A, Tonn T, Busch DH, Grigoleit GU, Einsele H, Odendahl M, Germeroth L, Ringhoffer M, Ringhoffer S, Wiesneth M, Greiner J, Michel D, Mertens T, Rojewski M, Marx M, von Harsdorf S, Dohner H, Seifried E, Bunjes D, Schmitt M. Adoptive transfer and selective reconstitution of streptamer-selected cytomegalovirus-specific CD8+ T cells leads to virus clearance in patients after allogeneic peripheral blood stem cell transplantation. Transfusion. 2011;51:591–599. doi: 10.1111/j.1537-2995.2010.02940.x. [DOI] [PubMed] [Google Scholar]
- 16.Blyth E, Clancy L, Simms R, Ma CKK, Burgess J, Deo S, Byth K, Dubosq M-C, Shaw PJ, Micklethwaite KP. Donor-derived CMV-specific T cells reduce the requirement for CMV-directed pharmacotherapy after allogeneic stem cell transplantation. Blood. 2013;121:3745–3758. doi: 10.1182/blood-2012-08-448977. [DOI] [PubMed] [Google Scholar]
- 17.Cobbold M, Khan N, Pourgheysari B, Tauro S, McDonald D, Osman H, Assenmacher M, Billingham L, Steward C, Crawley C, Olavarria E, Goldman J, Chakraverty R, Mahendra P, Craddock C, Moss PA. Adoptive transfer of cytomegalovirus-specific CTL to stem cell transplant patients after selection by HLA-peptide tetramers. J. Exp. Med. 2005;202:379–386. doi: 10.1084/jem.20040613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papadopoulou A, Gerdemann U, Katari UL, Tzannou I, Liu H, Martinez C, Leung K, Carrum G, Gee AP, Vera JF, Krance RA, Brenner MK, Rooney CM, Heslop HE, Leen AM. Activity of broad-spectrum T cells as treatment for AdV, EBV, CMV, BKV, and HHV6 infections after HSCT. Sci. Transl. Med. 2014;6:242ra283. doi: 10.1126/scitranslmed.3008825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holtappels R, Pahl-Seibert MF, Thomas D, Reddehase MJ. Enrichment of immediate-early 1 (m123/pp89) peptide-specific CD8 T cells in a pulmonary CD62L(lo) memory-effector cell pool during latent murine cytomegalovirus infection of the lungs. J. Virol. 2000;74:11495–11503. doi: 10.1128/jvi.74.24.11495-11503.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holtappels R, Thomas D, Podlech J, Reddehase MJ. Two antigenic peptides from genes m123 and m164 of murine cytomegalovirus quantitatively dominate CD8 T-cell memory in the H-2d haplotype. J. Virol. 2002;76:151–164. doi: 10.1128/JVI.76.1.151-164.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karrer U, Sierro S, Wagner M, Oxenius A, Hengel H, Koszinowski UH, Phillips RE, Klenerman P. Memory inflation: continuous accumulation of antiviral CD8+ T cells over time. J. Immunol. 2003;170:2022–2029. doi: 10.4049/jimmunol.170.4.2022. [DOI] [PubMed] [Google Scholar]
- 22.Ouyang Q, Wagner WM, Voehringer D, Wikby A, Klatt T, Walter S, Muller CA, Pircher H, Pawelec G. Age-associated accumulation of CMV-specific CD8+ T cells expressing the inhibitory killer cell lectin-like receptor G1 (KLRG1). Exp. Gerontol. 2003;38:911–920. doi: 10.1016/s0531-5565(03)00134-7. [DOI] [PubMed] [Google Scholar]
- 23.Sierro S, Rothkopf R, Klenerman P. Evolution of diverse antiviral CD8+ T cell populations after murine cytomegalovirus infection. Eur. J. Immunol. 2005;35:1113–1123. doi: 10.1002/eji.200425534. [DOI] [PubMed] [Google Scholar]
- 24.Sylwester AW, Mitchell BL, Edgar JB, Taormina C, Pelte C, Ruchti F, Sleath PR, Grabstein KH, Hosken NA, Kern F, Nelson JA, Picker LJ. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J. Exp. Med. 2005;202:673–685. doi: 10.1084/jem.20050882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Munks MW, Cho KS, Pinto AK, Sierro S, Klenerman P, Hill AB. Four distinct patterns of memory CD8 T cell responses to chronic murine cytomegalovirus infection. J. Immunol. 2006;177:450–458. doi: 10.4049/jimmunol.177.1.450. [DOI] [PubMed] [Google Scholar]
- 26.Komatsu H, Inui A, Sogo T, Fujisawa T, Nagasaka H, Nonoyama S, Sierro S, Northfield J, Lucas M, Vargas A, Klenerman P. Large scale analysis of pediatric antiviral CD8+ T cell populations reveals sustained, functional and mature responses. Immun. Ageing. 2006;3:11. doi: 10.1186/1742-4933-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snyder CM, Cho KS, Bonnett EL, van Dommelen S, Shellam GR, Hill AB. Memory inflation during chronic viral infection is maintained by continuous production of short-lived, functional T cells. Immunity. 2008;29:650–659. doi: 10.1016/j.immuni.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith CJ, Turula H, Snyder CM. Systemic Hematogenous Maintenance of Memory Inflation by MCMV Infection. PLoS Pathog. 2014;10:e1004233. doi: 10.1371/journal.ppat.1004233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Appay V, Dunbar PR, Callan M, Klenerman P, Gillespie GM, Papagno L, Ogg GS, King A, Lechner F, Spina CA, Little S, Havlir DV, Richman DD, Gruener N, Pape G, Waters A, Easterbrook P, Salio M, Cerundolo V, McMichael AJ, Rowland-Jones SL. Memory CD8+ T cells vary in differentiation phenotype in different persistent virus infections. Nat. Med. 2002;8:379–385. doi: 10.1038/nm0402-379. [DOI] [PubMed] [Google Scholar]
- 30.van Leeuwen EM, Gamadia LE, Baars PA, Remmerswaal EB, ten Berge IJ, van Lier RA. Proliferation requirements of cytomegalovirus-specific, effector-type human CD8+ T cells. J. Immunol. 2002;169:5838–5843. doi: 10.4049/jimmunol.169.10.5838. [DOI] [PubMed] [Google Scholar]
- 31.Gamadia LE, van Leeuwen EM, Remmerswaal EB, Yong SL, Surachno S, Wertheim-van Dillen PM, Ten Berge IJ, Van Lier RA. The size and phenotype of virus-specific T cell populations is determined by repetitive antigenic stimulation and environmental cytokines. J. Immunol. 2004;172:6107–6114. doi: 10.4049/jimmunol.172.10.6107. [DOI] [PubMed] [Google Scholar]
- 32.van Leeuwen EM, de Bree GJ, Remmerswaal EB, Yong SL, Tesselaar K, ten Berge IJ, van Lier RA. IL-7 receptor alpha chain expression distinguishes functional subsets of virus-specific human CD8+ T cells. Blood. 2005;106:2091–2098. doi: 10.1182/blood-2005-02-0449. [DOI] [PubMed] [Google Scholar]
- 33.Waller EC, McKinney N, Hicks R, Carmichael AJ, Sissons JG, Wills MR. Differential costimulation through CD137 (4-1BB) restores proliferation of human virus-specific “effector memory” (CD28 CD45RAHI) CD8+ T cells. Blood. 2007;110:4360–4366. doi: 10.1182/blood-2007-07-104604. [DOI] [PubMed] [Google Scholar]
- 34.Wallace DL, Masters JE, De Lara CM, Henson SM, Worth A, Zhang Y, Kumar SR, Beverley PC, Akbar AN, Macallan DC. Human cytomegalovirus- specific CD8+ T- cell expansions contain long- lived cells that retain functional capacity in both young and elderly subjects. Immunology. 2011;132:27–38. doi: 10.1111/j.1365-2567.2010.03334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rufer N, Zippelius A, Batard P, Pittet MJ, Kurth I, Corthesy P, Cerottini JC, Leyvraz S, Roosnek E, Nabholz M, Romero P. Ex vivo characterization of human CD8+ T subsets with distinct replicative history and partial effector functions. Blood. 2003;102:1779–1787. doi: 10.1182/blood-2003-02-0420. [DOI] [PubMed] [Google Scholar]
- 36.Price DA, Brenchley JM, Ruff LE, Betts MR, Hill BJ, Roederer M, Koup RA, Migueles SA, Gostick E, Wooldridge L. Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses. J. Exp. Med. 2005;202:1349–1361. doi: 10.1084/jem.20051357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iancu EM, Corthesy P, Baumgaertner P, Devevre E, Voelter V, Romero P, Speiser DE, Rufer N. Clonotype selection and composition of human CD8 T cells specific for persistent herpes viruses varies with differentiation but is stable over time. J. Immunol. 2009;183:319–331. doi: 10.4049/jimmunol.0803647. [DOI] [PubMed] [Google Scholar]
- 38.Gattinoni L, Lugli E, Ji Y, Pos Z, Paulos CM, Quigley MF, Almeida JR, Gostick E, Yu Z, Carpenito C. A human memory T cell subset with stem cell-like properties. Nat. Med. 2011;17:1290–1297. doi: 10.1038/nm.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Graef P, Buchholz VR, Stemberger C, Flossdorf M, Henkel L, Schiemann M, Drexler I, Höfer T, Riddell SR, Busch DH. Serial Transfer of Single-Cell-Derived Immunocompetence Reveals Stemness of CD8+ Central Memory T Cells. Immunity. 2014;41:116–126. doi: 10.1016/j.immuni.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 40.Berger C, Jensen MC, Lansdorp PM, Gough M, Elliott C, Riddell SR. Adoptive transfer of effector CD8+ T cells derived from central memory cells establishes persistent T cell memory in primates. J. Clin. Invest. 2008;118:294–305. doi: 10.1172/JCI32103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheinberg P, Melenhorst JJ, Brenchley JM, Hill BJ, Hensel NF, Chattopadhyay PK, Roederer M, Picker LJ, Price DA, Barrett AJ. The transfer of adaptive immunity to CMV during hematopoietic stem cell transplantation is dependent on the specificity and phenotype of CMV-specific T cells in the donor. Blood. 2009;114:5071–5080. doi: 10.1182/blood-2009-04-214684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wagner M, Jonjic S, Koszinowski UH, Messerle M. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J. Virol. 1999;73:7056–7060. doi: 10.1128/jvi.73.8.7056-7060.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Snyder CM, Cho KS, Bonnett EL, Allan JE, Hill AB. Sustained CD8+ T cell memory inflation after infection with a single-cycle cytomegalovirus. PLoS Pathog. 2011;7:e1002295. doi: 10.1371/journal.ppat.1002295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farrington LA, Smith TA, Grey F, Hill AB, Snyder CM. Competition for antigen at the level of the APC is a major determinant of immunodominance during memory inflation in murine cytomegalovirus infection. J. Immunol. 2013;190:3410–3416. doi: 10.4049/jimmunol.1203151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Turula H, Smith CJ, Grey F, Zurbach KA, Snyder CM. Competition between T cells maintains clonal dominance during memory inflation induced by MCMV. Eur. J. Immunol. 2013;43:1252–1263. doi: 10.1002/eji.201242940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Welten SPM, Redeker A, Franken KL, Benedict CA, Yagita H, Wensveen FM, Borst J, Melief CJM, van Lier RAW, van Gisbergen KPJM. CD27-CD70 Costimulation Controls T Cell Immunity during Acute and Persistent Cytomegalovirus Infection. J. Virol. 2013;87:6851–6865. doi: 10.1128/JVI.03305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J. Bioconductor: open software development for computational biology and bioinformatics. Genome biology. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26:2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Irizarry RA, Hobbs B, Collin F, Beazer- Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 50.MacDonald JW. mogene10sttranscriptcluster.db: Affymetrix mogene10 annotation data (chip mogene10sttranscriptcluster). R package version 8.1.0.
- 51.Smyth GK. Bioinformatics and computational biology solutions using R and Bioconductor. Springer; 2005. Limma: linear models for microarray data. pp. 397–420. [Google Scholar]
- 52.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Molec. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 53.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hertoghs KML, Moerland PD, van Stijn A, Remmerswaal EBM, Yong SL, van de Berg PJEJ, van Ham SM, Baas F, ten Berge IJM, van Lier RAW. Molecular profiling of cytomegalovirus-induced human CD8+ T cell differentiation. J. Clin. Invest. 2010;120:4077–4090. doi: 10.1172/JCI42758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thimme R, Appay V, Koschella M, Panther E, Roth E, Hislop AD, Rickinson AB, Rowland-Jones SL, Blum HE, Pircher H. Increased expression of the NK cell receptor KLRG1 by virus-specific CD8 T cells during persistent antigen stimulation. J. Virol. 2005;79:12112–12116. doi: 10.1128/JVI.79.18.12112-12116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hutchinson S, Sims S, O'Hara G, Silk J, Gileadi U, Cerundolo V, Klenerman P. A dominant role for the immunoproteasome in CD8 T cell responses to murine cytomegalovirus. PLoS One. 2011;6:e14646. doi: 10.1371/journal.pone.0014646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kern F, Khatamzas E, Surel I, Frommel C, Reinke P, Waldrop SL, Picker LJ, Volk HD. Distribution of human CMV-specific memory T cells among the CD8pos. subsets defined by CD57, CD27, and CD45 isoforms. Eur. J. Immunol. 1999;29:2908–2915. doi: 10.1002/(SICI)1521-4141(199909)29:09<2908::AID-IMMU2908>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 59.Tomiyama H, Takata H, Matsuda T, Takiguchi M. Phenotypic classification of human CD8+ T cells reflecting their function: inverse correlation between quantitative expression of CD27 and cytotoxic effector function. Eur. J. Immunol. 2004;34:999–1010. doi: 10.1002/eji.200324478. [DOI] [PubMed] [Google Scholar]
- 60.Jabbari A, Harty JT. Secondary memory CD8+ T cells are more protective but slower to acquire a central-memory phenotype. J. Exp. Med. 2006;203:919–932. doi: 10.1084/jem.20052237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Masopust D, Ha SJ, Vezys V, Ahmed R. Stimulation history dictates memory CD8 T cell phenotype: implications for prime-boost vaccination. J. Immunol. 2006;177:831–839. doi: 10.4049/jimmunol.177.2.831. [DOI] [PubMed] [Google Scholar]
- 62.Wirth TC, Xue HH, Rai D, Sabel JT, Bair T, Harty JT, Badovinac VP. Repetitive antigen stimulation induces stepwise transcriptome diversification but preserves a core signature of memory CD8(+) T cell differentiation. Immunity. 2010;33:128–140. doi: 10.1016/j.immuni.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fraser KA, Schenkel JM, Jameson SC, Vezys V, Masopust D. Preexisting High Frequencies of Memory CD8+ T Cells Favor Rapid Memory Differentiation and Preservation of Proliferative Potential upon Boosting. Immunity. 2013;39:171–183. doi: 10.1016/j.immuni.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kumar D, Chernenko S, Moussa G, Cobos I, Manuel O, Preiksaitis J, Venkataraman S, Humar A. Cell-mediated immunity to predict cytomegalovirus disease in high-risk solid organ transplant recipients. Am. J. Transplant. 2009;9:1214–1222. doi: 10.1111/j.1600-6143.2009.02618.x. [DOI] [PubMed] [Google Scholar]
- 65.Harvala H, Stewart C, Muller K, Burns S, Marson L, MacGilchrist A, Johannessen I. High risk of cytomegalovirus infection following solid organ transplantation despite prophylactic therapy. J. Med. Virol. 2013;85:893–898. doi: 10.1002/jmv.23539. [DOI] [PubMed] [Google Scholar]
- 66.Manuel O, Husain S, Kumar D, Zayas C, Mawhorter S, Levi ME, Kalpoe J, Lisboa L, Ely L, Kaul DR. Assessment of cytomegalovirus-specific cell-mediated immunity for the prediction of cytomegalovirus disease in high-risk solid-organ transplant recipients: a multicenter cohort study. Clin. Infect. Dis. 2013;56:817–824. doi: 10.1093/cid/cis993. [DOI] [PubMed] [Google Scholar]
- 67.Ljungman P, Hakki M, Boeckh M. Cytomegalovirus in hematopoietic stem cell transplant recipients. Hematol. Oncol. Clin. North. Am. 2011;25:151–169. doi: 10.1016/j.hoc.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jonjic S, Pavic I, Polic B, Crnkovic I, Lucin P, Koszinowski UH. Antibodies are not essential for the resolution of primary cytomegalovirus infection but limit dissemination of recurrent virus. J. Exp. Med. 1994;179:1713–1717. doi: 10.1084/jem.179.5.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cush SS, Flaño E. KLRG1+ NKG2A+ CD8 T cells mediate protection and participate in memory responses during γ-herpesvirus infection. J. Immunol. 2011;186:4051–4058. doi: 10.4049/jimmunol.1003122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Appay V, Van Lier RAW, Sallusto F, Roederer M. Phenotype and function of human T lymphocyte subsets: consensus and issues. Cytometry Part A. 2008;73:975–983. doi: 10.1002/cyto.a.20643. [DOI] [PubMed] [Google Scholar]
- 71.Ciocca ML, Barnett BE, Burkhardt JK, Chang JT, Reiner SL. Cutting edge: asymmetric memory T cell division in response to rechallenge. J. Immunol. 2012;188:4145–4148. doi: 10.4049/jimmunol.1200176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kurz SK, Rapp M, Steffens HP, Grzimek NK, Schmalz S, Reddehase MJ. Focal transcriptional activity of murine cytomegalovirus during latency in the lungs. J. Virol. 1999;73:482–494. doi: 10.1128/jvi.73.1.482-494.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kurz SK, Reddehase MJ. Patchwork pattern of transcriptional reactivation in the lungs indicates sequential checkpoints in the transition from murine cytomegalovirus latency to recurrence. J. Virol. 1999;73:8612–8622. doi: 10.1128/jvi.73.10.8612-8622.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Snyder CM. Buffered memory: a hypothesis for the maintenance of functional, virus-specific CD8(+) T cells during cytomegalovirus infection. Immunol. Res. 2011;51:195–204. doi: 10.1007/s12026-011-8251-9. [DOI] [PubMed] [Google Scholar]
- 75.Araki K, Turner AP, Shaffer VO, Gangappa S, Keller SA, Bachmann MF, Larsen CP, Ahmed R. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460:108–112. doi: 10.1038/nature08155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.