Abstract
The flight muscles (DLM1) of the Hawkmoth, Manduca sexta are synchronous, requiring a neural spike for each contraction. Stress/strain curves of skinned DLM1 showed hysteresis indicating the presence of titin-like elastic proteins. Projectin and kettin are titin-like proteins previously identified in Lethocerus and Drosophila flight muscles. Analysis of Manduca muscles with 1% SDS-agarose gels and western blots showed two bands near 1 MDa that cross-reacted with antibodies to Drosophila projectin. Antibodies to Drosophila kettin cross-reacted to bands at ~500 and ~700 kDa, but also to bands at ~1.6 and ~2.1 MDa, that had not been previously observed in insect flight muscles. Mass spectrometry identified the 2.1 MDa protein as a product of the Sallimus (sls) gene. Analysis of the gene sequence showed that all 4 putative Sallimus and kettin isoforms could be explained as products of alternative splicing of the single sls gene. Both projectin and Sallimus isoforms were expressed to higher levels in ventrally located DLM1 subunits, primarily responsible for active work production, as compared to dorsally located subunits, which may act as damped springs. The different expression levels of the 2 projectin isoforms and 4 Sallimus/kettin isoforms may be adaptations to the specific requirements of individual muscle subunits.
Keywords: Insect Flight Muscle, passive tension, elastic proteins
Introduction
The flight muscles in many flying insect species are regulated by delayed stretch activation[1] so that contraction is asynchronous with neural stimulation. Unlike these muscles, and like vertebrate striated muscles, contraction in the dorso-longitudinal flight muscles (DLM1) of the Hawk moth, Manduca sexta, as in other Lepidoptera, is synchronous with the neural input pulses [2]. The shape of the M. sexta DLM1 length tension curves is similar to mammalian cardiac muscle in that the muscles operate in the ascending part of length-tension curves in vivo [3].
Insect flight muscles need to bear rapid oscillatory contractions; therefore, the stiffness of the muscle is an important physiological adaptation that enables the storage and release of elastic strain energy [4]. While overall muscle stiffness incorporates a number of factors, with contributions from both passive and active components including actin-myosin crossbridges [5, 6], much of the passive tension and elastic force is based on elastic proteins that act in concert with the thick and thin filaments with which they interact. Such large extensible proteins, including titin (in vertebrate striated muscle, ~3 MDa), projectin (in insects, ~900 kDa, often called mini-titin), and Sallimus (Sls, also in insects, ~700kDa to 2 MDa), constitute elastic myofilaments that help maintain the structural stability of the sarcomere by providing an elastic restoring force to keep the A-bands centered in the sarcomeres and to prevent overstretching [7]. Passive tension generated by elastic proteins also appears to be an important component of delayed stretch activation in asynchronous insect flight muscles [8]. In vertebrate muscle, titin is anchored at both the Z-band and the M-line spanning half a sarcomere. The extensible PEVK and tandem Ig domains in the I-band region of the titin filaments can straighten out sequentially in response to stress [9, 10]. Through the expression of different titin isoforms, myofibril stiffness —shorter isoforms being generally stiffer than longer isoforms— and compliance can be tuned to the needs of the particular type of muscle [11]. In particular, variable lengths of the PEVK region found in different muscle types are associated with significant differences in the passive tension that a muscle can develop [11–13]. For example, cardiac muscles, that undergo repetitive stretch-activated contractions, are stiffer than skeletal muscle and, consequently have shorter, cardiac specific isoforms of titin [13, 14]. In asynchronous flight muscles (IFMs), such as those of Lethocerus indicus or Drosophila melanogaster, projectin and Sallimus proteins have been proposed to play similar roles to vertebrate titin [15, 16]. In IFMs, Sls and projectin are found within the so called elastic connecting C-filaments, which extend from the Z-line across the I-band and anchor on the A-band region but do not reach the M line [17, 18]. These proteins are much smaller, and hence the filament is shorter, than vertebrate skeletal or even cardiac muscle titin providing the high stiffness needed for the high frequency of oscillatory contractions associated with asynchronous mode and stretch activation [17]. mRNA transcripts for both proteins are highly differentially spliced generating multiple isoforms, which are often muscle type-specific. In D. melanogaster, the sallimus (sls) gene has the potential to encode many different polypeptides with the longer isoforms being collectively known as Sallimus (abbreviated to Sls). Zormin and kettin represent shorter polypeptides from the sls gene, containing only sequences from the NH2-terminus. The sequences of kettin and zormin can also be included in the longer Sallimus isoforms [19]. The asynchronous IFMs of L. indicus and D. melanogaster contain predominantly kettin, zormin, as well as short isoforms of projectin and Sallimus which represent a truncation or even a total loss of the elastic PEVK regions whereas body and leg synchronous muscles in these same insects additionally contain the longer isoforms from both genes [15, 19–21].
There are a number of known physiological differences between the synchronous M. sexta DLM1 and the prototype asynchronous flight muscles found in L. indicus and D. melanogaster. These asynchronous flight muscles extend and relax by only a few percent, consistent with their short I band region [17], and possibly their short PEVK elastic region. In contrast, synchronous M. sexta DLM1 can extend 8–10% in vivo [3] and the projectin PEVK region expressed in the M. sexta flight muscle is larger than the one expressed in D. melanogaster IFMs [22]. Other studies have also recently reported that the DLM1 in M. sexta show a gradient in temperature of 6°C from the cooler dorsal part to the warmer ventral part [23] when stimulated at 25 Hz. When the muscles operate at their physiological temperatures, the warmer ventral part produces positive power output and the cooler dorsal part produces negative power output, indicating that different subunits of DLM1 have different roles in powering the down stroke of the wing [4]. A large fraction of the differences in the apparent muscle elasticity between dorsal and ventral muscle during contraction, i.e. in active muscle, has been attributed to the different behavior of myosin crossbridges in the two regions [5, 23], but the passive properties of these muscles have not been investigated. An open question is whether the dorsal and ventral subunits are structurally and biochemically the same and the functional effects are due only to temperature, or, alternatively, whether the dorsal and ventral muscles have different structural and biochemical adaptations to their variable operating temperatures.
This study aimed to identify and perform preliminary characterization of proteins that might be responsible for the unusual mechanical properties of M. sexta flight muscle, the extent to which they differ in dorsal and ventral muscles, as well as compare and contrast their properties to those of other insect flight muscles. Here we show that M. sexta muscles not only contains projectin and kettin, similar in size to those of D. melanogaster and L. indicus IFM, but also two larger Sallimus (Sls) isoforms, not previously observed in insect flight muscle. Both projectin and the large Sls isoforms contain extensive PEVK elastic domains consistent with the ability of Manduca flight muscle to be stretched in vivo up to ~9% of their length [3].
Methods
Solutions
Relaxing solution contained: 20mM MOPS, 5mM NaN3, 5mM MgAc2.4H2O, 5mM Na2ATP, 5mM EGTA and 1X protease inhibitor (Roche). The muscle skinning wash solution contained: 100mM KCL, 10mM MOPS, 5mM EGTA, 20mM BDM (2,3-butanedione monoxime),9mM MgCl2, and 4mM Na2ATP and 1% TritonX-100. The above solutions were adjusted to pH 6.8 at 22°C by adding KOH. Urea protein sample buffer contained: 8 M urea, 2M thiourea, 3%SDS, 0.03% bromophenol blue and 0.05M Tris, pH 6.8. 75mM DTT was added freshly before extraction of the protein from sample. Glycerol buffer contained: 50% glycerol, 1mM PMSF, 0.02mM E64, and 0.08mM leupeptin. Tris extraction buffer contained: 50mM Tris pH 6.8, 2.5%SDS, 1.0mM DTT, 10% glycerol, 1μg/ml leupeptin, 1μg/ml pepstatin A, 1mM PMSF, and 5μg/ml aprotinin.
Skinned Muscle Preparations
Flight muscles (DLM1) were dissected from the mesothorax of M. sexta provided by Prof. T. Daniel from a colony maintained at the University of Washington. The dorso longitudinal muscles (DLM1) are divided into five subunits. The dorsal samples were taken from the top two subunits and ventral samples were taken from the bottom two subunits. The middle subunit was discarded. IFM fibers from wild type D. melanogaster thoraces were dissected in cold 1X relaxing solution with protease inhibitors. Muscle fibers were transferred into skinning solution containing 1% Triton X-100, and left at 4°C for 8–12 hours. After skinning, the muscle fibers were washed with 1X skinning wash solution with protease inhibitors three times, 5–10 minutes each, and were then used for mechanical experiments or for biochemical analysis.
Electrophoresis and Staining
For electrophoresis, M. sexta DLM1 and D. melanogaster flight muscle were used directly after dissection without skinning and the proteins extracted in urea buffer as described above. The L. indicus indirect flight muscle samples (a gift of M.K. Reedy, Duke University) were previously skinned and preserved in 75% glycerol solution. The glycerol was washed out and the muscles otherwise treated the same as the other insect muscle samples.
For detection of high molecular weight proteins, muscles were solubilized and electrophoresed using 1% agarose gels using as described [24] with the modification that the buffer to tissue ratio in the solubilization step was 15:1(v/w) instead of 40:1. Myofibrillar proteins extracted from bovine left ventricular heart (gift from H. Granzier, University of Arizona) along with Invitrogen HiMark™ high molecular gel markers were used as the molecular size references. The logarithm of the molecular weights of titin N2BA, N2B, myosin heavy chain (MHC) and high molecular weight gel markers were plotted vs. the relative mobility to calibrate molecular weight. The prominent bands from bovine heart sample are from N2BA titin isoform at 3.35MDa, the N2B titin isoform at 2.97 MDa and MHC at 0.205MDa [11]. The Invitrogen markers provided bands ranging from 31–460 kDa. Band intensities were analyzed using Carestream molecular imaging software (version 5.0) and normalized to that of MHC. Gel loading was optimized for visualizing the high molecular weight proteins in the 1% agarose gels. Under these conditions, the myosin heavy chain bands appear to be overloaded and are irregularly shaped. Nonetheless, when the background is subtracted and the integrated intensity/per unit area is plotted against loading, the relationship was linear up to 100 μg of total protein loading, which was not exceeded in our studies. The resolving agarose gel was 13 cm high and was typically run for 3.5 hours followed by Coomassie blue staining allowing resolution of the 460 kDa and 268 kDa marker proteins.
For separation and detection of the majority of other myofibrillar proteins, 4–15% Tris-glycine polyacrylamide gels were used as described by the manufacturer (Invitrogen). Gels were stained with Coomassie brilliant blue or with Pro-Q Diamond phosphoprotein gel stain and SYPRO ruby protein gel stain. The images were captured by a Carestream Gel Logic 2200 PRO imaging system.
Western blots
Immediately after electrophoresis, the agarose gels were placed in transfer buffer (190mM glycine, 24.76mM Tris-base and 20% methanol) for 5 min equilibration. PVDF (0.2μm thick) membranes were used for all transfers with a Bio Rad Mini Trans-Blot apparatus. Transfers were done at 4°C at 200 mA for 2 hours. Post-transfer, agarose gels were stained with Coomassie blue to confirm that most of the high molecular bands were transferred to the membrane. Membranes were stored in TBST (50mM Tris-HCL (pH 7.5), 200mM NaCl, 0.05% Tween 20) or air-dried. The membranes were blocked with 5% w/v non-fat milk in TBST at room temperature for one hour or at 4°C with shaking overnight. The blots were then rinsed with TBST 3 times for 10 min each. The primary antibodies (Table 1) and secondary antibodies (Table 2) were diluted in TBST. Primary antibodies were incubated overnight at 4°C or one hour at room temperature with shaking. Blots were washed in TBST3 times for 10 minutes each, between the first and secondary antibody treatment. All the secondary antibodies were incubated 1 to 2 hours then washed with TBST for 10 min, repeated 3 times, followed by development using the ECL plus kit (GE Healthcare).
Table 1.
Primary antibodies.
| Clone | Protein | Host | Dilution | Source | Source |
|---|---|---|---|---|---|
| MAC147 | Myosin | Rat | 1:5000 | Monoclonal IgG | B |
| 9D10 | Titin | Mouse | 1:300 | Hybridoma bank IgM | I |
| MAC155 | Kettin | Rat | 1:50 | Hybridoma bank supernatant | B |
| MAC 150 | Projectin | Rat | 1:3000 | IgG 7.9mg/ml | B |
Sources: B: Belinda Bullard, York University. I: Development al Studies Hybridoma Bank, Department of Biological Sciences, University of Iowa, Iowa City
Table 2.
Secondary antibodies
| Name | Host | Dilution | Conjugation | Company | |
|---|---|---|---|---|---|
| Anti-Rat | Goat | IgG | 1:5000 | HRP | GE |
| Anti-Mouse | Goat | IgM | 1:3000 | HRP | Invitrogen |
| Anti-Mouse | Goat | IgG | 1:5000 | HRP | Millipore |
| Anti-Goat | Rabbit | IgG | 1:5000 | HRP | Millipore |
Mechanical Experiments
Muscle fibers were attached to hooks with glue made from cellulose nitrate dissolved in acetone. One hook was attached to a dual-mode force/length servo-motor (Aurora Scientific Inc., model 300C) mounted on an X-Y-Z positioner. The other hook was attached to another X-Y-Z positioner that is adjusted to take up the slack in the fiber and then remains fixed during force measurements. Length was controlled and force measured using a National Instruments 16 bit digital/analog controller system controlled by custom software written in Labview (National Instruments Inc.). Sarcomere lengths were measured using laser diffraction with a helium-neon laser with a wavelength of 663nm.
Quantitative Analysis of Expression Levels for Elastic Proteins
M. sexta dorsal and ventral muscles were treated with skinning buffer containing 1% Triton X-100 for 8–12 hours. Samples were then washed with skinning buffer without Triton X-100 three times, 5–10 min each. Samples were weighed and 8M urea buffer was added (sample: buffer ratio 1:15 w/v) to extract proteins. The protein concentrations were determined using the 2-D quant kit (GE Healthcare). The concentration of both ventral and dorsal samples were adjusted by dilution to 3 mg/ml. Equal amounts of dorsal and ventral sample were mixed to be used as a reference standard to identify the range of loadings where OD of the gel bands is a linear function of concentration. For analysis of the 1.6 MDa protein, 2μL, 4μL 6μL 8μl and 10μL of the mixture was loaded as standards, and 5μl and 10μL of both ventral and dorsal samples were loaded on the same gel. For analysis of the 2.1 MDa protein, 5μL, 10μL, 15μL, 20μL and 25μl of the mixture were loaded as a reference standard, and 10μl and 20μL of each of ventral and dorsal samples was loaded on the same gel along with bovine left ventricle proteins as molecular size marker [11]. Each gel was run at 400V, 15mA and 20W for 3.5 hours. The gels were then fixed with 20% methanol and 10% acetic acid for 30 minutes, stained with Coomassie blue overnight and de-stained with 25% methanol. Images of the gels were recorded using a Carestream Gel Logic 2200 PRO imaging system and band intensities analyzed using Carestream Molecular Imaging Software v. 5.0.
Mass Spectrometry
Tandem mass spectrometry coupled to liquid chromatography (LC-MS/MS) analysis of trypsin-digested protein gel bands following 1D SDS-PAGE [25] was carried out using a LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific, San Jose, CA) equipped with an Advion nanomate ESI source (Advion, Ithaca, NY), following ZipTip (Millipore, Billerica, MA) C18 sample clean-up according to the manufacturer’s instructions. Peptides were eluted from a C18 precolumn (100-μm id × 2 cm, Thermo Fisher Scientific) onto an analytical column (75-μm ID × 10 cm, C18, Thermo Fisher Scientific) using a 5–10% gradient of solvent B (acetonitrile, 0.1% formic acid) over 5 minutes, followed by a 10–35% gradient of solvent B over 35 minutes, 35–50% gradient of solvent B over 20 minutes, 50–95% gradient of solvent B over 5minutes, and finally by a 95% solvent B held for another 5 minutes. All flow rates were at 400 nl/min. Solvent A consisted of water and 0.1% formic acid. Data dependent scanning was performed by the Xcalibur v 2.1.0 software[26] using a survey mass scan at 60,000 resolution in the Orbitrap analyzer scanning m/z 400–1600, followed by collision-induced dissociation (CID) tandem mass spectrometry (MS/MS) of the fourteen most intense ions in the linear ion trap analyzer. Precursor ions were selected by the monoisotopic precursor selection (MIPS) setting with selection or rejection of ions held to a +/− 10 ppm window. Dynamic exclusion was set to place any selected m/z on an exclusion list for 45 seconds after a single MS/MS. All MS/MS spectra were searched against a variety of protein databases using Thermo Proteome Discoverer 1.3 (Thermo Fisher Scientific, San Jose, CA). All MS/MS spectra were searched against the D. melanogaster protein database downloaded July 30, 2012 from Uniprot (http://www.uniprot.org/uniprot/?query=organism:7227+keyword:1185) using Thermo Proteome Discoverer 1.3 (Thermo Fisher Scientific). Proteins were identified at 99% confidence with XCorr score cut-offs [27] as determined by a reversed database search. The results were also validated using X!Tandem, another search engine[28] and displayed with Scaffold v 3.6.1 (Proteome Software Inc., Portland OR), a program that relies on various search engine results (i.e.: Sequest, X!Tandem, MASCOT) and uses Bayesian statistics to reliably identify more spectra [29, 30,31]. Matching to the M. sexta sequence was done using BLAST2p.
Genomic analysis
M. sexta genomic contigs containing the Sallimus (sls) or projectin genes were isolated from the M. sexta Whole Genome Sequence (WGS) database available on GenBank following tblastn searches using a series of fragments from D. melanogaster sls or projectin genes respectively. The tblastn algorithm compares a query protein sequence to the six frame-translation of a DNA sequence [32]. The exon-intron pattern was initially predicted over most of the two genes by performing translation in all three frames and aligning on the D. melanogaster amino acid sequences using LaLign [33]. In Drosophila, it is conventional to use italics to refer to genes whose names are derived from mutant phenotypes and non-italic uppercase for the corresponding proteins (i.e sls gene and Sls protein). We will follow this convention in the remainder of this report (http://flybase.org/static_pages/docs/nomenclature/nomenclature3.html). Several RNA-seq databases available from various stages or tissues of M. sexta were also searched and resulting cDNA sequences were aligned on the genomic sequences to predict the initial exon-intron gene structure.
cDNA analysis
M. sexta were purchased as larvae and/or pupae from Educational Science (TX) and reared up to emergence of the imago at which point they were sacrificed. Total RNA was purified from whole animals, isolated body parts (legs, heads, thoraces), and from dissected flight muscles using Bio-Rad Aurum total RNA kit. Further splice analysis was conducted by performing RT-PCR reactions with different RNA preparations and primer sets spanning the entire genes as described before [34]. DNA fragments were isolated after agarose gel electrophoresis and sequenced (Genewiz Inc.) either directly or after subcloning into the pGEM-T easy shuttle vector (Promega, Inc.). cDNA sequences were manually aligned with the genomic sequence to confirm splice sites and identify alternative splicing pathways. Protein sequences and molecular weights were predicted using translation and protein analysis software on the ExPasy website (http://www.expasy.org/). Repeat analyses were performed using Dotlet and CLUSTAL-W algorithm [35, 36].
Results
Passive Force/Extension curves from Manduca flight muscle
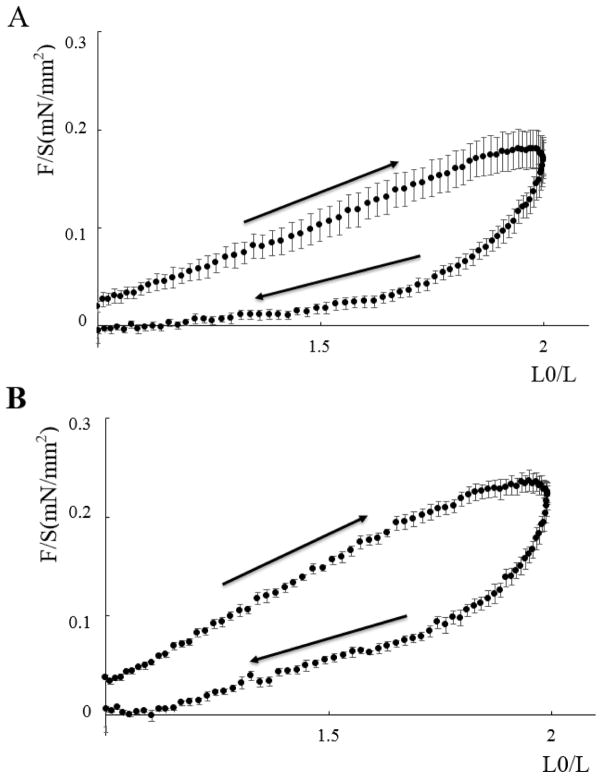
Figure 1 shows total passive force (normalized to cross sectional area) as a function of muscle length (as a fraction of slack length: L0) for Triton-skinned M. sexta DLM1. Remarkably, the muscles can be extended and released reproducibly up to 200% of slack length in repeated lengthening/release cycles (at least 4) that follow directly after each other. The shapes of the curves appear identical between dorsal and ventral muscle and show substantial hysteresis, resembling the behavior of vertebrate striated muscle [37]. This pattern indicates that the proteins responsible for passive tension quickly recover after release from the structural changes happening during stretch, and the recovery process is responsible for the hysteresis seen on release prior to restretch. Sarcomere length at slack length in both dorsal and ventral skinned muscle were not significantly different being 3.67±0.02μm (n=12) and 3.67±0.02μm (n=14) respectively.
Figure 1. Stress-strain curves for skinned Manduca sexta flight muscles.
Passive force as a function of muscle length from skinned ventral and dorsal M. sexta DLM1. A Dorsal flight muscle. B Ventral flight muscle. Length was normalized to that at slack length. (L0). Arrows indicate the stretch and release portions of the cycle. The small force offset at the beginning of the stretch cycle was due to manually taking up the slack in the muscle before starting the protocol. Error bars are the standard errors of the mean.
High molecular weight proteins in insect flight muscles
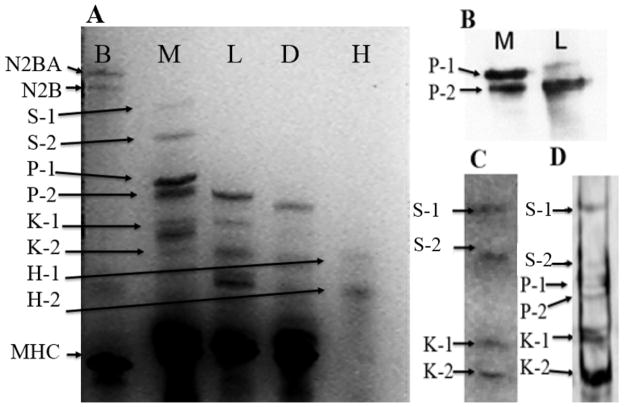
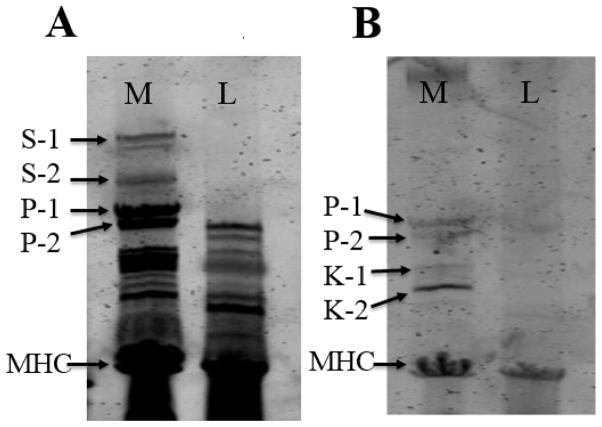
In order to separate proteins larger than 250 kDa, 1% agarose vertical gels were used with muscles from different insect species in order to compare their high molecular weight protein components (Figure 2). The relation between relative mobility and the logarithm of the molecular weight in these 1% agarose gels was linear (R2 =0.99) between molecular weights of ~ 205 kDa to 3350 kDa. This relationship was used to calculate the sizes of the large proteins. On this scale, the four largest proteins in the M. sexta sample were 2.1 MDa, 1.6 MDa, 1.05 MDa and 960 kDa while the largest protein detected in L. indicus was 912 kDa and in D. melanogaster, 864 kDa (Figure 2A, Table 3). The 1.05 MDa and 960 kDa proteins were tentatively assigned as isoforms of projectin (P1 and P2) based on the size similarity with that protein in L. indicus muscles while the 2.1 MDa and 1.6 MDa proteins (S1 and S2) were initially unidentified. Protein extracts from flight muscles of the cricket, cockroach, two species of butterflies and four other moth species were also analyzed for large proteins. Results are summarized in Table 3. The high molecular weight proteins S1 and S2 were only found in M. sexta and Bombyx mori flight muscles.
Figure 2. High Molecular Weight Proteins in Manduca sexta flight muscle.
Panel A) 1 % Vertical agarose gel electrophoresis (VAGE) gel. Lane B is from bovine left ventricular myocardium, lane M is from Manduca flight muscle, lane L is from Lethocerus flight muscle, lane D is from Drosophila flight muscle, and lane H is the protein size markers (see methods). S-1 is the larger isoform of sallimus protein, S-2 is the smaller isoform of sallimus protein. P-1 is the larger projectin isoform, P-2 is the smaller projectin isoform, K-1 is the larger kettin isoform and K-2 the smaller kettin isoform, 2. Panel B) Western blots against the projectin antibody. Panel C) Western blots against the kettin antibody. Panel D) Western blots against the 9D10 titin antibody.
Table 3.
Calculated sizes of high molecular weight proteins in various insect flight muscles
| M. sexta (Hawkmoth) | L. indicus (waterbug) | D. melana (fruit fly) | B. mori (Silk moth) | S. cynthia (Cynthia moth) | C. promethia (Promethea silkmoth) | A. luna (Luna moth) | V. virginiensis (Painted Lady Butterfly) | D. plexippus (Monarch Butterfly) | G. campestris (Cricket) | P. americana (cockroach) |
|---|---|---|---|---|---|---|---|---|---|---|
| (S-1) 2100 | 2100 | |||||||||
| (S-2) 1600 | 1600 | |||||||||
| (P-1) 1050 | 1090 | 1060 | 1040 | 1050 | 990 | 1040 | ||||
| (P-2) 960 | 910 | 860 | 930 | 950 | 930 | 890 | 940 | 890 | 920 | |
| (K-1) 710 | 700 | *700 | 730 | 800 | 720 | 750 | 740 | 780 | 810 | |
| (O) 610 | 670 | 600 | 670 | 640 | 660 | |||||
| (K-2) 490 | 500 | *540 | 610 | 570 | 520 | 540 | 520 | 500 |
From Kulke et al. [21]
Antibodies directed against projectin (Table 1) cross-react with M. sexta proteins showing two strong bands with molecular weights around 960 kDa and 1.05 MDa respectively (Figure 2B). We excluded the possibility that the lower molecular weight band was from a proteolytic product of the higher molecular weight protein by “aging” the muscle at room temperature for 1–2 days to allow endogenous proteases to degrade the proteins. Both putative projectin bands decreased at the same rate indicating that one is not a proteolytic product of the other. Manduca flight muscle therefore appears to have two proteins that cross-react with Lethocerus projectin antibody. The Sls antibody directed against the kettin isoform (Table 1) showed two bands in the Lethocerus flight muscle sample but four bands in M. sexta flight muscle (S1, S2, K1, K2 in Figure 2C). The K1 Sls isoform in M. sexta is similar in size to the largest of the Lethocerus Sls isoforms both at about 700 kDa whereas K2 is 500 kDa. Based on size and antibody reactivity, the 700 and 500 kDa Manduca proteins were identified as kettin isoforms of Sls (hence K1 and K2). Similarly, we also assigned the 2.1 (S1) MDa and 1.6 (S2) MDa proteins as two large isoforms of the Sls protein (hence S1 and S2). The 9D10 titin antibody [38] is immunospecific for mammalian cardiac muscle and targets the PEVK region of titin; however, our results showed that it cross-reacts with the putative M. sexta projectin bands and L. indicus projectin, which are known to contain PEVK regions ([15]; see below), as well as the 700 KDa and 500 kDa kettin isoforms in both insects (Figure 2D). The 9D10 antibody also cross-reacted with the 2.1 MDa (S1) and 1.6 MDa (S2) proteins, which also contain PEVK regions (see below).
Comparison of elastic protein expression levels in dorsal and ventral muscles
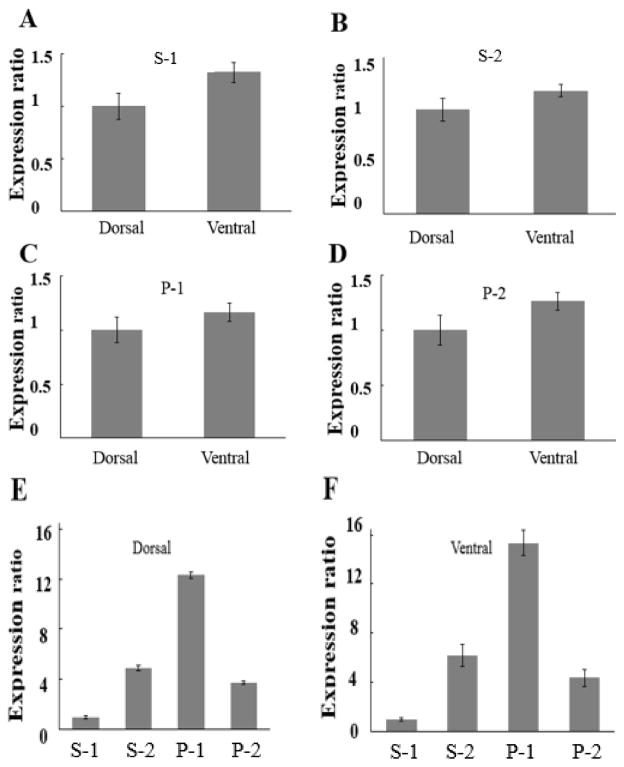
Because of the reported functional effects of the temperature gradient [4], one of the goals of this study was to see if we could detect differences between dorsal DLM1 and ventral DLM1 in terms of their elastic protein compositions. Figure 3 shows normalized expression ratios for the four largest previously unidentified proteins of M. sexta flight muscles, denoted S1, S2, P1, and P2. Ventral muscle has about 1.3 times more of the S1 protein than dorsal muscle (Figure 3A; p=0.0131 in paired t-tests). Similarly ventral muscle appeared to express about 1.2 times more of the S2 protein than dorsal muscle (Figure 3B but this was not significant). Figures 3C and 3D show normalized expression ratios for the P1 and P2 proteins with an increase of 1.2 (p=0.04, paired t-test) and 1.3 (p=0.02, paired t-test) respectively in ventral muscle compared to dorsal muscle. Normalization in panels A–D was achieved by dividing the measured banding intensities (in arbitrary units) by the loading volume and the means and standard errors of the mean calculated. For convenience of presentation, the larger mean number was divided by the smaller mean number so that the smaller mean number was then effectively normalized to 1. Figure 3 E and F show the relative amounts of proteins in dorsal and ventral muscle calculated using myosin heavy chain as a standard (see Materials and Methods). On this scale, myosin heavy chain would have a value of 80 ± 0.8. It can be seen that in both dorsal and ventral muscle, the larger isoform of Projectin (P1: 1050 kDa) is the most abundant of the elastic proteins.
Figure 3. Expression levels of elastic proteins comparing the dorsal and ventral regions of the DLM1 of Manduca sexta.
A expression of the larger Sallimus (Sls) isoform S-1 in dorsal and ventral DLM1. B expression of the smaller Sallimus (Sls) isoform protein S-2 in dorsal and ventral DLM1. C expression of projectin isoform P-1 in dorsal and ventral DLM1. D expression of projectin isoform P-2 in dorsal and ventral DLM1. Expression ratios were calculated by dividing the larger amount by the smaller amount, normalizing the smaller amount to 1. E & F relative expression of elastic proteins in dorsal (E) and ventral DLM1 (F). Expression is normalized to that of myosin heavy chain (see methods). Error bars represent standard errors of the mean.
Phosphorylation of myofilament proteins in insect flight muscles
Figure 4 shows the relative abundance of cytoskeletal proteins in 1% agarose gels stained with SYPRO-Ruby (Figure 4 A) and stained with Pro-Q Diamond (Figure 4 B) to show which proteins are phosphorylated. The Pro-Q image showed five phosphorylation band signals in the M. sexta sample located at about 1050, 960, 710, 490 and 230 kDa. The 1050 and 960 kD bands were located at the projectin isoform signal positions. The 710 and 490 kDa bands were located at the kettin isoform signal positions and the 230 kDa band is from myosin heavy chain.
Figure 4.
Phosphorylation assay for giant proteins from M. sexta. SYPRO Ruby-(SR) and Pro-Q Diamond-(PQ) stained 1 % agarose gels were used to determine protein phosphorylation states. SR, stained gel for relative abundance of protein; PQ, stained gel specific for relative abundance of phosphorylated proteins.
Projectin gene and protein structure in Manduca sexta
For the projectin annotation, the entire gene sequence was retrieved from the M. sexta WGS database as part of one contig (scaffold # #00026) and manually annotated as described in Materials and Methods. All splice sites were verified by either RNA-sequencing data or RT-PCR sequences. The analyses of the NH2-terminus and PEVK sequences of M. sexta projectin have been described elsewhere together with the complete gene structure for the Monarch butterfly (Danaus plexippus [22, 40]). The overall domain organization of projectin is identical to all the projectin proteins characterized so far [15, 20, 22]. The gene characteristics for M. sexta projectin are summarized in Table 4A and compared with two other Lepidoptera (D. plexippus and B. mori), as well as D. melanogaster [15, 22]. The three Lepidoptera genes are similar in overall gene size and number of exons representing an increase in exon number and average intron length as compared to the D. melanogaster gene (Table 4A). The protein size difference between the largest D. melanogaster and M. sexta isoforms is mostly due to the length of the full PEVK segment, which is longer in D. melanogaster (Table 4A). Known alternative splice variants within the PEVK and NH2-terminal regions would generate several isoforms in M. sexta with molecular weights ranging from 931 kDa to 974 kDa [22] (see Table 4A). These sizes are consistent with the two large proteins identified by projectin antibody staining as P1 and P2 (Figure 2B). All would contain a PEVK region consistent with the 9D10 antibody data presented in Figure 2D.
Table 4.
Comparison of projectin genes (A) and protein isoforms (B). Genes from three Lepidoptera species- Bombyx mori (B. mori), Danaus plexippus (D. plexippus) and Manduca sexta (M. sexta) are compared to the homolog in D melanogaster (D.mel) and Apis mellifera (A. mel). CDS size represents the transcript from start to stop codons. Gene size also includes 5′ and 3′ UTR sequences when known. The different isoforms found in M. sexta and D. melanogaster are listed to predict expected protein sizes and have been described before (Ayme-Southgate et al, 2011, 2013a,2013b).
| A) | ||||
|---|---|---|---|---|
| D. mel | B. mori | D. ple | M. sexta | |
| gene size (bp) | 51,073 | >116,150 | >72,600 | 104,668 |
| CDS size (bp) | 27,154 | 26,211 | 26,199 | 26,714 |
| # exons | 46 | 124 | ~122 | 125 |
| largest exon (bp) | 9,462 | 4,989 | 5,015 | 4,989 |
| # amino acids | 8,936 | 8,737 | ~8, 732 | 8,736 |
| Largest isoform (kDa) | 1,000,122 | 973,530 | 973,000 | 974,000 |
| FM isoform (kDa) | 941,232 | NA | NA | 955,455 |
| B) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Manduca | Drosophila | ||||||||
| splicing | size (kDa) | PEVK size | muscle type | splicing | size (kDa) | PEVK size | muscle type | ||
| flight | leg | flight | leg | ||||||
| Full isoform | 974.00 | 377 aa | Y | Y | Full isoform | 1,000.12 | 530 aa | Y | Y |
| short PEVK | 955.46 | 205 aa | Y | Y | short PEVK | 941.23 | 75 aa | Y | Y |
| delta Ig 3–4 (Δ) | 949.80 | 377 aa | N | Y | |||||
| short PEVK + Δ | 931.26 | 205 aa | N | Y | |||||
| % short/long PEVK | 54.4 | % short/long PEVK | 14.2 | ||||||
Sallimus gene (sls) and protein structure in Manduca sexta
Five non-overlapping genomic contigs (accession # AIXA01012021, AIXA01012022, AIXA01012023, AIXA01009211, and AIXA01009212) were isolated by tblastn search of the M. sexta WGS database. Despite our efforts, four gaps remain in the genomic sequence as repeated searches fail to return contigs overlapping these gaps, and we assumed that these sequences were not obtained during the original genome sequencing (Figure 5A). The genomic searches were complemented by mining several M. sexta RNA-seq databases available on GenBank, as well as sequencing RT-PCR fragments (see Materials and Methods for details). All the contigs, except for the two smaller contigs (#3 and 4) were connected as one gene by RNA-seq data or cDNA products (data not shown). The amino acid sequence encoded by the two smaller contigs is consistent with their inclusion into the sls gene, however their respective order relative to contigs #2 and 5 is unknown (see below).
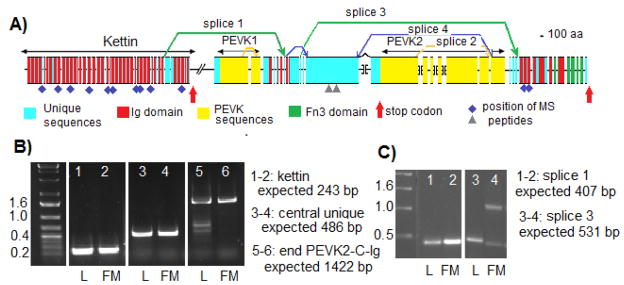
Figure 5.
Exon-intron structure, splicing alternative and domain organization of the sls gene in Manduca sexta. A) D iagram presenting the gene structure and identifying the characterized alternative splicing patterns. The extent of different protein regions are indicated by double head arrows above the map. Introns are not to scale. Brackets indicate gaps in the genomic sequences between individual contigs. B) Gel analysis of RT-PCR products for representative exons. Lanes 1–2 represent the terminal exon of kettin with the reverse primer placed after the stop codon. Lanes 3–4 represent a section of the main large exon for the central unique domain. Lanes 5–6 represent the splice from the last PEVK2 exon to the first Ig domain at the COOH terminus. L: RNA extracted from legs, FM: RNA extracted from flight muscles C) Gel analysis of RT-PCR products for specific splicing pattern. Lanes 1–2 product generated from splice 1 between NH2-terminal Ig domain and beginning of central unique region. Lanes 3–4 product generated from splice 3 between beginning of central unique region and beginning of first Ig domain at the COOH terminus. L: RNA extracted from legs, FM: RNA extracted from flight muscles. Purple diamonds and grey triangles: locations of the peptides identified from mass spectrometry with perfect or strong matches respectively.
Manual annotation of the initial contigs was performed as described in Materials and Methods. RNA sequencing and RT-PCR information were used to verify the splice site positions predicted from the annotation. The map in Figure 5A represents the current exon-intron structure of the sls gene, as well as the domain organization for the longest Sls isoform, which is very similar to the one in D. melanogaster [19]. M. sexta Sls protein contains a total of 47 Ig domains in three stretches, together with two separate and distinct PEVK segments and one large unique sequence region. The number of tandem Ig domains is the same in D. melanogaster and M. sexta at both the NH2 and COOH termini, but differs in the intermediate Ig segment, which contains only 3 Ig domains in M. sexta rather than 5 in D. melanogaster. The sls gene in all species for which it is available is more compact that the projectin gene, containing a comparatively smaller number of exons (Table 5A). The exons are, on average, much larger than in the projectin genes and their number is not very different between the Lepidoptera group and the Diptera (Table 5A). The longest M. sexta isoform (including all currently known coding exons) has a molecular weight of 1,826.822 kDa (1.827 MDa, Table 5A). Because of the genomic gaps, this is a minimum size as some exons are expected to be longer and some may still be missing (see Figure 5A).
Table 5.
Comparison of kettin/Sallimus genes (A) and protein isoforms (B). Genes from two Lepidoptera species- Danaus plexippus (D plexippus) and Manduca sexta (M. sexta) are compared to the homolog in D melanogaster (D.mel) and Apis mellifera (A. mel). CDS size represents the transcript from start to stop codons. Gene size also includes 5′ and 3′ UTR sequences when known. The different potential isoforms in M. sexta are listed with their expected protein sizes and compared to known D. melanogaster isoforms
| A) | |||
|---|---|---|---|
| D. mel | D. ple | M. sexta | |
| gene size (bp) | 75,870 | 68,393 | >88,175 |
| CDS size (bp) | 56,341 | 43,215 | 48,562 |
| # exons | 40 | 49 | 50 |
| # amino acids | 18,447 | 14,404 | 16,106 |
| Longest isoform kDa | 1,900 | 1,640 | 1,827 |
| B) | ||||||
|---|---|---|---|---|---|---|
| Manduca | Drosophila | |||||
| size (kDa) | composition | splicing | size (kDa) | composition | flight | thorax |
| 1,826.82 | PEVK1 and 2 | Full isoform | 1,900.00 | PEVK1 and 2 | N | Y |
| 1,501.28 | PEVK1 + 1/2 PEVK2 | splice 2 | 1,700.00 | PEVK1 +PEVK2, partial central unique | N | Y |
| 1,511.78 | full PEVK2, no PEVK1 | splice 1 | 1,500.00 | PEVK1, partial PEVK2, partial central unique | N | Y |
| 1,185.88 | 1/2 PEVK2, no PEVK1 | splice 1+2 (predicted) | ||||
| 1,032.04 | full PEVK1, no PEVK2 or central unique | splice 3 | ||||
| 988.64 | short PEVK2, no PEVK1 | splice 1+4 (predicted) | 1,000.00 | partial PEVK2, no PEVK1, partial central unique | N | Y |
| 716.65 | no PEVK, NH2 + COOH regions only | splice 1+3 (predicted) | 700.00 | no PEVK, NH2 + COOH regions only | Y | N |
| 542.75 (kettin) | NH2-terminus only | to 1st stop | 500 (kettin) | N-terminus only | Y | Y |
| Zormin | not found | na | 338/324 | Zormin | Y | Y |
The predicted Sls protein from D plexippus is available in GenBank as part of the genome’s initial annotation (accession # EHJ65548.1). Comparison of the domain organization and overall amino acid sequence of the two Lepidopteran Sls proteins confirms an identical domain organization, including the smaller intermediate Ig stretch of 3 domains only. The large unique sequence has a conserved length of 2010 amino acids between M. sexta and D. plexippus, but only 32% amino acid identity based on alignment generated by Lalign (data not shown) [33]. The full D. plexippus Sls protein is 1,640.245 kDa, 200 kDa smaller than the predicted M. sexta full length Sls.
All the major peptides from the Mass Spectroscopy analysis (see Materials and Methods) match perfectly to the NH2 terminal Ig domains except for a few peptides found within the Ig domains at the COOH-terminus (purple diamonds in Figure 5A). There are also some imperfect matches (~90%) to several sequences within the central unique domain (grey triangles in Figure 5A). This unique region is poorly conserved between D. plexippus and M. sexta (only 32%) and could be a highly polymorphic sequence even within a species.
The current M. sexta sls gene has the potential to encode kettin and several longer Sallimus isoforms, but not zormin. Not only the most NH2-terminal contig where kettin is found does not include any DNA sequence corresponding to zormin, but no sequence homologous to zormin could be retrieved from the M. sexta WGS or RNA-seq databases. There are three possible reasons – 1) the similarity to the D. melanogaster zormin is too low to enable isolation by BLAST searches, 2) the zormin genomic region was actually not sequenced or assembled in a contig during the initial M. sexta genome project, and 3) the coding region for zormin does not exist in M. sexta. Similarly, zormin was not included in the predicted D. plexippus sls gene and could not be isolated from the D. plexippus genome database by BLAST searches. In contrast, the A. mellifra sls gene contains a zormin sequence similar to the one in D. melanogaster (AAS, unpublished observation). For comparison purposes, the zormin segment was not included in the D. melanogaster parameters listed in Table 5A.
PEVK segments in Sallimus
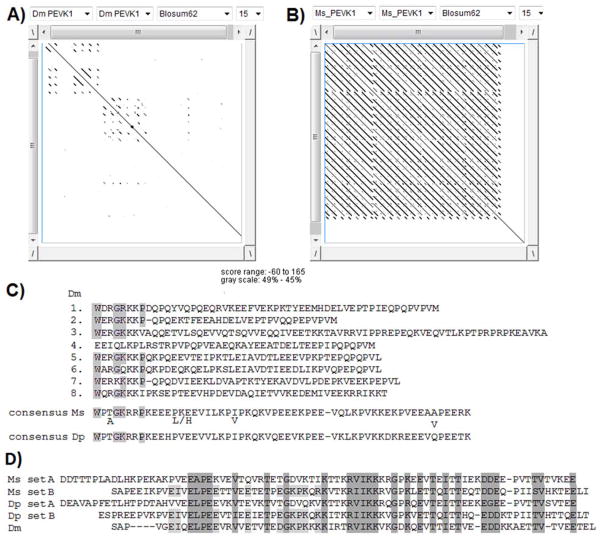
The PEVK1 segment (see Figure 5A) is 1636 amino acids in length in M. sexta and 1380 in D. plexippus. Both PEVK1 regions are mostly composed of highly conserved tandem repeats. In the Dotlet analysis presented in Figures 6A and 6B, alignments of the PEVK1 segment to itself reveal a striking difference between D. melanogaster and M. sexta. The PEVK1 region in D. melanogaster Sls contains 7 poorly conserved repeats of approximately 48 residues and a longer one of 91 residues, which are identified as extra short lines on either sides of the diagonal in Figure 5A. The D. melanogaster repeats have only a 58% P, E, V, and K amino acid content (Figure 6C, [19, 41]). By contrast the PEVK1 segment in M. sexta Sls is highly modular with 20 tandem repeats (Figure 6B), 16 with a 56 amino acid-length and four longer ones (between 81 and 107 amino acids). Each repeat has a P, E, V, and K content of approximately 80%, with the consensus sequence presented in Figure 6C. Similarly the PEVK1 region of D. plexippus has 22 highly conserved repeats, which are very similar in length and sequence to the PEVK1 repeats in M. sexta (Figure 6C). The consensus PEVK1 sequences are 84% conserved between M. sexta and D. plexippus whereas there is only a very short homology between the PEVK1 repeats of M. sexta and D. melanogaster (Figure 6C).
Figure 6.
Analysis of the the PEVK segments in the Sallimus gene.
A and B: dotlet analysis showing the repeat structure of PEVK1 in D. melanogaster (Dm) (A) and M. sexta (Ms) (B). C: Clustal-W alignment of the D. melanogaster (Dm) PEVK1 repeats and Clustal-generated consensus sequence for M. sexta (Ms) and D. plexippus (Dp) PEVK1 repeats. D) Clustal-W-generated consensus for the PEVK2 repeats in D. melanogaster (Dm), M. sexta (Ms) and D. plexippus (Dp).
The M. sexta PEVK2 segment is 4795 amino acid-long of which 4323 amino acids are organized as 57 tandem repeats with an average PEVK content of 51%. The two small contigs (#3 and 4) that could not be bridged encode several of these PEVK2 repeats, which is why these two contigs are considered bona fide parts of the sls gene. The nucleotide sequences of the two small contigs have several nucleotide differences to guarantee that there are indeed separate sequences and do not overlap with each other or contigs #2 or #5. This PEVK2 region could potentially be longer because of the remaining gaps. The tandem repeat region is followed by a non-repeated sequence of 472 amino acids with a 45% P, E, V, and K content. The complete PEVK2 region has an average P, E, V, and K content of 50.5% together with another unusual amino acid bias — threonine at 16%. The equivalent PEVK2 region in D. plexippus has a similar architecture; however it is shorter with only 3850 amino acids and 41 tandem repeats. The overall homology with M. sexta PEVK 2 is 58.6%. In both Lepidoptera the tandem repeats can be organized into two related sets of 74 (set A) and 94 (set B) amino acids, that alternate over most of the repeated segment (consensus in Figure 6D). By comparison the D. melanogaster PEVK 2 contains only 29 highly homologous (>90% identity) repeats of 71 residues and a shorter one of 45 residues with a 53% PEVK content overall [16, 19, 41]. The D. melanogaster repeats are related to both Lepidoptera set A and set B (Figure 6D).
Sallimus gene (sls) expression and alternative splicing in muscles of M. sexta
Several alternative splicing events and early termination codons would generate isoforms of different molecular weights and domain composition. At the NH2-terminus of the molecule, the sls gene contains a series of 35 Ig domains interspersed with short unique sequences. The skipping of the splice site marked by the first red arrow on Figure 5A leads to an early stop codon and generate the kettin isoform with a molecular weight of 543 kDa. (Table 5B). The expression of the kettin splice form was verified by using a primer set where the reverse primer is after the skipped splice site and stop codon in a RT-PCR amplification. The kettin splice form is expressed in both flight and leg muscle (Figure 5B, lanes 1–2). Splice variants containing the central unique region, as well as isoforms containing the end of PEVK2 and COOH-terminus Ig stretch can be amplified in RT-PCR reactions and are expressed in both leg and flight muscles (Figure 5B, lanes 3–4 and 5–6 respectively). So both leg and flight muscles would contain both the two shorter kettin variants and longer Sls isoforms.
The initial array of Ig domains can be differentially spliced to various downstream regions to create different Sls isoforms (Figure 5A). The splicing pattern #1 removes some Ig domains and the entire PEVK1 region, whereas splice pattern #3 removes most of the central unique region and the PEVK2. Splice pattern #4 removes most of the PEVK2 domain. These splicing patterns were predicted based on exon-intron boundaries and knowledge of D. melanogaster splicing patterns [19]. They were verified by RT-PCR reactions using primers from exons before and after the predicted alternate splice sites, and the amplification products were compared for size to the sequence-based predictions. Variants containing splice 1 or splice 3 are found in both leg and flight RNA (Figure 5C). The 1 kb amplified product visible in lanes 3 and 4 of Figure 5C could represent an alternative splice within one of the PEVK2 exons, but could not be verified further because the repeated nature of the PEVK2 region prevents the design of unique primers. Splice patterns 2 and 4 were recovered from RNA-seq data, but could not be confirmed because of the repetitive nature of the PEVK2 segment. The splice variant linking the NH2 region of kettin directly to the COOH Ig region (splices 1 and 3) is also expressed in both leg and flight muscle samples and would lead to a 700 kDa isoform (data not shown).
The amino acid sequences of different splice variant combinations were predicted, and the calculated sizes for the potential Sls isoforms are presented in Table 5B where they are compared to the known D. melanogaster variants [19].
Discussion
Even though synchronous flight muscles in various insects such as crickets and Lepidoptera show different levels of muscle stiffness, they display lower passive tension as compared to asynchronous muscles [42, 43]. The synchronous M. sexta DLM1 can undergo ~8–10% amplitude strains with a 25Hz wing beat [3] compared to the lower frequency 16Hz wing beat in locusts [1] and the higher frequency wing beats (~200 Hz) found in asynchronous D. melanogaster IFM [1]. Synchronous muscles also generally have longer sarcomere lengths with longer I-bands. We tested whether the more extensible and less stiff synchronous DLM1 muscle of M. sexta would contain longer isoforms of the elastic proteins, projectin and Sls.
Our gel analysis identified six high molecular weight molecules at 490 kDa, 710 kDa, 960 kDa, 1.05 MDa, 1.6 MDa and 2 MDa in M. sexta DLM1 that could be candidates for elastic proteins in this synchronous flight muscle. Our Western blot and genomic analyses allowed us to assign candidate homologues to all of these proteins.
A projectin antibody raised against L. indicus confirms that the 960 kDa (close to the L. indicus 800 kDa band) and 1.05 MDa bands are from projectin homologues present in M. sexta flight muscle. Similarly, the 9D10 antibody raised against the PEVK region in the titin sequence [38], cross-reacted with protein bands in the expected projectin and Sls positions in both M. sexta and L. indicus samples
Sequence analysis showed that projectin has a highly conserved sequence order (FnIII and Ig domains) between different insect species with an extensible PEVK region flanked by two Ig-tandem segments located in the NH2-terminus, which is anchored at the Z-line [44]. In M. sexta flight muscles several isoforms were characterized by molecular analysis [22]. The predicted sizes for these isoforms are consistent with the bands identified as projectin by Western blot analysis and confirm that neither the 1.6 nor 2.0 MDa can be projectin isoforms. This is also supported by the molecular analysis of projectin in B. mori and D. plexippus [22, 40]. Rotary shadowing of a 600 kDa projectin isoform from locust showed an elongated molecule measuring 260nm in length [45]. M. sexta flight muscle has a resting sarcomere length of ca. 3.5 microns with longer I–band than synchronous locust flight muscle (ca. 2.5 microns). The larger projectin isoform of M. sexta (1.05 Mda), if it is present as an extended molecule, could accommodate for the additional length of the I-band. The PEVK segment in the M. sexta synchronous flight isoforms is 205 amino acids long, which represents 70% of the total PEVK length and is much longer than the shortest PEVK variant expressed in D. melanogaster IFM with only 75 amino acids (Table 4B; [21,44]). Therefore the 1.05 MDa isoform could also provide for more extensible and less stiff sarcomeres and muscles, as compared to the shorter D. melanogaster isoform found in very short, stiff sarcomeres with almost non-existent I-bands.
Projectin has been shown to have a kinase domain at the COOH-terminus. The projectin kinase substrate is proposed to be troponin I, which might regulate the calcium sensitivity of actomyosin ATPase. Work in D. melanogaster has also shown that projectin is able to autophosphorylate in vitro [46,47]. Our results indicate that both projectin isoforms are phosphorylated in vivo in the M. sexta flight muscles.
We identified two bands at ~700 and 500 kDa reacting to the kettin antibody in M. sexta DLM1 and we assigned these proteins as homologs of the kettin isoform of Sls. Both of these kettin isoforms in M. sexta were found to also be phosphorylated. While D. melanogaster expresses only a 527 kDa kettin and 700 kDa Sls in its IFMs, it expresses larger Sls isoforms in other muscles. Non-flight muscles have 1500 and 2000 kDa isoforms and legs have a 1000 kDa isoform. All contain different combinations of PEVK and unique segments flanked by the kettin sequence at the NH2-terminal end and tandem-Ig domains at the COOH-terminus [19]. Our molecular analysis of the M. sexta sls gene indicates a mostly conserved gene structure with multiple possible alternative splice forms. The longest isoform encoded by the M. sexta sls gene would be 1.8 MDa containing two PEVK segments. Our current molecular analysis (Figure 5B) supports the possibility that the larger splice forms could be expressed in the flight muscles and could correspond to the 2 MDa band identified by the gel analysis. Exons included in the larger isoforms are clearly expressed in mRNA extracted from flight muscles; however the lengths of the transcripts preclude an analysis of splicing combinations. Availability of single molecule next generation sequencing [48] should enable us to further dissect the Sls isoform composition in M. sexta flight muscles. In D. melanogaster the larger Sls isoforms are only expressed in non-flight synchronous muscle, and it could be that M. sexta flight muscle expresses larger isoforms of Sls consistent with the longer sarcomere length with longer I-bands and lower stiffness [49].
One of the major differences between D. melanogaster Sls and Lepidopteran Sls is the absence of the zormin portion. Zormin in D. melanogaster can be expressed as at the NH2-terminal end of the longest Sls isoform or as an individual protein, in which case it is localized at both the Z band and M line. Zormin can bind to actin and to thick filaments and is proposed in D. melanogaster to form a complex with Rols7 and α-actinin during assembly of the Z-disc [19, 50]. Zormin however is not necessary for Z band anchoring as individual kettin molecules, without the zormin portion, can still anchor at the Z-band.
The other major difference is found within the first PEVK segment. In D. melanogaster, Sls PEVK 1 is 1254 amino acid in length with seven poorly conserved repeats (Figure 6A). In contrast M. sexta PEVK1 is not only longer (1636 amino acids), but more notably composed of 24 almost perfect repeats (Figure 6B). The M. sexta PEVK1 is more similar in structure to the PEVK2 segment, which in both insects is composed of almost perfect tandem repeats. The effect on extensibility of this different PEVK1 domain will be interesting to investigate and could contribute to the increased extensibility, lower passive tension, and recoil properties of the DLM1 muscles.
Elastic proteins larger than 1 MDa have not previously been identified in an insect flight muscle, either synchronous or asynchronous and were not present in a sampling of other insect species with synchronous flight muscles except for the silk moth Bombyx mori. The question then arises as to why there are so many different elastic proteins expressed in M. sexta DLM1? So far we have identified four isoforms of Sls including kettin and two of projectin. In vitro protein elasticity measurements showed that the Ig domain in kettin molecules could extend at low stretching force and unfold at high force, while projectin refolds very quickly even under high force conditions. This result indicates that both kettin and projectin can constitute folding-based springs [51] but with different properties. Working M. sexta DLM1 is known to have a temperature gradient from the cooler dorsal region to the warmer ventral region resulting in different power outputs from the different muscles. At physiological operating temperatures only the warmer ventral muscles have positive power output, suggesting that the cooler dorsal muscles operate mainly as energy storing and restoring damped springs. Therefore different portions appear to play different roles in DLM1. However, the detailed mechanism is still unknown. In our experiments, the ventral part expressed higher level of both projectin isoforms (Figure 3C and D). Similarly, the expression level of the 2 and 1.6 MDa Sls isoforms (Figure 3A and B) is higher in ventral than in dorsal muscle. This disparity in expression levels is consistent with the notion [4, 22] that dorsal muscle acts more like a damped spring (smaller amounts of elastic proteins i.e. presumably stiffer) and ventral muscle as an actuator (larger amounts of elastic proteins i.e. presumably more compliant). Location of the various proteins and insights into the role and exact composition of the different Sls isoforms could be revealed by future immunolocalization studies.
Conclusions
This study provides the first discussion of elastic protein composition for the flight muscle of M. sexta and helps provide a starting point for further research to explore the detailed molecular mechanisms of muscle contraction in this, in many ways, unique model system. We showed that M. sexta flight muscle contains both projectin and Sls proteins homologous to those in D. melanogaster and L. indicus IFM. The two larger proteins have been identified as alternative splice variants of the sls gene based on protein size, mass spectroscopy data and gene expression analysis. The larger 1 MDa projectin isoform in M. sexta is predominantly expressed in both dorsal and ventral subunits. However, the ventral subunits express higher total levels of projectin as well as Sls, perhaps adapting these muscle subunits to their proposed different physiological roles.
Acknowledgments
We would like to thank Prof. T. Daniel (U. Washington) for providing Manduca sexta samples. Research was supported by NSF IOS-1022058 (Irving). Mass spectrometry and proteomics data were acquired by the Arizona Proteomics Consortium supported by NIEHS grant ES06694 to the SWEHSC, NIH/NCI grant CA023074 to the AZCC and by the BIO5 Institute of the University of Arizona. The Thermo Fisher LTQ OrbitrapVelos mass spectrometer was provided by grant 1S10 RR028868-01 from NIH/NCRR. AAS and SF were supported by the College of Charleston.
References
- 1.Josephson RK, Malamud JG, Stokes DR. J Exp Biol. 2000;203:2713–2722. doi: 10.1242/jeb.203.18.2713. [DOI] [PubMed] [Google Scholar]
- 2.Stevenson RD, Josephson RK. Journal of Experimental Biology. 1990;149:61–78. [Google Scholar]
- 3.Tu MS, Daniel TL. J Exp Biol. 2004;207:2455–2464. doi: 10.1242/jeb.01039. [DOI] [PubMed] [Google Scholar]
- 4.George NT, Sponberg S, Daniel TL. J Exp Biol. 2012;215:471–479. doi: 10.1242/jeb.062901. [DOI] [PubMed] [Google Scholar]
- 5.George NT, Irving TC, Williams CD, Daniel TL. Science. 2013;340:1217–1220. doi: 10.1126/science.1229573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Granzier HL, Irving TC. Biophysical Journal. 1995;68:1027–1044. doi: 10.1016/S0006-3495(95)80278-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horowits R. Biophysical Journal. 1992;61:392–398. doi: 10.1016/S0006-3495(92)81845-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perz-Edwards RJ, Irving TC, Baumann BAJ, Gore D, Hutchinson DC, Kržic U, Porter RL, Ward AB, Reedy MK. Proc Natl Acad Sci U S A. 2011;108:120–125. doi: 10.1073/pnas.1014599107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gautel M, Goulding D. FEBS Lett. 1996;385:11–14. doi: 10.1016/0014-5793(96)00338-9. [DOI] [PubMed] [Google Scholar]
- 10.Linke WA, Ivemeyer M, Olivieri N, Kolmerer B, Ruegg JC, Labeit S. J Mol Biol. 1996;261:62–71. doi: 10.1006/jmbi.1996.0441. [DOI] [PubMed] [Google Scholar]
- 11.Cazorla O, Freiburg A, Helmes M, Centner T, McNabb M, Wu Y, Trombitas K, Labeit S, Granzier H. Circ Res. 2000;86:59–67. doi: 10.1161/01.res.86.1.59. [DOI] [PubMed] [Google Scholar]
- 12.Granzier H, Labeit S. J Physiol. 2002;541:335–342. doi: 10.1113/jphysiol.2001.014381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Granzier HL, Labeit S. Adv Protein Chem. 2005;71:89–119. doi: 10.1016/S0065-3233(04)71003-7. [DOI] [PubMed] [Google Scholar]
- 14.Linke WA, Rudy DE, Centner T, Gautel M, Witt C, Labeit S, Gregorio CC. J Cell Biol. 1999;146:631–644. doi: 10.1083/jcb.146.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ayme-Southgate AJ, Southgate RJ, Philipp RA, Sotka EE, Kramp C. J Mol Evol. 2008;67:653–669. doi: 10.1007/s00239-008-9177-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bullard B, Burkart C, Labeit S, Leonard K. J Muscle Res Cell Motil. 2005;26:479–485. doi: 10.1007/s10974-005-9032-7. [DOI] [PubMed] [Google Scholar]
- 17.Bullard B, Garcia T, Benes V, Leake MC, Linke WA, Oberhauser AF. Proc Natl Acad Sci U S A. 2006;103:4451–4456. doi: 10.1073/pnas.0509016103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trombitas K. Adv Exp Med Biol. 2000;481:1–23. doi: 10.1007/978-1-4615-4267-4_1. [DOI] [PubMed] [Google Scholar]
- 19.Burkart C, Qiu F, Brendel S, Benes V, Haag P, Labeit S, Leonard K, Bullard B. J Mol Biol. 2007;367:953–969. doi: 10.1016/j.jmb.2007.01.059. [DOI] [PubMed] [Google Scholar]
- 20.Ayme-Southgate A, Philipp RA, Southgate RJ. Insect Mol Biol. 2011;20:347–356. doi: 10.1111/j.1365-2583.2011.01069.x. [DOI] [PubMed] [Google Scholar]
- 21.Kulke M, Neagoe C, Kolmerer B, Minajeva A, Hinssen H, Bullard B, Linke WA. J Cell Biol. 2001;154:1045–1057. doi: 10.1083/jcb.200104016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ayme-Southgate AJ, Turner L, Southgate RJ. J Insect Sci. 2013;13 doi: 10.1673/031.013.8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.George NT, Daniel TL. J Exp Biol. 2011;214:894–900. doi: 10.1242/jeb.047969. [DOI] [PubMed] [Google Scholar]
- 24.Siegman MJ, Butler TM, Mooers SU, Davies RE. Science. 1976;191:383–385. doi: 10.1126/science.128819. [DOI] [PubMed] [Google Scholar]
- 25.Warren CM, Krzesinski PR, Greaser ML. Electrophoresis. 2003;24:1695–1702. doi: 10.1002/elps.200305392. [DOI] [PubMed] [Google Scholar]
- 26.Shevchenko A, Wilm M, Vorm O, Mann M. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 27.Andon NL, Hollingworth S, Koller A, Greenland AJ, Yates JR, 3rd, Haynes PA. Proteomics. 2002;2:1156–1168. doi: 10.1002/1615-9861(200209)2:9<1156::AID-PROT1156>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 28.Qian WJ, Liu T, Monroe ME, Strittmatter EF, Jacobs JM, Kangas LJ, Petritis K, Camp DG, 2nd, Smith RD. J Proteome Res. 2005;4:53–62. doi: 10.1021/pr0498638. [DOI] [PubMed] [Google Scholar]
- 29.Craig R, Beavis RC. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 30.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 31.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 32.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 33.Huang X, Miller W. Advances in Applied Mathematics. 1991;12:337–357. [Google Scholar]
- 34.Southgate R, Ayme-Southgate A. J Mol Biol. 2001;313:1035–1043. doi: 10.1006/jmbi.2001.5115. [DOI] [PubMed] [Google Scholar]
- 35.Junier T, Pagni M. Bioinformatics. 2000;16:178–179. doi: 10.1093/bioinformatics/16.2.178. [DOI] [PubMed] [Google Scholar]
- 36.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. Nuc clustal Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nedrud J, Labeit S, Gotthardt M, Granzier H. Biophys J. 2011;101:1385–1392. doi: 10.1016/j.bpj.2011.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trombitas K, Greaser M, French G, Granzier H. J Struct Biol. 1998;122:188–196. doi: 10.1006/jsbi.1998.3984. [DOI] [PubMed] [Google Scholar]
- 39.Contompasis JL, Nyland LR, Maughan DW, Vigoreaux JO. J Mol Biol. 2010;395:340–8. doi: 10.1016/j.jmb.2009.11.021. [DOI] [PubMed] [Google Scholar]
- 40.Ayme-Southgate A, Crowe M, Southgate RJ. J Proteomics and Genome Res. 2012;1:21–33. [Google Scholar]
- 41.Machado C, Andrew DJ. J Cell Biol. 2000;151:639–652. doi: 10.1083/jcb.151.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pringle JW. Proc R Soc Lond B Biol Sci. 1978;201:107–130. doi: 10.1098/rspb.1978.0035. [DOI] [PubMed] [Google Scholar]
- 43.Dudley R. The Biomechanics of Insect Flight:Form, Function, Evolution. Princeton: Princeton University Press; 2000. [Google Scholar]
- 44.Ayme-Southgate A, Saide J, Southgate R, Bounaix C, Cammarato A, Patel S, Wussler C. J Muscle Res Cell Motil. 2005;26:467–477. doi: 10.1007/s10974-005-9031-8. [DOI] [PubMed] [Google Scholar]
- 45.Nave R, Weber K. J Cell Sci. 1990;95:535–544. doi: 10.1242/jcs.95.4.535. [DOI] [PubMed] [Google Scholar]
- 46.Weitkamp B, Jurk K, Beinbrech G. J Biol Chem. 1998;273:19802–19808. doi: 10.1074/jbc.273.31.19802. [DOI] [PubMed] [Google Scholar]
- 47.Ayme-Southgate A, Southgate R, Saide J, Benian GM, Pardue ML. J Cell Biol. 1995;128:393–403. doi: 10.1083/jcb.128.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roberts RJ, Carneiro MO, Schatz MC. Genome Biol. 2013;14:405. doi: 10.1186/gb-2013-14-7-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bullard B, Linke WA, Leonard K. J Muscle Res Cell Motil. 2002;23:435–447. doi: 10.1023/a:1023454305437. [DOI] [PubMed] [Google Scholar]
- 50.Kreiskother N, Reichert N, Buttgereit D, Hertenstein A, Fischbach KF, Renkawitz-Pohl R. J Muscle Res Cell Motil. 2006;27:93–106. doi: 10.1007/s10974-006-9060-y. [DOI] [PubMed] [Google Scholar]
- 51.Bullard B, Leake M, Leonard K. Nature’s Versatile Engine: Insect Flight Muscle Inside and Out. Springer US; 2006. pp. 177–186. [Google Scholar]