Abstract
Purpose
MicroRNAs are small, endogenous noncoding RNAs that modulate posttranscriptional gene expression. Although the contribution of microRNAs to the pathogenesis of glaucomatous damage is unknown, supporting evidence from central nervous system (CNS) research suggests they may play a role. It was therefore hypothesized that microRNAs known to be altered in CNS injury are also altered in experimental glaucoma.
Methods
Intraocular pressure (IOP) was elevated in rats by unilateral injection of hypertonic saline and IOP monitored for 5 weeks. After rats were killed, retrobulbar optic nerve sections were graded for damage. MicroRNA was extracted from whole retinae of eyes with advanced nerve damage (n = 8) and from normal, noninjected control eyes (n = 8). Quantitative PCRs were performed using a panel of 17 microRNAs, reported from CNS research to be implicated in mechanisms also linked to glaucomatous damage. Computationally and experimentally derived gene targets were identified for the differentially expressed microRNAs. These were then integrated with existing gene array data. Functional interpretation was performed using the Molecular Signatures Database and DAVID Functional Annotation Clustering.
Results
Eight microRNAs were significantly downregulated in glaucomatous retinae compared with controls (miR-181c, miR-497, miR-204, let-7a, miR-29b, miR-16, miR106b, and miR-25); miR-27a was significantly upregulated. Enrichment of targets associated with extracellular matrix/cell proliferation, immune system, and regulation of apoptosis were observed. Cholesterol homeostasis and mTORC-1 pathways showed reduced expression.
Conclusions
MicroRNAs are differentially expressed in retinae of eyes with advanced glaucomatous damage compared with normal controls. Integrating microRNA with gene expression data may improve understanding of the complex biological responses produced by chronically elevated IOP.
Keywords: microRNA, glaucoma model, retina
Glaucoma is a leading cause of irreversible worldwide blindness, characterized by progressive loss of retinal ganglion cells (RGCs) and optic nerve degeneration that is usually secondary to elevated IOP.1–3 Current treatments for glaucoma are predominantly restricted to the modulation of IOP through pharmacological and surgical approaches. However, the effectiveness of this strategy can be limited in many patients by poor medication efficacy, intolerance, or surgical failure.
The primary insult leading to glaucomatous injury is thought to occur to RGC axons at the optic nerve head (ONH).4–7 In addition, patients with established glaucomatous damage may still suffer progressive loss of vision despite maximal IOP lowering. This progressive susceptibility of glaucomatous eyes to further damage may occur as a consequence of the molecular and structural mechanisms in the ONH and/or retina that occur during the disease process. An improved understanding of the biological processes in both locations may help direct future research toward the development of novel interventions to attenuate further loss of visual function in patients with glaucoma.
Experimental paradigms to model glaucomatous damage have been developed in a variety of species. In models ranging from rodents to primates, chronic elevation of IOP leads to loss of RGCs in a similar manner to that observed in human glaucoma,8 with secondary cell death thought to ultimately occur due to apoptosis.9–12 Studies of gene expression within the whole retina in experimental glaucoma highlight the complex and dynamic nature of the changes that occur following the induction of glaucomatous damage, which serve to modulate both positive and negative regulatory pathways.13–19 These studies have reported common themes, including upregulation of genes associated with cell death, cell proliferation, glial activation, TNF-α signaling, immune and inflammatory responses, and downregulation of genes associated with lipid biosynthesis and cytoskeleton and light sensation.13–19
However, with the initial IOP-related insult affecting RGC axons at the ONH, the impact of the primary injury would be expected to predominantly affect RGCs within the inner ganglion cell layer of the retina. Laser capture microdissection (LCM) has been used to isolate gene expression changes within the RGC layer following induction of experimental glaucoma,16,20 allowing further refinement of both the altered functional gene classes and the magnitude of any change, by reducing the potential dilution effect from gene responses in other retinal layers. Understanding the contribution of these various responses to the development of glaucomatous damage requires a detailed understanding of the variety of signals that may further impinge on their function.
MicroRNAs are small (∼18–22 nt), endogenous noncoding oligoribonucleotides that are highly conserved across species and modulate the posttranscriptional silencing of gene expression.21 They function through recognition of specific sequences in their target mRNAs and predominantly act to reduce expression of their mRNA targets.22 Understanding how these molecules modulate the mechanisms associated with glaucomatous damage may facilitate future targeting of microRNAs to attenuate glaucomatous injury.
Although the influence of microRNAs on the biological processes that occur within the glaucomatous ONH and retina is not currently known, there is an emerging body of evidence in central nervous system (CNS) research that suggests they may play an important role. Experimental studies of brain and spinal cord injury have demonstrated that microRNAs influence several pathways that are also known to be modulated within the glaucomatous retina. These include apoptotic cell death,23–30 ischemia,31–39 inflammatory and immune responses,30,40–45 extracellular matrix (ECM) remodeling27,46–48 and TGF-β signaling.43,49,50 The interactions of these biological responses are highly complex. In isolation, interpretation and simplification should be performed with great caution. However, examining the pattern of microRNA expression in conjunction with gene expression changes may help in our interpretation and understanding of the mechanisms that develop following glaucomatous injury.
Eyes with established severe injury are likely to demonstrate maximal responses to the ongoing primary IOP-related stimulus, in addition to the secondary mechanisms that will develop throughout the whole retina and not just restricted to the RGCs. In this study, we therefore tested the hypothesis that microRNAs altered in models of CNS injury are also altered within the glaucomatous retina with advanced damage.
Methods
All animals were treated in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research with approval from the Oregon Health and Science University (OHSU) for use in all the experiments.
Glaucoma Model
Eight-month-old Brown Norway rats were housed in constant low-level light according to the standard protocol in our laboratory. This approach was adopted to attenuate circadian fluctuations of IOP8,51 and to enable meaningful comparison with our previously published data using the same experimental glaucoma model housed in identical light conditions.8,16,20,52 Aqueous outflow obstruction was created by injecting hypertonic saline unilaterally into episcleral veins, resulting in sustained elevation of IOP.52 The IOP was measured in conscious animals at least three times per week using a handheld tonometer (Tono-Pen XL; Medtronic, Minneapolis, MN, USA). A weighted mean IOP elevation was calculated for each injected eye by dividing the cumulative IOP by the number of postinjection days. The cumulative IOP was determined as the area under the curve of the plot of days postinjection versus IOP measurement.
Tissues were harvested for further analysis 5 weeks after injection. Optic nerve injuries resulting from IOP elevation were evaluated using the grading method as previously described.8,53 This approach correlates strongly with total axon counting of optic nerve sections examined by transmission electron microscopy (TEM).8 Briefly, optic nerves were removed and postfixed in buffered 2.5% glutaraldehyde/2% paraformaldehyde and embedded in Spurr's plastic. Nerve cross sections were then graded from 1 (no injury) to 5 (active degeneration involving the whole nerve area) by five masked observers. Tissue from eyes with injury grades greater than 4.9 were used for these studies (n = 8; glaucoma model) along with noninjected control eyes with no injury (n = 8). Previous comparisons of nerves with similar advanced injury to actual axonal counts of TEM images from the same nerves, showed that a grade 5 injury is equivalent to a total intact axon count less than or equal to 45% of that within a normal optic nerve.8,53 Therefore, this study represents the contrast between microRNA expression in retinae with extensive IOP-induced degeneration of at least 50% of optic nerve axons and that in fellow eyes with no detectable axon degeneration.
Isolation and Assessment of MicroRNA
Whole retinae were quickly dissected from enucleated eyes and frozen at −80°C for subsequent analysis. Following evaluation of the extent of optic nerve injury, the mirVana microRNA isolation kit (Ambion; Thermo Fisher Scientific, Grand Island, NY, USA) was used to isolate total RNA (including microRNA). Whole frozen retinae were disrupted by immediate sonication in ice-cold lysis/binding buffer. The manufacturer's protocol was modified to include a second aqueous phase extraction during the phenol-chloroform purification step, so as to maximize microRNA yield.54 The DNAse treatment was performed following RNA elution (TURBO DNase, Ambion; Thermo Fisher Scientific). Quality of RNA was confirmed on the Agilent Bioanalyzer using the RNA 6000 Nanochip and the Eukaryotic Total RNA program (Agilent Technologies, Santa Clara, CA, USA).
Reverse Transcription, Pre-amplification, and Quantitative Real-Time PCR
TaqMan MicroRNA Assays (Thermo Fisher Scientific) were used to detect 17 mature microRNA sequences identified from recent CNS research to influence biological pathways known to be modulated within the glaucomatous retina, and a panel of three endogenous controls (Table 1). A custom RT primer pool was prepared for these 20 individual probes according to the manufacturer's instructions. Reverse transcription was performed in 15-μL reactions containing 222 ng of RNA/4 μL nuclease-free water, 6 μL RT primer pool, 0.3 μL 100 mM dNTPs with dTTP, 3 μL multiscribe reverse transcriptase (50 U/μL), 1.5 μL 10× RT buffer, and 0.2 μL RNAse Inhibitor (20 U/μL) for 30 minutes at 16°C, 30 minutes at 42°C, and 5 minutes at 85°C in a thermal cycler (GeneAmp PCR System 9700; Thermo Fisher Scientific).
Table 1.
Taqman MicroRNA Probes Used for Quantitative RT-PCR

A custom pre-amplification primer pool was prepared for 20 individual probes by combining each TaqMan MicroRNA Assay (containing a mix of forward and reverse primers) according to the manufacturer's instructions. Pre-amplification was performed in 25-μL reactions containing 2.5 μL RT product, 12.5 μL 2× TaqMan PreAmp Master Mix, 3.75 μL pre-amplification primer pool, and 6.25 μL nuclease-free water. These were incubated for 10 minutes at 95°C, 2 minutes at 55°C, 2 minutes at 72°C, and 5 minutes at 85°C, followed by 12 cycles of 95°C for 15 seconds and 60°C for 4 minutes, with a final 10-minute incubation at 99.9°C in the thermal cycler.
A total of 175 μL 0.1× Tris-EDTA buffer was added to each reaction to form the diluted pre-amplification product used for PCR reactions. This was subjected to quantitative PCR performed in 20-μL reactions and performed in triplicate for each sample (1 μL 20× TaqMan MicroRNA Assay, 0.2 μL diluted pre-amplification product, 10 μL 2× TaqMan Universal Master Mix II, No Amperase UNG, and 8.8 μL nuclease-free water) by incubation in a thermal cycler (Chromo4; Bio-Rad, Hercules, CA, USA) at 95°C for 10 minutes followed by 40 cycles of 95°C for 15 seconds and 60°C for 60 seconds.
Fluorescence data were analyzed (Opticon Monitor 3 software; Bio-Rad) with a five-point dilution standard curve. A baseline was subtracted by using the average-over-cycle range method (2–20 cycles) and threshold-adjusted to give r2 = 0.9 to 1.0 (as close to 1.0 as possible). Differential microRNA expression between control and glaucomatous retinae was calculated by relative quantification using the comparative CT method with normalization performed using a panel of three endogenous controls (U6 snRNA, snoRNA234, and snoRNA202). Each experimental group consisted of eight biological replicates. Data were expressed as the change (x-fold) in glaucomatous retinae compared with retinae from control eyes by dividing the mean normalized expression value in experimental retinae by the mean value in control specimens. If this number (the relative quantity) was less than 1, the (negative) reciprocal was listed (e.g., 0.5, or a decrease of 50% compared with the control, is reported as −2-fold change). Statistical analysis was performed by comparing the normalized expression between glaucomatous and control retinae using an unpaired two-tailed t-test with significance considered for values of P less than 0.05.
Determination of Gene Targets of Differentially Expressed MicroRNAs
Experimentally validated gene targets for each differentially expressed microRNA in retinae from eyes with advanced glaucomatous damage were obtained by searching the freely available curated online database, miRWalk (v2.0) (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/index.html).55 Due to the highly conserved nature of microRNAs, we combined validated targets for both human and rat. In the context of this study, it is important to emphasize that these gene targets were not validated in retina, but within other cells or tissues derived from the either human or rat.
These were then augmented by including predicted gene targets for the rat using computational approaches that evaluate the interaction of the first seven nucleotides of the mature microRNA strand (seed sequence) with the 3′- untranslated region (UTR) of mRNA sequences, with prediction based on factors such as the degree of complementarity and thermodynamic stability of the putative bond. Four freely available algorithms were used (miRWalk 2.0 [http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/index.html],56 RNAhybrid [http://bibiserv.techfak.uni-bielefeld.de/rnahybrid],57 miRanda [http://www.microrna.org/microrna/],58 and Targetscan 6.2 [www.targetscan.org])59 so as to identify potential gene targets.
The statistical basis of the miRWalk algorithm involves modeling the probability distribution of random matches between the 5′ end of the microRNA sequence and the 3′-UTR of the mRNA sequence with a Poisson distribution. Longer predicted matches to the so-called seed sequence (a seven-nucleotide sequence comprising 5′-bases 2 to 8) of a microRNA would be associated with a more effective microRNA-mRNA interaction and lower probability. The algorithm output reports predicted genes with P less than 0.05.56 The RNAhybrid reports targets based on an adjusted P less than 0.05, which incorporates extreme value statistics of normalized minimum free energies of the microRNA-mRNA bond, a Poisson estimation of multiple binding sites, and a calculation of the effective numbers of orthologous targets from comparative studies of multiple organisms.60 The miRanda-predicted targets are ranked according to the mirSVR regression model, which generates a weighted sum based on features of the predicted microRNA-mRNA duplex.58 The output was adjusted to report predicted targets with an mirSVR score of −1.0 or lower, which represent the top 7% of predicted targets, with 35% or more probability of having a log expression change of less than or equal to −1.0 (−2.7-fold change), and 50% or more probability of a log expression changes of at least −0.5 (−1.65-fold change).58 Targetscan evaluates base-pairing complementarity within the seed sequence of a microRNA, the thermodynamic free energy of the putative microRNA-mRNA interaction, and identifies and ranks targets according to the predicted efficacy of targeting (context + scores) and the probability of conserved targeting across multiple genomes.61,62 Targetscan has an estimated sensitivity of 82% with a false-positive rate of approximately 21%.63 To mitigate the risk of including excessive false positives and excluding false negatives, genes were accepted as potential targets only when predicted by two or more of the above algorithms.
Combination of Gene Targets With Existing Microarray Data and Functional Interpretation
Having identified potential gene targets for each differentially expressed microRNA in retinae from eyes with advanced glaucomatous damage, these were then compared against published gene array data of whole retinae from the same experimental model with an identical injury grade.16 Based on the assumption that the dominant action of microRNAs is to decrease target mRNA levels,22 we looked for significantly upregulated genes (≥1.3-fold change) on the gene array where the microRNA was downregulated, and looked for downregulated genes (≥1.3-fold change) on the gene array where the microRNA was upregulated. This comparison enabled the generation of lists of potential targets for each individual microRNA, which also showed significant change in the gene array analysis of whole retinae from eyes with advanced glaucomatous damage.
Two approaches were used to derive functional interpretations of the above lists of potential gene targets generated for each individual microRNA. First, significant overlaps between each microRNA-specific list of gene targets and so called “Hallmark Gene Sets” (developed at the Broad Institute, Cambridge, MA, USA) were investigated by using the Molecular Signatures Database v5.0 (http://www.broadinstitute.org/gsea/msigdb/index.jsp).64 These “Hallmark Gene Sets” summarize and represent specific, well-defined biological processes and have been derived using a computational approach so as to reduce noise and redundancy for use in the formal gene set enrichment analysis of genomic data.64 The investigation of gene sets uses a hypergeometric distribution of overlapping genes using the whole human genome as a background, so as to generate a significance value (P value) with an associated false discovery rate analogue to correct for multiple hypothesis testing (q-value < 0.00001).
Database for Annotation, Visualization, and Integrated Discovery (DAVID), an established online resource (https://david.ncifcrf.gov/home.jsp) that provides a comprehensive set of functional annotations for large gene lists, was also used.65,66 The DAVID functional annotation clustering module67 was used to identify related annotation groups that were enriched for each list of microRNA-specific gene targets. This approach evaluates functional classification by examining relationships among annotation terms to cluster heterogeneous yet similar annotations into functional groups. This was performed for each microRNA-specific gene list by using annotations from the Gene Ontology (GO) consortium (http://geneontology.org), namely biological process, cell component, and molecular function, and those derived from the KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway database (http://www.genome.jp/kegg/pathway.html). A list of array probes included on the microarray platform (Supplementary Table S1) was uploaded as the background for the clustering analyses. The EASE method65 was used to identify individual overrepresented GO categories or KEGG pathways by calculating a modified Fisher's Exact P value to demonstrate enrichment, where annotations showing P less than 0.05 were considered to be more enriched than by random chance. An enrichment score65 for each cluster was calculated to rank the overall biological importance (enrichment) of the gene groups. Briefly, this represents the geometric mean of all enrichment EASE scores for each annotation term, with a negative log transformation applied to emphasize its nature as a relative score. Higher enrichment scores indicate that annotation term members play a more significant role. A score of 1.3 is equivalent to 0.05 on a nonlogarithmic scale and it is suggested that more attention is given to groups with scores of 1.3 or higher, although annotation groups with lower scores may also be of potential interest.
Results
History of IOP and Evaluation of Experimental Glaucoma
Whole retinae from eight unoperated control eyes and eight experimental eyes were analyzed in this study. The mean/maximum IOP (± SD) in control eyes was 28.2 ± 0.2/29.0 ± 0.3 mm Hg compared with 39.2 ± 3.3/51.7 ± 1.0 mm Hg in experimental eyes (Fig. 1A). The cumulative IOP profile of all eyes by group across the duration of the experiment is shown in Figure 1B. Evaluation of optic nerve sections confirmed advanced glaucomatous damage in experimental eyes with a mean grade of 4.98, compared with a mean grade of 1.0 in control eyes (i.e., no evidence of injury).
Figure 1.
(A) Mean/maximum IOP of control eyes and those with experimental glaucoma over the duration of the experiment. Error bars represent ± SD. (B) Cumulative IOP profile of control and experimental eyes over the duration of the experiment.
Differential MicroRNA Expression Within Glaucomatous Retinae
Nine of the studied microRNAs showed significant differential expression in glaucomatous retinae when compared with noninjected controls (Table 2). Of these, miR-181c, miR-497, miR-204, let-7a, miR-29b, miR-16, miR106b, and miR-25 were significantly downregulated and miR-27a was the only microRNA found to be significantly upregulated. No significant change in expression of miR-21, miR-22, miR-23a, miR-145, miR-200b, miR-223, or miR-424 was observed.
Table 2.
Differentially Expressed MicroRNAs in Whole Retinae of Eyes With Advanced Glaucomatous Damage

Integration of Potential Gene Targets of Differentially Expressed MicroRNAs With Gene Array Data
A mean ± SD of 7602 ± 1320 experimentally validated targets were identified using the miRWalk database for each differentially expressed microRNA. Combining these genes with the additional predicted targets (identified by two or more of the computational algorithms) resulted in potential targets comprising 9502 ± 728 genes per individual microRNA (Supplementary Table S2).
Comparing these potential targets with published gene array data16 identified 112 ± 7 (mean ± SD) genes per microRNA that were upregulated on the array that were therefore either predicted or validated targets of the respective downregulated microRNAs (Supplementary Table S3). Interestingly, there was remarkable consistency among the genes targeted by the downregulated microRNAs identified, with 66 genes upregulated on the array being potential targets of all eight downregulated microRNAs. Forty-three of these shared gene targets were also significantly upregulated on a gene array of the RGC layer obtained by LCM from the same experimental model with an identical injury grade.16 Twenty-nine (78.4%) of 37 shared gene targets showed a significantly greater degree of change within the RGC layer when compared with the whole retina (unpaired two-tailed t-test, P < 0.008) (Table 3).
Table 3.
Of the 66 Altered Genes Within Whole Retina That Are Potentially Targeted By All Eight Downregulated MicroRNAs, 37 Were Also Significantly Upregulated in a Separate Gene Array Analysis of Retinal Ganglion Cell Layer (RGCL) Samples Obtained by Laser Capture Microdissection Using the Same Experimental Model With an Identical Injury Grade.16 All 37 Genes Were Experimentally Validated Targets (in Cells/Tissues Other Than Retina, Derived From Either Human or Rat) of All Eight Downregulated MicroRNAs, Although Computational Prediction of These Genes Varied Considerably as Shown in the Table. 29/37 Genes Identified Showed a Significantly Greater Degree of Change in the RGCL When Compared to Whole Retina (P < 0.008). The Extent of Change in the 8/37 Genes Showing a Greater Fold Change in Whole Retina Than the RGCL Did Not Differ Significantly

Eighty-six downregulated genes identified on the array were either predicted or validated targets of the single upregulated microRNA, miR-27a (Supplementary Table S3).
Functional Interpretation of Altered Genes Targeted by Differentially Expressed MicroRNAs
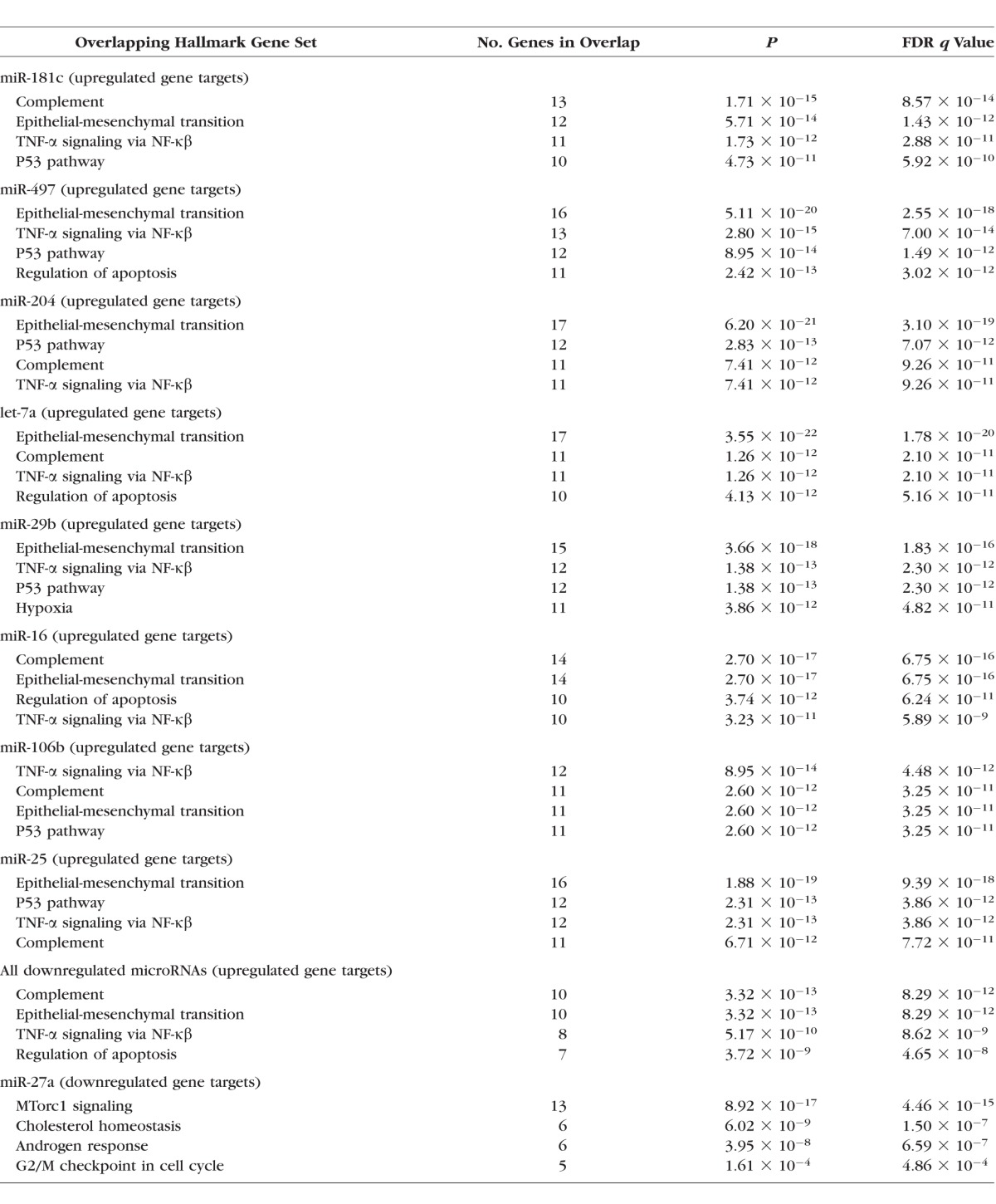
The leading Hallmark Gene Sets showing significant overlap with genes that were either predicted or validated targets of the respective differentially expressed microRNAs, which also changed on the microarray, are shown in Table 4. Enriched gene sets potentially influenced by all eight downregulated microRNAs included those defining epithelial-mesenchymal transition (e.g., wound healing, fibrosis) and genes regulated by NF-κβ in response to TNF-α signaling. Six downregulated microRNAs were associated with enrichment of genes encoding components of the complement system and p53 pathways and networks, and others associated with genes regulating programmed cell death and genes upregulated in response to hypoxia. Analysis of the gene targets common to all eight downregulated microRNAs showed significant enrichment of gene sets associated with the complement system, epithelial-mesenchymal transition, genes regulated by NF-κβ in response to TNF-α signaling, and the regulation of apoptosis. The single upregulated microRNA, miR-27a, was associated with reduced expression of gene sets relating to mTORC-1 (“mechanistic target of rapamycin complex”) signaling and cholesterol homeostasis.
Table 4.
Hallmark Gene Sets Showing Significant Overlap With Targets That Were Also Altered on the Microarray (i.e., Upregulated Genes) for Each Downregulated MicroRNA; List of Hallmark Gene Sets Showing Significant Overlap With Targets Common to All Eight Downregulated MicroRNAs (i.e., Upregulated Genes); Hallmark Gene Sets Showing Significant Overlap With Targets That Also Altered on the Microarray (i.e., Downregulated Genes) for the Single Upregulated MicroRNA
Summaries of the major DAVID functional annotation clusters associated with predicted or validated targets of the respective differentially expressed microRNAs, which also changed on the microarray are shown in Table 5. Detailed information about specific annotation terms and the relevant genes associated with each cluster are presented in Supplementary Table S4. Analysis of the gene targets that showed increased expression on the microarray for each individual downregulated microRNA showed robust enrichment of annotation clusters associated with ECM/cell proliferation and the immune system. Several downregulated microRNAs were associated with strong enrichment of annotation clusters associated with signal transduction/protein kinase cascades, with four microRNAs also associated with the regulation of apoptosis, although with lower enrichment scores. Analysis of the 66 gene targets common to all eight downregulated microRNAs demonstrated robust enrichment of clusters associated with ECM/cell proliferation, the immune system, and signal transduction/protein kinase cascades. The single upregulated microRNA, miR-27a, was associated with reduced expression of functional clusters relating cholesterol homeostasis, light responses, and axonal development.
Table 5.
Summary of the Major Functional Annotation Clusters by Enrichment Score for Each Downregulated MicroRNA Obtained Using the DAVID Functional Annotation Clustering Module (Full Details in Supplementary Table S4), Using MicroRNA-Specific Gene Targets That Were Also Altered on the Microarray (i.e., Upregulated Genes); Summary of the Major Functional Annotation Clusters by Enrichment Score Using Upregulated Gene Targets Common to All Eight Downregulated MicroRNAs (Full Details in Supplementary Table S4); Summary of the Major Functional Annotation Clusters by Enrichment Score for the Single Upregulated MicroRNA (Full Details in Supplementary Table S4), Using MicroRNA-Specific Gene Targets That Were Also Altered on the Microarray (i.e., Downregulated Genes)

Discussion
The results of this study demonstrate for the first time that a range of microRNAs known to be altered in CNS injury are also altered within the retinae of rats with advanced glaucomatous damage. The integration and analysis of both differential microRNA and gene expression data also help provide a better understanding of the complex responses that are known to occur in retinae with advanced injury. Despite recent advances, the potential significance of microRNAs in modulating the pathogenesis of glaucomatous damage within the ONH and retina has yet to be investigated. However existing knowledge regarding the role of microRNAs in modulating the sequelae of experimental CNS injury may provide a suitable paradigm for approaching these unanswered questions.
Although not providing evidence of a direct causal relationship in glaucomatous retinae, our experimental observations provide correlations between microRNA and gene expression data that are consistent with experimentally validated responses in other tissues. It is also prudent to highlight that our findings reflect responses to chronically elevated IOP that occur across the entire retina. Most cells within the retina survive this insult, with cell loss predominantly restricted to RGCs.1 Improved understanding of the complex interplay between the responses to glaucomatous injury throughout the retina would be a significant advance, as future neuroprotective strategies for glaucoma may well involve the modulation of surviving retinal glia and neurons. Also, the findings of this study describe a snapshot of events within the advanced glaucomatous retina and therefore do not fully reflect the dynamic nature of mechanisms that occur during the early onset and progression of glaucomatous damage.
Upregulation of genes associated with apoptosis within the retina has been observed in a variety of experimental glaucoma models.9–12 Although apoptosis is known to occur in glaucoma, the observed alterations in the microRNA signature would suggest an attempt to shift toward an opposite, more protective environment (Fig. 2A). Upregulation of let-7a,23 miR-16,23 miR-29b,29 and mir-49724 have been observed following both CNS trauma and ischemia. Let-7a exerts its proapoptotic activity by inducing neurodegeneration through Toll-like receptor (TLR)-7 activity27 and miR-16, miR-29b, and mir-497 promote cell death through negative regulation of the antiapoptotic protein Bcl-2.23,24,29 In addition, a reduction of miR-27a expression following traumatic brain injury leads to upregulation of proapoptotic molecules, whereas administration of exogenous miR-27a results in a reduction in lesion volume and neuronal loss following injury.28 Our observed downregulation of these four proapoptotic microRNAs (miR-16, miR-29b, miR-497, and let-7a) and upregulation of the antiapoptotic miR-27a within the glaucomatous retina suggest that the microRNA expression may be working to adjust the microenvironment in a protective manner at this advanced stage of the disease, by attempting to attenuate the extent of apoptotic cell death throughout the retina.
Figure 2.
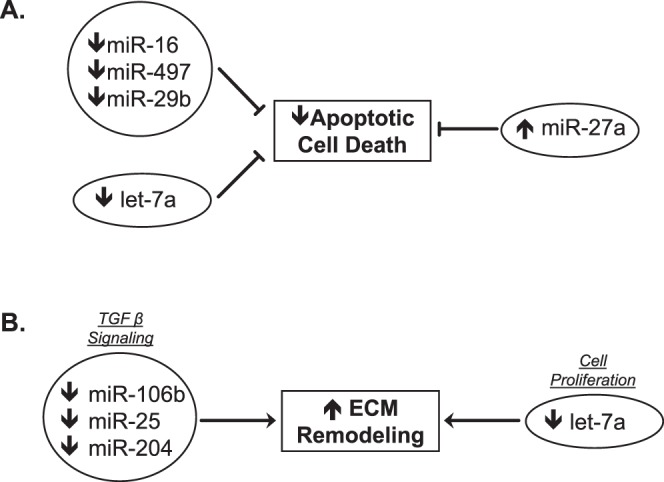
Biological interpretations of differential microRNA expression in retinae with advanced glaucomatous damage. (A) Reduced expression of proapoptotic microRNAs (miR-16, miR-497, miR-29b, and let 7-a) would aim to reduce apoptotic cell death. Increased expression of the antiapoptotic miR-27a would also enhance this effect. These changes in the microRNA signature are suggestive of a protective shift to limit apoptosis. (B) Reduced expression of several microRNAs (miR-106b, miR25, and miR-204) is associated with enhanced TGF-β signaling and a reduction in let-7a expression is also known to enhance cell proliferation. These changes in the MicroRNA signature within glaucomatous retina are consistent with extensive ECM remodeling.
Signaling of TNF-α, classified as a component of the immune response, has complex and contrasting roles in the response to elevated IOP, and may act to further modulate the equilibrium between RGC death and survival.68 On one hand, this pathway plays a role in mediating RGC death within the glaucomatous human retina,69 with recent clinical reports even suggesting an association between systemic anti-TNF therapy and reduced incidence of glaucoma.70 However, through activation of NF-κβ pathways, although often considered as initiators of inflammation, TNF-α may also exert a protective effect by resolving inflammation and initiating tissue repair.71
Our findings appear to lend some clarity to the role of TNF-α signaling within the glaucomatous retina. We identified several downregulated microRNAs (miR-106b, miR-29b, miR-497, miR-181c, and let-7a) that were associated with increased TNF-α–mediated NF-κβ signaling. These included upregulation of NF-κβ1 and AP-1 complex components (cJun and cFos),72 which both function in an antiapoptotic manner. This also suggests an apparent protective shift in the cellular responses following advanced injury. In contrast, retinal glia may contribute a proinflammatory effect through these downregulated microRNAs and their correspondingly enriched genes. Rodent microglia activated by hypoxic culture conditions showed reduced miR-181c expression, which was associated not only with enhanced NF-κβ1 expression but also with increased expression of proinflammatory mediators, including TLR-4.45 In addition, miR-181c knockdown led to an enhanced proinflammatory response in primary astrocyte cultures, with knockin studies resulting in increased production of the anti-inflammatory cytokine IL-10.40 These observations suggest that the different constituent glia within the retina may have discrete roles in the modulation of the inflammatory responses to elevated IOP. Future investigation of the glaucomatous retinal milieu using in situ techniques may help to identify the specific microRNA and gene expression responses of the individual retinal cell types.
In this study, we observed that downregulation of several microRNAs, in particular miR-181c and miR-16, were associated with enriched expression of genes encoding components of the complement and immune system. Enhanced expression of the complement cascade and immune response has also been observed within the retina across a spectrum of experimental glaucoma models.13,14,16,18,73,74 As a part of the innate immune response, complement pathways are also intrinsically linked with the process of glial activation in the pathogenesis of glaucomatous damage within the retina.13,75 Aside from its role on apoptotic pathways, reduced expression of miR-16 has also been shown to enhance susceptibility to damage from TNF-α–related pathways in human retinal vascular endothelial cell culture.76 This effect, in conjunction with proinflammatory effects of miR-181c,45 may lead to promote complement-mediated cell death in the context of retinae with advanced injury from chronically elevated IOP.
The responses of retinal glia to a glaucomatous insult are complex, and may also change as the disease progresses from early to advanced injury.77 Responses during this time may be protective to both RGC axons and the glia themselves, such as regulation of ionic imbalances and limitation of glutamate-induced excitotoxicity by Müller glia or astrocytes.78 They may also be injurious, including mechanisms leading to axonal atrophy, RGC involution, or RGC death. Retinae from our eyes with advanced glaucomatous damage demonstrate a marked shift toward mechanisms associated with ECM remodeling and repair (Fig. 2B), as shown by the enrichment of gene sets associated with the ECM and cell proliferation. These genes are potentially influenced by all eight downregulated microRNAs, and would be consistent with a potential causal relationship. Extracellular matrix remodeling has been demonstrated within the ONH79,80 and optic nerve itself81 in response to glaucomatous injury. Similar changes have been described within the glaucomatous retina,16,82 although this area has been less intensively studied.
The let-7a also has been established as a master regulator of cell proliferation pathways48 and plays a crucial role in neural progenitor cell differentiation.83 These characteristics are consistent with our observed reduction in let-7a expression leading to enhanced expression of genes associated with remodeling and cellular proliferation within the glaucomatous retina. Signaling of TGF-β is integral to the CNS response to injury, with blockade of this pathway shown to reduce glial scarring.84 Several of the microRNAs we observed to be downregulated in glaucomatous retinae influence TGF-β signaling, with repression of miR-204,49 miR-106b,43,50 and miR-2550 known to stimulate downstream components of this pathway.
The only microRNA observed to show increased expression in glaucomatous retinae was miR-27a. Functional analysis of its altered gene targets demonstrated repression of genes associated with mTORC-1 signaling, lipid biosynthesis, and axonal development. Autophagy is implicated in the pathogenesis of several neurodegenerative diseases,85 and its complex role in the pathogenesis of glaucomatous damage is becoming a focus of emerging research.86,87 Signaling of mTORC-1 is a major regulator of autophagy, and observations that in vivo inhibition may attenuate damage in animal models of neurodegeneration88 has led to considerable interest in modulating this signaling pathway as a potential therapeutic intervention. Further investigation of the interactions between miR-27a and autophagy in the context of experimental glaucoma models is an area that merits further investigation.
There is significant interplay between microRNAs and their gene targets, in that an individual microRNA may influence multiple targets and, conversely, an individual gene target may be modulated by multiple microRNAs. This makes the discovery of biologically significant microRNA targets a challenging process, as different microRNAs may regulate individual targets in a cooperative manner, but with differing hierarchy in terms of relative effect. This concept may explain why we observed no significant change in expression of several microRNAs anticipated to be altered in the glaucomatous retina on the basis of existing CNS research.
It is also worth noting the consistency of genes targeted by all the downregulated microRNAs identified, with 66 shared potential targets showing a significant increase in expression on the gene array of whole retina. It is also important to emphasize that almost two-thirds of these shared targets were also significantly upregulated within the RGC layer of glaucomatous eyes16 (Table 3). Interestingly, a closer comparison of the relative upregulation of these genes between the RGC layer and whole retina suggests that analysis of whole retina most likely represents a dilution of a larger change in gene/microRNA expression that is occurring within the RGC layer.16 It is therefore reasonable to propose that the downregulated microRNAs identified within our study are likely to modulate the specific gene expression changes that occur within the RGC layer of eyes with advanced glaucoma. An improved understanding of the hierarchical role of these and other microRNAs may be achieved by the assay of additional microRNAs within this tissue in the future. The behavior of individual microRNAs may also vary across tissue subtypes, leading to variations in biological function even within tissues of similar developmental origin, such as the retina and CNS.
It may be that animals housed in constant light are not physiologically normal, due to circadian cycle disruption.89 In this study, both groups were housed in identical light conditions, therefore controlling for any differences attributable to the use of constant light. This approach may well minimize the fluctuation of microRNAs known to have circadian or diurnal variations90−92; however, these specific targets were not investigated in this study. Nevertheless, the groups examined differ only in the extent of IOP-related damage as characterized by the extent of optic nerve injury, and are therefore able to successfully differentiate microRNA and gene expression between two groups of animals differing only in the extent of glaucomatous damage.
In the context of human glaucoma, the differential microRNA expression observed in this study may be comparable to what could be expected in a head-to-head comparison between the retinae of patients with refractory advanced glaucoma and healthy controls. These observations provide further insight into the regulation of the retinal microenvironment in the context of advanced glaucomatous damage. The integration of both microRNA and gene expression data also offers further understanding of the complex biological responses that occur in response to chronically elevated IOP.
The therapeutic application of microRNA biology to other aspects of human disease is a rapidly emerging field, with early clinical trials currently under way.93 Further research in experimental glaucoma models may potentially lead to future opportunities to explore novel neuroprotective approaches using microRNA-targeted techniques aimed toward modulating the retinal milieu during advanced disease. However, it is important to emphasize that glaucomatous changes within the retina are secondary to a primary insult at the ONH. Detailed study of the dynamic nature of alterations in microRNA and gene expression during the early responses to an elevated IOP insult, in the ONH as well as the retina, would enhance our understanding of the mechanisms responsible for the initiation and development of RGC damage. This may provide a real opportunity to correlate changes in both microRNA and gene expression in response to elevated IOP at early time points and provide a basis for future translational research toward attenuating the impact of the mechanisms associated with the development of glaucomatous damage.
Supplementary Material
Acknowledgments
Supported by the US-UK Fulbright Commission in conjunction with Fight for Sight, The Special Trustees of Moorfields Eye Hospital (HJ), National Institutes of Health Grants R01EY010145 (JCM) and P30EY010572 (OHSU Core Grant), and an unrestricted grant from Research to Prevent Blindness, Inc (RPB). JCM is an RPB Senior Investigator.
Disclosure: H. Jayaram, Allergan (R); W.O. Cepurna, None; E.C. Johnson, None; J.C. Morrison, None
References
- 1. Quigley HA. Neuronal death in glaucoma. Prog Retin Eye Res. 1999; 18: 39–57. [DOI] [PubMed] [Google Scholar]
- 2. Weinreb RN,, Khaw PT. Primary open-angle glaucoma. Lancet. 2004; 363: 1711–1720. [DOI] [PubMed] [Google Scholar]
- 3. Tham YC,, Li X,, Wong TY,, Quigley HA,, Aung T,, Cheng CY. Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology. 2014; 121: 2081–2090. [DOI] [PubMed] [Google Scholar]
- 4. Buckingham BP,, Inman DM,, Lambert W,, et al. Progressive ganglion cell degeneration precedes neuronal loss in a mouse model of glaucoma. J Neurosci. 2008; 28: 2735–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Howell GR,, Libby RT,, Jakobs TC,, et al. Axons of retinal ganglion cells are insulted in the optic nerve early in DBA/2J glaucoma. J Cell Biol. 2007; 179: 1523–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Quigley HA,, Addicks EM,, Green WR,, Maumenee AE. Optic nerve damage in human glaucoma. II. The site of injury and susceptibility to damage. Arch Ophthalmol. 1981; 99: 635–649. [DOI] [PubMed] [Google Scholar]
- 7. Quigley HA,, Hohman RM,, Addicks EM,, Massof RW,, Green WR. Morphologic changes in the lamina cribrosa correlated with neural loss in open-angle glaucoma. Am J Ophthalmol. 1983; 95: 673–691. [DOI] [PubMed] [Google Scholar]
- 8. Morrison JC,, Johnson EC,, Cepurna W,, Jia L. Understanding mechanisms of pressure-induced optic nerve damage. Prog Retin Eye Res. 2005; 24: 217–240. [DOI] [PubMed] [Google Scholar]
- 9. Quigley HA,, Nickells RW,, Kerrigan LA,, Pease ME,, Thibault DJ,, Zack DJ. Retinal ganglion cell death in experimental glaucoma and after axotomy occurs by apoptosis. Invest Ophthalmol Vis Sci. 1995; 36: 774–786. [PubMed] [Google Scholar]
- 10. Hanninen VA,, Pantcheva MB,, Freeman EE,, Poulin NR,, Grosskreutz CL. Activation of caspase 9 in a rat model of experimental glaucoma. Curr Eye Res. 2002; 25: 389–395. [DOI] [PubMed] [Google Scholar]
- 11. Johnson EC,, Deppmeier LM,, Wentzien SK,, Hsu I,, Morrison JC. Chronology of optic nerve head and retinal responses to elevated intraocular pressure. Invest Ophthalmol Vis Sci. 2000; 41: 431–442. [PubMed] [Google Scholar]
- 12. Reichstein D,, Ren L,, Filippopoulos T,, Mittag T,, Danias J. Apoptotic retinal ganglion cell death in the DBA/2 mouse model of glaucoma. Exp Eye Res. 2007; 84: 13–21. [DOI] [PubMed] [Google Scholar]
- 13. Ahmed F,, Brown KM,, Stephan DA,, Morrison JC,, Johnson EC,, Tomarev SI. Microarray analysis of changes in mRNA levels in the rat retina after experimental elevation of intraocular pressure. Invest Ophthalmol Vis Sci. 2004; 45: 1247–1258. [DOI] [PubMed] [Google Scholar]
- 14. Steele MR,, Inman DM,, Calkins DJ,, Horner PJ,, Vetter ML. Microarray analysis of retinal gene expression in the DBA/2J model of glaucoma. Invest Ophthalmol Vis Sci. 2006; 47: 977–985. [DOI] [PubMed] [Google Scholar]
- 15. Yang Z,, Quigley HA,, Pease ME,, et al. Changes in gene expression in experimental glaucoma and optic nerve transection: the equilibrium between protective and detrimental mechanisms. Invest Ophthalmol Vis Sci. 2007; 48: 5539–5548. [DOI] [PubMed] [Google Scholar]
- 16. Guo Y,, Cepurna WO,, Dyck JA,, Doser TA,, Johnson EC,, Morrison JC. Retinal cell responses to elevated intraocular pressure: a gene array comparison between the whole retina and retinal ganglion cell layer. Invest Ophthalmol Vis Sci. 2010; 51: 3003–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Howell GR,, Walton DO,, King BL,, Libby RT,, John SW. Datgan a reusable software system for facile interrogation and visualization of complex transcription profiling data. BMC Genomics. 2011; 12: 429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Miyahara T,, Kikuchi T,, Akimoto M,, Kurokawa T,, Shibuki H,, Yoshimura N. Gene microarray analysis of experimental glaucomatous retina from cynomologous monkey. Invest Ophthalmol Vis Sci. 2003; 44: 4347–4356. [DOI] [PubMed] [Google Scholar]
- 19. Jakobs TC. Differential gene expression in glaucoma. Cold Spring Harb Perspect Med. 2014; 4: a020636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guo Y,, Johnson EC,, Cepurna WO,, Dyck JA,, Doser T,, Morrison JC. Early gene expression changes in the retinal ganglion cell layer of a rat glaucoma model. Invest Ophthalmol Vis Sci. 2011; 52: 1460–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bartel DP. MicroRNAs: genomics biogenesis, mechanism, and function. Cell. 2004; 116: 281–297. [DOI] [PubMed] [Google Scholar]
- 22. Guo H,, Ingolia NT,, Weissman JS,, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010; 466: 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu G,, Keeler BE,, Zhukareva V,, Houle JD. Cycling exercise affects the expression of apoptosis-associated microRNAs after spinal cord injury in rats. Exp Neurol. 2010; 226: 200–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yin KJ,, Deng Z,, Huang H,, et al. miR-497 regulates neuronal death in mouse brain after transient focal cerebral ischemia. Neurobiol Dis. 2010; 38: 17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Han Z,, Chen F,, Ge X,, Tan J,, Lei P, Zhang J. miR-21 alleviated apoptosis of cortical neurons through promoting PTEN-Akt signaling pathway in vitro after experimental traumatic brain injury. Brain Res. 2014; 1582: 12–20. [DOI] [PubMed] [Google Scholar]
- 26. Jovicic A, Zaldivar Jolissaint JF, Moser R, Silva Santos Mde F, Luthi-Carter R. MicroRNA-22 (miR-22) overexpression is neuroprotective via general anti-apoptotic effects and may also target specific Huntington's disease-related mechanisms. PLoS One. 2013; 8: e54222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lehmann SM,, Kruger C,, Park B,, et al. An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat Neurosci. 2012; 15: 827–835. [DOI] [PubMed] [Google Scholar]
- 28. Sabirzhanov B,, Zhao Z,, Stoica BA,, et al. Downregulation of miR-23a and miR-27a following experimental traumatic brain injury induces neuronal cell death through activation of proapoptotic Bcl-2 proteins. J Neurosci. 2014; 34: 10055–10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shi G,, Liu Y,, Liu T,, et al. Upregulated miR-29b promotes neuronal cell death by inhibiting Bcl2L2 after ischemic brain injury. Exp Brain Res. 2012; 216: 225–230. [DOI] [PubMed] [Google Scholar]
- 30. Zhang L,, Dong LY,, Li YJ,, Hong Z, Wei WS. miR-21 represses FasL in microglia and protects against microglia-mediated neuronal cell death following hypoxia/ischemia. Glia. 2012; 60: 1888–1895. [DOI] [PubMed] [Google Scholar]
- 31. Buller B,, Liu X,, Wang X,, et al. MicroRNA-21 protects neurons from ischemic death. FEBS J. 2010; 277: 4299–4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chang CY,, Lui TN,, Lin JW,, et al. Roles of microRNA-1 in hypoxia-induced apoptotic insults to neuronal cells [published online ahead of print September 20, 2014] Arch Toxicol.doi:10.1007/s00204-014-1364-x. [DOI] [PubMed]
- 33. Khanna S,, Rink C,, Ghoorkhanian R,, et al. Loss of miR-29b following acute ischemic stroke contributes to neural cell death and infarct size. J Cereb Blood Flow Metab. 2013; 33: 1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moon JM,, Xu L,, Giffard RG. Inhibition of microRNA-181 reduces forebrain ischemia-induced neuronal loss. J Cereb Blood Flow Metab. 2013; 33: 1976–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ouyang YB,, Lu Y,, Yue S,, et al. miR-181 regulates GRP78 and influences outcome from cerebral ischemia in vitro and in vivo. Neurobiol Dis. 2012; 45: 555–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ouyang YB,, Xu L,, Lu Y,, et al. Astrocyte-enriched miR-29a targets PUMA and reduces neuronal vulnerability to forebrain ischemia. Glia. 2013; 61: 1784–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Peng Z,, Li J,, Li Y,, et al. Downregulation of miR-181b in mouse brain following ischemic stroke induces neuroprotection against ischemic injury through targeting heat shock protein A5 and ubiquitin carboxyl-terminal hydrolase isozyme L1. J Neurosci Res. 2013; 91: 1349–1362. [DOI] [PubMed] [Google Scholar]
- 38. Roshan R,, Shridhar S,, Sarangdhar MA,, et al. Brain-specific knockdown of miR-29 results in neuronal cell death and ataxia in mice. RNA. 2014; 20: 1287–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhao H,, Wang J,, Gao L,, et al. MiRNA-424 protects against permanent focal cerebral ischemia injury in mice involving suppressing microglia activation. Stroke. 2013; 44: 1706–1713. [DOI] [PubMed] [Google Scholar]
- 40. Hutchison ER,, Kawamoto EM,, Taub DD,, et al. Evidence for miR-181 involvement in neuroinflammatory responses of astrocytes. Glia. 2013; 61: 1018–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jadhav SP,, Kamath SP,, Choolani M,, Lu J, Dheen ST. microRNA-200b modulates microglia-mediated neuroinflammation via the cJun/MAPK pathway. J Neurochem. 2014; 130: 388–401. [DOI] [PubMed] [Google Scholar]
- 42. Murray AR,, Chen Q,, Takahashi Y,, Zhou KK,, Park K,, Ma JX. MicroRNA-200b downregulates oxidation resistance 1 (Oxr1) expression in the retina of type 1 diabetes model. Invest Ophthalmol Vis Sci. 2013; 54: 1689–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang H,, Liu J,, Zong Y,, et al. miR-106b aberrantly expressed in a double transgenic mouse model for Alzheimer's disease targets TGF-beta type II receptor. Brain Res. 2010; 1357: 166–174. [DOI] [PubMed] [Google Scholar]
- 44. Zhang L,, Dong LY,, Li YJ,, Hong Z,, Wei WS. The microRNA miR-181c controls microglia-mediated neuronal apoptosis by suppressing tumor necrosis factor. J Neuroinflammation. 2012; 9: 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang L,, Li YJ,, Wu XY,, Hong Z,, Wei WS. MicroRNA-181c negatively regulates the inflammatory response in oxygen-glucose-deprived microglia by targeting Toll-like receptor 4. J Neurochem. 2015; 132: 713–723. [DOI] [PubMed] [Google Scholar]
- 46. Hong P,, Jiang M,, Li H. Functional requirement of dicer1 and miR-17-5p in reactive astrocyte proliferation after spinal cord injury in the mouse. Glia. 2014; 62: 2044–2060. [DOI] [PubMed] [Google Scholar]
- 47. Wang CY,, Yang SH,, Tzeng SF. MicroRNA-145 as one negative regulator of astrogliosis. Glia. 2015; 63: 194–205. [DOI] [PubMed] [Google Scholar]
- 48. Johnson CD,, Esquela-Kerscher A,, Stefani G,, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007; 67: 7713–7722. [DOI] [PubMed] [Google Scholar]
- 49. Wang FE,, Zhang C,, Maminishkis A,, et al. MicroRNA-204/211 alters epithelial physiology. FASEB J. 2010; 24: 1552–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. De Santis G,, Ferracin M,, Biondani A,, et al. Altered miRNA expression in T regulatory cells in course of multiple sclerosis. J Neuroimmunol. 2010; 226: 165–171. [DOI] [PubMed] [Google Scholar]
- 51. Lozano DC,, Hartwick AT,, Twa MD. Circadian rhythm of intraocular pressure in the adult rat. Chronobiol Int. 2015; 32: 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Morrison JC,, Moore CG,, Deppmeier LM,, Gold BG,, Meshul CK,, Johnson EC. A rat model of chronic pressure-induced optic nerve damage. Exp Eye Res. 1997; 64: 85–96. [DOI] [PubMed] [Google Scholar]
- 53. Jia L,, Cepurna WO,, Johnson EC,, Morrison JC. Patterns of intraocular pressure elevation after aqueous humor outflow obstruction in rats. Invest Ophthalmol Vis Sci. 2000; 41: 1380–1385. [PubMed] [Google Scholar]
- 54. Burgos KL,, Javaherian A,, Bomprezzi R,, et al. Identification of extracellular miRNA in human cerebrospinal fluid by next-generation sequencing. RNA. 2013; 19: 712–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dweep H, Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods. 2015; 12: 697. [DOI] [PubMed] [Google Scholar]
- 56. Dweep H,, Sticht C,, Pandey P, Gretz N. miRWalk—database: Prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform. 2011; 44: 839–847. [DOI] [PubMed] [Google Scholar]
- 57. Kruger J,, Rehmsmeier M. RNAhybrid: microRNA target prediction easy fast and flexible. Nucleic Acids Res. 2006; 34: W451–W454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Betel D,, Koppal A,, Agius P,, Sander C,, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010; 11: R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lewis BP,, Burge CB,, Bartel DP. Conserved seed pairing often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005; 120: 15–20. [DOI] [PubMed] [Google Scholar]
- 60. Rehmsmeier M,, Steffen P,, Hochsmann M,, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004; 10: 1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Friedman RC,, Farh KK,, Burge CB,, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009; 19: 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Grimson A,, Farh KK,, Johnston WK,, Garrett-Engele P,, Lim LP,, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007; 27: 91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhang Y,, Verbeek FJ. Comparison and integration of target prediction algorithms for microRNA studies. J Integr Bioinform. 2010; 7: 127. [DOI] [PubMed] [Google Scholar]
- 64. Subramanian A,, Tamayo P,, Mootha VK,, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005; 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009; 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 66. Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009; 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Huang DW,, Sherman BT,, Tan Q,, et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007; 8: R183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tezel G. TNF-alpha signaling in glaucomatous neurodegeneration. Prog Brain Res. 2008; 173: 409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tezel G,, Li LY,, Patil RV,, Wax MB. TNF-alpha and TNF-alpha receptor-1 in the retina of normal and glaucomatous eyes. Invest Ophthalmol Vis Sci. 2001; 42: 1787–1794. [PubMed] [Google Scholar]
- 70. Stein JD,, Talwar N,, Kang JH,, Okereke OI,, Wiggs JL,, Pasquale LR. Bupropion use and risk of open-angle glaucoma among enrollees in a large U.S. managed care network. PLoS One. 2015; 10: e0123682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ghosh S,, Hayden MS. New regulators of NF-kappaB in inflammation. Nat Rev Immunol. 2008; 8: 837–848. [DOI] [PubMed] [Google Scholar]
- 72. Shaulian E,, Karin M. AP-1 in cell proliferation and survival. Oncogene. 2001; 20: 2390–2400. [DOI] [PubMed] [Google Scholar]
- 73. Howell GR,, Macalinao DG,, Sousa GL,, et al. Molecular clustering identifies complement and endothelin induction as early events in a mouse model of glaucoma. J Clin Invest. 2011; 121: 1429–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Stevens B,, Allen NJ,, Vazquez LE,, et al. The classical complement cascade mediates CNS synapse elimination. Cell. 2007; 131: 1164–1178. [DOI] [PubMed] [Google Scholar]
- 75. Soto I,, Howell GR. The complex role of neuroinflammation in glaucoma. Cold Spring Harb Perspect Med. 2014; 4: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ye EA, Steinle JJ. miR-15b/16 protects primary human retinal microvascular endothelial cells against hyperglycemia-induced increases in tumor necrosis factor alpha and suppressor of cytokine signaling 3. J Neuroinflammation. 2015; 12: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Nickells RW. From ocular hypertension to ganglion cell death: a theoretical sequence of events leading to glaucoma. Can J Ophthalmol. 2007; 42: 278–287. [PubMed] [Google Scholar]
- 78. Johnson EC,, Morrison JC. Friend or foe? Resolving the impact of glial responses in glaucoma. J Glaucoma. 2009; 18: 341–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Johnson EC,, Jia L,, Cepurna WO,, Doser TA,, Morrison JC. Global changes in optic nerve head gene expression after exposure to elevated intraocular pressure in a rat glaucoma model. Invest Ophthalmol Vis Sci. 2007; 48: 3161–3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Hernandez MR. The optic nerve head in glaucoma: role of astrocytes in tissue remodeling. Prog Retin Eye Res. 2000; 19: 297–321. [DOI] [PubMed] [Google Scholar]
- 81. Selles-Navarro I,, Ellezam B,, Fajardo R,, Latour M,, McKerracher L. Retinal ganglion cell and nonneuronal cell responses to a microcrush lesion of adult rat optic nerve. Exp Neurol. 2001; 167: 282–289. [DOI] [PubMed] [Google Scholar]
- 82. Guo L,, Moss SE,, Alexander RA,, Ali RR,, Fitzke FW,, Cordeiro MF. Retinal ganglion cell apoptosis in glaucoma is related to intraocular pressure and IOP-induced effects on extracellular matrix. Invest Ophthalmol Vis Sci. 2005; 46: 175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Meza-Sosa KF,, Pedraza-Alva G, Perez-Martinez L. microRNAs: key triggers of neuronal cell fate. Front Cell Neurosci. 2014; 8: 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Moon LD,, Fawcett JW. Reduction in CNS scar formation without concomitant increase in axon regeneration following treatment of adult rat brain with a combination of antibodies to TGFbeta1 and beta2. Eur J Neurosci. 2001; 14: 1667–1677. [DOI] [PubMed] [Google Scholar]
- 85. Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006; 443: 780–786. [DOI] [PubMed] [Google Scholar]
- 86. Wang Y,, Huang C,, Zhang H,, Wu R. Autophagy in glaucoma: crosstalk with apoptosis and its implications. Brain Res Bull. 2015; 117: 1–9. [DOI] [PubMed] [Google Scholar]
- 87. Davis CH,, Kim KY,, Bushong EA,, et al. Transcellular degradation of axonal mitochondria. Proc Natl Acad Sci U S A. 2014; 111: 9633–9638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Sarkar S,, Rubinsztein DC. Small molecule enhancers of autophagy for neurodegenerative diseases. Mol Biosyst. 2008; 4: 895–901. [DOI] [PubMed] [Google Scholar]
- 89. McMahon DG,, Iuvone PM,, Tosini G. Circadian organization of the mammalian retina: from gene regulation to physiology and diseases. Prog Retin Eye Res. 2014; 39: 58–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wang Q,, Bozack SN,, Yan Y,, Boulton ME,, Grant MB,, Busik JV. Regulation of retinal inflammation by rhythmic expression of MiR-146a in diabetic retina. Invest Ophthalmol Vis Sci. 2014; 55: 3986–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Kinoshita C,, Aoyama K,, Matsumura N,, Kikuchi-Utsumi K,, Watabe M,, Nakaki T. Rhythmic oscillations of the microRNA miR-96-5p play a neuroprotective role by indirectly regulating glutathione levels. Nat Commun. 2014; 5: 3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Cheng HY,, Obrietan K. Revealing a role of microRNAs in the regulation of the biological clock. Cell Cycle. 2007; 6: 3034–3035. [DOI] [PubMed] [Google Scholar]
- 93. Shibata C,, Otsuka M,, Kishikawa T,, et al. Current status of miRNA-targeting therapeutics and preclinical studies against gastroenterological carcinoma. Mol Cell Ther. 2013; 1: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



