Abstract
Heart failure is a complex clinical syndrome and has become the most common reason for adult hospitalization in developed countries. Two subtypes of heart failure, ischemic heart disease (ISCH) and dilated cardiomyopathy (DCM), have been studied using microarray platforms. However, microarray has limited resolution. Here we applied RNA sequencing (RNA-Seq) to identify gene signatures for heart failure from six individuals, including three controls, one ISCH and two DCM patients. Using genes identified from this small RNA-Seq dataset, we were able to accurately classify heart failure status in a much larger set of 313 individuals. The identified genes significantly overlapped with genes identified via genome-wide association studies for cardiometabolic traits and the promoters of those genes were enriched for binding sites for transcriptions factors. Our results indicate that it is possible to use RNA-Seq to classify disease status for complex diseases such as heart failure using an extremely small training dataset.
Keywords: RNA-seq, Heart Failure, disease classification
Introduction
Heart failure, defined as the inability of the heart to pump sufficient blood to meet the body’s demands, is a syndrome associated with high morbidity and mortality. An estimated five million Americans are diagnosed with heart failure every year, causing more than 250,000 deaths annually. Heart failure is a complex disease that involves multiple genetic and environmental factors. Two of the most common subtypes of heart failure include ischemic heart disease (ISCH), which is caused by reduced blood supply to heart muscle, and dilated cardiomyopathy (DCM) in which the heart becomes weakened and enlarged despite normal blood flow [1]. Although ISCH and DCM can lead to similar symptoms of heart failure, emerging evidence suggest that the two subtypes may produce different structural and/or functional phenotypes and may respond differently to therapy [1–3]. In addition, patients with ISCH generally have reduced survival compared to those with DCM [1, 2].
Most human genomic studies in heart failure are limited by insufficient clinical samples from patients with advanced heart failure [1]. As such, researchers have used animal models in combination with functional genomics to study the molecular underpinnings of heart failure [4, 5]. Attempts have also been made to link gene signatures in human blood with heart failure outcomes [6, 7]. Recently, several studies have been published based on human myocardium. Tan et al. [8] showed that end-stage heart failure is associated with an increase in expression levels for genes encoding for matrix/cytoskeletal and proteolysis/stress proteins based on a comparison of eight hearts from patients with end-stage heart failure and seven non-failing controls. Kittlesonet al. [2] used microarray with a machine learning approach to distinguish patients with histological evidence of ischemic injury from those without a history of myocardial infarction, revascularization, or coronary artery disease. Using a much larger dataset derived from 185 failing and 14 non-failing hearts, Margulies et al. [9] identified 3,088 differentially expressed transcripts with only a small subset demonstrating improvements that was correlated to the favorable remodeling observed during mechanical circulatory support. Using this dataset, Hannenhalli et al. [3] explored transcription factors that are associated with heart failure.
All of the aforementioned studies were based on microarrays. Although microarrays have been the predominant method for gene expression studies due to their ability to measure thousands of transcripts simultaneously, they are subject to biases in hybridization strength, and potential for cross-hybridization to probes with similar sequences. Additionally, they are unable to identify novel genes or novel splicing events because of their reliance on existing gene models. RNA sequencing (RNA-Seq) is a newer approach for transcriptome profiling [10–12]. It is the first sequencing-based method that allows an unbiased survey of the entire transcriptome in a high-throughput manner. Briefly, RNA-Seq involves fragmenting poly-A selected RNA molecules into small fragments and converting into a cDNA library with adaptors attached to cDNA fragments. The cDNA library is then sequenced to obtain short sequences, which are subsequently aligned to a reference genome and/or transcriptome or assembled de novo without the reference sequence. The expression level for a gene is determined by counting the number of reads that are mapped to it. With RNA-Seq data, transcripts spanning multiple exons can be directly observed. Moreover, RNA-Seq has a greater dynamic range than microarrays, which suffer from non-specific hybridization and saturation biases [13].
Motivated by the advantages of RNA-Seq technology for gene expression profiling, we sequenced the transcriptomes of six human individuals’ left ventricle tissue to identify genes that are associated with heart failure. Our study includes one ISCH patient, two DCM patients and three individuals with non-failing hearts (NF). Based on these six individuals, we identified genes that were differentially expressed between ISCH and NF, DCM and NF, and ISCH and DCM. A remarkable finding of our study is that using genes identified from this small RNA-Seq dataset, we were able to classify a much larger set of 313 individuals with failing or non-failing hearts. Our results suggest that, with highly accurately measured gene expression levels using RNA-Seq, it is possible to classify disease status for complex diseases such as heart failure using an extremely small training dataset.
Materials and Methods
Sample collection
Samples of cardiac tissue (n = 6 for RNA-Seq, n = 313 for microarrays) were acquired from subjects from the MAGNet consortium (http://www.med.upenn.edu/magnet/). The heart was perfused with cold cardioplegia prior to cardiectomy to arrest contraction and prevent ischemic damage. Left ventricular free-wall tissue was harvested and snap frozen with liquid nitrogen at the time of cardiac surgery from subjects with heart failure undergoing transplantation and from unused donor hearts. Cause of heart failure (ISCH or DCM) was determined by medical history and pathological examination of the explanted hearts. All the samples were stored in −80°C freezer until analyses. This study was approved by the University of Pennsylvania Institutional Review Board and the Cleveland Clinic Institutional Review Board. All participants were 18 years or older and provided written informed consent.
RNA extraction, library preparation and sequencing
RNAs for six selected individuals were extracted using RNeasy Lipid Tissue total RNA mini kit (Qiagen, Valencia, CA). Extracted RNA samples underwent quality control (QC) assessment using the Agilent Bioanalyzer (Agilent, Santa Clara, CA) and all RNA samples submitted for sequencing had an RNA Integrity Number (RIN) > 6, with a minimum of 1μg input RNA. Poly-A library preparation and RNA sequencing were performed at the Penn Genome Frontiers Institute’s High-Throughput Sequencing Facility per standard protocols. Briefly, we generated first-strand cDNA using random hexamer-primed reverse transcription, followed by second-strand cDNA synthesis using RNase H and DNA polymerase, and ligation of sequencing adapters using the TruSeq RNA Sample Preparation Kit (Illumina, San Diego, CA). Fragments of ~350 bp were selected by gel electrophoresis, followed by 15 cycles of PCR amplification. The prepared libraries were then sequenced using Illumina’sHiSeq 2000 with four RNA-seq libraries per lane (2×101 bp paired-end reads).
Analysis of RNA-Seq data
The RNA-Seq data were aligned to the hg19 reference genome using Tophat with default options [14]. In order to eliminate mapping errors and reduce potential mapping ambiguity due to homologous sequences, several filtering steps were applied. Specifically, we required (1) the mapping quality score of each read is 30, (2) reads from the same pair were mapped to the same chromosome with expected orientations and the mapping distance between the read pair was < 500,000 bp, and (3) each read was uniquely mapped to the genome. All subsequent analyses were based on filtered alignment files.
Transcripts were assembled using Cufflinks [15, 16]. For each gene, we compared the expression levels between two individuals for each of the three categories, including ISCH vs. NF, DCM vs. NF, and ISCH vs. DCM. To test for differential expression, Cufflinks first computes the logarithm of the ratio of Fragments Per Kilobase of exon per Million fragments mapped (FPKMs) between the two subjects, and then uses delta method to estimate the variance of the log ratio. The test statistic is log ratio of the FPKMs divided by the standard deviation of the log ratio. It is possible to estimate the standard deviation based on a single subject because of the availability of multiple reads per subject. To ensure reliable expression estimates, we required the FPKM value to be greater than or equal to 3 for at least one of the two individuals under comparison [17]. A gene was considered differentially expressed if the FDR adjusted p-value was < 0.05.
For differentially expressed genes, we carried out functional annotation analysis using DAVID [18, 19]. Differentially expressed genes were used as input gene list, and all human genes that were expressed in heart were used as the background. We looked for enrichment for genetic association with disease class, KEGG pathways, and biological processes in Gene Ontology (GO). Multiple testing was adjusted using Benjamini approach, and enrichment was declared if Benjamini adjusted p-value was less than 0.05.
To search for evidence of over representation of transcription factor binding sites in heart failure, we used a computational approach previously developed by Hannenhalli et al. [3]. First, a set of cardiac genes was determined from RNA-Seq data by selecting genes with FPKM > 3. Each cardiac gene was then mapped to its corresponding promoter region sequence, defined as the 5 kb of genomic sequence upstream from the transcription start site, based on the RefSeq annotation. Transcription factor binding sites were determined within these promoters with the TRANSFAC database [20] of vertebrate transcription factor binding sites, with a focus on promoter regions that show human-mouse evolutionary sequence conservation. We then determined which binding sites were statistically over represented among genes that showed altered expression in heart failure.
For differentially expressed genes, we further examined whether they were more likely to overlap with GWAS findings. Our analysis was based on all GWAS signals summarized in the NHGRI GWAS catalogue (http://www.genome.gov/gwastudies). We only considered GWAS signals for cardiometabolic traits (Supplementary Table 6). Enrichment analysis was investigated using Fisher’s exact test.
RNA preparation and processing of microarray data
RNAs for 313 subjects, including 95 ISCH patients, 82 DCM patients, and 136 individuals with normal hearts were hybridized with Affymetrix Human Exon ST1.1 arrays using manufacturer instructions. The resulting CEL files were normalized with the robust multiarray analysis using Bioconductor to generate transcript-level intensity estimates [21]. To remove residual batch effect, expression values were further adjusted using ComBat [22, 23], an empirical Bayes method that estimates parameters for location and scale adjustment of each batch for each gene independently. Probe sets were removed if the log2-transformed expression values were less than four on all arrays. This filtering yielded sets of genes present well above background levels in the human heart. For the remaining probe sets, their Affymetrix probe annotations were cross checked by mapping probe sequences to the hg19 reference genome. Only uniquely mapped probes with no mismatches were kept for subsequent analysis.
Classification of disease status using gene signatures identified from RNA-Seq
Our goal was to use those differentially expressed genes identified from RNA-Seq as feature vectors to classify disease status for the 313 individuals with microarray data. In order to classify the ISCH/NF (n = 231) individuals, we used genes that were differentially expressed in all pairwise comparisons (defined as globally differentially expressed) of ISCH vs. NF in RNA-Seq as the feature vector. Similarly, globally differentially expressed genes were used as the feature vectors to classify DCM/NF individuals (n = 218), and ISCH/DCM individuals (n = 177). After the feature vectors were determined, the K-means clustering algorithm implemented in R’s “amap: Another Multidimentional ” package was used to classify the individuals into two groups, and Pearson correlation distance metric was used in the clustering with a maximum of 50 iterations.
Data Access
RNA-Seq and microarray data have been deposited in the Gene Expression Omnibus (GEO) database (accession number GSE57345).
Results
RNA-Seq data alignment
The RNA-Seq data were aligned and filtered as described in Methods. We obtained a high mapping rate with 76–83% of reads mapped to the reference genome, and 66–71% were uniquely mapped, properly filtered, and used in subsequent analysis (Supplementary Table 1). All RNA-Seq samples passed FastQC’s basic statistics test (Supplementary Figure 1).
Analysis of differential expression using RNA-Seq
First, we compared the gene expression profiles of the ISCH and NF individuals. Our RNA-Seq experiment includes one ISCH patient and three individuals with non-failing hearts, yielding three possible pairwise comparisons for differential expression analysis. Using Cufflinks, we identified 492, 522 and 418 differentially expressed genes in the three pairs, respectively (Table 1; Supplementary Table 2A–C). Union of these gene lists gave 983 genes that were differentially expressed in at least one of the three pairs, among which 531 (54%) had higher expression levels in ISCH and 452 (46%) had higher expression levels in NF (Supplementary 3A). By intersecting differentially expressed genes across all three pairs, 70 genes were retained and we call these genes as globally differentially expressed and used them as feature vector for the K-means clustering of the 231 ISCH/NF individuals with microarray data (Supplementary Table 4A).
Table 1.
Summary of differentially expressed (DE) genes for comparisons of ISCH vs. NF, DCM vs. NF, and ISCH vs. DCM.
| Comparison | Pair | No. of DE genes | No. of overlapping DE genes | No. of union DE genes |
|---|---|---|---|---|
| ISCH vs. NF | 234 vs. 1207 | 522 | 70 | 983 |
| 234 vs. 1256 | 418 | |||
| 234 vs. D111 | 492 | |||
| DCM vs. NF | 333 vs. 1207 | 491 | 12 | 1,109 |
| 333 vs. 1256 | 482 | |||
| 333 vs. D111 | 343 | |||
| X2182 vs. 1207 | 361 | |||
| X2182 vs. 1256 | 393 | |||
| X2182 vs. D111 | 491 | |||
| ISCH vs. DCM | 234 vs. 333 | 484 | 129 | 825 |
| 234 vs. X2182 | 492 |
Next, we compared DCM and NF individuals. With two DCM and three NF individuals, there were six possible pairwise comparisons for differential expression analysis. Using Cufflinks, we identified 361, 393, 491, 482, 343 and 491 differentially expressed genes, respectively, for the six pairs (Table 1; Supplementary Table 2D–I). Union of these gene lists gave 1,109 genes that were differentially expressed in at least one of the six pairs (Supplementary 3B). Among these genes, 844 (76%) had higher expression levels in DCM and 265 (24%) had higher expression levels in NF. By intersecting differentially expressed genes across all six pairs, we identified 12 genes that were globally differentially expressed (Supplementary 4B). These genes were used as feature vector in the K-means clustering of the 218 DCM/NF individuals with microarray data.
We also compared the two subtypes of heart failure. Two possible combinations were considered based on one ISCH and two DCM individuals. We found 484 and 492 differentially expressed genes in the two pairs (Table 1; Supplementary Table 2J–K), respectively, yielding a total of 825 differentially expressed genes in at least one pair, including 476 (58%) with higher expression levels in ISCH and 349 (42%) with higher expression levels in DCM (Supplementary Table 3C). Interaction of the gene lists yielded 129 genes that were differentially expressed in both pairs and they were used as the feature vector in the K-means clustering of the 177 ISCH/DCM individuals with microarray data (Supplementary Table 4C).
Categories of differentially expressed genes
To investigate what categories of genes were differentially expressed, we carried out functional annotation analysis using DAVID. For each set of differential expression analysis, genes that were expressed (FPKM 3) in at least one individual under comparison were used as the background. These include 9,919 genes for ISCH vs. NF comparison, 10,462 genes for DCM vs. NF comparison, and 10,190 for ISCH vs. DCM comparison.
ISCH vs. NF comparison
For genes that had higher expression levels in ISCH, they were enriched and only enriched for CARDIOVASCULAR (p-value = 0.0028) in disease class and ECM-receptor interaction pathway (p-value = 0.000152) in KEGG. For Gene Ontology (GO), these genes were significantly enriched for extracellular matrix formation processes (p-value < 10−25) (Supplementary Figure 2 (A)–(C)). ECM-receptor interaction and their formation related processes had been shown to play critical roles in ischemic heart remodeling.[24] For genes that had higher expression levels in the NF individuals, they were enriched for CARDIOVASCULAR (p-value = 0.011) and RENAL (p-value = 0.0024) in disease class, but no significant enrichment was found in KEGG. For GO, these genes were significantly enriched for terms related to system development (p-value = 8.68×10−8) and organ development (p-value = 9.34×10−6) (Supplementary Figure 2 (D)).
DCM vs. NF comparison
For genes that had higher expression levels in DCM, they were enriched for CARDIOVASCULAR (p-value = 0.00022) in disease class. They were also enriched for focal adhesion pathway (p-value = 0.029) in KEGG. For GO, these genes were significantly enriched for terms related to plasma membrane (p-value < 10−10) and extracellular region (p-value < 10−6) (Supplementary Figure 2 (E)–(G)). For genes that had higher expression levels in NF, no significant enrichment was found in disease class and KEGG, but for GO, these genes were significantly enriched for terms related to extracellular matrix (Supplementary Figure 2 (H)).
ISCH vs. DCM comparison
Genes that had higher expression levels in ISCH were enriched for CARDIOVASCULAR (p-value = 0.0015) and IMMUNE (p-value = 0.0001) in disease class. In KEGG, only the ECM-receptor interaction pathway was significantly enriched (p-value = 1.85×10−6). For GO, terms related to extracellular matrix formation processes (p-value < 10−28), response to external stimulus (p-value = 2.06×10−14) and inflammatory response (p-value = 6.99×10−12) were significantly enriched (Supplementary Figure 2 (I)–(K)). Extracellular matrix formation process was again found highly enriched, indicating that extracellular matrix related genes were not only differentially expressed in ISCH vs. NF, but also differentially expressed in subtypes of heart failure. No significant enrichment was found for genes that had higher expression levels in DCM.
Overrepresented transcription factor binding sites in heart failure
To investigate whether there is a discrete set of cardiac transcription factors potentially driving the observed gene expression changes in heart failure cases relative to controls, we examined whether certain transcription factor binding sites are over- or under-represented among those that showed altered gene expression patterns in heart failure using a computational approach developed by Hannenhalli et al. [3]. Specifically, we determined enrichment of TRANSFAC motifs by counting the frequency with which a given binding site was present in the promoters of differentially expressed genes relative to the frequency in the reference set of genes (i.e., genes expressed in heart). We performed analysis separately for genes that were up-regulated or down-regulated in heart failure cases.
For the comparison of ISCH vs. NF, the binding sites of NKX2-5, MAZ, and MZF1 were over-represented in down-regulated genes in all three pairs. Further examination of the gene expression of these transcription factors suggests that gene that encodes NKX2-5 was differentially expressed between the ISCH subject 234 and the NF subject 1256 (p-value = 0.0062), with subject 234 showing lower expression level than subject 1256, which is consistent with the fact that the target genes of NKX2-5 were down-regulated in ISCH. The expression level of subject 234 was also lower than the other two NF subjects, 1207 and D111, but the gene expression difference was not statistically significant (Supplementary Table 5). We did not find evidence of differential expression in genes that encode MAZ and MZF1. Several factors may affect the lack of differential expression in transcription factor genes. First, transcription factor genes are generally expressed at very low levels, and this affects the statistical power in detecting their differential expression. Second, transcription factors are often regulated at post-translational levels and are not expected to exhibit differential expression [26]. Third, a large fraction of the observed differences in target gene expression is likely due to genotype differences in cis-elements and not differential expression of the transcription factor regulators. Fourth, it is entirely possible that certain transcription factors can both activate certain genes and repress others, depending on the genomic context and also whether the binding site is polymorphic between cases and controls and the effect of the corresponding polymorphisms. We performed similar enrichment analysis for up-regulated genes in the ISCH vs. NF comparison and both up- and down-regulated genes for the DCM vs. NF comparison, but did not identify transcription factor motifs that were significantly enriched in all pairs.
Overlap of differentially expressed genes with GWAS loci for cardiometabolic traits
We queried our differentially expressed genes against published GWAS loci for cardiometabolic traits as derived from the NHGRI GWAS catalogue (Supplementary Table 6). Compared to genes that were not differentially expressed, we found a statistically significant overlap with GWAS loci for cardiometabolic traits for the ISCH vs. NF comparison (p-value = 0.00029), and the DCM vs. NF comparison (p-value = 0.0012). The overlap was less significant for the ISCH vs. DCM comparison (p-value = 0.0093) (Table 2). Our results suggest that the identified differentially expressed genes might play a specific role for cardiometabolic disease.
Table 2.
Overlap of differentially expressed (DE) genes with GWAS loci for cardiometabolic traits.
| Comparison | Category | Total | Overlap with GWAS loci | % Overlap | P-value |
|---|---|---|---|---|---|
| ISCH vs. NF | DE genes | 983 | 72 | 7.32 | 0.0013 |
| Non-DE genes | 8,936 | 408 | 4.57 | ||
| DCM vs. NF | DE genes | 1,109 | 78 | 7.03 | 0.0041 |
| Non-DE genes | 9,353 | 438 | 4.68 | ||
| ISCH vs. DCM | DE genes | 825 | 57 | 6.90 | 0.017 |
| Non-DE genes | 9,365 | 448 | 4.78 |
RNA-Seq selected genes classify heart failure status in samples with microarray data
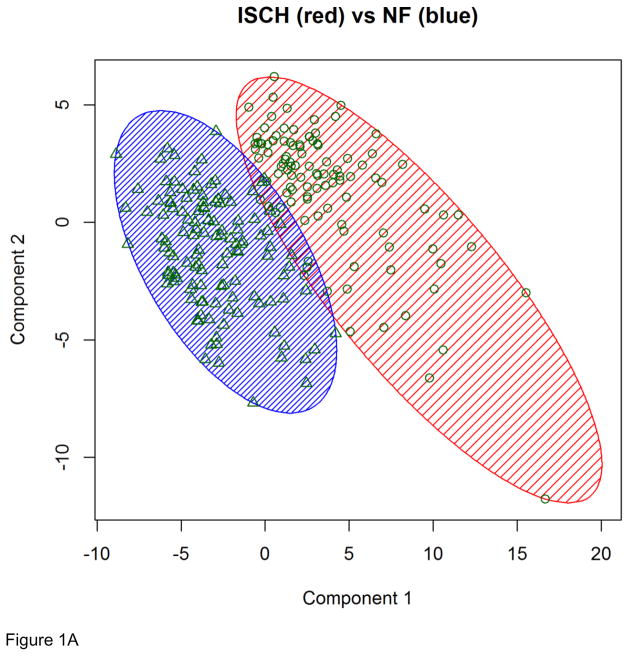
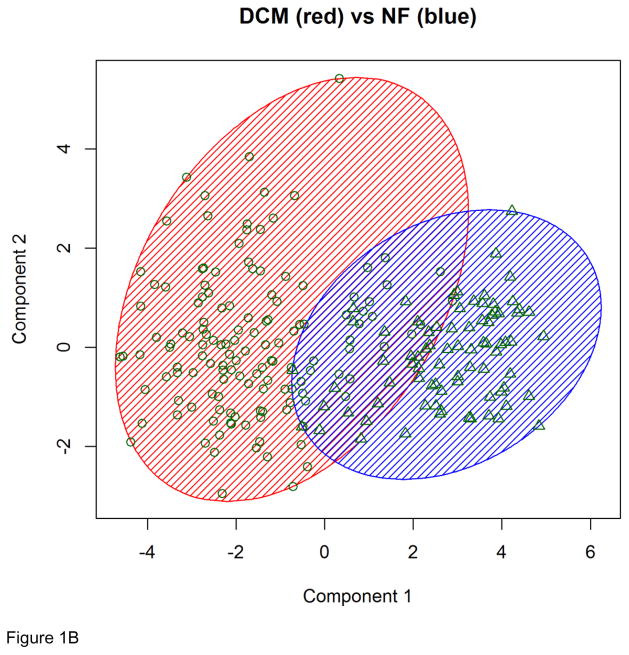
Genes that were globally differentially expressed were used as feature vectors to classify the 313 individuals with microarray data, which include 95 with ISCH, 82 with DCM, and 136 with NF. In the classification of the 231 ISCH/NF individuals, the K-means clustering algorithm using the RNA-Seq determined feature vector with 70 genes correctly classified 216 of the individuals, yielding a misclassification rate of 6.5% (Figure 1 (A)). In the classification of the 218 DCM/NF individuals, the RNA-Seq determined feature vector with 12 genes was used in the K-means clustering algorithm and this led to the correct classification of 194 individuals, yielding a misclassification rate of 11.0% (Figure 1 (B)). Notably, all six individuals who had both RNA-Seq and microarray data were correctly classified into the right group. Our results suggest that by using feature vectors determined from a training set with six individuals only, we were able to correctly classify nearly 200 individuals in the testing dataset into the correct clinical phenotype category. It is remarkable to achieve such high classification accuracy with datasets of extremely small training/testing ratio (4:231 for ISCH/NF and 5:218 for DCM/NF) (Table 3A).
Figure 1. K-means clustering results based on RNA-Seq determined feature vectors.
(A) Clustering results for the 231 ISCH/NF individuals. (B) Clustering results for the 218 DCM/NF individuals.
Table 3.
Misclassification rates using K-means clustering for RNA-Seq determined feature vectors (A), and microarray determined feature vectors (B).
| (A)
| ||||||
|---|---|---|---|---|---|---|
| Groups | No. genes in feature vector | No. of individuals in group 1 | No. of individuals in group 2 | Total | No. of misclassified individuals | Misclassification rate |
| ISCH + NF | 70 | 95 | 136 | 231 | 15 | 6.49% |
| DCM + NF | 12 | 82 | 136 | 218 | 24 | 11.01% |
| ISCH + DCM | 129 | 95 | 82 | 177 | 80 | 45.20% |
| (B)
| ||||||
|---|---|---|---|---|---|---|
| Groups | No. genes in feature vector | No. of individuals in group 1 | No. of individuals in group 2 | Total | No. of misclassified individuals | Misclassification rate |
| ISCH + NF | 70 | 95 | 136 | 231 | 29 | 12.55% |
| DCM + NF | 12 | 82 | 136 | 218 | 101 | 46.33% |
| ISCH + DCM | 129 | 95 | 82 | 177 | 79 | 44.63% |
As a comparison, we repeated the classification analysis based on feature vectors determined from the microarray data. We focused on the six individuals that had both RNA-Seq and microarray data, but the genes were selected by mean fold change of expression levels based on the microarray data. We used a different method to select signature genes because there is only a single intensity measure per probe, which disallows pairwise comparisons due to the lack of variance estimate for gene expression. For the ISCH/NF classification, we used the top 70 genes that had the largest fold change on gene expression as the feature vector. The clustering algorithm with this feature vector gave a misclassification rate of 12%, which is about twice as high as the misclassification rate when the RNA-Seq determined feature vector was used (Supplementary Figure 3). Similarly, for the DCM/NF classification, the clustering algorithm with microarray determined feature vector gave a misclassification rate of 46%, which is about four times as high as that for RNA-Seq determined feature vector (Supplementary Figure 4). The reduced misclassification rates using RNA-Seq determined feature vectors indicate the much higher accuracy of RNA-Seq in quantifying gene expression levels than microarray (Table 3B).
To evaluate the confidence of our classification results, we performed random sampling. For the classification of the ISCH/NF individuals, we randomly selected 70 genes from the union of the 9,919 expressed genes obtained from RNA-Seq and used this list of genes as the feature vector for classification. We repeated this process 100 times and obtained an empirical distribution of the misclassification rate, which ranges from 8.2% to 49%. Similar analysis was carried out for the classification of the DCM/NF individuals, and the range of the misclassification rate was 16% to 49%. Results from these analyses suggest that the low misclassification rates observed in our original analysis are unlikely due to random variation.
We also attempted to classify the ISCH/DCM individuals. However, with the 129 RNA-Seq selected differentially expressed genes serving as the feature vector, the accuracy for classification was only slightly better than random (misclassification rate was 45%, Supplementary Figure 5 (A)). The misclassification rate based on genes selected from microarray based on mean fold change of expression was also 45% (Supplementary Figure 5 (B)). In the case of heart failure subtype classification, the differentially expressed genes could not distinguish between ISCH and DCM in an independent dataset. This failure of classification might be due to several reasons: 1) all of the ISCH and DCM individuals had end-stage heart failure, and this had obviated their initial differences [27]; 2) the relative small sample size of the training dataset; 3) gene expression levels in the testing dataset were not accurately measured by microarray. A recent study also reported the difficulty of discriminating between cardiomyopathies of different causes [17].
Discussion
Heart failure results from abnormalities in multiple biological processes that contribute to cardiac dysfunction. In this study, we tested the hypothesis that a small set of genes with distinct expression patterns between failing and non-failing hearts can accurately classify disease status for complex diseases such as heart failure. Using RNA-Seq data on six individuals, we identified genes that were differentially expressed between ISCH and NF, DCM and NF, and ISCH and DCM individuals. A remarkable finding of our study is that using the gene signatures identified from this small RNA-Seq dataset, we were able to classify a much larger set of 313 individuals with failing or non-failing hearts, and the misclassification rates for the classification of ISCH/NF and DCM/NF individuals were one to three times lower than those obtained from microarray data. Such remarkable results are likely due to the highly accurate gene expression measurements obtained from RNA-Seq and careful selection of feature vectors in classification.
The unbiased RNA-Seq approach as we employed in this study identified genes that were differentially expressed between individuals with heart failure and those with non-failing hearts. Typical differential expression analysis involves group-wise comparison, i.e., comparing gene expression levels between two groups (each with multiple biological replicates), and searching for genes with different mean expression levels. Instead of searching for such overall differentially expressed genes between heart failure cases and controls, we did pairwise comparisons between every two individuals that have different disease status. We then took the intersection of the identified genes and used them as feature vectors for downstream clustering of microarray data. By so doing, we achieved a high accuracy with extremely small training: testing ratio (4:231 for ISCH/NF and 5:218 for DCM/NF); in other words, using ~2% of the samples, we correctly classified disease status for the remaining ~98% of the samples.
The advantage of the pairwise comparison lies in its ability to identify sets of genes that were differentially expressed in all pairs, i.e., globally differentially expressed, and this minimizes the contribution of less informative genes in the classification. To demonstrate this point, we compared the misclassification rates from pairwise comparisons with those obtained from group-wise comparisons (Supplementary Table 7). As expected, the group-wise comparisons identified more differentially expressed genes; for the ISCH vs. NF comparison, 50 more differentially expressed genes were identified but the misclassification rate increased from 6.5% to 13.4%, which almost doubled the misclassification rate obtained from pairwise comparison; for the DCM vs. NF comparison, the number of differentially expressed genes increased more than four times and the misclassification rate reduced slightly from 11% to 7.8%; for the ISCH vs. DCM comparison, the number of differentially expressed genes was almost doubled, but the misclassification rate was only 1% lower than that from pairwise comparison. Taken together, these results indicate that pairwise comparisons significantly reduced the number of signature genes but achieved similar level or even better classification accuracy than group-wise comparisons.
In our comparison with microarray, we have used the same number of individuals (n = 6) in the training set as RNA-Seq. Since the cost of microarray is lower than RNA-Seq on a per-sample basis, it is of interest to compare the performance of RNA-Seq with microarray when larger number of training individuals is used in microarray data analysis. We randomly selected half of the microarray samples for the training set and used the other half for testing. The misclassification rates for each comparison were shown in Supplementary Table 8. The number of signature genes identified from this analysis was much larger than that from the pairwise RNA-Seq analysis (~8 times more for the ISCH vs. NF comparison and ~48 times more for the DCM vs. NF comparison). Although using much larger number of genes in the feature vector, the misclassification rate for the ISCH vs. NF comparison was still higher than that from RNA-seq: 7.9% vs. 6.5%; for the DCM vs. NF comparison, microarray had slightly lower misclassification rate: 7.4% vs. 11.0%. However, it is worth noting that the cost of generating microarray data for 165 subjects is much higher than that for six RNA-Seq samples. Therefore, the 3.6% reduction in misclassification rate represents a modest improvement.
Since we only had one ISCH patient, to assess the robustness of our conclusion, we analyzed two recently sequenced ISCH subjects from our ongoing MAGNet study. We repeated our analyses using these two new subjects, and obtained misclassification rates of 6.5% and 7.8%, respectively, which are comparable to the 6.5% misclassification rate obtained from the original ISCH subject. Although preliminary, this result suggests that our conclusion is robust to the choice of different ISCH patients.
Although RNA-Seq has demonstrated its superior power in studying the complexity of eukaryotic transcriptomes, this approach is still relatively new for cardiovascular genomics research. Here we report an RNA-Seq study of advanced heart failure together with a microarray study of the same population. Most studies reporting gene expression variations in heart failure have focused on small numbers of samples with advanced heart failure. Because of the relative small sample size, the resulting genes have frequently failed to be replicated. Our study represents the largest heart failure transcritpomic study reported to date. An important implication of our findings is the identification of myocardial genes associated with heart failure in humans.
RNA-Seq is a recently developed approach for transcriptome profiling that uses deep-sequencing technologies. Studies using this approach have already altered our view of the extent and complexity of eukaryotic transcriptomes. As shown by our results, RNA-Seq provides a far more precise measurement of levels of gene expression than microarray. In this study, we focused on gene expression quantification. However, using RNA-Seq, we can also quantify gene expression at the isoform level [28]. Additionally, we can examine differences of alternative splicing between two conditions, and integrate with DNA sequence data to examine allelic imbalance and RNA editing. In contrast to gene expression quantification, these analyses require a much higher sequencing depth to yield reliable results [29]. We will explore these various aspects of transcriptomic variations as we generate RNA-Seq data with higher sequencing depths.
In conclusion, we have utilized the RNA-Seq technology to identify genes with distinct expression patterns between failing and non-failing hearts. Our study demonstrates how knowledge gained from a small set of samples with accurately measured gene expressions using RNA-Seq and creative selection of classifier genes can be leveraged as a complementary strategy to discern the genetics of complex diseases. We note that analysis methods for RNA-Seq data are continuing to evolve. Additional studies employing improved analytical methods hold the potential to reveal a more complete picture of the genetic architecture of heart failure.
Supplementary Material
Highlights.
We applied RNA-Seq to identify gene signatures for heart failure from six individuals. Using genes identified from pairwise differential expression analysis, we accurately classified heart failure status in a much larger set of 313 individuals. Our results indicate that it is possible to use RNA-Seq to classify disease status for complex diseases such as heart failure using an extremely small training dataset.
Acknowledgments
This project was supported by the U.S. National Institutes of Health: R01 HL105993 to K.B.M., R01HL089847 to K.B.M. and T.P.C., R01HL088577 to T.P.C., R01GM108600, R01HL113147, R01HG006465, R01GM097505 to M.L., and R01HL103931 to W.H.T.
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest
Authors’ contributions
M.L., Y.L. and T.C. conceived and designed the study. K.B., T.C., E.A., W.H.T., and C.M. collected the data. Y.L., M.M., S.H., Y.H. and M.L. analyzed the data. M.L. and Y.L. wrote the paper. J.B. performed the microarray and RNA-Seq experiments. K.B. and T.C. critically reviewed the manuscript. All authors read and approved the final manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Yichuan Liu, Email: yichuan.edwdard.liu@gmail.com.
Michael Morley, Email: mmorley@mail.med.upenn.edu.
Jeffrey Brandimarto, Email: bjeff@mail.med.upenn.edu.
Sridhar Hannenhalli, Email: sridhar@umiacs.umd.edu.
Yu Hu, Email: huyu1@mail.med.upenn.edu.
Euan A. Ashley, Email: euan@stanford.edu.
W.H. Wilson Tang, Email: TANGW@ccf.org.
Christine S. Moravec, Email: MORAVEC@ccf.org.
Kenneth B. Margulies, Email: Kenneth.Margulies@uphs.upenn.edu.
Thomas P. Cappola, Email: thomas.cappola@uphs.upenn.edu.
Mingyao Li, Email: mingyao@mail.med.upenn.edu.
References
- 1.Liew CC, Dzau VJ. Molecular genetics and genomics of heart failure. Nature reviews Genetics. 2004;5(11):811–825. doi: 10.1038/nrg1470. [DOI] [PubMed] [Google Scholar]
- 2.Kittleson MM, Ye SQ, Irizarry RA, Minhas KM, Edness G, Conte JV, Parmigiani G, Miller LW, Chen Y, Hall JL, et al. Identification of a gene expression profile that differentiates between ischemic and nonischemic cardiomyopathy. Circulation. 2004;110(22):3444–3451. doi: 10.1161/01.CIR.0000148178.19465.11. [DOI] [PubMed] [Google Scholar]
- 3.Hannenhalli S, Putt ME, Gilmore JM, Wang J, Parmacek MS, Epstein JA, Morrisey EE, Margulies KB, Cappola TP. Transcriptional genomics associates FOX transcription factors with human heart failure. Circulation. 2006;114(12):1269–1276. doi: 10.1161/CIRCULATIONAHA.106.632430. [DOI] [PubMed] [Google Scholar]
- 4.Weinberg EO, Mirotsou M, Gannon J, Dzau VJ, Lee RT, Pratt RE. Sex dependence and temporal dependence of the left ventricular genomic response to pressure overload. Physiological genomics. 2003;12(2):113–127. doi: 10.1152/physiolgenomics.00046.2002. [DOI] [PubMed] [Google Scholar]
- 5.Tang Z, McGowan BS, Huber SA, McTiernan CF, Addya S, Surrey S, Kubota T, Fortina P, Higuchi Y, Diamond MA, et al. Gene expression profiling during the transition to failure in TNF-alpha over-expressing mice demonstrates the development of autoimmune myocarditis. Journal of molecular and cellular cardiology. 2004;36(4):515–530. doi: 10.1016/j.yjmcc.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 6.Vanburen P, Ma J, Chao S, Mueller E, Schneider DJ, Liew CC. Blood gene expression signatures associate with heart failure outcomes. Physiological genomics. 2011;43(8):392–397. doi: 10.1152/physiolgenomics.00175.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whitney AR, Diehn M, Popper SJ, Alizadeh AA, Boldrick JC, Relman DA, Brown PO. Individuality and variation in gene expression patterns in human blood. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(4):1896–1901. doi: 10.1073/pnas.252784499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan FL, Moravec CS, Li J, Apperson-Hansen C, McCarthy PM, Young JB, Bond M. The gene expression fingerprint of human heart failure. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(17):11387–11392. doi: 10.1073/pnas.162370099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Margulies KB, Matiwala S, Cornejo C, Olsen H, Craven WA, Bednarik D. Mixed messages: transcription patterns in failing and recovering human myocardium. Circ Res. 2005;96(5):592–599. doi: 10.1161/01.RES.0000159390.03503.c3. [DOI] [PubMed] [Google Scholar]
- 10.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456(7218):53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nature biotechnology. 2008;26(5):541–547. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 13.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roberts A, Pimentel H, Trapnell C, Pachter L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics. 2011;27(17):2325–2329. doi: 10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
- 16.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang KC, Yamada KA, Patel AY, Topkara VK, George I, Cheema FH, Ewald GA, Mann DL, Nerbonne JM. Deep RNA Sequencing Reveals Dynamic Regulation of Myocardial Noncoding RNA in Failing Human Heart and Remodeling with Mechanical Circulatory Support. Circulation. 2014 doi: 10.1161/CIRCULATIONAHA.113.003863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang da W, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic acids research. 2007;35(Web Server issue):W169–175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 20.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic acids research. 2006;34(Database issue):D108–110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome biology. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 23.Chen C, Grennan K, Badner J, Zhang D, Gershon E, Jin L, Liu C. Removing batch effects in analysis of expression microarray data: an evaluation of six batch adjustment methods. PloS one. 2011;6(2):e17238. doi: 10.1371/journal.pone.0017238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallagher GL, Jackson CJ, Hunyor SN. Myocardial extracellular matrix remodeling in ischemic heart failure. Frontiers in bioscience : a journal and virtual library. 2007;12:1410–1419. doi: 10.2741/2157. [DOI] [PubMed] [Google Scholar]
- 25.Andersen ME, Clewell HJ, Carmichael PL, Boekelheide K. Can case study approaches speed implementation of the NRC report: “toxicity testing in the 21st century: a vision and a strategy?”. ALTEX. 2011;28(3):175–182. doi: 10.14573/altex.2011.3.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Everett LJ, Jensen ST, Hannenhalli S. Transcriptional regulation via TF-modifying enzymes: an integrative model-based analysis. Nucleic Acids Res. 2011;39(12):e78. doi: 10.1093/nar/gkr172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steenman M, Chen YW, Le Cunff M, Lamirault G, Varro A, Hoffman E, Leger JJ. Transcriptomal analysis of failing and nonfailing human hearts. Physiological genomics. 2003;12(2):97–112. doi: 10.1152/physiolgenomics.00148.2002. [DOI] [PubMed] [Google Scholar]
- 28.Hu Y, Liu Y, Mao X, Jia C, Ferguson JF, Xue C, Reilly MP, Li H, Li M. PennSeq: accurate isoform-specific gene expression quantification in RNA-Seq by modeling non-uniform read distribution. Nucleic Acids Res. 2014;42(3):e20. doi: 10.1093/nar/gkt1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y, Ferguson JF, Xue C, Silverman IM, Gregory B, Reilly MP, Li M. Evaluating the Impact of Sequencing Depth on Transcriptome Profiling in Human Adipose. PLoS One. 2013;8(6):e66883. doi: 10.1371/journal.pone.0066883. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.