Abstract
Importance
The association of rare copy number variants (CNVs) with complex disorders is almost exclusively evaluated using clinically ascertained cohorts. As a result, the contribution of these genetic variants to cognitive phenotypes in the general population remains unclear.
Objectives
- To investigate the clinical features of genomic disorders in adult carriers without clinical pre-selection.
- To assess the genome-wide burden of rare CNVs on carriers’ educational attainment and intellectual disability prevalence in the general population.
Design, Setting, and Participants
The population biobank of Estonia (EGCUT) contains 52,000 participants, or 5% of the Estonian adults, enrolled in 2002-2010. General practitioners examined participants and filled out a questionnaire of health- and lifestyle-related questions, as well as reported diagnoses. As EGCUT is representative of the country's population, we investigated a random sample of 7877 individuals for CNV analysis and genotype-phenotype associations with education and disease traits.
Main Outcomes and Measures
Phenotypes of genomic disorders in the general population, prevalence of autosomal CNVs, and association of the latter variants with decreased educational attainment and increased prevalence of intellectual disability.
Results
We identified 56 carriers of genomic disorders. Their phenotypes are reminiscent of those described for carriers of identical rearrangements ascertained in clinical cohorts. We also generated a genome-wide map of rare (frequency ≤0.05%) autosomal CNVs and identified 10.5% of the screened general population (n=831) as carriers of CNVs ≥250kb. Carriers of deletions ≥250kb or duplications ≥1Mb show, compared to the Estonian population, a greater prevalence of intellectual disability (P=0.0015, OR=3.16, (95%CI: 1.51-5.98); P=0.0083, OR=3.67, (95%CI: 1.29-8.54), respectively), reduced mean education attainment (a proxy for intelligence; P=1.06e-04; P=5.024e-05, respectively) and an increased fraction of individuals not graduating from secondary school (P=0.005, OR=1.48 (95%CI: 1.12-1.95); P=0.0016, OR=1.89 (95%CI: 1.27-2.8), respectively). The deletions show evidence of enrichment for genes with a role in neurogenesis, cognition, learning, memory and behavior. Evidence for an association between rare CNVs and decreased educational attainment was confirmed by analyses in adult cohorts of Italian (HYPERGENES) and European American (Minnesota Center for Twin and Family Research) individuals, as well as in the Avon Longitudinal Study of Parents and Children (ALSPAC) birth cohort.
Conclusions and Relevance
Our results challenge the assumption that carriers of known syndromic CNVs identified in population cohorts are asymptomatic. They also indicate that individually rare but collectively common intermediate-size CNVs contribute to the variance in educational attainment. Refinements of these findings in additional population groups is warranted given the potential implications of this observation for genomics research, clinical care, and public health.
Keywords: genomic disorders, CNV, 16p11.2, population biobanks, education, intelligence, EGCUT, ALSPAC
Introduction
Recent studies showed that two human individuals differ on approximately 0.8% of their genome1. The Database of Genomic Variants catalogs ~2.4 million DNA copy number variants (CNVs), i.e. stretches of DNA that display altered copy-numbers, mapping to ~200,000 unique loci that cover 72% of the human genome2. With such an extent of genomic sequences concerned, CNVs contribute to inter-individual variation3-6. Large recurrent CNVs were found to be associated with complex disorders, particularly developmental delay and intellectual disability7,8 characterized by limited intellectual functioning and impaired adaptive behavior in everyday life. These CNVs are listed in DECIPHER, a database for genomic variants and phenotypes in humans9 and are often regrouped under the term “genomic disorder”8.
Since associations of large rare CNVs with pathologies were almost exclusively evaluated using clinically ascertained, often pediatric, cohorts, it is unclear whether these structural variants affect health in the general adult population. For example, the reports of seemingly asymptomatic (reviewed in10), but not fully evaluated, control individuals and transmitting parents underscored their possible incomplete penetrance. Here we investigated the phenotypic profiles of adult carriers of known pathological CNVs who were not clinically pre-selected and assessed the burden of rare intermediate-size autosomal CNVs on educational attainment and intellectual disability.
Methods
EGCUT cohort
The Estonian Genome Centre of the University of Tartu (EGCUT) cohort is a population biobank containing 5% of the Estonian adult population11. Samples have been collected in all 15 Estonian counties and diverse social groups by 454 general practitioners (GPs; i.e. 56% of the GPs in the registry of the Estonian Health Board). The age, sex and geographical distribution of the 52,000 participants closely reflect those of the Estonian adult population. The detailed description of the EGCUT cohort was previously published 11. At baseline, GPs performed a standardized objective examination of the participants and filled out a questionnaire that encompassed >1000 health- and lifestyle-related questions, as well as provided the diagnoses of diseases present in the medical history of the participating individual using the format of the WHO international classification of diseases (WHO ICD-10)11 (see details in eMethods). The data are continuously updated through periodic linking to national electronic health registries. The wide range of phenotypes, ages and social groups makes the cohort ideally suited to population-based studies. See eFigure 1 and eMethods for details on the EGCUT phenotype data. EGCUT is conducted according to the Estonian Human Genes Research Act and managed in conformity with the standard ISO 9001:2008. The Ethics Review Committee on Human Research of the University of Tartu approved the project. Written informed consent was obtained from all participants for the baseline and follow-up investigations.
The relevant phenotype traits of EGCUT individuals identified as carriers of DECIPHER-listed syndromic CNVs (eTable 1) were obtained from the baseline questionnaire and compared with the reviewed characteristics of corresponding syndromes (eTable 2). To further investigate the clinical features of adult carriers not clinically pre-selected, we invited back all 16p11.2 600kb BP4-BP5 (breakpoint) deletion and reciprocal duplication carriers identified in EGCUT for follow-up investigations. These CNVs were selected because of their relatively high prevalence and variable phenotype. These carriers were phenotyped using the standardized clinical and neuropsychological protocol we developed previously to specifically study 16p11.2 syndrome patients ascertained through clinical cohorts10,12. In agreement with the known population prevalence of 16p11.2 600kb BP4-BP5 CNVs12, we identified 4 deletion (0.05%) and 7 duplication carriers (0.09%) in the EGCUT set.
The EGCUT cohort (and Estonian population in general) is an outbred population with no substantial regional or ethnic differences. SNP allele frequencies and linkage disequilibrium patterns are similar to those found in populations with European ancestry13. We did not find small series of non-recurrent CNVs and/or inflation of recurrent rearrangements typical of founder effects14,15 (eMethods). Accordingly, EGCUT samples have been successfully used to discover or replicate hundreds of SNP associations, which are vulnerable to population frequencies and stratification differences (e.g.16-18). See eMethods and eFigure 2 for details on the Estonian population makeup and stratification.
CNV calling
The genomic DNA of 8110 subjects (7020 for discovery and 1090 for replication cohort; eTable 3), randomly selected among the 52,000 EGCUT participants, was subjected to CNV analysis. A third cohort of 1066 individuals (“high-functioning replication cohort”) was used to further assess the significance of the signal obtained regarding education attainment. SNP-genotyping and CNV calling were performed using Illumina platforms and the Hidden Markov Model-based software PennCNV according to the manufacturer's and developer's protocols 19, respectively. The 6819 discovery, 1058 replication and 993 “high-functioning” replication samples that passed the quality control parameters were retained (see eMethods for details).
Genotype-phenotype correlations
We analyzed the difference of studied phenotypes between CNV carriers and population. A two-sided Fisher's exact test and Welch two-sample t-test were used for statistical analysis in The R Project for Statistical Computing environment (http://www.r-project.org, R version 3.0.2). Odds ratios (OR), 95% confidence intervals and P-values were calculated; a threshold of P≤0.05 was set to indicate statistical significance. See eMethods for assessment of phenotype and determination of the prevalence of recurrent genomic syndromes. Briefly, intellectual disability (F70-79 of the WHO ICD-10) is defined in the DSM-IV as a deficit in overall cognitive functioning along with limitations in adaptive behavior. All diagnoses, including intellectual disability, were diagnosed according to diagnostic standards throughout the participant medical history and reported to the EGCUT database by the participant's GP at the moment of recruitment. Intellectual disability prevalence is estimated at 1-3% in developed countries20, which is consistent with the prevalence found in the EGCUT discovery cohort (1.7%).
Education levels were uniformly coded at the time of enrollment according to the Estonian education curriculum from 1 to 7, i.e. from less than primary school to scientific degree, respectively (details in eMethods). In both discovery and replication cohorts the mean education attainment (MEA) corresponded to secondary education (MEA=4.09 and 4.0, respectively) in agreement with the country's MEA. See eMethods for details on the Estonian population religiousness, school curriculum organization and education system performance.
Function of CNV-embedded genes
We used three previously published datasets to functionally annotate genes embedded in rare CNVs and assess if we could use those characteristics to predict CNV deleteriousness (eMethods): (i) the neurodevelopmental gene list21,22; (ii) the haploinsufficiency scores (HiS), i.e. the probability that a given gene maintains its normal function with only one functional copy23, and (iii) the list of ohnologs, i.e. paralogous genes resulting from ancestral whole-genome duplication events24. Since a CNV may preserve a gene's integrity yet indirectly affect it through changes in the copy-number of its regulatory elements4,5,25, we also tested the potential contributions of the latter by stratifying CNVs using the number of encompassed regulatory elements identified in 26 (eMethods). To further assess the functions of imbalanced genes we used Thomson Reuters MetaCore™, an integrated software suite for data-mining and pathway analysis based on a manually-curated biological knowledge database (eMethods).
ALSPAC cohort
The Avon Longitudinal Study of Parents and Children (ALSPAC) is a birth cohort based in Bristol, UK 27. 14,541 pregnant women with expected delivery dates between 1st April 1991 and 31st December 1992 were initially enrolled. 13,988 children who were alive at 1 year of age, and additional families were enrolled in later phases. Detailed phenotypic information on the children and their parents were collected during clinic visits and by completion of questionnaires, as well as from linkage with external data sources (eMethods). Ethical approval for the study was obtained from the ALSPAC Ethics and Law Committee and the Local Research Ethics Committees.
The Illumina HumanHap550 Quad platform was used to genotype 9912 children in ALSPAC. CNVs were called with PennCNV19. After quality control (eMethods), the subset of 5218 unrelated individuals with education information were retained for analysis (eTable 3). Log R Ratio (LRR) and B Allele Frequency (BAF) metrics were derived from raw data using published guidelines 28.
Within ALSPAC, educational attainment was assessed using data from the UK-based Key Stage 3 National Curriculum Tests in English and Mathematics, taken at age 13-14 years, also known as Standard Assessment Tests (SATs). A discrete level is awarded for these tests, but to further account for i) the exact mark received, and ii) the fact that the maximum and minimum level achievable for Mathematics was dependent upon the tier of examination for which the child was entered, results were scaled and adjusted as described previously 29,30. Due to non-normal distribution of the data, these two variables were then inverse-rank normal transformed, and then standardized. Furthermore, tertiles of the untransformed English and Mathematics scores were created (eTable 4). Differences in means of educational attainment according to rare CNV carrier status (frequency ≤0.05%) were compared using a Welch two-sided t-test. This was performed separately for each of the inverse-rank transformed, standardized English and Mathematics educational attainment scores. To obtain an interpretable estimate of effect univariable logistic regression models were assessed separately for English and Mathematics. The top tertile was coded as the reference group, and the bottom tertiles as the risk group. Separate odds ratios were estimated for membership of the risk group, comparing CNV carriers corresponding to increasing size groups against baseline (no large CNVs at a given frequency, 0.0005). The binary educational outcome was then regressed against CNV carrier status as an ordered variable, including all four size-categories, and the P value was reported as an assessment of trend.
MCTFR cohort
Participants from two studies conducted by the Minnesota Center for Twin and Family Research (MCTFR) were used as replication samples: the Sibling Interaction and Behavior Study (SIBS), and the Minnesota Twin Family Study (MTFS). MTFS is a longitudinal study of a community-based sample of same-sex twins born between 1972 and 1994 in the State of Minnesota (USA) and their parents 31. SIBS is an adoption study of sibling pairs and their parents32; its community-based sample contains families where both siblings are adopted, where both are biologically related to the parents, or where one is adopted and one is biologically related. In the current analyses, only a single random individual was selected for inclusion in analyses in order to create a dataset of unrelated participants (n=2390, eTable 3). The collection, genotyping, and analysis of DNA samples for both studies were approved by the University of Minnesota Institutional Review Board's Human Subjects Committee. Written informed assent or consent was obtained from all participants; parents provided written consent for their minor children.
Genotyping was performed using the Illumina 660W-Quad array. Whole-blood extracted DNA samples were only analyzed if the participant was i) white non-Hispanic and the standard deviation of the GC-corrected 33 autosomal log R ratios was less than 0.20. CNVs were called using PennCNV and then processed and filtered. Adjacent CNVs were merged if they had the same copy number and if the number of markers in the intervening gap was less than 20% of the number of total markers spanning the called CNVs. Rare (frequency ≤0.05%) deletions ≥250kb and duplications ≥1Mb were retained in the burden analysis.
Full-Scale IQ (FSIQ) was estimated using an abbreviated form of either the Wechsler Intelligence Scale for Children-Revised (WISC-R; for children 16 years and younger) or the Wechsler Adult Intelligence Scale-Revised (WAISR; for individuals older than 16). The short forms consisted of two Performance subtests (Block Design and Picture Arrangement) and Verbal subtests (Information and Vocabulary) and were prorated to determine FSIQ. Estimates from this short form have been shown to correlate 0.94 with FSIQ from the complete test 34. Samples with multiple FSIQ measurements were averaged together for analysis (mean=104.52±14.27; range=67-150).
Italian HYPERGENES cohort
The Italian follow-up is based on 451 individuals belonging to the cohort ascertained as controls for genome-wide association studies of hypertension (HYPERGENES)35 (eTable 3). Years of Schooling were defined in accordance with the ISCED 1997 classification, leading to seven categories of educational attainment that are internationally comparable (see details in eMethods). SNPs were genotyped using Illumina Human 1M-Duo BeadChips and CNVs called with PennCNV as for the discovery cohort. Differences in means of educational attainment were compared using a Welch two samples one-tailed t-test and Wilcoxon rank-sum test in R. Both tests returned comparable results.
Results
Prevalence and phenotypes of pathological CNVs in EGCUT
To investigate the medical burden of rare CNVs in the general population we opted for a genotype-first approach and analyzed a random sample cohort from EGCUT. Within a combined discovery and replication sample of 7877 unrelated individuals, we identified 56 carriers of known recurrent autosomal genomic disorders (0.7%; eTable 1). While the prevalence of each genomic disorder is lower than previously reported in clinical cohorts36,37, it is only slightly lower than the 67 individuals expected according to the reported population prevalence of the 57 autosomal syndromes listed in the DECIPHER database of genomic disorders9 (eTable 2, eTable 5 and eMethods). EGCUT is depleted (6 observed carriers/17 expected, P=0.03, OR=0.35, CI95%(0.11; 0.94)) of the most deleterious CNVs (graded 1-2 by DECIPHER), while the frequency of CNVs graded 3 and ungraded is as expected (50/50, P=1, OR=1, CI95%(0.66; 1.51)).
The clinical features of EGCUT carriers of DECIPHER-listed CNVs are comparable to those reported in disease cohorts. 31 (55%; including only formal diagnosis) and 39 out of 56 (70%; including self-reported problems) carriers recruited from the general population with no prior awareness of their genetic disorder present phenotypes previously associated with their genomic lesion in the literature (see eTable 1 for the phenotypes identified in the 56 EGCUT carriers and eTable 2 for phenotypes associated with DECIPHER-listed CNVs). For example, carriers of the 16p11.2 600kb BP4-BP5 deletions and reciprocal duplications identified in clinical cohorts show opposite phenotypes on body weight, head size and volume of specific cortico-striatal structures. They exhibit reduced full-scale intellectual quotient (FSIQ), as well as neuro-psychiatric problems and congenital abnormalities10,12,38-44. Correspondingly, the baseline questionnaires of the 4 deletion (cases no 41-44 in eTable 1) and 7 duplication (no 45-51) carriers identified in EGCUT indicated high and low body mass indexes, respectively, as well as neuropsychiatric traits, learning and developmental problems. The follow-up evaluation of these carriers uncovered additional similarities in the spectrum and severity distribution of phenotypic features found in 16p11.2 BP4-BP5 rearrangement carriers identified through pan-European recruitment via clinical genetics center (eTable 6).
Rare intermediate size CNVs and educational attainment
We then generated the genome-wide map of rare autosomal CNVs in the discovery set of 6819 individuals (eTable 3) and identified a total of 216 deletion and 509 duplication carriers (≥250kb with carrier frequency ≤0.05%; eTable 7). The underrepresentation of deletions compared to duplications (P=2.2e-16) is consistent with previous reports and concordant with the hypothesis that the former are more deleterious1,14. We found evidence for an association between carrier status and prevalence of intellectual disability (3.2% (n=23) in rare CNV carriers vs 1.7% (n=114) in EGCUT; P=0.007, OR=1.93 (95%CI: 1.17-3.06)). This effect was mainly driven by deletions (5.1% of intellectual disability, n=11; P=0.0015; OR=3.16 (95%CI: 1.51-5.98)) and remained even after exclusion of carriers of DECIPHER-listed CNVs (2.8% n=19; P=0.05, OR=1.64 (95%CI: 0.95-2.71), 8.9% of which were diagnosed with intellectual disability (n=4, P=0.0072, OR=5.74 (95%CI: 1.47-16.22)).
We next assessed the correlation between CNV size and intellectual disability, as it was previously reported that in comparison to controls, cohorts of affected patients show an excess of CNVs and that this excess is larger for longer CNVs7. The frequency of intellectual disability increases with CNV size (4.3% (n=6, P=0.033, OR=2.65 (95%CI 0.94-6.11)) in 250-500kb versus 8.3% for ≥1Mb deletions (n=36, P=0.023, OR=5.34 (95%CI: 1.03-17.42)), while associations with duplications are only detectable when rearrangements exceed 1Mb in size (5.9% n=102; P=0.0083, OR=3.67; (95%CI: 1.29-8.54); Table 1). Smaller deletions (125kb≤CNV<250kb; n=275) had no apparent impact on this prevalence (2.5% (n=7, P=0.24, OR=1.5 (95%CI: 0.59-3.28)).
Table 1.
Prevalence of intellectual disability diagnosis in EGCUT
| Cohort | Sample size | Intellectual disability prevalence (nb of cases) | OR (CI95%) | P-value |
|---|---|---|---|---|
| EGCUT all samples | 6819 | 1.7% (114) | ||
| DECIPHER CNV carriers | 45 | 8.9% % (4)** | 5.74 (1.47; 16.22) | 0.0072 |
| DEL carriers ≥ 1 Mb | 36 | 8.3% (3)* | 5.34 (1.03; 17.42) | 0.023 |
| DEL carriers ≥ 500 kb | 77 | 6.5% (5)** | 4.08 (1.26; 10.25) | 0.01 |
| DEL carriers ≥ 250 kb | 216 | 5.1% (11)** | 3.16 (1.51; 5.98) | 0.0015 |
| 500 kb ≤ DEL carriers < 1 Mb | 41 | 4.9% (2) | 3.02 (0.35; 11.9) | 0.1522 |
| 250 kb ≤ DEL carriers < 500 kb | 139 | 4.3% (6)* | 2.65 (0.94; 6.11) | 0.0326 |
| DUP carriers ≥ 1 Mb | 102 | 5.9% (6)** | 3.67 (1.29; 8.54) | 0.0083 |
| DUP carriers ≥ 500 kb | 235 | 3.4% (8) | 2.07 (0.86; 4.29) | 0.066 |
| DUP carriers ≥ 250 kb | 509 | 2.4% (12) | 1..42 (0.71; 2.6) | 0.285 |
| 500 kb ≤ DUP carriers < 1 Mb | 133 | 1.5% (2) | 0.87 (0.11; 3.38) | 1.0000 |
| 250 kb ≤ DUP carriers < 500 kb | 274 | 1.5% (4) | 0.87 (0.23; 2.32) | 1.0000 |
Carriers of rare deletions are indicated as DEL and rare duplications as DUP. The results are presented as cumulative or as size-separated groups. DECIPHER CNV correspond to all CNV listed within the DECIPHER database (see text for details).
Significant results are highlighted in bold; P-values ≤ 0.05, ≤ 0.01 and ≤ 0.001 are pinpointed by *, ** and *** respectively.
1For both deletions and duplications, ‘Controls’ are those individuals carrying neither a deletion nor a duplication ≥250kb.
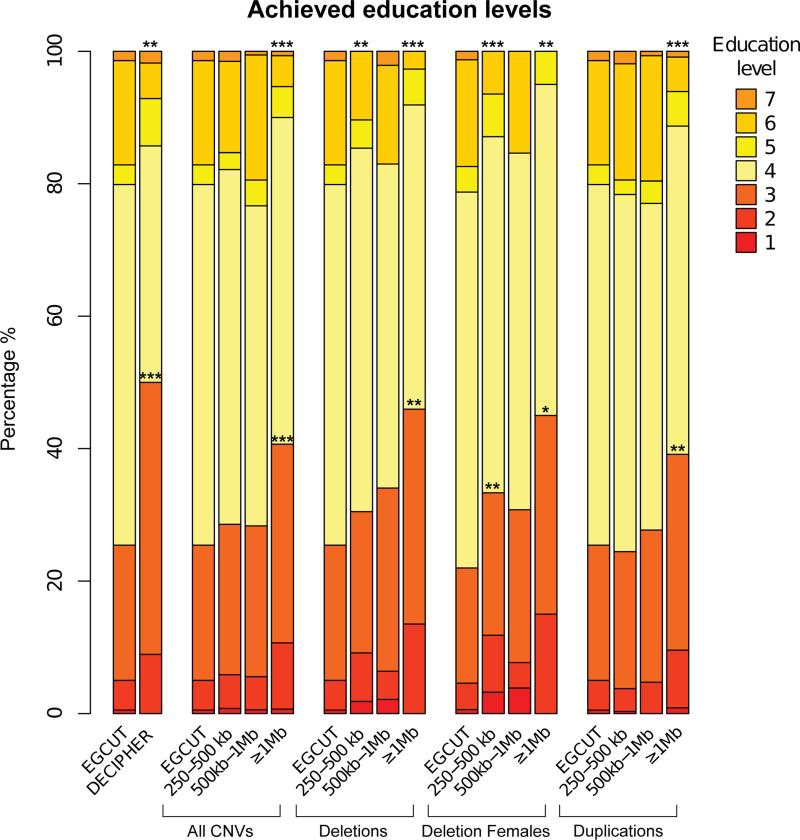
The diagnosis of intellectual disability is binary; thus to assess with greater granularity the effect of rare CNVs, we investigated if their occurrence and size are related to achieved education levels, a proxy for global cognition45,46. For this purpose we used the scale of seven sublevels of the Estonian education curriculum (eMethods). While 25.3% (n=1729) of sampled EGCUT individuals fail to complete secondary school (level 4; EGCUT mean education attainment (MEA)=4.09, similar to the Estonian population11), this proportion is higher in carriers of DECIPHER-listed genomic disorders with 48.9% (n=22) of them only reaching elementary or basic education (P=0.0008, OR=2.8 (95%CI: 1.49-5.3); MEA=3.71, P=0.028; Figure 1). The fraction of carriers that fail to reach secondary education increases with CNV size (e.g. ≥1Mb CNV carriers have a MEA=3.65 (P=4.6e-07) and 40.6% (n=56) of them do not complete secondary school (P=0.0001, OR=2.01 (95%CI: 1.40-2.87)); Figure 1). Deletions are responsible for the bulk of the outcome with MEA decreasing to 3.5 (P=0.0004) and 47.2% (n=17) of 1Mb deletion carriers not completing secondary education (P=0.006, OR=2.63 (95%CI: 1.28-5.36); Figure 1). A decrease is already seen in the 250-500kb CNV carrier group with MEA=3.86 (P=0.017) and 29.5% (n=41) of carriers not graduating from secondary school (P=0.28, OR=1.23 (95%CI: 0.83-1.80)). In agreement with the intellectual disability results, smaller deletions (125kb≤CNV<250kb: n=275) were not associated with changes in education attainment (MEA=4.11 (P=0.80), less than secondary education 26.2% (n=72, P=0.78, OR=1.04 (95%CI: 0.78-1.38))), while duplications were associated only with rearrangements ≥1Mb (MEA=3.71; P=0.00015 and 38.2% (n=39) of carriers failing to complete secondary school; P=0.0042, OR=1.82 (95%CI: 1.19-2.77); Figure 1).
Figure 1. Rare intermediate-size CNVs are associated with decreased education metrics.
The education attainment decreases with CNV size. The different panels compare the distribution of achieved education levels of the assessed general population (EGCUT) with carriers of DECIPHER-listed rearrangements (DECIPHER) or with carriers of CNVs (frequency ≤ 0.05%), deletions, deletion females and duplications segregated by size. Asterisks placed above the stacked columns specify different distributions, while the ones positioned at the boundary between basic and secondary education levels (levels 3 and 4, respectively) denote differences in the fraction of individuals who reached at least secondary education. P-values ≤ 0.05, ≤ 0.01 and ≤ 0.001 are indicated by *, ** and ***, respectively. The actual P-values are mentioned in the main text and Table 3. Education levels are coded according to the Estonian education curriculum: 1 - less than primary; 2 - primary; 3 – basic; 4 – secondary; 5 – professional higher/college; 6 – university/academic degree; 7 – scientific degree (see eMethods for details). Note that while 21.2% of EGCUT females (n=855) hold college or academic degrees, the presence of a rare deletion is associated with a decreased ability to achieve these highest education levels (del 250-499kb: 12.9% (n=12) of female with levels 5-7; P=0.05, OR=0.55 (95%CI:0.27-1.02)). For example, only one (a carrier of the 17p12 deletion causative for HNPP peripheral neuropathy, OMIM #162500) of 20 females carrying deletions ≥1Mb reached an education level above secondary school.
EGCUT ancestry principal components are not associated with CNV burden (eFigure 2), indicating that genetic stratification is likely not confounding the association with educational attainment. Likewise, differences in education possibilities due to religion or ethnicity could not account for the observed associations as the OECD “Program for International Student Assessment” and “for International Assessment of Adult Competencies” surveys showed that the “free education for all” Estonian system is among the best in the world in term of results and equal opportunity (eMethods).
Estonian replication
We conducted a replication of the education analysis on a non-overlapping random set of 1058 unrelated EGCUT individuals recruited similarly (eTable 3), but sampled at a different time-point and genotyped using a different array platform (replication cohort: MEA=4.00, 25.6% (n=271) failing to complete secondary school). In agreement with the discovery cohort, we noted a diminished education attainment in 250kb≤CNV<500kb deletion carriers (MEA=3.68, P=0.056; 36% (n=9) with only basic education or less, P=0.25, OR=1.63 (95%CI: 0.63-3.98)) and ≥1Mb duplication carriers (MEA=3.54, P=0.15; 46.2% (n=6), P=0.11, OR=2.49 (95%CI: 0.68-8.72). The joint analyses of these two random cohorts confirmed the negative effect of rare deletions ≥250kb (MEA=3.81; P=1.06e-04; less than secondary 33.5% (n=83); P=0.005, OR=1.48 (95%CI: 1.12-1.95) and duplications ≥1Mb (MEA=3.69; P=5.024e-05; less than secondary 39.1% (n=45); P=0.0016, OR=1.89 (95%CI: 1.27-2.8) on educational attainment (see full details in Figure 1 and Table 2). To challenge our results further, we then used a non-overlapping set of 993 unrelated individuals that, due to different ascertainment criteria (eTable 3 and eMethods), were biased towards higher than average socio-cognitive functioning (high-functioning replication cohort: MEA=4.77, lower than secondary education 9.4% (n=93)). Even in this group that is probably partially depleted of severe impact CNVs, we observe a lowering of the MEA of 250kb≤CNV<500kb deletion and ≥1Mb duplication carriers of the same order of magnitude (MEA=4.36, Δ=−0.41 and 4.44, Δ=−0.33, respectively). Combining both independent replication cohorts confirmed our results (replication cohorts MEA=4.36; 250kb≤CNV<500kb deletion carriers MEA=3.91 (P=0.004); ≥1Mb duplication: MEA=3.79 (P=0.057)); the same holds true if all three Estonian cohorts were analyzed together (eTable 8).
Table 2.
Education attainment in EGCUT (joint analysis of discovery and replication cohorts)
| Cohort | Sample size | MEA | P-value | Fraction not reaching secondary education (nb of case) | OR (CI95%) | P-value |
|---|---|---|---|---|---|---|
| EGCUT all samples | 7877 | 4.08 | 25.4% (2000) | |||
| DECIPHER CNV carriers | 56 | 3.64** | 0.003 | 50% (28)*** | 2.94 (1.67; 5.16) | 8.334e-05 |
| DEL carriers ≥ 1 Mb | 37 | 3.51*** | 0.0004 | 46% (17)** | 2.5 (1.23; 5.03) | 0.0072 |
| DEL carriers ≥ 500 kb | 84 | 3.75** | 0.0057 | 39.3% (33)** | 1.9 (1.18; 3.01) | 0.0054 |
| DEL carriers ≥ 250 kb | 248 | 3.81*** | 1.06-e04*** | 33.5% (83)** | 1.48 (1.12; 1.95) | 0.005 |
| 500 kb ≤ DEL carriers < 1 Mb | 47 | 3.93 | 0.383 | 34.0% (16) | 1.52 (0.77; 2.87) | 0.1803 |
| 250 kb ≤ DEL carriers < 500 kb | 164 | 3.84** | 0.0041 | 30.5% (50) | 1.29 (0.9; 1.82) | 0.1474 |
| DUP carriers ≥ 1 Mb | 115 | 3.69*** | 5.024e-05 | 39.1% (45)** | 1.89 (1.27; 2.8) | 0.0016 |
| DUP carriers ≥ 500 kb | 264 | 3.91* | 0.0174 | 32.6% (86)** | 1.42 (1.08; 1.86) | 0.01 |
| DUP carriers ≥ 250 kb | 583 | 4.04 | 0.493 | 28.1% (164) | 1.15 (0.95; 1.39) | 0.1536 |
| 500 kb ≤ DUP carriers < 1 Mb | 149 | 4.10 | 0.8186 | 28.1% (43) | 1.19 (0.81; 1.72) | 0.343 |
| 250 kb ≤ DUP carriers < 500 kb | 319 | 4.14 | 0.2951 | 24.5% (78) | 0.95 (0.72; 1.24) | 0.743 |
Carriers of rare deletions are indicated as DEL and rare duplications as DUP. The results are presented as cumulative or as size-separated groups. DECIPHER CNV correspond to all CNV listed within the DECIPHER database (see text for details).
Significant results are highlighted in bold; P-values ≤ 0.05, ≤ 0.01 and ≤ 0.001 are pinpointed by *, ** and *** respectively. Abbreviations: MEA – mean education attainment
ALSPAC, HYPERGENES and MCTFR follow-up
We sought to strengthen the inference from our results using the ALSPAC birth cohort and SATs scores at the age of 13-14 years as an alternative measure of education attainment (n=5218; eTable 3). When mean education attainment was studied using the transformed variables, Mathematics scores were decreased in carriers of rare intermediate-size deletions compared to controls (250kb≤CNV<500kb: Welch two-sided t-test comparing means P=0.019), and English scores were decreased in carriers of large deletions (≥1Mb, Welch two-sided t-test comparing means P=0.020)(eTable 9). Mean educational attainment in English and Mathematics was decreased in those who carried large duplications (≥1Mb; P=0.020 and P=0.049 respectively, Welch two-sided t-test). These results confirm the association between education attainment and rare CNVs using a different education metrics in a geographically distinct and differently ascertained cohort of adolescents.
Larger CNV size increased the odds of individuals belonging to the lowest tertile of SATs score (compared to the top, reference tertile) for both English and Mathematics. This was apparent both for carriers of deletions (English: 250kb≤CNV<500kb, OR 1.26 [95%CI 0.81, 1.95]; 500kb≤CNV<1Mb, OR 1.69 [95%CI 0.88-3.30]; ≥1Mb, OR 4.18 [95%CI 1.48, 14.87]; trend [p=0.002]; Mathematics: 250kb≤CNV<500kb, OR 1.42 [95%CI 0.91, 2.21]; 500kb≤CNV<1Mb, OR 2.21 [95%CI 1.01, 5.06]); ≥1Mb, OR 3.69 [95%CI 1.51, 10.29], trend [p=0.0002]) and duplications, albeit in this analysis, substantive evidence for an association of duplications and educational attainment was only observed for English results (English: 250kb≤CNV<500kb, OR 1.14 [95%CI 0.81, 1.61]; 500kb≤CNV<1Mb, OR 1.19 [95%CI 0.76,1.87]; ≥1Mb, OR 2.22 [95%CI 1.07, 4.84]; trend [p=0.035]; Mathematics: 250kb≤CNV<500kb, OR 1.10 [95%CI 0.78, 1.54]; 500kb≤CNV<1Mb, OR 1.03 [95%CI 0.68, 1.55]; ≥1Mb, OR 1.54 [95%CI 0.80, 3.01]; trend [p=0.273]) (Table 3).
Table 3.
Univariable logistic regression models for performance in the SATs assessment in ALSPAC CNV carriers
| Type of CNV | Exposure1 | English | Mathematics | ||||||
|---|---|---|---|---|---|---|---|---|---|
| OR | 95% LCI | 95% UCI | P value (trend) | OR | 95% LCI | 95% UCI | P value (trend) | ||
| Deletion | 250kb≤deletion<500kb vs controls | 1.26 | 0.81 | 1.95 | 0.002 | 1.42 | 0.91 | 2.21 | 0.0002 |
| 500kb≤deletion<1Mb vs controls | 1.69 | 0.88 | 3.30 | 2.21 | 1.01 | 5.06 | |||
| ≥1Mb deletion vs controls | 4.18 | 1.48 | 14.87 | 3.69 | 1.51 | 10.29 | |||
| Duplication | 250kb≤duplication<500kb vs controls | 1.14 | 0.81 | 1.61 | 0.035 | 1.10 | 0.78 | 1.54 | 0.273 |
| 500kb≤duplication<1Mb vs controls | 1.19 | 0.76 | 1.87 | 1.03 | 0.68 | 1.55 | |||
| ≥1Mb duplication vs controls | 2.22 | 1.07 | 4.84 | 1.54 | 0.80 | 3.01 | |||
Univariable logistic regression models for performance in the SATs assessment according to CNV status (carrier frequency ≤ 0.05%). Results are shown separately for English and Mathematics. For each subject, the top tertile was coded as the reference tertile, and the bottom tertile as the risk tertile. The exposure was CNV carrier status, divided into four size groups1. For each of the deletion and duplication analyses, odds ratios and 95% confidence intervals are presented, where each binary exposure is each CNV size group separately, in comparison to the control CNV group.1 In addition, a P-value estimating trend is presented. It was calculated by fitting an univariable logistic model, estimating odds ratio of the binary educational outcome per increase in CNV size group.1 OR=Odds ratio; 95% LCI=lower bound of 95% confidence interval, 95% UCI=upper bound of 95% confidence interval.
Our results were followed-up in two separate cohorts of healthy individuals with normal cognitive functioning (eMethods). Consistent with this ascertainment, both Italians and European Americans recruited for the HYPERGENES and MCTFR cohort, respectively, showed evidence of paucity of DECIPHER-listed CNVs [1 observed vs. 4 expected (P=0.37; OR=0.25, CI95% 0.005-2.53) and 14 vs. 20 (P=0.39, OR=0.7, 95%CI 0.32-1.46, respectively (eTable 2)]. Of note, the HYPERGENES analysis was restricted by small sample size (n=451; eTable 3) resulting in both a limited statistical power and limited CNV frequency calculation (≥0.25%). At this 5-fold higher level of prevalence, the MEA was reduced in carriers of deletion 500kb≤CNV<1Mb (ΔMEA=−0.26; P=0.39, Wilcoxon test) and the ≥1Mb duplications (ΔMEA=−0.66; P=0.11, Wilcoxon test)(eTable 10). A consistent, but similarly underpowered, decrease of FSIQ was found in MCTFR carriers of rare deletions (500kb≤CNV<1Mb: Δ=−4.23 IQ points, P=0.43; ≥1Mb: Δ=−13.82 IQ points, P=0.09) and duplications (500kb≤CNV<1Mb: Δ=−5.56, P=0.01; ≥1Mb: Δ=−6.03, P=0.16) (eTable 11).
Female mutation burden
In contrast to duplication carriers (male:female ratio=1.06 (303:285), we observe an excess of female carriers in every deletion size class ≥250kb separately and together within the combined Estonian discovery and replication cohort [M:F=0.78 (109:139); P=0.14, OR=1.22 (95%CI: 0.94-1.59)]. Female deletion carriers also show a more severe decrease in MEA than males in EGCUT [Female MEA=4.13 vs. Female del 250kb≤CNV<500kb MEA=3.71 (P=0.0003); Male MEA=4.02 vs Male del 250kb≤CNV<500kb MEA=4.00 (P=0.847); Figure 1). The joint analysis of the three Estonian cohorts confirmed that female deletion carriers are responsible for the majority of the decrease in education attainment (EGCUT combined female MEA=4.22, 250kb≤CNV<500kb deletion female MEA=3.71, P=3.9e-08; less than secondary education 20.3% (n=920), deletion female 33.6% (n=49), P=0.00024, OR=1.99 (95%CI: 1.37-2.85) (eTable 8). Consistent with the Estonian results in the MCTFR cohort deletions ≥500kb had a stronger effect on FSIQ in females (Δ=−13.73; P=0.03) compared to males (Δ=−0.12; p=0.98; eTable 11).
Assessment of CNV deleteriousness and function
Investigating the functions of the 642 protein-coding genes encompassed in the identified rare ≥250kb deletions, we found evidence of enrichment for genes with a role in neurogenesis, cognition, learning, memory and behavior (29 out of the top 50 GO processes with strongest evidence; all with FDR<2.45e-05; eTable 12). We then assessed if we could use gene characteristics to more accurately predict CNV deleteriousness. We stratified CNVs by the number of embedded i) protein-coding and non-coding genes, ii) neurodevelopmental (ND) genes22, iii) ohnologs24, or by iv) the sum of imbalanced genes’ probability score for haploinsufficiency (HiS)23, and v) the highest HiS in the CNV. A decrease of cognitive abilities was present in carriers of deletions encompassing ≥2 genes (MEA=3.82, P=0.003) and duplications including ≥11 genes (MEA=3.74, P=0.0003) (eFigure 3). When genes were present in the rearranged interval, deleteriousness was associated with the presence of at least one protein-coding gene (intellectual disability prevalence 5.3% (n=8), P=0.0046; OR=3.31 (95%CI: 1.37-6.93); MEA=3.79, P=0.0014; 33.3% (n=50) not reaching secondary education, P=0.029, OR=1.47 (95%CI: 1.02-2.1) in agreement with the observation that the majority of Mendelian pathogenic mutations disrupt coding sequences 47. Prevalence of intellectual disability was best correlated with the presence of at least one ND-gene in the deleted interval (prevalence 8.8% (n=6), P=0.001, OR=5.69 (95%CI: 1.97-13.47); MEA=3.76, P=0.03) and the sum of HiS (Highest quartile of HiS sums: 8.9% (n=4), P=0.0072, OR=5.74 (95%CI: 1.46-16.22; MEA=3.91, P=0.27). Presence of an ohnolog in the deletion is associated with a higher prevalence of intellectual disability, however to a lesser degree (5.9% (n=6), P=0.008, OR=3.7 (95%CI: 1.29-8.54). Neither separately nor together did the numbers of promoters, enhancers, transcriptional elements and insulators within a CNV correlate with intellectual disability and educational attainment.
Discussion
While various large pathogenic CNVs are known, the vast majority of rare CNVs of intermediate size (250-500kb) were thought to be non-deleterious. In the current report we show that the presence of both recurrent syndromic and rare intermediate-size non-recurrent CNVs, which are cumulatively frequent in the general population (10.5%), correlates positively with prevalence of intellectual disability and negatively with educational attainment. Our results are likely to be underestimated through i) exclusion of the most severely affected patients, ii) inclusion of patients with CNVs known to have no impact on cognition and iii) incorrect inclusion of carriers of large somatic/tumorigenic genomic lesions.
The link between impaired cognitive functioning and lower academic achievement in CNV carriers parallels the recognized correlation between health and education48. This health-education gradient was postulated to result from the combination of i) heritable factors impacting both traits, ii) poor early-life health that affects learning, and iii) health-related behaviors being modulated by education. While recurrent CNVs conferring risk of autism spectrum disorders or schizophrenia were associated with a decrease in IQ of individuals from the general population49 and phenotype mining of carriers of genomic variants in the Northern Finland 1966 Birth Cohort revealed an excess of lower IQ, school grade retention before age 14 and impaired hearing among individuals carrying deletions >500kb previously implicated in neurodevelopmental disorders14, both studies failed to recognize that other CNVs, in particular non-recurrent ones, were also associated with decreases in cognitive capabilities.
Although 40-80% of the variance in intelligence and 20-40% in educational attainment are explained by genetic factors 50-53 studies failed to find major contributors to this heritability. For example, three individual SNPs each with an approximate effect size of one month of schooling per allele have been identified in a GWAS encompassing >126,000 individuals (largest estimated effect = 0.02%) 17 and only a polygenic model including ~300,000 common SNPs genome-wide explained 28-29% of variation in general cognition 54. While earlier studies failed to identify common CNVs as major contributors to the above heritabilities 55-58, our results suggest that rare structural variants ≥250kb for deletions and ≥1Mb for duplications are associated with complex social-science traits in population cohorts. About 2% of the analyzed biobank participants carry a rare CNV ≥1Mb. Even without considering other health problems, a fifth of them appear to be linked with decreased life quality as the fraction reaching secondary education level is lowered by 15% when comparing CNV carriers to the general population. This reduction results in a MEA that is half a level lower. If we add to this fraction of rare ≥1Mb CNVs both the smaller intermediate-size CNVs associated with decreased educational attainment identified in this report (at least 0.2% of the population), and the highly pathogenic anomalies absent from EGCUT (0.15%), the life quality of 1 of 40 people might be negatively affected by rare CNVs. These variants may account for a sizable portion of the heritability of the complex “educational attainment” measure52.
The observed excess of females carrying rare genomic deletions supports the recently described female-biased mutational burden 21,59. Females appear “protected” from neurodevelopmental disorders. This potentially allows females to be enrolled in general population cohorts despite the fact that they carry rare CNVs, while their male counterparts who likely present more severe phenotypes are excluded from such recruitment. Consequently and corroboratively, female deletion carriers mostly drove the signal on education attainment.
While intellectual disability prevalence was increased with presence of a neurodevelopmental or ohnolog gene in the deleted interval or a high haploinsufficiency score of imbalanced genes, none of the assessed evaluators correctly capture the variation in education attainment, possibly because they are limited to protein-coding genes. Investigation of the function of the encompassed protein-coding genes revealed that they were enriched for genes involved in neurogenesis, cognition, learning, memory and behavior. This is consistent with the hypothesis that these rearrangements are rare because they impact genes important for neurodevelopment and thus are rapidly purged from the population.
While none of the carriers of known syndromic CNVs identified in EGCUT were previously diagnosed with a genetic disease, many suffered from major clinical problems (e.g. intellectual disability, congenital anomalies, neuropathies, neuropsychiatric disturbances, extreme obesity and reproductive problems). As the latter are most likely caused by the newly-found genetic alterations, it suggests that these individuals have escaped the attention of the medical genetics system and thus not received proper examination and counseling.
Conclusions
Our results suggest that population carriers of known syndromic CNVs identified in the general population are not asymptomatic. They also indicate that individually rare, but collectively common, intermediate-size CNVs negatively contribute to the variance in educational attainment. Validation of this finding in additional population groups is warranted given the potential implications of this observation for genomics research, clinical care, and public health.
Acknowledgments
Funding/Support: Katrin Männik is a grantee of a scholarship from the Swiss Scientific Exchange NMS Program. Anna Guyatt is funded by a PhD studentship from the Wellcome Trust [102433/Z/13/Z]. Sébastien Jacquemont is a Bursary Professor of the Swiss National Science Foundation (SNSF). This study is supported by two SNSF grants (Reymond, Kutalik), a specific 16p11.2 SNSF Sinergia grant (Reymond), the Simons Foundation Autism Research Initiative (SFARI274424)(Reymond), Leenaards Foundation Prizes (Jacquemont, Reymond, Kutalik), European Commission FP7 grants (278913, 306031, 313010) (Metspalu), Center of Excellence in Genomics (EXCEGEN) and University of Tartu (SP1GVARENG) (Metspalu), Estonian Research Council Grant IUT20-60 (Metspalu), USPHS Grants from the National Institute on Alcohol Abuse and Alcoholism (AA09367 and AA11886) (Pankratz), the National Institute on Drug Abuse (DA05147, DA13240, and DA024417) (Pankratz), and the National Institute on Mental Health (MH066140) (Pankratz). Nicholas Timpson, Tom Gaunt and Hashem Shihab work within the MRC Integrative Epidemiology Unit, supported by the Medical Research Council [MC_UU_12013/1-9].
Role of the Funder/Sponsor: None of the funders or sponsors had any role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
Author contributions: Alexandre Reymond had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Männik, Reymond
Acquisition of data: Männik, Mägi, Cole, Guyatt, Shihab, Maillard, Alavere, Kolk, Reigo, Mihailov, Leitsalu, Nõukas, Teumer, Salvi, Cusi, McGue, Iacono, Gaunt, Jacquemont, Pankratz, Timpson
Analysis or interpretation of data: Männik, Mägi, Macé, Cole, Guyatt, Shihab Ferreira, Reymond
Statistical analysis: Männik, Mägi, Macé, Cole, Guyatt, Shihab, Mihailov, Kutalik, Pankratz, Timpson
Drafting of the manuscript: Männik, Reymond
Critical revision of the manuscript for important intellectual content: Männik, Macé, Mägi, Cole, Guyatt, Shihab, Maillard, Reigo, Beckmann, Jacquemont, Kutalik, Pankratz, Timpson, Reymond,
Obtained funding: Metspalu, Reymond
Administrative, technical, or material support: Mihailov, Alavere, Metspalu, Reymond
Study supervision: Metspalu, Reymond
Conflict of Interest Disclosure: All authors have completed and submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. The authors declare no competing interests.
Additional Contributions: We express our gratitude to EGCUT participants. We thank the EGCUT personnel for their compensated assistance in recruiting, phenotyping, routing samples, genotyping and administrative responsibilities especially Mari Nelis, PhD, Lili Milani, PhD, Viljo Soo, Kairit Mikkel, Mari-Liis Tammesoo, MSc and Steven Smit. We are grateful to Sven Bergmann, PhD, University of Lausanne for comments on the manuscript. We are extremely grateful to all the families who took part in the ALSPAC study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses. We are grateful to Mary Ward for her advice on the use of the ALSPAC education data. The UK Medical Research Council and the Wellcome Trust (Grant ref: 102215/2/13/2) and the University of Bristol provide core support for ALSPAC. This publication is the work of the authors, who will serve as guarantors for the contents of this paper. GWAS data was generated by Sample Logistics and Genotyping Facilities at the Wellcome Trust Sanger Institute and LabCorp (Laboratory Corporation of America) using support from 23andMe. ALSPAC analyses were completed using computational facilities of the Advanced Computing Research Centre, University of Bristol - http://www.bris.ac.uk/acrc.
References
- 1.Conrad DF, Pinto D, Redon R, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010 Apr 1;464(7289):704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Macdonald JR, Ziman R, Yuen RK, Feuk L, Scherer SW. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic acids research. 2014 Jan 1;42(1):D986–992. doi: 10.1093/nar/gkt958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaignat E, Yahya-Graison EA, Henrichsen CN, et al. Copy number variation modifies expression time courses. Genome research. 2011 Jan;21(1):106–113. doi: 10.1101/gr.112748.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henrichsen CN, Chaignat E, Reymond A. Copy number variants, diseases and gene expression. Human molecular genetics. 2009 Apr 15;18(R1):R1–8. doi: 10.1093/hmg/ddp011. [DOI] [PubMed] [Google Scholar]
- 5.Henrichsen CN, Vinckenbosch N, Zollner S, et al. Segmental copy number variation shapes tissue transcriptomes. Nature genetics. 2009 Apr;41(4):424–429. doi: 10.1038/ng.345. [DOI] [PubMed] [Google Scholar]
- 6.Stranger BE, Forrest MS, Dunning M, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007 Feb 9;315(5813):848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper GM, Coe BP, Girirajan S, et al. A copy number variation morbidity map of developmental delay. Nature genetics. 2011 Sep;43(9):838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lupski JR. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends in genetics : TIG. 1998 Oct;14(10):417–422. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- 9.Swaminathan GJ, Bragin E, Chatzimichali EA, et al. DECIPHER: web-based, community resource for clinical interpretation of rare variants in developmental disorders. Human molecular genetics. 2012 Oct 15;21(R1):R37–44. doi: 10.1093/hmg/dds362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zufferey F, Sherr EH, Beckmann ND, et al. A 600 kb deletion syndrome at 16p11.2 leads to energy imbalance and neuropsychiatric disorders. Journal of medical genetics. 2012 Oct;49(10):660–668. doi: 10.1136/jmedgenet-2012-101203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leitsalu L, Haller T, Esko T, et al. Cohort Profile: Estonian Biobank of the Estonian Genome Center, University of Tartu. International journal of epidemiology. 2014 Feb 11; doi: 10.1093/ije/dyt268. [DOI] [PubMed] [Google Scholar]
- 12.Jacquemont S, Reymond A, Zufferey F, et al. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature. 2011 Oct 6;478(7367):97–102. doi: 10.1038/nature10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelis M, Esko T, Magi R, et al. Genetic structure of Europeans: a view from the North-East. PloS one. 2009;4(5):e5472. doi: 10.1371/journal.pone.0005472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pietilainen OP, Rehnstrom K, Jakkula E, et al. Phenotype mining in CNV carriers from a population cohort. Human molecular genetics. 2011 Jul 1;20(13):2686–2695. doi: 10.1093/hmg/ddr162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walters RG, Coin LJ, Ruokonen A, et al. Rare genomic structural variants in complex disease: lessons from the replication of associations with obesity. PloS one. 2013;8(3):e58048. doi: 10.1371/journal.pone.0058048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perry JR, Day F, Elks CE, et al. Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature. 2014 Oct 2;514(7520):92–97. doi: 10.1038/nature13545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rietveld CA, Medland SE, Derringer J, et al. GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science. 2013 Jun 21;340(6139):1467–1471. doi: 10.1126/science.1235488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wood AR, Esko T, Yang J, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nature genetics. 2014 Nov;46(11):1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang K, Li M, Hadley D, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome research. 2007 Nov;17(11):1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maulik PK, Mascarenhas MN, Mathers CD, Dua T, Saxena S. Prevalence of intellectual disability: a meta-analysis of population-based studies. Research in developmental disabilities. 2011 Mar-Apr;32(2):419–436. doi: 10.1016/j.ridd.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 21.Jacquemont S, Coe BP, Hersch M, et al. A higher mutational burden in females supports a “female protective model” in neurodevelopmental disorders. Am J Hum Genet. 2014 Mar 6;94(3):415–425. doi: 10.1016/j.ajhg.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krumm N, O'Roak BJ, Karakoc E, et al. Transmission disequilibrium of small CNVs in simplex autism. Am J Hum Genet. 2013 Oct 3;93(4):595–606. doi: 10.1016/j.ajhg.2013.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang N, Lee I, Marcotte EM, Hurles ME. Characterising and predicting haploinsufficiency in the human genome. PLoS genetics. 2010 Oct;6(10):e1001154. doi: 10.1371/journal.pgen.1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makino T, McLysaght A. Ohnologs in the human genome are dosage balanced and frequently associated with disease. Proceedings of the National Academy of Sciences of the United States of America. 2010 May 18;107(20):9270–9274. doi: 10.1073/pnas.0914697107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reymond A, Henrichsen CN, Harewood L, Merla G. Side effects of genome structural changes. Current opinion in genetics & development. 2007 Oct;17(5):381–386. doi: 10.1016/j.gde.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 26.Ernst J, Kheradpour P, Mikkelsen TS, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011 May 5;473(7345):43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boyd A, Golding J, Macleod J, et al. Cohort Profile: the 'children of the 90s'- -the index offspring of the Avon Longitudinal Study of Parents and Children. International journal of epidemiology. 2013 Feb;42(1):111–127. doi: 10.1093/ije/dys064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peiffer DA, Le JM, Steemers FJ, et al. High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome research. 2006 Sep;16(9):1136–1148. doi: 10.1101/gr.5402306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ward ME, McMahon G, St Pourcain B, et al. Genetic variation associated with differential educational attainment in adults has anticipated associations with school performance in children. PloS one. 2014;9(7):e100248. doi: 10.1371/journal.pone.0100248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levăcić R, Jenkins A, Vignoles A, Steele F, Allen R. Estimating the Relationship Between School Resources and Pupil Attainment at Key Stage 3. In: Skills DfEa., editor. DFES Research Report RR679. London: 2005. [Google Scholar]
- 31.Iacono WG, Carlson SR, Taylor J, Elkins IJ, McGue M. Behavioral disinhibition and the development of substance-use disorders: findings from the Minnesota Twin Family Study. Development and psychopathology. 1999;11(4):869–900. doi: 10.1017/s0954579499002369. Fall. [DOI] [PubMed] [Google Scholar]
- 32.McGue M, Keyes M, Sharma A, et al. The environments of adopted and non-adopted youth: evidence on range restriction from the Sibling Interaction and Behavior Study (SIBS). Behavior genetics. 2007 May;37(3):449–462. doi: 10.1007/s10519-007-9142-7. [DOI] [PubMed] [Google Scholar]
- 33.Diskin SJ, Li M, Hou C, et al. Adjustment of genomic waves in signal intensities from whole-genome SNP genotyping platforms. Nucleic acids research. 2008 Nov;36(19):e126. doi: 10.1093/nar/gkn556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sattler JM. Assessment of Children (Revised) W. B. Saunders Company; Philadelphia: 1974. [Google Scholar]
- 35.Salvi E, Kutalik Z, Glorioso N, et al. Genomewide association study using a high-density single nucleotide polymorphism array and case-control design identifies a novel essential hypertension susceptibility locus in the promoter region of endothelial NO synthase. Hypertension. 2012 Feb;59(2):248–255. doi: 10.1161/HYPERTENSIONAHA.111.181990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dittwald P, Gambin T, Szafranski P, et al. NAHR-mediated copy-number variants in a clinical population: mechanistic insights into both genomic disorders and Mendelizing traits. Genome research. 2013 Sep;23(9):1395–1409. doi: 10.1101/gr.152454.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaminsky EB, Kaul V, Paschall J, et al. An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities. Genetics in medicine : official journal of the American College of Medical Genetics. 2011 Sep;13(9):777–784. doi: 10.1097/GIM.0b013e31822c79f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bochukova EG, Huang N, Keogh J, et al. Large, rare chromosomal deletions associated with severe early-onset obesity. Nature. 2010 Feb 4;463(7281):666–670. doi: 10.1038/nature08689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walters RG, Jacquemont S, Valsesia A, et al. A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature. 2010 Feb 4;463(7281):671–675. doi: 10.1038/nature08727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shinawi M, Liu P, Kang SH, et al. Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. Journal of medical genetics. 2010 May;47(5):332–341. doi: 10.1136/jmg.2009.073015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weiss LA, Shen Y, Korn JM, et al. Association between microdeletion and microduplication at 16p11.2 and autism. The New England journal of medicine. 2008 Feb 14;358(7):667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 42.McCarthy SE, Makarov V, Kirov G, et al. Microduplications of 16p11.2 are associated with schizophrenia. Nature genetics. 2009 Nov;41(11):1223–1227. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Golzio C, Willer J, Talkowski ME, et al. KCTD13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11.2 copy number variant. Nature. 2012 May 17;485(7398):363–367. doi: 10.1038/nature11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maillard AM, Ruef A, Pizzagalli F, et al. The 16p11.2 locus modulates brain structures common to autism, schizophrenia and obesity. Molecular psychiatry. 2014 Nov 25; doi: 10.1038/mp.2014.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brody N. Intelligence, Schooling, and Society. Am. Psychol. 1997;52(10):1046–1050. [Google Scholar]
- 46.Matarazzo JD, Herman DO. Relationship of Education and IQ in the WAISR Standardization Sample. Journal of Consulting and Clinical Psychology. 1984;52(4):631–634. [Google Scholar]
- 47.Bamshad MJ, Ng SB, Bigham AW, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nature reviews. Genetics. 2011 Nov;12(11):745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 48.Deary IJ. Intelligence. Annual review of psychology. 2012;63:453–482. doi: 10.1146/annurev-psych-120710-100353. [DOI] [PubMed] [Google Scholar]
- 49.Stefansson H, Meyer-Lindenberg A, Steinberg S, et al. CNVs conferring risk of autism or schizophrenia affect cognition in controls. Nature. 2014 Jan 16;505(7483):361–366. doi: 10.1038/nature12818. [DOI] [PubMed] [Google Scholar]
- 50.Deary IJ, Penke L, Johnson W. The neuroscience of human intelligence differences. Nature reviews. Neuroscience. 2010 Mar;11(3):201–211. doi: 10.1038/nrn2793. [DOI] [PubMed] [Google Scholar]
- 51.Devlin B, Daniels M, Roeder K. The heritability of IQ. Nature. 1997 Jul 31;388(6641):468–471. doi: 10.1038/41319. [DOI] [PubMed] [Google Scholar]
- 52.Flint J, Munafo M. Genetics. Herit-ability. Science. 2013 Jun 21;340(6139):1416–1417. doi: 10.1126/science.1240684. [DOI] [PubMed] [Google Scholar]
- 53.Vinkhuyzen AA, van der Sluis S, Maes HH, Posthuma D. Reconsidering the heritability of intelligence in adulthood: taking assortative mating and cultural transmission into account. Behavior genetics. 2012 Mar;42(2):187–198. doi: 10.1007/s10519-011-9507-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davies G, Armstrong N, Bis JC, et al. Genetic contributions to variation in general cognitive function: a meta-analysis of genome-wide association studies in the CHARGE consortium (N=53 949). Molecular psychiatry. 2015 Feb;20(2):183–192. doi: 10.1038/mp.2014.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kirkpatrick RM, McGue M, Iacono WG, Miller MB, Basu S, Pankratz N. Low-Frequency Copy-Number Variants and General Cognitive Ability: No Evidence of Association. Intelligence. 2014 Jan 1;42:98–106. doi: 10.1016/j.intell.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McRae AF, Wright MJ, Hansell NK, Montgomery GW, Martin NG. No association between general cognitive ability and rare copy number variation. Behavior genetics. 2013 May;43(3):202–207. doi: 10.1007/s10519-013-9587-9. [DOI] [PubMed] [Google Scholar]
- 57.Need AC, Attix DK, McEvoy JM, et al. A genome-wide study of common SNPs and CNVs in cognitive performance in the CANTAB. Human molecular genetics. 2009 Dec 1;18(23):4650–4661. doi: 10.1093/hmg/ddp413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bagshaw AT, Horwood LJ, Liu Y, Fergusson DM, Sullivan PF, Kennedy MA. No effect of genome-wide copy number variation on measures of intelligence in a New Zealand birth cohort. PloS one. 2013;8(1):e55208. doi: 10.1371/journal.pone.0055208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Desachy G, Croen LA, Torres AR, et al. Increased female autosomal burden of rare copy number variants in human populations and in autism families. Molecular psychiatry. 2015 Feb;20(2):170–175. doi: 10.1038/mp.2014.179. [DOI] [PubMed] [Google Scholar]