Abstract
Different engineered organisms have been used to produce L-lactate. Poor yields of lactate at low pH and expensive downstream processing remain as bottlenecks. Aspergillus niger is a prolific citrate producer and a remarkably acid tolerant fungus. Neither a functional lactate dehydrogenase (LDH) from nor lactate production by A. niger is reported. Its genome was also investigated for the presence of a functional ldh. The endogenous A. niger citrate synthase promoter relevant to A. niger acidogenic metabolism was employed to drive constitutive expression of mouse lactate dehydrogenase (mldhA). An appraisal of different branches of the A. niger pyruvate node guided the choice of mldhA for heterologous expression. A high copy number transformant C12 strain, displaying highest LDH specific activity, was analyzed under different growth conditions. The C12 strain produced 7.7 g/l of extracellular L-lactate from 60 g/l of glucose, in non-neutralizing minimal media. Significantly, lactate and citrate accumulated under two different growth conditions. Already an established acidogenic platform, A. niger now promises to be a valuable host for lactate production.
Introduction
Lactic acid is a versatile organic acid used in various industrial applications. It is a commonly used acidulant and preservative in food, leather, textile industries and helps in controlled drug delivery. Demand for lactic acid has increased in recent years due to two emerging products—polylactide (a biodegradable plastic) and ethyl lactate (an environment friendly solvent). The L(+) form of lactate is used in food and pharma industries as humans can assimilate this isomer [1]. Industrial scale biosynthesis of L-lactic acid is achieved through fermentation of microorganisms belonging to Lactobacillus, Bacillus and Rhizopus genera. Lactobacillus species were the first promising candidate for lactic acid production. They are fastidious with respect to growth requirements and cannot utilize starchy raw materials. Hence the pretreatment of raw materials raises the cost of production. Their growth is inhibited at low pH during lactate fermentation and the addition of neutralizing agents makes downstream processing difficult [1]. Amongst filamentous fungi Rhizopus oryzae is known to naturally produce lactate. But lactate production using this fungus requires near neutral pH conditions and yields are compromised due to formation of ethanol and fumarate as by-products [2,3]. Different organisms have been engineered to improve process parameters and increase lactate yields. Among fungi, Saccharomyces cerevisiae [4–6], Kluyveromyces lactis [7,8], R. oryzae [9], Pichia stipites [10], Candida utilis [11] and Candida sonorensis [12] were genetically modified to produce L-lactate. Lactic acid production by most engineered hosts is hampered by the formation of high levels of by-products like ethanol (in yeast) and fumarate (in Rhizopus), compromised growth due to gene deletions, low pH tolerance, long fermentation time and inability to utilize different raw materials [1,13]. Thus, development of other production platforms to overcome some of these limitations continues to attract attention.
Aspergillus niger produces high levels of citrate and its acidogenic fermentation parameters are well established. Its saprophytic mode of nutrition allows for utilization of various raw materials and grows well at pH 3.0 or lower. These advantages along with GRAS (Generally Regarded as Safe) status make this organism an industrial favorite to produce citrate and gluconate [14]. With the advent of different genetic tools, A. niger has also been manipulated to produce oxalate [15], succinate [16] and itaconate [17–20]. A. niger is not a natural L-lactate producer and a functional lactate dehydrogenase (LDH) has not been reported in this fungus. Recently, two different Aspergillus species, albeit with indistinct acidogenic capacities, were engineered to produce lactate [21,22]. However, A. niger is a well known acid producer with better understood metabolic profile and a strong glycolytic flux [23]. The pyruvate so formed could be diverted to form L-lactate by expressing a suitable ldh gene. We chose and expressed the mouse LDH using a homologous strong constitutive A. niger citrate synthase promoter [24]. Such a transformant was capable of producing L-lactate aerobically on minimal media (Indian Patent Application No. 2077/MUM/2014). The advantages of A. niger as a lactate production platform are highlighted in this report.
Material and Methods
Strains and media
A. niger NCIM 565 (National collection of Industrial Microorganisms, National Chemical Laboratory-Pune, India) was the parent strain for this study. Different A. niger strains were maintained on potato dextrose (PDA) agar or yeast nitrogen base (without amino acids and ammonium sulfate), dextrose and ammonium nitrate (YDA) agar supplemented with DL- phosphinothricin (PPT) (YDA+PPT agar for bar transformants). The PPT (technical grade sample from Bayer Crop-Science; supplied as 45–50% aqueous solution, w/v) at 1.0 mg/ml of final concentration was used for selection of transformants. The bar marker from plasmid pCB1265 was employed to select A. niger transformants. A. niger mycelia were harvested from shake flask cultures grown in minimal medium (MM) [25] for protein extraction and lactate fermentation. The MM contained 20.0 g/l glucose, 3.0 g/l KH2PO4, 6.0 g/l Na2HPO4, 0.5 g/l MgSO4.7H2O, 2.25 g/l NH4NO3, 10 mg/l ZnSO4.7H2O, 3.0 mg/l MnSO4.7H2O, 1.5 mg/l Na2MoO4.H2O, 20.0 mg/l FeCl3.6H2O and 1.0 mg/l CuSO4.H2O. The medium pH was adjusted with 0.1 N HCl to 5.5–6.0. For citrate production, A. niger growth was carried out in acidogenic medium (AM) containing 140.0 g/l sucrose, 1.0 g/l KH2PO4, 0.1 g/l Fe(NH4)2(SO4)2, 2.25 g/l NH4NO3 and 0.25 g/l MgSO4.7H2O for eight days. A. niger strains were also grown in MM under conditions of hypoxia according to Terabayashi et al. [26].
Construction of heterologous and homologous ldh expression vectors
Mouse ldhA cDNA (NCBI accession number BC094019) was PCR amplified from a cDNA clone (pCMV from OriGene, USA), using the primers ldhNcF1 and ldhXbR1 (Table 1). The amplicon (999 bp DNA encoding a 332 residue protein) was cloned between NcoI and XbaI sites in pCBΔXCE [24] by replacing the EGFP CDS. The resultant plasmid pCBΔXCmldh consisted of PcitA-mldhA expression cassette along with the bar selection marker. The putative A. niger ldh sequence (corresponding to An04g08220 of A. niger CBS strain) was PCR amplified (primers AnldhNcF1 and AnldhXR1; Table 1) from A. niger NCIM 565 genomic DNA. This amplicon was cloned between NcoI and XbaI sites of pCBΔXCmldh thereby replacing the mouse ldhA cDNA to obtain pCBΔXCAnldh.
Table 1. List of primers used in this study.
| Primer | Sequence (5'-3') a | Description |
|---|---|---|
| ldhNcF1 | cccaccatggcaaccctcaaggacc | Mouse ldhA specific forward primer (NcoI site) |
| ldhXbR1 | ggtctagattagaactgcagctccttctgg | Mouse ldhA specific reverse primer (XbaI site) |
| AnldhNcF1 | gagaccatggcaacaatcgccttaatcg | A. niger ldhA specific forward primer (NcoI site) |
| AnldhXR1 | gctctagactacgaccctccagcatca | A. niger ldhA specific reverse primer (XbaI site) |
| CitF4 | ctaccgtgtcctgatataccc | A. niger PcitA specific forward primer |
| mldhR4 | caactgtaatcttgttctggg | Mouse ldhA reverse primer |
| actAF | ttgcggtacagcctccattg | A. niger actA forward primer |
| actAR | cgcttggactgtgcctcatc | A. niger actA reverse primer |
aRestriction enzyme recognition sites are in bold.
PCR reactions in 100 μl contained—3.5mM MgCl2, 250 μM dNTP’s, 0.5 μM primers and 5 units of Pfu polymerase (MBI Fermentas, St. Leon-Rot, Germany). Cloning and propagation of plasmids was done in Escherichia coli XL1-Blue (Stratagene, CA, USA), according to standard procedures [27].
A. niger transformation and characterization of transformants
A. niger was transformed and the A. niger transformants were selected on YDA+PPT agar [24]. Genomic DNA was isolated from mycelia using QIAGEN DNeasy Plant Mini kit. PcitA-mldhA copy number in the genome of A. niger mldh transformants was determined by qPCR (Stratagene Mx3000P; Agilent Technologies). The PcitA-mldhA fragment was amplified with CitF4 and mldhR4 primer pair (Table 1). The single copy A. niger actin gene served as a reference and was amplified with actAF and actAR primers (Table 1). Amplification efficiencies were determined using serial dilutions (0.1–10 ng/μl) of genomic DNA. The qPCR reactions (in 20 μl) were performed in triplicates and each reaction mixture contained 10 μl 2× Brilliant III Ultra fastSYBR Green master mix (Agilent Technologies), 3 μl suitably diluted genomic DNA and 50 nM each of forward and reverse primers. The qPCR conditions were as follows: initial denaturation at 95°C for 3 min followed by 40 cycles of 95°C for 20 s, 61°C for 30 s. Melt curve analysis was performed to check for the presence of primer dimers and/or nonspecific products. The copy number was calculated according to Skulj et al. [28].
cDNA preparation
Total RNA from A. niger was prepared using QIAGEN RNeasy Plant Mini kit. A. niger ldhA cDNA was prepared from total RNA using Transcription first strand cDNA synthesis kit (Roche Diagnostics India Pvt. Ltd.). Oligo(dT) primers were used to generate cDNA which was subsequently purified using cDNA purification column (Invitrogen). The amplification of A. niger ldhA cDNA was done with gene specific primers AnldhNcF1 and AnldhXR1.
Preparation of cell-free extract and lactate dehydrogenase assay
A. niger mycelial proteins were extracted with 2 volumes (cell wet weight:volume) of ice-cold extraction buffer (200 mM Tris-HCl pH 8.0, 1 mM 2-mercaptoethanol and 1 mM phenylmethanesulfonylfluoride). The cell-free extract was clarified at 12400×g for 20 min in a refrigerated centrifuge. For LDH assays, the final supernatant was desalted (on Sephadex G-25 column) before use.
The LDH activity was determined in the direction of lactate oxidation, by measuring the NADH formed at 340 nm. The assay mixture composed of 100 mM Tris-HCl buffer (pH 8.0), 200 mM sodium L-lactate, 2.5 mM NAD+ and suitably diluted A. niger cell-free extract. One unit of enzyme activity corresponds to the amount of enzyme that generates 1.0 nmole of NADH per min. The LDH activity was also monitored in the direction of pyruvate reduction (1.0 mM sodium pyruvate, 0.1 mM NADH in the same buffer as mentioned above, but at pH 7.4), when required. The LDH activity staining protocol was essentially as that for glutamate dehydrogenase [25], but using lactate and NAD+ as substrates.
Analytical methods
L-Lactate, citrate and glucose were estimated enzymatically. The assay mixture for lactate estimation consisted of 1.0 M glycine buffer (pH 9.5), 0.4 M hydrazine, 2.5 mM NAD+, 10 units of rabbit muscle L-LDH (Sigma Aldrich) and a suitable aliquot of the lactate sample. The mixture was incubated at 37°C for 60 min and absorbance at 340 nm was recorded. Appropriate enzyme blanks were included and L-lactate concentration was calculated using a standard curve. The lactate concentration was determined in millimolar (mM) units and 1 mM corresponds to 0.09 g/l. Citrate was estimated using citrate lyase (from Aerobacter aerogenes; Roche Diagnostics India Pvt. Ltd.) based on established procedure [29]. After an initial 15 min incubation with phenylhydrazine, the citrate sample was incubated for further 15 min with citrate lyase assay buffer. The absorbance of oxaloacetate phenylhydrazone was measured at 330 nm. Appropriate enzyme blanks were used to account for endogenous oxaloacetate contribution. Glucose was estimated using glucose oxidase-peroxidase reagent (Biolab Diagnostics Pvt. Ltd., India).
For intracellular lactate and citrate estimations, the A. niger mycelia were harvested by filtering through muslin cloth, washed with 3–5 volumes of distilled water, blotted dry and frozen immediately in liquid nitrogen. The whole operation was concluded within couple of minutes after harvest. The mycelia were crushed in liquid nitrogen and suitable buffer to obtain cell-free extracts. The cell-free extracts were treated with perchloric acid: methanol: water (8:40:52) mixture (10% of sample volume) to precipitate proteins and the precipitate cleared by centrifugation (10000×g for 5 min). Clear supernatants were neutralized with 1.0 M KOH before estimating the lactate/citrate present. The protocol employed here takes a few minutes and may not rapidly quench the mycelial metabolism [30]; to this extent, intracellular lactate and citrate measurements may be biased. However, their extracellular levels were better assessed as the samples were quickly filtered to remove mycelia and frozen. Intracellular concentration of lactate/citrate was calculated assuming an intracellular volume of 0.233 ml/g wet weight of A. niger mycelia [31].
Unless otherwise mentioned, all enzyme activity data and the metabolite measurements are representative of at least three separate experiments (performed in duplicates). Error bars represent the standard deviation.
Shake flask culture for lactate fermentation
The A. niger NCIM 565 and ldhA transformants were grown in MM supplemented with different glucose concentrations (1–10 percent). CaCO3 (30 g/l) was added to MM for growth under neutralizing conditions. Different A. niger strains were inoculated (108 spores/100 ml) in 200 ml medium (in 1000 ml Erlenmeyer flasks) and the lactate levels were monitored for 7 days.
Results and Discussion
Search for a functional ldh locus in A. niger
Use of A. niger as a host to produce lactate requires that an endogenous or heterologous ldh gene be expressed. Presence of a functional LDH is not reported in A. niger. Marginal but reproducible lactate oxidation activity was observed in the parent strain suggesting that an endogenous LDH-like enzyme may be present in A. niger. Lactate (in μM range) was also detected in the culture media under hypoxic growth of a few Aspergilli [26]. Putative ldh loci were identified from in silico analysis of published genome sequences of Aspergilli, including A. niger [32,33]. In this context, functional annotation of A. niger ldh and exploring its role in L-lactate formation is important. Five out of 272 hits in A. niger ATCC 1015 JGI genome database (v4.0) are annotated as putative NAD-dependent lactate/malate dehydrogenase (Table 2); others are either putative cytochrome-dependent dehydrogenases or hypothetical proteins [34]. A blastp analysis, with three different ldhA sequences (R. oryzae, AAF74436.1; A. nidulans, AN5842; mouse, BC094019) as queries, also retrieved one of these five putative ldh from JGI genome database (JGI protein id: 1094595, Table 2). Further, this sequence corresponds to An04g08220 locus (putative ldhA) in A. niger CBS genome [32,33]. This putative ldhA sequence contains one intron and shows around 30% amino acid sequence similarity with R. oryzae LDH (isoform A). The NADH binding sites are conserved in this putative ldh from A. niger; but the LDH signature sequence at the active site (VGVRDSES) is not well conserved. The catalytically important H residue is replaced by an R residue. This active site His (H193 of mouse LDH) is conserved in all functional LDHs [35]. This residue is also conserved in putative LDH from other Aspergilli except for A. niger and A. fumigatus (S1 Fig). The other putative NAD-dependent lactate/malate dehydrogenases (Table 2) do contain this H residue. However, a comparison of their full length protein sequences suggests they may be malate dehydrogenases and hence they were not considered.
Table 2. Putative ldh sequences found in A. niger ATCC 1015 JGI genome database. a .
| JGI protein id | Location | Genbank (CBS strain) | Putative role | Similarity | |
|---|---|---|---|---|---|
| R. oryzae LDH | Mouse LDH | ||||
| 1143375 | chr_304:32072–33915 (-) | CAK42176.1 | NAD/NADP lactate/malate dehydrogenase | Score: 50.1 Identity: 24% Coverage: 93% | Score: 66.2 Identity: 27%Coverage: 96% |
| 48047 | chr_701:1791847–1793109 (-) | CAK40769.1 | NAD/NADP lactate/malate dehydrogenase | Score: 43.9 Identity: 24% Coverage: 68% | Score: 43.5 Identity: 24% Coverage: 87% |
| 1094595 | chr_603:61034–62047 (-) | CAK44788.1 | NAD/NADP lactate/malate dehydrogenase | Score: 165 Identity: 36% Coverage: 91% | Score: 105 Identity: 27% Coverage: 90% |
| 1143842 | chr_401:461235–462893 (-) | CAK39307.1 | NAD/NADP lactate/malate dehydrogenase | Score: 61.6 Identity: 24% Coverage: 96% | Score: 55.5 Identity: 25% Coverage: 91% |
| 1156951 | chr_301:246807–247963 (+) | CAK39136.1 | NAD/NADP malate/lactate dehydrogenase | Score: 15.4 Identity: 31% Coverage: 19% | Score: 16.2 Identity: 57% Coverage: 1% |
a JGI protein models were identified by searching the A. niger ATCC 1015 JGI genome database (v4.0) with the keyword ‘L-lactate dehydrogenase’ and the putative NAD-dependent lactate/malate dehydrogenase were identified based on NAD/NADP binding domains.
A functional annotation of the putative ldhA ORF (JGI protein id: 1094595) was attempted. The genomic clone, containing a predicted 90 nt intron and encoding a 308 residue protein, was fused to the A. niger citrate synthase promoter (PcitA) (pCBΔXCAnldh) for constitutive expression in A. niger. Around twenty transformants (through bar selection) were obtained and the integration of PcitA-Anldh cassette was confirmed by genomic PCR. None of these transformants displayed significant increase in their LDH activity (activity was comparable to that of the parent strain). The functionality of this ORF was also tested by expressing the corresponding cDNA in E. coli. For this, cDNA was prepared from one of the A. niger transformants (the intron was spliced as expected), cloned in pET-28a and expressed in E. coli BL21 (DE3). However, the expressed protein was not active on different substrate combinations like L-lactate, D/L-lactate, ethanol, NAD+ and NADP+ (data not shown). An R174H site directed mutant was constructed to see if this made the putative A. niger LDH functional. This mutant protein also did not show any LDH activity. The putative ldhA in A. niger (JGI protein id: 1094595) therefore does not code for a functional LDH in E. coli.
Expression of heterologous mouse ldhA (mldhA) in A. niger
Since attempts to identify functional ldh gene(s) from A. niger genome were unsuccessful, a heterologous ldh sequence was expressed to divert the carbon flux from pyruvate towards lactate in A. niger. A range of ldh sources have been evaluated for fungal lactate production since the optimal carbon distribution at the pyruvate node is dependent upon the properties of LDH employed [12,36,37]. In the earlier two reports with Aspergilli, bovine and R. oryzae ldhA genes were used to achieve LDH expression [21,22]. However, the A. oryzae strains constructed to express R. oryzae ldhA genes did not accumulate detectable amounts of lactate [21]. Expressing LDH with appropriate kinetic features is desirable for better lactate yields. Different branches of the A. niger pyruvate node were scrutinized and the kinetic and regulatory properties of different enzymes at this node were analyzed. With micromolar intracellular pyruvate levels (40 μM, [30]) and correspondingly low Km values for different enzymes (Fig 1), choice of an LDH with low Km for pyruvate and high Ki for lactate was obvious. Accordingly, mouse ldhA (mldhA) having low K m for pyruvate (0.13 mM) was chosen for expression. Promoters of glycolytic pathway enzymes namely, Ppki and PgpdA were employed earlier to manipulate oxalate and itaconate production in A. niger, respectively [15,20]. However, ldh expression for lactate production in fungi was earlier achieved through various promoters like yeast PDC1 [11,36], A. oryzae sodM [21] and A. nidulans gpdA [22] genes. The fact that A. niger is a major citrate producer with a high glycolytic flux makes its own citrate synthase promoter (PcitA) an ideal pick. The strong constitutive A. niger PcitA was also selected for as it was active throughout growth on MM and AM [24]. The compact PcitA (0.5 kb) was used successfully to express both homologous and heterologous CDSs in A. niger [24,38]. Further, it is comparable in strength to the well established promoters like PglaA and Pgpd [39]. Not surprisingly, the choice of mldhA in combination with PcitA for LDH expression resulted in good titers of lactic acid (see below).
Fig 1. Metabolic fates of pyruvate in A. niger.
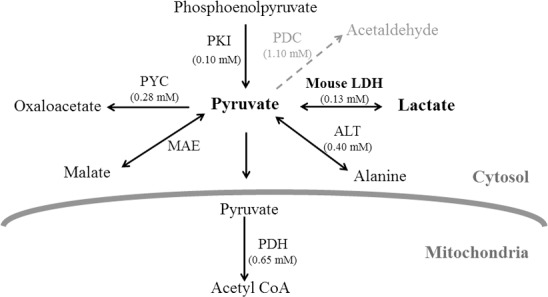
All enzymes catalyzing the reactions with pyruvate (as substrate or product) are depicted and the pathway shown with dotted arrow is from A. nidulans. ALT- alanine transaminase, LDH- lactate dehydrogenase [40], MAE- malic enzyme [41], PDC- pyruvate decarboxylase, PDH- pyruvate dehydrogenase complex, PKI- pyruvate kinase [42] and PYC- pyruvate carboxylase [43]. Respective K m values for the enzymes are given in brackets. These are either taken from Aspergillus literature or for the corresponding yeast enzymes from BRENDA (www.brenda-enzymes.org).
The plasmid pCBΔXCmldh (bearing PcitA-mldhA gene construct, see Material and methods section) was linearized and used to transform A. niger NCIM 565. Most of the bar + transformants displayed significantly higher LDH specific activity (in cell-free extracts) than the parent strain. No extracellular (spent medium) LDH activity was found in any of these transformants. The pyruvate reduction rates were always higher than the corresponding lactate oxidation rates; lactate oxidation is a better measure of specific LDH activity. Negligible yet consistent lactate oxidation activity was observed in the parent strain; that the rudimentary activity may be due to endogenous LDH cannot be excluded at this point. That A. niger may contain an endogenous LDH (or LDH-like) activity is discussed below.
Six bar + transformants displaying a range of LDH specific activities (two highest- D5, C12; two intermediate- C2, C7; and two basal- C16, C3) were selected for further analysis (Table 3). The enzyme activity staining results were consistent with the corresponding LDH specific activities measured for these six transformants (Fig 2A). The integration of PcitA-mldhA DNA into the genome of the transformants was confirmed by genomic PCR. A 1.0 kb amplicon (with primers citF4 and ldhXR1) was obtained for all the transformants except C3 (Fig 2B); this is consistent with the corresponding LDH activity data (Table 3). Integrated DNA could also be picked up in Southern blots (not shown). Since the DNA integration events are random, the observed differences in LDH specific activity of the transformants may reflect either copy number effects or locus specific transcriptional effects. Therefore, PcitA-mldhA copy number in the genome of these A. niger transformants was determined by qPCR (Fig 2C). All the transformants (except C3 strain, which was however bar +) had at least one copy of PcitA-mldhA cassette integrated in their genome (Table 3). There was no linear correlation between the copy number integrated and LDH specific activity. Transformants with only one copy of PcitA-mldhA integrated (C7 and C16 strains) had different LDH specific activities. The D5 transformant with 4 copies of PcitA-mdlhA showed higher LDH specific activity than C12 and C2 transformants having 11 and 13 copies of PcitA-mdlhA, respectively. This could possibly be the effect of integration context on expression levels. While marginal LDH (lactate oxidation) activity was observed in the parent strain, as expected, it was devoid of the PcitA-mldhA cassette. Similar analysis in the case of A. oryzae ldh transformants is not available [21]. The LDH activity in A. brasiliensis ldh transformants increased with ldhA gene copy number up to 6 copies. But LDH activity gradually decreased as the gene copy increased above 6 [22].
Table 3. LDH specific activity of the A. niger mldhA transformants.
| Strain | Activity (U) a | Specific activity (U/mg protein) | mldhA copy number |
|---|---|---|---|
| D5 | 360.5 ± 35.7 | 171.7 ± 15.0 | 4 |
| C12 | 302.7 ± 14.4 | 137.0 ± 14.0 | 11 |
| C2 | 180.2 ± 31.2 | 81.3 ± 6.5 | 13 |
| C7 | 86.7 ± 16.8 | 40.0 ± 2.0 | 1 |
| C16 | 18.8 ± 6.9 | 8.3 ± 3.1 | 1 |
| C3 | 2.0 ± 1.1 | 0.9 ± 0.7 | 0 |
| Parent | 4.8 ± 4.6 | 2.3 ± 1.5 | 0 |
a LDH was assayed in the direction of lactate oxidation.
Fig 2. Analysis of mldhA transformants by activity staining, genomic PCR and qPCR: a—Activity staining for LDH from A. niger and six of its transformants.
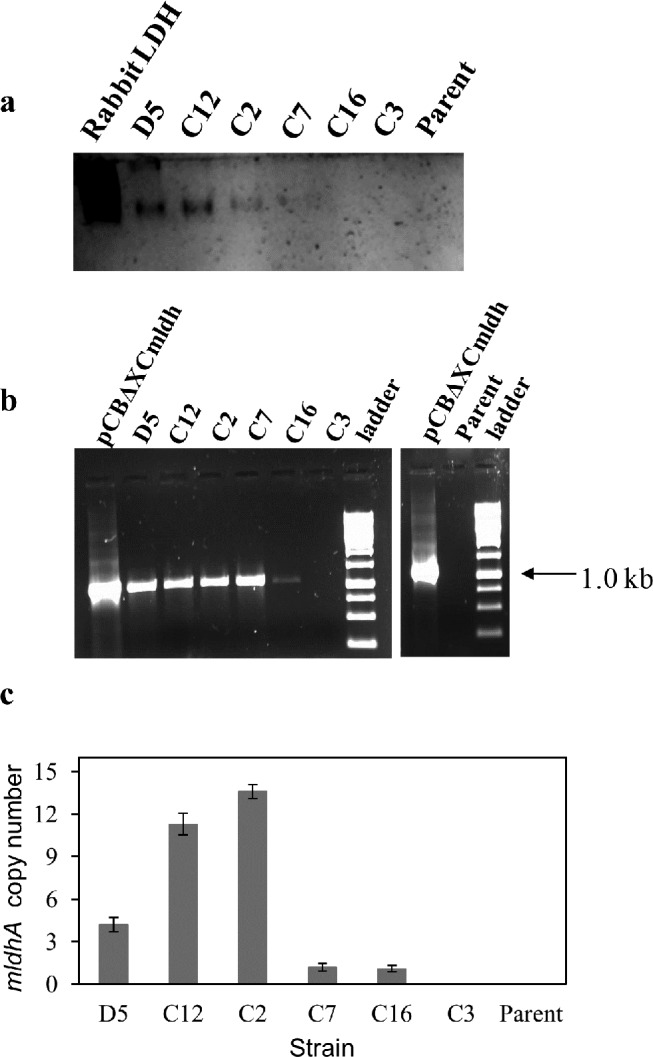
Desalted cell-free extract (5 μg protein each) was loaded on polyacrylamide gels. Purified rabbit skeletal muscle LDH served as positive control. b—PCR amplification of integrated PcitA- mldhA DNA from the transformants. Parent A. niger genomic DNA and pCBXCmldh served as negative and positive controls, respectively.c—PcitA-mldhA copy number determination in transformants by qPCR. The mldhA copy number was determined using the single copy A. niger actin gene as reference. The data are plotted as mean values obtained from different concentrations of genomic DNA. Error bars represent the standard deviation.
Lactate formation in mldhA transformants
The parent A. niger NCIM 565 strain is incapable of making L-lactic acid and if any, sub-micromolar levels of lactate are detected in its cell-free extracts. With the exception of C3 strain, L-lactate was detected in the spent medium (extracellular) of all the transformants grown in MM (without pH regulation) for 24 h (Fig 3). Intracellular lactate (in the cell-free extracts) was also detected in these transformants and the levels were invariably higher than those found in the spent medium. The levels of citrate in all the transformants however remain unchanged. In general, lactate levels were positively correlated with the LDH specific activity (Fig 3), and not the mldhA copy number (see Table 3), of the transformants. The transformant D5 is an exception in that, despite the highest LDH specific activity (albeit with only 4 gene copies integrated), it had lower lactate levels when compared with C12 strain. Strain D5 also conidiated poorly on PDA plates. Increasing LDH specific activity in transgenic S. cerevisiae [37], C. sonorensis [12] and A. brasiliensis [22] had a positive impact on lactate yield. The C12 transformant strain of A. niger showed maximal extracellular lactate levels on MM containing 1.0 percent glucose (Fig 3). This transformant is deposited in the Microbial Culture Collection at NCCS, Pune, India (Accession number MCC0019). The C12 strain was further tested under different growth conditions for lactate production.
Fig 3. LDH specific activity and lactate and citrate levels in mldhA transformants: Extracellular (Panel a) and intracellular (Panel b) concentrations of L-lactate (filled circles) and citrate (open circles) are shown.
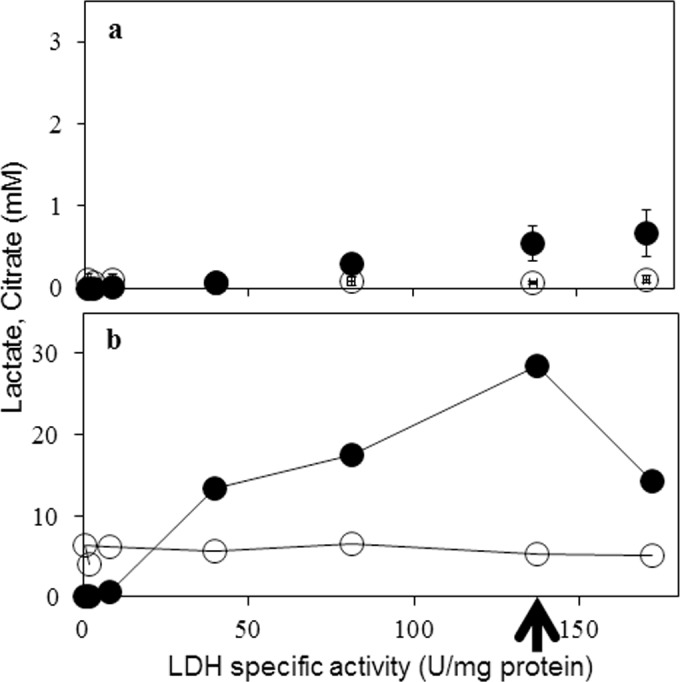
Data in panel b is an average of two measurements. Each LDH specific activity data point represents one individual transformant (see Table 3). Arrow indicates the data for C12 strain.
Lactate formation by the transgenic A. niger C12 strain
Growth on different glucose concentrations
The effect of increasing initial glucose concentrations on lactate production by the C12 strain was studied. The LDH specific activity in this strain was not significantly different (range 130–180 U/mg protein) when grown for 24 h on MM with different glucose amounts (1 to 10 percent); whereas the corresponding LDH specific activity of the parent strain ranged between 8–17 U/mg of protein. Lactate was detected both in the spent medium (extracellular) and in the cell-free extracts (intracellular) of C12 strain (Fig 4). The lactate levels were unaffected by increased glucose in the growth medium. Intracellular lactate levels after 24 h growth were invariably higher (between 30–40 mM) than those found in the spent medium (around 1.5 mM). It was also instructive to measure citrate levels as A. niger is a recognized citrate producer. While the intracellular citrate remained steady (between 7–9 mM), its levels increased in the spent medium with increasing glucose; similar results for citrate were obtained with the parent A. niger strain (Fig 4). Detection of TCA cycle intermediates (malate and succinate) were reported with transgenic A. oryzae and it was suggested that excess glucose may induce pyruvate run-off into the TCA cycle [21].
Fig 4. Effect of increasing medium glucose on the formation of lactate and citrate by A. niger C12 strain: Extracellular (Panel a) and intracellular (Panel b) concentrations of the two acids from C12 strain (circles) and the parent (squares) are shown.
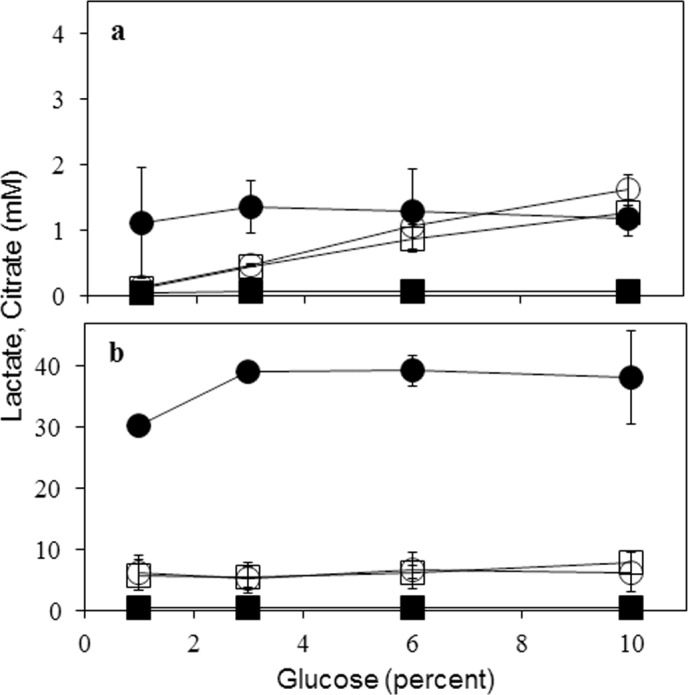
Filled symbols represent L-lactate and open symbols represent citrate. Data are for 24 h growth.
Lactate fermentation on MM
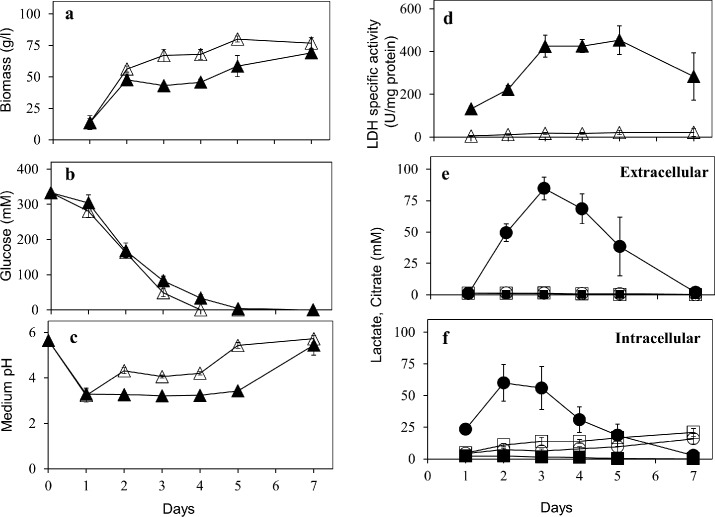
An extended time course of lactate formation by C12 strain was monitored by growth on MM supplemented with 60 g/l of glucose. While their glucose consumption was comparable (Fig 5B), the C12 strain formed less biomass than the parent strain during the lactate production phase (day 2 to day 4; Fig 5A). The LDH activity in C12 strain was detected throughout the seven days of growth (Fig 5D). The L-lactate levels and LDH specific activity (as also the total LDH activity) peaked on day three. By this time, C12 strain produced 85 mM (7.7 g/l) of extracellular L-lactate from 333 mM (60.0 g/l) of glucose; with an overall yield of 13 percent (g lactate/g glucose) (Fig 5E). The pH of the spent medium decreased to around 3.0 (Fig 5C) upon lactate accumulation. This may not be an issue as A. niger is tolerant to and capable of growth in acidic pH particularly during acidogenesis [23]. However, lactate production by engineered yeasts is adversely affected in non-neutralizing media; addition of CaCO3 significantly improved the lactate yield [10,12,36]. Therefore, lactate secretion by C12 strain was also monitored under neutralizing conditions—on MM buffered with CaCO3 (30 g/l). The medium pH was stable between pH 6.0–6.3 during the course of fermentation; with a maximum extracellular lactate concentration of 70 mM on day four (not shown). While addition of CaCO3 in MM had no effect on lactate secretion by C12 strain, extracellular lactate did not disappear (see below) at later growth stages. Addition of CaCO3 had no significant effect on lactate titers and conversion yields in the case of A. brasiliensis ldh transformant. However, the nature of nitrogen source in the medium affected the lactate titers [22]. The recent report on lactate fermentation by engineered A. oryzae provides data only with CaCO3-supplemented media [21]. While CaCO3 could be added to production media, undesirable costs to downstream processing of lactate may accrue [1].
Fig 5. Lactate formation from glucose by A. niger C12 strain: The shake flask growth over 7 days was performed and biomass (Panel a; wet weight), medium glucose (Panel b), medium pH (Panel c), LDH specific activity (Panel d) and extracellular (Panel e) and intracellular (Panel f) concentrations of the two acids are shown.
In panels a, b, c and d, filled triangles represent data for C12 strain while open triangles represent data for the parent strain. The symbols in panels e and f mean the same as in Fig 4.
Intracellular lactate concentrations have been reported only in C. sonorensis engineered yeast [12]. Lactate concentrations inside C12 strain were comparable to that secreted in the non-neutralizing MM (Fig 5F). Nevertheless, intracellular lactate levels were invariably high at all the glucose concentrations tested (Fig 4). Lactate is known to be actively transported and hence could be a bottleneck. Over-expression of lactate permease (encoded by JEN1) improved lactate production by an engineered S. cerevisiae [37]. Presence of a functional lactate transporter (lacA) was also demonstrated in R. oryzae [44]. A. niger genome harbors a putative lactate transporter gene (NCBI accession number CAK42727), which is homologous to JEN1 of yeast. Engineering a functional lactate transporter might augment lactate secretion in A. niger.
Since A. niger is acid tolerant and a prolific citrate producer [23], a comparison of lactate and citrate formation by the C12 strain was of interest. Both A. brasiliensis and A. niger belong to black Aspergilli but the effects on citrate levels due to ldhA expression in the former are not known [22]. L-Lactate levels were always far higher than citrate levels of C12 strain (Fig 5E and 5F). Intracellular citrate in C12 strain remained largely steady whereas extracellular citrate levels were very low. This pattern of citrate levels (both extracellular and intracellular) was comparable to that of the parent strain under all growth conditions tested (Figs 4, 5E and 5F). Therefore, inducing lactate formation by mldhA expression does not appear to perturb citrate levels in A. niger. This is an advantage in lactate fermentation in MM with minimal interference of citrate.
Lactate fermentation on AM
Citric acid fermentation conditions are well established in A. niger. Inducing lactate formation by mldhA expression in C12 strain does not appear to influence citrate levels when grown in MM. This was tested further by growing C12 strain in AM (conducive to citrate production). Despite the presence of active LDH throughout the course of fermentation, both extracellular and intracellular lactate levels in the C12 strain were negligible (Table 4). However, both parent and C12 strain of A. niger displayed high citrate levels. Citrate synthase activity is present throughout the acidogenic growth of A. niger [23]. In the present study, citrate synthase promoter was employed to express mouse ldhA [24]. It is interesting that minimal lactate was produced by the C12 strain grown on AM. Pyruvate availability may be a factor as pyruvate carboxylase is also active during acidogenesis [23].
Table 4. LDH specific activity, lactate and citrate levels in A. niger grown on AM.
| Strain | LDH specific activity (U/mg protein) | Lactate (mM) | Citrate (mM) | ||
|---|---|---|---|---|---|
| Extracellular | Intracellular | Extracellular | Intracellular | ||
| C12 | 128.5 ± 29.0 | 0.23 ± 0.10 | 0.85 ± 0.25 | 9.7 ± 4.8 | 2.7 ± 1.1 |
| Parent | 4.25 ± 1.8 | 0.02 ± 0.01 | 0.20 ± 0.14 | 16.0 ± 1.0 | 5.0 ± 1.3 |
LDH activity and lactate/citrate was estimated after eight days of growth.
Lactate fermentation under hypoxic and nitrogen starvation
The conversion of glucose to L-lactate by pre-grown mycelia of C12 strain was assessed under hypoxic conditions and on MM devoid of nitrogen source. After 48 h incubation in these two conditions, no significant increase in biomass was observed whereas extracellular lactate levels remained around 5 mM. In a similar study, lactate yield by the parent strain was negligible (10–15 μM). Lactate formation in C12 strain therefore appears to be a growth associated phenomenon.
Among all growth conditions tested, maximum lactate secretion by C12 strain was obtained on non-neutralizing MM with 6 percent glucose. Lactate was secreted within 3 days of fermentation which is less compared to 4 and 10 days required by engineered A. brasiliensis and A. oryzae, respectively (Table 5). Although the lactate yield obtained was low, it is comparable to other engineered fungi grown on non-neutralizing media (Table 5). Further improvements can be achieved through media and growth optimization. More interestingly, lactate and citrate production could be separated based on growth on two different media. Exploring lactate production by C12 strain on other carbon sources also merits further attention.
Table 5. L-Lactate production by engineered fungi.
| Fungus a | Source of ldhA | Glucose (g/l) | Lactate (g/l) | Fermentation time (days) | Reference |
|---|---|---|---|---|---|
| S. cerevisiae (Δpdc1Δadh1) | Bovine | 100 | 74.0 | 2 | [45] |
| K. lactis (ΔKlPDC1) | Bovine | 50 | 12.0 | 4 | [7] |
| C. utilis (ΔCuPDC1) | Bovine | 110 | 103.3 | 1.5 | [11] |
| C. sonorensis * (Δpdc1Δpdc2) | L. helveticus | 50 | 10.0 | 5 | [12] |
| B. megaterium | 50 | 5.0 | 5 | [12] | |
| R. oryzae | 50 | 4.0 | 5 | [12] | |
| P. stipites * | L. helveticus | 55 | 8.0 | 2 | [10] |
| A. oryzae (Δ871) | Bovine | 100 | 45.0 | 10 | [21] |
| A. brasiliensis * | R. oryzae | 50 | 13.9 | 5.5 | [22] |
| A. niger * | Mouse | 60 | 7.7 | 3 | This study |
a Respective gene deletions are shown in the brackets.
* Growth was carried out in non-neutralized medium.
Lactate utilization by A. niger C12 strain
Very little is known about lactate metabolism in Aspergilli [46]. A. niger was unable to utilize and grow on L-lactate as sole carbon source, both on liquid and solid MM. Expression of a functional LDH was expected to support the growth of C12 strain on L-Lactate; however, this was not the case. Besides unfavorable kinetic features of mouse LDH for lactate oxidation, uptake of lactate could be an issue. Efforts to feed lactate at lower pH values (pH 3.0 and pH 5.7) and also as methyl lactate were unsuccessful. The C12 strain (as well as the parent strain) grew very poorly on other three-carbon compounds like L-alanine, glycerol or glycerol plus lactate. A. niger is incapable of utilizing ethanol as a carbon source [32]. An inefficient gluconeogenesis in A. niger is one possibility. Interestingly, pre-formed intra and extracellular lactate levels decreased on prolonged growth of C12 strain in non-neutralizing MM (see Fig 5E and 5F). Disappearance of lactate only after glucose exhaustion suggests its reutilization by this strain. Such a lactate utilization/ reutilization possibility was not examined in studies with A. oryzae and A. brasiliensis [21,22]. Both C12 and parent strains were fed with additional lactate (20 mM) after 5 days of growth on MM (6 percent glucose, without pH buffering). Surprisingly, decrease in added lactate was observed for both the parent and the C12 strain within 24 h of further growth. These results point to utilization of lactate through an endogenous enzyme of the parent strain (and not via the expressed mouse LDH). However a mechanism for the observed lactate utilization in A. niger (Fig 5E and 5F) remains to be elucidated. Extracellular lactate decreased after glucose consumption, both in S. cerevisiae [47] and C. sonorensis [12]. It was proposed that such lactate reutilization may be a strategic response to weak acid stress. S. cerevisiae utilizes L-lactate via a mitochondrial L(+)-lactate:cytochrome c oxidoreductase (encoded by CYB2) and disruption of CYB2 improved the lactate production [48]. Lactate yield marginally improved in A. oryzae when a putative ldh (AO090023000871) locus was disrupted [21]. Putative NAD- and cytochrome-dependent ldh(s) are also known in A. niger and these need further attention.
Conclusions
The high acidogenic potential and glycolytic flux make A. niger an attractive target for the production of lactic acid. Its genome was analyzed for potential ldhA genes for functional expression. While A. niger may contain an endogenous LDH (or LDH-like) activity, an ldhA gene coding for such an activity could not be identified. Considering the metabolic features of the A. niger pyruvate node, the LDH expression construct was designed based on sequence comparisons and using an endogenous promoter relevant to its acidogenic metabolism. The integration of PcitA-mldhA DNA led to LDH expression and resulted in transformants that gave significant lactic acid titers. Lactate formation was analyzed as a function of different growth media, hypoxia and nitrogen starvation. Although A. niger does not utilize lactate as a sole carbon source, it was able to re-utilize secreted lactate subsequent to glucose consumption. Interestingly, the formation of lactate and citrate by the C12 strain could be segregated by choosing different growth media. Already an industrial favorite, A. niger promises to be a good platform to produce lactate as yet another organic acid.
Supporting Information
The relevant sequence alignment region covering the LDH active site signature (underlined) is shown.
(TIF)
(DOC)
Acknowledgments
Bayer Crop-Science is gratefully acknowledged for providing glufosinate ammonium (PPT).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Khyati Dave received Council of Scientific and Industrial Research (CSIR) research fellowship.
References
- 1. Wang Y, Tashiro Y, Sonomoto K. Fermentative production of lactic acid from renewable materials: Recent achievements, prospects, and limits. J Biosci Bioeng. 2015; 119: 10–18. 10.1016/j.jbiosc.2014.06.003 [DOI] [PubMed] [Google Scholar]
- 2. Soccol CR, Stonoga VI, Raimbault M. Production of L-lactic acid by Rhizopus species. World J Microbiol Biotechnol. 1994; 10: 433–5. 10.1007/BF00144467 [DOI] [PubMed] [Google Scholar]
- 3. Zhan ZY, Jin B, Kelly JM. Production of lactic acid from renewable materials by Rhizopus fungi. Biochem Eng J. 2007; 35: 251–63. [Google Scholar]
- 4. Ishida N, Saitoh S, Onishi T, Tokuhiro K, Nagamori E, Kitamoto K, et al. The effect of pyruvate decarboxylase gene knockout in Saccharomyces cerevisiae on L-lactic acid production. Biosci Biotechnol Biochem. 2006; 70: 1148–53. [DOI] [PubMed] [Google Scholar]
- 5. Saitoh S, Ishida N, Onishi T, Tokuhiro K, Nagamori E, Kitamoto K, et al. Genetically engineered wine yeast produces a high concentration of L-lactic acid of extremely high optical purity. Appl Environ Microbiol. 2005; 71: 2789–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Skory CD. Lactic acid production by Saccharomyces cerevisiae expressing a Rhizopus oryzae lactate dehydrogenase gene. J Ind Microbiol Biotechnol. 2003; 30: 22–7. [DOI] [PubMed] [Google Scholar]
- 7. Porro D, Bianchi MM, Brambilla L, Menghini R, Bolzani D, Carrera V, et al. Replacement of a metabolic pathway for large-scale production of lactic acid from engineered yeasts. Appl Environ Microbiol. 1999; 65: 4211–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bianchi MM, Brambilla L, Protani F, Liu CL, Lievense J, Porro D. Efficient homolactic fermentation by Kluyveromyces lactis strains defective in pyruvate utilization and transformed with the heterologous LDH gene. Appl Environ Microbiol. 2001; 67: 5621–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Skory CD, Ibrahim AS. Native and modified lactate dehydrogenase expression in a fumaric acid producing isolate Rhizopus oryzae 99–880. Curr Genet. 2007; 52: 23–33. [DOI] [PubMed] [Google Scholar]
- 10. Ilmen M, Koivuranta K, Ruohonen L, Suominen P, Penttila M. Efficient production of L-lactic acid from xylose by Pichia stipitis . Appl Environ Microbiol. 2007; 73: 117–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ikushima S, Fujii T, Kobayashi O, Yoshida S, Yoshida A. Genetic engineering of Candida utilis yeast for efficient production of L-lactic acid. Biosci Biotechnol Biochem. 2009; 73: 1818–24. [DOI] [PubMed] [Google Scholar]
- 12. Ilmen M, Koivuranta K, Ruohonen L, Rajgarhia V, Suominen P, Penttila M. Production of L-lactic acid by the yeast Candida sonorensis expressing heterologous bacterial and fungal lactate dehydrogenases. Microb Cell Fact. 2013; 12: 53 10.1186/1475-2859-12-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Okano K, Tanaka T, Ogino C, Fukuda H, Kondo A. Biotechnological production of enantiomeric pure lactic acid from renewable resources: recent achievements, perspectives, and limits. Appl Microbiol Biotechnol. 2010; 85: 413–423. 10.1007/s00253-009-2280-5 [DOI] [PubMed] [Google Scholar]
- 14. Magnuson JK, Lasure LL. Organic acid production by filamentous fungi Tkacz JS and Lange L, editors. Advances in fungal biotechnology for industry, agriculture and medicine, Kluwer Academic/Plenum Publishers, New York; 2004. pp. 307–340. [Google Scholar]
- 15. Kobayashi K, Hattori T, Honda Y, Kirimura K. Oxalic acid production by citric acid-producing Aspergillus niger overexpressing the oxaloacetate hydrolase gene oahA . J Ind Microbiol Biotechnol. 2014; 41: 749–56. 10.1007/s10295-014-1419-2 [DOI] [PubMed] [Google Scholar]
- 16. Meijer S, Otero J, Olivares R, Andersen MR, Olsson L, Nielsen J. Overexpression of isocitrate lyase-glyoxylate bypass influence on metabolism in Aspergillus niger . Metab Eng. 2009; 11: 107–16. [DOI] [PubMed] [Google Scholar]
- 17. Li A, van Luijk N, ter Beek M, Caspers M, Punt P, van der Werf M. A clone-based transcriptomics approach for the identification of genes relevant for itaconic acid production in Aspergillus . Fungal Genet Biol. 2011; 48: 602–11. 10.1016/j.fgb.2011.01.013 [DOI] [PubMed] [Google Scholar]
- 18. Li A, Pfelzer N, Zuijderwijk R, Punt P. Enhanced itaconic acid production in Aspergillus niger using genetic modification and medium optimization. BMC Biotechnol. 2012; 12: 57 10.1186/1472-6750-12-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Blumhoff ML, Steiger MG, Mattanovich D, Sauer M. Targeting enzymes to the right compartment: metabolic engineering for itaconic acid production by Aspergillus niger . Metab Eng. 2013; 19: 26–32. 10.1016/j.ymben.2013.05.003 [DOI] [PubMed] [Google Scholar]
- 20. van der Straat L, Vernooij M, Lammers M, van den Berg W, Schonewille T, Cordewener J, et al. Expression of the Aspergillus terreus itaconic acid biosynthesis cluster in Aspergillus niger . Microb Cell Fact. 2014; 13:11 10.1186/1475-2859-13-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wakai S, Yoshie T, Asai-Nakashima N, Yamada R, Ogino C, Tsutsumi H, et al. L-lactic acid production from starch by simultaneous saccharification and fermentation in a genetically engineered Aspergillus oryzae pure culture. Bioresour Technol. 2014; 173: 376–83. 10.1016/j.biortech.2014.09.094 [DOI] [PubMed] [Google Scholar]
- 22. Liaud N, Rosso MN, Fabre N, Crapart S, Herpoel-Gimbert I, Sigoillot JC, et al. L-lactic acid production by Aspergillus brasiliensis overexpressing the heterologous ldhA gene from Rhizopus oryzae . Microb Cell Fact. 2015; 14: 66 10.1186/s12934-015-0249-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Papagianni M. Advances in citric acid fermentation by Aspergillus niger: biochemical aspects, membrane transport and modeling. Biotechnol Adv. 2007; 25: 244–63. [DOI] [PubMed] [Google Scholar]
- 24. Dave K, Punekar NS. Utility of Aspergillus niger citrate synthase promoter for heterologous expression. J Biotechnol. 2011; 155: 173–7. 10.1016/j.jbiotec.2011.06.012 [DOI] [PubMed] [Google Scholar]
- 25. Noor S, Punekar NS. Allosteric NADP-glutamate dehydrogenase from Aspergilli: purification, characterization and implications for metabolic regulation at the carbon-nitrogen interface. Microbiology. 2005; 151: 1409–19. [DOI] [PubMed] [Google Scholar]
- 26. Terabayashi Y, Shimizu M, Kitazume T, Masuo S, Fujii T, Takaya N. Conserved and specific responses to hypoxia in Aspergillus oryzae and Aspergillus nidulans determined by comparative transcriptomics. Appl Microbiol Biotechnol. 2012; 93: 305–17. 10.1007/s00253-011-3767-4 [DOI] [PubMed] [Google Scholar]
- 27. Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbour Laboratory Press, New York; 2001. [Google Scholar]
- 28. Skulj M, Okrslar V, Jalen S, Jevsevar S, Slanc P, Strukelj B, et al. Improved determination of plasmid copy number using quantitative real-time PCR for monitoring fermentation processes. Microb Cell Fact. 2008; 7:6 10.1186/1475-2859-7-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Petrarulo M, Facchini P, Cerelli E, Marangella M, Linari F. Citrate in urine determined with a new citrate lyase method. Clin Chem. 1995; 41: 1518–21. [PubMed] [Google Scholar]
- 30. Ruijter GJG, Visser J. Determination of intermediary metabolites in Aspergillus niger . J Microbiol Meth. 1996; 25: 295–302. [Google Scholar]
- 31. Legisa M, Kidric J. Initiation of citric acid accumulation in the early stages of Aspergillus niger growth. Appl Microbiol Biotechnol. 1989, 31: 453–457. [Google Scholar]
- 32. Flipphi M, Sun J, Robellet X, Karaffa L, Fekete E, Zeng AP, et al. Biodiversity and evolution of primary carbon metabolism in Aspergillus nidulans and other Aspergillus spp . Fungal Genet Biol. 2009; 46:S19–S44. [DOI] [PubMed] [Google Scholar]
- 33. Andersen MR, Nielsen ML, Nielsen J. Metabolic model integration of the bibliome, genome, metabolome and reactome of Aspergillus niger . Mol Syst Biol. 2008; 4:178 10.1038/msb.2008.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Andersen MR, Salazar MP, Schaap PJ, van de Vondervoort PJ, Culley D, Thykaer J, et al. Comparative genomics of citric-acid-producing Aspergillus niger ATCC 1015 versus enzyme-producing CBS 513.88. Genome Res. 2011; 21:885–97. 10.1101/gr.112169.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Clarke AR, Wilks HM, Barstow DA, Atkinson T, Chia WN, Holbrook JJ. An investigation of the contribution made by the carboxylate group of an active site histidine-aspartate couple to binding and catalysis in lactate dehydrogenase. Biochemistry. 1988; 27: 1617–22. [DOI] [PubMed] [Google Scholar]
- 36. Ishida N, Saitoh S, Tokuhiro K, Nagamori E, Matsuyama T, Kitamoto K, et al. Efficient production of L-Lactic acid by metabolically engineered Saccharomyces cerevisiae with a genome-integrated L-lactate dehydrogenase gene. Appl Environ Microbiol. 2005; 71: 1964–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Branduardi P, Sauer M, De GL, Zampella G, Valli M, Mattanovich D, et al. Lactate production yield from engineered yeasts is dependent from the host background, the lactate dehydrogenase source and the lactate export. Microb Cell Fact. 2006; 5:4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kumar S, Tejaswani S, Punekar NS. Novel route for agmatine catabolism in Aspergillus niger involves 4-guanidinobutyrase. Appl Environ Microbiol. 2015; 81: 03987–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Punekar NS, Dave K, inventors; Novel recombinant protein expression system involving a strong, constitutive promoter of citrate synthase from Aspergillus. India patent 2542/MUM/2009. 2009.
- 40. Hawtrey CO, Goldberg E. Some kinetic aspects of sperm specific lactate dehydrogenase in mice. J Exp Zool. 1970; 174: 451–61. [DOI] [PubMed] [Google Scholar]
- 41. Jernejc K, Legisa M. The influence of metal ions on malic enzyme activity and lipid synthesis in Aspergillus niger . FEMS Microbiol Lett. 2002; 217: 185–90. [DOI] [PubMed] [Google Scholar]
- 42. Meixner-Monori B, Kubicek CP, Rohr M. Pyruvate kinase from Aspergillus niger: a regulatory enzyme in glycolysis? Can J Microbiol. 1984; 30: 16–22. [DOI] [PubMed] [Google Scholar]
- 43. Feir HA, Suzuki I. Pyruvate carboxylase of Aspergillus niger: kinetic study of a biotin-containing carboxylase. Can J Biochem. 1969; 47: 697–710. [DOI] [PubMed] [Google Scholar]
- 44. Skory CD, Hector RE, Gorsich SW, Rich JO. Analysis of a functional lactate permease in the fungus Rhizopus . Enzyme Microb Technol. 2010; 46: 43–50. [Google Scholar]
- 45. Tokuhiro K, Ishida N, Nagamori E, Saitoh S, Onishi T, Kondo A, et al. Double mutation of the PDC1 and ADH1 genes improves lactate production in the yeast Saccharomyces cerevisiae expressing the bovine lactate dehydrogenase gene. Appl Microbiol Biotechnol. 2009; 82: 883–90. 10.1007/s00253-008-1831-5 [DOI] [PubMed] [Google Scholar]
- 46. Hynes M. Gluconeogenesis In: Borkovich KA, Ebbole DJ, editors. Cellular and molecular biology of filamentous fungi. Washington: ASM press; 2010. pp. 312–24. [Google Scholar]
- 47. Pacheco A, Talaia G, Sa-Pessoa J, Bessa D, Goncalves MJ, Moreira R, et al. Lactic acid production in Saccharomyces cerevisiae is modulated by expression of the monocarboxylate transporters Jen1 and Ady2 . FEMS Yeast Res. 2012; 12: 375–81. 10.1111/j.1567-1364.2012.00790.x [DOI] [PubMed] [Google Scholar]
- 48. Ookubo A, Hirasawa T, Yoshikawa K, Nagahisa K, Furusawa C, Shimizu H. Improvement of L-lactate production by CYB2 gene disruption in a recombinant Saccharomyces cerevisiae strain under low pH condition. Biosci Biotechnol Biochem. 2008; 72: 3063–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The relevant sequence alignment region covering the LDH active site signature (underlined) is shown.
(TIF)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.