Abstract
Background
Pregnancy induces drastic biological changes systemically, and has a beneficial effect on some autoimmune conditions such as rheumatoid arthritis (RA). However, specific systemic changes that occur as a result of pregnancy have not been thoroughly examined in healthy women or women with RA. The goal of this study was to identify genes with expression patterns associated with pregnancy, compared to pre-pregnancy as baseline and determine whether those associations are modified by presence of RA.
Results
In our RNA sequencing (RNA-seq) dataset from 5 healthy women and 20 women with RA, normalized expression levels of 4,710 genes were significantly associated with pregnancy status (pre-pregnancy, first, second and third trimesters) over time, irrespective of presence of RA (False Discovery Rate (FDR)-adjusted p value<0.05). These genes were enriched in pathways spanning multiple systems, as would be expected during pregnancy. A subset of these genes (n = 256) showed greater than two-fold change in expression during pregnancy compared to baseline levels, with distinct temporal trends through pregnancy. Another 98 genes involved in various biological processes including immune regulation exhibited expression patterns that were differentially associated with pregnancy in the presence or absence of RA.
Conclusions
Our findings support the hypothesis that the maternal immune system plays an active role during pregnancy, and also provide insight into other systemic changes that occur in the maternal transcriptome during pregnancy compared to the pre-pregnancy state. Only a small proportion of genes modulated by pregnancy were influenced by presence of RA in our data.
Background
Pregnancy is known to have beneficial effects on rheumatoid arthritis (RA) in a number of ways. New onset of RA is rare during the gestational period, suggesting that pregnancy may be protective against development of RA [1–3]. Pregnancy even appears to protect against RA onset beyond the gestational period in the form of vaccine-like protection [4]. Further, women with RA often experience a natural and dramatic improvement in disease activity during pregnancy [5–8]. Some pregnancy-related factors that have been associated with the pregnancy-induced improvement of RA disease activity include maternal-fetal HLA disparity [9] and microchimerism [10]. The mechanism(s) behind the protective effects of pregnancy are, however, not entirely clear. While there is no doubt that pregnancy induces drastic biological changes systemically, the specific systemic changes that occur as a result of pregnancy and their effects on maternal health have not been thoroughly examined. Hence, as a first step in examining the influence of pregnancy on RA, it is important that we gain a better understanding of systemic biological changes associated with healthy pregnancy and that we determine whether pregnancy-induced biological changes are altered by the presence of RA, irrespective of disease activity levels.
Global gene expression studies are well-suited to inform us on such changes that occur during pregnancy. However, surprisingly little is known about pregnancy-induced systemic changes in gene expression among healthy women because most gene expression studies in pregnancy have focused on the maternal-fetal interface to understand how fetal tolerance is established and maintained [11–13]. There have been very few gene expression studies on RA pregnancy [14–17], all of which were focused on pregnancy-induced changes in disease activity. Thus, it still remains to be determined whether systemic biological changes associated with pregnancy can be altered by the presence of RA, irrespective of disease activity levels.
In the present study, we have examined the hypothesis that the maternal immune system is active during pregnancy among both healthy women and women with RA. We have examined global gene expression profiles among 5 healthy women and 20 women with RA from pre-pregnancy to the third trimester, to identify genes that exhibit pregnancy-induced changes in expression. We also examined whether the presence of RA (irrespective of level of disease activity) influences pregnancy-associated gene expression.
Results
Study subjects
Overall, age at conception was similar among healthy women and women with RA: healthy (mean±SD): 31.4 ± 5.8 years; RA: 31.6 ± 4.6 years (p = 0.99). The women with RA had the disease for a mean duration of 5.1 ± 3.3 years.
Medication use: Among the women with RA who had complete follow up (n = 13), 2 did not take any medications while in the study, 7 took prednisolone and/or sulfasalazine before and/or during pregnancy, and 4 took anti-TNF agents before pregnancy and in the first trimester. Of the women on prednisolone and/or sulfasalazine, one took methotrexate before pregnancy, and one was on hydroxychloroquine throughout pregnancy. The 7 women with incomplete follow up took prednisolone or sulfasalazine (n = 6) or were not on medication (n = 1).
Data quality
The average number of paired-end reads across all samples was 73 million. Of these, an average of 40% mapped to the reference transcriptome (S1 Fig). No significant differences were observed in number of mapped reads across time points (ANOVA, p = 0.93). Sample replicates processed in separate batches achieved a Pearson correlation of at least 0.97, following correction for batch effects (S2 Fig). After filtering out genes with low expression across all samples and genes differentially expressed between replicate samples across batches, a total of 13,655 genes (39%) remained for downstream analyses.
Genes modulated by pregnancy in healthy women and women with RA
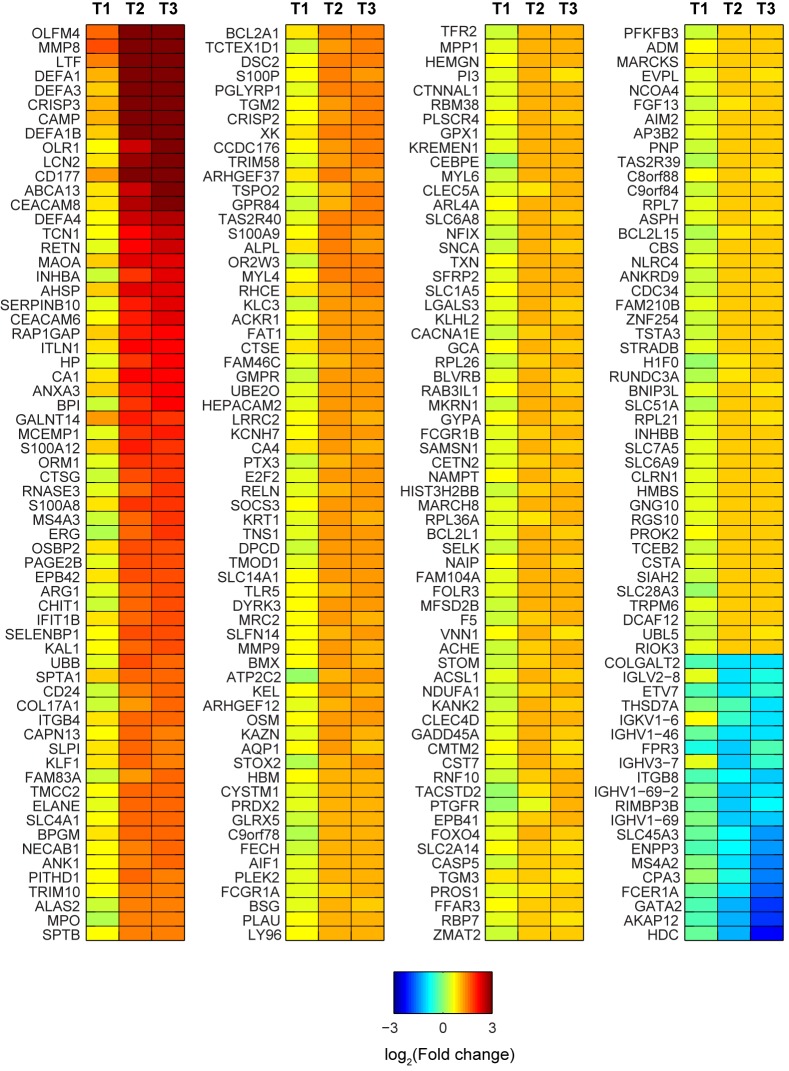
In the GEE models, of 13,655 genes analyzed, 4,710 had expression levels associated with pregnancy status over time (q<0.05), irrespective of whether a woman did or did not have RA. Pathway analysis indicated that many of these genes were enriched pathways for: genetic information processing, metabolism, signal transduction, cellular processes and organismal systems (immune, endocrine, excretory, nervous), development, and disease pathways (Table 1). Among the genes with pregnancy-associated expression, 256 genes showed greater than two-fold change in expression levels during pregnancy compared to pre-pregnancy levels among healthy women. These had distinct temporal trends, with most being up-regulated as pregnancy progressed and others showing reduced expression with advancing pregnancy stages (Fig 1). Significant changes in the expression profiles compared to pre-pregnancy were observed during second trimester and were maintained during third trimester. Examples of genes involved in immune system processes and defense response that displayed a marked pregnancy-related up-regulation included: OLFM4, MMP8, LTF, DEFA1, DEFA3, DEFA1B, CRISP3, CAMP, OLR1, LCN2, CD177, ABCA13 and CEACAM8. Furthermore, genes involved in mast cell activation and immunoglobulin binding were significantly downregulated in the second and third trimesters. Two such significant transcripts were the Fc fragment of immunoglobulin Epsilon Receptor (FCER1A) and Membrane Spanning 4-domains subfamily A member 2 (MS4A2).
Table 1. Significantly enriched KEGG pathways among 4,710 genes associated with pregnancy.
| KEGG pathways | Genes identified | q value* | |||
|---|---|---|---|---|---|
| (Genes in pathway) | |||||
| Pathways enriched in genes up-regulated during pregnancy | |||||
| Genetic information processing | |||||
| Translation | |||||
| Ribosome | 51 (89) | 7x10-24 | |||
| Folding, sorting and degradation | |||||
| Ubiquitin mediated proteolysis | 30 (135) | 2x10-3 | |||
| Protein processing in endoplasmic reticulum | 30 (165) | 0.02 | |||
| Proteasome | 11 (44) | 0.03 | |||
| Replication and repair | |||||
| Non-homologous end joining | 5 (13) | 0.04 | |||
| Transcription | |||||
| Spliceosome | 24 (127) | 0.03 | |||
| Metabolism | |||||
| Metabolism | |||||
| Metabolic pathways | 169 (1130) | 5x10-4 | |||
| Energy metabolism | |||||
| Oxidative phosphorylation | 31 (132) | 6x10-4 | |||
| Metabolism of other amino acids | |||||
| Glutathione metabolism | 13 (50) | 0.02 | |||
| Carbohydrate metabolism | |||||
| Amino sugar and nucleotide sugar metabolism | 14 (48) | 4x10-3 | |||
| Inositol phosphate metabolism | 14 (57) | 0.02 | |||
| Galactose metabolism | 8 (27) | 0.03 | |||
| Pentose phosphate pathway | 8 (27) | 0.03 | |||
| Signal transduction | |||||
| Signal transduction | |||||
| MAPK signaling pathway | 53 (268) | 4x10-4 | |||
| mTOR signaling pathway | 14 (52) | 8x10-3 | |||
| Jak-STAT signaling pathway | 30 (155) | 0.01 | |||
| Phosphatidylinositol signaling pathway | 16 (78) | 0.04 | |||
| Cellular processes | |||||
| Transport and catabolism | |||||
| Phagosome | 39 (152) | 2x10-5 | |||
| Lysosome | 27 (121) | 3x10-3 | |||
| Endocytosis | 46 (201) | 4x10-5 | |||
| Regulation of autophagy | 9 (34) | 0.04 | |||
| Cell motility | |||||
| Regulation of actin cytoskeleton | 41 (212) | 3x10-3 | |||
| Cell growth and death | |||||
| Apoptosis | 20 (87) | 8x10-3 | |||
| Organismal processes | |||||
| Immune system | |||||
| Fc gamma R-mediated phagocytosis | 29 (94) | 1x10-5 | |||
| Toll-like receptor signaling pathway | 30 (102) | 2x10-5 | |||
| NOD-like receptor signaling pathway | 17 (58) | 1x10-3 | |||
| B cell receptor signaling pathway | 19 (75) | 3x10-3 | |||
| Leukocyte transendothelial migration | 24 (116) | 0.01 | |||
| Fc epsilon RI signaling pathway | 18 (79) | 0.01 | |||
| Hematopoietic cell lineage | 19 (88) | 0.02 | |||
| T cell receptor signaling pathway | 21 (108) | 0.03 | |||
| Chemokine signaling pathway | 32 (183) | 0.04 | |||
| Natural killer cell mediated cytotoxicity | 24 (136) | 0.05 | |||
| Development | |||||
| Osteoclast differentiation | 44 (128) | 2x10-10 | |||
| Endocrine system | |||||
| Insulin signaling pathway | 33 (138) | 3x10-4 | |||
| Adipocytokine signaling pathway | 18 (68) | 3x10-3 | |||
| Excretory system | |||||
| Collecting duct acid secretion | 10 (27) | 3x10-3 | |||
| Nervous system | |||||
| Neurotrophin signaling pathway | 28 (127) | 3x10-3 | |||
| Long-term potentiation | 15 (70) | 0.03 | |||
| Disease pathways | |||||
| Infectious diseases: Bacterial | |||||
| Epithelial cell signaling in Helicobacter pylori infection | 19 (68) | 1x10-3 | |||
| Vibrio cholerae infection | 13 (54) | 0.02 | |||
| Shigellosis | 14 (61) | 0.03 | |||
| Bacterial invasion of epithelial cells | 15 (70) | 0.03 | |||
| Infectious diseases: Parasitic | |||||
| Leishmaniasis | 21 (72) | 4x10-4 | |||
| Chagas disease | 25 (104) | 1x10-3 | |||
| Toxoplasmosis | 29 (132) | 2x10-3 | |||
| Cancers | |||||
| Renal cell carcinoma | 20 (70) | 7x10-4 | |||
| Pathways in cancer | 55 (326) | 7x10-3 | |||
| Endometrial cancer | 13 (52) | 0.02 | |||
| Acute myeloid leukemia | 14 (57) | 0.02 | |||
| Non-small cell lung cancer | 13 (54) | 0.02 | |||
| Glioma | 15 (65) | 0.02 | |||
| Chronic myeloid leukemia | 16 (73) | 0.03 | |||
| Pancreatic cancer | 15 (70) | 0.03 | |||
| Pancreatic cancer | 15 (70) | 0.03 | |||
| Thyroid cancer | 8 (29) | 0.04 | |||
| Neurodegenerative diseases | |||||
| Alzheimer's disease | 40 (167) | 5x10-5 | |||
| Huntington's disease | 41 (183) | 2x10-4 | |||
| Amyotrophic lateral sclerosis | 15 (53) | 4x10-3 | |||
| Parkinson’s disease | 27 (130) | 7x10-3 | |||
| Immune diseases | |||||
| Rheumatoid arthritis | 18 (91) | 0.04 | |||
| Endocrine and metabolic diseases | |||||
| Type II diabetes mellitus | 11 (48) | 0.05 | |||
| Pathways enriched in genes downregulated during pregnancy | |||||
| Metabolism | |||||
| Glycam biosynthesis and metabolism | |||||
| N-Glycan biosynthesis | 17 (49) | 2x10-3 | |||
| Disease pathways | |||||
| Immune diseases | |||||
| Primary imunodeficiency | 19 (35) | 2x10-7 | |||
* q value refers to the FDR-adjusted p value associated with the KEGG pathways enriched in pregnancy-associated genes. Pathways are grouped into broad functional categories.
Fig 1. Heatmap showing temporal trends in expression among genes with pregnancy-associated expression, compared to pre-pregnancy levels in healthy women.
Log-transformed (log2) values of the average fold change in expression compared to pre-pregnancy levels are plotted. Only genes that exhibited 2-fold or higher change in expression (compared to healthy pre-pregnancy baseline levels) in any trimester are shown. These patterns were similar in healthy women and in women with RA.
Genes with pregnancy-associated expression modified by presence of RA
In contrast to the genes with pregnancy-associated expression irrespective of the presence or absence of RA, another 98 genes exhibited expression patterns during pregnancy that were dependent on both pregnancy status and presence or absence of RA; these were identified through the interaction term (RA×Pregnancy) in the GEE models. Functional classification using the GO database revealed that these genes were involved in a variety of biological processes including regulation of immune system (Table 2). Of these, thirteen genes displayed greater than two-fold change in expression before and/or during pregnancy, compared to pre-pregnancy levels among healthy women (Fig 2).
Table 2. Functional categories of genes with expression patterns differentially associated with pregnancy in presence or absence of RA.
| Biological process / Molecular function | Genes |
|---|---|
| Regulation of immune system process | HLA-DRB3, IL15 |
| Metabolic process | ACAD11, DECR2, LCAT, PKD2, PNPLA2, IL15, CAMKK1 |
| Regulation of catalytic activity | PKD2, IFI27, CAMKK1 |
| Hormone transport | HLA-DRB3, CACNA1C |
| Cellular response to osmotic stress | PKD2, RCSD1 |
| Neurogenesis | MAP1S, CTHRC1, CACNA1C, USH2A, ARHGDIA |
| Tissue morphogenesis | PKD2, CTHRC1, CCM2 |
| Voltage-gated calcium channel activity | PKD2, CACNA1C |
| Endothelial cell migration | EPHB4, HDAC5 |
| Protein binding | TNNI2, CACNA1C, RPH3AL, USH2A, DAAM2, RCSD1, MAP1S, PKD2, FAM101B, MYO7B, SH2D4A, SYTL2 |
| Cofactor binding | SQLE, ACAD11, HSD11B2, MTO1 |
| Hydrolase activity | CES2, HDAC5, LCAT, PNPLA2 |
Genes up-regulated in healthy pregnancy are shown in normal font, genes downregulated in healthy pregnancy are shown in italics, genes up-regulated in RA pregnancy are not underlined and genes downregulated in RA pregnancy are underlined.
Fig 2. Heatmap showing genes with pregnancy-associated expression patterns that are altered by presence of RA.
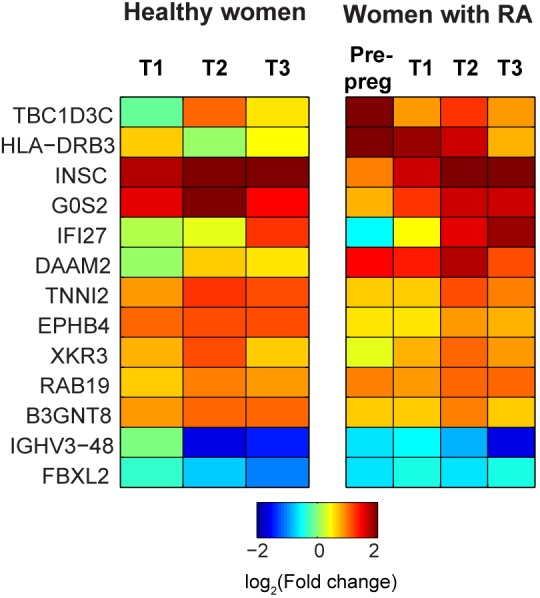
Log-transformed (log2) values of the average fold change in expression compared to baseline (i.e. pre-pregnancy levels among healthy women) are plotted for healthy women and for women with RA. Only genes that exhibited 2-fold or higher change in expression (compared to healthy pre-pregnancy baseline levels) in any trimester are shown.
Genes with expression levels associated with presence of RA and/or medication use
One hundred and eleven genes had expression levels that were associated with presence of RA in the GEE models (q<0.05), irrespective of pregnancy status. The pathways and/or biological processes that these genes are involved in are shown in Table 3. Our analyses also detected genes with expression levels influenced by medication use; these were involved in immune response, defense response, RNA binding, and transferase activity.
Table 3. KEGG pathway and Gene Ontology Biological process categories of the genes showing association with RA.
| Pathway / biological process | Genes |
|---|---|
| mTOR signaling pathway | ULK1, AKT1 |
| Toll-like receptor signaling pathway | IKBKG, AKT1, TOLLIP |
| Metabolic pathways | MAN2A2, GAA, GALC, CSGALNACT1, MAOA, NDUFV3, ABAT, LIPC, DBT, PISD |
| Fatty acid metabolic process | FADS1, FADS2, FADS3, ABAT, AKT1, LIPC, SCAP |
| Cellular polysaccharide metabolic process | GAA, AKT1, PYGM, CSGALNACT1 |
| Intracellular transport of viral material | VPS37C, VPS37B |
| Neurotransmitter catabolic process | MAOA, ABAT |
| t-RNA binding | SLFN11, IGHMBP2 |
| DNA helicase activity | IGHMBP2, HELB |
| Catalytic activity | GAA, TUBB2A, PYGM, SPTSSB, ACAP1, ULK1, ERCC4, RNF43, IGHMBP2, ABAT, GRK6, KIF21B, PISD, SPPL2A, FADS3, ECHDC3, TPST1, NDUFV3, DBT, UBE2M, CARS2, DCTN1, PLK3, FADS1, FADS2, AKT1, ADCK3, GALC, HELB, UFSP1, PPM1F, USP10, PGLYRP1, LIPC, ALK, MAN2A2, PRDX5, MAOA, CSGALNACT1, DTD1, TRIM36 |
| GTPase activator activity | ARHGAP9, GMIP, ARHGAP27, ACAP1, RAP1GAP2 |
Up-regulated genes are shown in normal font and downregulated genes are in italics.
Discussion
Using samples from an ethnically homogeneous pregnancy cohort followed in real time from pre-pregnancy, we have examined changes in global gene expression among healthy women and women with RA using RNA-seq technology. Given that pregnancy-induced gene expression could not previously be examined relative to the pre-pregnancy state, our findings are novel. We have identified genes that demonstrate altered expression during pregnancy compared to the pre-pregnancy baseline, and shown how their temporal patterns of expression change throughout pregnancy. These include several potentially novel genes that are at least two-fold differentially expressed during pregnancy compared to before pregnancy. In addition, we have also identified genes that demonstrate different temporal patterns of expression through pregnancy between healthy women and women with RA; that is, their pregnancy-associated expression patterns were altered by whether a woman was healthy or had RA.
Human pregnancy is known to induce extensive physiological changes in the mother, involving almost every system. Our results at the gene expression level fit well in this context, indicating systemic changes in maternal transcriptome spanning multiple cellular and organismal systems, pathways and biological processes during pregnancy. Among the pathways that we identified as being enriched in genes with pregnancy-associated expression, several were related to immune function. This is especially relevant since pregnancy is known to be associated with immunological changes and challenges, not just locally at the maternal-fetal interface, but also at the systemic level. The pathways identified relate to natural killer cell mediated cytotoxicity, and signaling pathways involving Toll-like receptor, NOD-like receptor, T cell receptor, and B cell receptor, suggesting that both innate and adaptive immune responses play a role in pregnancy. Further, several of the genes that were the most highly up-regulated during pregnancy are expressed in neutrophils, in line with previous reports of neutrophil activation during pregnancy [18–20]. An overexpression of neutrophil-related genes, namely OLFM4, MMP8, DEFA1 and CEACAM8, during pregnancy has also been reported by Heung et. al. [21]. Our findings thus add support to mounting evidence that the maternal immune system is active, and not generally immuno-suppressed during pregnancy [22].
Previous studies of systemic gene expression changes that occur in the mother during healthy pregnancy have been few and the findings inconsistent. Some studies reported no significant differences between pregnancy and post-partum expression profiles of healthy women [14, 17]. Other reports share some degree of overlap in genes/pathways that we have identified as being modulated by pregnancy. For example, a study of peripheral blood mononuclear cells (PBMC) profiles using microarray data identified genes differentially expressed during and after pregnancy [16]. Pathways that overlapped with our findings included those for: apoptosis, cancer, Fc gamma R-mediated phagocytosis, natural killer cell-mediated cytotoxity, and signaling pathways involving MAPK, T cell receptor, Toll-like receptor, and adipocytokine. In a longitudinal study of 11 women followed from the first trimester through 6 weeks post-partum, pairwise analyses of microarray and RNA-seq data from cell-free plasma RNA identified 16 genes in common with our findings [23]. Of interest, several genes such as MMP8, S100P, LIN7A, ZNF438, TCN1, B3GNT5, PLEK2 and PAPPA showed similar temporal changes in expression as pregnancy progressed as observed in our study. A small (n = 4) study of cell-free plasma comparing RNA-seq data from before and after delivery identified several genes with pregnancy-associated expression that were also found in our data [24].
The inconsistencies in findings between these previous studies and our study may be accounted for by a number of factors. First, whilst these studies used either cell-free sera or PBMC as the source of total RNA, our source of total RNA included both the cellular and cell-free fractions of whole blood. Second, these few studies have identified pregnancy-related changes using postpartum data as baseline. However, the postpartum state is itself associated with major changes such as lactation, increased risk of some cancers [25], depression [26, 27], and significant immunologic changes as reflected by increased risk of some autoimmune diseases [1, 28, 29]. Therefore, the appropriate baseline when identifying pregnancy-specific changes is the pre-pregnancy state, which was not available in these studies. Third, we used GEE models to make the most of the data available at all time points. Pairwise analyses of data available from the same subjects across time points [23, 24] limit the power of the analyses. Additionally, in cases where data on each woman was available at only one time-point (i.e. cross-sectional data) [16], changes identified between time points may have been in part due to between-subject variability. Fourth, while several of the previous studies were based on microarray data [17, 23], we used the more accurate and reliable RNA-seq technology [30, 31].
Our findings relating to genes that have pregnancy-related patterns altered by presence of RA are novel. Several of the genes identified have previously been implicated in RA. For example, HLA-DRB3 and G0/G1 Switch 2 (G0S2) expression have been reported to predict response to anti-TNF therapy [32, 33], while Interferon, Alpha-Inducible Protein 27 (IFI27) and Troponin I type 2 (TNNI2) appear to be involved in RA onset or progression [34, 35]. To our knowledge, it had thus far not been demonstrated that these genes can be modulated by both RA and pregnancy status. It is not clear yet whether and how the remaining genes that exhibited pregnancy-related patterns altered by presence of RA, may be involved in the disease.
Our study does have strengths as well as limitations. The sample size was small, but longitudinal samples available from the majority of subjects enabled us to use GEE models to enhance power and to eliminate noise due to between-subject variability and time-stable confounders. The ethnic homogeneity of our study population also was an advantage. We cannot eliminate the possibility of technical bias and/or batch effects having been introduced in the data. However, we randomized sample order prior to any sample processing, used a block design for sequencing, and at the data processing step, we used sample replicates to assess and mitigate batch effects. We also adjusted for any residual batch effects in the statistical models. Although we adjusted for medication use in the model, we did not adjust for specific medications that may have an effect on the immune system and/or dosage due to the heterogeneity in medication use. Because our goal was to identify overall systemic gene expression changes resulting either from altered expression of specific genes or from changes in cell proportions, we also did not examine whether proportions of different cell types in blood samples changed across time points. We nevertheless adjusted for medication use in the analysis, which should have corrected for drug-induced changes in cell type proportions. The use of total RNA from whole blood may also mean that expression profiles of neutrophils may have dominated a large part of the observed expression patterns. Some genes expressed by neutrophils such as MMP8, LTF, CRISP3, CD177 and DEFA4 did exhibit significant changes in expression during the course of pregnancy since neutrophils appear to have an active role in later stages of pregnancy and in labor [36]. However, the sensitivity of RNA-seq technology enabled us to also detect transcripts that were not neutrophil-specific, including those specific to cell types present in low proportions in blood. We have not attempted to separate RA-associated gene expression from disease activity-associated gene expression because the women with RA showed a broad range of disease activity, ranging from remission to high disease activity, both before and during pregnancy. Since the pregnancy-related gene expression changes were obtained by averaging over all RA patients, we do not expect them to have been induced by specific disease activity, but by presence of the disease in general.
Conclusions
In summary, our findings support the hypothesis that the maternal immune system is active during pregnancy, as depicted by the significant changes in expression of immune response genes in the global transcriptome of both healthy women and women with RA. We have identified several genes that demonstrate pregnancy-associated expression patterns that were similar in healthy women and in women with RA, with only few genes showing altered expression in the presence of RA. These findings broaden our understanding of pregnancy-associated systemic changes in the maternal transcriptome among healthy women and women with RA. While it is not yet known how these changes in gene expression might influence the decreased risk of RA onset during pregnancy or contribute to pregnancy-induced amelioration of RA, these results, though still exploratory, may represent a first step towards elucidating the beneficial influence of pregnancy on RA.
Methods
Study subjects
Twenty five women of Danish descent, 5 healthy and 20 with RA, were recruited and enrolled in a pregnancy cohort in Denmark. All 5 healthy women and 14 of the women with RA were enrolled before pregnancy; another 6 women with RA were enrolled after they conceived, in the first (n = 2), second (n = 1) and third (n = 3) trimesters. One of the 14 women with RA enrolled before pregnancy was not followed after she conceived because she had a miscarriage. Study subjects were followed prospectively from the time of enrollment through the third trimester. Subjects with RA fulfilled the 1987 revised American College of Rheumatology criteria for RA [37]. The study was approved by the Ethics Committee for Region Hovedstaden (Denmark), the Danish Data Protection Agency, and the Children’s Hospital Oakland Research Institute Institutional Review Board. All subjects provided written informed consent prior to enrolment.
Sample collection and processing
Blood samples were drawn into PAXgene RNA tubes at 4 time points: before conception and once every trimester during pregnancy (gestational weeks 6–8, 24 and 32). Data on medication use during the 3 months prior to the blood draw were also collected at the same time points from the subjects with RA. Total RNA was extracted from frozen blood samples using the PAXgene Blood RNA Isolation kit according to the manufacturer’s protocol. RNA integrity was assayed using 2100 Bioanalyzer and D1000 ScreenTape (Agilent Technologies). The Illumina TruSeq RNA sample preparation kit was used to generate barcoded cDNA libraries depleted of ribosomal RNAs (rRNAs) and globin mRNAs. Pooled libraries were sequenced on an Illumina HiSeq2500 instrument to generate on average 60 million paired-end reads of 100 bp length. cDNA library preparation and sequencing were performed in two batches. Sample replicates were included in both batches to assess and correct for batch effects. To minimize confounding from experimental effects introduced by variations in library preparation, flow cells or lanes, sample order was randomized prior to library preparation and a blocking design [38] was adopted for sequencing.
Bioinformatic analyses
The raw sequence reads (FASTQ format) were aligned to cDNA sequences of the human GRCh38 reference assembly available in Ensembl using the Bowtie2 (v2.2.5) algorithm [39]. The reads were mapped as concordant paired reads with stringent read and reference gap penalties (—rdg 6,5—rfg 6,5—score-min L,-.6,-.4) to report all alignments. The resulting alignments in SAM format were converted to BAM format using SAMtools [40]. Transcript abundances were calculated from the BAM files using eXpress (v1.5.1) with default settings [41]. BioMart [42] annotations were used to map the transcript Ensembl identifiers to their associated gene loci using in-house python scripts. Gene-level quantifications were determined by combining the transcript-level quantifications onto their associated gene loci. To remove unwanted variation in the transcriptome profiles associated with batch effects, genes which showed at least 10-fold difference in expression (FDR-adjusted p-value < 0.05) across two batches in replicate samples were filtered out, and correlations in expression levels between batches were computed. Genes with very low read counts across all samples were also removed. To adjust for variable sequencing depths between samples, the raw gene counts were normalized using a weighted trimmed mean of the log expression ratios (Trimmed Mean of M values [TMM] algorithm) as implemented in the edgeR Bioconductor package [43, 44].
Statistical analyses
Longitudinal regression models
To examine associations between repeated measures of gene expression levels over 4 time points corresponding to different pregnancy status (pre-pregnancy, first, second and third trimesters), Generalized Estimating Equations (GEE) models were fitted, with robust estimation, using normalized gene counts as the outcome variable and pregnancy status as the main explanatory temporal variable. Covariates included in the models were presence of RA (present/absent), medication use before/during pregnancy (yes/no), and sequencing batch (1/2). To determine whether associations between gene expression levels and pregnancy status were modified by the presence of RA, an interaction term (RA×Pregnancy) was also included in the model. These models allowed within-person correlations in the repeated measures of the data to be adjusted for. A negative binomial link function was used to handle the over-dispersion in RNA-seq gene counts. Both independent and exchangeable correlation structures were tested. Correction for multiple testing was achieved using the Benjamini-Hochberg False Discovery Rate (FDR) method [45]. We refer to the FDR-adjusted p values as q values [46] henceforth. A threshold of q<0.05 was used to assess statistical significance of associations tested. All statistical computations were performed using STATA version 13.1 software.
Temporal patterns in expression levels
For genes demonstrating statistical association with pregnancy status, temporal variations, i.e. from pre-pregnancy to the third trimester, in gene expression levels (log-transformed normalized gene counts) among healthy women and women with RA were examined using pre-pregnancy expression levels in healthy women as reference. The ratio of average expression levels at each time point relative to the average expression levels in the reference group was evaluated as the mean fold change, and these were plotted as a heat map.
Gene ontology and pathway analyses
Sets of genes demonstrating evidence of association with pregnancy status were analyzed for over-representation of biological processes and pathways terms in the Gene Ontology (GO) and KEGG databases using a hypergeometric test implemented in WebGestalt [47]. A q value<0.05 was used to define significant enrichment.
Supporting Information
Bar plot showing total number of mapped reads for each sample from healthy women and women with RA.
(TIF)
The plots and correlations shown are representative of 3 independent sets of sample replicates included in the 2 batches of samples, to correct for batch effects. In the left panel, log-transformed* gene-level counts from technical replicates of the same biological sample (sample 1) are plotted against each other. These technical replicates represent independent cDNA libraries prepared within a single batch of samples (batch 1). The Pearson correlation between the within-batch replicates was 0.99. In the right panel, the log-transformed gene-level counts are plotted for sample 1/batch 1 on the x-axis and for a technical replicate of sample 1 prepared as part of a separate batch of samples (replicate 2/batch 2) on the y-axis, after correction for batch effects. The Pearson correlation for these between-batch replicates was 0.98. (* To accommodate genes which had a read count of zero, log(counts+1) was used.)
(TIF)
Acknowledgments
We are grateful to the study subjects for their participation in the study.
The Rheumatology departments at the following hospitals in Denmark facilitated collection of data and samples: Rigshospitalet (Glostrup), Odense Universitetshospital, Kong Christian X's Gigthospital (Gråsten), Århus University Hospital NBG and Regionshospitalet Viborg.
We thank Anne-Grethe Rasmussen, Charlotte Schön Frengler, Dorte Heide, Randi Petersen, Tove Thorup Rasmussen, Lone Thomasen, Britta Hvidberg Nielsen, Teresa Rozenfeldt, Kirsten Junker, Lis Kastberg Schubert, Lis Lund, and Jette Barlach for their contribution with data and sample collection, and Rikke Godtkjær Andersen, Mie Rasmussen and Tashnia Hossain for management of data and samples.
We also greatly appreciate valuable assistance provided by Majbritt Norman Nielsen, and DANBIO personnel.
Dr. Hanne Kjærgaard passed away before the submission of the final version of this manuscript. Dr. Damini Jawaheer accepts responsibility for the integrity and validity of the data collected and analyzed.
Abbreviations
- ANOVA
analysis of variance
- bp
base pair
- cDNA
complementary DNA
- FDR
False Discovery Rate
- GEE
Generalized Estimating Equations
- GO
gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- mRNA
messenger RNA
- RA
rheumatoid arthritis
- RNA-seq
RNA sequencing
- rRNA
ribosomal RNA
- TMM
Trimmed Mean of M-values
Data Availability
Data are governed by Danish privacy laws. The authors are legally forbidden from publicly sharing data under the terms of their agreement with the Danish Data Protection Agency. Data are available upon request to the corresponding author, after approval is granted by the Danish Data Protection Agency.
Funding Statement
This work was supported in part by funds from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS, http://www.niams.nih.gov/), USA (grant R21AR057931); and Gigtforeningen (https://www.gigtforeningen.dk/), Denmark (grant R87-A1477-B512). Author DJ received the funding. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Silman A, Kay A, Brennan P. Timing of pregnancy in relation to the onset of rheumatoid arthritis. Arthritis Rheum. 1992;35(2):152–5. [DOI] [PubMed] [Google Scholar]
- 2. Lansink M, de Boer A, Dijkmans BA, Vandenbroucke JP, Hazes JM. The onset of rheumatoid arthritis in relation to pregnancy and childbirth. Clin Exp Rheumatol. 1993;11(2):171–4. [PubMed] [Google Scholar]
- 3. Oka M. Effect of pregnancy on the onset and course of rheumatoid arthritis. Ann Rheum Dis. 1953;12(3):227–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guthrie KA, Dugowson CE, Voigt LF, Koepsell TD, Nelson JL. Does pregnancy provide vaccine-like protection against rheumatoid arthritis? Arthritis Rheum. 2010;62(7):1842–8. 10.1002/art.27459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hench PS. The ameliorating effect of pregnancy on chronic atrophic (infectious rheumatoid) arthritis, fibrositis, and intermittent hydrarthrosis. Mayo Clin Proc. 1938;13:161–7. [Google Scholar]
- 6. Nelson JL, Ostensen M. Pregnancy and rheumatoid arthritis. Rheum Dis Clin North Am. 1997;23(1):195–212. [DOI] [PubMed] [Google Scholar]
- 7. de Man YA, Dolhain RJ, van de Geijn FE, Willemsen SP, Hazes JM. Disease activity of rheumatoid arthritis during pregnancy: results from a nationwide prospective study. Arthritis Rheum. 2008;59(9):1241–8. 10.1002/art.24003 [DOI] [PubMed] [Google Scholar]
- 8. Barrett JH, Brennan P, Fiddler M, Silman AJ. Does rheumatoid arthritis remit during pregnancy and relapse postpartum? Results from a nationwide study in the United Kingdom performed prospectively from late pregnancy. Arthritis Rheum. 1999; 42(6):1219–27. [DOI] [PubMed] [Google Scholar]
- 9. Nelson JL, Hughes KA, Smith AG, Nisperos BB, Branchaud AM, Hansen JA. Maternal-fetal disparity in HLA class II alloantigens and the pregnancy-induced amelioration of rheumatoid arthritis. N Engl J Med. 1993; 329(7):466–71. [DOI] [PubMed] [Google Scholar]
- 10. Adams KM, Nelson JL. Microchimerism: an investigative frontier in autoimmunity and transplantation. JAMA. 2004;291(9):1127–31. [DOI] [PubMed] [Google Scholar]
- 11. Winn VD, Haimov-Kochman R, Paquet AC, Yang YJ, Madhusudhan MS, Gormley M et al. Gene expression profiling of the human maternal-fetal interface reveals dramatic changes between midgestation and term. Endocrinology. 2007;148(3):1059–79. 10.1210/en.2006-0683 [DOI] [PubMed] [Google Scholar]
- 12. Mikheev AM, Nabekura T, Kaddoumi A, Bammler TK, Govindarajan R, Hebert MF et al. Profiling gene expression in human placentae of different gestational ages: an OPRU Network and UW SCOR Study. Reprod Sci. 2008;15(9):866–77. 10.1177/1933719108322425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Matjila M, Millar R, van der Spuy Z, Katz A. The differential expression of Kiss1, MMP9 and angiogenic regulators across the feto-maternal interface of healthy human pregnancies: implications for trophoblast invasion and vessel development. PLoS One. 2013;8(5):e63574 10.1371/journal.pone.0063574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Haupl T, Ostensen M, Grutzkau A, Radbruch A, Burmester GR, Villiger PM. Reactivation of rheumatoid arthritis after pregnancy: increased phagocyte and recurring lymphocyte gene activity. Arthritis Rheum. 2008;58(10):2981–92. 10.1002/art.23907 [DOI] [PubMed] [Google Scholar]
- 15. Haupl T, Ostensen M, Grutzkau A, Burmester GR, Villiger PM. Interaction between rheumatoid arthritis and pregnancy: correlation of molecular data with clinical disease activity measures. Rheumatology (Oxford). 2008;47 Suppl 3:iii19–22. 10.1093/rheumatology/ken157 [DOI] [PubMed] [Google Scholar]
- 16. Weix J, Forger F, Haupl T, Surbek D, Ostensen M, Villiger PM. Influence of pregnancy on the adipocytokine and peroxisome proliferator-activated receptor pathways in peripheral blood mononuclear cells from healthy donors and rheumatoid arthritis patients. Arthritis Rheum. 2012;64(7):2095–103. 10.1002/art.34375 [DOI] [PubMed] [Google Scholar]
- 17. Weix J, Haupl T, Raio L, Villiger PM, Forger F. The physiologic increase in expression of some type I IFN-inducible genes during pregnancy is not associated with improved disease activity in pregnant patients with rheumatoid arthritis. Transl Res. 2013;161(6):505–12. 10.1016/j.trsl.2013.02.007 [DOI] [PubMed] [Google Scholar]
- 18. Wang Y, Gu Y, Philibert L, Lucas M. Neutrophil activation induced by placental factors in normal and pre-eclamptic pregnancies in vitro. Placenta. 2001;22(6):560–5. [DOI] [PubMed] [Google Scholar]
- 19. Luppi P, Haluszczak C, Trucco M, Deloia J. Normal pregnancy is associated with peripheral leukocyte activation. American Journal of Reproductive Immunology. 2002;47(2):72–81. [DOI] [PubMed] [Google Scholar]
- 20. Sacks GP, Studena K, Sargent IL, Redman CW. Normal pregnancy and preeclampsia both produce inflammatory changes in peripheral blood leukocytes akin to those of sepsis. American journal of obstetrics and gynecology. 1998;179(1):80–6. [DOI] [PubMed] [Google Scholar]
- 21. Heung M, Jin S, Tsui N, Ding C, Leung TY, Lau TK et al. Placenta-derived fetal specific mRNA is more readily detectable in maternal plasma than in whole blood. PLoS One. 2009;4(6):e5858 10.1371/journal.pone.0005858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mor G, Cardenas I, Abrahams V, Guller S. Inflammation and pregnancy: the role of the immune system at the implantation site. Ann N Y Acad Sci. 2011;1221:80–7. 10.1111/j.1749-6632.2010.05938.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Koh W, Pan W, Gawad C, Fan HC, Kerchner GA, Wyss-Coray T et al. Noninvasive in vivo monitoring of tissue-specific global gene expression in humans. Proc Natl Acad Sci U S A. 2014;111(20):7361–6. 10.1073/pnas.1405528111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tsui NB, Jiang P, Wong YF, Leung TY, Chan KC, Chiu RW et al. Maternal plasma RNA sequencing for genome-wide transcriptomic profiling and identification of pregnancy-associated transcripts. Clin Chem. 2014;60(7):954–62. 10.1373/clinchem.2014.221648 [DOI] [PubMed] [Google Scholar]
- 25. Lyons TR, Schedin PJ, Borges VF. Pregnancy and breast cancer: when they collide. J Mammary Gland Biol Neoplasia. 2009;14(2):87–98. 10.1007/s10911-009-9119-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. O'Hara MW, Wisner KL. Perinatal mental illness: definition, description and aetiology. Best Pract Res Clin Obstet Gynaecol. 2014;28(1):3–12. 10.1016/j.bpobgyn.2013.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Scrandis DA, Sheikh TM, Niazi R, Tonelli LH, Postolache TT. Depression after delivery: risk factors, diagnostic and therapeutic considerations. ScientificWorldJournal. 2007;7:1670–82. 10.1100/tsw.2007.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Amino N, Tada H, Hidaka Y. Postpartum autoimmune thyroid syndrome: a model of aggravation of autoimmune disease. Thyroid. 1999;9(7):705–13. [DOI] [PubMed] [Google Scholar]
- 29. Poser S, Poser W. Multiple sclerosis and gestation. Neurology. 1983;33(11):1422–7. [DOI] [PubMed] [Google Scholar]
- 30. Fu X, Fu N, Guo S, Yan Z, Xu Y, Hu H et al. Estimating accuracy of RNA-Seq and microarrays with proteomics. BMC Genomics. 2009;10:161 10.1186/1471-2164-10-161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. 10.1038/nrg2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Julia A, Erra A, Palacio C, Tomas C, Sans X, Barcelo P et al. An eight-gene blood expression profile predicts the response to infliximab in rheumatoid arthritis. PLoS One. 2009;4(10):e7556 10.1371/journal.pone.0007556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim TH, Choi SJ, Lee YH, Song GG, Ji JD. Gene expression profile predicting the response to anti-TNF treatment in patients with rheumatoid arthritis; analysis of GEO datasets. Joint, bone, spine: revue du rhumatisme. 2014;81(4):325–30. 10.1016/j.jbspin.2014.01.013 [DOI] [PubMed] [Google Scholar]
- 34. Yarilina A, Park-Min KH, Antoniv T, Hu X, Ivashkiv LB. TNF activates an IRF1-dependent autocrine loop leading to sustained expression of chemokines and STAT1-dependent type I interferon-response genes. Nat Immunol. 2008;9(4):378–87. 10.1038/ni1576 [DOI] [PubMed] [Google Scholar]
- 35. Kwan Tat S, Pelletier JP, Amiable N, Boileau C, Lajeunesse D, Duval N et al. Activation of the receptor EphB4 by its specific ligand ephrin B2 in human osteoarthritic subchondral bone osteoblasts. Arthritis Rheum. 2008;58(12):3820–30. 10.1002/art.24029 [DOI] [PubMed] [Google Scholar]
- 36. Gomez-Lopez N, StLouis D, Lehr MA, Sanchez-Rodriguez EN, Arenas-Hernandez M. Immune cells in term and preterm labor. Cell Mol Immunol. 2014;11(6):571–81. 10.1038/cmi.2014.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31(3):315–24. [DOI] [PubMed] [Google Scholar]
- 38. Fang Z, Cui X. Design and validation issues in RNA-seq experiments. Brief Bioinform. 2011;12(3):280–7. 10.1093/bib/bbr004 [DOI] [PubMed] [Google Scholar]
- 39. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9(4):357–9. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Roberts A, Pachter L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nature methods. 2013;10(1):71–3. 10.1038/nmeth.2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Biomart. Available: http://wwwbiomartorg/biomart/martview.
- 43. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society, Series B. 1995;57:289–300. [Google Scholar]
- 46. Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440–5. 10.1073/pnas.1530509100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic acids research. 2005;33(suppl 2):W741–W8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bar plot showing total number of mapped reads for each sample from healthy women and women with RA.
(TIF)
The plots and correlations shown are representative of 3 independent sets of sample replicates included in the 2 batches of samples, to correct for batch effects. In the left panel, log-transformed* gene-level counts from technical replicates of the same biological sample (sample 1) are plotted against each other. These technical replicates represent independent cDNA libraries prepared within a single batch of samples (batch 1). The Pearson correlation between the within-batch replicates was 0.99. In the right panel, the log-transformed gene-level counts are plotted for sample 1/batch 1 on the x-axis and for a technical replicate of sample 1 prepared as part of a separate batch of samples (replicate 2/batch 2) on the y-axis, after correction for batch effects. The Pearson correlation for these between-batch replicates was 0.98. (* To accommodate genes which had a read count of zero, log(counts+1) was used.)
(TIF)
Data Availability Statement
Data are governed by Danish privacy laws. The authors are legally forbidden from publicly sharing data under the terms of their agreement with the Danish Data Protection Agency. Data are available upon request to the corresponding author, after approval is granted by the Danish Data Protection Agency.