Abstract
Toxicological assays measuring mortality are routinely used to describe insecticide response, but sub-lethal exposures to insecticides can select for resistance and yield additional biological information describing the ways in which an insecticide impacts the insect. Here we present the Wiggle Index (WI), a high-throughput method to quantify insecticide response by measuring the reduction in motility during sub-lethal exposures in larvae of the vinegar fly Drosophila melanogaster. A susceptible wild type strain was exposed to the insecticides chlorantraniliprole, imidacloprid, spinosad, and ivermectin. Each insecticide reduced larval motility, but response times and profiles differed among insecticides. Two sets of target site mutants previously identified in mortality studies on the basis of imidacloprid or spinosad resistance phenotypes were tested. In each case the resistant mutant responded significantly less than the control. The WI was also able to detect a spinosad response in the absence of the primary spinosad target site. This response was not detected in mortality assays suggesting that spinosad, like many other insecticides, may have secondary targets affecting behaviour. The ability of the WI to detect changes in insecticide metabolism was confirmed by overexpressing the imidacloprid metabolizing Cyp6g1 gene in digestive tissues or the central nervous system. The data presented here validate the WI as an inexpensive, generic, sub-lethal assay that can complement information gained from mortality assays, extending our understanding of the genetic basis of insecticide response in D. melanogaster.
Introduction
Insect pests are major vectors of human infectious diseases and also impose massive production losses and control costs in agriculture [1]. Chemical insecticides are often used as a weapon of choice to control or eliminate pests. While insecticides are potent in the short term, long term efficacy is often stymied by the evolution and spread of resistance [2,3]. There are a limited number of chemicals, targeting an even smaller number of insect target proteins, meaning it is critical to manage or even prevent resistance to current generation insecticides [4]. A deeper understanding of the insect’s biological response to an insecticide, and thus the array of resistance options available to insect pests, underpins rational resistance management strategies. The vinegar fly Drosophila melanogaster, while not a pest species, has proven to be powerful model that can be manipulated to interrogate the insect: insecticide interface [5–7].
There are many genetically determined biological mechanisms that influence how an insect responds to an insecticide. Among these, two that are particularly intensively studied because of their large contribution to insecticide resistance both in field and laboratory settings. The overexpression of genes encoding drug metabolizing enzymes (DMEs) such as cytochrome P450s (CYPs), carboxylesterases (CoEs) glutathione S transferases (GSTs) and glucuronosyltransferases (UGTs), which have the capacity to modify the chemical structure of insecticides, can cause resistance [8]. For example, the overexpression Cyp6cm1 in Bemisia tabaci and Cyp6g1 in D. melanogaster is sufficient to confer resistance to a range of insecticide classes [9–11]. The modification or loss of insecticide target sites is another commonly found and well-studied resistance mechanism. In D. melanogaster a variety of mutations in Dα1 or Dβ2 nicotinic acetylcholine receptor (nAChR) subunit genes confer imidacloprid resistance [12], while knockouts and amino acid substitutions in the Dα6 subunit confer resistance to spinosad [13,14]. These findings were mirrored by studies in other insects, which found nAChR modifications linked to imidacloprid resistance in Nilaparvata lugens [15] and M. persicae [16] and to spinosad resistance in Plutella xylostella and Frankinella occidentalis [17,18].
Much of what is currently known about the mechanisms by which insects respond to insecticides comes from mortality assays [19], but sub-lethal insecticide response is an economically relevant phenotype that can describe additional information about insecticide biology [20,21]. Pest populations are frequently subjected to sub-lethal exposures in field settings [22,23], which have been shown to provide selection pressure [24]. For example, sub-lethal exposures to insecticides have been shown to significantly alter the fecundity [25], feeding behaviour [26] and locomotion [27] of targeted pest species. Each of these phenotypes have fitness costs and are often elicited by concentrations far below the LC50 value of a compound [28]. Considering these sub-lethal effects on pests may also give new insights into the mechanisms that influence insecticide resistance. Following the ingestion of an insecticide there is a cascade of biological processes which determine the response of the insect, with death as one possible endpoint. Observing the impact of sub-lethal exposures on living insects could focus on processes earlier in this cascade and the potential mechanisms of resistance that may arise from them. These points can also be extended to non-pest insects, which often undergo the same sub-lethal exposures [20]. Special attention has recently been given to the sub-lethal neonicotinoid exposures in honeybees that have been suggested to contribute to colony collapse disorder [29], although this is controversial [30].
Several methods to measure the sub-lethal effects of xenobiotics have been developed. For example, the crawling speed and pattern of D. melanogaster larvae from sensitive and control backgrounds were found to differ during acute exposure to ethanol [31]. More recently, several video tracking softwares such as Ethovision (Nodulus) and VideoTrack (ViewPoint Life Sciences) have been used to more precisely measure insect motility. These kinds of software have been used to describe sub-lethal insecticide response in mosquitos [27] and beetles [32] with a range of different insecticides. Other studies have measured alterations in sub-lethal phenotypes discussed above such as fecundity, feeding behaviour and locomotion after sub-lethal xenobiotic exposures [25–27].
Here, we describe an additional sub-lethal behavioural assay the Wiggle Index (WI; 38), which uses an open source ImageJ macro to quantify the temporal motility response of D. melanogaster larvae to sub-lethal insecticide exposures. Larvae from a wild type susceptible strain were tested with the WI with insecticides from four mode of action (MoA) classes. The response curve of the larvae differed depending on the insecticide used. The capacity of WI to detect insecticide resistance was tested using several pairs of resistant and matching control (susceptible) genotypes previously characterized using mortality assays. In each case resistant and susceptible genotypes were distinguishable. Additional biological information was also gained by observing motility response to the insecticide spinosad in a highly resistant target site mutant, suggesting additional target(s) for this compound. Using the wealth of genetic resources available in D. melanogaster, the WI can be used to investigate the role of individual genes in sublethal insecticide response.
Methods
Fly Stocks
All stocks were maintained at room temperature and under constant light. Armenia14 is an isofemale line derived from Armenia60 (Drosophila Genomics Resource Center #103394), and was used as a susceptible wild type for all dose response experiments. Three additional sets of genotypes consisted of lines previously associated with either imidacloprid or spinosad resistance in D. melanogaster, and had genetically matched control backgrounds (Table 1). Additionally, each of these sets had relatively well characterized resistance mechanisms that have been elaborated on in other insect species [17] or with in vitro studies [33,34]. Set one contains nAChR subunit mutants Dα1 M4 and Dβ2 L351Q that were generated with EMS mutagenesis of Armenia14 and are resistant to imidacloprid in mortality assays [12]. Dα1 M4 produces a truncated Dα1 protein product that is cut short in the transmembrane M4 domain, and Dβ2 L351Q contains a single amino acid replacement in the Dβ2 subunit. Set two includes two genotypes also generated with EMS mutagenesis and selection of Armenia14 carrying mutations in the Dα6 nAChR subunit (Dα6 nx, Dα6 W337*); each confers spinosad resistance [7]. Dα6 W337* is truncated 13 residues after the third transmembrane domain, while Dα6 nx has no detectable Dα6 expression. Set three used the HR_GAL4 to drive the expression of Cyp6g1 in the midgut, Malpighian tubules, fat body (referred to as digestive tissues here) by crossing it to the UAS_Cyp6g1 line [6]. A matched control for this genotype was generated by crossing the HR_GAL4 driver line to the Φ86FB genotype, which has the same genetic background as the UAS_Cyp6g1 line [35]. Previous studies linked overexpression of Cyp6g1 in these digestive tissues with resistance to multiple classes of insecticide [36]. Finally, in set four the overexpression of Cyp6g1 was achieved in the central nervous system (CNS) using the Elav-GAL4 (Bloomington Stock Center # 458) driver line. Unlike the previous genotype sets, the overexpression of Cyp6g1 in the CNS has not been previously linked to resistance.
Table 1. Genotypes Used in the Present Study.
| Name | Background | Stock # | Resistance |
|---|---|---|---|
| Armenia 14 | Armenia | DGRC (103394) | Susceptible |
| Dα1 M4 | Armenia 14 | Batterham Lab Stock | Imidacloprid |
| Dβ2 L351R | Armenia 14 | Batterham Lab Stock | Imidacloprid |
| Dα6 nx | Armenia 14 | Batterham Lab Stock | Spinosad |
| Dα6 W337* | Armenia 14 | Batterham Lab Stock | Spinosad |
| Φ86FB | Φ86FB | Bischoff et. al 2007 | Susceptible |
| UAS_Cyp6g1 | Φ86FB | Batterham Lab Stock | Imidacloprid |
| Elav-GAL4 | Elav-GAL4 | Bloomington (458) | Susceptible |
All genotypes used in the current study are displayed along with their backgrounds and previously reported resistance status.
Obtaining Third Instar Larvae
For all isogenic lines, sixty 2–5 day old females and twenty males were collected and transferred onto maize meal medium (S1 Table) in vials sprinkled with dried yeast. For all crosses, the same procedure was followed with virgin females. In each case flies were left undisturbed for one day at 25°C for oviposition and then cleared from the vial. The vials were kept at 25°C for 68 additional hours to generate a population of early third instar larvae, which were recovered from the food using sucrose extraction [37]. Briefly, 30mL of 20% w/v sucrose (non-Analytical Reagent) in distilled H20 was poured onto the food. The top layer of the food was then gently disrupted with a metal rod in order to release the larvae, which float in the sucrose solution. The solution was then carefully poured onto a fine cloth mesh to isolate the larvae, which were then dried and transferred onto grape juice agar plates (S1 Table). Third Instar larvae (~5mm in length) were then individually picked, 25 per well, into a NUNC cell culture treated 24 well plate (Thermo-Scientific) preloaded with 200μL Analytical Reagent 5% w/v sucrose (Chem Supply) in distilled H20. This volume of 5% sucrose was chosen because it is the minimum amount needed to cover the 1.8cm2 bottom of the well. In this environment the larvae were able to translocate, but stationary gyrations represented the majority of their activity. To dose the larvae, 50μL of 5x insecticide stock solution was added to each well in order to bring the final concentration to the desired dosage. After mixing, 50μL of the mixed solution was removed in order to bring the final volume back to 200μL.
Insecticide Dilution
Chlorantraniliprole (10 gL-1 Coragen®; Du Pont), Imidacloprid (200 gL-1 Confidor®; Bayer Crop Science), and Spinosad (10 gL-1 Success®; Yates) were all purchased commercially and diluted to 5,000ppm stocks using distilled water. On the day of exposure, 5x stocks were generated for each dose being used (Chlorantraniliprole: 60, 30, 15, 7.5, 3.75, 0ppm; Imidacloprid: 240, 120, 60, 30, 15, 0ppm; Spinosad: 240, 60, 30, 15, 7.5, 3.75, 0 ppm) by diluting the 5,000ppm stock in 5% Analytical Reagent sucrose (Chem Supply). A similar procedure was followed for Ivermectin (Sigma), and a 10,000ppm stock was generated using DMSO as a solvent. 5x stocks were generated (120, 60, 30, 15, 7.5, 0 ppm) by dissolving the original stock in 5% sucrose, and the highest concentration of DMSO used in dosing was added to the 0ppm solution in order to control for solvent effects, none of which were observed.
Experimental Design
To assess the effect of different insecticides on movement, the susceptible control Armenia14 was tested with a range of 5 doses for imidacloprid (3ppm-48ppm), ivermectin (1.5ppm-24ppm), spinosad (1.5ppm-24ppm), and chlorantraniliprole (.75ppm-12ppm). Pilot studies were used to determine the best range of doses to be used for each insecticide. Set one mutants (Dα1 M4 and Dβ2 L351Q) were tested at 12 and 48ppm imidacloprid and the same doses were used for set two mutants (Dα6 nx and Dα6 W337*) with spinosad. Set three and four genotypes expressing Cyp6g1 in the three digestive tissues or CNS were tested at 48ppm imidacloprid.
Filming
Ten second videos were taken at 5,10,15, 30, 45, 60, 120 and 240 minutes after the addition of insecticide and compared to videos taken immediately before the addition of the insecticide (time = 0). Each video captured larval activity in 4 individual wells that were processed separately. All filming was done using a Panasonic 3CCD Ultra-Compact™ Digital Palmcorder® against a LED light box (Huion L4S Led Light Pad). As some of the insecticides examined here are light sensitive [38,39], larvae were kept in darkness except for a brief period before and during filming. 15 seconds was allowed for larvae to acclimate to light conditions on the LED pad, before filming began. Because each 24 well plate required 6 separate videos to be taken, larvae were exposed to the light for approximately 120 seconds for each time point.
Video Processing and Analysis of Larval Movement
Raw videos were named in bulk via a renaming script written in R (R: A language and Environment for Statistical Computing). Named videos were split into jpeg image sequences using the free software Video Jpg Converter (DVDVideoSoft), and a ten second video produced 250 frames for analysis. Image Sequences were then uploaded to a server on the NeCtar research cloud for further processing. There, the Fiji distribution of ImageJ [40] was used to run the WI script (Fig 1; S1 Script).
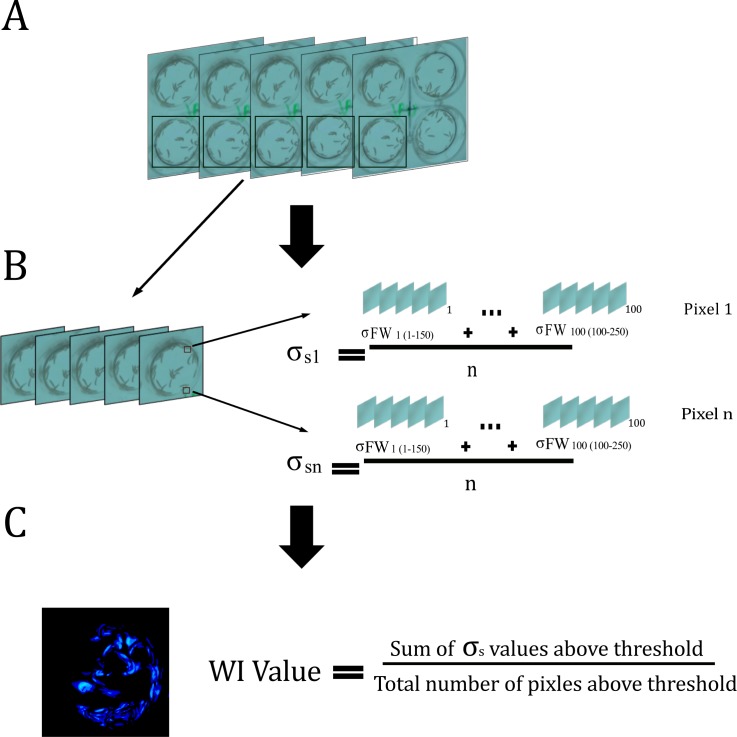
Fig 1. The Wiggle Index.
The Wiggle Index measures total motility in a given well A) Individual wells are cropped out of a sequence of 250.jpeg images B) σS values are calculated for each pixel by calculating the standard deviation of all σFW values over the entire sequence. C) σS values from each pixel are filtered based on a threshold value and then averaged to yield the Wiggle Index (WI) value.
The WI script analysed individual image sequences and calculated the total motility of the larvae in a given well. Firstly, the script crops one of the four wells from the image sequence and converts it into stacks of.tiff format (Fig 1A). An ImageJ algorithm was then performed similar to that of Preston et. al [41], which is briefly outlined here. Prior to any calculations a Gaussian Blur of 2 pixels was applied to each.tiff stack in order to minimize the impact of artefacts due to video compression. Then for each pixel, a series of frame window standard deviations (σFW) was calculated by measuring the standard deviation of the pixel’s light intensity for 150 frame windows in a rolling manner starting with frame 1 (F1-F150) and continuing until the end of the stack (F101-F250). The formula is:
where W denotes the number of windows, F denotes the frame window size, μW denotes the average frame window intensity and xf i denotes the frame intensity value. A second intermediate variable (σS) was then calculated for each pixel by taking the standard deviation of the series of previously calculated series of σFW values (Fig 1B). The formula for these calculations is:
where μδW is the frame window intensity average.
Using the σS values for each pixel, WI values were calculated, which represent the total motility of the larvae in the ten second video. This was accomplished by averaging the σS values for every pixel above a cut-off threshold (Fig 1C). The formula for WI value is:
where T is the threshold cut off, TA is the number of pixels above the threshold and Σσs≥T σs is the sum of values above the threshold. The cut-off threshold, in this case 30, was applied to remove any background noise due to global movement of the plate or fluctuations of the liquid.
Visualization and Statistics
The WI script generates heat maps (Fig 1C) and reports numeric estimates of total motility (WI values) for each well at a given time. Two independent corrections were applied after the calculation of WI values to account for differences in initial motility between genotypes and to isolate the organisms’ responses to the insecticide.
The first correction divided WI values at a given time point by the WI value of the same larvae prior to the addition of the insecticide and generated relative movement ratio values (RMR values; Fig 2). The RMR correction normalized the motility of larvae to those same larvae in the absence of insecticide. For example, a genotype with a WI value of 10 before the addition of an insecticide and 3 at a later time point would have a RMR value of 3/10 = .3 at that time point. Mean corrected RMR values were plotted together with their associated 95% confidence intervals to visualize the response of different genotypes for a given dose or the response to different doses for a given genotype. Visualizing the data allowed for the description of several aspects of the insecticide response. The Response Time was defined as the time needed for a treatment to significantly and irreversibly reduce the RMR value less than one based on a 95% confidence interval. The End Point RMR value was defined as the final RMR value recorded (240 minutes) and was compared among doses or genotypes using Tukey’s Honestly Significant Difference (HSD) pairwise test (P≤.05).
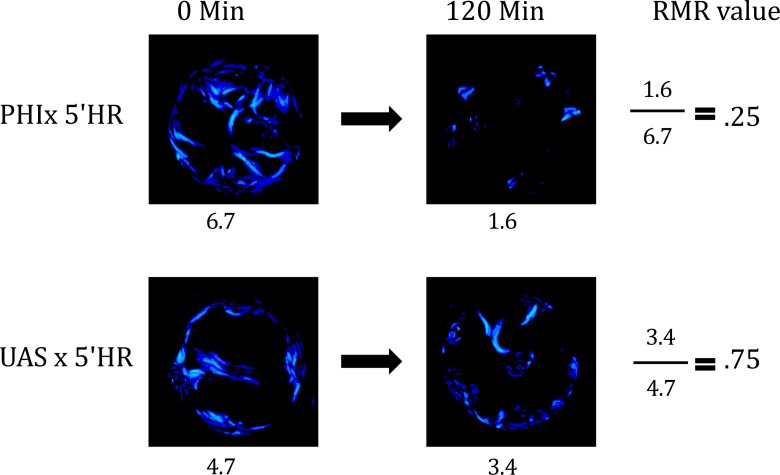
Fig 2. Wiggle Index Output and RMR Calculation.
The Wiggle Index produced heat maps and numeric estimates of total larval motility in each well at a given time. RMR values were calculated by dividing the WI value at time = x from the WI value from the same well at time = 0.
The second correction fits a generalized linear model (GLM) to the response curve based on WI values from a particular well over time. In the case of spinosad, no transformation of the data was needed, but for imidacloprid, a log10 transformed time scale was used as it better fit the data based on adjusted R squared values. The inverse of the slopes of these regression lines (β values) were used as quantitative measurements of overall response. β values of each genotype were compared to control lines using a Student’s t test (P≤.05) and mean β values were plotted with 95% confidence intervals. An interactive R script WI_Analysis was written that accepts raw data from the WI script and generates a series of descriptive graphs including RMR curves and GLM analysis (available on GitHub https://github.com/shanedenecke/WI_Analysis).
Results
The Response of Wild Type to Different Insecticides
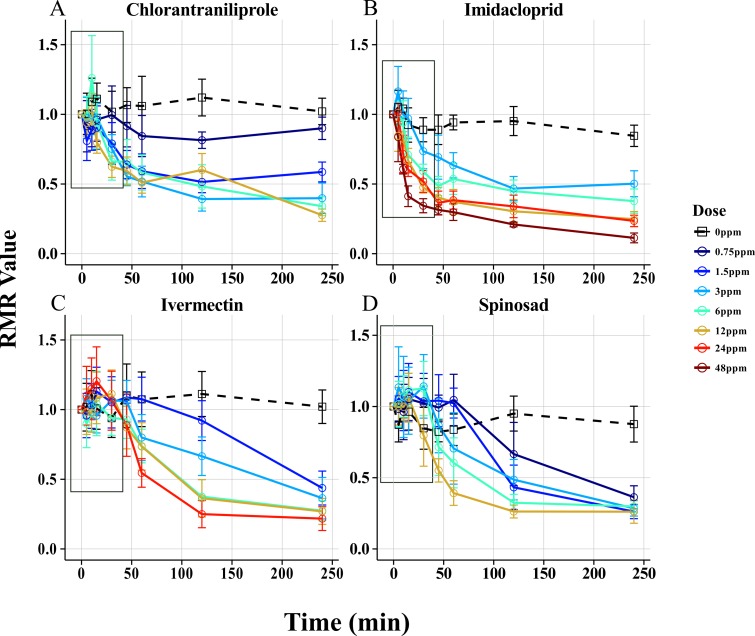
To observe the effect of different classes of insecticides on motility, dose response profiles were generated for Armenia14 with four insecticides from distinct (MoA) classes (Fig 3; Fig 4). Regardless of dose or insecticide, each chemical reduced the motility of larvae decreased as exposure time increased, but two distinct response types were observed depending on the insecticide used, fast acting and slow acting. Fast Acting responses were characterized by rapid, dose dependent Response Times and dose dependent End Point RMR values, while the Slow Acting response displayed delayed, dose dependent Response Times but with similar End Point RMR values. Imidacloprid and chlorantraniliprole were both Fast Acting, with Response Times of ten minutes or less at the highest doses tested followed by End Point RMR values that were significantly different among doses (Fig 3A and 3B; Fig 4A and 4B). Spinosad and Ivermectin were Slow Acting, with Response Times of 45 minutes or greater. The End Point RMR values for these Slow Acting insecticides were not significantly different as determined by Tukey’s HSD (Fig 3C and 3D; Fig 4C and 4D).
Fig 3. Armenia14 Dose Response Curves.
Armenia14 was tested using the Wiggle Index at 5 doses of 4 insecticides. Chlorantraniliprole (A) and Imidacloprid (B) each show rapid response times and dose dependent long term RMR values. Ivermectin (C) and Spinosad (D) responded differently, displaying delayed response times and similar long term RMR values. All points correspond to the mean RMR values with 95% confidence intervals. Boxes in each plot indicate values between 0 and 30 minutes magnified in Fig 4.
Fig 4. Armenia14 Short time Point Dose Response Curves.
Armenia14 was tested using the Wiggle Index at 5 doses of Chlorantraniliprole (A), Imidacloprid (B), Ivermectin (C), and Spinosad (D). All points correspond to the mean RMR values with 95% confidence intervals. All measurements in this figure were observed between 0 and 30 minutes.
Set One: Imidacloprid Resistant nAChR Alleles
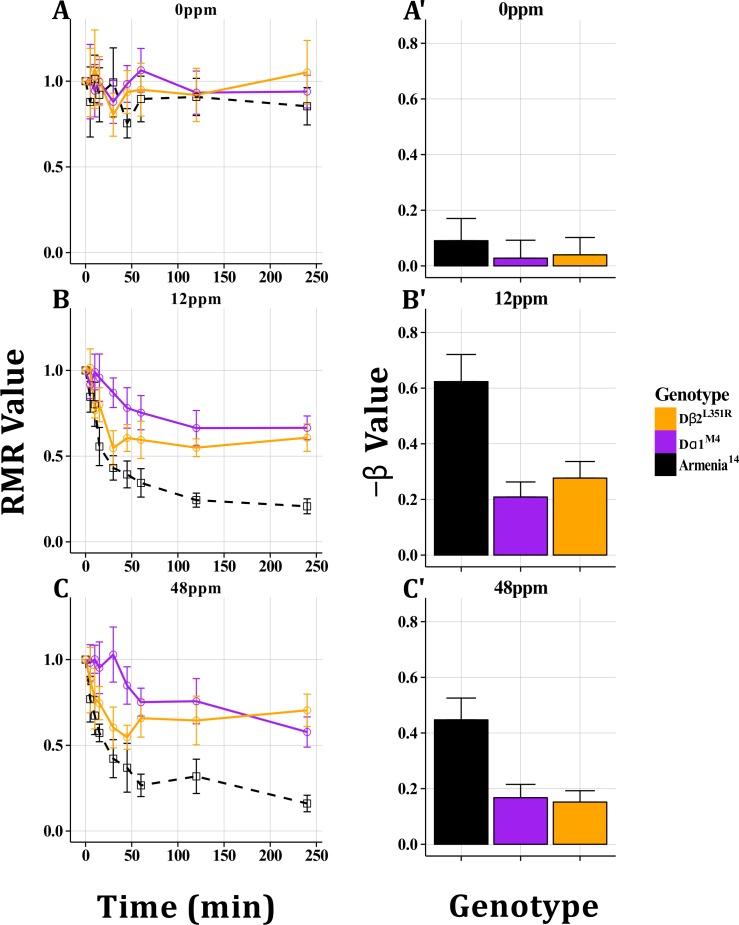
The motility of previously characterized imidacloprid resistant Dα1 and Dβ2 mutants was examined at two different concentrations of imidacloprid (12, 48ppm). Armenia14 showed a greater response to imidacloprid than either of the resistant mutants in the GLM analysis (Fig 5B’ and 5C’) and End Point RMR values (Fig 5B and 5C). Dα1 M4 had the longest Response Time and the highest End Point RMR value. Dβ2 L351Q displayed an intermediate Response Time and End Point RMR Value compared to the other genotypes. In the absence of imidacloprid, there were no significant differences among genotypes in the GLM or RMR analysis (Fig 5A and 5A’).
Fig 5. Imidacloprid Resistance in nAChR Mutants.
Armenia14, Dα1 M4 and Dβ2 L351R were tested using the Wiggle Index. At 0ppm (A, A’), no significant differences were observed between genotypes. As measured by Response Time, End Point RMR values and GLM analysis at 12 and 48ppm imidacloprid (B, B’, C, C’) Dα1 M4 responded the least, Dβ2 L351R displayed an intermediate phenotype and Armenia14 responded the most. All plots display mean RMR or β values with 95% confidence intervals.
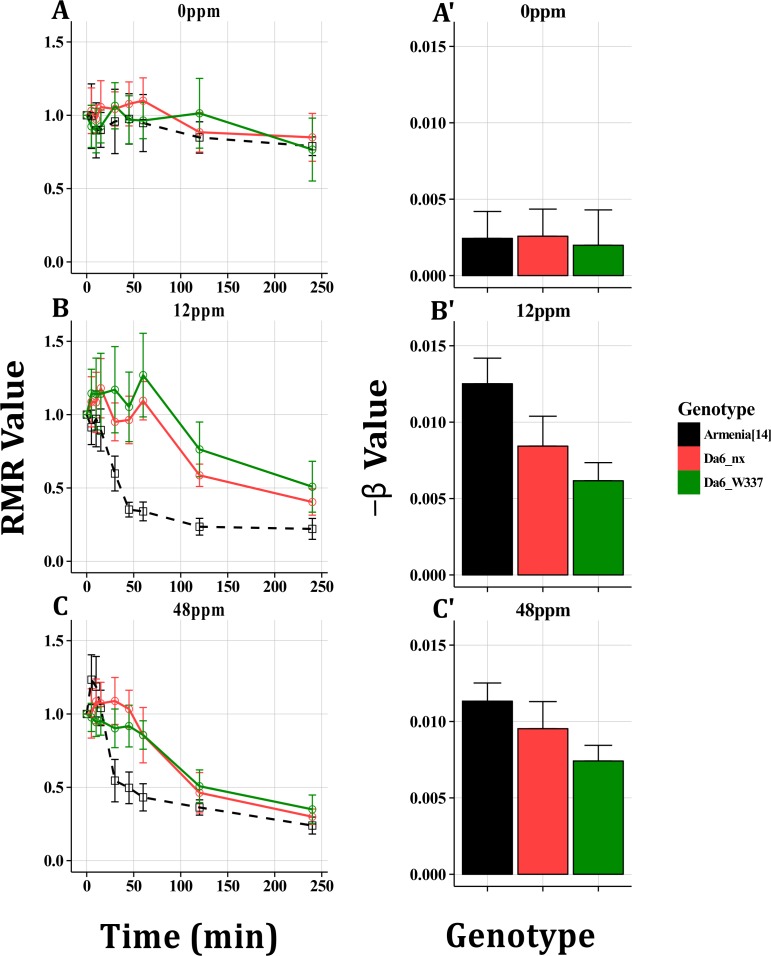
Set Two: Spinosad Resistant Dα6 Alleles
Dα6 mutant genotypes, Dα6 nx and Dα6 W337*, were screened at 2 concentrations of spinosad (12, 48ppm). At each dose both Dα6 mutants showed delayed Response Times compared to Armenia14. At 12ppm Dα6 W337* had a larger End Point RMR value when compared to Armenia14, but the Dα6 nx End Point RMR value was not statistically different. At 48ppm there were no differences in End Point RMR values among any of genotypes (Fig 6B and 6C). GLM analysis indicated small but significant differences between Armenia14and each Dα6 mutant at 12ppm, but at 48ppm only Dα6 W337* was significantly different from the control (Fig 6B’ and 6C’). Each genotype displayed near identical response profiles at 0ppm, indicating that there were no motility differences in the absence of insecticide (Fig 6A and 6A’).
Fig 6. Spinosad Resistance in nAChR Mutants.
Armenia14, Dα6 W337* and Dα6 nx were tested using the Wiggle Index at 0ppm (A, A’), no significant differences were observed between genotypes. At 12 ppm (B, B’) both Dα6 mutants displayed significantly delayed response times and responded less in the GLM analysis. However, while Dα6 337* differed from Armenia14 in End Point RMR value, Dα6 nx did not. At 48ppm (C, C’), while Response Times were still delayed in Dα6 mutants, End Point RMR values were not significantly different and GLM analysis indicated that only Dα6 337* responded more than Armenia14. All plots display mean RMR or β values with 95% confidence intervals.
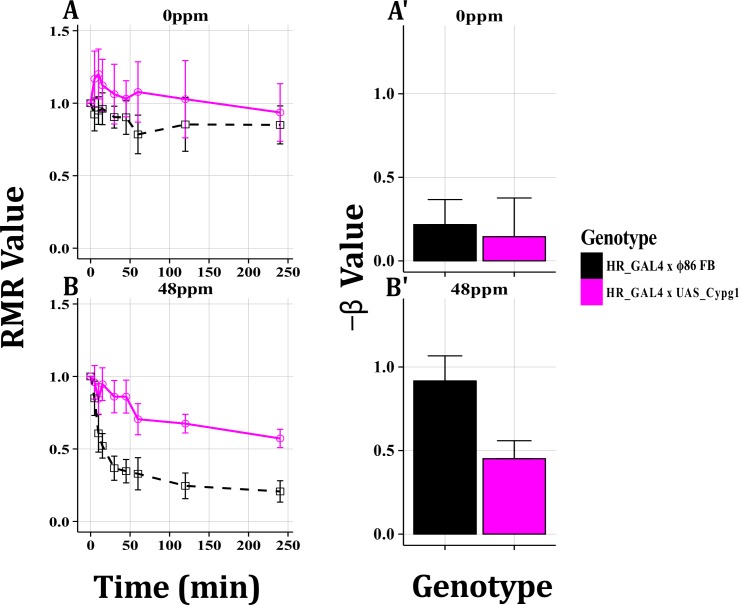
Set Three and Four: Cyp6g1 Overexpression
Cyp6g1 was overexpressed in the midgut, fat body and Malpighian tubules and tested at 48ppm imidacloprid. Lines that overexpressed Cyp6g1, showed a slightly delayed Response Time to imidacloprid than the matched control and showed a significantly higher End Point RMR value, which was supported by GLM analysis (Fig 7B and 7B’). No significant difference was observed at 0ppm (Fig 7A and 7A’).
Fig 7. Cyp6g1 Expression in the Digestive Tissues.
Cyp6g1 overexpression was achieved in the midgut, Malpighian tubule and the fat body using the GAL4-UAS system. At 0ppm (A, A’), no significant differences were observed between genotypes. At 48ppm imidacloprid the overexpression line responded less as measured by Response Time, End Point RMR values and GLM analysis. All plots display mean RMR or β values with 95% confidence intervals.
Cyp6g1 overexpression in the CNS (Elav x UAS_Cyp6g1 c.f. Elav x Φ86FB) was tested at 48ppm imidacloprid. Elav x UAS_Cyp6g1 displayed a 5 minute delay in Response Time and had an End Point RMR value significantly higher than the control (Fig 8B). GLM analysis also found differences between overexpression and control genotypes (Fig 8B’). Overexpression of Cyp6g1 in the CNS had less of an impact on all phenotypes than overexpression in the digestive tissues.
Fig 8. Cyp6g1 Expression in the Central Nervous System.
Cyp6g1 overexpression was achieved in the CNS using the GAL4-UAS system. At 0ppm (A, A’), no significant differences were observed between genotypes. At 48ppm imidacloprid the overexpression line responded less as measured by Response Time, End Point RMR values and GLM analysis. All plots display mean RMR or β values with 95% confidence intervals.
Discussion
Armenia14 Dose Response Profiles
Insect pests impose significant costs on agriculture and human health, but lack of descriptive phenotypes have hindered a deeper understanding of how insects respond and interact with insecticides. Here, a novel method of assessing sub lethal insecticide response in the model insect D. melanogaster is described, in part, by testing a wild type strain with insecticides of different MoA classes. This strain (Armenia14) responded in two distinct ways depending on which of the 4 insecticides was used. Although each of these insecticides belongs to a distinct MoA class, characteristics common between classes may partially explain the types of response profiles observed. Imidacloprid and spinosad target distinct nicotinic acetylcholine receptor subtypes found in the CNS, ivermectin targets ligand gated chloride channels in the same tissue [42], and chlorantraniliprole binds to the ryanodine receptor in muscle cells [43]. Because, imidacloprid, ivermectin, and spinosad all target receptors in the CNS, they must therefore cross the blood-brain barrier. The molecular weight of xenobiotics has been shown to influence their rates of diffusion [44] and transport [45] into the CNS. This could explain the differences in Response Time observed in the WI. Spinosad and ivermectin have molecular weights of 732 Da and 875 Da respectively, while imidacloprid is relatively smaller with a weight of 255 Da. Chlorantraniliprole, though of intermediate size (430 Da), would not be affected by this phenomenon due to its targeting of the more easily accessible ryanodine receptor in muscle cells [43].
Target Site Mutants
For each set of target site mutants used in this study, significant differences were detected in insecticide response for resistant mutants compared to their matched controls using the WI. Following the addition of imidacloprid, there was always a reduction in RMR values. Dα1 and Dβ2 mutants displayed significantly higher End Point RMR values and lower β values than did Armenia14 (Fig 5). In contrast, Dα6 mutants tested with spinosad displayed similar End Point RMR values to Armenia14, but their Response Time was significantly delayed (Fig 6). These same mutants were previously found to be more resistant in mortality assays, and it was hypothesized that this was due to a decreased ability of the insecticide to bind its primary target [7,12]. While both mortality and motility are complex phenotypes, these data do link specific alleles to insecticide response phenotypes. Resistance levels observed in mortality assays, however, do not correlate perfectly with the data presented here. For example, Dα1 M4 showed less response to imidacloprid than Dβ2 L351Q as measured by all three WI phenotypes even though the latter has higher resistance in mortality assays [46]. The partial overlap in the mortality and motility phenotypes for these mutants suggest that different aspects of insect/insecticide interactions are being detected by each assay. These differences could be partially explained by differences in the concentrations of insecticides, stages of development and exposure methods in the two assays. The mortality assays discussed here, were performed on first instar larvae reared to adulthood on solid media dosed with insecticide with much of the mortality occurring at the first instar stage. In contrast, the WI used third instar larvae, exposed for a short time (240 minutes) in liquid media.
The Dα6 nx mutant, which has no detectable Dα6 expression, still responds to spinosad in the WI and has similar End Point RMR values when compared to Armenia14 and Dα6 W337* even though the two Dα6 mutants have far higher mortality LC50 value (Fig 5; Fig 8). To date Dα6 orthologues are the only genes for which a role in spinosad target site resistance has been demonstrated [17,47]. The Dα6 nx data presented here suggest that spinosad may also bind to at least one additional target impacting motility. Early mode of action studies indicated that other ligand gated chloride channels may also be spinosad targets [48]. No mutations were found in the Dα6 orthologue in a spinosad resistant strain of Musca domestica [49], but there is evidence that spinosad can bind to M. domestica α6 [7]. The WI has allowed us to make this significant observation, not detected using classical mortality studies.
There is the potential that baseline differences in motility in the absence of insecticide may confound any motility responses measured by the WI. In the current study the only genotypes to show such differences were the imidacloprid target site mutants (Dα1 M4 Dβ2 L351Q ) with each mutant moving significantly less than Armenia14 at time point 0 (S1 Fig). Despite lower initial WI values, in the longer term the WI values of the mutants were higher than Armenia14. Therefore, even in the absence of any correction for motility differences in the insecticide free control, the WI was still able to differentiate target site mutants. However, only matched genetic backgrounds were tested in the current study. It is therefore possible that comparing field populations may restrict the WI to describing alleles of large effect as is the case with mortality testing [24]. However, quantitative genetics approaches including genome wide association studies may help overcome this obstacle (See section 4.4).
Cyp6g1
The impact of imidacloprid on motility was reduced significantly when Cyp6g1 was overexpressed in the digestive tissues (Fig 6). These data correlate with those previously reported for mortality assays [10]. As imidacloprid targets nAChRs in the CNS, the response of larvae may be an indicator of the levels of imidacloprid and/or toxic metabolites in the CNS at a given time. Hence, the data presented here could be explained if the overexpression of Cyp6g1 reduces the effective concentration of imidacloprid in the CNS. This hypothesis is supported by data from metabolic studies conducted under the same exposure conditions employed here (3rd instar larvae exposed to imidacloprid in 5% sucrose solution). Hoi et al. [50] used Twin Ion Mass Spectrometry to show that increased in vivo expression of Cyp6g1 in the digestive tissues leads to a large increase in the metabolism of imidacloprid to toxic metabolites that are rapidly excreted. As a consequence, when Cyp6g1 is overexpressed in the metabolic tissues, significantly lower concentrations of imidacloprid and metabolites are retained in the body with access to the CNS.
The impact of imidacloprid on motility was also significantly reduced when Cyp6g1 was overexpressed in the CNS (Fig 7). While the capacity of Cyp6g1 to provide neuroprotection has not been previously reported, a role has been proposed for a Cytochrome P450, Cyp6cm1, in Tribolium castaneum [9].
One advantage of using the WI in combination with the GAL4-UAS system is that it provides the capacity to determine the route(s) that an insecticide follows to arrive at its target. It is theoretically possible that under the conditions used for the WI, that the insecticides tested may have entered the body orally, reaching the CNS via the digestive system, or through the cuticle, gaining direct access to the CNS via the hemolymph. Cuticular thickness has been cited as a resistance mechanism in adult D. melanogaster [51]. The large difference in RMR values at early time points (5 minutes; Fig 7B) when Cyp6g1 is overexpressed in digestive tissues indicates that a significant portion of imidacloprid reaches the CNS via the digestive system, probably due to oral ingestion. The path that insecticides and their metabolites follow in larvae could be further studied by using the WI in combination with Twin Ion Mass Spectrometry [50] and the GAL4-UAS system to examine the role of different families of genes such as those encoding transporter or cuticular proteins.
The Utility of the WI
While only the response of D. melanogaster was tested in this study, this method offers several insights into insecticide biology in pest species. D. melanogaster has been frequently used to elaborate on resistance mechanisms in matched genetic backgrounds. For example, the contribution of individual pest genes to resistance has been measured by driving the expression of genes encoding metabolic enzymes and targets in D. melanogaster [6,9,52]. Beyond the transgenic expression of pest genes in D. melanogaster, there is also the possibility that the methods used in this study may be modified for use in some other insect species. A similar exposure method has already been used to study drug responses in the parasitic nematode Haemonchus contortus (38). While the exposure method described here (24 well plates, chemical concentrations etc.) may be D. melanogaster specific, the ImageJ algorithm itself only measures the average change in light intensity of pixels in a video. Thus, in theory a variety of different exposure conditions may be used so long as the organism under investigation can be differentiated from its background.
Several technical aspects of the WI make it an attractive option for assessing an insecticide response. Analysing motility is a relatively cheap, high-throughput process when compared with other methods. The WI setup requires only an inexpensive video camera, a source of even light and free downloadable software (See Materials and Methods). Typical mortality assays are measured in adults by analysing how many adults survive insecticide exposure after a given interval or in larvae by measuring what proportion of individuals develop to adulthood [19,53]. The WI measures insecticide response earlier in the life cycle of the organism and significant differences can be observed between genotypes in as little as 5 minutes. The ease of sampling (filming) allows for a nearly unlimited number of time points that can be analysed in a single run. While in this study three parameters were used to quantify insecticide response, many other features of the response profile could be analysed. These factors make the WI suitable for screening large numbers of genotypes.
One of the distinct advantages of using D. melanogaster is the ability to implicate individual genes in a phenotype by using previously generated genotypes from stock centres such as Vienna Drosophila RNAi Centre [54], the Bloomington Deficiency Kit [55] and the MiMic library [56]. Each stock in these collections contains a defined genetic change in a common background, much like the genotypes discussed in the current study. However, other Drosophila specific resources use genetic variation similar to those observed in field settings to implicate alleles in complex traits. The Drosophila Genetic Reference Panel is a collection of fully sequenced homozygous stocks derived from one field population [57,58]. By phenotyping the collection and performing a subsequent genome wide association study, alleles of potentially very small effect can be associated with a complex trait, such as sub-lethal insecticide response, as has been done for other traits [59,60]. A similar resource, the Drosophila Synthetic Population Resource, relies on a synthetic populations derived from several fully sequenced founder individuals [61] and has previously been used to describe the genetic basis for nicotine resistance [62]. The use of these resources, in combination with the WI may lead to the identification of previously undescribed genes influencing insecticide ingestion, metabolism, transport, excretion and those encoding targets.
Conclusion
The WI quantifies insecticide response by measuring motility and is a valuable addition to the current array of bioassays. By measuring the motility of larvae as they are being exposed, a more complete understanding of this response can emerge. Using insecticides of distinct classes and validation with previously characterized metabolic and target site resistant D. melanogaster, we have demonstrated that this assay is both sensitive and rapid, while providing novel biological information.
Supporting Information
The uncorrected responses (WI values) for imidacloprid resistant alleles during exposure to 48ppm imidacloprid over A) 240 minutes and B) 30 minutes. Despite lower starting values, the WI Value prior to correction are still clearly capable of discriminating between the known resistant alleles and susceptible strain (Armenia14).
(TIF)
Written in imagej macro language, this script will iterate over a folder of image sequences and will return a sub-folder full of heat maps and a table of WI values. These images and values correspond to the motility of larvae in one well in each image sequence in the folder.
(TXT)
Recipes for both maize meal media (A) and grape juice plates (B) used in this study.
(XLSX)
Acknowledgments
The authors would like to thank the Basler lab, the Bloomington stock centre and the Drosophila Genome Resource Center for the provision of D. melanogaster stocks and the Australian Drosophila Biomedical Research Support Facility for handling the quarantine of imported flies. This research was supported by funding from the Australian Research Council (Discovery Project Grant DP130102414) awarded to PB.
Data Availability
All data is contained within the manuscript and its Supporting Information files except for the WI_Analysis.R script. It is available at the URL https://github.com/shanedenecke/WI_Analysis
Funding Statement
This research was supported by the Australian Research Council (Discovery Project Grant DP130102414), http://www.arc.gov.au/. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Flood J. The importance of plant health to food security. Food Secur. 2010;2: 215–231. 10.1007/s12571-010-0072-5 [DOI] [Google Scholar]
- 2. Palumbi SR. Evolution—Humans as the world's greatest evolutionary force. Science (80-). 2001;293: 1786–1790. [DOI] [PubMed] [Google Scholar]
- 3. Feyereisen R, Dermauw W, Van Leeuwen T. Genotype to phenotype, the molecular and physiological dimensions of resistance in arthropods. Pestic Biochem Physiol. Elsevier Inc.; 2015; 1–17. 10.1016/j.pestbp.2015.01.004 [DOI] [PubMed] [Google Scholar]
- 4. McKenzie J a, Batterham P. Resistance. Genetica. 1994;9. [Google Scholar]
- 5. Ffrench-Constant RH. The molecular genetics of insecticide resistance. Genetics. 2013;194: 807–15. 10.1534/genetics.112.141895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Daborn PJ, Lumb C, Harrop TWR, Blasetti A, Pasricha S, Morin S, et al. Using Drosophila melanogaster to validate metabolism-based insecticide resistance from insect pests. Insect Biochem Mol Biol. Elsevier Ltd; 2012;42: 918–24. 10.1016/j.ibmb.2012.09.003 [DOI] [PubMed] [Google Scholar]
- 7. Perry T, Somers J, Yang YT, Batterham P. Expression of insect α6-like nicotinic acetylcholine receptors in Drosophila melanogaster highlights a high level of conservation of the receptor:spinosyn interaction. Insect Biochem Mol Biol. Elsevier Ltd; 2015; 1–10. 10.1016/j.ibmb.2015.01.017 [DOI] [PubMed] [Google Scholar]
- 8. Li X, Schuler M a, Berenbaum MR. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu Rev Entomol. 2007;52: 231–53. 10.1146/annurev.ento.51.110104.151104 [DOI] [PubMed] [Google Scholar]
- 9. Zhu F, Parthasarathy R, Bai H, Woithe K, Kaussmann M, Nauen R, et al. A brain-specific cytochrome P450 responsible for the majority of deltamethrin resistance in the QTC279 strain of Tribolium castaneum . Proc Natl Acad Sci U S A. 2010;107: 8557–8562. 10.1073/pnas.1000059107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Daborn PJ, Yen JL, Bogwitz MR, Le Goff G, Feil E, Jeffers S, et al. A single p450 allele associated with insecticide resistance in Drosophila . Science. 2002;297: 2253–6. 10.1126/science.1074170 [DOI] [PubMed] [Google Scholar]
- 11. Schmidt JM, Good RT, Appleton B, Sherrard J, Raymant GC, Bogwitz MR, et al. Copy Number Variation and Transposable Elements Feature in Recent, Ongoing Adaptation at the Cyp6g1 Locus. PLoS Genet. Public Library of Science; 2010;6: e1000998 10.1371/journal.pgen.1000998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Perry T, Heckel DG, McKenzie J a, Batterham P. Mutations in Dα1 or Dβ2 nicotinic acetylcholine receptor subunits can confer resistance to neonicotinoids in Drosophila melanogaster . Insect Biochem Mol Biol. Elsevier Ltd; 2008;41: 520–528. 10.1016/j.ibmb.2007.12.007 [DOI] [PubMed] [Google Scholar]
- 13. Perry T, McKenzie J a, Batterham P. A Dα6 knockout strain of Drosophila melanogaster confers a high level of resistance to spinosad. Insect Biochem Mol Biol. 2007;37: 184–188. 10.1016/j.ibmb.2006.11.009 [DOI] [PubMed] [Google Scholar]
- 14. Watson GB, Chouinard SW, Cook KR, Geng C, Gifford JM, Gustafson GD, et al. A spinosyn-sensitive Drosophila melanogaster nicotinic acetylcholine receptor identified through chemically induced target site resistance, resistance gene identification, and heterologous expression. Insect Biochem Mol Biol. Elsevier Ltd; 2010;40: 376–84. 10.1016/j.ibmb.2009.11.004 [DOI] [PubMed] [Google Scholar]
- 15. Liu Z, Williamson MS, Lansdell SJ, Denholm I, Han Z, Millar NS. A nicotinic acetylcholine receptor mutation conferring target-site resistance to imidacloprid in Nilaparvata lugens (brown planthopper). Proc Natl Acad Sci U S A. 2005;102: 8420–5. 10.1073/pnas.0502901102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bass C, Puinean AM, Andrews M, Cutler P, Daniels M, Elias J, et al. Mutation of a nicotinic acetylcholine receptor β subunit is associated with resistance to neonicotinoid insecticides in the aphid Myzus persicae. BMC Neurosci. BioMed Central Ltd; 2011;12: 51 10.1186/1471-2202-12-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Puinean AM, Lansdell SJ, Collins T, Bielza P, Millar NS. A nicotinic acetylcholine receptor transmembrane point mutation (G275E) associated with resistance to spinosad in Frankliniella occidentalis . J Neurochem. 2013;124: 590–601. 10.1111/jnc.12029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Baxter SW, Chen M, Dawson A, Zhao JZ, Vogel H, Shelton AM, et al. Mis-spliced transcripts of nicotinic acetylcholine receptor α6 are associated with field evolved spinosad resistance in Plutella xylostella (L.). PLoS Genet. 2010;6 10.1371/journal.pgen.1000802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.IRAC [Insecticide Resistance Action Committee] (2015) Test Methods. Available: http://www.irac-online.org/methods/. Accessed 1 October 2015.
- 20. Desneux N, Decourtye A, Delpuech J-M. The sublethal effects of pesticides on beneficial arthropods. Annu Rev Entomol. 2007;52: 81–106. 10.1146/annurev.ento.52.110405.091440 [DOI] [PubMed] [Google Scholar]
- 21. Guedes RNC, Smagghe G, Stark JD, Desneux N. Pesticide-Induced Stress in Arthropod Pests for Optimized Integrated Pest Management Programs. Annu Rev Entomol. 2016;61: annurev–ento–010715–023646. 10.1146/annurev-ento-010715-023646 [DOI] [PubMed] [Google Scholar]
- 22. Remucal CK. The role of indirect photochemical degradation in the environmental fate of pesticides: a review. Environ Sci Process Impacts. 2014;16: 628–53. 10.1039/c3em00549f [DOI] [PubMed] [Google Scholar]
- 23. Bonmatin JM, Giorio C, Girolami V, Goulson D, Kreutzweiser DP, Krupke C, et al. Environmental fate and exposure; neonicotinoids and fipronil. Environ Sci Pollut Res. 2014; 35–67. 10.1007/s11356-014-3332-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McKenzie JA, Batterham P. Predicting insecticide resistance : mutagenesis, selection and response. 1998; 1729–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wei H, Wang J, Li HS, Dai HG, Gu XJ. Sub-lethal effects of fenvalerate on the development, fecundity, and juvenile hormone esterase activity of diamondback moth, Plutella xylostella (L.). Agric Sci China. Chinese Academy of Agricultural Sciences; 2010;9: 1612–1622. 10.1016/S1671-2927(09)60258-3 [DOI] [Google Scholar]
- 26. Cameron R, Lang EB, Annan IB, Portillo HE, Alvarez JM. Use of Fluorescence, a Novel Technique to Determine Reduction in Bemisia tabaci (Hemiptera: Aleyrodidae) Nymph Feeding When Exposed to Benevia and Other Insecticides. J Econ Entomol. 2013;106: 597–603. 10.1603/Ec12370 [DOI] [PubMed] [Google Scholar]
- 27. Tomé HV, Pascini T V, Dângelo RA, Guedes RN, Martins GF. Survival and swimming behavior of insecticide-exposed larvae and pupae of the yellow fever mosquito Aedes aegypti . Parasit Vectors. 2014;7: 195 10.1186/1756-3305-7-195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hellou J. Behavioural ecotoxicology, an “early warning” signal to assess environmental quality. Environ Sci Pollut Res. 2011;18: 1–11. 10.1007/s11356-010-0367-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fairbrother A, Purdy J, Anderson T, Fell R. Risks of neonicotinoid insecticides to honeybees. Environ Toxicol Chem. 2014;33: 719–731. 10.1002/etc.2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cresswell JE, Desneux N, vanEngelsdorp D. Dietary traces of neonicotinoid pesticides as a cause of population declines in honey bees: An evaluation by Hill’s epidemiological criteria. Pest Manag Sci. 2012;68: 819–827. 10.1002/ps.3290 [DOI] [PubMed] [Google Scholar]
- 31. Wolf FW, Rodan AR, Tsai LT, Heberlein U. High-Resolution Analysis of Ethanol-Induced Locomotor Stimulation in Drosophila. 2002;22: 11035–11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tooming E, Merivee E, Must A, Sibul I, Williams I. Sub-lethal effects of the neurotoxic pyrethroid insecticide Fastac® 50EC on the general motor and locomotor activities of the non-targeted beneficial carabid beetle Platynus assimilis (Coleoptera: Carabidae). Pest Manag Sci. 2013; 10.1002/ps.3636 [DOI] [PubMed] [Google Scholar]
- 33. Bai D, Lummis SCR, Leicht W, Breerc H, Sattelleat DB. Actions of Imidacloprid and a Related Nitromet hylene on Cholinergic Receptors of an Identified Insect Motor Neurone. 1991; 197–204. [Google Scholar]
- 34. Joußen N, Heckel DG, Haas M, Schuphan I. Metabolism of imidacloprid and DDT by P450 CYP6G1 expressed in cell cultures of Nicotiana tabacum suggests detoxification of these insecticides in Cyp6g1-overexpressing strains of Drosophila melanogaster, leading to resistance. 2008;73: 65–73. 10.1002/ps [DOI] [PubMed] [Google Scholar]
- 35. Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific φC31 integrases. Proc Natl Acad Sci U S A. 2007;104: 3312–3317. 10.1073/pnas.0611511104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Daborn PJ, Lumb C, Boey A, Wong W, Ffrench-Constant RH, Batterham P. Evaluating the insecticide resistance potential of eight Drosophila melanogaster cytochrome P450 genes by transgenic over-expression. Insect Biochem Mol Biol. 2007;37: 512–9. 10.1016/j.ibmb.2007.02.008 [DOI] [PubMed] [Google Scholar]
- 37. Nichols CD, Becnel J, Pandey UB. Methods to assay Drosophila behavior. J Vis Exp. 2012; 3–7. 10.3791/3795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cleveland CB, Bormett G a., Saunders DG, Powers FL, McGibbon AS, Reeves GL, et al. Environmental fate of spinosad. 1. Dissipation and degradation in aqueous systems. J Agric Food Chem. 2002;50: 3244–3256. 10.1021/jf011663i [DOI] [PubMed] [Google Scholar]
- 39. Liu Z, Dai Y, Huang G, Gu Y, Ni J, Wei H, et al. Soil microbial degradation of neonicotinoid insecticides imidacloprid, acetamiprid, thiacloprid and imidaclothiz and its effect on the persistence of bioefficacy against horsebean aphid Aphis craccivora Koch after soil application. Pest Manag Sci. 2011;67: 1245–52. 10.1002/ps.2174 [DOI] [PubMed] [Google Scholar]
- 40. Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9: 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Preston S, Jabbar A, Nowell C, Joachim A, Ruttkowski B, Baell J, et al. Low cost whole-organism screening of compounds for anthelmintic activity. Int J Parasitol. 2015;45: 1–27. 10.1016/j.ijpara.2015.01.007 [DOI] [PubMed] [Google Scholar]
- 42. Kane NS, Hirschberg B, Qian S, Hunt D, Thomas B, Brochu R, et al. Drug-resistant Drosophila indicate glutamate-gated chloride channels are targets for the antiparasitics nodulisporic acid and ivermectin. Proc Natl Acad Sci U S A. 2000;97: 13949–13954. 10.1073/pnas.240464697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lahm GP, Cordova D, Barry JD. New and selective ryanodine receptor activators for insect control. Bioorganic Med Chem. Elsevier Ltd; 2009;17: 4127–4133. 10.1016/j.bmc.2009.01.018 [DOI] [PubMed] [Google Scholar]
- 44. Banks W a. Characteristics of compounds that cross the blood-brain barrier. BMC Neurol. 2009;9 Suppl 1: S3 10.1186/1471-2377-9-S1-S3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Begley DJ. ABC transporters and the blood-brain barrier. Curr Pharm Des. 2004;10: 1295–1312. 10.2174/1381612043384844 [DOI] [PubMed] [Google Scholar]
- 46. Perry T, Chan JQ, Batterham P, Watson GB, Geng C, Sparks TC. Effects of mutations in Drosophila nicotinic acetylcholine receptor subunits on sensitivity to insecticides targeting nicotinic acetylcholine receptors. Pestic Biochem Physiol. Elsevier Inc.; 2012;102: 56–60. 10.1016/j.pestbp.2011.10.010 [DOI] [Google Scholar]
- 47. Hsu JC, Feng HT, Wu WJ, Geib SM, Mao CH, Vontas J. Truncated transcripts of nicotinic acetylcholine subunit gene Bdα6 are associated with spinosad resistance in Bactrocera dorsalis . Insect Biochem Mol Biol. Elsevier Ltd; 2012;42: 806–815. 10.1016/j.ibmb.2012.07.010 [DOI] [PubMed] [Google Scholar]
- 48. Watson GB. Actions of Insecticidal Spinosyns on γ-Aminobutyric Acid Responses from Small-Diameter Cockroach Neurons. Pestic Biochem Physiol. 2001;71: 20–28. 10.1006/pest.2001.2559 [DOI] [Google Scholar]
- 49. Gao JR, Deacutis JM, Scott JG. The nicotinic acetylcholine receptor subunit Mdα6 from Musca domestica is diversified via post-transcriptional modification. Insect Mol Biol. 2007;16: 325–334. 10.1111/j.1365-2583.2007.00730.x [DOI] [PubMed] [Google Scholar]
- 50. Hoi KK, Daborn PJ, Battlay P, Robin C, Batterham P, O’Hair R a J, et al. Dissecting the insect metabolic machinery using twin ion mass spectrometry: A single P450 enzyme metabolizing the insecticide imidacloprid in vivo. Anal Chem. 2014;86: 3525–3532. 10.1021/ac404188g [DOI] [PubMed] [Google Scholar]
- 51. Strycharz JP, Lao A, Li H, Qiu X, Lee SH, Sun W, et al. Resistance in the highly DDT-resistant 91-R strain of Drosophila melanogaster involves decreased penetration, increased metabolism, and direct excretion. Pestic Biochem Physiol. Elsevier Inc.; 2013;107: 207–217. 10.1016/j.pestbp.2013.06.010 [DOI] [Google Scholar]
- 52. Bass C, Zimmer CT, Riveron JM, Wilding CS, Wondji CS, Kaussmann M, et al. Gene amplification and microsatellite polymorphism underlie a recent insect host shift. Proc Natl Acad Sci U S A. 2013;110: 19460–5. 10.1073/pnas.1314122110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bloomquist J, Roush R, ffrench-Constant R. Reduced Neuronal Sensitivity to Dieldrin and Picrotoxinin in a Cyclodiene-Resistant Strain of Drosophila melanogaster (Meigen). 1992;25: 17–25. [DOI] [PubMed] [Google Scholar]
- 54. Dietzl G, Chen D, Schnorrer F, Su K-C, Barinova Y, Fellner M, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila . Nature. 2007;448: 151–6. 10.1038/nature05954 [DOI] [PubMed] [Google Scholar]
- 55. Roote J, Russell S. Toward a complete Drosophila deficiency kit. Genome Biol. 2012;13: 149 10.1186/gb4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Venken KJT, Schulze KL, Haelterman N a, Pan H, He Y, Evans-Holm M, et al. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat Methods. 2011;8: 737–743. 10.1038/nmeth.1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mackay TFC, Richards S, Stone E a, Barbadilla A, Ayroles JF, Zhu D, et al. The Drosophila melanogaster Genetic Reference Panel. Nature. 2012;482: 173–8. 10.1038/nature10811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Huang W, Massouras A, Inoue Y, Peiffer J, Ràmia M, Tarone AM, et al. Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Res. 2014;24: 1193–1208. 10.1101/gr.171546.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Garlapow ME, Huang W, Yarboro MT, Peterson KR, Mackay TFC (2015) Quantitative Genetics of Food Intake in Drosophila melanogaster . PLoS ONE 10(9): e0138129 10.1371/journal.pone.013812960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ivanov DK, Escott-Price V, Ziehm M, Magwire MM, Mackay TFC, Partridge L, et al. Longevity GWAS Using the Drosophila Genetic Reference Panel. Journals Gerontol Ser A Biol Sci Med Sci. 2015; 1–9. 10.1093/gerona/glv047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. King EG, Macdonald SJ, Long AD. Properties and power of the Drosophila synthetic population resource for the routine dissection of complex traits. Genetics. 2012;191: 935–949. 10.1534/genetics.112.138537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Marriage TN, King EG, Long AD, Macdonald SJ. Fine-mapping nicotine resistance Loci in Drosophila using a multiparent advanced generation inter-cross population. Genetics. 2014;198: 45–57. 10.1534/genetics.114.162107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The uncorrected responses (WI values) for imidacloprid resistant alleles during exposure to 48ppm imidacloprid over A) 240 minutes and B) 30 minutes. Despite lower starting values, the WI Value prior to correction are still clearly capable of discriminating between the known resistant alleles and susceptible strain (Armenia14).
(TIF)
Written in imagej macro language, this script will iterate over a folder of image sequences and will return a sub-folder full of heat maps and a table of WI values. These images and values correspond to the motility of larvae in one well in each image sequence in the folder.
(TXT)
Recipes for both maize meal media (A) and grape juice plates (B) used in this study.
(XLSX)
Data Availability Statement
All data is contained within the manuscript and its Supporting Information files except for the WI_Analysis.R script. It is available at the URL https://github.com/shanedenecke/WI_Analysis